Abstract
Objectives:
Among women initiating 1st-line non-nucleoside-reverse-transcriptase-inhibitor-(NNRTI)-based-ART with-and-without a history of single-dose-nevirapine-(sdNVP)±zidovudine±lamivudine (ZDV±3TC) for prevention-of-mother-to-child-HIV-transmission (PMTCT), we hypothesized that pre-ART HIV-drug-resistance would be associated with virologic failure
Design/Methods:
In a prospectively enrolled study, three genotypic drug-resistance assays (oligonucleotide-ligation-assay (OLA), consensus sequencing, and next-generation-sequencing by Illumina) were retrospectively performed to detect pre-ART drug-resistance. Minority or majority drug-resistant variants identified in pre-ART RNA and/or DNA, a history of antiretrovirals for PMTCT, and other risk factors were assessed for association with virologic failure.
Results:
Failure occurred in 38/169 (22.5%) women, and was associated with pre-ART drug-resistance detected by any assay (OLA of plasma or PBMC, consensus sequencing of PBMC and/or plasma, and next-generation-sequencing of PBMC at frequencies ≥10% and as minority variants; all P<0.0001). Failure was also associated with PMTCT using sdNVP+ZDV±3TC, but not sdNVP-only; however, the longer time-interval between PMTCT and ART-initiation observed for sdNVP-only women showed no interaction with failure. Viral loads and OLA of PBMC in longitudinal specimens demonstrated rapid failure and emergence of drug-resistance, particularly among sdNVP+ZDV±3TC-experienced women with pre-ART drug-resistant minority variants by next-generation-sequencing but without drug-resistance by OLA or consensus sequencing.
Summary/Conclusions:
Pre-ART drug-resistance was detected similarly by OLA of PBMC or plasma and by consensus sequencing, and was associated with virologic failure soon after initiation of 1st-line NVP-based ART. A history of sdNVP+ZDV±3TC for PMTCT or minority variants detected by next-generation-sequencing identified additional women with failure. These findings emphasize the value of assessing individual antiretroviral history, particularly non-suppressive antiretrovirals with ≥2 drug classes, and testing for pre-ART drug-resistance, including minority variants.
Keywords: HIV drug resistance, ART, prevention of mother to child transmission, quasispecies, nevirapine, women
Introduction
Access to antiretroviral-therapy (ART) for human immunodeficiency virus type-1 (HIV)-infected persons has increased markedly in resource-limited settings (RLS) [1]. While ART decreases morbidity and mortality [2] and new infections [3], access to ART is associated with increased “acquisition” of drug-resistance and transmission of drug-resistance [4]. Pre-ART drug-resistance is rising in Africa [5–7], especially in young Kenyan women [8]. Increases in virologic failure associated with pre-ART drug-resistance [6,9–10] warrants expectant management to maintain ART benefits and to minimize forward transmission of drug-resistant variants.
Peripartum single-dose nevirapine (sdNVP) has frequently been used in resource-limited settings to prevent mother-to-child-transmission (PMTCT). While moderately effective [11], sdNVP can select drug-resistance that impacts subsequent ART efficacy [10,12]. Pre-partum zidovudine (ZDV) added to sdNVP [13], and later combined with a 5–7 day postpartum “tail” of ZDV and lamivudine (3TC) [14] to prevent drug-resistance selection in women, dramatically reduced MTCT [15–16], but was complex to administer and contributed to growing rates of pre-ART drug-resistance [17]. Furthermore, NVP-resistant viruses demonstrate cross-resistance to other non-nucleoside reverse-transcriptase inhibitors (NNRTIs) including efavirenz (EFV) [18], which became the WHO-preferred 1st-line ART regimen for individuals ≥2 years of age in 2013, including pregnant women [19].
To evaluate the role of pre-ART drug-resistance in virologic failure of women, adult Kenyan females with and without a history of sdNVP who qualified for initial 1st-line NNRTI-based-ART were prospectively enrolled and followed for one year. Pre-ART drug-resistance was identified retrospectively by consensus sequencing, and a relatively low-cost oligonucleotide-ligation-assay (OLA) previously validated to detect transmitted or sdNVP-selected pre-ART drug-resistance associated with failure [6,20]. Our study included primary, secondary, and post-hoc hypotheses; Primary: Pre-ART drug-resistance detected by OLA at four codons associated with resistance to N/NRTIs would predict virologic failure as well or better than consensus sequencing; Secondary #1: sdNVP-experienced women without pre-ART drug-resistance by OLA and consensus sequencing would rapidly select N/NRTI-associated mutations and experience failure (detected by frequent monitoring of plasma viral load) within a few months of ART initiation; Secondary #2: Archived mutations not detected as pre-ART drug-resistance by OLA that contribute to failure can be identified by longitudinal OLA genotyping of PBMC; and Post-hoc #1: sdNVP-experienced women who failed without pre-ART drug-resistance by OLA and consensus sequencing would harbor pre-ART drug-resistant minority variants detectable by next-generation-sequencing
Methods
HIV-infected women ≥18 years-old initiating 1st-line-ART entered the study at Coptic Hope Center for Infectious Diseases, an HIV treatment clinic in Nairobi [21] (Figure 1). ART-experienced, seriously ill, or individuals planning to move within a year study were excluded. Participants provided informed consent, as approved by Human Subjects’ Ethics Boards (Seattle #:11676 and Kenya #:P209/08/2008). Socio-demographic and clinical data from prior to ART initiation were collected: age, CD4 T-lymphocyte counts, and prior exposure to peripartum antiretrovirals for PMTCT (by pharmacy records and self-report), including sdNVP, 3rd-trimester ZDV and/or post-partum ZDV±3TC (sdNVP±ZDV±3TC). Women were prescribed 1st-line ART containing NVP or EFV plus ZDV+3TC; EFV was used when NVP was contraindicated or not tolerated. Blood plasma and peripheral blood mononuclear cells (PBMC) were collected prior to ART initiation and every two months through month-12 of ART.
