Abstract
Direct lineage reprogramming involves the remarkable conversion of cellular identity. Single-cell technologies aid in deconstructing the considerable heterogeneity that emerges during lineage conversion. However, lineage relationships are typically lost during cell processing, complicating trajectory reconstruction. Here, we present ‘CellTagging’, a combinatorial cell indexing methodology, permitting the parallel capture of clonal history and cell identity, where sequential rounds of cell labelling enable the construction of multi-level lineage trees. CellTagging and longitudinal tracking of fibroblast to induced endoderm progenitor (iEP) reprogramming reveals two distinct trajectories: one leading to successfully reprogrammed cells, and one leading to a ‘dead-end’ state, paths determined in the earliest reprogramming stages. We find that expression of a putative methyltransferase, Mettl7a1, is associated with the successful reprogramming trajectory, where its addition to the reprogramming cocktail increases the yield of iEPs. Together, these results demonstrate the utility of our lineage tracing method to reveal dynamics of direct reprogramming.
Direct lineage reprogramming bypasses pluripotency to convert cell identity between somatic states, yielding clinically valuable cell types1. However, these conversion strategies are generally inefficient, producing incompletely converted, and developmentally immature cells that fail to fully recapitulate target cell identity2,3. This considerable heterogeneity arising during reprogramming has concealed the molecular mechanisms underlying lineage conversion. In this respect, single-cell RNA-sequencing (scRNA-seq) has enabled fully converted cells to be distinguished from partially reprogrammed intermediates4,5, although these analytical approaches typically result in the loss of spatial, temporal, and lineage information. While, elegant computational approaches can infer missing observations6,7, reconstruction of true reprogramming trajectories using these tools remains challenging. Although sophisticated lineage tracing solutions to connect cell history with fate are emerging, these protocols are either not compatible with high-throughput scRNA-seq8–11, or require genome editing strategies that are not readily deployed in some systems12–15.
To enable simultaneous single-cell profiling of cell identity and clonal history, we have developed a straightforward, high-throughput cell tracking method, ‘CellTagging’. Sequential lentiviral delivery of heritable random unique molecular indexes, CellTags, permits the construction of multi-level lineage trees. Here, we apply CellTagging to transcription factor (TF)-induced direct lineage reprogramming of mouse embryonic fibroblasts (MEFs) to induced endoderm progenitors (iEPs), a valuable self-renewing cell type that possesses both hepatic and intestinal potential3,16. iEP generation represents a prototypical cell fate engineering methodology, reflecting the inefficiency and infidelity of many reprogramming protocols2,3. CellTagging and tracking over 100,000 cells converting to iEPs reveals two distinct trajectories: one, a route toward successfully reprogrammed cells, and an alternate path into a putative ‘dead-end’ state, marked by re-expression of fibroblast genes. Although few cells successfully reprogram, clonally-related cells tend to follow the same trajectories, suggesting that their reprogramming outcome may be determined from the earliest stages of conversion. These clonal dynamics and lineages can be explored on our companion website, CellTag Viz: http://www.celltag.org/. In later stages of conversion, our analyses reveal expression of a putative methyltransferase, Mettl7a1, along the successful reprogramming trajectory. Addition of this factor to the reprogramming cocktail increases the yield of successfully converted iEPs. Together, these findings demonstrate the utility of CellTagging for lineage reconstruction, providing molecular insights into reprogramming that serve to improve the outcome of this generally inefficient process.
CellTagging: combinatorial indexing of cells to trace their clonal history
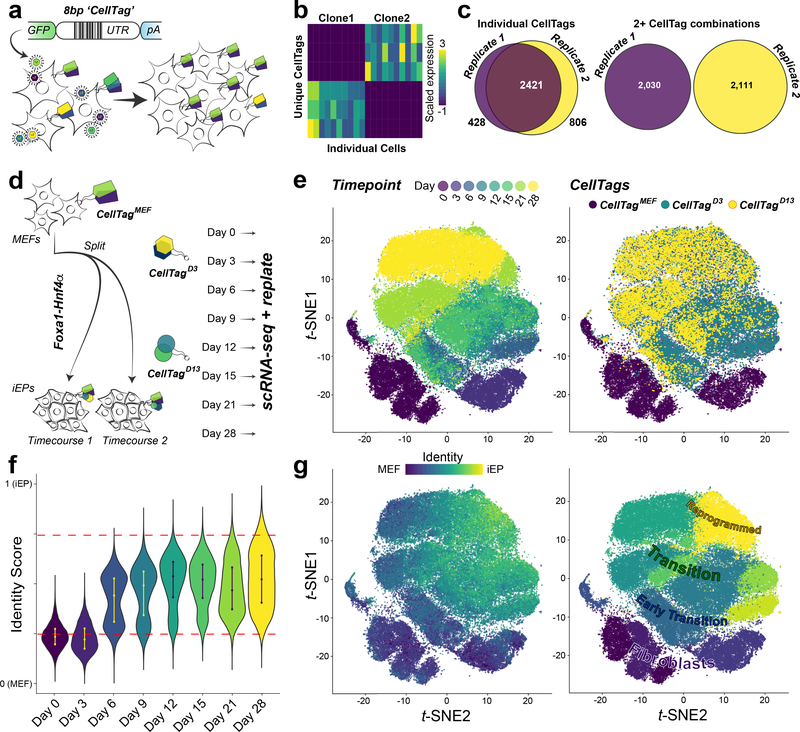
CellTagging is a lentiviral-based approach to uniquely label individual cells with heritable barcode combinations. CellTags are highly expressed and readily captured within each single-cell transcriptome, allowing clonal history to be recorded over time, in parallel with cell identity (Fig. 1a). Recovery of CellTag expression, followed by filtering and error-correction, ensures sensitive and specific identification of clonally-related cells (Extended Data Fig. 1a-g). The efficacy of this approach to label cells with distinguishing barcode combinations is demonstrated via CellTagging a ‘species-mix’ of genetically distinct human 293T cells and MEFs (Extended Data Fig. 1h-j). This is further supported by labelling two independent biological replicates with the same CellTag library: while individual CellTags appear in both pools of cells, no signatures of 2 or more CellTags are shared between replicates, confirming that clones are derived from uniquely-labelled cells (n=8,326 cells expressing 3±4×10−4 (mean+s.e.m.) CellTags per cell, Fig. 1b,c). Finally, CellTagging does not perturb cell physiology or reprogramming efficiency (Extended Data Fig. 2). Together, these data validate the utility of CellTagging to deliver unique, heritable labels into cells, permitting clonal relationships to be tracked longitudinally, with a high degree of confidence.
Figure 1. CellTagging: clonal tracking applied to reprogramming.
(a) CellTagging workflow: A lentiviral construct contains an 8bp random ‘CellTag’ barcode in the 3’UTR of GFP, followed by an SV40 polyadenylation signal. Transduced cells express unique CellTag combinations, resulting in distinct, heritable signatures, enabling tracking of clonally-related cells. (b) Representative CellTag expression in two clones, defined by unique combinations of three CellTags (n=10 cells per clone). (c) Left: Overlap of individual CellTags in two independent biological replicates tagged with the same CellTag library. Right: CellTag signatures are not shared between the two replicates (replicate 1: n=8,535 cells; replicate 2: n=11,997 cells). (d) Experimental approach: Mouse Embryonic Fibroblasts (MEFs) are tagged with the CellTagMEF library, expanded for two days and then split for reprogramming in two independent biological replicates. Additional tagging was performed at 3 days (CellTagD3) and 13 days (CellTagD13) post-initiation of reprogramming. Every 3–7 days, cells were harvested for scRNA-seq with the remainder replated. (e) Visualization of scRNA-seq data: projection of timepoint onto t-distributed stochastic neighbor embedding plot (t-SNE, timecourses 1 and 2: n=85,010 cells. (f) Scoring single-cell identity via quadratic programming, cells scoring >0.75 (upper red line) classify as iEPs, cells scoring <0.25 (lower red line) classify as fibroblasts (n=85,010 cells). (g) Left: Projection of identity scores onto the t-SNE plot. Right: t-SNE cluster designations: Fibroblast, Early Transition, Transition, and Reprogrammed.
We next applied CellTagging to direct reprogramming of fibroblasts to iEPs, driven by Foxa1 and Hnf4α TF overexpression, in four independent biological replicates. To enable lineage reconstruction, we devised a sequential CellTagging scheme where fibroblasts were transduced with an initial CellTag library, CellTagMEF. Following a 48hr expansion period, these cells were split into independent biological replicates for reprogramming. Tagging with a second library (CellTagD3) was performed at the end of the 3-day period of retroviral TF-delivery, followed by a third round (CellTagD13) 13 days after reprogramming initiation, coinciding with the phenotypic emergence of iEPs. Each CellTagging round is demultiplexed via a short motif preceding the random CellTag region. Cells were harvested every 3–7 days over the 28-day timecourse, methanol preserving portions for high-throughput Drop-seq17 and 10x Genomics18 scRNA-seq, re-plating the remaining cells to permit clonal growth and lineage reconstruction (Fig. 1d). In total, 96,356 high-quality single-cell transcriptomes were captured. Downstream analysis focused on data captured using the 10x Genomics platform (85,010 single-cell transcriptomes, merging timecourses 1 and 2; Fig. 1e; Extended Data Fig. 3a-c; Supplementary Table 1). Canonical correlation analysis19 demonstrates consistent replication across technologies and biological replicates (Extended Data Fig. 3d,e).
