Abstract
Background:
OBI-3424 is a highly selective prodrug that is converted by aldo-keto reductase family 1 member C3 (AKR1C3) to a potent DNA alkylating agent. OBI-3424 has entered clinical testing for hepatocellular carcinoma and castrate resistant prostate cancer, and it represents a potentially novel treatment for acute lymphoblastic leukemia (ALL).
Experimental Procedures:
We assessed AKR1C3 expression by RNA-Seq and immunoblotting, and evaluated the in vitro cytotoxicity of OBI-3424. We investigated the pharmacokinetics of OBI-3424 in mice and non-human primates, and assessed the in vivo efficacy of OBI-3424 against a large panel of patient-derived xenografts (PDXs).
Results:
AKR1C3 mRNA expression was significantly higher in primary T-ALL (n=264) than B-ALL (n=1,740) (P<0.0001) and OBI-3424 exerted potent cytotoxicity against T-ALL cell lines and PDXs. In vivo, OBI-3424 significantly prolonged the event-free survival (EFS) of 9/9 ALL PDXs by 17.1–77.8 days (Treated/Control values 2.5–14.0), and disease regression was observed in 8/9 PDXs. A significant reduction (P<0.0001) in bone marrow infiltration at Day 28 was observed in 4/6 evaluable T-ALL PDXs. The importance of AKR1C3 in the in vivo response to OBI-3424 was verified using a B-ALL PDX that had been lentivirally transduced to stably overexpress AKR1C3. OBI-3424 combined with nelarabine resulted in prolongation of mouse EFS compared with each single agent alone in 2 T-ALL PDXs.
Conclusion:
OBI-3424 exerted profound in vivo efficacy against T-ALL PDXs derived predominantly from aggressive and fatal disease, and therefore may represent a novel treatment for aggressive and chemoresistant T-ALL in an AKR1C3 biomarker-driven clinical trial.
Keywords: Leukemia, Patient-Derived Xenografts, Biomarkers, OBI-3424, AKR1C3
INTRODUCTION
Five-year survival rates for children diagnosed with acute lymphoblastic leukemia (ALL), the most common pediatric malignancy, have continually improved since the 1970s and are currently approximately 90%.(1–3) Pediatric ALL can be broadly divided into B-lineage ALL (B-ALL) and T-lineage ALL (T-ALL).(4) Disease-free survival and overall survival are comparable between children with B-ALL and T-ALL treated on contemporary risk-adapted regimens,(5) although outcome is poor for children with T-ALL following relapse.(6)
Aldo-keto reductases (AKRs) are a superfamily of NAD(P)(H)-dependent oxidoreductases that primarily catalyze the reduction of aldehydes and ketones to their corresponding alcohols.(7) AKR family I member C3 (AKR1C3), is a hydroxysteroid dehydrogenase involved in the synthesis of steroid hormones and prostaglandins(8) and is expressed in a range of normal human tissues at varying levels.(9) AKR1C3 is also expressed in pediatric and adult ALL, with significantly higher mRNA levels detected in T-ALL than B-ALL.(10) Recently, elevated expression and activity of AKR1C3 and other subfamily members were shown in diagnostic samples from pediatric T-ALL patients who went on to respond poorly to treatment when compared to treatment responders, suggesting a relationship between AKR1C3 expression and sensitivity of T-ALL cells to conventional drugs including daunorubicin, vincristine, and L-asparaginase.(11) Moreover, the activity of AKR1C3 and its subfamily members affects the in vitro sensitivity of primary T-ALL cells and cell lines to vincristine.(11) Therefore, AKR1C3 overexpression in pediatric T-ALL is a potential therapeutic target as well as a possible biomarker for treatment resistance.
A potential innovative therapeutic approach is to exploit the enzymatic activity of AKR1C3 to activate novel prodrugs. The hypoxia-activated nitrogen mustard pre-prodrug PR-104 is hydrolyzed in vivo to PR-104A, which then undergoes activation under hypoxic conditions to DNA cross-linking metabolites by 1-electron NADPH:cytochrome P450 oxidoreductase (CYPOR) and related flavoproteins.(12,13) While PR-104 was initially developed as a hypoxia-activated prodrug, it was subsequently discovered to also be activated by AKR1C3 under aerobic conditions.13 Despite exhibiting promising preclinical activity against T-ALL patient-derived xenografts (PDXs),(14) PR-104 showed limited efficacy in a phase I/II clinical trial in adult patients with ALL or acute myeloid leukemia, and myelosuppression was the major dose-limiting toxicity (DLT).(15,16) Moreover, PR-104 also exhibits considerable bystander effects in its mechanism of action, which is also likely to contribute to both its efficacy and toxicity.(17)
ALL cells are known to be preferentially sensitive to DNA-damaging agents. The most effective DNA alkylating agent used to treat pediatric ALL, cyclophosphamide, is itself a prodrug converted by liver enzymes to active metabolites that then enter the circulation to reach their target cells. Myelosuppression is also the major DLT of cyclophosphamide.(18) Theoretically, a potent DNA-alkylating prodrug that is both activated and retained within the target cancer cell may exhibit favorable properties of safety, selectivity and efficacy compared with both cyclophosphamide and PR-104. OBI-3424 (previously TH-3424) is a prodrug that is selectively activated by AKR1C3 to a potent DNA alkylating agent that is then retained within the cell in which it is activated. It has entered clinical testing for hepatocellular carcinoma and castrate resistant prostate cancer (NCT03592264). The goal of this study was to evaluate the efficacy of OBI-3424 against preclinical models of pediatric ALL with reference to AKR1C3 expression.
MATERIALS AND METHODS
Cell lines and PDXs for in vitro and in vivo studies
The non-small-cell lung cancer H460 cell line was purchased from ATCC. All cell line studies were outsourced to HD Biosciences (Shanghai, China). All experimental work was performed with approvals from the respective institutional review boards and animal ethics committees of each institution. Experiments used continuous PDXs established previously in 20 to 25 g female non-obese diabetic/severe combined immunodeficient (NOD.CB17-Prkdcscid/SzJ, NOD/SCID) or NOD/SCID/interleukin-2 receptor γ–negative (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJAusb, NSG) mice, as described elsewhere.(19) The development of lentivirally-transduced ALL-11 PDXs [empty vector (EV) and AKR1C3 overexpressing] has been described previously.(10) The patient demographics and passage numbers of the ALL PDXs used in this study are shown in Supplementary Table S2, and genomic characterization can be accessed at https://pedcbioportal.org and in Supplementary Table S3. The ALL-11/EV and ALL-11/AKR1C3 PDXs were used at 5° passage. OBI-3424 was provided by Threshold Pharmaceuticals, Inc. and is being developed by OBI Pharmaceuticals, Inc. (ex-Asia) and Ascentawits Pharmaceuticals, Ltd. (Asia).
