Abstract
MicroRNAs (miRNAs) play a great contribution to the development of diabetic nephropathy (DN). The aim of this study was to explore potential miRNAs-genes regulatory network and biomarkers for the pathogenesis of DN using bioinformatics methods.
Gene expression profiling data related to DN (GSE1009) was obtained from the Gene Expression Omnibus (GEO) database, and then differentially expressed genes (DEGs) between DN patients and normal individuals were screened using GEO2R, followed by a series of bioinformatics analyses, including identifying key genes, conducting pathway enrichment analysis, predicting and identifying key miRNAs, and establishing regulatory relationships between key miRNAs and their target genes.
A total of 600 DEGs associated with DN were identified. An additional 7 key DEGs, including 6 downregulated genes, such as vascular endothelial growth factor α (VEGFA) and COL4A5, and 1 upregulated gene (CCL19), were identified in another dataset (GSE30528) from glomeruli samples. Pathway analysis showed that the down- and upregulated DEGs were enriched in 14 and 6 pathways, respectively, with 7 key genes mainly involved in extracellular matrix–receptor interaction, PI3K/Akt signaling, focal adhesion, and Rap1 signaling. The relationships between miRNAs and target genes were constructed, showing that miR-29 targeted COL4A and VEGFA, miR-200 targeted VEGFA, miR-25 targeted ITGAV, and miR-27 targeted EGFR.
MiR-29 and miR-200 may play important roles in DN. VEGFA and COL4A5 were targeted by miR-29 and VEGFA by miR-200, which may mediate multiple signaling pathways leading to the pathogenesis and development of DN.
Keywords: bioinformatics, diabetic nephropathy, differentially expressed genes, microRNAs
1. Indroduction
Diabetic nephropathy (DN) is one of the most common and severe microvascular complications of diabetes. Approximately, 30% to 40% of all patients with diabetes develop DN.[1] DN has become the main cause of end-stage renal disease in developed countries.[2] As the lack of effective treatment for DN, expensive medical cost status is persisting across the globe. From 1990 to 2012, the number of deaths caused by DN increased by 94%.[3] Therefore, investigating the pathogenic mechanisms of DN and early screening of patients with DN are important to prevent the occurrence and development of DN.
The role of miRNAs in the physiological mechanisms and therapeutic interventions of DN have received increasing attention.[4] The accumulating data and new discoveries are expected to generate new insight into the complex pathogenesis of DN.[5,6] Regulating miRNAs expression could become a very attractive measure for preventing DN. Although a number of miRNAs have recently been identified related to DN,[7] more efforts are needed to discover new targets for DN treatment and to validate these targets through in vitro and in vivo experiments and large-scale population studies.
The recent rise of bioinformatics has shed new light on studies of diabetic kidney disease. Researchers have used microarray data from mouse DN models in the Gene Expression Omnibus (GEO) database to analyze molecular mechanisms and genetic factors related to DN.[8] Previous bioinformatics analyses using human DN-related gene chip data (GSE1009) from the GEO database found that the CXCR2, CASR, CSF2, VEGFA, and CD2AP genes played important roles in maintaining kidney functions in DN and that miR-1 targeting VEGFA and miR-25 targeting CD2AP might affect the occurrence and development of DN.[9] Cui et al[10] identified several important genes associated with DN using GSE30122 chip data, including CD44, FN1, CCL5, and CXCR4; these genes were mainly enriched in the extracellular matrix (ECM)-receptor and cytokine–cytokine receptor interaction pathways. At the same time, 3 potential miRNA (miR-17-5p, miR-20a, and miR-106a) were discovered. However, the regulatory network involving these miRNA-DEGs and the related signaling pathways remain unknown.
In the present study, the differentially expressed genes (DEGs) between DN glomerular tissue and normal glomerular tissue were screened, and the results were combined with the data from the GSE30122 microarray data about glomerular tissue samples. Notably, the regulatory relationship between miRNAs and genes were constructed and validated using 5 target gene prediction databases. In addition, the regulatory network of miRNAs and target genes were connected with their corresponding signaling pathways to reveal potential regulatory mechanisms of DN pathogenesis and to promote the discovery of novel DN markers and therapeutic targets.
2. Methods
2.1. Acquisition of microarray data
The gene expression profiles associated with DN were obtained from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). The following search terms were used: “diabetic nephropathy or diabetic kidney disease,” “Homo sapiens,” and “Expression profiling by array.” After careful screening, the microarray data (GSE1009) submitted by Baelde et al[11] were selected for subsequent analysis. The experimental platform GPL8300 used in the above study was the Affymetrix HG_U95Av2 Array (Affymetrix GeneChip U95 Version 2 Array). The dataset included a total of 6 samples: 3 DN glomerular tissue samples and 3 normal glomerular tissue samples. Another set of microarray data related to diabetic kidney disease (DKD) (GSE30122) contained 69 samples,[12] including 9 DKD glomerular tissue samples, 26 normal control glomerular tissue samples, 10 DKD renal tubular tissue samples, and 24 normal control renal tubular tissue samples, of which 35 glomerular tissue samples were selected for analysis. Ethical approval was not necessary in this study because the gene expression profiles were downloaded from GEO, a public database, and we do not conduct new experiments in patients or animals.
2.2. Screening of DEGs
The online analysis tool GEO2R[13] was used to preprocess and analyze the microarray data and screen for DEGs associated with DN. The screening conditions were P value <.01 and |logFC| ≥1.5.
2.3. Identification of key DEGs
To analyze the interactions among the encoded proteins, the Search Tool for the Retrieval of Interacting Genes (STRING10.5)[14] database was used to construct a protein–protein interaction network. The CytoHubba plugin[15] was installed in the visualization software Cytoscape,[16] and the hubba values of each DEG were calculated according to the 12 topological algorithms by the CytoHubba plugin. The data were downloaded and sorted, and the top 15 hub nodes were selected. The common hub nodes calculated by ≥5 topological algorithms were eventually designated the hub genes. The intersections between the hub genes and the DEGs identified in the GSE30122 microarray data were used to determine the key genes for DN.
2.4. Gene ontology functional enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analysis
The gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization, and Integration Discovery (DAVID 6.8).[17] The P value <.05 and gene count ≥2 were set as the cutoff criteria.
