Abstract
Objective:
Researchers have evaluated the associations between mitochondrial DNA (mtDNA) 4977 bp deletion and presbycusis. This study aimed to assess the differences of mtDNA 4977 bp deletion between presbycusis patients and controls by conducting a meta-analysis of published studies.
Methods:
Databases, including PubMed, Embase, Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang Data were searched to collect case–control studies on the correlation between mitochondrial DNA 4977 bp deletion and presbycusis. The research findings of related articles were collected according to the inclusion criteria. Pooled odds ratios (ORs) and corresponding confidence intervals (CIs) were calculated. Meanwhile, subgroup analysis was performed to examine the source of heterogeneity. Revman 5.3 and Stata 12.0 software were used for data synthesis.
Results
: Eight English and Chinese studies were included in the meta-analysis, the results of which showed that mitochondrial DNA 4977 bp deletion could increase the risk of presbycusis (OR = 8.16, 95% CI: 3.51–18.99), and the difference was statistically significant (P <. 01). Analysis of the polled OR showed the incidence of mtDNA 4977 bp deletion was 8.50 times higher in Asians with presbycusis than in the control group. And the OR in the studies of occidentals was 7.24. Sample source analysis was also performed with the sample source divided by temporal bone source and other sources (hair and blood). The OR was 4.18 and 22.36 for the temporal bone and other sources, respectively.
Conclusion:
Mitochondrial DNA 4977 bp deletion could increase the risk of presbycusis.
Keywords: mitochondrial DNA, mtDNA 4977 bp deletion, presbycusis
1. Introduction
Aging is an unavoidable process of a society's development. Although some elderly people retain good hearing, presbycusis, also known as age-related hearing loss (AHL), accumulates with age and has become a severe health and social problem. Presbycusis sufferers develop a high-toned hearing loss first, which plays a major adverse role in communication, particularly in reverberant, and/or noisy listening situations. Epidemiologically, some health problems related to presbycusis have been reported, such as social isolation, cognitive decline, depression, and dementia. However, the etiology of the disease remains unknown.[1–8]
Several factors contribute to presbycusis. Oxidative stress caused by mitochondrial dysfunction plays a causal role in presbycusis.[8–10] Mitochondria are the most important energy-producing organelles in the eukaryotic cells. Mitochondrial DNA (MtDNA) is more sensitive to damage than nuclear DNA is, because there are not enough enzymes in mitochondria to repair DNA damage and its lack of protective histones.[11] Sometic mtDNA deletions have been reported to be associated with the process of aging in many tissues, and mtDNA is particularly susceptible to large-scale deletions in the sections characterized by the appearance of flanking repeats. The most abundant and prevalent pathogenic mtDNA rearrangements are common deletion of 4977-bp. Mitochondrial 4977 bp deletion occurs frequently in tissues in an aging-related manner.[12–14]
Therefore, it is not surprising that a significant number of clinical studies have researched the effects of 4977 bp deletion on aging. However, researchers have not always indicated a positive association between 4977 bp deletion and aging. Although some authors reported an association between 4977 bp deletion and aging,[12,13,15,16] several studies in humans have shown an association between presbycusis and mtDNA deletion.[17–19] It is still unknown how relevant it is and whether mtDNA 4977 can be used as a susceptibility gene for presbycusis. The primary aim of the current meta-analysis is to explore the effects of 4977 bp deletion and presbycusis by performing a quantitative analysis of available published data about this subject.
2. Materials and methods
2.1. Search strategy and selection criteria
This systematic review and meta-analysis are developed and performed per the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklists.[20] Ethical approval was unnecessary in this study because it was a meta-analysis analyzing existing articles and did not need handle individual patient data.
Relevant literature search was performed in:
-
(1)
PubMed,
-
(2)
Embase,
-
(3)
Web of Science,
-
(4)
China National Knowledge Infrastructure (CNKI), and
-
(5)
Wanfang Chinese databases.
The medical subject headings (MeSH) and free terms were all included in our search terms, which are listed as follows: “Presbycusis”, “Age-Related Hearing Impairment”, “age-related hearing loss”, “mitochondrial DNA 4977-bp deletion”, “common deletion”. Our search logic in the PubMed database was listed as follows: (presbycusis[Mesh] OR presbycuses OR Age-Related Hearing Impairment OR age-related hearing loss) AND ((((common deletion) OR 4977 bp deletion) OR 4977 bp) OR mitochondrial DNA 4977-bp deletion). All research we searched was reported before June 5, 2018. We also manually checked all articles listed in the reference lists of the retrieved literature. The detailed steps of the literature search are shown in Figure 1.