In vitro cytotoxicity assays
H460 and leukemia cell lines (Supplementary Table S1) were suspended in RPMI medium supplemented with fetal bovine serum (Biosera, Shanghai, China) while ALL PDXs cells were cultured in QBSF medium (Quality Biological Inc, Gaithersburg MD, USA) supplemented with Flt-3 ligand (20 ng/ml, Bionovus Life Sciences, Cherrybrook, NSW, Australia) or IL-7 (10–20 ng/ml, Jomar Life Research, Melbourne, VIC, Australia). Cells were plated according to optimal cell density(14,20) and incubated for 3 h or overnight (37°C, 5% CO2). H460 cells were pre-treated with 3 μM TH-3021 (SN336384), a potent and specific inhibitor of AKR1C3,(21) for 2 h, co-treated with OBI-3424 for 2 h, washed and incubated in fresh medium for 48 h. PDX cells and leukemia cell lines were treated with OBI-3424 (10 μM - 1 pM) or vehicle control for 48 h or 72 h, respectively. Viability was determined using Alamar Blue reduction assay,(14,22) or Cell Titer-Glo Luminescent Cell Viability Assay (Promega, Madison, WI). The half-maximal inhibitory concentration (IC50) was calculated by interpolation of non-linear regression curves calculated by GraphPad Prism 7 software.
RNA Sequencing (RNA-Seq) analysis
For analysis of AKR1C3 expression in primary patient aspirates patients were stratified into B-ALL and T-ALL and their relevant subgroups as previously described.(23,24) Paired-end reads were mapped to the GRCh37 human genome reference by STAR(25) (version 2.5.1b) through the recommended two pass mapping pipeline with default parameters and the Picard MarkDuplicates module was used to mark the duplication rate. Gene annotation files were downloaded from Ensembl (http://www.ensembl.org/) and used for STAR mapping and subsequent gene expression level evaluation. To evaluate gene expression profiles, read counts for annotated genes were called by HTSeq(26) (version 0.6.0) and processed by DESeq2 R package(27) to normalize gene expression into regularized log2 values (rlog).
For analysis of PDX samples, Illumina paired-end RNA-Seq data were aligned to the human genome assembly (build hg38) using STAR (version 2.5) with quantMode parameters set to TranscriptomeSAM for alignments translated into transcript coordinates. Alignments were run through RSEM (version 1.2.31) command rsem-calculate-expression to calculate raw gene counts, TPM, FPKM and isoform expression. All RNA-Seq values are expressed as fragments per kilobase million (FPKM). FPKM data were log2 transformed.
Immunoblotting and real-time quantitative reverse transcription PCR (RT-qPCR)
Procedures for immunoblotting and RT-qPCR have been described previously(14,28,29) and are detailed in the Supplementary Methods, and data were quantified relative to HeLa cells.
Single cell gel electrophoresis assays
Single cell alkali gel electrophoresis to calculate the interstrand crosslink (ICL) Index was performed using the Trevigen CometAssay® Kit (Bioscientific, Kirrawee, NSW, Australia) and is described in detail in the Supplementary Methods.(30)
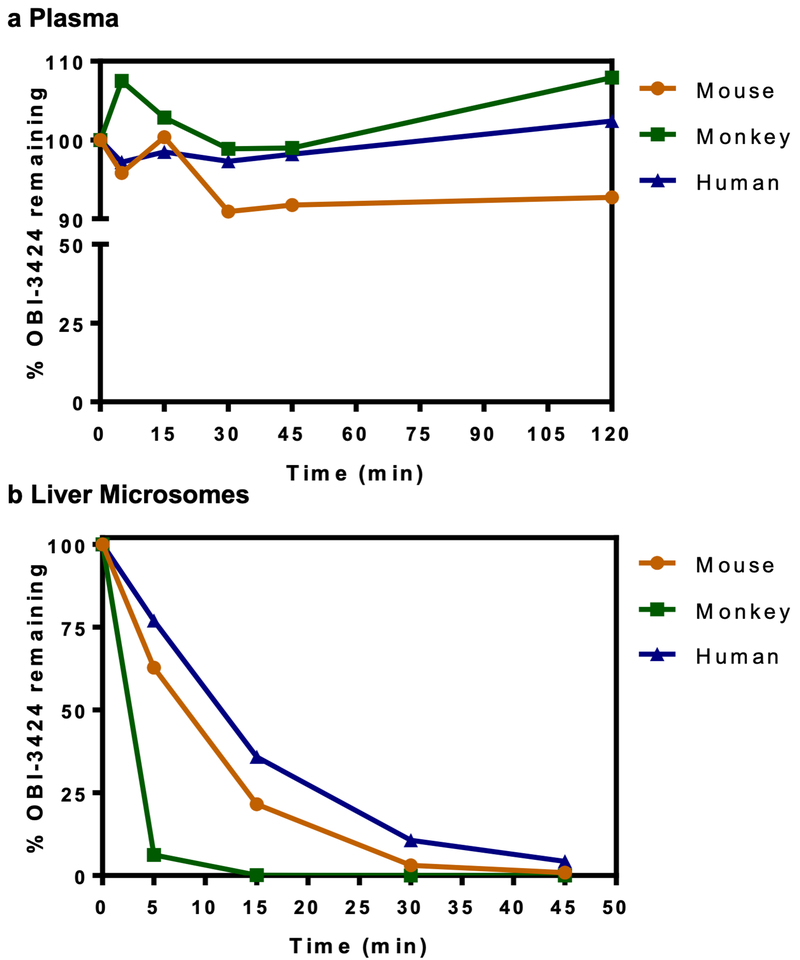
Activation and stability of OBI-3424 in mouse, monkey and human cytosol, plasma and liver microsomes.
Samples of plasma, liver cytosol (1 mg/mL, in 100 mM PBS, 2 mM NADPH) and liver microsomes (0.5 mg/mL, in 100 mM PBS, 2 mM NADPH) were each obtained from mouse, monkey and human. For each sample, OBI-3424 was incubated at a final concentration of 5 μM (plasma, liver cytosol) or 1 μM (liver microsomes) at 37°C for 120 min (plasma), 60 min (liver cytosol) or 45 min (liver microsomes). Progesterone (5 μM) and Midazolam (1 μM) were used as positive controls for liver cytosol and liver microsomes, respectively. Additionally, liver cytosol samples were tested ± 3 μM TH-3021.(21) Reactions were terminated by adding acetonitrile containing propranolol as an internal standard. After centrifugation at 4°C, the supernatants were analyzed by liquid chromatography tandem mass spectrometry (LC/MS-MS).
Pharmacokinetic study of OBI-3424 in mice and monkeys
Procedures for the evaluation of OBI-3424 pharmacokinetics and toxicity in nude mice and cynomolgus monkeys are detailed in the Supplementary Methods.
Assessment of in vivo drug efficacy
Leukemia engraftment and progression were assessed in groups of 8 female 20–25 g NSG mice following intravenous inoculation of PDX cells by weekly flow cytometric enumeration of the proportion of human versus mouse CD45+ (%huCD45+) cells in the peripheral blood (PB). Individual mouse event-free survival (EFS) was calculated as the number of days from treatment initiation until the %huCD45+ reached 25%, computed by interpolating between bleeds directly preceding and following events, assuming log-linear growth. Efficacy of drug treatment was evaluated by the difference between median EFS of vehicle control (C) and drug-treated (T) cohorts, as well as T/C values, and by an objective response measure (ORM), as described previously (see Supplementary Methods and Supplementary Table S4).(31) Leukemic infiltration was also assessed in the femoral bone marrow, spleen and blood at Day 0 (n=3 mice), Day 28 (n=4 mice) or event, whichever occurred first. OBI-3424 (or vehicle control) was administered via intraperitoneal (IP) injection once weekly for 3 weeks. For combination studies, mice were treated with OBI-3424 administered on the above schedule plus nelarabine at 150 mg/kg via IP injection once daily for 5 days and repeated at day 14.