2.5. Prediction of key miRNAs
Web-based Gene Set Analysis Toolkit (WebGestalt)[18] online datebase was used to predict miRNAs that targeted the DEGs associated with DN. The differentially expressed miRNAs were selected according to the screening criterion of P value <.01. The results were exported into the Cytoscape software for analysis. The miRNA–protein interaction networks were constructed through network topology characteristics. The node degree was calculated using the Network analysis plugin, and miRNAs with a node degree >12 were selected as the key miRNAs.
2.6. Exploration of regulatory relationships between the key miRNAs and their target genes
The regulatory relationships between the key miRNAs and their target genes of DN were established based on Cytoscape and verified through 5 miRNA target gene prediction databases, including TargetScan (http://www.targetscan.org/vert_71/), starBase (http://starbase.sysu.edu.cn/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/), microRNA.org (http://34.236.212.39/microrna/home.do), and miRDB (http://Mirdb.org/). The potential regulatory network for the pathogenesis and development of DN were established by combining these findings with the KEGG pathway analysis results.
3. Results
3.1. Identification of DEGs
Overall, 600 DEGs were identified when DN glomerular tissue samples versus normal glomerular tissue samples from GSE1009, of which 257 were downregulated and 343 were upregulated. The same method was used to analyze and identify 301 DEGs between DKD and normal glomerular tissues in the GSE30122 microarray data, with 253 downregulated genes and 48 upregulated genes.
3.2. Screening of key genes of DN
The 15 downregulated hub genes (eg, VEGFA, EGFR, and ITGA3) and 15 upregulated hub genes (eg, AGT, CXCR2, and CCL19) were selected (Table 1). By intercrossing with the glomerular tissue-related DEGs in GSE30122, the 7 key genes for DN were identified, including 6 downregulated genes (VEGFA, ITGA3, ITGB5, COL4A3, COL4A5, and CBLB) and 1 upregulated gene (CCL19) (Fig. 1).
Table 1.
Hub genes of diabetic nephropathy.
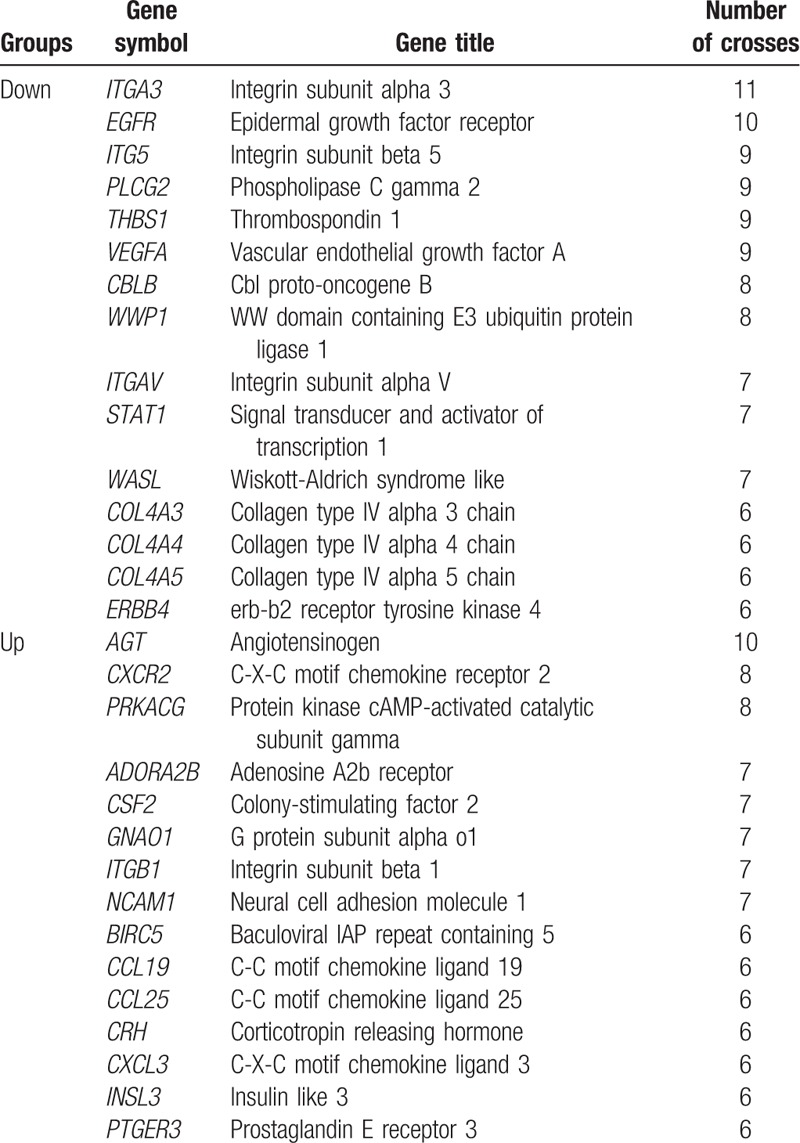
Figure 1.
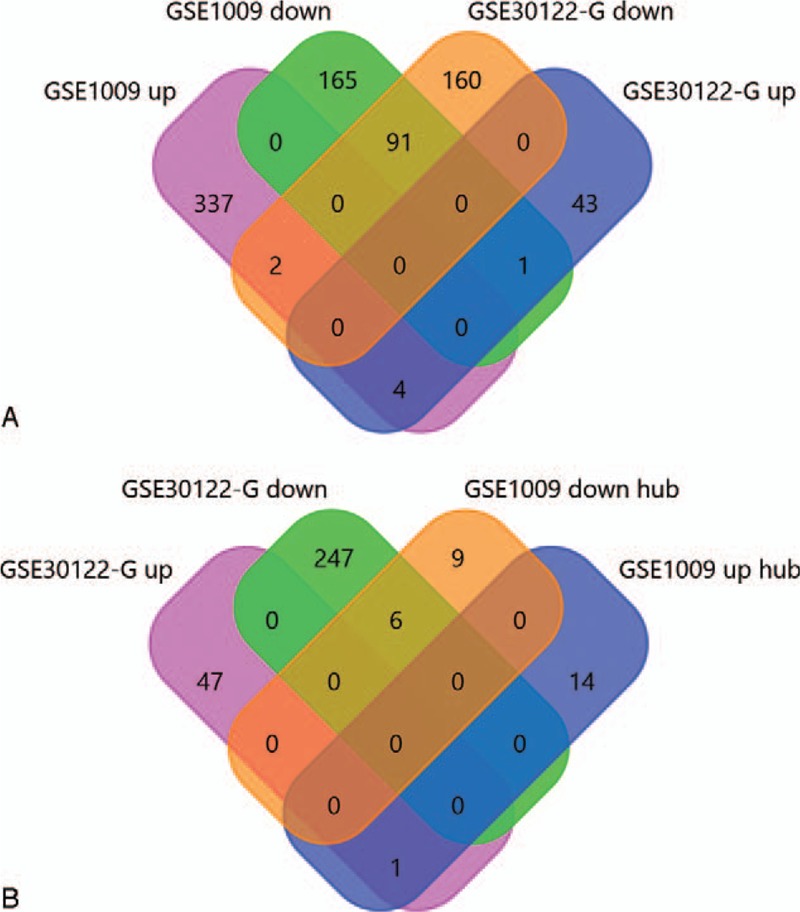
Differentially expressed genes (DEGs) common to GSE1009 and GSE30122. (A) Venn diagram of the up- and downregulated DEGs in GSE1009 and GSE30122. (B) Venn diagram of the hub genes identified in GSD1009 and the DEGs in glomerular tissues identified in GSE30122.