Figure 1.
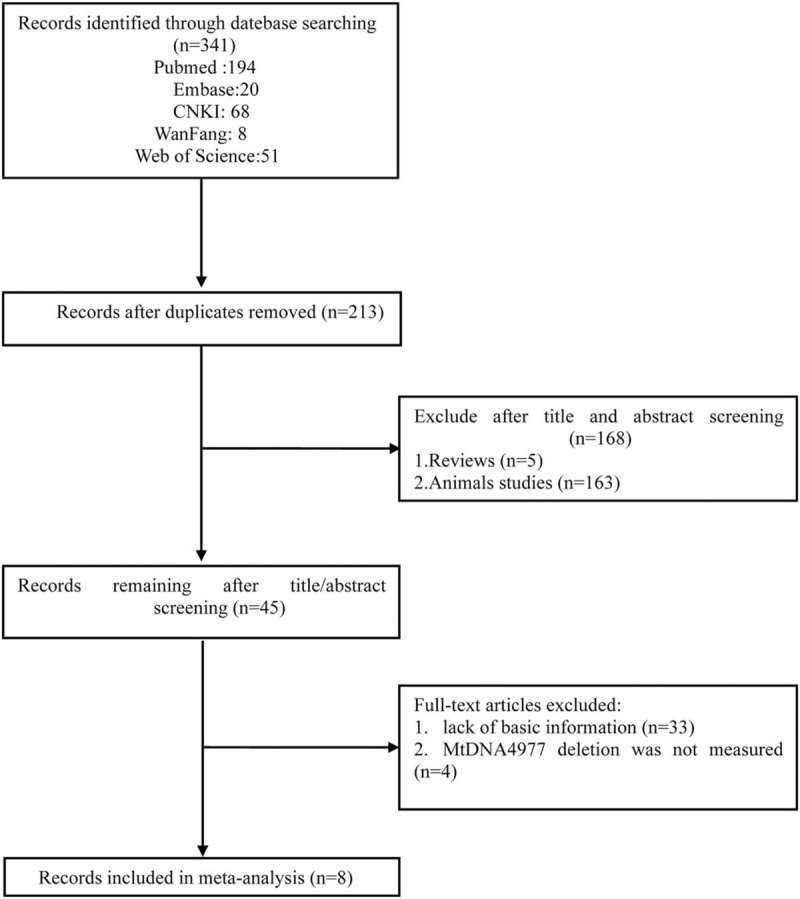
Flow chart of the study selection process. After careful discussion among the 3 reviewers, 8 studies were included to perform the meta-analysis.
2.2. Inclusion criteria
The following inclusion criteria were applied to select studies in the meta-analysis:
-
(1)
articles evaluating the relationship of the mtDNA 4977 bp deletion and presbycusis;
-
(2)
the object of study was human;
-
(3)
a case–control study;
-
(4)
published in either English or Chinese;
-
(5)
cases were diagnosed as presbycusis, according to clinical diagnostic criteria, and the control group had normal hearing;
-
(6)
the frequency distribution of mtDNA 4977 bp deletions in the case and control groups could be provided directly or indirectly.
2.3. Exclusion criteria
The exclusion criteria were as follows:
-
(1)
duplicate publications,
-
(2)
incomplete information,
-
(3)
insufficient data,
-
(4)
no control group,
-
(5)
review articles or conference literature.
2.4. Statistical methods
The odds ratio (OR) of mtDNA 4977 bp deletion in presbycusis compared with those without hearing loss was calculated with a random-effects model or a fixed-effects model, using between-study heterogeneity derived from the Mantel–Haenszel model. The effects model was applied, depending on the P value of the chi-squared statistic. When the I2 value >50% and P was <0.05, the random-effects model was applied, and when the I2 value <50%, the fixed-effects model was applied to combine effect size. The extent of homogeneity was assessed by I-squared statistics (I2 <25%, no heterogeneity; I2 = 25%–50%, moderate heterogeneity; I2 = 50%–75%, moderate heterogeneity, I2 >75%, high heterogeneity). To explore the origin of homogeneity, subgroup analysis was performed for ethnic groups (Asian and Occidental) and organ (auditory organ and other organs). Sensitivity analysis was used to evaluate the stability of the result of the meta-analysis. We conducted meta-regression to identify the possible sources of heterogeneity. The forest plot was computer-generated. Publication bias was statistically assessed by 2 formal methods: by Begg rank correlation and Egger regression test. We also employed the trim-and-fill method to identify and correct for funnel plot asymmetry arising from publication bias. Two-sided P value <.05 was considered statistically significant except for the test for publication bias, where P <.10 was used. Statistical calculations were performed by using Stata version 12.0 and Review Manager 5.3.