Statistical analysis
All statistical methods used in this study are described in detail in the Supplementary Methods.
RESULTS
Structure and in vitro anti-leukemic efficacy of OBI-3424
OBI-3424 (previously TH-3424) was developed as a highly potent DNA alkylating prodrug that is selectively activated by AKR1C3 (Figure 1a). In the presence of NADPH, OBI-3424 is reduced by AKR1C3 to an intermediate that spontaneously hydrolyzes to OBI-2660 (Figure 1a), which has a structure reminiscent of the DNA alkylating drug thioTEPA (N,N’,N”-triethylenethiophosphoramide).(32) OBI-3424 exerted potent cytotoxicity against the H460 lung cancer cell line (IC50 4.0 nM); in contrast the IC50 of OBI-2660 was >330 μM (Figure 1b). The cytotoxicity of OBI-3424 was highly AKR1C3 dependent (Figure 1b); OBI-3424 IC50 values were 4.0 nM and 6.3 μM in the absence or presence, respectively, of 3 μM TH-3021.
Figure 1.
In vitro activity of OBI-3424. (a) chemical structure and mechanism of activation of OBI-3424. (b) Cytotoxicity of OBI-3424 against H460 cells ± TH-3021, a specific inhibitor of AKR1C3. (c-e) Cytotoxicity of OBI-3424 against a panel of 6 B-ALL (c), 7 T-ALL (d) and 6 ETP-ALL (e) PDXs. Data in c-e are displayed as the mean ± SEM of biological triplicates.
In order to assess the potential anti-leukemic activity of OBI-3424, in vitro cytotoxicity assays were carried out on a broad range of leukemia cell lines. OBI-3424 exhibited potent cytotoxicity, in particular against cell lines derived from T-ALL with high AKR1C3 expression, with IC50 values in the low nM range (Supplementary Table S1). The difference in IC50 values between cell lines with high/medium AKR1C3 expression and those with low expression was statistically significant (P = 0.0016). In vitro cytotoxicity assays were then carried out against a panel of 19 PDXs representative of B-ALL, T-ALL and early T-cell precursor ALL (ETP-ALL) (Supplementary Table S2). Similar to the results obtained with leukemia cell lines, OBI-3424 exerted potent cell killing against ALL PDXs (Figure 1c–e). The median IC50 values were 60.3 nM (range 3.2 nM - >10 μM) for B-ALL (Figure 1c, Supplementary Table S2), 9.7 nM (1.1 – 745 nM) for T-ALL (Figure 1d, Supplementary Table S2) and 31.5 nM (4.0 – 130 nM) for ETP-ALL (Figure 1e, Supplementary Table S2). When cell survival relative to vehicle-treated controls was compared at 100 nM OBI-3424, T-ALL PDXs were more sensitive than B-ALL and ETP-ALL PDXs (Supplementary Figure S1).
AKR1C3 expression in pediatric ALL biopsy specimens and PDXs
We next assessed AKR1C3 expression in diverse pediatric ALL subtypes with respect to OBI-3424 sensitivity. AKR1C3 expression was significantly higher in diagnosis bone marrow aspirates from pediatric T-ALL patients (n=264) compared with B-ALL (n=1,740) (P < 0.0001, Figure 2a). AKR1C3 expression in B-ALL was relatively low, especially in cases harboring DUX4 and MEF2D rearrangements (Supplementary Figure S2a). In contrast, AKR1C3 expression in T-ALL was generally high, with the exception of subtypes harboring TLX1/3 rearrangements (Supplementary Figure S2b). AKR1C3 expression did not differ significantly between ETP-ALL and T-ALL (Supplementary Figure S2c). There was no evidence of genomic alterations or mutations of AKR1C3 as potential factors influencing differential expression levels of AKR1C3 between B- and T-ALL.
Figure 2.
AKR1C3 expression in primary ALL and PDX cells and quantification of DNA ICLs. (a-b) RNA-Seq data of primary (a) and PDX (b) B-ALL versus T-ALL cells expressed as FPKM on a log2 scale. (c-d) AKR1C3 mRNA (c) and protein (d) expression levels in a panel of 6 B-ALL, 6 T-ALL and 6 ETP-ALL PDXs. A representative immunoblot is shown in (d). (e-f) Correlations of PDX cell survival following treatment with OBI-3424 and AKR1C3 protein expression at 100 nM (e) or 10 nM (f) OBI-3424. (g) Comet assay results showing DNA ICL Indices in 3 ALL PDXs (ALL-8, ETP-2, ALL-19) following OBI-3424 treatment (4 h). (h) Representative comet images from G are shown for ALL-8. In (a) and (b) bars represent the median and interquartile range. Data in c, d and g are displayed as the mean ± SEM of biological triplicates. Solid and dashed lines in e and f represent linear regression and 95% confidence interval, respectively.
Analysis of RNA-seq data from 90 ALL PDXs, including 25 derived from patients at relapse, confirmed significantly higher AKR1C3 expression in T-ALL (n=25) versus B-ALL (n=65) (P < 0.0001, Figure 2b, Supplementary Figure S2d), and was confirmed by qRT-PCR (Figure 2c) and immunoblotting (Figure 2d). AKR1C3 expression was also significantly higher in T-ALL than most normal tissues (Supplementary Figure S3), with the exception of several tissues with comparable or higher expression (adipose tissue, colon, kidney, and liver). There was a significant correlation between AKR1C3 mRNA and protein expression (R = 0.58; P = 0.0003) (Supplementary Figure S4). Moreover, AKR1C3 protein expression showed significant inverse correlations with the in vitro cell survival of the 18 ALL PDXs at OBI-3424 concentrations of both 100 nM (Figure 2e) and 10 nM (Figure 2f). Of note, an AKR1C3high B-ALL PDX (ALL-7) exhibited relative sensitivity to OBI-3424, while an AKR1C3low T-ALL PDX (ALL-42) was OBI-3424 resistant, highlighting the importance of AKR1C3 expression, rather than ALL lineage, in sensitivity to OBI-3424 (Supplementary Table S2).
OBI-3424 also induced a concentration-dependent increase in the ICL Index in ALL-8 and ETP-2, but not ALL-19 (AKR1C3low, Figure 2c and d, Supplementary Table S2), as assessed by comet assay (Figure 2g and h). These results, along with the relative resistance of ALL-19 to the cytotoxic effects of OBI-3424 (Figure 1c) are consistent with AKR1C3-dependent activation of OBI-3424 into a DNA alkylator resulting in cytotoxic DNA ICLs. The higher AKR1C3 expression in primary T-ALL cells and PDXs compared with B-ALL suggests that T-ALL may be particularly susceptible to targeting in vivo with OBI-3424.