Simultaneous capture of reprogramming and clonal dynamics
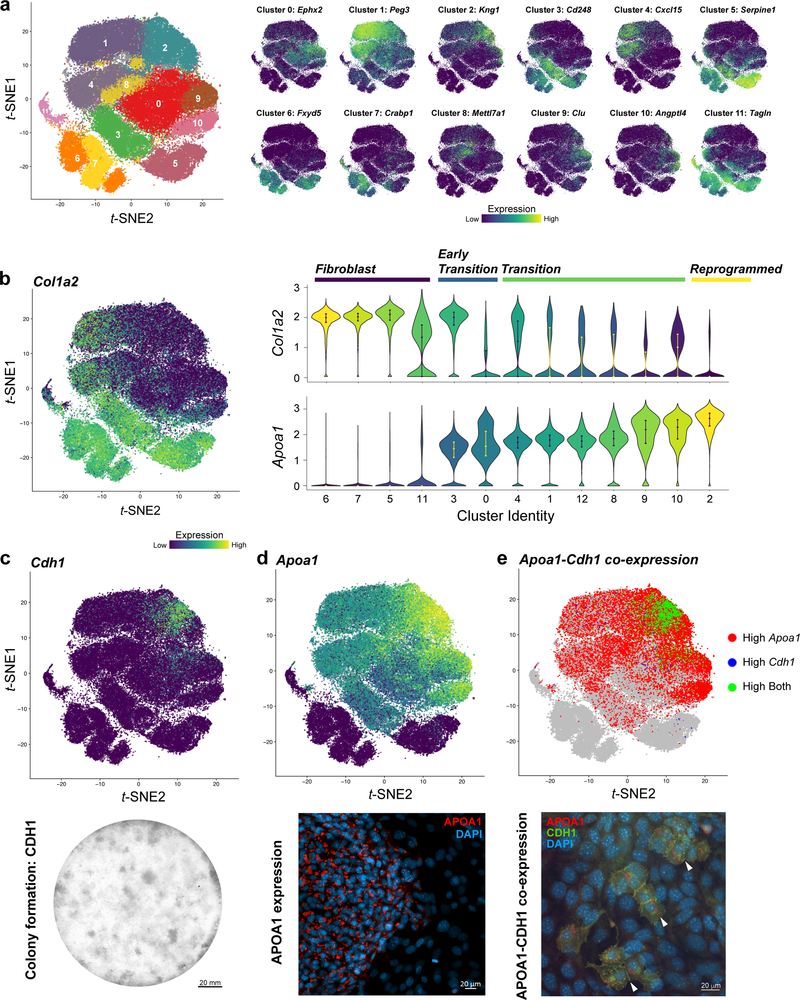
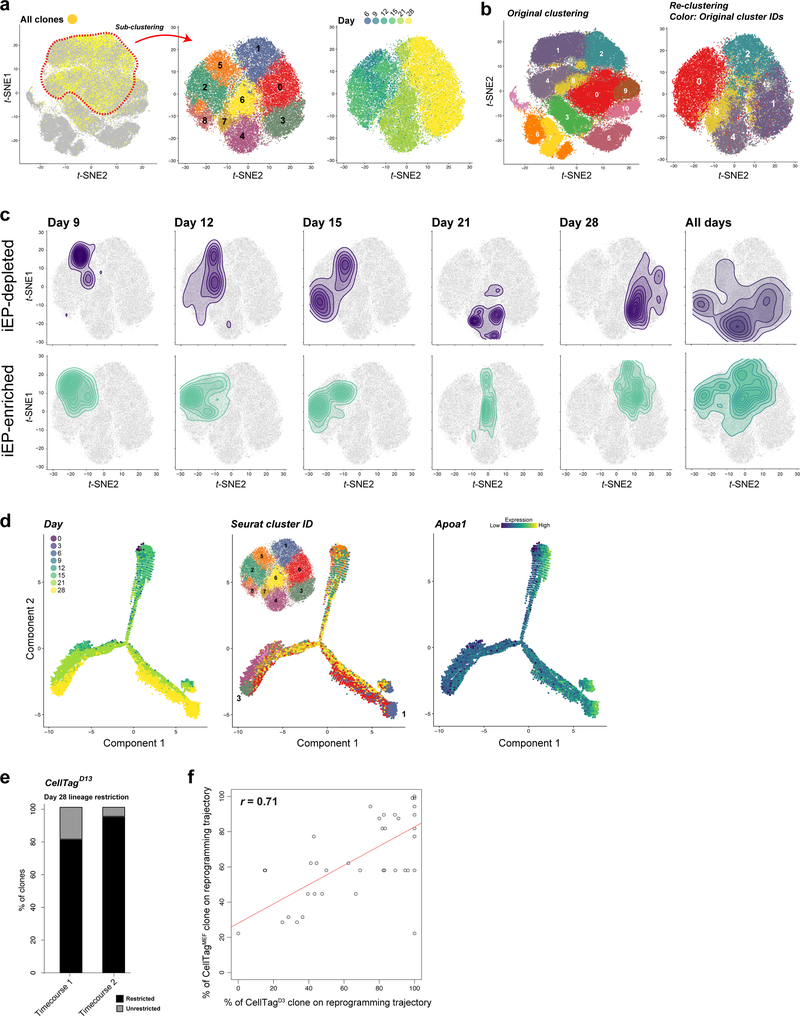
Using t-distributed stochastic neighbor embedding6 (t-SNE), the 28-day reprogramming process resolves into 13 clusters of transcriptionally distinct cells (Extended Data Fig. 3f,g). CellTag expression is detected in 99% of cells, with CellTagMEF expression detected across all timepoints, CellTagD3 from day 6, and CellTagD13 from day 15 (Fig. 1e). 65% of cells sequenced pass the ≥2 CellTag threshold required for tracking (n=55,571/85,010 cells, Extended Data Fig. 4). To investigate reprogramming dynamics, we first performed gene expression analysis for each cluster, revealing progressive silencing of fibroblast identity (Extended Data Fig. 5a,b; Supplementary Tables 2,3). To track iEP emergence, we employed quadratic programming5 to score individual cell identities as a fraction of starting and target cell types, revealing that iEP identity is progressively gained from day 6 of reprogramming (Fig. 1f). Projection of identity scores onto the t-SNE plot localizes iEPs to cluster 2, coinciding with reprogramming days 21 and 28 (Fig. 1e,g). Further examination of this iEP-harboring cluster identifies new markers, including Apolipoprotein A1 (Apoa1; Extended Data Fig. 5a,b; Supplementary Table 3), where immunostaining for APOA1 demonstrates protein-level co-expression with the canonical iEP marker, CDH1 (E-Cadherin)3,16 (Extended Data Fig. 5c-e). Although previous studies show that only ~1% of cells successfully reprogram3,16, we observe a high proportion of cells expressing Apoa1, commencing from day 6 (62.5±5.5%; Extended Data Fig. 5b,d,e). Together, these observations suggest that many cells initiate reprogramming but few complete the transition to iEPs, broadly partitioning the process into four phases: Fibroblast, Early Transition, Transition, and Reprogrammed (Fig. 1g; Extended Data Fig. 5b).
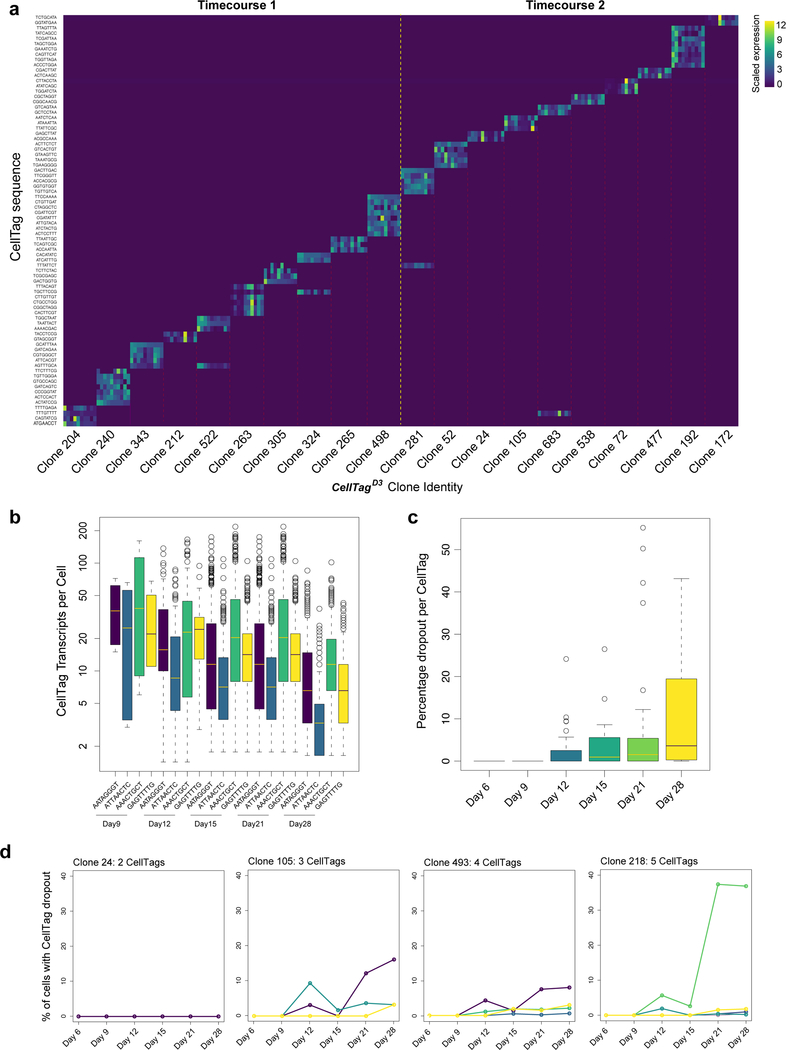
We next integrated clonal relationships into this single-cell landscape: from a total of 55,571 cells passing the ≥2 CellTag threshold to support clone-calling, we identified 27,020 cells possessing clonal relatives, based on shared CellTag signatures. Defining a clone as three or more related cells, we identified 706 CellTagMEF clones and 884 CellTagD3 clones. Since CellTagD13 clones had less time to expand, we also included related cell pairs for this later labelling, resulting in 561 clones (Supplementary Table 4). In agreement with our above validation experiments, examination of 10 major clones from CellTagD3-labelled replicates shows that CellTag combinations used to identify clonally-related cells are unique (Extended Data Fig. 6a). CellTags are reliably detected over a 10-week period; although their expression slightly diminishes over time, they are not silenced (Extended Data Fig. 4c; 6b-d). Together, this demonstrates the advantage of our CellTag combinatorial indexing method to reliably label cells and track them over extended periods.
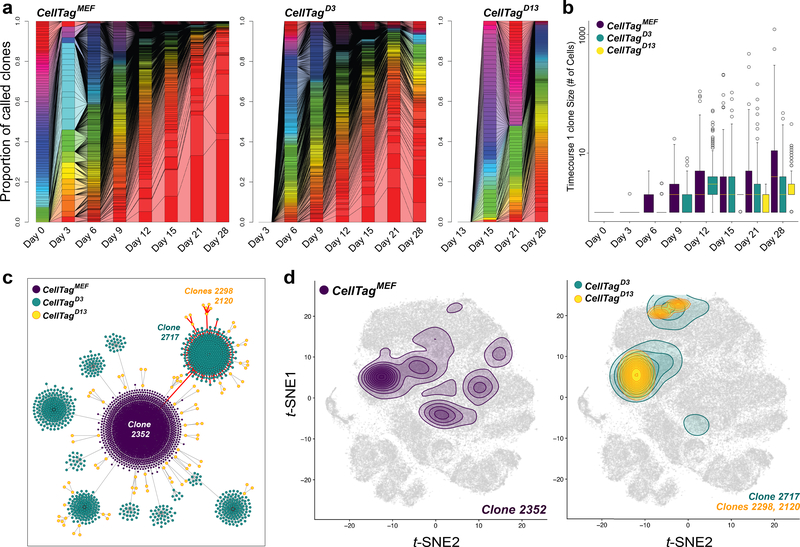
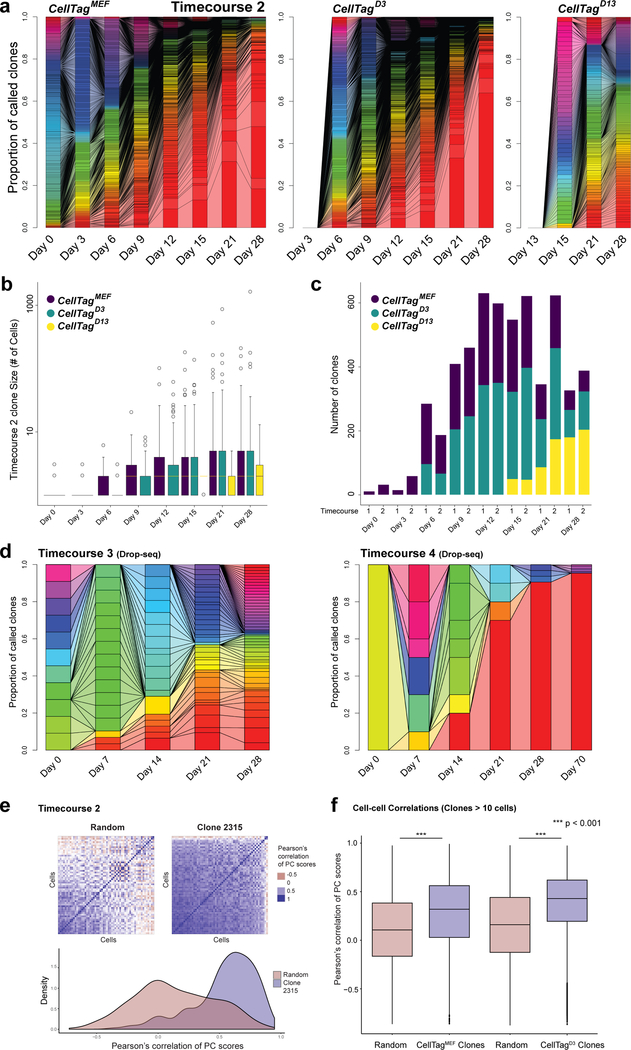
During reprogramming, we observe extensive clonal growth: CellTagMEF clones reach an average size of 47±22 cells per clone by day 28. (Fig. 2a,b; Extended Data Fig. 7a-d). Expanding at a similar rate, CellTagD3 clones are first detected from day 6, whereas smaller clones arise from CellTagD13-labelling (Fig. 2a,b). In some instances, we observe dramatic growth of an individual clone during reprogramming (Extended Data Fig. 7d). This could not be reconciled with viral integration analysis (Supplementary Table 5), suggesting that the clonal growth we observe is associated with iEPs entering a self-renewing, progenitor-like state. As a consequence of this growth, iEPs are derived from a handful of clones. We next sought to connect these clonal relationships over time, to trace the origins of successfully reprogramming cells. In this approach, we assume that the identity or state of each cell we capture is representative of its collective clone. Indeed, we do find that gene expression is highly correlated between clonally-related cells, suggesting that family members are likely to behave in a similar manner, sharing reprogramming outcomes (Extended Data Fig. 7e,f).
Figure 2. Tracking reprogramming clonal dynamics and constructing lineage trees.
(a) Connected barplots showing individual clones as a proportion of all clones over reprogramming, for each CellTagging round (Timecourse1, n=12,932 cells, 1,031 clones). (b) Average number of cells per clone, per timepoint, for each round of CellTagging (n=1,031 clones). (c) Reconstruction and visualization of lineages via force-directed graph drawing. Each node represents an individual cell, and edges represent clonal relationships between cells: Purple=CellTagMEF, Blue=CellTagD3, Yellow=CellTagD13 clones. (d) Contour plots, representing cell density of each clone, projected onto the t-SNE, for the red highlighted lineage in (c) (n=2,199 cells). All lineages and clone distributions can be explored via CellTag Viz (http://www.celltag.org/).