3. Results
3.1. Search results and characteristics of the eligible studies
Figure 1 shows details of the literature search. Two of our investigators (Sun HY and Han BA) independently abstracted the initial data. Discrepancies were resolved by consensus, and 341 articles were preliminarily selected. Screening by title and abstract was conducted according to information given in the title and abstract. After a thorough discussion, 45 articles were found to be related to this study. These articles then were subjected to second-stage review. Finally, 8 studies were included for the meta-analysis, covering data from a total of 458 samples. The information of authors, publication year, national sources, materials, ages, and sample size of each study are listed in Table 1 and Table 2.
Table 1.
Characteristics of the trials included in the meta-analysis.
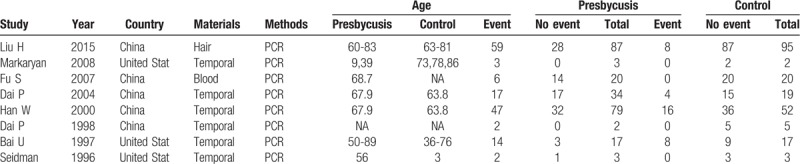
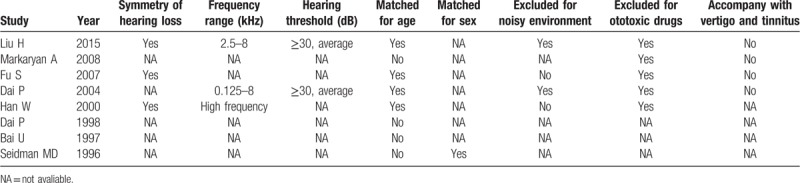
Table 2.
Characteristics of included studies.
3.2. Pooled analysis
The meta-analysis exhibited that the overall pooled OR of mtDNA 4977 bp deletion in presbycusis compared with the control group was 8.16 (95% confidence interval [CI]: 3.51 to 18.99, P <.01). The value of I2 was 51%, indicating that the studies were moderately heterogeneous. Therefore, the random-effects model was used to combine effect size (Fig. 2).
Figure 2.
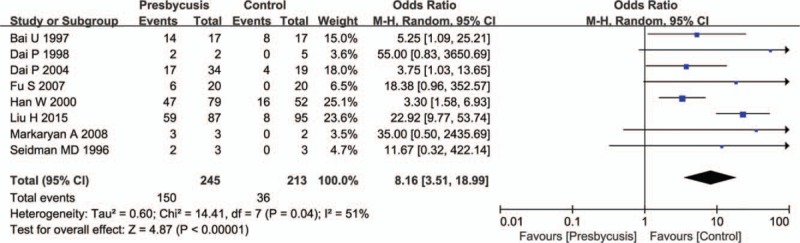
Comparison of mtDNA 4977 bp deletion between presbycusis group and control group in the 8 included studies. Calculation based on random-effects model. Results are expressed as OR and 95% CI. Meta-analysis showed that the overall pooled OR of mtDNA 4977 bp deletion in presbycusis compared with control group was 8.16. CI = confidence intervals, OR = odds ratio.
3.3. Subgroup analysis: Ethnic group
Asian: Analysis of the polled OR showed the incidence of mtDNA 4977 bp deletion was 8.50 times higher in Asians with presbycusis than in the control group [OR = 8.50 (95% CI: 2.71 to 26.68, P <.01, Fig. 3A)]. Occidental: the OR in the studies of occidentals was 7.24 (95% CI: 1.90–27.55, P <.05, Fig. 3B).
Figure 3.