OBI-3424 stability, preclinical pharmacokinetics and toxicology
Since there is no functional murine equivalent of AKR1C3,(33) mouse liver cytosolic fractions are unable to activate OBI-3424. In contrast, monkey and human liver cytosolic fractions rapidly activate OBI-3424 in an AKR1C3-dependent fashion (Supplementary Figure S5). Therefore, prior to evaluation of OBI-3424 in immune-deficient murine preclinical experimental models it was important to assess the stability, pharmacokinetics and toxicity of OBI-3424 in relevant preclinical model systems.
OBI-3424 retained >90% stability in mouse, monkey and human plasma for at least 2 h (Figure 3a), although it was most unstable in hepatic microsomes from monkeys and more stable in those from mice and humans (Figure 3b). OBI-3424 was well tolerated in mice at doses >10 mg/kg and OBI-3424 plasma pharmacokinetics in female H460 tumor-bearing mice (Nu-Foxn 1nu NU/NU, 5 mg/kg) and cynomolgus monkeys (2 mg/kg) are shown in Supplementary Tables S5 and S6, respectively. In a non-GLP toxicology study in cynomolgus monkeys, OBI-3424 was administered by 30 min IV infusion every 7 days × 2 for 2 cycles with a one week break between cycles. No toxicity was observed at 0.32 mg/kg, with mean white blood cell count (WBC) remaining within reference intervals for the duration of the study (Supplementary Figure S6), while all animals receiving 1 mg/kg OBI-3424 were humanely killed on Days 10, 12 or 13 due to severe diarrhea. Microscopic examination showed correlative severe pathology in the small intestine with villous atrophy, crypt loss, and reactive hyperplasia with necrotic cell debris.
Figure 3.
In vitro stability of OBI-3424. (a) Stability of OBI-3424 in plasma. (b) Stability of OBI-3424 in liver microsomes. OBI-3424 was assayed by LC/MS-MS. Data represent the mean ± relative SD (RSD). Data in a and b are displayed as the mean ± relative standard deviation (RSD, coefficient of variation).
No OBI-3424-related clinical signs were observed in animals administered 0.32 mg/kg OBI-3424, with no marked changes in body weight, hematology, clinical chemistry or urinalysis parameters (Supplementary Tables S7–S12 and data not shown). OBI-3424-related clinical chemistry changes were limited to increases in creatine kinase. On Days 8 (13.6-fold; Female 2101) and 10 (10.9- fold; Female 2102 at euthanasia) in animals treated with 1 mg/kg O2BI-3424 creatine kinase values increased relative to Day 1 baseline values. A similar elevation was observed (15.3-fold baseline) on Day 43 in one female (1101) treated with 0.32 mg/kg OBI-3424, and a minor increase (3.8-fold baseline) in the same animal at Day 8. Therefore, OBI-3424 was well tolerated in cynomolgus monkeys at the level of 0.32 mg/kg/dose.
Extrapolation of the AUC values between mouse and monkey in Supplementary Tables S5 and S6 indicates that a dose of 0.32 mg/kg IV in the cynomolgus monkey approximates to 2.5 mg/kg IP in the mouse in terms of equivalent plasma drug exposure levels. While this dose is well below the mouse maximum tolerated dose (MTD), the dose of 2.5 mg/kg administered every 7 days IP was selected as the maximum dose for all subsequent experiments to assess the in vivo efficacy of OBI-3424 against ALL PDX models in immune-deficient mice.
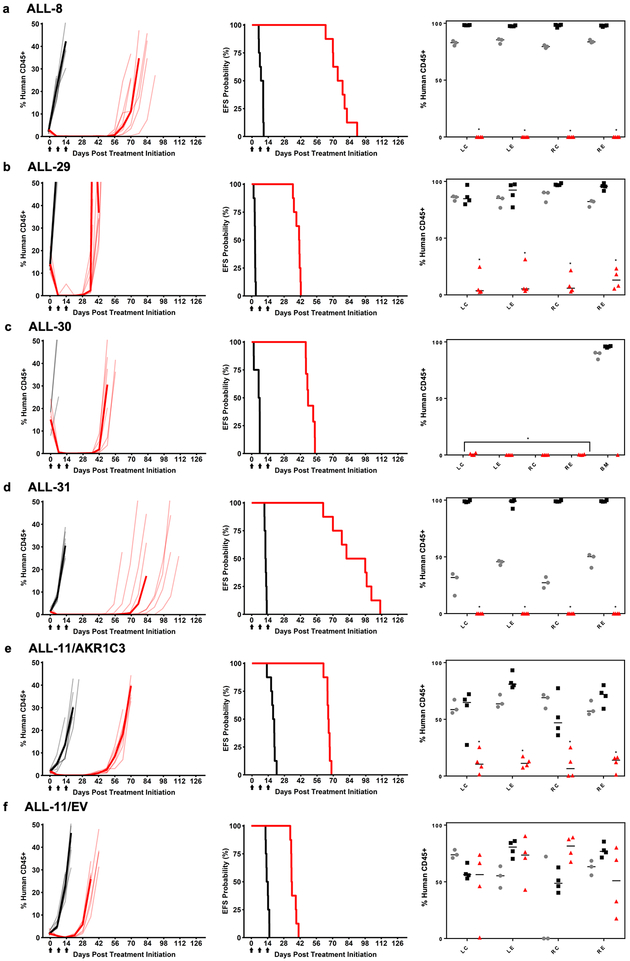
In vivo efficacy of OBI-3424 against PDX models of pediatric ALL
The in vivo efficacy of OBI-3424 was evaluated against 7 pediatric ALL PDXs (6 × T-ALL, 1 × B-ALL, Table 1), as well as a B-ALL PDX (ALL-11) that had previously been lentivirally transduced to overexpress AKR1C3 (ALL-11/AKR1C3) or empty vector control (ALL-11/EV).(14) OBI-3424 administered as a single agent every 7 days for only 3 doses significantly delayed the progression of all PDXs tested by between 17.1 and 77.8 days, including those derived from T-ALL patients who experienced fatal disease (Figure 4, Table 1, Supplementary Figure S7, Supplementary Table S13). Moreover, the EFS treated/control (T/C) values were 3.9 – 14.0 for T-ALL PDXs and 2.5 for the 2 B-ALL PDXs (ALL-28 and ALL-11/EV). The T/C value (3.5) for ALL-11/AKR1C3 was more consistent with the T-ALL PDXs, confirming the importance of AKR1C3 in the in vivo sensitivity of ALL PDXs to OBI-3424. OBI-3424 also induced regressions in 8/9 PDXs tested, with 2 PDXs achieving CRs and 6 MCRs (Table 1, Supplementary Table S13). Of note, only 2/72 mice treated with OBI-3424 were euthanized for toxicity due to reaching the pre-defined endpoint of ≥20% weight loss (Supplementary Table S13).
Table 1.