Lineage reconstruction and identification of reprogramming trajectories
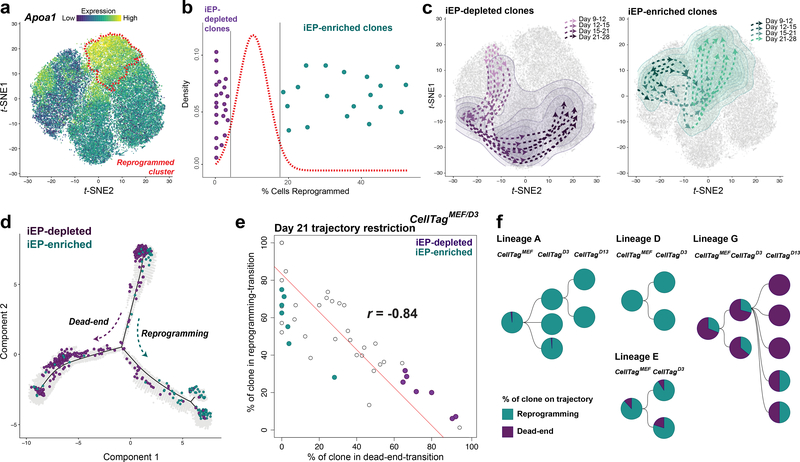
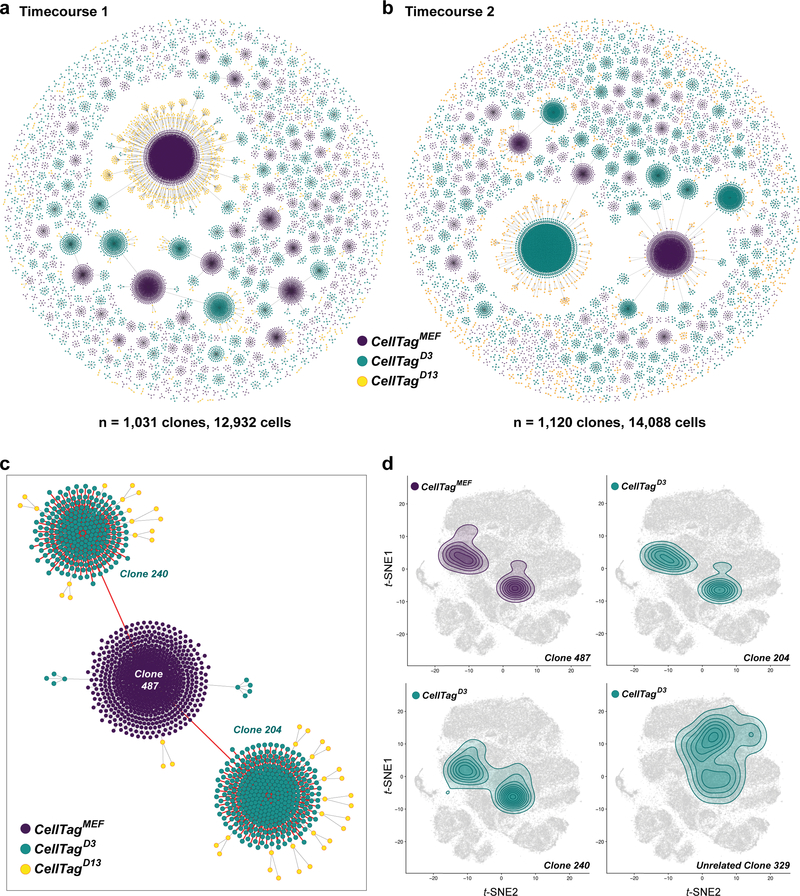
Sequential CellTagging enables the reconstruction of lineage trees and reprogramming trajectories. First, we apply force-directed graphing to construct hundreds of multi-level lineages (Extended Data Fig. 8a,b), which can be explored on our companion website, CellTag Viz: http://www.celltag.org/. Fig. 2c shows a representative lineage stemming from one CellTagMEF clone, branching into CellTagD3 and CellTagD13 descendants. Next, to visualize clonally-related cell distribution, we use contour plotting in combination with t-SNE plotting. This reveals considerable overlap of clones belonging to the same lineage, supporting the above observations that clonally-related cells are transcriptionally similar (Fig. 2d; Extended Data Fig. 8c,d). From these analyses, we observed enrichment or depletion of iEPs within many lineages. To quantify this, we re-clustered cells in the later stages of reprogramming, providing high-coverage clone information. Within this subset, 8% of cells classify as fully-reprogrammed iEPs (Fig. 3a; Extended Data Fig. 9a,b). We then performed randomized testing to identify major clones significantly enriched for, or depleted of iEPs, yielding 20 iEP-enriched clones in which ~20–50% cells are fully reprogrammed. In contrast, we found 24 iEP-depleted clones in which less than 3% of cells classify as iEPs (Fig. 3b).
Figure 3. Mapping reprogramming trajectories and timing of cell fate commitment.
(a) Apoa1 expression in a subset of cells from timecourses 1 and 2 (n=48,515 cells); fully reprogrammed iEPs outlined in red (cluster 1). (b) Density plot of the mean proportion of reprogrammed cells for groups of randomly-selected cells (defined by cluster 1 occupancy, n=59 groups, 14,987 cells). Randomized testing of 59 CellTagMEF/D3 clones (>35 cells per clone, n=10,259 cells) identifies iEP-enriched clones (n=20 clones, 6,128 cells, p<0.05) and iEP-depleted clones (n=24 clones, 3,117 cells, p<0.05). (c) Clones spanning all timepoints were selected for further analysis: trajectories showing connections between areas of highest clonal density across each day of reprogramming, for iEP-depleted (left, n=7 clones, 2,270 cells) and iEP-enriched clones (right, n=7 clones, 1,037 cells). (d) Pseudo-temporal ordering of the timecourse 1 and 2 subset (a), with overlay of individual cells belonging to iEP-enriched and iEP-depleted clones, defining reprogramming and dead-end trajectories (n=14 clones, 3,307 cells). (e) Proportions of clones occupying clusters 6 and 7 (reprogramming-transition) or cluster 4 (dead-end-transition) at reprogramming day 21 (r =−0.84, Pearson’s correlation, n=44 clones, 9,624 cells). (f) Lineage trees of related clones, with the proportion of each clone contributing to reprogramming or dead-end trajectories shown (n=1,185 cells).
Via contour plotting, we find that iEP-enriched/depleted clones are clearly segregated, suggesting the existence of discrete reprogramming trajectories, supported by orthogonal pseudotemporal ordering analysis (Fig. 3c,d; Extended Data Fig. 9c,d). Quantification of these trajectories reveals a bifurcation at day 21 where successfully reprogramming clones transition through clusters 6 and 7, leading to the reprogrammed state at day 28. Conversely, these transition clusters are bypassed on the iEP-depleted-trajectory, where clones traverse cluster 4 on day 21, entering a putative reprogramming ‘dead-end’ by day 28 (Fig. 3e, r=−0.84). To investigate the timing of trajectory commitment, we quantified CellTagD13-labelled cell occupancy of reprogrammed and putative dead-end states (cluster1 and 3, respectively) at day 28. The distribution of clonally-related cells between these states reveals their restriction to one of the two states indicating that reprogramming outcome is determined by day 13 (88±8% restricted clones; Extended Data Fig. 9e). These divergent routes appear to be rooted in distinct transcriptional states as early as day 9 (Fig. 3c), suggesting their establishment early in the reprogramming process.
The existence of early-labelled clones that are biased in their reprogramming outcome, in addition to the shared transcriptional signatures we observe between clonally-related cells, suggests that cells do not reprogram in a stochastic manner. Here, sequential CellTagging and quantification of reprogramming outcome for each clone within a lineage allows us to probe the probability with which cells will successfully generate iEPs. To explore this, we identified lineages of CellTagD3-labelled clones arising from common CellTagMEF-labelled ancestors. For each clone within the lineage, we calculated the proportion of cells occupying reprogramming and dead-end trajectories. In a stochastic model of reprogramming, we would expect the post-reprogramming-induction, CellTagD3-labelled clones from a common ancestor to reprogram with different efficiencies. On the contrary, Fig. 3f shows that CellTagD3-descendant clones reprogram with similar efficiencies to each other, and to their CellTagMEF-labelled parent, particularly for those lineages reprogramming at high efficiency (r=0.71, Extended Data Fig. 9f). This suggests that reprogramming outcome may be determined from early stages. It is a possibility that an elite cell type, predisposed to reprogram, exists in the highly heterogeneous fibroblast starting population. To explore this possibility, cells were first tagged and then split for reprogramming in two biological replicates. We identify 84 clones appearing across both replicates, where only 4 clones both reprogram, with different efficiencies (Supplementary Table 6), arguing against the existence of an elite reprogramming cell type in the fibroblast population.
A putative methyltransferase, Mettl7a1, delineates successful reprogramming
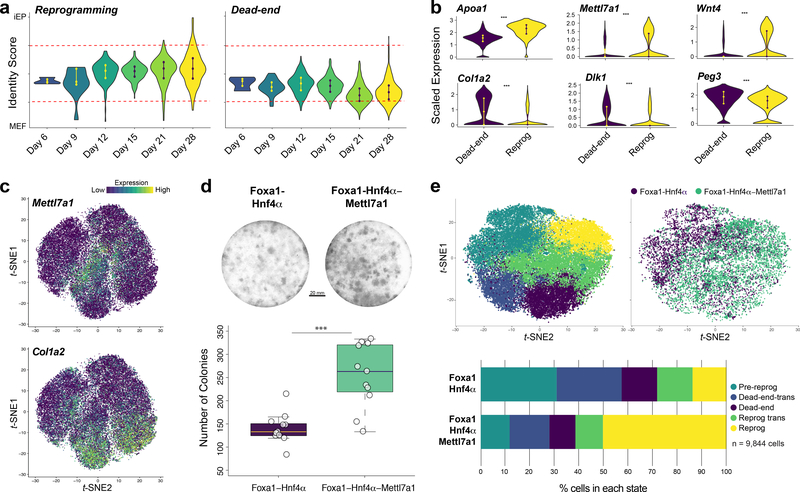
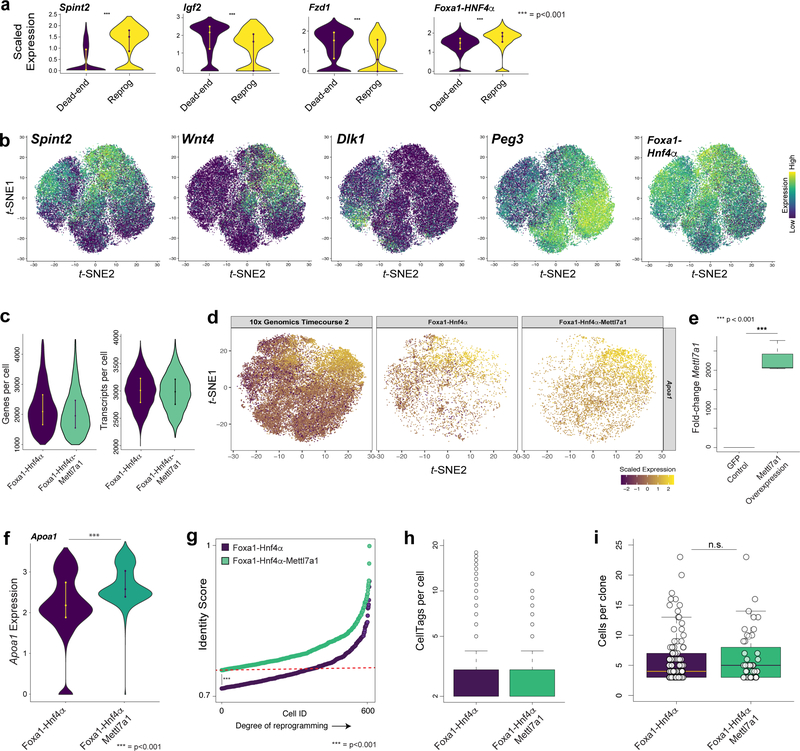
To investigate the molecular characteristics underpinning the distinct reprogramming paths revealed here, we compared cells between reprogramming and dead-end trajectories (n=2,047 cells). Along the reprogramming trajectory, iEP identity scores gradually increase over time. In contrast, partial fibroblast identity is re-established with progression down the dead-end trajectory, suggesting that this is indeed represents a reprogramming impasse (Fig. 4a). Significant changes in gene expression between these two trajectories are apparent, including key elements of Wnt, Igf2, and HGF signaling pathways. The dead-end trajectory is enriched for imprinted gene expression (Dlk1 and Peg3), in concert with reactivation of fibroblast gene expression and reprogramming transgene silencing. Many of these gene expression differences are evident from day 6, including significant upregulation of Apoa1 and concomitant downregulation of Col1a2 on the reprogramming trajectory, supporting our observations that these outcomes are established from early stages. We do not detect significant differences in transgene expression between the two trajectories at these early stages, suggesting that this is not a bifurcation driver (Fig. 4b,c; Extended Data Fig. 10a,b; Supplementary Table 7).