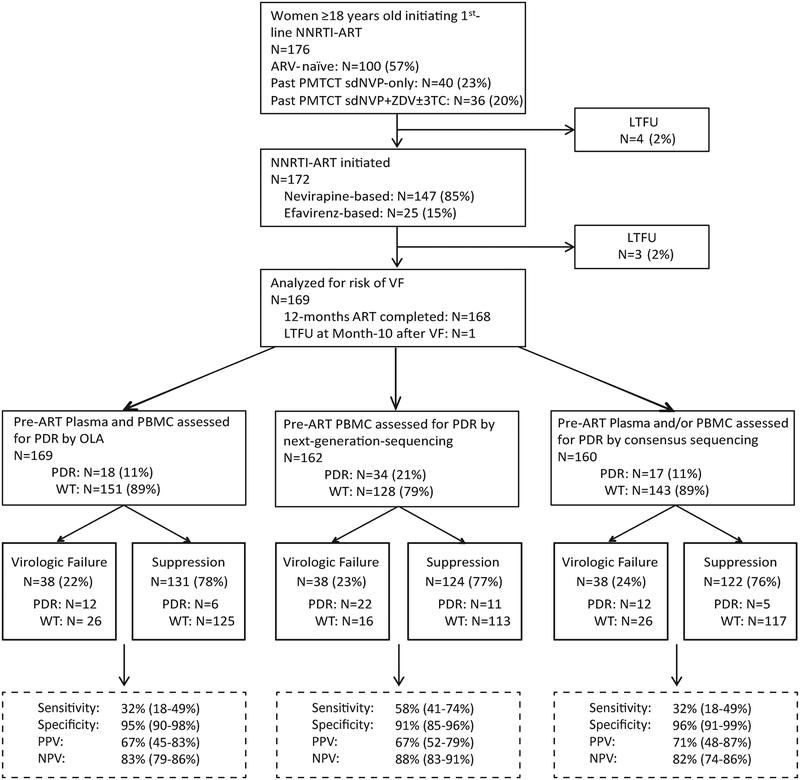
Figure 1. Schema of study of women initiating 1st-line non-nucleoside reverse transcriptase inhibitor (NNRTI)-based ART with retrospective assessments of pre-ART HIV drug-resistance (PDR), and virologic outcome at month-12 of study follow-up.
Enrolled participants either had no history of antiretroviral (ARV) treatment or a history of single-dose nevirapine (sdNVP) with or without prepartum zidovudine (ZDV) and/or post-partum ZDV ± lamivudine (3TC) for prevention-of-mother-to-child-transmission (PMTCT). Specimens were retrospectively assessed for PDR by an oligonucleotide ligation assay (OLA), consensus sequencing and next-generation-sequencing using the Illumina Miseq platform. Study outcomes, virologic failure (plasma viral load ≥400 HIV RNA copies/mL) or virologic suppression (plasma viral load <400 copies/mL) are shown, with contributions of participants with PDR vs. wild-type (WT) by each method of drug-resistance genotyping shown for each outcome. Diagnostic sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) have been calculated and presented for OLA, consensus sequencing, and next-generation-sequencing. Participants lost-to-follow-up (LTFU) are shown.
Pre-ART drug-resistance was assessed in plasma and PBMC by OLA, in plasma and/or PBMC by consensus sequencing [22] and in PBMC by next-generation-sequencing using the Illumina MiSeq platform. PBMC DNA and plasma RNA had HIV templates quantified by a CLIA-certified in-house quantitative PCR (qPCR) targeting the HIV gag [23]. An estimated 100–300 amplifiable copies were submitted to OLA designed to detect mutations at pol reverse transcriptase codons 103, 181, 184, and 190. OLA standard curves allowed quantification of mutant frequencies within participants’ viral quasispecies. If multiple mutant codons were detected within an individual’s quasispecies, the value from the codon with the highest mutant frequency was considered “peak” and used for analysis. The amplicon from RNA and/or DNA also underwent consensus sequencing. Nucleotide sequences are available in the NCBI Genbank under accession numbers MH509760-MH509936.
Next-generation-sequencing, as described [8], evaluated pre-ART drug-resistant minority variants in HIV pol below the limit-of-detection by OLA (2%) and consensus sequencing (15–25%). Briefly, two regions of pol encoding reverse transcriptase were amplified from PBMC DNA by nested PCR with a high-fidelity enzyme and 2nd-round primers containing Illumina sequencing adapters. These amplicons were purified, pooled, and sequenced bidirectionally over 300 base-pairs on an Illumina Miseq. Raw sequence reads were processed by a custom variant-calling analysis pipeline (downloadable here), resulting in tables containing amino acid variants and associated frequencies. PCR and sequencing error rates at each base were assessed by an in-house Perl script to estimate genuine pre-ART drug-resistant populations (downloadable here). The average mismatch error rate ranged from 0.58–0.68%, suggesting a conservative 1% cut-off frequency. To exclude minority variants due to Illumina “index hopping”, all minority variants were evaluated by phylogenetic clustering to participants’ consensus sequences, and non-clustering variants were discarded. Next-generation-sequencing datasets are available in the NCBI Sequence Read Archive under accession number SRP154562.
Pre-ART drug-resistance, hypermutation, and HIV subtypes were defined by the Stanford HIV Database [24]. Next-generation-sequencing pre-ART drug-resistant mutants encoded by hypermutation were excluded from analysis. When pre-ART drug-resistance was not detected by any assay or conferred a Stanford penalty score <10, the participant’s HIV was defined as wild-type. Subtypes were determined from consensus sequences, or next-generation-sequencing data when consensus sequences were unavailable.
Plasma viral load (RealTime HIV, Abbott Molecular) at month-12 of ART or final specimen identified subjects with virologic failure, defined as ≥400 HIV RNA copies (c)/mL plasma. In women with failure and/or pre-ART drug-resistance by OLA, longitudinal plasma RNA and PBMC DNA specimens collected at 2-month intervals between months 0–12 had plasma viral load measured and OLA performed, respectively, to identify the onset of failure and detect emerging drug-resistance. Consensus sequencing of plasma at failure assessed accumulated drug-resistance.
Data Analysis
Amongst participants stratified by prior ARV experience for PMTCT, Welch two-sample t-tests compared continuous enrollment characteristics (CD4 count, enrollment plasma viral load, the interval between PMTCT and ART initiation), while chi-square tests compared binary variables (pre-ART drug-resistance prevalence by OLA). Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated for each genotypic resistance assay using the MedCalc Diagnostic test evaluation calculator for Windows, version 15.0 (MedCalc Software, Ostend, Belgium). Pre-ART risk factors including plasma viral load, CD4 T-cell count, prior antiretroviral-experience for PMTCT, NNRTI (NVP or EFV) used for ART, and pre-ART drug-resistance by OLA, consensus sequencing, and next-generation-sequencing, were assessed for associations with virologic failure using both univariable and multivariable Cox Proportional-Hazards regression with robust variance estimates. Pre-ART drug-resistance was assessed as both a binary and categorical independent variable, the latter stratified by mutant frequency for pre-ART drug-resistance by OLA and next-generation-sequencing. Multivariable regression analysis included the covariates plasma viral load, prior antiretroviral-experience, pre-ART drug-resistance (by OLA, consensus sequencing, or next-generation-sequencing), and NNRTI used for ART. Kaplan-Meier survival analyses show time from ART initiation to failure among all women stratified by prior antiretroviral-experience, among women with next-generation-sequencing results stratified by pre-ART drug-resistant mutant frequency, and in a second analysis with women with minority variants further stratified by exposure to sdNVP+ZDV±3TC. The survival distributions were compared across these groups using pairwise log-rank tests with Benjamini-Hochberg adjustment for multiple comparisons. All statistical analyses were performed in RStudio (version 1.0.44).