Results of in vivo OBI-3424 efficacy testing against a panel of 9 ALL PDXs
| PDX | N | Na | EFS T-C (Days) | EFS T/C | p-value | Min CD45 | Median Response | Mean BM %huCD45+ (day*) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | OBI-3424 | p-value | ||||||||
| ALL-8 | 8 | 8 | 67.3 | 8.7 | <0.001 | 0.0 | MCR | 97.9 (15) | 0 (28) | <0.0001 |
| ALL-27 | 8 | 4 | 70.6 | 11.4 | 0.008 | 0.0 | MCR | 49.8 (8,14) | 0 (28) | NS |
| ALL-29 | 8 | 8 | 38.1 | 14.0 | <0.001 | 0.013 | CR | 92.3 (8) | 10.7 (28) | <0.0001 |
| ALL-30 | 8 | 7 | 42.1 | 7.5 | <0.001 | 0.0 | MCR | 95.7 (7) | 0.25 (28) | <0.0001 |
| ALL-31 | 8 | 8 | 77.8 | 7.5 | <0.001 | 0.0 | MCR | 98.7 (14,21) | 0 (28) | <0.0001 |
| ALL-32 | 8 | 8 | 17.1 | 3.9 | <0.001 | 1.1 | SD | 94.4 (8,15) | 96.3 (22,28) | NS |
| ALL-28 | 8 | 8 | 58.2 | 2.5 | <0.001 | 0.0 | MCR | 69.5 (28) | 0.69 (28) | 0.05 |
| ALL-11/EV | 8 | 8 | 21.1 | 2.5 | <0.001 | 0.13 | CR | 66.2 (19) | 61.7 (28) | NS |
| ALL-11/1C3 | 8 | 8 | 47.2 | 3.5 | <0.001 | 0.0 | MCR | 65.8 (20,25) | 11.2 (28) | <0.0001 |
N, total number of mice entering experiment; Na, number of mice in analysis; EFS T – C, difference in median time-to-event (days) between T and C groups; EFS T/ C, ratio of median time-to-event (days) between T and C groups; P-value, between C and T EFS by Gehan-Wilcoxon test; Min CD45, average minimum huCD45% for treated group; Median response, median response evaluation (see Supplementary Methods for definitions); BM, bone marrow;
, days post treatment initiation on which BM samples were harvested. For complete in vivo response data see Supplementary Table S13.
Figure 4.
In vivo efficacy of OBI-3424 against ALL PDXs. Mice engrafted with (a) ALL-8, (b) ALL-29, (c) ALL-30, (d) ALL-31, (e) ALL-11/AKR1C3, and (f) ALL-11/EV were treated with OBI-3424 (2.5 mg/kg IP once weekly × 3 weeks, red lines) or vehicle control (black lines). The lighter hued lines represent individual mice for each treatment (vehicle or OBI-3424), whereas the darker line represents the median values for each group. Left panels, engraftment of each PDX, showing the %huCD45+ over time. Middle panels, mouse EFS (see Table 1 and Supplementary Table S13 for P-values). Arrows on the x axes represent the 3 treatments. Right panels, % infiltration of femoral bone marrow pre-treatment (gray circles), control mice at event (black squares) or OBI-3424-treated mice at Day 28 post treatment initiation (red triangles). BM, whole bone marrow; LC, left central region; LE, left endosteal region; RC, right central region; RE, right endosteal region. *P < 0.0001 comparing OBI-3424-treated and control mice at event.
While OBI-3424 induced substantial and prolonged regressions of disease as assessed by the surrogate marker of leukemic blasts in the peripheral blood (left and middle panels of Figure 4 and Supplementary Figure S7), of particular note was the observation that OBI-3424 caused profound reductions in bone marrow infiltration of the disease at Day 28 (14 days after the last OBI-3424 treatment) in 6/9 ALL PDXs (right panels of Figure 4 and Supplementary Figure S7, Table 1). Specifically, OBI-3424 caused significant reductions in bone marrow infiltration to <5% human versus mouse CD45+ cells in all bone marrow regions analyzed for 6 PDXs (ALL-8, −28, −29, −30, −31 and ALL-11/AKR1C3). Notable exceptions were ALL-27 (in which most mice were lost due to mouse-related lymphoma), ALL-32 (a PDX derived from a T-ALL patient at relapse harboring a NUP214-ABL1 translocation, Supplementary Table S3), and ALL-11/EV (a B-ALL PDX).
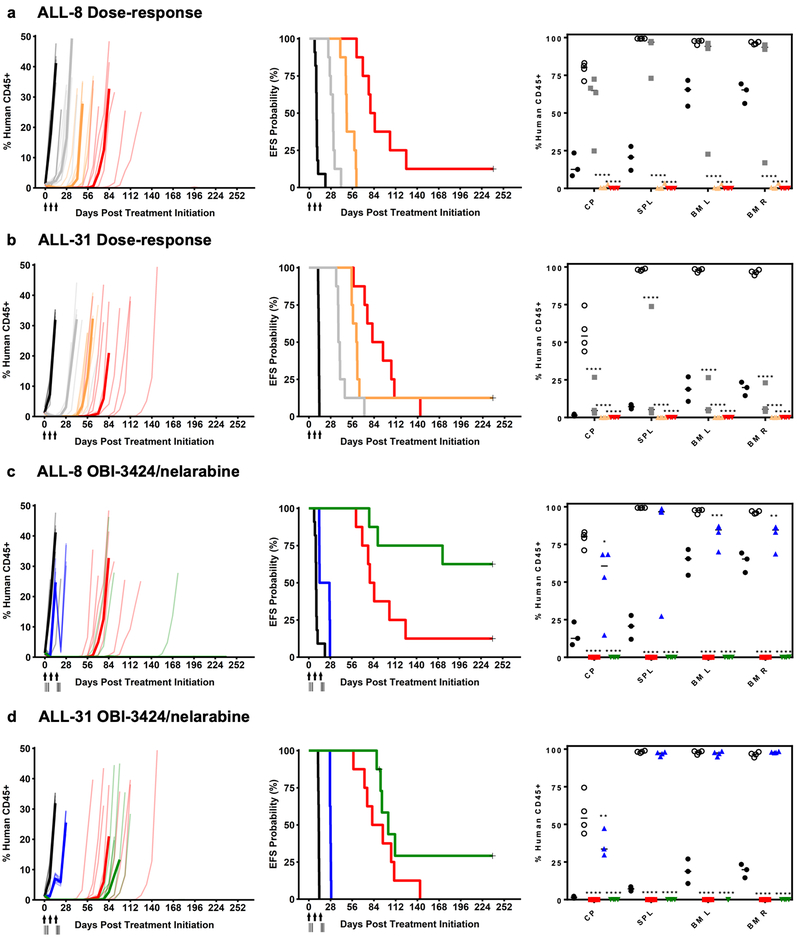
We next assessed the in vivo efficacy OBI-3424 against 3 clinically relevant criteria: (1) the possible development of drug resistance; (2) efficacy across a broad dose range; and (3) efficacy in combination with standard-of-care-drugs. First, mice engrafted with all 9 PDXs were subjected to re-treatment with OBI-3424 after each mouse had reached event (>25% huCD45+ in the PB). Despite the repeat course of treatment occurring at a much higher disease burden than the initial treatments, 7/9 PDXs achieved objective responses, with median %huCD45+ in the PB <1% for at least one week (Supplementary Figure S8, Supplementary Table S14). Second, 2 T-ALL PDXs derived from patients who experienced aggressive and fatal disease (ALL-8 and −31) were tested over a 5-fold reduction in OBI-3424 dose (0.5, 1.0 and 2.5 mg/kg). OBI-3424 at all 3 doses significantly delayed the progression of both PDXs (Figure 5a and b left and middle panels, Supplementary Table S13) and induced objective responses in ALL-31, while objective responses in ALL-8 were elicited at the 2 highest doses (1.0 and 2.5 mg/kg). The responses of both PDXs at the highest dose of OBI-3424 were remarkably similar between separate experiments (compare Figure 5a with 4a, and 5b with 4d). Moreover, significant reductions in blood (cardiac puncture), spleen and bone marrow infiltration were observed at Day 28 for ALL-31 at all 3 dose levels (Figure 5b right panel), and in ALL-8 at the 2 highest doses (Figure 5a right panel).