Figure 4. Molecular hallmarks of reprogramming trajectories.
(a) Identity scores of cells on the reprogramming (left, n=7 clones, 1,037 cells) and dead-end trajectories (right, n=7 clones, 1,037 cells, random downsampling from 2,270 cells) from reprogramming days 6 to 28. Cells scoring >0.75 (upper red line) classify as iEPs, cells scoring <0.25 (lower red line) classify as fibroblasts. (b) Violin plots of significantly different (p<0.001, permutation test, one-sided) gene expression between reprogramming and dead-end trajectories (n=14 clones, 2,074 cells). (c) Projection of Mettl7a1 and Col1a2 expression onto the t-SNE plot (n=48,515 cells). (d) Colony formation assay (CDH1/E-cadherin immunohistochemistry) for cells reprogrammed with Foxa1-Hnf4α, or Foxa1-Hnf4α-Mettl7a1. Scale bar=20mm. Blinded and automated colony quantification, (n=22 technical replicates, 3 independent biological replicates, p=8×10−5, t-test, one-sided). (e) Upper: scRNA-seq analysis of 6,559 Foxa1-Hnf4α reprogrammed cells and 6,559 (10,161 cells prior to random downsampling) Foxa1-Hnf4α-Mettl7a1 reprogrammed cells, 14 days post-reprogramming initiation. Lower: quantification of Foxa1-Hnf4α-Mettl7a1 reprogrammed cell distribution across reprogramming stages, represented by fold-change in distribution relative to Foxa1-Hnf4α cell distribution.
Focusing on later stages of reprogramming, we performed differential expression analysis of the trajectory bifurcation at day 21 (Supplementary Table 7). Mettl7a1, an as yet uncharacterized putative methyltransferase is transiently and significantly upregulated along the successful reprogramming trajectory (Fig. 4b,c). A related methyltransferase-like gene, METTL3 catalyzes N6-methyl-adenosine (m6A) modification of mRNAs, regulating stem cell differentiation and reprogramming to pluripotency20,21, thus we focused on Mettl7a1 in the context of enhancing reprogramming efficiency. Addition of Mettl7a1 to the standard Foxa1-Hnf4α reprogramming cocktail results in a two-fold increase in iEP colony formation (Fig. 4d). In agreement, scRNA-seq of Foxa1-Hnf4α and Foxa1-Hnf4α-Mettl7a1 reprogrammed cells shows that addition of Mettl7a1 to the reprogramming cocktail results in 2.5-fold more cells entering the fully reprogrammed state
(Fig. 4e,Extended Data Fig. 10c-g). Inclusion of CellTags in these reprogramming experiments show that under both control and Mettl7a1 conditions, average clone size did not significantly differ between the two conditions (Extended Data Fig. 10h-i). Thus, Mettl7a1, rather than expanding existing iEPs, promotes a true increase in reprogramming efficiency.
Discussion
Here, we have developed and validated a combinatorial indexing strategy, CellTagging, enabling the simultaneous analysis of clonal history and cell identity at single-cell resolution. Our longitudinal dissection of Foxa1-Hnf4α-mediated direct lineage reprogramming to iEPs reveals two distinct conversion trajectories: one to successful reprogramming, and one to a ‘dead-end’ state. We observe striking parallels between direct lineage reprogramming and induction of pluripotency: For instance, almost all cells initiate reprogramming, although transition to a fully pluripotent state is rare. This is characterized by two waves where in the second phase, a subset of cells are able to stably maintain the core pluripotency network4,22. In this context, the later bifurcation leading to the iEP state may parallel this second phase of reprogramming to pluripotency. Our identification of Mettl7a1 as a pro-reprogramming factor suggests that it may be an important molecular player in the stabilization of iEP identity in later stages of lineage conversion.
Fibroblast to iEP conversion also shares a common feature with reprogramming to pluripotency in terms of inefficiency. Based on the low frequency of pluripotent cell generation, earlier studies have suggested that the initiation and early phases of reprogramming are stochastic processes4,23. Here, our method of sequential CellTagging and lineage reconstruction is powerful in that it enables the probability of successful reprogramming to be quantified. Tracking reprogramming outcome of clones derived from a shared ancestor strongly suggests that the trajectory of cell conversion is determined from the outset in many cases. If these early stages of reprogramming were stochastic, we would expect to see heterogeneity in reprogramming outcome between clones of the same lineage. On the contrary, we observe that clones of the same lineage follow similar reprogramming trajectories. In agreement with earlier studies23, our CellTag and split approach shows that clonally-related cells, split into independent biological replicates, do not share reprogramming outcome, arguing against the existence of an elite cell type that is primed to reprogram. It is important to note here that although we control reprogramming factor stoichiometry, we do not control copy number or location of integration, which may produce a variable outcome between biological replicates.
Together, the evidence we present here suggests the existence of a privileged cell state in which reprogramming potential is predetermined. This is supported by several recent studies from reprogramming to pluripotency, suggesting the existence of a privileged state, or that cells can be coaxed into such a state via transient factor expression24–28. Furthermore, DNA barcode-based clonal analyses support a deterministic model of reprogramming29. Finally, scRNA-seq in combination with computational trajectory reconstruction suggests that reprogramming outcome can be predicted as early as two days following initiation via factor expression30. The next challenge will be to uncover the molecular hallmarks of this permissive state, enabling further future improvements in reprogramming cells toward any desired cell identity with high efficiency and fidelity.
Methods
Experimental Methods
Mice and derivation of mouse embryonic fibroblasts.
Mouse Embryonic Fibroblasts (MEFs) were derived from E13.5 C57BL/6J embryos. (The Jackson laboratory: 000664). Heads and visceral organs were removed and the remaining tissue was minced with a razor blade and then dissociated in a mixture of 0.05% Trypsin and 0.25% Collagenase IV (Life Technologies) at 37°C for 15 minutes. After passing the cell slurry through a 70μM filter to remove debris, cells were washed and then plated on 0.1% gelatin-coated plates, in DMEM supplemented with 10% FBS (Sigma-Aldrich), 2mM L-glutamine, and 50mM β-mercaptoethanol (Life Technologies). All animal procedures were based on animal care guidelines approved by the Institutional Animal Care and Use Committee.
Lenti- and Retrovirus Production.
Lentiviral particles were produced by transfecting 293T-17 cells (ATCC: CRL-11268) with the pSMAL-CellTag construct (see below), along with packaging constructs pCMV-dR8.2 dvpr (Addgene plasmid 8455), and pCMV-VSVG (Addgene plasmid 8454). Constructs were titered by serial dilution on 293T cells. Hnf4α-t2a-Foxa1 and Mettl7a1 were cloned into the pGCDN-Sam Retroviral construct and packaged with pCL-Eco (Imgenex), titered on fibroblasts. We opted to generate a bicistronic Hnf4α-Foxa1 construct, based on the T2A sequence to increase the consistency of reprogramming via maintenance of exogenous transcription factor stoichiometry. Virus was harvested 48hr and 72hr after transfection and applied to cells immediately following filtering through a low-protein binding 0.45μM filter.
CellTagging methodology.
To generate CellTags, we introduced an 8bp variable region into the 3’UTR of GFP in the pSMAL lentiviral construct31, using a gBlock gene fragment (Integrated DNA Technologies) and megaprimer insertion. This approach relies on the presence of 60bp ‘arms’ in the gene fragment that are homologous to the desired plasmid insertion site. The fragments were then introduced into the plasmid via PCR, followed by DpnI (New England Biolabs) treatment to digest non-modified plasmid. All the recovery from bacterial transformation (Stellar Competent Cells, Takara Biosciences) was grown overnight in liquid culture, followed by maxi-prep extraction of the plasmid DNA. This complex library of CellTag constructs was used to generate lentivirus (above) which was then used to transduce fibroblasts at a multiplicity of infection of ~3–4. For CellTag versions 2 and 3, a short 6bp sequence was also included, just upstream of the variable CellTag region. For CellTag version 2, this sequence motif is “GTGATG”. For CellTag version 3, this sequence motif is “TGTACG”. For both Drop-seq and 10x Genomics-based experiments, the starting fibroblast population was transduced with CellTag version 1 (denoted as CellTagMEF) for 24hr, followed by washing and culture for a further 48hr. At this point, cells were split with one portion taken for Drop-seq/10x and two portions replated for reprogramming to iEPs in two biological replicates. For 10x Genomics-based experiments, cells were tagged again, just following the 72hr of reprogramming, with CellTag version 2 (denoted as CellTagD3). One further round of CellTagging followed at day 13 post-initiation of reprogramming with CellTag version 3 (denoted as CellTagD13). Pooled CellTag libraries are deposited at Addgene: pSMAL-CellTag-V1 (https://www.addgene.org/115643); pSMAL-CellTag-V2 (https://www.addgene.org/115644); pSMAL-CellTag-V3 (https://www.addgene.org/115645).
Generation and collection of iEPs.
Early passage MEFs (< passage 6) were reprogrammed as in Sekiya and Suzuki, 201116. We modified this protocol, transducing cells every 12hr for 3 days, with fresh Hnf4α-t2a-Foxa1 retrovirus in the presence of 4mg/ml Protamine Sulfate (Sigma-Aldrich). These transduced cells were then cultured on 0.1% gelatin-treated plates for 1 week in hepato-medium: DMEM:F-12, supplemented with 10% FBS, 1 mg/ml insulin (Sigma-Aldrich), 100nM dexamethasone (Sigma-Aldrich), 10mM nicotinamide (Sigma-Aldrich), 2mM L-glutamine, 50mM β-mercaptoethanol (Life Technologies), and penicillin/streptomycin, containing 20 ng/ml hepatocyte growth factor (Sigma-Aldrich), and 20 ng/ml epidermal growth factor (Sigma-Aldrich). After 7 days of culture, the cells were transferred onto plates coated with 5μg/cm2 Type I rat collagen (Gibco, A1048301). For Drop-seq based experiments (two independent biological replicates), with a cell capture rate of 5%, 2×106 cells were initially seeded and cells harvested every seven days. At each harvest, cells were gently dissociated in TrypLE Express (Gibco), and 1.5×106 cells were collected for Drop-seq, replating and culturing the remaining cells. For 10x Genomics-based experiments, with a cell encapsulation rate of up to 60%, 5×105 cells were initially seeded and harvested every 3–7 days. At each cell harvest, 3×105 dissociated cells were fixed in methanol, replating and culturing the remaining cells. Methanol fixation was performed as previously described in32. Briefly, cells were collected and washed in Phosphate Buffered Saline (PBS), followed by resuspension in ice-cold 80% Methanol in PBS, with gentle vortexing. These cells were stored at −80°C for up to three months, and processed in the same batch on the 10x Genomics platform (below). iEP lines at the end of reprogramming tested negative for mycoplasma.
Immunostaining.
iEP cells were grown in 4-Chamber Culture Slides (Falcon #354114) and fixed in 4% paraformaldehyde (PFA). Cells were permeabilized in 0.1% Triton-X100, followed by blocking in 10% fetal bovine serum in PBS (blocking buffer). Primary antibody, goat anti-Apolipoprotein A-I antibody (1:100, Novus Biologicals, NB600–609, lot: 30506) or mouse anti-E-Cadherin (1:50, BD Biosciences, 610181, Clone: 36/E-Cadherin, lot: 7187865) in blocking buffer was applied overnight before washing and applying secondary antibody, Alexa Fluor 555 rabbit anti-goat IgG (1:1000, Invitrogen A-21431) or Alexa Fluor 488 goat anti-mouse IgG (1:1000, Invitrogen A-32723), diluted in blocking buffer. Nuclear staining was performed using 300nM DAPI in PBS. Slides were mounted with ProLong Gold antifade reagent (Invitrogen P36930). Images were captured using a Zeiss Axio Imager Z2 fluorescent microscope.
Mettl7a1 reprogramming and colony formation assay.