3.3. GO enrichment analysis of DEGs
GO functional enrichment analysis was performed on the DEGs associated with DN from the aspects of molecular function (MF), cellular component (CC), and biological process (BP).
The GO analysis results for the downregulated DEGs showed that most of these genes had MF such as protein-binding, integrin-binding and calcium ion-binding. These DEGs were mainly concentrated in the extracellular exosome, cell surface, actin cycoskeleton and plasama membrane, as revealed by the GO cellular component analysis. GO biological process analysis showed these genes were mainly involved in positive regulation of cell migration, lung development, glomerulus development, and glomerular basement membrane development (Fig. 2A).
Figure 2.
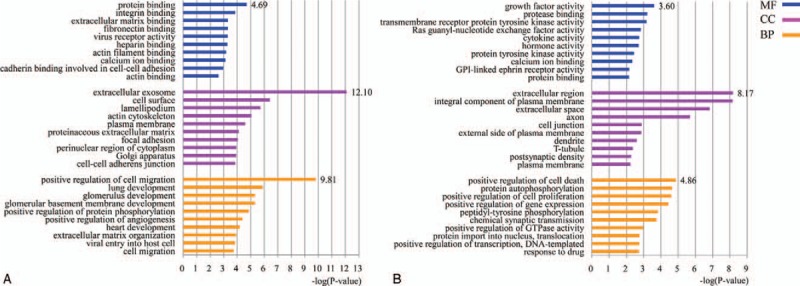
(A) GO analysis of the downregulated DEGs in DN (top 10). (B) GO analysis of the upregulated DEGs in DN (top 10). BP = biological process, CC = cellular component, DEGs = differentially expressed genes, DN = diabetic nephropathy, GO = gene ontology, MF = molecular function.
The GO analysis results for the upregulated DEGs showed that these DEGs mainly had MF such as growth factor and cytokine activity, protease-binding, and calcium ion-binding. These DEGs were mainly concentrated in the extracellular region, integral component plasma membrane, cell junction, and axon, as revealed by the GO cellular component analysis. GO biological process analysis showed these genes were mainly involved in positive regulation of cell death and proliferation, protein autophosphorylation, and GTPase activity (Fig. 2B).
3.4. KEGG pathway analysis of DEGs
KEGG pathway analysis of DEGs associated with DN revealed that the downregulated DEGs were involved in a total of 14 pathways (P < .05), including ECM–receptor interaction, cancer, focal adhesion, complement and coagulation cascades, PI3K/Akt and Rap1 signaling pathways. KEGG pathway analysis of the upregulated DEGs revealed 6 pathways (P < .05), including neuroactive ligand–receptor interaction, cytokine–cytokine receptor interaction, renin secretion, and PI3K/Akt signaling pathways (Table 2). Among these pathways, the 7 key genes were mainly involved in the PI3K/Akt, focal adhesion, ECM–receptor interaction, and Rap1 signaling pathways (Table 3).
Table 2.
KEGG analysis of the differentially expressed genes.
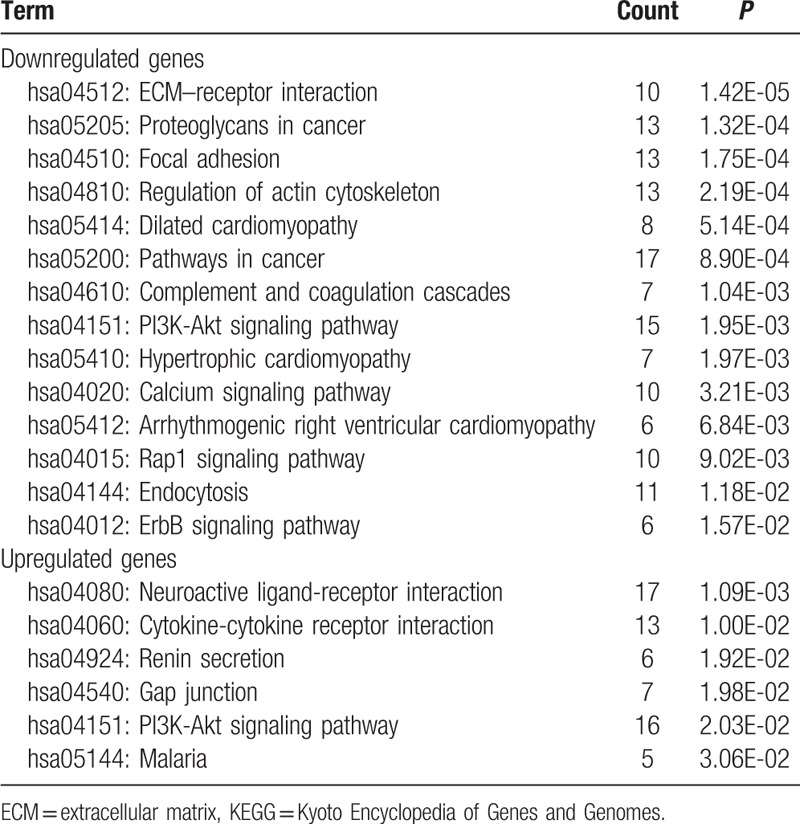
Table 3.