Figure 5.
In vivo dose response of OBI-3424 and in combination with nelarabine. (a) ALL-8 and (b) ALL-31 dose response in mice treated with vehicle control (black lines) or OBI-3424 (0.5 mg/kg, gray lines; 1.0 mg/kg, orange lines; 2.5 mg/kg, red lines). (c) ALL-8 and (d) ALL-31 OBI-3424 in combination with nelarabine in mice treated with vehicle control (black lines), nelarabine alone (blue lines), OBI-3424 alone (red lines) or OBI-3424 plus nelarabine (green lines). (a-d) Left panels show engraftment of each PDX as the %huCD45+ over time, while middle panels show the mouse EFS (see Table 1 and Supplementary Table S13 for P-values). The lighter hued lines in the left panels represent individual mice while the darker lines represent the median values for each group. Arrows indicate OBI-3424 treatment times, while vertical lines indicate nelarabine treatments. Right panels show the %huCD45+ in blood (cardiac puncture, CP), spleen (SPL) and femoral bone marrow pre-treatment (black circles), control mice at event (open circles), and Day 28 post treatment initiation in mice treated with OBI-3424 at 0.5 mg/kg (gray squares), 1.0 mg/kg (orange triangles), 2.5 mg/kg (red squares or triangles), nelarabine (blue triangles), and OBI-3424 plus nelarabine (green triangles). BM L, left bone marrow; BM R, right bone marrow. *P < 0.05, ** P < 0.01, *** P < 0.0001, **** P < 0.0001 comparing OBI-3424 treated or OBI-3424 plus nelarabine treated and control mice at event.
Third, we tested the in vivo efficacy of OBI-3424 in combination with the nucleoside analog nelarabine.(34) At its MTD nelarabine significantly delayed the progression of ALL-8 and −31 (Figure 5c and d left and middle panels, Supplementary Table S13), but did not significantly decrease organ infiltration (Figure 5c and d right panels). The single-agent efficacy of OBI-3424 was consistent with that reported above, and OBI-3424 combined with nelarabine further delayed the progression of ALL-8 (by >156 days) and ALL-31 (by 14 days) compared with OBI-3424 alone, and compared with nelarabine alone (ALL-8, >218 days; ALL-31, 75 days) (Figure 5c and d, left and middle panels, Supplementary Table S13). The OBI-3424/nelarabine combination also profoundly decreased organ infiltration of both PDXs at Day 28 compared with nelarabine alone or vehicle control (Figure 5c and d, right panels). However, since pharmacokinetic analysis was not carried out, it was not possible to exclude the possibility that the enhanced in vivo effects of the OBI-3424/nelarabine combination were due to drug-drug interactions increasing the exposure to one or both of the drugs.
DISCUSSION
OBI-3424 is a novel prodrug that selectively releases a bis-functional DNA alkylating agent upon reduction by AKR1C3 in the presence of NADPH. The precise nature of the DNA crosslinks induced by OBI-3424 are unknown, but are likely to be similar to other ethylene imine-based DNA alkylating drugs such as thioTEPA, which can alkylate the N-7 position of guanosine and the N-3 or N-7 position of adenosine to form monofunctional and bifunctional DNA alkylations leading to DNA strand breaks, ICLs and intrastrand crosslinks.(32) Release of the aziridine moiety may also result in the formation of stable adducts with guanosine. The potent induction of DNA ICLs by low nM concentrations of OBI-3424 in 2 AKR1C3high PDXs but not in an AKR1C3low PDX is consistent with AKR1C3-dependent DNA crosslinking being the major mode of cytotoxicity induced by OBI-3424 in T-ALL cells.
While AKR1C3 is known to be overexpressed in several adult malignancies,(35,36) in particular liver(13)and prostate(37,38) cancer, its relatively high expression in pediatric T-ALL indicates that OBI-3424 also represents a targeted therapy for T-ALL. While the contribution of cyclophosphamide to improving patient outcomes in pediatric ALL cannot be understated, the long-term effects of treatment with this DNA alkylating agent include reproductive problems, infertility and secondary neoplasms. Both cyclophosphamide and OBI-3424 are prodrugs, although the former is activated by hepatic enzymes with reactive metabolites disseminating into the general circulation. In contrast, OBI-3424 is activated intracellularly to produce a potent DNA alkylating compound OBI-2660, which exists as a polar salt at pH 7.4 and cannot penetrate the plasma membrane, and this property has the potential to result in reduced systemic and/or bystander toxicity.
In this study using a cohort of >2,000 primary pediatric ALL cases, AKR1C3 mRNA expression was significantly higher in T-ALL compared with B-ALL, a difference that was confirmed in a separate cohort of 90 pediatric ALL PDXs. The mechanistic basis for differential regulation of AKR1C3 expression between B-ALL and T-ALL is currently unknown, but may involve additional genes regulated by the Keap1-Nrf2-Antioxidant Response Element signaling pathway including AKR1C1, AKR1C2, AKR1C4 and NQO1.(13)
OBI-3424 exerted potent in vitro cytotoxicity against T-ALL cell lines and in vitro-cultured T-ALL PDXs, with IC50 values generally in the low nM range. When tested on the same cohort of pediatric T-ALL PDXs this degree of in vitro potency was approximately 1,000-fold greater than the hypoxia-activated DNA alkylating pre-prodrug PR-104, which is also activated under aerobic conditions by AKR1C3.(13,14) Moreover, the level of AKR1C3 expression appeared to be a more important determinant of in vitro OBI-3424 sensitivity than cell lineage, since the B-ALL PDX ALL-7 (AKR1C3high) and the T-ALL PDX ALL-42 (AKR1C3low) exhibited relative sensitivity and resistance, respectively, to OBI-3424. This inference was further supported by the significant correlation between AKR1C3 protein expression and OBI-3424 sensitivity exhibited by pediatric ALL PDXs.