Mouse Mettl7a1 (NM 027334, Origene: MC205948) was sub-cloned into the retroviral vector, pGCDNSam16, and retrovirus produced as above. For comparative reprogramming experiments, MEFs (1.2×105 cells per 6cm plate, in 3 independent biological replicates) were serially transduced over 72hr (as above), followed by splitting and seeding at 4×104 cells per well of a 6-well plate to generate technical replicates. In control experiments, virus produced from an empty vector control expressing only GFP was added to the Hnf4α-Foxa1 reprogramming cocktail. In Mettl7a1 experiments, virus produced from the Mettl7a1-IRES-GFP construct was added to Hnf4α and Foxa1. Mettl7a1 overexpression was confirmed by harvesting RNA from Hnf4α-Foxa1 and Hnf4α-Foxa1-Mettl7a1- transduced cells (RNeasy kit, Qiagen). Following cDNA synthesis (Maxima cDNA synthesis kit, Life Tech), qPCR was performed to quantify Mettl7a1 overexpression (TaqMan Probe: Mm03031185_sH, TaqMan qPCR Mastermix, Applied Biosystems). Cells were reprogrammed for two weeks, at which point the cells of some wells were dissociated and methanol fixed for 10x Genomics-based single-cell analysis (details below). The remaining wells were processed for colony formation assays: cells were fixed on the plate with 4% PFA, permeabilized in 0.1% Triton-X100 then blocked with Mouse on Mouse Elite Peroxidase Kit (Vector PK-2200). Primary antibody, mouse anti-E-Cadherin (1:100, BD Biosciences) was applied for 30 min before washing and processing with the VECTOR VIP Peroxidase Substrate Kit (Vector SK-4600). Colonies were visualized on a flatbed scanner, adding heavy cream to each well in order to increase image contrast. Colonies were counted, using automated counting via an ImageJ plugin (https://imagej.nih.gov/ij/plugins/colony-counter.html). These analyses were blinded.
Drop-seq procedure.
Cells were dissociated using TrypLE Express (Gibco), washed in PBS containing 0.01% BSA and diluted to 100 cells/μl, then processed by Drop-seq within 15 minutes of their harvest. Drop-seq was performed as previously described17 (http://mccarrolllab.com/dropseq/). Briefly, cells and beads were diluted to an estimated co-occupancy rate of 5% upon co-encapsulation: 1×105 cells/ml and 1.2×105 beads/ml. Two independent lots of beads (Macosko201110, ChemGenes Corporation, Wilmington MA) were used: 091615 (Timecourse 3), 032516B (Timecourse 4). Emulsions were collected and broken using 1ml of Perfluorooctanol (Sigma) for 15ml of emulsion, followed by washing in 6X Saline-sodium citrate (SSC) buffer to recover beads. Reverse transcription was then performed using the Maxima H Minus Reverse Transcriptase kit (EP0752, Life Tech). After treatment with 2000U/ml of ExonucleaseI (New England Biolabs), aliquots of 2,000 beads (representing ∼100 single-cell transcriptomes for a cell: bead co-encapsulation rate of 5%) were amplified by PCR for 14 cycles, using Kapa HiFi Hotstart Readymix (Kapa Biosystems). The PCR product resulting from this reaction was purified by addition of 0.6x AMPure XP beads (Agencourt). 600 pg of this purified cDNA product from an estimated 5,000 cells was tagmented by Nextera XT according to the manufacturer’s protocol (Illumina). The resulting cDNA library was again purified using 0.6x AMPure XP beads, followed by 1x AMPure XP beads. cDNA concentrations were assessed by Tapestation (Agilent) analysis. Libraries were sequenced on an Illumina HiSeq 2500, with custom priming (Read1CustSeqB Drop-seq primer).
10x Genomics procedure.
For single-cell library preparation on the 10x Genomics platform, we used: the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (PN-120237), Chromium Single Cell 3′ Chip kit v2 (PN-120236) and Chromium i7 Multiplex Kit (PN-120262), according to the manufacturer’s instructions in the Chromium Single Cell 3′ Reagents Kits V2 User Guide. Just prior to cell capture, methanol-fixed cells were placed on ice, then spun at 3000rpm for 5 minutes at 4°C, followed by resuspension and rehydration in PBS, according to32. 17,000 cells were loaded per lane of the chip, aiming for capture of 10,000 single-cell transcriptomes. All samples were processed in parallel, on the same day. Resulting cDNA libraries were quantified on an Agilent Tapestation and sequenced on an Illumina HiSeq 3000.
Viral integration analysis.
Genomic DNA was harvested from control mouse embryonic fibroblasts and iEPs derived from clone 1 (Timecourse 4), using the DNeasy Blood & Tissue kit (Qiagen). Sample quality was assessed by Qubit DNA Assay Kit and gel electrophoresis. Library construction was carried out using the Nextera XT Library prep kit (Illumina) following the manufacturer’s recommendations. The lentivirus integration boundary sequence was enriched by amplifying using primers specific for lentivirus LTR and the Nextera XT adapter sequence. Two separate PCR reactions were performed for each sample, one for 3’LTR and another for 5’LTR. The final PCR was performed to add Illumina sequencing adapters with unique barcodes for each sample. The libraries for each sample were pooled into 1 final library and assessed by Qubit DNA assay, Agilent Bioanalyzer, and qPCR. The library was sequenced on the NextSeq 500 system using the 150 Cycle High Output flow cell. Fastq data was extracted from the NextSeq system using bcl2fastq and the QC of the data was performed using FastQC. Fastq reads were aligned to the mouse reference genome (GRCm38) using BWA MEM. Deduplication was performed using Samtools. Peak calling and comparison between two samples for putative lentivirus integration site was performed using MACS2.
Library preparation and sequencing of CellTag plasmid libraries for whitelist generation.
Library construction was carried out using the Nextera XT Library prep kit (Illumina), following the manufacturer’s recommendations. The CellTag region was enriched by amplifying using primers specific for the pSMAL lentivirus GFP UTR and the Nextera XT adapter sequence. A final PCR was performed to add Illumina sequencing adapters. The libraries for each CellTag version were pooled into 1 final library and assessed by Tapestation (Agilent). The library was sequenced on an Illumina MiSeq. Reads that contained the CellTag “motif” were identified (below). A 90% percentile cutoff in terms of reads reported for each CellTag was used to select CellTags for inclusion on the whitelist.
Computational methods
10x Genomics and Drop-seq alignment, digital gene expression matrix generation.
The Cell Ranger v2.1.0 pipeline (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest) was used to process data generated using the 10x Chromium platform. This pipeline was used in conjunction with a custom reference genome, created by concatenating the sequences corresponding to the Hnf4α-t2a-Foxa1 transgene and the GFP-CellTag transgene as new chromosomes to the mm10 genome. The unique UTRs in the Hnf4α-t2a-Foxa1 and GFP-CellTag transgene constructs allowed us to monitor transgene expression. To create Cell Ranger compatible reference genomes, the references were rebuilt according to instructions from 10x Genomics (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references). To achieve this, we first created a custom gene transfer format (GTF) file, containing our transgenes, followed by indexing of the FASTA and GTF files, using Cell Ranger ‘mkgtf’ and ‘mkref’ functions. Following this step, the default Cell Ranger pipeline was implemented, with the filtered output data used for downstream analyses. For Drop-seq analysis, raw reads were processed, filtered, and aligned as previously described17, including correction of barcode synthesis errors. This process, and the required tools, are further outlined online in the Drop-seq Alignment Cookbook (http://mccarrolllab.com/dropseq/). Again, in an effort to facilitate downstream analyses the reference genome used during alignment was modified to include the three transgenic sequences, above. Processed reads were aligned to a custom genome build, using STAR. Across all experiments, the mean number of confidently mapped reads per cell was 38,259 (Table S1).
Following alignment, digital gene expression (DGE) matrices were generated for each timepoint, for all time courses. Drop-seq DGEs were aggregated using a custom R script. Merged 10x Genomics DGE files were generated using the aggregation function of the Cell Ranger pipeline. We then performed initial filtering of these DGE files as a quality control step. We first removed cells with a low number (<200) of unique detected genes. We then removed cells for which the total number of unique molecules (UMIs, after log10 transformation) was not within three standard deviations of the mean. This was followed by the removal of outlying cells with an unusually high or low number of UMIs given their number of reads by fitting a loess curve (span = 0.5, degree = 2) to the number of UMIs with number of reads as predictor (after log10 transformation), removing cells with a residual more than three standard deviations away from the mean. This process was also used to remove cells for with unusually high or low number of genes given their number of UMIs. Finally, we removed cells in which the proportion of the UMI count attributable to mitochondrial genes was greater than 10% (for Drop-seq-based experiments) or 20% (for 10x Genomics-based experiments).
Data normalization and scoring of cell cycle phase.
Following DGE filtering, cell cycle scores were generated for each cell and data normalized. Cell cycle scores were generated using a pre-defined classifier to assign cell cycle phase for each cell. This classifier was built from training data by identifying pairs of genes where the difference in expression within each pair changed sign across phases. Via examination of the sign of the difference in test data, cell cycle phase is assigned to each cell. After calculating the cell cycle scores, the data was normalized using the “deconvolution” method. This method pools cells and combines the expression values of the cells in a pool. The pooled expression values are used to calculate size-factors for normalization. These pool-based normalization factors can then be deconvoluted into cell-specific normalization factors, which are then used to normalize each cell’s expression. This deconvolution normalization method is an attempt to address the abundance of zero counts that is prevalent to scRNA-seq. The cell cycle scores and data normalization was facilitated by the Scater package33, available on Bioconductor.
CellTag demultiplexing.
Reads containing the CellTag sequence were extracted from the processed, filtered, and unmapped reads BAM files produced in intermediate steps of the 10x Genomics and Drop-seq pipelines. Reads that contained the CellTag “motif” were identified as follows: CellTagV1 (CellTagMEF): “CCGGTNNNNNNNNGAATTC”, CellTagV2 (CellTagD3): “GTGATGNNNNNNNNGAATTC”, CellTagV3 (CellTagD13): “TGTACGNNNNNNNNGAATTC”. Following extraction of reads from the BAM file, a custom gawk script was utilized to parse the output, capturing the Read ID, Sequence, Cell Barcode, UMI, CellTag Sequence, and Aligned Genes for each read. This parsed output was then used to construct a Cell Barcode x CellTag UMI matrix. CellTags were grouped by Cell Barcodes and then the number of unique UMIs for each Cell Barcode, CellTag pair was counted. The matrix was then filtered to remove any cell barcodes not found in the filtered Cell Ranger and Drop-seq output files. Finally, the CellTags were filtered to remove any that were represented by ≤1 UMI. The construction and filtering of the CellTag UMI matrix was accomplished using a custom R script. Using this matrix, an error-correction step was then performed to remove errors generated via PCR and sequencing errors: CellTags one edit-distance apart were collapsed on a cell-by-cell basis, using Starcode34, an algorithm to determine which sequence pairs lie within a given Levenshtein distance, merging matched pairs into clusters of similar sequences. This filtered CellTag UMI count matrix was then utilized for all downstream clone and lineage analysis.
CellTag filtering and clone calling.
The CellTag matrix was initially filtered by removing CellTags that do not appear on the whitelists generated for each CellTag plasmid library (above). CellTags appearing in >5% of cells in the first timepoint were also removed as it would suggest dominance of the library by individual CellTags that would interfere with accurate clone-calling. The requirement for this filtering was rare. Cells expressing more than 20 CellTags (likely to correspond to cell multiplets), and less than 2 CellTags per cell were filtered out. To identify clonally-related cells, Jaccard analysis using the R package, Proxy was employed to calculate the similarity of CellTag signatures between cells. A Jaccard score of >0.7 was used as a cutoff to identify cells highly likely to be related, based on our experimental findings. We found this cutoff to be stringent enough for unrelated cells not to be connected, but in a small number of instances, we found related cells that were not connected, likely due to CellTag errors that were not corrected or CellTag dropout – these related cells were united as part of lineage construction, below. Clones were defined as groups of 3 or more related cells (for CellTagMEF, CellTagD3), or 2 or more related cells (for CellTagD13) identified using a custom R script. Clones were visualized using the Corrplot package with hierarchical clustering, contour plotting using GGPlot2, or via force-directed network graphs (see below). Clones were called on cells pre-filtered for numbers of genes, UMIs and mitochondrial RNA content.
Seurat, Monocle, and quadratic programming analyses.
After filtering and normalization, the R package, Seurat6 was used to cluster and visualize cells. As the data was previously normalized, it was loaded into Seurat without normalization, scaling, or centering. Along with the expression data, metadata for each cell is collected, including information such as clone identity, cell cycle phase, and timepoint (Supplementary Table 4). Seurat was used to remove unwanted variation, regressing out number of UMIs, proportion of mitochondrial UMIs, and cell cycle scores. Next, highly variable genes were identified and used as input for dimensionality reduction via principal component analysis (PCA). The resulting PCs and the correlated genes were examined to determine the number of components to include in downstream analysis. These PCs were then used as input to cluster the cells, visualizing these clusters using t-distributed stochastic neighbor embedding (t-SNE). Semi-supervised Monocle27 analysis was used to order cells in pseudotime, based on expression of the fibroblast marker, Col1a2, and the iEP marker, Apoa1. Quadratic programming, previously described in5, was employed to score fibroblast and iEP identity. This approach was modified to use bulk expression data of MEF and iEP collected previously16 and whole transcriptome profiles of the two cell types were used for identity score calculation. The R Package, QuadProg was used for quadratic programming to generate cell identity scores.