Key genes-related signaling pathways.
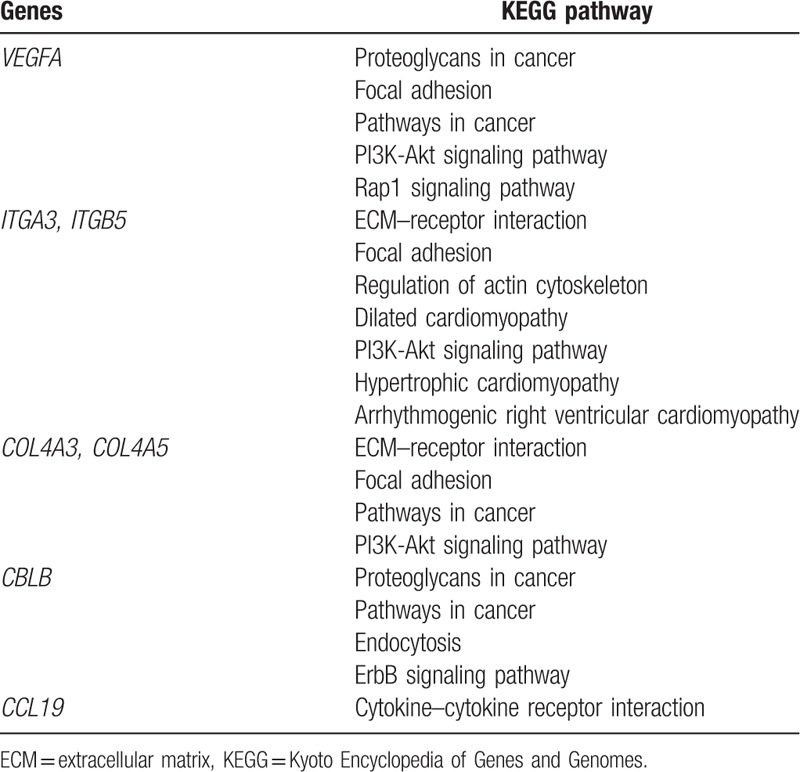
3.5. Prediction of miRNAs targeting DEGs
A total of 9 groups of key miRNAs that targeted the downregulated DEGs were predicted: miR-506, miR-23a/b, the miR-200 family (miR-200b/c/429), miR-124a, the miR-181 family (miR-181a/b/c/d), the miR-25/32/92/363/367 cluster, the miR-17 family (miR-17-5p, miR-20a/b, miR-106a/b, and miR-519d), miR-27a/b, and the miR-29 family (miR-29a/b/c) (Fig. 3A). Only 2 groups of key miRNAs targeted the upregulated DEGs were found (the miR-17 family and the miR-200 family) (Fig. 3B).
Figure 3.
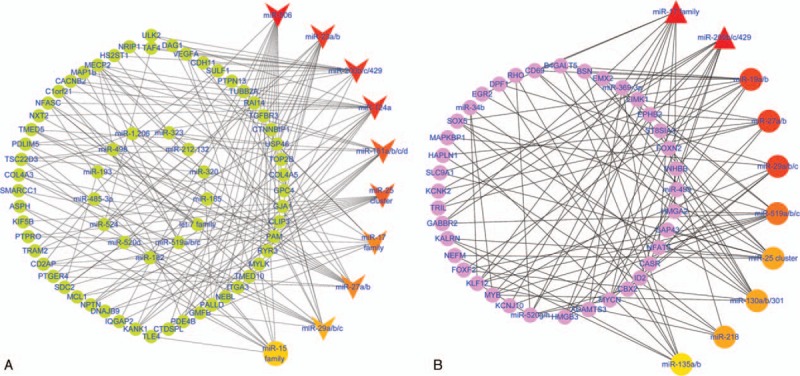
(A) MiRNA–protein interaction network between targeting miRNAs and the downregulated DEGs. The V-shaped nodes represent the key miRNAs (degree >12). The color gradient represents the degree (top 10 miRNAs). The round nodes represent the downregulated genes related to the key miRNAs or other miRNAs. (B) MiRNA–protein interaction network between targeting miRNAs and the upregulated DEGs. The triangle nodes represent the key miRNAs (degree >12). The color gradient represents the degree (top 10 miRNAs). The round nodes represent the upregulated genes related to the key miRNAs or other miRNAs. DEGs = differentially expressed genes, miRNAs = microRNAs.
3.6. Exploration of relationships between the key miRNAs and their target genes
The regulatory relationships between the miRNAs and their target genes were established using Cytoscape, which showed that the single gene was regulated by multiple miRNAs and that the single miRNA could target multiple genes (Fig. 4). Subsequently, miR-29 targeting VEGFA and COL4A5 and miR-200 targeting VEGFA were verified in all 5 databases. A total of 4 databases showed that miR-29 targeted COL4A3, the miR-17 family targeted VEGFA, and miR-181 targeted CBLB. Only 3 databases showed that miR-17 family targeted COL4A3, miR-23 targeted COL4A5, and miR-506 targeted ITGA3, and only 2 databases showed that miR-181 targeted ITGA3 (Table 4). Integrating with the result of KEGG pathway analysis, it was indicated that these key miRNA-target genes were mainly involved in the ECM–receptor interaction, PI3K/Akt signaling pathways, and focal adhesion to influence the pathogenesis and development of DN (Fig. 5).
Figure 4.