When tested as a single agent in vivo OBI-3424 exerted potent efficacy, not only in the peripheral blood, but also in dramatically reducing bone marrow infiltration of T-ALL PDXs that were for the most part derived from aggressive disease. In some instances, leukemic infiltration of the femoral bone marrow was reduced to almost undetectable levels, even at 14 days following the cessation of OBI-3424 treatment. This level of single-agent efficacy compares highly favorably alongside >50 novel agents previously tested by the Pediatric Preclinical Testing Program (PPTP).(39) The importance of AKR1C3 in the in vivo response to OBI-3424 was also reinforced by utilizing a B-ALL PDX (ALL-11) that had been lentivirally transduced to stably express AKR1C3.
In contrast to the vast majority of targeted single agents initially tested by the PPTP,(39) OBI-3424 was initially tested at a dose well below its MTD. Since mice lack a functional equivalent of human AKR1C3,(33) the dose of OBI-3424 selected for our in vivo efficacy studies was extrapolated from pharmacokinetic data derived from mice and cynomolgus monkeys to mimic a dose that will be well tolerated in humans. The selected dose for NSG mice (2.5 mg/kg) was well tolerated by all treated mice, and was 220-fold less than the initial (MTD) dose used in the preclinical in vivo efficacy testing of PR-104 against ALL PDXs,(40) with both drugs being administered weekly.
Since the development of drug resistance remains a significant barrier to cancer cure, we also tested the OBI-3424 responses of previously treated ALL PDXs following relapse of the disease. While the second cycle of treatment occurred at a much higher leukemic burden than the initial treatment, OBI-3424 was able to induce remissions in the re-treated PDXs, suggesting that acute drug resistance was not induced by the original schedule of OBI-3424 treatment. Furthermore, the combination of OBI-3424 and nelarabine was also well tolerated in NSG mice and induced disease regression in 2 T-ALL PDXs that were derived from aggressive and fatal disease. Since nelarabine is FDA approved for relapsed T-ALL,(34) the combination of OBI-3424 and nelarabine could also be considered for future clinical evaluation in relapsed/refractory T-ALL. Should OBI-3424 show activity in the relapsed/refractory setting, it may be an attractive candidate for evaluation in newly diagnosed patients (e.g. by substituting it for cyclophosphamide or by adding it to nelarabine treatment blocks).
The OBI-3424 in vivo preclinical data presented in this study should only be used to inform of potential efficacy, and both its safety and efficacy are currently being evaluated in a phase ½ clinical trial for patients with hepatocellular carcinoma or castrate-resistant prostate cancer (ClinicalTrials.gov NCT03592264). Nevertheless, the cross-species pharmacokinetic comparisons (efficacy in mice, toxicity in non-human primates, and allometric extrapolation to humans) suggest that there will indeed be a therapeutic window. This premise is also supported by the vastly different preclinical toxicity profile of OBI-3424 (gastrointestinal, with no significant effects on WBC) compared with the clinical DLT of myelosuppression for both cyclophosphamide and PR-104. Given the potent in vivo efficacy of OBI-3424 against preclinical models of pediatric T-ALL, its efficacy over a broad dose range, and the excellent correlation between AKR1C3 mRNA and protein expression levels, OBI-3424 represents a promising candidate for a biomarker-driven clinical trial in relapsed/refractory T-ALL.
Supplementary Material
Statement of Translational Relevance.
In this study we investigated the efficacy of a novel prodrug, OBI-3424, in preclinical models of pediatric acute lymphoblastic leukemia (ALL). OBI-3424 is activated by the enzyme aldo-keto reductase 1C3 (AKR1C3) into a potent DNA alkylating agent. Using a large cohort of primary patient samples and patient-derived xenografts (PDXs) we showed that AKR1C3 expression was significantly higher in T-ALL than B-ALL and almost all normal human tissues. In addition, OBI-3424 exerted potent in vitro cytotoxicity against T-ALL cell lines and PDX cells. When tested in vivo using PDX models in immune-deficient mice, OBI-3424 at drug exposure levels likely to be achieved in humans profoundly reduced the bone marrow infiltration of pediatric T-ALL PDXs, and significantly delayed disease progression. OBI-3424 also caused prolongation of mouse survival when combined with nelarabine in T-ALL PDXs and represents a potential novel treatment for aggressive and chemoresistant T-ALL.
ACKNOWLEDGEMENTS
The authors thank Ms Sara Danielli for assistance in setting up the comet assay. The X-RAD 320 Biological Irradiator was provided by the Biological Resource Imaging Laboratory, Mark Wainwright Analytical Centre, UNSW Sydney. Children’s Cancer Institute Australia is affiliated with UNSW Sydney and The Sydney Children’s Hospitals Network.
Financial Support: This research was funded by grants from the National Cancer Institute (CA199222 and CA199000), and the National Health and Medical Research Council of Australia (NHMRC Fellowships APP1059804 and APP1157871 to RBL).
Footnotes
Conflict of Interests Disclosure Statement: J.D., F.M. and D.J. are employees of Ascentawits Pharmaceuticals, Ltd.; C-S.S. is an employee of OBI Pharma, Inc.; Ti.P. is a consultant of OBI Pharma, Inc.; all other authors declare no competing financial interests.
REFERENCES
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Eng J Med 2015;373(16):1541–52 doi 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 2.Pui C-H, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, et al. Childhood acute lymphoblastic leukemia: progress through collaboration. J Clin Oncol 2015;33(27):2938–48 doi 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MA, Altekruse SF, Adamson PC, Reaman GH, Seibel NL. Declining childhood and adolescent cancer mortality. Cancer 2014;120(16):2497–506 doi 10.1002/cncr.28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwig WD, Reiter A, Loffler H, Gokbuget, Hoelzer D, Riehm H, et al. Immunophenotypic features of childhood and adult acute lymphoblastic leukemia (ALL): experience of the German Multicentre Trials ALL-BFM and GMALL. Leuk Lymphoma 1994;13 Suppl 1:71–6 doi 10.3109/10428199409052679. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol 2003;21(19):3616–22 doi 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 6.Locatelli F, Schrappe M, Bernardo ME, Rutella S. How I treat relapsed childhood acute lymphoblastic leukemia. Blood 2012;120(14):2807–16 doi 10.1182/blood-2012-02-265884. [DOI] [PubMed] [Google Scholar]