Results
Study Population
In total, 176 Kenyan women enrolled and 169 completed the study. Seven (4%) women total were lost-to-follow-up, including four prior to ART initiation and three during the study (at study months 2, 2, and 4); these were excluded from analyses (Figure 1). NVP- and EFV-based ART were prescribed to 144 and 25 women, respectively. The 169 women analyzed were infected with HIV subtypes A (n=64, 37.9%), CRF01_AE (n=39, 23.1%), C (n=14, 8.3%), or D (n=40, 23.7%). sdNVP was used for PMTCT by 72/169 (43%) women at a median of 424 days (IQR 169–784) prior to ART initiation. Among the sdNVP-experienced women, pre-partum ZDV or stavudine (d4T) was taken by 34/72 (47%) women for a median duration of 90 days (IQR 60–90), and post-partum ZDV±3TC was taken by 14 of these women for a median duration of 7 (IQR 7–7) days. sdNVP±ZDV±3TC-experienced vs. antiretroviral-naïve women had lower enrollment age (mean (95% CI) years, 30.3 (29.1–31.5) vs. 36.9 (35.1–38.7), P<0.0001) and higher enrollment CD4 T-cell count (mean (95% CI) cells/uL, 188.8 (168.9–208.7) vs. 159.6 (143.9–175.3), P=0.02), while enrollment plasma viral load (log10 c/mL, 4.8 (4.6–5.0) vs. 4.7 (4.4–4.9), P=0.44) were similar. The interval between PMTCT and ART initiation was greater among those who took sdNVP alone vs. sdNVP+ZDV±3TC (mean (95% CI) days, 635 (510–761) vs. 349 (241–457), P<0.001).
Pre-ART drug-resistance by OLA, consensus sequencing and next-generation-sequencing
OLA assessed pre-ART drug-resistance on an estimated median of 471 (IQR 121–551) and 131 (IQR 108–195) HIV RNA (quantified by gag qPCR in only sdNVP-experienced; otherwise estimated from plasma viral load) and DNA templates (all quantified by gag qPCR), respectively. OLA detected pre-ART drug-resistance at codons K103N, Y181C, M184V, and/or G190A in the plasma and/or PBMC samples of 18/169 (10.7%) women (Supplemental Table 1). Pre-ART drug-resistance was more prevalent in sdNVP-experienced vs. antiretroviral-naïve women: 12/72 (16.7%) vs. 6/97 (6.2%), prevalence-ratio (PR) (95% CI) = 2.69 (1.06–6.84), P=0.03. Among sdNVP-experienced women, the prevalence of pre-ART drug-resistance by OLA was lower in those who received sdNVP+ZDV±3TC (4/34, 11.8%) vs. sdNVP alone (8/38, 21.1%), but this difference was not statistically significant, PR (95% CI) = 0.56 (0.18–1.69), P=0.29).
In women with pre-ART drug-resistance by OLA, the median pre-ART mutant frequency was 100% (IQR 17.5–100) in plasma and 85% (IQR 37–92) in PBMC. The six antiretroviral-naïve women with pre-ART drug-resistance had a median frequency of 100% (IQR 89–100) in plasma and 81% (IQR 60–88.5) in PBMC and none had peak pre-ART drug-resistance at frequencies 2–9%. In contrast, 2/12 (16.7%) sdNVP-experienced women had peak mutants of frequencies 2–9%.
Consensus sequencing detected 22/26 (84.6%) pre-ART drug-resistance mutations identified by OLA in 15/18 women but missed four mutations with a median OLA frequency of 7% (range 2–14). In the 18 women with pre-ART drug-resistance by OLA, consensus sequencing detected 7 additional mutant codons associated with intermediate-to-high-level resistance to N/NRTIs, including K70R, L74I, A98G, K101E, and T215Y. Consensus sequencing identified one additional participant with intermediate-to-high-level pre-ART drug-resistance (M230I), and 9 women with polymorphisms conferring potential or low-level resistance.
Next-generation-sequencing was performed on pre-ART PBMC DNA, a compartment more likely to harbor archived drug-resistance [25], from 162/169 (95.9%) women (seven had insufficient specimen/amplification), including all 38 participants that experienced virologic failure. A median of 1,147 (IQR 793–1,441) HIV DNA templates per specimen were sequenced, as estimated by gag qPCR, producing a median of 129 (IQR 90–190) sequence reads per input viral template. Next-generation-sequencing confirmed 25/26 (96.2%) DR mutations identified by OLA and 30/30 detected by consensus sequencing (Supplemental Table 1). The single mutant population detected by OLA but not next-generation-sequencing was Y181C at a frequency of 4%. Next-generation-sequencing detected 26 minority variants not detected by consensus sequencing or OLA, including 6/26 (26%) at OLA codons: K103N (n=3), M184V (n=2), and G190A (n=1)). These 26 minority variants conferred low-to-high-level resistance in 22 women, including 14/22 (64%) who were wild-type by OLA and consensus sequencing. Notably, 9 of the latter women were sdNVP-experienced, and six also ZDV±3TC-experienced. The minority variants had a median frequency of 2.0% (IQR 1.3–3.8) within participants’ viral quasispecies. One of these variants exceeded the 1–9% threshold (K70R=24%) but was considered minority as it was not detected by consensus sequencing.
Pre-ART Risk Factors associated with virologic failure
Among 169 participants, 38 (22.5%) experienced virologic failure by study month-12, including 34/144 (24%) prescribed NVP- and 4/25 (16%) prescribed EFV-based ART (P=0.34). At study enrollment, women with and without failure were similar in age, plasma viral load, and CD4 T-cell counts (Table 1). In univariable Cox regression analyses, pre-ART drug-resistance in plasma and/or PBMC by OLA, consensus sequencing or next-generation-sequencing were associated with increased risk of failure; Hazard Ratio (HR) (95% CI): 7.11 (3.32–15.23), 7.59 (3.50–16.46), 9.50 (4.95–18.22), respectively, all P<0.0001). Mutations K103N and/or G190A identified all women with pre-ART drug-resistance by OLA that experienced failure. Pre-ART drug-resistance by OLA at frequencies ≥10% in plasma and/or PBMC were associated with increased risk of failure compared to wild-type; HR (95% CI): 9.28 (4.20–20.45), P<0.0001. Only two participants had pre-ART drug-resistance by OLA at peak frequencies between 2–9% and neither had failure. In contrast, detection of minority variants by next-generation-sequencing (Table 1) with Stanford mutation penalty scores ≥10 was associated with increased risk of failure compared to wild-type, HR (95% CI): 7.05 (3.21–15.48), P<0.0001.
Table 1.