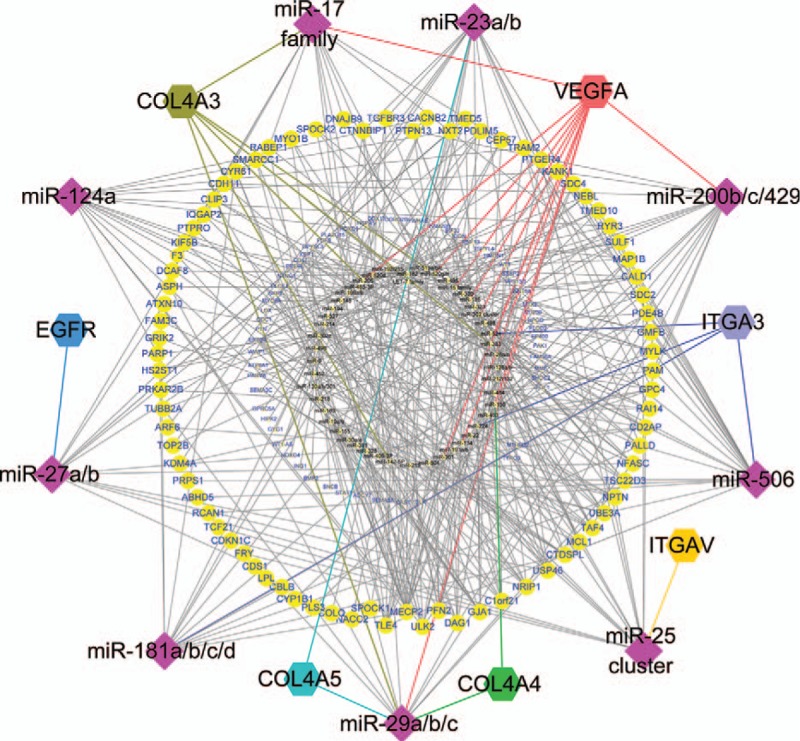
Global network diagram of the interactions between the key miRNAs and their target genes. The diamond nodes represent the 9 key miRNAs. The hexagonal nodes represent the 5 hub genes (ITGB5 and CCL19 did not correspond to the miRNAs). The large round nodes represent genes associated with the 9 key miRNAs, and the small round nodes represent the other upregulated genes and miRNAs. miRNAs = microRNAs.
Table 4.
Verification of potential miRNAs-target genes regulatory relationships using 5 databases.

Figure 5.
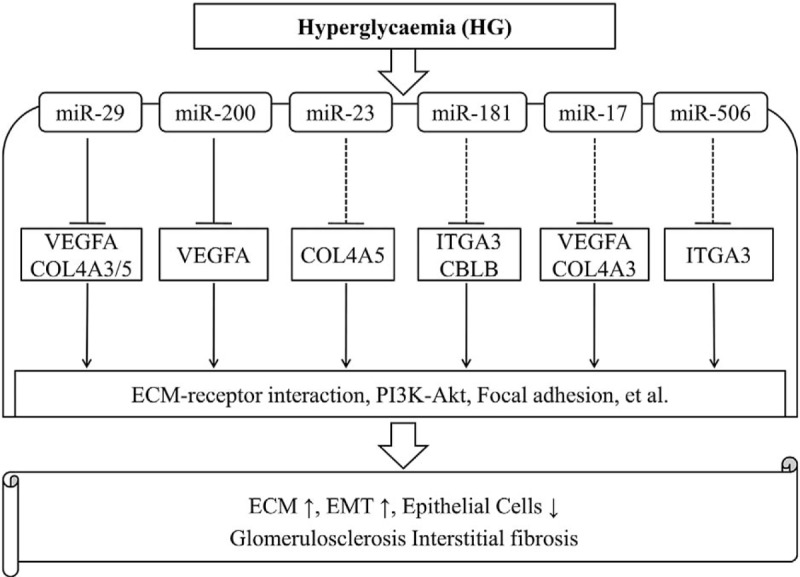
Schematic diagram of potential miRNAs-genes regulatory network in DN. DN = diabetic nephropathy, miRNAs = microRNAs.
4. Discussion
The recent rise of bioinformatics has shed new light on studies of multiple diseases including autoimmune disorders, neoplastic disease, and DN among others. Especially, these disease-related miRNAs were successfully identified. For example, the differentially expressed miRNAs including miR-371a-5p and let-7a-5p may be potential diagnosis biomarkers to illuminate pathogenic mechanisms in systemic lupus erythematosus[19–21] and lupus nephritis.[22,23] In addition, many miRNAs were predicted using the WebGestalt toolkit[24] then validated to be important in acute ischemic stroke.[25–27] Bioinformatics analysis was also performed to explore the underlying functions, pathways, and networks of the differentially expressed miRNAs and targeted genes in various types of cancer such as hepatocellular carcinoma,[28] gastric carcinoma,[29] breast cancer,[30] prostate cancer,[31] and so on. Many miRNAs identified as potential biomarkers based on bioinformatics analysis were further confirmed by reverse transcription quantitative PCR and other functional experiments.
The bioinformatics technology has also provided new perspectives for researchers to study the potential molecular mechanisms and regulatory targets of DN. In the present study, a total of 600 DEGs associated with DN were screened. VEGFA, ITGA3, ITGB5, COL4A3, COL4A5, CBLB, and CCL19 were identified as hub genes based on a network topology. The key miRNAs related to DN were also predicted, including miR-200b/c, miR-29a/b/c, miR-25, miR-27, miR-23, miR-181, miR-17, miR-506, and miR-124a. In the present study, we made an appropriate significance level cutoff (the screening criterion of P value <.01 and network node degree >12) for more detailed investigation of the data and narrowed down the screening range of key miRNAs based on WebGestalt software tool[18,32] and previous screening cutoff point.[9,24,33] Strikingly, this study suggested the potential regulatory network of the miRNAs-genes and signaling pathways. For example, miR-29 targeted VEGFA and COL4A5, and miR-200 targeted VEGFA, which mediated ECM–receptor interaction and PI3K/Akt signaling pathways as well as other signaling pathways to initiate the pathogenesis of DN.
VEGFA is the most important vascular endothelial growth factor and plays an important role in angiogenesis, tumor growth, and ischemic diseases. The up- and downregulation of the podocyte VEGFA levels during renal development led to glomerular disease.[34] This study identified that VEGFA was a key gene that was downregulated in DN, consistent with previous studies. Sivaskandarajah et al[35] found that selective knockout of VEGFA in glomerular podocytes of diabetic mice resulted in a significant increase in proteinuria with severe glomerular scar formation and increased apoptosis. However, Veron et al[36] found that overexpression of the VEGFA isoform (VEGF164) in the podocytes of adult transgenic mice resulted in proteinuria, glomerular hyperplasia, glomerular basement membrane thickening, and podocyte disappearance, which might be related to different types of diabetes models or the length of studies by different research institutes.