- 7.Penning TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys 2007;464(2):241–50 doi 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penning TM. The aldo-keto reductases (AKRs): Overview. Chem Biol Interact 2015;234:236–46 doi 10.1016/j.cbi.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penning TM, Burczynski ME, Jez JM, Hung CF, Lin HK, Ma H, et al. Human 3alpha-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem J 2000;351(Pt 1):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamieson SM, Gu Y, Manesh DM, El-Hoss J, Jing D, Mackenzie KL, et al. A novel fluorometric assay for aldo-keto reductase 1C3 predicts metabolic activation of the nitrogen mustard prodrug PR-104A in human leukaemia cells. Biochem Pharmacol 2014;88(1):36–45 doi 10.1016/j.bcp.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Bortolozzi R, Bresolin S, Rampazzo E, Paganin M, Maule F, Mariotto E, et al. AKR1C enzymes sustain therapy resistance in paediatric T-ALL. Br J Cancer 2018;118(7):985–94 doi 10.1038/s41416-018-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson AV, Ferry DM, Edmunds SJ, Gu Y, Singleton RS, Patel K, et al. Mechanism of action and preclinical antitumor activity of the novel hypoxia-activated DNA cross-linking agent PR-104. Clin Cancer Res 2007;13(13):3922–32 doi 10.1158/1078-0432.CCR-07-0478. [DOI] [PubMed] [Google Scholar]
- 13.Guise CP, Abbattista MR, Singleton RS, Holford SD, Connolly J, Dachs GU, et al. The bioreductive prodrug PR-104A is activated under aerobic conditions by human aldo-keto reductase 1C3. Cancer Res 2010;70(4):1573–84 doi 10.1158/0008-5472.CAN-09-3237. [DOI] [PubMed] [Google Scholar]
- 14.Moradi Manesh D, El-Hoss J, Evans K, Richmond J, Toscan CE, Bracken LS, et al. AKR1C3 is a biomarker of sensitivity to PR-104 in preclinical models of T-cell acute lymphoblastic leukemia. Blood 2015;126(10):1193–202 doi 10.1182/blood-2014-12-618900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konopleva M, Thall PF, Yi CA, Borthakur G, Coveler A, Bueso-Ramos C, et al. Phase I/II study of the hypoxia-activated prodrug PR104 in refractory/relapsed acute myeloid leukemia and acute lymphoblastic leukemia. Haematologica 2015;100(7):927–34 doi 10.3324/haematol.2014.118455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKeage MJ, Gu Y, Wilson WR, Hill A, Amies K, Melink TJ, et al. A phase I trial of PR-104, a pre-prodrug of the bioreductive prodrug PR-104A, given weekly to solid tumour patients. BMC Cancer 2011;11:432 doi 10.1186/1471-2407-11-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foehrenbacher A, Patel K, Abbattista MR, Guise CP, Secomb TW, Wilson WR, et al. The Role of Bystander Effects in the Antitumor Activity of the Hypoxia-Activated Prodrug PR-104. Front Oncol 2013;3:263 doi 10.3389/fonc.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ. Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet 1991;20(3):194–208 doi 10.2165/00003088-199120030-00002. [DOI] [PubMed] [Google Scholar]
- 19.Lock RB, Liem N, Farnsworth ML, Milross CG, Xue C, Tajbakhsh M, et al. The nonobese diabetic/severe combined immunodeficient (NOD/SCID) mouse model of childhood acute lymphoblastic leukemia reveals intrinsic differences in biologic characteristics at diagnosis and relapse. Blood 2002;99(11):4100–8. [DOI] [PubMed] [Google Scholar]
- 20.Dolai S, Sia KC, Robbins AK, Zhong L, Heatley SL, Vincent TL, et al. Quantitative phosphotyrosine profiling of patient-derived xenografts identifies therapeutic targets in pediatric leukemia. Cancer Res 2016;76:2766–77 doi 10.1158/0008-5472.CAN-15-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanagan JU, Atwell GJ, Heinrich DM, Brooke DG, Silva S, Rigoreau LJ, et al. Morpholylureas are a new class of potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase (AKR1C3). Bioorg Med Chem 2014;22(3):967–77 doi 10.1016/j.bmc.2013.12.050. [DOI] [PubMed] [Google Scholar]
- 22.Khaw SL, Suryani S, Evans K, Richmond J, Robbins A, Kurmasheva RT, et al. Venetoclax responses of pediatric ALL xenografts reveal sensitivity of MLL-rearranged leukemia. Blood 2016;128(10):1382–95 doi 10.1182/blood-2016-03-707414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49(8):1211–8 doi 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9 doi 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106 doi 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res 2007;67(9):4482–90 doi 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- 29.Suryani S, Carol H, Chonghaile TN, Frismantas V, Sarmah C, High L, et al. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clin Cancer Res 2014;20(17):4520–31 doi 10.1158/1078-0432.CCR-14-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spanswick VJ, Craddock C, Sekhar M, Mahendra P, Shankaranarayana P, Hughes RG, et al. Repair of DNA interstrand crosslinks as a mechanism of clinical resistance to melphalan in multiple myeloma. Blood 2002;100(1):224–9. [DOI] [PubMed] [Google Scholar]
- 31.Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer 2007;49(7):928–40 doi 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 32.van Maanen MJ, Smeets CJ, Beijnen JH. Chemistry, pharmacology and pharmacokinetics of N,N’,N” -triethylenethiophosphoramide (ThioTEPA). Cancer Treat Rev 2000;26(4):257–68 doi 10.1053/ctrv.2000.0170. [DOI] [PubMed] [Google Scholar]
- 33.Velica P, Davies NJ, Rocha PP, Schrewe H, Ride JP, Bunce CM. Lack of functional and expression homology between human and mouse aldo-keto reductase 1C enzymes: implications for modelling human cancers. Mol Cancer 2009;8:121 doi 10.1186/1476-4598-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunsmore KP, Devidas M, Linda SB, Borowitz MJ, Winick N, Hunger SP, et al. Pilot study of nelarabine in combination with intensive chemotherapy in high-risk T-cell acute lymphoblastic leukemia: a report from the Children’s Oncology Group. J Clin Oncol 2012;30(22):2753–9 doi 10.1200/JCO.2011.40.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller VL, Lin HK, Murugan P, Fan M, Penning TM, Brame LS, et al. Aldo-keto reductase family 1 member C3 (AKR1C3) is expressed in adenocarcinoma and squamous cell carcinoma but not small cell carcinoma. Int J Clin Exp Pathol 2012;5(4):278–89. [PMC free article] [PubMed] [Google Scholar]
- 36.Rizner TL, Smuc T, Rupreht R, Sinkovec J, Penning TM. AKR1C1 and AKR1C3 may determine progesterone and estrogen ratios in endometrial cancer. Mol Cell Endocrinol 2006;248(1–2):126–35 doi 10.1016/j.mce.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Fung KM, Samara EN, Wong C, Metwalli A, Krlin R, Bane B, et al. Increased expression of type 2 3alpha-hydroxysteroid dehydrogenase/type 5 17beta-hydroxysteroid dehydrogenase (AKR1C3) and its relationship with androgen receptor in prostate carcinoma. Endocr Relat Cancer 2006;13(1):169–80 doi 10.1677/erc.1.01048. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Lou W, Zhu Y, Yang JC, Nadiminty N, Gaikwad NW, et al. Intracrine androgens and AKR1C3 activation confer resistance to enzalutamide in prostate cancer. Cancer Res 2015;75(7):1413–22 doi 10.1158/0008-5472.CAN-14-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones L, Carol H, Evans K, Richmond J, Houghton PJ, Smith MA, et al. A review of new agents evaluated against pediatric acute lymphoblastic leukemia by the Pediatric Preclinical Testing Program. Leukemia 2016;30(11):2133–41 doi 10.1038/leu.2016.192. [DOI] [PubMed] [Google Scholar]
- 40.Houghton PJ, Lock R, Carol H, Morton CL, Phelps D, Gorlick R, et al. Initial testing of the hypoxia-activated prodrug PR-104 by the pediatric preclinical testing program. Pediatr Blood Cancer 2011;57(3):443–53 doi 10.1002/pbc.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.