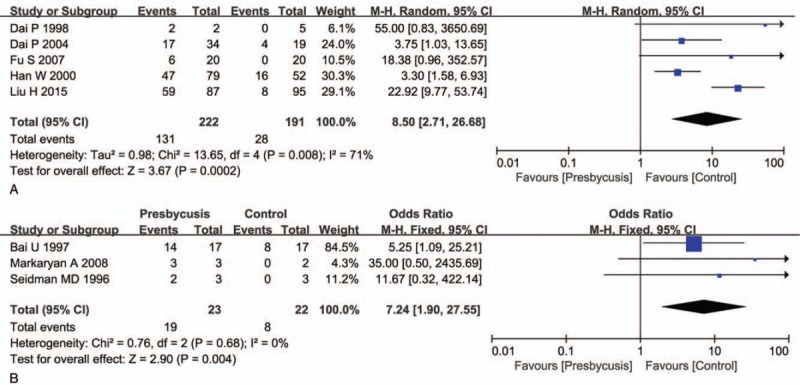
Subgroup analysis in Asian group and Occidental group. A shows the results on Asian group, and B shows the Occidental group results. A: Calculation based on random-effects model. Results are expressed as OR and 95% CI. The OR value in the studies was 8.50 (95% CI: 2.71–26.68, P <.01). B: Calculation based on fixed-effects model. Results are expressed as OR and 95% CI. The OR value in the studies was 7.24 (95% CI: 1.90–27.55, P <.05). CI = confidence intervals, OR = odds ratio.
3.4. Subgroup analysis: Sample source
Analysis was also performed with the sample source divided by temporal bone source and other sources (hair and blood). The OR was 4.18 (Fig. 4A) and 22.36 (Fig. 4B) for the groups, temporal bone, and other sources, respectively.
Figure 4.
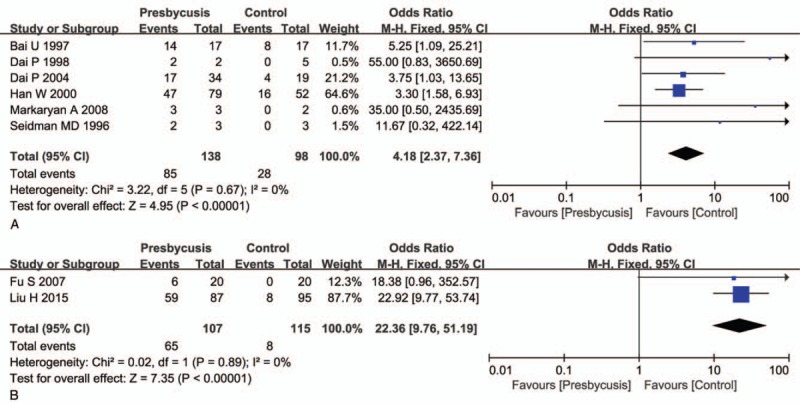
Subgroup analysis in temporal bone source and other sources. Analysis was also according to sample source. A shows the results on the temporal bone source group, and B shows the other sources group. A: Calculation based on fixed-effects model. Results are expressed as OR and 95% CI. The OR value in the studies was 4.10 (95% CI: 2.37–7.36, P <.01). B: Calculation based on fixed-effects model. Results are expressed as odds ratio (OR) and 95% CI. The OR value in the studies was 22.36 (95% CI: 9.76–51.19, P <.01). CI = confidence intervals, OR = odds ratio.
3.5. Sensitivity analysis
The stability of the pooled ORs of the mtDNA 4977 bp deletion was estimated by eliminating each of the included studies in turn. Sensitivity analysis showed that no significant alteration of the pooled ORs was found as any of the included studies was omitted (Fig. 5). These results indicate that the corresponding ORs were relatively robust.
Figure 5.
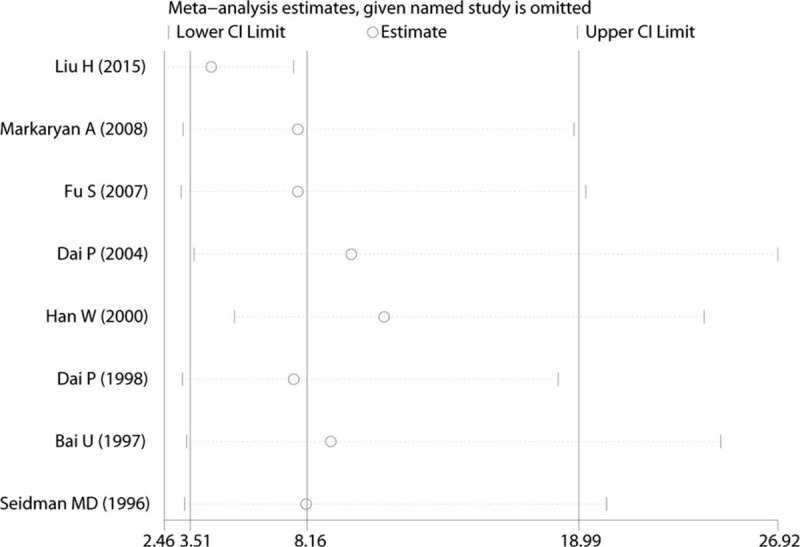
Sensitivity analysis of the pooled effective size on mtDNA 4977 bp deletion between presbycusis and control groups.