This study showed that VEGFA was regulated by multiple miRNAs, including miR-29 and miR-200. Previous studies have confirmed that VEGFA is a target of miR-29[37,38] and that miR-200 suppresses VEGFA expression.[39,40] The loss of miR-29a/b is thought to be closely related to progressive kidney damage in diabetes. MiR-29a and miR-29b promote resistance to renal fibrosis and prevent podocyte apoptosis.[41,42] MiR-29c can induce apoptosis and increase the accumulation of extracellular matrix proteins, whereas knockdown of miR-29c prevented high glucose-induced apoptosis.[43] Several studies have shown that miR-200 family members can regulate the epithelial-to-mesenchymal transition (EMT) and its reversal process, and that the overexpression of miR-200 family members can prevent the TGF-β-induced EMT.[44,45] The EMT plays an important role in renal fibrosis, and renal interstitial fibrosis is a common feature of the progression of various kidney diseases developing into end-stage renal failure,[46] suggesting that miR-200 can affect the development of DN.
The KEGG pathway analysis showed that VEGFA was mainly enriched in the PI3K/Akt and focal adhesion pathways. Currently, studies in vivo and in vitro have indicated that the PI3K/Akt signaling pathway is involved in DN tubular cell apoptosis, mesangial cell hyperplasia, and podocyte dysfunction. High glucose induces increased Akt expression and accelerates the transdifferentiation of renal tubular epithelial cells, leading to renal fibrosis.[47–49] Previous studies reported that miR-200 ultimately promoted glomerular hypertrophy and renal fibrosis through the PI3K/Akt signaling pathway.[50,51] These studies indicated that the mechanism of miR-200 inducing DN by targeting VEGFA through the PI3K/Akt signaling pathway was feasible. Thus, miR-200 may be a prospective target for the prevention or treatment of renal fibrosis in diabetic patients. This mechanism needs to be further verified in vivo and in vitro experiments as well as large-scale clinical trials in the future.
This study also found that COL4A3 and COL4A5 were downregulated in DN glomeruli. COL4A3 was regulated by miR-29 and miR-17, and COL4A5 was regulated by miR-29 and miR-23. Collagen type IV (COL4) is the main collagen component of the ECM and plays an important role in the renal pathological changes of DN.[52] The accumulation of ECM is considered the key to glomerulosclerosis and renal failure in DN.[53] Zhao et al[54] revealed that miR-23b was downregulated in DN and promoted renal fibrosis and proteinuria via the PI3K/Akt signaling pathway.[55] COL4A3 and COL4A5, which are subtypes of type IV collagen, may be regulated by multiple miRNAs, including miR-29 and miR-23, and participate in the pathogenesis of DN through the ECM–receptor interaction and PI3K/Akt signaling pathways.
The hub genes found in this study also include ITGA3, ITGB5, CBLB, and CCL19. ITGA3 encodes integrin α3, and ITGB5 encodes integrin β5, which have been found to be differentially expressed in the glomeruli and tubules of DN patients.[12] Integrin α3β1 is mainly expressed in podocytes and is thought to play a key role in podocytes attachment to the glomerular basement membrane (GBM)[56]; a decrease in integrins leads to the detachment of podocytes from the GBM, which causes proteinuria and promotes renal damage.[57,58] However, Sawada et al[59] found that ITGA3 expression was upregulated in early DN podocytes. Therefore, the roles of ITGA3 and ITGB5 in DN require further investigation. Currently, chemokines and their receptors play a beneficial role in deepening understanding of the inflammatory mechanism of DN. CCL19, which is a member of the chemokine family, was identified as a key factor in this study and might be involved in kidney damage in diabetic conditions. CCL19 expression was increased in DN patients. Many previous studies have confirmed the roles of CCL2, CXCL9, CXCL16, and other chemokines and their receptors in DN and have verified that high levels of chemokines mediate renal injury.[60–62] At present, blocking RAAS is the main clinical strategy for treating the inflammatory process of DN. Newer strategies based on chemokine receptor antagonists or immunosuppressive therapy are still in the experimental stages. Moreover, there are lack of relevant animal experiments and clinical studies for confirming the effect of CCL19 in DN. Thus, CCL19 may be a new research target. Notably, miR-181 targeted both ITGA3 and CBLB, whereas ITGA3 was regulated by miR-506, which have not been previously reported in the field of DN and may be the starting point for future studies.
Other key miRNAs identified in this study were miR-25, miR-27, and miR-124a. Previous studies have reported that miR-25 downregulation plays an important part in diabetic kidney injury, such as high glucose-induced podocyte apoptosis, persistent proteinuria, and secondary hypertension.[63–65] In addition, high glucose stimulation has been shown to increase miR-27a expression, leading to the worsening of renal function and increasing tubulointerstitial fibrosis in DN patients.[66–68]
Previous studies based on bioinformatics analysis have mostly analyzed either gene expression profiling or noncoding RNA profiling, but not conducted between miRNAs, genes, and signaling pathway. In the present study, we made a perfect combination of miRNAs-genes-signaling pathway and identified several important miRNAs-genes regulatory network in DN such as miR-200-VEGFA-PI3K/Akt pathway. Furthermore, a few novel miRNA-target pairs (eg, miR-23 targeting COL4A5, miR-181 targeting ITGA3, miR-17 targeting VEGFA and COL4A3, and miR-506 targeting ITGA3) were found to be involved in diabetic kidney injury. These key genes and miRNAs have potential value for the diagnosis and treatment of DN and may provide new breakthroughs for further studies focusing on the new biomarkers and therapeutic targets of DN. However, we only identified candidate aberrantly DEGs and predicted some potential miRNAs-genes regulatory network in DN based on bioinformatics analysis. Our study is just at the tip of the iceberg of understanding for the prevention, diagnosis, treatment, and prognosis of DN. Further molecular biological experiments are necessary to confirm the function of the identified DEGs as well as the interaction between microRNAs and genes in DN. In future studies, we also need to analyze more clinical samples to validate these results.
Acknowledgments
All the data contributors are sincerely appreciated for data submitted in the GEO and other databases.
Author contributions
Conceptualization: Li Yuan, Huiqing Li, Fengying Yang.
Data curation: Fengying Yang.
Formal analysis: Fengying Yang, Zhenhai Cui.
Funding acquisition: Li Yuan, Huiqing Li.
Investigation: Hongjun Deng, Ying Wang, Yang Chen.
Methodology: Fengying Yang, Huiqing Li.