Demographic, Immunologic, and Virologic Parameters at Enrollment Associated with Virologic Suppression vs. Failure (>400c/mL) at month-12 of ART
| Risk Factor | Plasma HIV RNA at 12-month ART | Univariable Cox Regression | Multivariable with PDR by OLA | Multivariable with PDR by CS | Multivariable with PDR by NGS | ||
|---|---|---|---|---|---|---|---|
| Total N= | <400 c/mL; N=131 |
≥400 c/mL; N=38 |
HR (95% CI), P value |
HR (95% CI), P value |
HR (95% CI), P value |
HR (95% CI), P value |
|
| Age, mean years (95% CI) | -- | 34 (33–36) | 33 (30–36) | 0.98 (0.94–1.02), 0.33 | -- | -- | -- |
| CD4 lymphocyte count, mean cell/uL (95% CI) | -- | 170 (156–184) | 179 (151–208) | 1.00 (1.00–1.00), 0.66 | -- | -- | -- |
| Plasma HIV RNA, mean log c/mL (95% CI) | -- | 4.70 (4.53–4.88) | 4.88 (4.66–5.10) | 1.17 (0.82–1.69), 0.38 | 1.27 (0.80–2.01), 0.31 | 1.12 (0.70–1.77), 0.64 | 1.44 (0.85–2.44), 0.18 |
| Time interval between PMTCT and ART, mean days (95% CI) | -- | 519 (418–620) | 454 (262–646) | 1.00 (1.00–1.00), 0.45 | -- | -- | -- |
| HIV Subtype | 159 | -- | -- | -- | -- | -- | -- |
| A | 64 | 48 (75%) | 16 (25%) | reference | -- | -- | -- |
| C | 14 | 12 (86%) | 2 (14%) | 0.52 (0.12–2.25), 0.38 | -- | -- | -- |
| D | 40 | 30 (75%) | 10 (20%) | 0.96 (0.44–2.09), 0.91 | -- | -- | -- |
| G | 2 | 1 (50%) | 1 (50%) | 2.30 (0.33–15.89), 0.40 | -- | -- | -- |
| CRF01_AE_ | 39 | 30 (77%) | 9 (23%) | 0.90 (0.40–2.05), 0.80 | -- | -- | -- |
| ART regimen prescribed | 169 | -- | -- | -- | -- | -- | -- |
| Nevirapine-based 1st-line ART, N (%) | 144 | 110 (76%) | 34 (24%) | reference | reference | reference | reference |
| Efavirenz-based 1st-line ART, N (%) | 25 | 21 (84%) | 4 (16%) | 0.64 (0.23–1.79), 0.34 | 1.09 (0.33–3.66), 0.88 | 1.12 (0.35–3.65), 0.85 | 1.05 (0.33–3.34), 0.94 |
| History of ARV | 169 | -- | -- | ||||
| Antiretroviral-naïve, N (%) | 97 | 80 (82%) | 17 (18%) | reference | reference | reference | reference |
| sdNVP±ZDV±3TC for PMTCT, N (%) | 72 | 51 (71%) | 21 (29%) | 1.87 (0.99–3.52), 0.06 | -- | -- | -- |
| History of sdNVP only, N (%) | 38 | 29 (76%) | 9 (24%) | 1.43 (0.64–3.18), 0.38 | 0.87 (1.15–2.19), 0.76 | 0.68 (0.26–1.74), 0.42 | 0.68 (0.28–1.65), 0.40 |
| History of sdNVP+ZDV±3TC, N (%) | 34 | 22 (65%) | 12 (35%) | 2.43 (1.15–5.14), 0.02 | 2.63 (1.26–5.47), <0.01 | 2.18 (1.04–4.54), 0.04 | 1.70 (0.80–3.62), 0.17 |
| Assay platforms to detect PDR | |||||||
| OLA test results (Plasma and/or PBMC) | 169 | -- | -- | -- | -- | -- | -- |
| WT: no PDR or frequency <2% quasispecies, N (%) | 151 | 125 (83%) | 26 (17%) | reference | reference | -- | -- |
| PDR at any frequency, 2–100%, N (%) | 18 | 6 (33%) | 12 (67%) | 7.11 (3.32–15.23), <0.0001 | 10.86 (5.31–22.20), <0.0001 | -- | -- |
| Low, 2–9%, N (%) | 2 | 2 (100%) | 0 (0%) | -- | -- | -- | -- |
| Moderate-to-high, 10–100%, N (%) | 16 | 4 (25%) | 12 (75%) | 9.28 (4.20–20.45), <0.0001 | -- | -- | -- |
| CS (Plasma and/or PBMC) | 160 | -- | -- | -- | -- | -- | -- |
| WT: no PDR mutations, N (%) | 143 | 117 (82%) | 26 (18%) | reference | -- | reference | -- |
| PDR: any codon with a Stanford score ≥10, N (%) | 17 | 5 (29%) | 12 (71%) | 7.59 (3.50–16.46), <0.0001 | -- | 13.03 (6.35–26.72), <0.0001 | -- |
| PDR at OLA codons, N (%) | 15 | 3 (20%) | 12 (80%) | 10.40 (4.61–23.44), <0.0001 | -- | -- | -- |
| PDR at non-OLA codons, N (%) | 2 | 2 (100%) | 0 (0%) | -- | -- | -- | -- |
| NGS test results (PBMC)a | 162 | -- | -- | -- | -- | -- | -- |
| WT: defined as Stanford score <10, N (%) | 129 | 113 (88%) | 16 (12%) | reference | -- | -- | reference |
| PDR: Stanford score ≥10, frequency 1–100% quasispecies, N (%) | 33 | 11 (33%) | 22 (67%) | 9.50 (4.95–18.22), <0.0001 | -- | -- | -- |
| Low, 1–9%, N (%) | 17 | 7 (41%) | 10 (59%) | 7.05 (3.21–15.48), <0.0001 | -- | -- | 7.57 (3.35–17.08), <0.0001 |
| Moderate-to-high, 10–100%, N (%) | 16 | 4 (25%) | 12 (75%) | 13.66 (5.90–31.70), <0.0001 | -- | -- | 16.14 (6.93–37.59), <0.0001 |
excludes sequences with evidence of G-to-A transitions indicative of APOBEC hypermutations statistically significant Cox Proportional-Hazard Ratios shown in bold; “--” indicates that not available or not performed
Abbreviations: ART=antiretroviral (ARV) therapy; PDR=pre-ART drug-resistance; OLA=oligonucleotide-ligation-assay; CS=consensus sequencing; NGS=next-generation-sequencing; c/mL=HIV RNA copies/milliliter of plasma; HR (95%CI)=Cox proportional hazards ratio (95% confidence interval); PMTCT=prevention of mother-to-child-transmission; sdNVP=single-dose nevirapine; ZDV=peripartum zidovudine; 3TC=peripartum lamivudine; PBMC=peripheral blood mononuclear cells; WT=wild-type.