3.6. Publication bias
The studies seem to have slight publication bias because the funnel plot was not perfectly symmetrical (Fig. 6), but Begg tests (P = .466) and Egger tests (P = 1.48) did not provide any evidence of publication bias in our study. Furthermore, the trim-and-fill method demonstrated that no research needed to be statistically corrected for funnel plot asymmetry. The methodological quality of each included study is shown in Figure 7.
Figure 6.
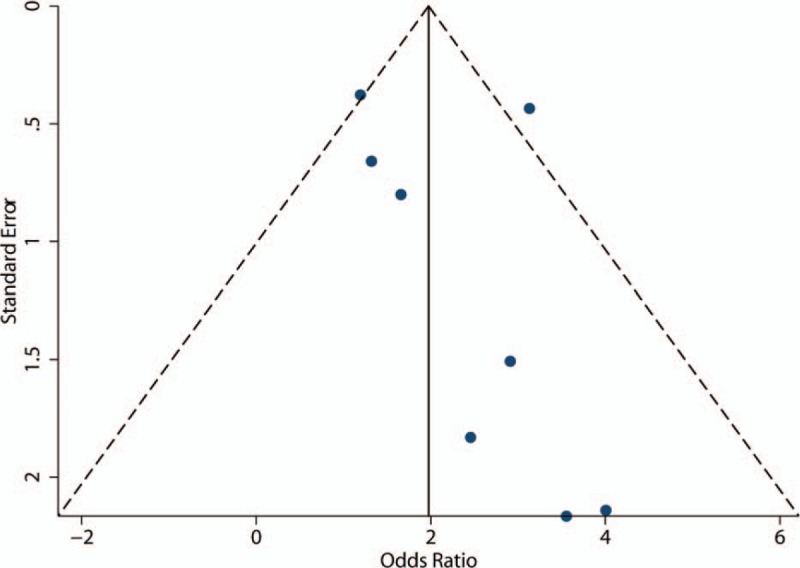
Funnel plot shows the possibility of a small publication bias.
Figure 7.
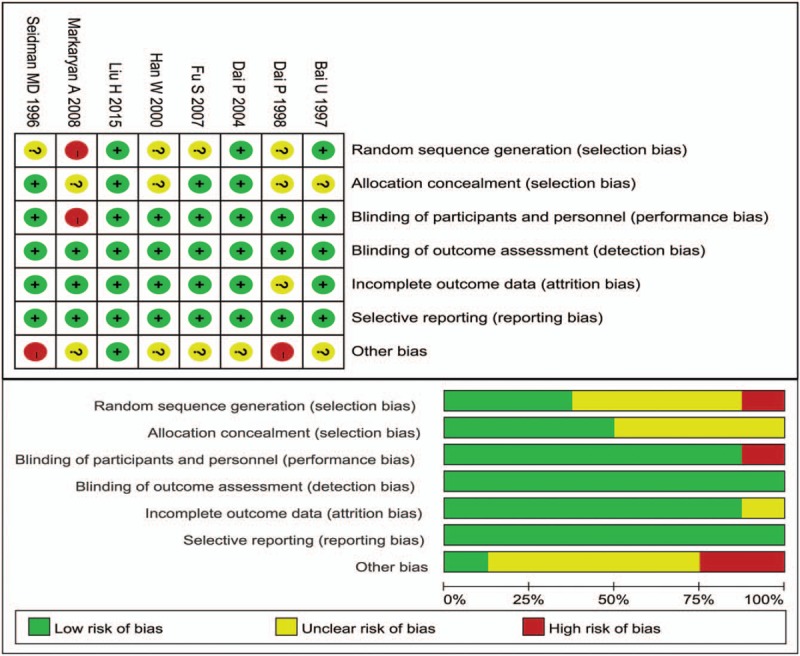
Risk of bias summary and graph: review authors’ judgements about each domain for each included study.
4. Discussion
The present research used meta-analytic methods to evaluate the difference in mitochondrial 4977 bp deletion between presbycusis and the control group. Our research demonstrated that individuals with mitochondrial 4977 bp deletion have a closer association with presbycusis than control subjects do. Begg tests (P = .466) and Egger tests (P = 1.48) did not provide evidence of publication bias in our study. Furthermore, although the value of I2 = 51% (I2 >50, P <.05), it indicated that this research contained moderate heterogeneity. Subgroup analysis was performed to evaluate the sources of heterogeneity, so the results of this research could represent the true relationship between 4977 bp deletion and presbycusis. Meanwhile, sensitivity analysis demonstrated that after any individual research was omitted, the overall conclusions and results still existed. Therefore, we are confident that the results of this meta-analysis show strong association between 4977 bp deletion and presbycusis.