Project administration: Li Yuan, Huiqing Li.
Software: Fengying Yang.
Supervision: Zhenhai Cui, Hongjun Deng.
Writing – review & editing: Li Yuan, Huiqing Li, Fengying Yang.
Writing – original draft: Fengying Yang.
fengying yang orcid: 0000-0002-1428-0395.
Footnotes
Abbreviations: DKD = diabetic kidney disease, DN = diabetic nephropathy, ECM = extracellular matrix, EMT = epithelial-to-mesenchymal transition, GEO = Gene Expression Omnibus, GO = gene ontology, KEGG = kyoto encyclopedia of genes and genomes, miRNAs = microRNAs, PI3K/Akt = phosphoinositide 3 kinese/protein kinase B, PPI = protein–protein interaction, VEGFA = vascular endothelial growth factor α, WebGestalt = WEB-based GEne SeT AnaLysis Toolkit.
FY and ZC contributed equally to this work.
Funding information: National Natural Science Foundation of China (grant no. 81370880), and the Health and Family planning Commission Project of Hubei Province of China (grant no. WJ2017M104).
The authors report no conflicts of interest.
References
- [1].Umanath K, Lewis JB. Update on diabetic nephropathy: core Curriculum 2018. Am J Kidney Dis 2018;71:884–95. [DOI] [PubMed] [Google Scholar]
- [2].Tuttle KR, Bakris GL, Bilous RW, et al. Diabetic kidney disease: a report from an ADA consensus conference. Am J Kidney Dis 2014;64:510–33. [DOI] [PubMed] [Google Scholar]
- [3].Doshi SM, Friedman AN. Diagnosis and management of type 2 diabetic kidney disease. Clin J Am Soc Nephrol 2017;12:1366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alvarez ML, DiStefano JK. Towards microRNA-based therapeutics for diabetic nephropathy. Diabetologia 2013;56:444–56. [DOI] [PubMed] [Google Scholar]
- [5].Simpson K, Wonnacott A, Fraser DJ, et al. MicroRNAs in diabetic nephropathy: from biomarkers to therapy. Curr Diab Rep 2016;16:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci 2015;1353:72–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu H, Kong L, Zhou S, et al. The role of microRNAs in diabetic nephropathy. J Diabetes Res 2014;2014:920134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang WN, Zhang WL, Zhou GY, et al. Prediction of the molecular mechanisms and potential therapeutic targets for diabetic nephropathy by bioinformatics methods. Int J Mol Med 2016;37:1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qu W, Han C, Li M, et al. Revealing the underlying mechanism of diabetic nephropathy viewed by microarray analysis. Exp Clin Endocrinol Diabetes 2015;123:353–9. [DOI] [PubMed] [Google Scholar]
- [10].Cui C, Cui Y, Fu Y, et al. Microarray analysis reveals gene and microRNA signatures in diabetic kidney disease. Mol Med Rep 2018;17:2161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baelde HJ, Eikmans M, Doran PP, et al. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis 2004;43:636–50. [DOI] [PubMed] [Google Scholar]
- [12].Woroniecka KI, Park AS, Mohtat D, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846–7. [DOI] [PubMed] [Google Scholar]
- [14].Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39(Database issue):D561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8Suppl 4:S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods in molecular biology (Clifton, NJ) 2011;696:291–303. [DOI] [PubMed] [Google Scholar]
- [17].Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41(Web Server issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tang D, Chen Y, He H, et al. Integrated analysis of mRNA, microRNA and protein in systemic lupus erythematosus-specific induced pluripotent stem cells from urine. BMC Genomics 2016;17:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Carlsen AL, Schetter AJ, Nielsen CT, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum 2013;65:1324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen N, Liang D, Tang Y, et al. MicroRNAs—novel regulators of systemic lupus erythematosus pathogenesis. Nature Rev Rheumatol 2012;8:701–9. [DOI] [PubMed] [Google Scholar]
- [22].Wang W, Mou S, Wang L, et al. Up-regulation of Serum MiR-130b-3p Level is Associated with Renal Damage in Early Lupus Nephritis. Sci Rep 2015;5:12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Y, Fang X, Li QZ. Biomarker profiling for lupus nephritis. Genom Proteomics Bioinformatics 2013;11:158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zeng Y, Liu JX, Yan ZP, et al. Potential microRNA biomarkers for acute ischemic stroke. Int J Mol Med 2015;36:1639–47. [DOI] [PubMed] [Google Scholar]
- [25].Li P, Teng F, Gao F, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol 2015;35:433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Peng G, Yuan Y, Wu S, et al. MicroRNA let-7e is a potential circulating biomarker of acute stage ischemic stroke. Transl Stroke Res 2015;6:437–45. [DOI] [PubMed] [Google Scholar]
- [27].Tiedt S, Prestel M, Malik R, et al. RNA-Seq identifies circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as potential biomarkers for acute ischemic stroke. Circul Res 2017;121:970–80. [DOI] [PubMed] [Google Scholar]
- [28].Zhang Y, Mo WJ, Wang X, et al. Microarraybased bioinformatics analysis of the prospective target gene network of key miRNAs influenced by long noncoding RNA PVT1 in HCC. Oncol Rep 2018;40:226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Q, Li J, Dai W, et al. Differential regulation analysis reveals dysfunctional regulatory mechanism involving transcription factors and microRNAs in gastric carcinogenesis. Artif Intell Med 2017;77:12–22. [DOI] [PubMed] [Google Scholar]
- [30].Zhang GM, Goyal H, Song LL. Bioinformatics analysis of differentially expressed miRNA-related mRNAs and their prognostic value in breast carcinoma. Oncol Rep 2018;39:2865–72. [DOI] [PubMed] [Google Scholar]
- [31].Li D, Hao X, Song Y. Identification of the key MicroRNAs and the miRNA-mRNA regulatory pathways in prostate cancer by bioinformatics methods. Biomed Res Int 2018;2018:6204128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server issue):W741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nat Methods 2012;9:1069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carranza K, Veron D, Cercado A, et al. Cellular and molecular aspects of diabetic nephropathy; the role of VEGF-A. Nefrologia 2015;35:131–8. [DOI] [PubMed] [Google Scholar]