Lineage visualization via construction of force-directed network graphs.
Network graphs were constructed by integrating all data for all rounds of CellTagging. In the graphs, each node represents an individual cell, and edges represent clonal relationships between cells. First, using a custom R-based script, cells were assembled into sub-clusters, according to CellTagMEF, CellTagD3, and CellTagD13 information. Then, these sub-clusters were connected to each other to build lineages of related cells, connected across the different rounds of CellTagging – i.e. two different CellTagD3 clones sharing the same CellTagMEF labels are part of the same lineage. Via this approach, we identified collisions in 4.5±1.1% of clones – where a collision is defined as one clone sharing two or more parents. In these cases, we inspected the CellTag signature for each clone and united any clones that had been split, reducing the collision rate to 0.9±0.6%. The resulting networks are visualized as force-directed network graphs via Cytoscape 3.6.0 and Allegro Layout. Allegro Spring-Electric was used as the layout protocol to render force-directed network graphs. Individual graphs for each lineage can be explored via our Shiny-based interactive platform, CellTag Viz: http://www.celltag.org/.
Trajectory discovery via randomized testing.
To identify clones with an enriched or depleted rate of iEP generation, we used randomized testing to evaluate if each clone (of at least 35 cells in size) possesses a similar percentage of fully reprogrammed cells, relative to a randomly selected population of the same size. Here, the percentage of reprogrammed cells is defined as the proportion of cells within each group found in the reprogrammed cluster, as defined by Seurat. Two groups, cells of the clone and that of the overall population, are compared with the null percentage calculated using the cells in each clone. Let N represent the number of cells in each clone and M represent the remaining cell population size. We pool the two groups of cells (size = N+M) and resample N random cells, without replacement, from the pooled cells for (N+M)/N times such that every possible separation with ending groups of size N and M can be sampled and captured. During this process, the percentage is calculated based on the N randomly sampled cells. With the percentage calculated, p-values can be evaluated based on the proportion of randomly sampled cells with a percentage greater than or equal to the null percentage. Using the p-value of <0.05 (>0.95 for the other tail), we identified clones with enriched or depleted for reprogrammed cells. These calculations were performed using a custom R-based script. Clones with at least 35 cells were selected to increase the statistical power of this analysis. For permutation testing to analyze differences in trajectory-specific gene expression, a custom Python-based script was used.
Code availability.
Code for processing of CellTag data, clone-calling, and construction of lineage trees is available on GitHub (https://github.com/morris-lab).
Data availability.
All source data, including sequencing reads and single-cell expression matrices, are available from the Gene Expression Omnibus (GEO) under accession code GSE99915.
Reagent availability.
Pooled CellTag libraries are deposited and available from Addgene: pSMAL-CellTag-V1 (https://www.addgene.org/115643); pSMAL-CellTag-V2 (https://www.addgene.org/115644); pSMAL-CellTag-V3 (https://www.addgene.org/115645).
Extended Data
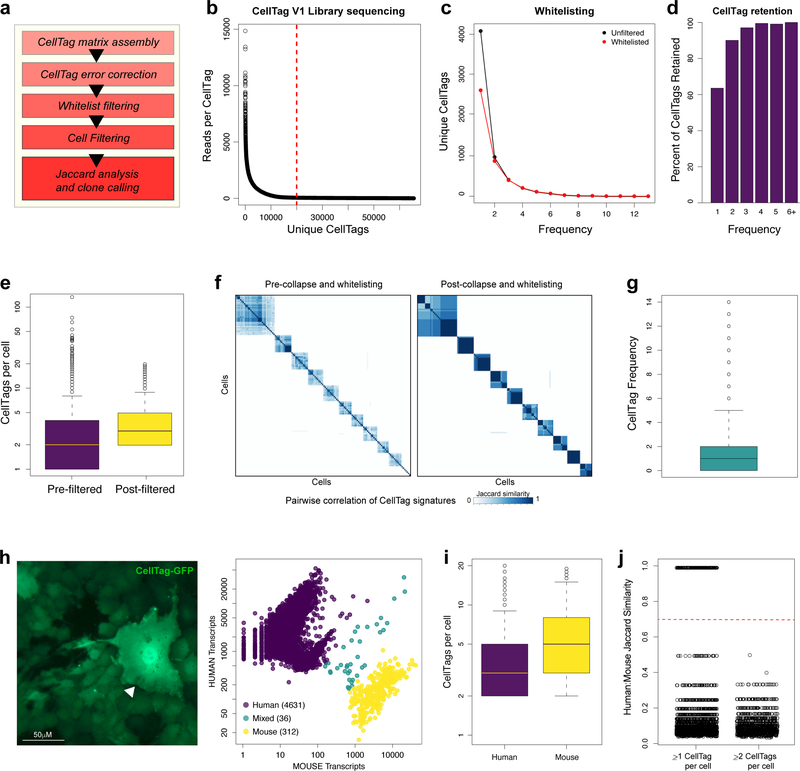
Extended Data Figure 1. Outline of the CellTag filtering pipeline, whitelisting and species mixing validations.
(a) Schematic of the CellTag processing and filtering pipeline: CellTag sequences are first extracted from aligned sequencing reads, followed by construction of a matrix of CellTag expression in each cell. To mitigate potential artifacts arising as a result of PCR and sequencing errors, we implemented an error-correction step via the collapse of similar barcodes one edit-distance apart, on a cell-by-cell basis. An initial filtering step removes any CellTags that do not appear on a ‘whitelist’ of CellTags that are confirmed to exist in the complex lentiviral library. A second filtering step removes cells expressing less than two, and more than 20 unique CellTags. Using this filtered dataset, Jaccard analysis is then applied (using the R package, Proxy) to identify related cells, based on CellTag signature similarity, allowing clones to be called. (b) Generation of the CellTag whitelist. Following CellTag lentiviral plasmid sequencing, CellTags were extracted from the raw fastq files via identification of the adjacent motifs described in the methods section. A 90th percentile cutoff in terms of reads reporting each CellTag was used to select CellTags for inclusion on the whitelist. Of a possible 65,536 unique combinations, we detected 19,973 sequences passing this 90th percentile of read counts. Data for CellTag version 1 (CellTagMEF) is shown here. Whitelist creation was also performed for CellTag versions 2 (CellTagD3) and 3 (CellTagD13). (c) CellTag frequency, i.e. how many times each CellTag is detected in a population of transduced cells, before (black data points) and after removal (red data points) of CellTags that do not feature on the whitelist. This ‘whitelisting’ predominantly results in the removal of CellTags appearing only once, singletons likely to arise due to sequencing and PCR errors. This is reflected in the histogram in (d), showing that only 60% of singleton CellTags detected are retained, whereas over 90% of CellTags appearing at a frequency of two or more are retained. (e) Mean CellTags per cell pre- and post-CellTag pipeline filtering. Cells in this figure correspond to the cells shown in Fig. 1b,c (replicate 1: n=8,535 cells; replicate 2: n=11,997 cells). (f) Pairwise correlation scores (Jaccard similarity) and hierarchical clustering of 10 major clones arising from this tag and trace experiment. Hierarchical clustering is based on each cell’s Jaccard correlation relationships with other cells, where each defined ‘block’ of cells represents a clone. Left panel: scoring and clustering of pairwise correlations, pre-whitelisting and filtering. Right panel: Post-whitelisting and filtering, pairwise correlations are stronger and more cells are detected within each clone (n=869 cells). (g) CellTag frequency metric: Each detected CellTag appears in less than two cells (n=9,072 cells in total) at the start of the experiment, on average, thus the library is not dominated by any abundant CellTags, which would potentially generate false positive results. (h) A species mixing experiment, consisting of a mixture of human 293T cells and mouse embryonic fibroblasts (left panel), labelled with ~3–5 CellTags per cell and expressing GFP as a result. A fibroblast (white arrow) is visible within a colony of 293T cells, scale bar=50μM. 72hr-post-transduction, cells were harvested and processed for scRNA-seq via Drop-seq. Right panel: following sequencing and alignment, cells were assigned to their corresponding species, revealing a low rate of doublet formation (n=4,631 human cells, 312 mouse cells, 36 mixed). (i) Mean CellTags per cell for human and mouse cells in the species mixing experiment. CellTag transcripts were detected in 70% of cells (n=3,493/4,979 cells). Of the tagged population, each cell expressed 5 CellTags on average: 3.8±0.002 (mean ± s.e.m.) in human cells, and 5.9±0.02 in mouse cells. (j) For each cell, CellTag signatures were extracted and Jaccard similarity analysis was performed to assess the frequency of CellTag signature overlap between the two species. To establish a false positive baseline, we initially compared CellTag overlap between mouse and human populations, as these cells are not related. From the analysis of 4,943 cells, we identified 200 instances of mouse:human cell pairings, out of a possible 1.5×107 pairs, sharing the same individual CellTags. This demonstrates that reliance on only one CellTag per cell does not uniquely label cells with high confidence. Excluding cells represented by only one CellTag removes this noise, resulting in no detection of cross-species CellTag signatures (Jaccard similarity index <0.7). This highlights the importance of combinatorial labelling, and the efficacy of our approach to uniquely label unrelated cells.
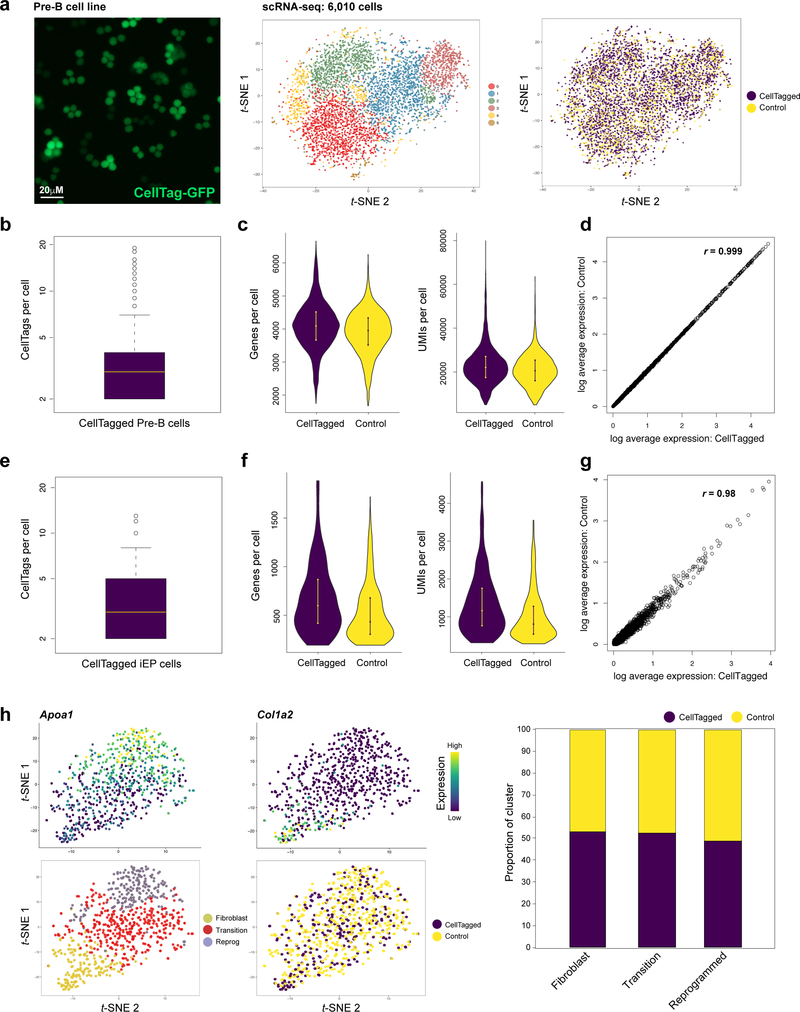
Extended Data Figure 2. CellTagging does not perturb cell physiology or reprogramming efficiency.