Numerous studies have reported that deletions of mtDNA play a pivotal role in aging and impairment of various human organs, such as the heart,[21] brain,[15,22] skin,[5,23] cornea,[24] and skeletal muscle.[25] MtDNA deletions are abundant in cochlear tissue, which have been shown, including mtDNA 10422, 13162, 7663, 4989, 7436, 4989, and 4977 bp deletion. The most common deletion is mtDNA 4977 bp deletion, and its absence will hinder mitochondrial oxidative phosphorylation.[26] Dai et al determined mtDNA 4977 bp deletion by using nested polymerase chain reaction (PCR) in temporal bone specimens. The results demonstrated that in the presbycusis group, the percentage of mtDNA 4977 bp deletion was as high as 50%, 0.0% in the young and middle-aged group, and 21% in the age-related control group.[27] Markaryan et al studied mtDNA 4977 bp deletion in the cochlea of senior individuals. They reported that the incidence of mtDNA 4977 bp deletion was 12 ± 2% in the normal hearing control group and 32 ± 14% in the presbycusis group.[28] A study by Hong Liu et al suggested a relationship between mtDNA 4977 bp deletion in the hair shaft and the severity of hearing loss in presbycusis.[17] These studies have therefore shown that mtDNA 4977 bp deletion is universally observed in patients with presbycusis.
Although many studies and this meta-analysis have shown a certain correlation between mtDNA 4977 deletion and presbycusis, its relevance and whether mtDNA 4977 can be used as a susceptibility gene for presbycusis is still unknown. Through extensive review of related data, this study eventually included 8 Chinese and foreign literature for meta-analysis.[17,27–32] The meta-analysis showed that the risk of mtDNA 4977 deletion in presbycusis increased (OR = 7.84, 95% CI: 3.63∼16.93). The OR values obtained after sensitivity analysis fluctuated between 4.57 and 10.63. The subgroup analysis results showed that mtDNA 4977 deletion had a strong association with presbycusis. From the results of this meta-analysis, we could conclude mtDNA 4977 participation in presbycusis pathogenesis.
However, the limitations of this meta-analysis should be considered. First, due to the rare related study materials included in the research, the sample size is not large enough, especially in the case of subgroup analysis of ethnic groups; the study is merely comparing a limited number of reports from China and the 2 reports from the United States. Thus, it seems inappropriate to draw a conclusion and group the ethnicity based on such little information.
Second, there is no stratified analysis of factors such as age, gender, and disease severity. The matching conditions of the control group are not completely consistent; there are objectively confounding factors that prevent accurate reflection of the true situation of both groups. In addition, presbycusis is caused by multiple factors such as the external environment and individual susceptibility. Some researches demonstrated that a functional deficit will not occur until the level of deletion in tissues accumulates a significant level.[33,34] Furthermore, publication bias is inevitable because studies are not likely to report negative findings. Even if publication bias was not statistically derived, its possibility could not be excluded. At present, the mechanism of mtDNA4977 deletion in presbycusis remains unclear. Large case–control studies to provide more evidence-based assessment of mtDNA 4977 bp deletion as a cause of genetic susceptibility to presbycusis is needed.
Although this study objectively has certain limitations, sensitivity analysis, and research bias analysis results may indicate that the combined effect value is reliable and stable; the combined meta-analytic results of this study can still provide certain strong evidence for the prevention and treatment of senile spasticity. The exact mechanism of mtDNA4977 leading to presbycusis remains to be further explored. Specific gerontological biological markers and effective prevention and treatment drugs need further study. The deep exploration of the correlation between presbycusis and mtDNA CD4977 not only helps elucidate the part of the mechanism of presbycusis at the molecular level but also provides new ideas for the prevention and treatment of presbycusis.
Author contributions
Data curation: Baoai Han, Yaqin Tu, Jie Yuan.
Formal analysis: Baoai Han, Yaqin Tu, Zuhong He.
Funding acquisition: Haiying Sun, Tao Zhou, Tian Wang.
Investigation: Haiying Sun, Tao Zhou.
Methodology: Baoai Han, Yaqin Tu.
Project administration: Yongqin Li.