- [35].Sivaskandarajah GA, Jeansson M, Maezawa Y, et al. Vegfa protects the glomerular microvasculature in diabetes. Diabetes 2012;61:2958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Veron D, Reidy KJ, Bertuccio C, et al. Overexpression of VEGF-A in podocytes of adult mice causes glomerular disease. Kidney Int 2010;77:989–99. [DOI] [PubMed] [Google Scholar]
- [37].Liu L, Bi N, Wu L, et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer 2017;16:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu Q, Liao F, Wu H, et al. Different expression of miR-29b and VEGFA in glioma. Artif Cells Nanomed Biotechnol 2016;44:1927–32. [DOI] [PubMed] [Google Scholar]
- [39].Mitra RN, Nichols CA, Guo J, et al. Nanoparticle-mediated miR200-b delivery for the treatment of diabetic retinopathy. J Control Release 2016;236:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu GT, Chen HT, Tsou HK, et al. CCL5 promotes VEGF-dependent angiogenesis by down-regulating miR-200b through PI3K/Akt signaling pathway in human chondrosarcoma cells. Oncotarget 2014;5:10718–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lin CL, Lee PH, Hsu YC, et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol 2014;25:1698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang B, Komers R, Carew R, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 2012;23:252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Long J, Wang Y, Wang W, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 2011;286:11837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kato M, Putta S, Wang M, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- [46].Zhao L, Wang X, Sun L, et al. Critical role of serum response factor in podocyte epithelial-mesenchymal transition of diabetic nephropathy. Diab Vasc Dis Res 2016;13:81–92. [DOI] [PubMed] [Google Scholar]
- [47].Liu L, Hu X, Cai GY, et al. High glucose-induced hypertrophy of mesangial cells is reversed by connexin43 overexpression via PTEN/Akt/mTOR signaling. Nephrol Dial Transplant 2012;27:90–100. [DOI] [PubMed] [Google Scholar]
- [48].Rane MJ, Song Y, Jin S, et al. Interplay between Akt and p38 MAPK pathways in the regulation of renal tubular cell apoptosis associated with diabetic nephropathy. Am J Physiol Renal Physiol 2010;298:F49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ha TS. High-glucose and advanced glycosylation end products increased podocyte permeability via PI3-K/Akt signaling. J Mol Med (Berl) 2010;88:391–400. [DOI] [PubMed] [Google Scholar]
- [50].Park JT, Kato M, Yuan H, et al. FOG2 protein down-regulation by transforming growth factor-beta1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem 2013;288:22469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hyun S, Lee JH, Jin H, et al. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009;139:1096–108. [DOI] [PubMed] [Google Scholar]
- [52].Watanabe H, Sanada H, Shigetomi S, et al. Urinary excretion of type IV collagen as a specific indicator of the progression of diabetic nephropathy. Nephron 2000;86:27–35. [DOI] [PubMed] [Google Scholar]
- [53].Kolset SO, Reinholt FP, Jenssen T. Diabetic nephropathy and extracellular matrix. J Histochem Cytochem 2012;60:976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhao B, Li H, Liu J, et al. MicroRNA-23b targets Ras GTPase-activating protein SH3 domain-binding protein 2 to alleviate fibrosis and albuminuria in diabetic nephropathy. J Am Soc Nephrol 2016;27:2597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu H, Wang X, Liu S, et al. Effects and mechanism of miR-23b on glucose-mediated epithelial-to-mesenchymal transition in diabetic nephropathy. Int J Biochem Cell Biol 2016;70:149–60. [DOI] [PubMed] [Google Scholar]
- [56].Cosio FG. Cell-matrix adhesion receptors: relevance to glomerular pathology. Am J Kidney Dis 1992;20:294–305. [DOI] [PubMed] [Google Scholar]
- [57].Pozzi A, Jarad G, Moeckel GW, et al. Beta1 integrin expression by podocytes is required to maintain glomerular structural integrity. Dev Biol 2008;316:288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wolf G, Chen S, Ziyadeh FN. From the periphery of the glomerular capillary wall toward the center of disease: podocyte injury comes of age in diabetic nephropathy. Diabetes 2005;54:1626–34. [DOI] [PubMed] [Google Scholar]
- [59].Sawada K, Toyoda M, Kaneyama N, et al. Upregulation of alpha3 beta1-integrin in podocytes in early-stage diabetic nephropathy. J Diabetes Res 2016;2016:9265074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Elewa U, Sanchez-Nino MD, Mahillo-Fernandez I, et al. Circulating CXCL16 in diabetic kidney disease. Kidney Blood Press Res 2016;41:663–71. [DOI] [PubMed] [Google Scholar]
- [61].Lin Z, Gong Q, Zhou Z, et al. Increased plasma CXCL16 levels in patients with chronic kidney diseases. Eur J Clin Invest 2011;41:836–45. [DOI] [PubMed] [Google Scholar]
- [62].Ruster C, Wolf G. The role of chemokines and chemokine receptors in diabetic nephropathy. Front Biosci 2008;13:944–55. [DOI] [PubMed] [Google Scholar]
- [63].Liu Y, Li H, Liu J, et al. Variations in microRNA-25 expression influence the severity of diabetic kidney disease. J Am Soc Nephrol 2017;28:3627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li H, Zhu X, Zhang J, et al. MicroRNA-25 inhibits high glucose-induced apoptosis in renal tubular epithelial cells via PTEN/AKT pathway. Biomed Pharmacother 2017;96:471–9. [DOI] [PubMed] [Google Scholar]
- [65].Oh HJ, Kato M, Deshpande S, et al. Inhibition of the processing of miR-25 by HIPK2-Phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Sci Rep 2016;6:38789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhou Z, Wan J, Hou X, et al. MicroRNA-27a promotes podocyte injury via PPARgamma-mediated beta-catenin activation in diabetic nephropathy. Cell Death Dis 2017;8:e2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hou X, Tian J, Geng J, et al. MicroRNA-27a promotes renal tubulointerstitial fibrosis via suppressing PPARgamma pathway in diabetic nephropathy. Oncotarget 2016;7:47760–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wu L, Wang Q, Guo F, et al. MicroRNA-27a induces mesangial cell injury by targeting of PPARgamma, and its in vivo knockdown prevents progression of diabetic nephropathy. Sci Rep 2016;6:26072. [DOI] [PMC free article] [PubMed] [Google Scholar]