Compared to OLA (31.6% (17.5%−48.7%)) and consensus sequencing (31.6% (17.5%−48.7%)), next-generation-sequencing appeared to detect pre-ART drug-resistance in women with virologic failure with greater sensitivity (57.9% (40.8%−73.7%)) (Figure 1). However, specificity of next-generation-sequencing (91.1% (84.7%−95.5%) was slightly less than OLA (95.4% (90.3–98.3%)) and consensus sequencing (95.9% (90.7–98.7%)). Overall, OLA, consensus sequencing, and next-generation-sequencing had similar positive (66.7% (44.6 – 83.3%) vs 70.6% (47.5–86.5%) vs 66.7% (51.7–78.9%), respectively) and negative (82.8% (79.4–85.7%) vs 81.8% (73.6–86.4%) vs 87.6% (82.9–91.2%), respectively) predictive values (Figure 1).
sdNVP-experienced vs. antiretroviral-naïve women had increased risk of virologic failure, but this was not statistcally significant, HR (95% CI): 1.87 (0.99–3.52), P=0.06 (Table 1). Compared to antiretroviral-naïve participants, a statistically significant increase in failure was observed in women who took sdNVP+ZDV±3TC but not in those who took sdNVP alone (HR (95% CI): 2.43 (1.15–5.14), P=0.02 and 1.43 (0.64–3.18), P=0.38, respectively. However, a comparison of the time interval between PMTCT and ART initiation found no difference in risk of failure between women exposed to sdNVP-only versus sdNVP+ZDV±3TC; HR (95% CI): 1.00 (1.00–1.00), P=0.45). Previous sdNVP+ZDV±3TC experience remained associated with failure in multivariable Cox regression analyses that included the covariates: enrollment plasma viral load, NVP or EFV-based ART, and pre-ART drug-resistance by OLA (HR (95% CI), 2.63 (1.26–5.47), P<0.01) or consensus sequencing (HR (95%CI) 2.18 (1.04–4.54), P=0.04). This indicated potential additional sources of risk of failure in sdNVP+ZDV±3TC-experienced women and precipitated the analysis of minority variants by next-generation-sequencing. Indeed, adjusting for pre-ART drug-resistance by next-generation-sequencing at frequencies 1–9% and 10–100% attenuated the association between prior sdNVP+ZDV±3TC-experience and risk of virologic failure (HR (95% CI), 1.70 (0.80–3.62), P=0.17), compared to models adjusting for pre-ART drug-resistance by OLA or consensus sequencing.
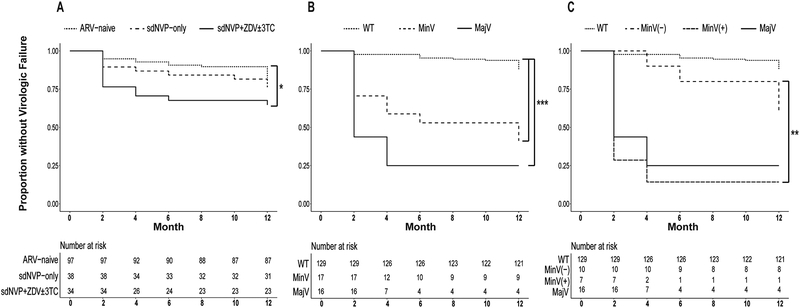
Kaplan-Meier survival analyses, showing the proportion of participants without virologic failure, are presented for all women stratified by history of antiretrovirals for PMTCT (Panel A), for women with next-generation-sequencing data available stratified by pre-ART drug-resistance category (Panel B), and for women with next-generation-sequencing data by pre-ART drug-resistance category with those with minority variants further stratified by exposure to ZDV±3TC for PMTCT (Panel C) (Figure 2). Increased virologic failure was observed in women with a history of sdNVP+ZDV±3TC for PMTCT (Panel A, P=0.05) and with pre-ART drug-resistance as both minority variants and majority variants (Panel B, both P<0.0001). While sample sizes were small, among women with minority variants exposure to ZDV±3TC was associated with increased failure compared to sdNVP-only and antiretroviral-naïve women, P=0.01 (Panel C).
Figure 2. Kaplan-Meier survival analysis.
Survival probabilities (i.e., proportion without virologic failure) during 12 months of non-nucleoside reverse transcriptase inhibitor-based ART (NNRTI-ART) of all women stratified by history of antiretroviral (ARV) treatment for prevention-of-mother-to-child-transmission (PMTCT) (Panel A), and by pre-ART drug-resistance category (wild-type (WT), minority variants (MinV), majority variants (MajV)) (Panel B). Women with minority variants were further stratified into those with (MinV(+)) and without (MinV(−)) exposure to sdNVP+ZDV±3TC. Benjamini-Hochberg-adjusted P values denote levels of significance for pairwise log-rank comparisons between survival curves: ‘*’ 0.05, ‘**’ 0.01, ‘***’ <0.0001. Risk tables denote total number of participants with outcome at each timepoint below survival plots.
Panel A: Increased virologic failure was observed in women with a history of sdNVP+ZDV±3TC for PMTCT compared to ARV-naïve women (P=0.05).
Panel B: next-generation-sequencing analyses found that minority variants (MinV) (1–9% of one or more mutant variants within an individual’s HIV quasispecies) and majority variants (MajV) (10–100% mutant) were associated with increased virologic failure compared to wild-type (WT) (<1% mutant) (both P<0.0001).
Panel C: Next-generation-sequencing analyses found that among women with minority variants, increased virologic failure was associated with sdNVP+ZDV±3TC-experience (MinV(+)) compared to ARV-naïve and sdNVP-only-experienced women (MinV(−)), (P=0.01).
Longitudinal OLA for Detection of Emergent DR in Women with failure
To evaluate if “archived” mutations not detected as pre-ART drug-resistance by OLA contributed to failure through selection by 1st-line ART, longitudinal OLA genotyping of PBMC and plasma viral load measurements were performed for 45 women with failure and/or pre-ART drug-resistance by OLA (Supplemental Figure 1). In women with failure but without pre-ART drug-resistance by OLA, longitudinal OLA genotyping of PBMC detected emergent drug-resistance in 9/11 (81.8%) antiretroviral-naïve and 12/14 (78.6%) sdNVP-experienced women, including all 9 sdNVP+ZDV±3TC-experienced women (Supplemental Figure 1, Panels B, D and F). Drug-resistance emerged prior to failure in only three women (two antiretroviral-naïve and one sdNVP-only-experienced), and at the onset of failure in 10 women. Failure occurred rapidly, by month-6 of ART, in 7/10 of these women, including 1/3 antiretroviral-naïve and 6/7 sdNVP-experienced women. Notably, all six sdNVP-experienced women had peripartum ZDV±3TC. Pre-ART drug-resistance composed of minority variants was detected by next-generation-sequencing in 5 of 6 of these women at both OLA (M184V) and non-OLA codons (D67N, K70R, V108I, K219Q) conferring Stanford penalty scores ≥10 to NVP, ZDV, and 3TC.
Discussion
In this study of Kenyan women, virologic failure within 12 months of initiating 1st-line NNRTI-based ART, primarily with NVP, was increased in association with pre-ART drug-resistance by OLA and consensus sequencing, minority and majority frequency variants by next-generation-sequencing, and a history of PMTCT. Consistent with past findings and our primary hypothesis, a more than 10-fold increase in failure was observed in women with pre-ART drug-resistance by OLA; particularly those who had previously taken sdNVP±ZDV±3TC for PMTCT [25–28], or if antiretroviral-naïve, in those with apparent transmitted drug-resistance [6–7]. The risk of failure from pre-ART drug-resistance appeared primarily conferred by majority frequency mutations detected similarly in participants’ plasma RNA and/or PBMC DNA HIV, by OLA and consensus sequencing. OLA detection of K103N and/or G190A codons identified all women with pre-ART drug-resistance by OLA who experienced failure, which generally occurred at or prior to month-4 of ART. However, a subset of women with majority K103N achieved and maintained ART-suppression, as has been reported [26].