Resources: Zuhong He.
Software: Baoai Han, Yaqin Tu, Zuhong He, Yongqin Li.
Supervision: Tian Wang.
Visualization: Tao Zhou.
Writing – original draft: Baoai Han, Yaqin Tu, Yongqin Li, Xiuping Yang.
Writing – review & editing: Haiying Sun, Tao Zhou.
Haiying Sun orcid: 0000-0001-8112-4803.
Footnotes
Abbreviations: CI = confidence interval, mtDNA = mitochondrial DNA, ORs = odds ratios.
BH and TZ contribute the equal work to this article.
This study was supported by the National Natural Science Foundation of China (No, 81600801, 81771005, 81502527, and 81670938). National Science Foundation of Hubei Province, China (No, 2015CFB527).
The authors have no conflicts of interest to disclose.
References
- [1].Vaisbuch Y, Santa Maria PL. Age-related hearing loss: innovations in hearing augmentation. Otolaryngol Clin North Am 2018;51:705–23. [DOI] [PubMed] [Google Scholar]
- [2].Patel R, McKinnon BJ. Hearing loss in the elderly. Clin Geriatr Med 2018;34:163–74. [DOI] [PubMed] [Google Scholar]
- [3].Servidoni AB, Conterno LO. Hearing loss in the elderly: is the hearing handicap inventory for the elderly - screening version effective in diagnosis when compared to the audiometric test. Int Arch Otorhinolaryngol 2018;22:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Su P, Hsu CC, Lin HC, et al. Age-related hearing loss and dementia: a 10-year national population-based study. Eur Arch Otorhinolaryngol Off J Eur Feder Otorhinolaryngol Soc 2017;274:2327–34. [DOI] [PubMed] [Google Scholar]
- [5].Pye A, Charalambous AP, Leroi I, et al. Screening tools for the identification of dementia for adults with age-related acquired hearing or vision impairment: a scoping review. Int Psychogeriatr 2017;29:1771–84. [DOI] [PubMed] [Google Scholar]
- [6].Golub JS, Luchsinger JA, Manly JJ, et al. Observed hearing loss and incident dementia in a multiethnic cohort. J Am Geriatr Soc 2017;65:1691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deal JA, Albert MS, Arnold M, et al. A randomized feasibility pilot trial of hearing treatment for reducing cognitive decline: Results from the Aging and Cognitive Health Evaluation in Elders Pilot Study. Alzheimers Dement 2017;3:410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tavanai E, Mohammadkhani G. Role of antioxidants in prevention of age-related hearing loss: a review of literature. Eur Arch Otorhinolaryngol Off J Eur Feder Otorhinolaryngol Soc 2017;274:1821–34. [DOI] [PubMed] [Google Scholar]
- [9].Bottger EC, Schacht J. The mitochondrion: a perpetrator of acquired hearing loss. Hearing Res 2013;303:12–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Menardo J, Tang Y, Ladrech S, et al. Oxidative stress, inflammation, and autophagic stress as the key mechanisms of premature age-related hearing loss in SAMP8 mouse Cochlea. Antioxid Redox Signal 2012;16:263–74. [DOI] [PubMed] [Google Scholar]
- [11].Sun HY, Hu YJ, Zhao XY, et al. Age-related changes in mitochondrial antioxidant enzyme Trx2 and TXNIP-Trx2-ASK1 signal pathways in the auditory cortex of a mimetic aging rat model: changes to Trx2 in the auditory cortex. FEBS J 2015;282:2758–74. [DOI] [PubMed] [Google Scholar]
- [12].Huang YH, Chen CM, Lee YS, et al. Detection of mitochondrial DNA with 4977 bp deletion in leukocytes of patients with ischemic stroke. PLoS One 2018;13:e0193175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salehi Z, Haghighi A, Haghighi S, et al. Mitochondrial DNA deletion Delta4977 in peptic ulcer disease. Mol Biol 2017;51:37–41. [DOI] [PubMed] [Google Scholar]
- [14].Zabihi Diba L, Mohaddes Ardebili SM, Gharesouran J, et al. Age-related decrease in mtDNA content as a consequence of mtDNA 4977 bp deletion. Mitochondrial DNA Part A, DNA Mapp Seq Anal 2016;27:3008–12. [DOI] [PubMed] [Google Scholar]