To assess the potential impact of CellTagging on cell physiology we performed scRNA-seq on CellTagged cells and unlabelled, control cells, 72hr post-tagging. (a) Left panel: Fluorescent image of CellTagged, GFP-expressing, pre-B cell line, HAFTL-1. Right panels: 10x Genomics-based scRNA-seq of CellTagged (n=3,943 cells) and non-tagged control cells (n=2,067 cells). Cells were clustered using Seurat, resulting in a t-SNE plot with 6 clusters of transcriptionally distinct cells. CellTagged and control cells were evenly distributed across these populations. (b) The CellTagged B-cell population expresses a mean of 3.5±0.02 CellTags per cell. (c) We detect no observable differences in numbers of genes or unique molecular identifiers (UMIs) per cell in either population. (d) Average gene expression values between CellTagged and control cells are highly correlated (r=0.999, Pearson’s correlation), demonstrating that our labelling approach does not induce significant changes in gene expression. These experiments were performed independently, twice, with similar results. (e) To assess the potential impact of CellTagging on reprogramming outcome, we induced lineage conversion of CellTagged cells in parallel with unlabelled, control cells, followed by three weeks of culture and processing on the Drop-seq platform (n=773 cells passing quality control). A mean of 3.3±0.09 CellTags per cell are expressed in a labelled reprogrammed cell population. (f) There are no observable differences in numbers of genes or UMIs per cell in either the labelled or unlabelled populations. (g) Average gene expression values between CellTagged and control cells are highly correlated (r =0.98, Pearson’s correlation), again demonstrating that our labelling approach does not induce significant changes in gene expression. (h) Seurat clustering of cells, where cells in fibroblast (Col1a2-high), transition, and fully-reprogrammed (Apoa1-high) states can be identified. Right panel: CellTagged and control cells are distributed fairly evenly across these reprogramming stages. Some variation is expected between these independent biological replicates. These experiments were performed independently, twice, with similar results.
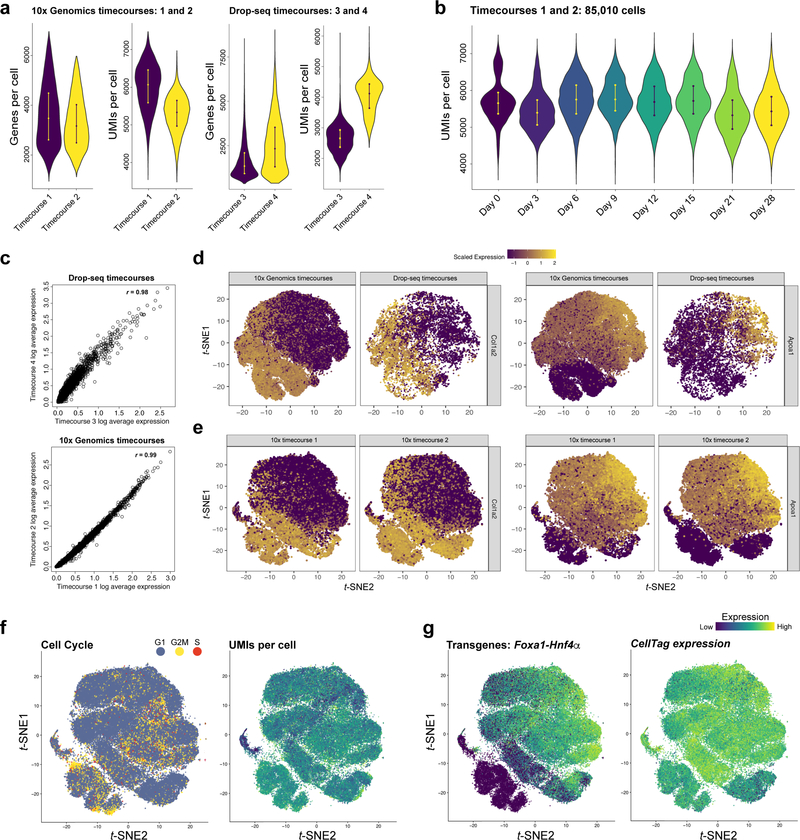
Extended Data Figure 3. scRNA-seq metrics and quality control of cell clustering.
(a) Numbers of genes and UMIs per cell for 10x Genomics-based (Timecourse 1, n=30,733 cells and timecourse 2: n=54,277 cells) and Drop-seq-based (Timecourse 3, n=5,932 cells and timecourse 4: n=5,414 cells) reprogramming timecourses. In these cross-platform comparisons, we apply more stringent filtering of Drop-seq data to include only those cells with 1000 or more UMIs. For Drop-seq experiments, with a cell capture rate of 5%, 2×106 MEFs were initially seeded for reprogramming. For 10x Genomics experiments, with a cell encapsulation rate of up to 60%, 5×105 MEFs were initially seeded for reprogramming. (b) Mean numbers of UMIs per cell (5,570±2.2), at each captured timepoint during reprogramming, in two independent biological replicates (10x Genomics, timecourses 1 and 2): Cells were captured at days 3, 6, 9, 12, 15, 21, and 28, along with the initial MEF population (day 0). (c) Average gene expression values between 10x Genomics replicates, and Drop-seq replicates are highly correlated at day 0, demonstrating technical consistency (r=0.99, and r=0.98, respectively, Pearson’s correlation). (d) Alignment of independent 10x Genomics replicates (Timecourses 1 and 2) with Drop-seq replicates (Timecourses 3 and 4) via canonical correlation analysis19. Left panels: Expression of MEF marker, Col1a2. Right panel: iEP marker, Apoa1. Overlay of data from these two sources demonstrates a high level of technical and biological consistency between the two technologies. (e) Alignment of 10x Genomics replicates (Timecourse 1 and 2) via canonical correlation analysis. Left panels: Expression of fibroblast marker, Col1a2. Right panel: iEP marker, Apoa1. Integration of these two replicates demonstrates a high level of technical and biological consistency. (f) Projections of cell cycle phase and UMIs per cell onto t-SNE alignment of timecourses 1 and 2 shows that clustering is independent of these factors. (e) Reprogramming factor expression (via detection of bicistronic Foxa1-t2a-Hnf4α transgene expression) and CellTag expression across timecourses 1 and 2.
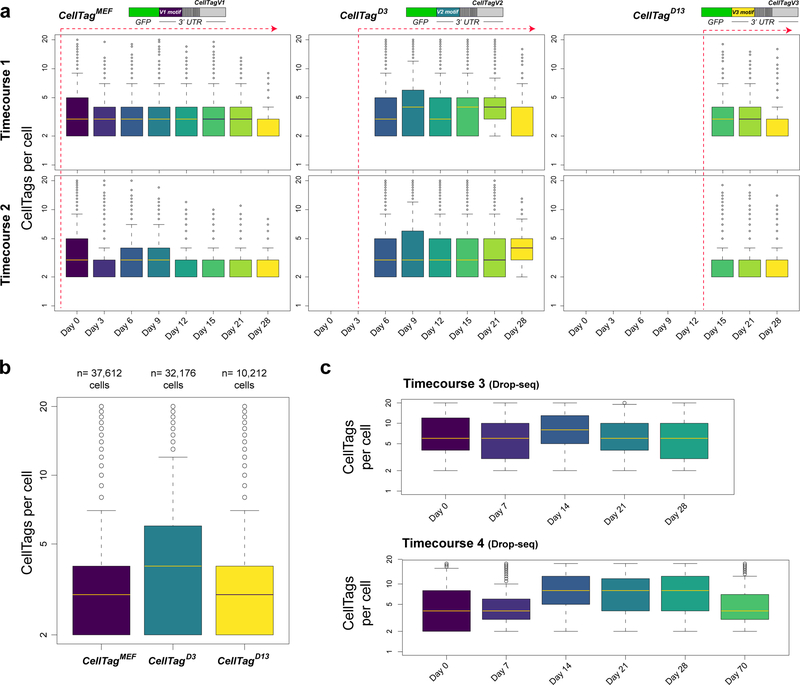
Extended Data Figure 4. CellTag expression metrics.
(a) Mean counts of CellTags expressed per cell, following whitelisting and filtering for timecourses 1 (n=19,581 cells passing filtering) and 2 (n=38,943 cells passing filtering), broken down by timepoint and CellTag version. Red dashed lines denote time of CellTag introduction. (b) Mean number of CellTags expressed per cell, post-whitelisting and filtering, for each round of CellTagging across timecourses 1 and 2. CellTagMEF: 3.4±0.01 CellTags per cell, n=37,612 cells. CellTagD3: 4.5±0.02 CellTags per cell, n=32,176 cells. CellTagD13: 3.2±0.02 CellTags per cell, n=10,212 cells. 65% of sequenced cells pass the ≥2 CellTag expression threshold to support tracking. (c) Mean CellTags per cell following whitelisting and filtering for both Drop-seq timecourses, broken down by timepoint. All cells with 200 or more genes were included in this analysis (Timecourse 1: n=10,038 cells, timecourse 2: n=9,839 cells). CellTags were introduced only in MEFs, prior to reprogramming in these experiments. In Drop-seq timecourses, we detected a mean of 7.8±0.07 CellTags per cell, across 61% of cells (12,086/19,877 cells) passing the tracking threshold.
Extended Data Figure 5. Assignment of cluster identities based on mRNA and protein expression.
(a) Top enriched gene expression associated with each cluster, projected onto the reprogramming t-SNE plot (n=85,010 cells). (b) Left panel: Expression of the fibroblast marker, Col1a2, projected onto the t-SNE plot. Upper right panel: Violin plot of Col1a2 expression levels in each cluster. Lower right panel: Violin plot of Apoa1 expression levels in each cluster, ordered by gain of expression over the course of reprogramming. Clusters are classified as one of four reprogramming stages: Clusters 5, 6, 7, 11 = Fibroblast. Clusters 0, 3 = Early Transition. Clusters, 1, 4, 8, 9,10,12 = Transition. Cluster 2 = Reprogrammed. Apoa1 is not expressed in the fibroblast clusters. (c) Upper panel: Expression of the previously reported iEP marker, Cdh1 (E-Cadherin)3,16, projected onto the t-SNE plot, highlighting the location of fully reprogrammed cells. Lower panel: Staining of CDH1 protein in iEP colonies emerging following three-weeks of reprogramming (control shown from Fig.4d). Scale bar=20mm. (d) Upper panel: Expression of the novel iEP marker, Apolipoprotein A1, Apoa1, projected onto the t-SNE plot. Lower panel: Immunofluorescent staining and imaging of APOA1 protein in an iEP colony, emerging following three-weeks of reprogramming. APOA1 (red) is localized to vesicles. This is a representative image selected from five independent biological replicates. Scale bar=20μM. (e) Upper panel: Co-expression of Apoa1 and Cdh1, within the same individual cells at the transcript level in the fully reprogrammed cluster confirms Apoa1 as a marker of iEP emergence. Lower panel: Immunofluorescent co-staining and imaging of APOA1 and CDH1 protein in iEPs. White arrows mark emerging iEP colonies co-expressing these two proteins. APOA1 expression (red) is found localized to vesicles of CDH1-positive cells (green), where the most intense CDH1 staining is observed at cell-cell junctions. This is a representative image selected from two independent biological replicates. Scale bar=20μM.
Extended Data Figure 6. Combinatorial CellTagging to identify clonally-related cells.
(a) Heatmap showing scaled expression of individual CellTags in 20 major clones called from CellTagD3-labelled cells (n=10 representative cells per clone, timecourses 1 and 2). Dashed yellow line marks separation between the two timecourses. Dashed red lines mark separation between independent clones. Although some CellTags are shared between these independent biological replicates, the combined CellTag signatures are unique. (b) Expression levels of individual CellTags per cell over three weeks in a representative clone labelled by 4 unique CellTags. Expression diminishes over time, but is not completely silenced. (c) To assess CellTag silencing we selected 10 major clones (n=6,728 cells), defining the intact CellTag signature for each clone at reprogramming day 6. We then assess loss, or ‘dropout’ of CellTags from each signature over the timecourse, to day 28. By week 4, expression of an individual CellTag ‘drops-out’ in 1 out of 10 cells - i.e. expected CellTag expression was not detected in 11±2% of cells. Conversely, CellTag expression is retained in almost 90% of cells by day 28. Later rounds of CellTagging (CellTagD13) are less prone to this effect, with CellTags dropping out in only 3±1.5% of cells. (d) We mapped CellTag expression across four representative clones, where expression of each CellTag is plotted over time. The y-axis denotes the percentage of cells within each clone where specific CellTag expression has dropped out. Typically, only one CellTag exhibits dropout, where expression of the other CellTags is maintained. We do not observe complete silencing, i.e. loss of expected CellTag expression in 100% of cells. This demonstrates the advantage of our CellTag combinatorial indexing method to reliably label cells and track them over an extended period of time. For example, reliance on the expression of a single, longer barcode would not be effective following integration into a region that later becomes silenced.
Extended Data Figure 7. Visualizing growth of clones and gene expression correlation within clones.