While evaluation of different participant subsets and amplicons prevented direct statistical comparison of assay sensitivity and specificity, next-generation-sequencing detection of pre-ART drug-resistance appeared to be more sensitive, identifying 10 more women with failure than both OLA and consensus sequencing. This potential advantage was offset slightly by a reduced specificity. However, the assays had similar positive and negative predictive values. Notably, none of the assays had sensitivity greater than 58%, with OLA and consensus sequencing sensitive to 32% only. This likely reflects the multiple pathways by which failure may occur, including poor adherence to ART, which was not assessed in this study.
Testing for pre-ART drug-resistance by OLA and consensus sequencing identified individuals at increased risk of failure, as in our previous studies [6,20,22]. However, because we anticipated that neither OLA nor consensus sequencing would detect all pre-ART drug-resistance in women who experienced failure, we explored additional strategies to identify at-risk women: (1) frequent plasma viral load monitoring and (2) testing PBMC by OLA at 2-month intervals during ART; and added post-hoc 3) testing for pre-ART drug-resistant minority variants by next-generation-sequencing.
Longitudinal plasma viral load testing identified viral rebound or failure-to-suppress viral replication within a few months of initiating ART in women with high-frequency pre-ART drug-resistance. Among those wild-type by OLA and consensus sequencing, failure among antiretroviral-naïve women often occurred after month-6 of ART, consistent with incomplete adherence [29]. In contrast, sdNVP±ZDV±3TC-experienced women without pre-ART drug-resistance by OLA or consensus sequencing experienced failure-to-suppress or rapid viral rebound, resembling those with high-frequency pre-ART drug-resistance, suggesting that frequent plasma viral load monitoring may benefit these women. Emergent drug-resistant variants in most were detected by OLA at or shortly after the onset of failure, and rarely before failure, indicating that OLA of PBMC did not effectively screen for ongoing selection of mutations during ART.
Cox proportional hazards regression and survival analyses indicated that sdNVP+ZDV±3TC-experience, but not sdNVP alone, conferred residual risk of failure among women without pre-ART drug-resistance by OLA and consensus sequencing. Other studies suggest an interaction between the interval of time between PMTCT and initiation of ART and risk of failure [10,20]. However, we did not observe such an interaction in our analysis, mostly likely because the median time intervals from sdNVP or sdNVP+ZDV±3TC to ART initiation in our subgroups were 2–4-fold longer than the 6-month interval previously analyzed. The longer interval in our study may have allowed decay of rare undetected mutant variants to clinically irrelevant levels [26]. Pre-ART drug-resistance genotyping by next-generation-sequencing was performed. Next-generation-sequencing provides sequence data with a greater sensitivity compared to consensus sequencing and a broader range of codons compared to the OLA. Next-generation-sequencing detected pre-ART drug-resistant minority variants associated with failure, especially among women who had taken sdNVP+ZDV±3TC for PMTCT. Notably, failure occurred by month-4 of ART in 6/6 sdNVP+ZDV±3TC-experienced women with minority variants by next-generation-sequencing who were wild-type by OLA and consensus sequencing. Minority variants were detected at codons associated with ZDV, 3TC or NNRTI resistance, including thymidine analogue mutations (TAMs), suggesting that mutants selected by multiple non-suppressive antiretrovirals for PMTCT were archived at levels below the limit of detection by consensus sequencing, and rapidly re-selected upon initiation of ART. Persistent minority variants following antiretrovirals for PMTCT have been linked to poor response to ART in women [26,30–31] and infants [32] in other studies, which combined with our observations emphasize the risks of serial antiretrovirals for PMTCT. While numbers were too small to draw statistical conclusions, antiretroviral-naïve women with minority variants in our study did not appear to experience rapid failure, in agreement with some [33–34] but not other reports [35–36], suggesting a complex interaction of multiple factors on this outcome. The median frequency of detected minority variants (2%, range 1–24%), suggests an OLA or allele-specific PCR could assay for clinically relevant mutants. While ZDV-monotherapy has been abandoned, individuals who fail pre-exposure-prophylaxis (PrEP) may harbor similarly relevant minority variants [37–40].
Our and others’ [20,30,41] findings demonstrate that past antiretroviral-experience can increase the risk of failure, and thus should be considered when choosing initial ART regimens. In such antiretroviral-experienced individuals, the use of a sensitive point-mutation or next-generation-sequencing genotypic-resistance assays could also provide helpful guidance. Our finding of TAMs among minority variants associated with failure suggests that OLA (or similar point-mutation assays) require timely tailoring for the detection of prevalent DR codons.
In 2017, the WHO included ART with dolutegravir (DTG) in 1st-line combination recommendations. While antiretroviral-naïve individuals given DTG-based ART in randomized clinical trials rarely had drug-resistance at virologic failure, DTG-monotherapy selects DR [42–44]. Whether DTG will suppress HIV replication in individuals with pre-ART drug-resistance or who are experiencing failure of 1st-line NNRTI-based ART remains uncertain. The high prevalence of the drug-resistant mutants observed at failure in our study, including M184V and TAMs, strongly suggests potential benefits from pre-ART drug-resistance testing to guide the choice of antiretroviral and prevent de facto DTG-monotherapy, especially among those with sequential antiretroviral-experience for PMTCT or PrEP. Additionally, past antiretroviral-use may inform selection of alternate ART options for successful viral suppression.
This study has several important limitations. First, and most salient, the participants were not randomly assigned to antiretrovirals for PMTCT. Thus, risk factors other than previous antiretroviral, such as adherence and access to care may be unmeasured contributors to virologic failure, a possibility that is further suggested by the high rate of failure among antiretroviral-naïve women relative to our previous study [7]. Second, small sample sizes reduced our ability to discern more nuanced associations between certain risk factors and failure; specifically, more precise comparisons of failure rates and PMTCT-associated minority variants between antiretroviral-naïve, sdNVP- and sdNVP+ZDV±3TC-experienced women. Third, the read length of the Illumina platform necessitated sequencing two separate regions, preventing linkage analyses of resistance mutations across these two amplicons on virologic outcome. Importantly, PCR primers for next-generation-sequencing were different from those used for consensus sequencing and OLA, and represent a potential source of bias across assays. Fourth, next-generation-sequencing of PBMC DNA precluded the use of “primer ID” methodology [45]. Thus, while the number of viral DNA templates sequenced was estimated using gag qPCR, the next-generation-sequencing mutant frequencies presented were not direct measurements from the participants’ HIV quasispecies. Similarly, the input number of viral templates into OLA, consensus sequencing, and next-generation-sequencing for a given participant likely differed, and could have resulted in variable mutant genotypes and frequencies across the three assays, particularly for minority variants. Fifth, nevirapine has been replaced by efavirenz in NNRTI-based ART, and recent studies suggest that pre-ART drug-resistance to multiple antiretroviral classes, but not single NNRTI mutations increase the risk of failure with EFV-based ART [46–47].