- [15].Dzinic T, Dencher NA. Oxygen concentration and oxidative stress modulate the influence of Alzheimer's disease abeta1-42 peptide on human cells. Oxid Med Cell Longev 2018;2018:7567959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Karimova A, Oltulu YM, Azakli H, et al. Lack of association between increased mitochondrial DNA(4977) deletion and ATP levels of sputum cells from chronic obstructive pulmonary disease patients versus healthy smokers. Mitochondrial DNA Part A, DNA Mapp Seq Anal 2017;28:361–9. [DOI] [PubMed] [Google Scholar]
- [17].Liu H, Han Y, Wang S, et al. Association between the mitochondrial DNA 4977 common deletion in the hair shaft and hearing loss in presbycusis. Mol Med Rep 2015;11:1127–31. [DOI] [PubMed] [Google Scholar]
- [18].Du Z, Yang Q, Liu L, et al. NADPH oxidase 2-dependent oxidative stress, mitochondrial damage and apoptosis in the ventral cochlear nucleus of D-galactose-induced aging rats. Neuroscience 2015;286:281–92. [DOI] [PubMed] [Google Scholar]
- [19].Du Z, Li S, Liu L, et al. NADPH oxidase 3associated oxidative stress and caspase 3dependent apoptosis in the cochleae of Dgalactoseinduced aged rats. Mol Med Rep 2015;12:7883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yoshitoku Y, Toyonori O. Practice guideline of evidence-based medicine: Preferred Reporting Items for Systematic Reviews and Meta-analyses (the PRISMA statement). J Info Proc Manag /Joho Kanri 2011;54:254–66. [Google Scholar]
- [21].Siqueira RG, Silva DA, Melo LD, et al. Common Deletion (CD) in mitochondrial DNA of irradiated rat heart. Anais Acad Brasileira de Ciencias 2014;0:0. [DOI] [PubMed] [Google Scholar]
- [22].Krishnan KJ, Ratnaike TE, De Gruyter HL, et al. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer's disease. Neurobiol Aging 2012;33:2210–4. [DOI] [PubMed] [Google Scholar]
- [23].Powers JM, Murphy G, Ralph N, et al. Polypharmacy and sun exposure: implications for mitochondrial DNA deletions in skin. J Photochem Photobiol B 2017;173:397–403. [DOI] [PubMed] [Google Scholar]
- [24].Gendron SP, Mallet JD, Bastien N, et al. Mitochondrial DNA common deletion in the human eye: a relation with corneal aging. Mech Ageing Dev 2012;133:68–74. [DOI] [PubMed] [Google Scholar]
- [25].Rocha MC, Rosa HS, Grady JP, et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann Neurol 2018;83:115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhong Y, Hu YJ, Chen B, et al. Mitochondrial transcription factor A overexpression and base excision repair deficiency in the inner ear of rats with D-galactose-induced aging. FEBS J 2011;278:2500–10. [DOI] [PubMed] [Google Scholar]
- [27].Dai P, Yang W, Jiang S, et al. Correlation of cochlear blood supply with mitochondrial DNA common deletion in presbyacusis. Acta Otolaryngol 2004;124:130–6. [DOI] [PubMed] [Google Scholar]
- [28].Markaryan A, Nelson EG, Hinojosa R. Detection of mitochondrial DNA deletions in the cochlea and its structural elements from archival human temporal bone tissue. Mutat Res 2008;640:38–45. [DOI] [PubMed] [Google Scholar]
- [29].Seidman MD, Bai U, Khan MJ, et al. Association of mitochondrial DNA deletions and cochlear pathology: a molecular biologic tool. Laryngoscope 1996;106:777–83. [DOI] [PubMed] [Google Scholar]
- [30].Han W, Han D, Jiang S. Mitochondrial DNA4977 deletions associated with human presbycusis. Zhonghua Er Bi Yan Hou Ke Za Zhi 2000;35:416–9. [PubMed] [Google Scholar]
- [31].Bai U, Seidman MD, Hinojosa R, et al. Mitochondrial DNA deletions associated with aging and possibly presbycusis: a human archival temporal bone study. Am J Otol 1997;18:449–53. [PubMed] [Google Scholar]
- [32].Dai P, Jiang S, Yang W. Extraction, amplification, recombination and sequencing of the mitochondrial DNA from celloidin embedded human temporal bone sections. Zhonghua Er Bi Yan Hou Ke Za Zhi 1998;33:206–9. [PubMed] [Google Scholar]
- [33].Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 2006;38:515–7. [DOI] [PubMed] [Google Scholar]
- [34].Kraytsberg Y, Kudryavtseva E, McKee AC, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 2006;38:518–20. [DOI] [PubMed] [Google Scholar]