(a) Connected barplots showing individual clones as a proportion of all clones called at each reprogramming timepoint for timecourse 2, for each round of CellTagging (n=14,088 cells across 1,120 clones). Connected bars denote clonal expansion and growth over time. (b) Average number of cells per clone, per timepoint, for each round of CellTag labelling (timecourse 2, n=1,120 clones). (c) Number of clones detected at each timepoint, for each round of CellTagging over reprogramming timecourse 1 (n=1,031 clones) and 2 (n=1,120 clones). The number of clones detected gradually increases over time as probability of capture increases with clonal growth. The number of clones then begins to decrease as the growth of some individual clones outcompetes other clones which are lost from the population over time. (d) Connected barplots showing individual clones as a proportion of all clones called at each reprogramming timepoint for Drop-seq replicates 1 (n=103 clones) and 2 (n=37 clones). In replicate 2, a single clone progressively dominates the culture over 10 weeks of growth. In our viral integration analyses shown in Supplementary Table 5, we detect three viral integration sites in the cells of this clone. We did not detect any differential expression of genes proximal to these integration sites. Similarly, analysis of gene expression enrichment in 12 dominant clones across two biological replicates does not reveal any common signature of these clones to explain their rapid expansion (data not shown). This suggests that the clonal growth we observe is a normal part of the iEP reprogramming process, where the cells enter a progenitor-like state. Even so, these analyses do not exclude the acquisition of genetic and epigenetic changes endowing these expanding clones with increased fitness. (e) Correlation of Principal Component Analysis (PCA) scores in clonally-related cells (clone 2315, n=58 cells), relative to a random sampling of cells. Correlation between PC scores was used as a proxy for transcriptional similarity between cells. Clonally related cells were much more closely correlated, relative to randomly selected cells. (f) Quantification of correlation analysis for all timecourse 2 clones consisting of 10 cells or more, for CellTagMEF (n=78 clones, 3,963 cells) and CellTagD3-labelled clones (n=109 clones, 6,265 cells). Mean correlation scores for clonally-related cells are significantly higher than random cell groupings (p<0.001, t-test, one-sided). We tagged cells both before and after the 72hr reprogramming window, expecting much heterogeneity to be introduced via serial viral transduction. On the contrary, there is only a slight but insignificant increase in PCA correlation between CellTagMEF and CellTagD3-labelled, clonally-related cells.
Extended Data Figure 8. Reconstruction and visualization of lineages via force-directed graph drawing.
(a) Force-directed graph of all clonally-related cells and lineages reconstructed from timecourse 1 (1,031 clones, 12,932 cells) and (b) timecourse 2 (1,120 clones, 14,088 cells). All lineages and clone distributions can be interactively explored via our companion website, CellTag Viz (http://www.celltag.org/). (c) In this tree, we follow CellTagMEF clone 487 from timecourse 1, and its descendants. Each node represents an individual cell, and edges represent clonal relationships between cells. Purple = CellTagMEF clone, Blue = CellTagD3 clones, Yellow = CellTagD13 clones. In the lineage highlighted in red, we follow the CellTagMEF clone (n=678 cells), branching into two CellTagD3 lineages (clone 204 (n=363 cells) and clone 240 (n=260 cells)). (d) Contour plots, representing cell density of each clone, projected onto the t-SNE plot, for the lineage shown in (a). Upper left: Cells belonging to clone 487 (CellTagMEF). Clones 204 and 240 (CellTagD3) descend from this first clone, exhibiting a high degree of overlap within 2D-space, on the t-SNE plot. An unrelated CellTagD3 clone, 329 (n=38 cells), does not overlap with this lineage, demonstrating the high similarity between cells belonging to the same lineage.
Extended Data Figure 9. Mapping reprogramming trajectories and timing of cell fate decisions.
(a) Projection of all clones (yellow, n=2,151 clones, 27,020 cells) across reprogramming timecourses 1 and 2 (n=85,010 cells). Clusters with the highest density of detected clones, outlined in red (clusters 0, 1, 2, 4, 8, and 12) were subsetted from this larger dataset and re-clustered to generate a higher-resolution t-SNE plot, focusing on reprogramming days 6 to 28 (n=48,515 cells). (b) Left panel: original cluster identities of all cells (n=85,010 cells). Right-panel: subset of 48,515 cells, colored by original cluster identity. (c) Contour plots of iEP-depleted clone distribution (upper panels, (n=7 clones, 1,037 cells) and iEP-enriched clone distribution (lower panels, (n=7 clones, 2,270 cells) broken down by reprogramming day, and across days 9–28 (right panels). These specific clones were selected from the larger iEP-depleted and -enriched group as they had cells distributed across all timepoints to enable definition of the trajectories. Via these distributions, clusters 8, 4 and 3 are iEP-depleted, thus representing the dead-end trajectory. Conversely, clusters 2, 6 and 1 are iEP-enriched, representing the reprogramming trajectory. These trajectories divide cluster 0 into two halves, although re-clustering does not offer any higher resolution (data not shown). Deeper sequencing of more cells may provide further insights into this cluster in future. (d) Monocle2 psuedotemporal ordering of subsetted cells (n=48,515 cells), colored by day of reprogramming (left panel), Seurat cluster ID (middle panel) and Apoa1 expression (right panel). Monocle2 uses dimension reduction to represent each single-cell in 2D space and effectively ‘connects-the-dots’ to construct a differentiation trajectory. In this analysis, we performed semi-supervised ordering using Col1a2 (marking fibroblast identity) expression as a start point and Apoa1 expression (marking iEP identity) as an endpoint. Here, the branched trajectory generated by monocle is in general agreement with our clonal analyses. (e) Restriction of CellTagD13 clones (timecourse 1, n=79 clones, 240 cells, timecourse 2, n=30 clones, 148 cells) to either the reprogrammed cluster (cluster 1), or the dead-end cluster (cluster 3) at day 28. 88±8% of clones from these two biological replicates exhibit adherence to one of these trajectories by day 13 of reprogramming. (f) We identified lineages where multiple CellTagD3-labelled clones share a common CellTagD0-labelled ancestor. The proportion of each clone on the reprograming trajectory (defined as occupancy of clusters 2, 6, and 1 on the subsetted t-SNE plot), and proportion of each clone on the dead-end trajectory (defined as occupancy of clusters 8, 4, and 3) was calculated. We then plotted the proportion of each CellTagD3-labelled clone on the reprogramming trajectory against that of its CellTagD3-labelled descendants (r=0.71, Pearson’s correlation, n=13 lineages, 57 clones, 6,035 cells).
Extended Figure 10. Reprogramming trajectory-enriched gene expression: Mettl7a1 expression promotes iEP generation.
(a) Violin plots of significantly different gene expression between reprogramming and dead-end trajectories (n=2,074 cells). Wnt4 and Spint2 expression is significantly upregulated along the reprogramming trajectory (p<0.001, permutation test, one-sided, n=1,037 cells). Dlk1 and Peg3 expression is significantly upregulated along the dead-end trajectory (p<0.001, permutation test, one-sided, n=1,037 cells). Expression of the Foxa1-Hnf4α transgene is significantly downregulated along the dead-end trajectory (p<0.001, permutation test, one-sided, n=1,037 cells). (b) Projection of gene expression onto the t-SNE plot (n=48,515 cells). (c) Mean numbers of genes and transcripts per cell following 10x Genomics-based scRNA-seq analysis: Foxa1-Hnf4α reprogrammed cells (n=6,559 cells) and Foxa1-Hnf4α-Mettl7a1 reprogrammed cells (n=10,161 cells), harvested 14 days after initiation of reprogramming. For subsequent analyses, the Foxa1-Hnf4α-Mettl7a1 experimental group was randomly downsampled for direct comparison to the Foxa1-Hnf4α experimental group (n=6,559 cells for both groups). (d) Via canonical correlation analysis19, the Foxa1-Hnf4α and Foxa1-Hnf4α-Mettl7a1 scRNA-seq datasets were merged with cells from timecourse 2, to help place these two experimental groups within these previously defined trajectories. Expression levels of Apoa1 are projected onto this t-SNE plot. (e) Confirmation of Mettl7a1 expression, by qPCR, following transduction of cells with Foxa1-Hnf4α-GFP vs. Foxa1-Hnf4α-Mettl7a1 retroviruses (**p=5.3×10−3, t-test, one-sided). (f) Violin plot of mean Apoa1 expression in Foxa1-Hnf4α, and Foxa1-Hnf4α-Mettl7a1 reprogrammed cells. Addition of Mettl7a1 to the reprogramming cocktail results in a significant increase in Apoa1 expression, supporting observations that this factor increases the yield of fully reprogrammed cells (p<0.001, permutation test, one-sided). (g) Plot of identity scores of Foxa1-Hnf4α (purple) and Foxa1-Hnf4α-Mettl7a1 (green) reprogrammed cells, where cells are ordered according to an increase in iEP identity. Red dashed line indicates a cutoff of 0.75, where above this score cells are considered as iEPs. 3-fold more Foxa1-Hnf4α-Mettl7a1 cells classify as iEPs, relative to Foxa1-Hnf4α cells, represented as a significant increase in iEP score (p<0.001, permutation test, one-sided). (h) Boxplot of mean CellTag expression between Foxa1-Hnf4α (3±0.05 CellTags per cell) and Foxa1-Hnf4α-Mettl7a1 (2.5±0.04 CellTags per cell) experimental groups. (i) Boxplot of cells per clone for Foxa1-Hnf4α and Foxa1-Hnf4α-Mettl7a1 experimental groups, following data processing via our CellTag demultiplexing and clone calling pipeline. Clone size does not significantly differ between these two groups: Foxa1-Hnf4α, 6±0.4 cells per clone (n=99 clones, 595 cells), Foxa1-Hnf4α-Mettl7a1: 6.3±0.65 cells per clone (n=43 clones, 277 cells), demonstrating that addition of Mettl7a1 enhances iEP yield via an increase in the number of unique reprogramming events. For comparison, average clone size at ~ day 14 for timecourse replicates 1 and 2 is ~ 8 cells per clone.
Supplementary Material
Acknowledgments.
We thank members of the Morris lab, and T.Druley and R.Mitra for critical discussions; S.McCarroll, E.Macosko and M.Goldman for advice establishing Drop-seq; B.Treutlein for quadratic programming assistance; J.Dick for the kind gift of pSMAL31; K.Kniepkamp for CellTag Viz assistance. This work was funded by National Institutes of Health (NIH) grants R01-GM126112, R21-HG009750; Silicon Valley Community Foundation, Chan Zuckerberg Initiative Grants HCA-A-1704–01646 and HCA2-A-1708–02799; The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital MI-II-2016–544. S.A.M is supported by a Vallee Scholar Award; B.A.B: NIH- T32HG000045–18; S.E.W: NIH-5T32GM007067–44; K.K: Japan Society for the Promotion of Science Postdoctoral Fellowship.
References
- 1.Vierbuchen T & Wernig M Direct lineage conversions: unnatural but useful? Nat. Biotechnol 29, 892–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahan P et al. CellNet: Network Biology Applied to Stem Cell Engineering. Cell 158, 903–915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris SA et al. Dissecting Engineered Cell Types and Enhancing Cell Fate Conversion via CellNet. Cell 158, 889–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buganim Y et al. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell 150, 1209–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treutlein B et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature (2016). doi: 10.1038/nature18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter SN, Baker LC, Mittelman D & Porteus MH Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 15, R75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Z et al. A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell Stem Cell 20, 120–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat. Biotechnol. (2018). doi: 10.1038/nbt.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. (2018). doi: 10.1038/nbt.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner DE et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science eaar4362 (2018). doi: 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekiya S & Suzuki A Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T et al. m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 16, 289–301 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Batista PJ et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polo JM et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell 151, 1617–1632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna J et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo S et al. Nonstochastic Reprogramming from a Privileged Somatic Cell State. Cell 156, 649–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babos KN et al. Balancing dynamic tradeoffs to drive cellular reprogramming. bioRxiv 393934 (2018). doi: 10.1101/393934 [DOI]
- 26.Rais Y et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502, 65–70 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Di Stefano B et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506, 235–239 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Di Stefano B et al. C/EBPα creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat. Cell Biol 18, 371–381 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Yunusova AM, Fishman VS, Vasiliev GV & Battulin NR Deterministic versus stochastic model of reprogramming: new evidence from cellular barcoding technique. Open Biol. 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiebinger G et al. Reconstruction of developmental landscapes by optimal-transport analysis of single-cell gene expression sheds light on cellular reprogramming. bioRxiv (2017).
- 31.van Galen P et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–72 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Alles J et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy DJ, Campbell KR, Lun ATL & Wills QF Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 347, btw777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zorita E, Cuscó P & Filion GJ Starcode: sequence clustering based on all-pairs search. Bioinformatics 31, 1913–1919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data, including sequencing reads and single-cell expression matrices, are available from the Gene Expression Omnibus (GEO) under accession code GSE99915.