In summary, we examined several strategies to identify women at high risk for virologic failure of 1st-line NNRTI (primarily NVP)-based ART, including diagnosis of pre-ART drug-resistance by multiple genotyping assays and early detection of emergent drug-resistance and failure. OLA and consensus sequencing had similar sensitivity in identifying individuals with pre-ART drug-resistance associated with failure. Testing for pre-ART drug-resistant minority variants by next-generation-sequencing and consideration of antiretroviral histories for past use of ZDV plus sdNVP for PMTCT identified additional women with failure not captured by OLA or consensus sequencing. Testing for detectable viral load soon after the initiation of ART identified women with apparent re-selection of pre-ART drug-resistant minority variants. Given the residual risk for virologic failure conferred by non-suppressive antiretrovirals for PMTCT, our findings endorse evaluating pre-ART drug-resistance in ZDV and sdNVP-experienced women of childbearing age who may be restricted from initiating DTG-based ART and treated instead with EFV-based ART [48]. In addition, it is uncertain whether DTG-based ART will suppress viral replication in those with drug-resistance due to past PMTCT, failed PrEP or virologic failure during NNRTI-based ART, warranting further studies among such individuals.
Supplementary Material
Acknowledgment:
The authors acknowledge the commitment of the women participants, study nurses, contributions of Steven Bii, visiting scientist Segundo Leon, and valuable advice from Joseph Fitzgibbon, PhD. Sequence analysis was performed with assistance of the Molecular Profiling and Computational Biology core of the UW CFAR [P30 AI027757].
Funding: This work was supported by grants from the National Institutes of Health awards [R01 AI058723, R01 AI100037 to LMF], including an American Recovery and Reinvestment Act supplement [R01 AI058723]. Support of the study was also provided by the Clinical Retrovirology [P30 AI027757 to RWC] core of the University of Washington Center for AIDS Research. The Coptic Hope Center for Infectious Diseases is supported by the President’s Emergency Plan for AIDS Relief through a cooperative agreement [U62/CCU024512 to MHC] from the Centers for Disease Control and Prevention.
Footnotes
These data were presented, in part, at the 2014 International Workshop on Antiviral Drug Resistance held June 3–7, 2014 in Berlin, Germany.
Authorship Statement: I.A.B., S.D., J.N.K., S.R.S., M.H.C., R.W.C., and L.M.F. conceived and designed the study; R.S.M, R.A.S., J.M., W.D., and T.R.S. performed data collection and analysis; R.S.M., R.A.S., and L.M.F., wrote the article. None of the authors declare a conflict of interest in this study, and all have reviewed and approved this manuscript.
References
- 1.WHO| HIV/AIDS Data and Statistics: Estimated numbers of people receiving antiretroviral therapy globally and by WHO Region and percentage coverage globally, 2000–2015. In: WHO Online; 2017. pp. Chart of HIV-infected persons. [Google Scholar]
- 2.Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining Morbidity and Mortality among Patients with Advanced Human Immunodeficiency Virus Infection. New England Journal of Medicine 1998; 338(13):853–860. [DOI] [PubMed] [Google Scholar]
- 3.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 Infection with Early Antiretroviral Therapy. New England Journal of Medicine 2011; 365(6):493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panichsillapakit T, Smith DM, Wertheim JO, Richman DD, Little SJ, Mehta SR. Prevalence of Transmitted HIV Drug Resistance Among Recently Infected Persons in San Diego, CA 1996–2013. J Acquir Immune Defic Syndr 2016; 71(2):228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DHJ, Gregson J, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. The Lancet; 380(9849):1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung MH, Beck IA, Dross S, Tapia K, Kiarie JN, Richardson BA, et al. Oligonucleotide ligation assay detects HIV drug resistance associated with virologic failure among antiretroviral-naive adults in Kenya. J Acquir Immune Defic Syndr 2014; 67(3):246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung MH, Silverman R, Beck IA, Yatich N, Dross S, McKernan-Mullin J, et al. Increasing HIV-1 pretreatment drug resistance among antiretroviral-naive adults initiating treatment between 2006 and 2014 in Nairobi, Kenya. Aids 2016; 30(10):1680–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silverman RA, Beck IA, Kiptinness C, Levine M, Milne R, McGrath CJ, et al. Prevalence of Pre-antiretroviral-Treatment Drug Resistance by Gender, Age, and Other Factors in HIV-Infected Individuals Initiating Therapy in Kenya, 2013–2014. In: J Infect Dis. United States; 2017. pp. 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamers RL, Schuurman R, Sigaloff KCE, Wallis CL, Kityo C, Siwale M, et al. Effect of pretreatment HIV-1 drug resistance on immunological, virological, and drug-resistance outcomes of first-line antiretroviral treatment in sub-Saharan Africa: a multicentre cohort study. The Lancet infectious diseases 2012; 12(4):307–317. [DOI] [PubMed] [Google Scholar]
- 10.Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med 2007; 356(2):135–147. [DOI] [PubMed] [Google Scholar]
- 11.Guay LA, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 1999; 354(9181):795–802. [DOI] [PubMed] [Google Scholar]
- 12.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, Bowonwatanuwong C, Kantipong P, Leechanachai P, et al. Intrapartum Exposure to Nevirapine and Subsequent Maternal Responses to Nevirapine-Based Antiretroviral Therapy. New England Journal of Medicine 2004; 351(3):229–240. [DOI] [PubMed] [Google Scholar]
- 13.Lallemant M, Jourdain G, Le Coeur S, Mary JY, Ngo-Giang-Huong N, Koetsawang S, et al. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N Engl J Med 2004; 351(3):217–228. [DOI] [PubMed] [Google Scholar]
- 14.McIntyre JA, Hopley M, Moodley D, Eklund M, Gray GE, Hall DB, et al. Efficacy of Short-Course AZT Plus 3TC to Reduce Nevirapine Resistance in the Prevention of Mother-to-Child HIV Transmission: A Randomized Clinical Trial. PLOS Medicine 2009; 6(10):e1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Micek MA, Blanco AJ, Carlsson J, Beck IA, Dross S, Matunha L, et al. Effects of short-course zidovudine on the selection of nevirapine-resistant HIV-1 in women taking single-dose nevirapine. J Infect Dis 2012; 205(12):1811–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler MG, Qin M, Fiscus SA, Currier JS, Flynn PM, Chipato T, et al. Benefits and risks of antiretroviral therapy for perinatal HIV prevention. New England Journal of Medicine 2016; 375(18):1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser A, Sewangi J, Mbezi P, Dugange F, Lau I, Ziske J, et al. Emergence of minor drug-resistant HIV-1 variants after triple antiretroviral prophylaxis for prevention of vertical HIV-1 transmission. PloS one 2012; 7(2):e32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melikian GL, Rhee SY, Varghese V, Porter D, White K, Taylor J, et al. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother 2014; 69(1):12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health O Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. 2013 In; 2013. [PubMed] [Google Scholar]
- 20.Jourdain G, Wagner TA, Ngo-Giang-Huong N, Sirirungsi W, Klinbuayaem V, Fregonese F, et al. Association between detection of HIV-1 DNA resistance mutations by a sensitive assay at initiation of antiretroviral therapy and virologic failure. Clin Infect Dis 2010; 50(10):1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung MH, Drake AL, Richardson BA, Reddy A, Thiga J, Sakr SR, et al. Impact of prior HAART use on clinical outcomes in a large Kenyan HIV treatment program. Current HIV research 2009; 7(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck IA, Deng W, Payant R, Hall R, Bumgarner RE, Mullins JI, et al. Validation of an oligonucleotide ligation assay for quantification of human immunodeficiency virus type 1 drug-resistant mutants by use of massively parallel sequencing. Journal of clinical microbiology 2014; 52(7):2320–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gantt S, Shetty AK, Seidel KD, Matasa K, Musingwini G, Woelk G, et al. Laboratory indicators of mastitis are not associated with elevated HIV-1 DNA loads or predictive of HIV-1 RNA loads in breast milk. J Infect Dis 2007; 196(4):570–576. [DOI] [PubMed] [Google Scholar]
- 24.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clinical infectious diseases 2006; 42(11):1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner TA, Kress CM, Beck I, Techapornroong M, Wittayapraparat P, Tansuphasawasdikul S, et al. Detection of HIV-1 drug resistance in women following administration of a single dose of nevirapine: comparison of plasma RNA to cellular DNA by consensus sequencing and by oligonucleotide ligation assay. Journal of clinical microbiology 2010; 48(5):1555–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coovadia A, Hunt G, Abrams EJ, Sherman G, Meyers T, Barry G, et al. Persistent Minority K103N Mutations among Women Exposed to Single-Dose Nevirapine and Virologic Response to Nonnucleoside Reverse-Transcriptase Inhibitor–Based Therapy. Clin Infect Dis 2009; 48(4):462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eshleman SH, Mracna M, Guay LA, Deseyve M, Cunningham S, Mirochnick M, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). Aids 2001; 15(15):1951–1957. [DOI] [PubMed] [Google Scholar]
- 28.Flys T, Nissley DV, Claasen CW, Jones D, Shi C, Guay LA, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. The Journal of infectious diseases 2005; 192(1):24–29. [DOI] [PubMed] [Google Scholar]
- 29.Harrigan PR, Hogg RS, Dong WWY, Yip B, Wynhoven B, Woodward J, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. The Journal of infectious diseases 2005; 191(3):339–347. [DOI] [PubMed] [Google Scholar]
- 30.Boltz VF, Zheng Y, Lockman S, Hong F, Halvas EK, McIntyre J, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A 2011; 108(22):9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowley CF, Boutwell CL, Lee EJ, MacLeod IJ, Ribaudo HJ, Essex M, et al. Ultrasensitive detection of minor drug-resistant variants for HIV after nevirapine exposure using allele-specific PCR: clinical significance. AIDS research and human retroviruses 2010; 26(3):293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLeod IJ, Rowley CF, Thior I, Wester C, Makhema J, Essex M, et al. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J Clin Virol 2010; 48(3):162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boltz VF, Bao Y, Lockman S, Halvas EK, Kearney MF, McIntyre JA, et al. Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis 2014; 209(5):703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzner KJ, Rauch P, Braun P, Knechten H, Ehret R, Korn K, et al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. Journal of Clinical Virology 2011; 50(2):156–161. [DOI] [PubMed] [Google Scholar]
- 35.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. Jama 2011; 305(13):1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson JA, Li J-F, Wei X, Lipscomb J, Irlbeck D, Craig C, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment–naïve populations and associate with reduced treatment efficacy. PLoS medicine 2008; 5(7):e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehman DA, Baeten JM, McCoy CO, Weis JF, Peterson D, Mbara G, et al. Risk of drug resistance among persons acquiring HIV within a randomized clinical trial of single-or dual-agent preexposure prophylaxis. The Journal of infectious diseases 2015; 211(8):1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant RM, Liegler T, Defechereux P, Kashuba ADM, Taylor D, Abdel-Mohsen M, et al. Drug resistance and plasma viral RNA level after ineffective use of oral pre-exposure prophylaxis in women. Aids 2015; 29(3):331–337. [DOI] [PubMed] [Google Scholar]
- 39.Hurt CB, Eron JJ Jr, Cohen MS. Pre-exposure prophylaxis and antiretroviral resistance: HIV prevention at a cost? Clinical Infectious Diseases 2011; 53(12):1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivay MV, Li M, Piwowar-Manning E, Zhang Y, Hudelson SE, Marzinke MA, et al. Characterization of HIV seroconverters in a TDF/FTC PrEP study: HPTN 067/ADAPT. JAIDS Journal of Acquired Immune Deficiency Syndromes 2017; 75(3):271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huntington S, Thorne C, Newell M, Anderson J, Taylor G, Pillay D, et al. Outcomes after 2 years on antiretroviral therapy (ART): comparison of women who were ART-naive and women who previously used short-course ART in pregnancy. In: 15th European AIDS Conference Barcelona; 2015. [Google Scholar]
- 42.Brenner BG, Thomas R, Blanco JL, Ibanescu R-I, Oliveira M, Mesplede T, et al. Development of a G118R mutation in HIV-1 integrase following a switch to dolutegravir monotherapy leading to cross-resistance to integrase inhibitors. Journal of Antimicrobial Chemotherapy 2016; 71(7):1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijting I, Rokx C, Boucher C, van Kampen J, Pas S, de Vries-Sluijs T, et al. Dolutegravir as maintenance monotherapy for HIV (DOMONO): a phase 2, randomised non-inferiority trial. The Lancet HIV 2017; 4(12):e547–e554. [DOI] [PubMed] [Google Scholar]
- 44.Trevillyan JM, Hoy JF. Dolutegravir monotherapy as maintenance ART bites the dust. The Lancet HIV 2017; 4(12):e531–e532. [DOI] [PubMed] [Google Scholar]
- 45.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proceedings of the National Academy of Sciences 2011; 108(50):20166–20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck IA, Levine M, Milne R, So I, Andersen N, Watling M, et al. Impact of pretreatment HIV-drug resistance on virologic outcome of first-line ART. [Abstract 490] 24th Conference on retroviruses and opportunistic infections (CROI) 13–16 February 2017. [Google Scholar]
- 47.Derache A, Iwuji CC, Baisley K, Danaviah S, Marcelin AG, Calvez V, et al. Impact of next generation sequencing defined HIV pre-treatment drug resistance on virological outcomes in the ANRS 12249 treatment as prevention trial. LID - 10.1093/cid/ciy881 [doi]. (1537–6591 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO. Updated Recommendations on First-Line and Second-Line Antiretroviral Regimens and Post-Exposure Prophylaxis and Recommendations on Early Infant Diagnosis of HIV. 2018. In; 2018
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.