Abstract
Background and Objectives:
Colonization of Pseudomonas aeruginosa in Cystic Fibrosis (CF) patients may lead to severe pulmonary disease and death. Different characteristics of P. aeruginosa from these patients were determined in the present study.
Materials and Methods:
Antimicrobial susceptibility and AmpC-overproduction were determined. The β-lactamase genes were detected by PCR and the oprD gene was sequenced in some of the carbapenem resistance isolates. Distribution of exo genes was determined by PCR. Cytotoxicity of Exo effector proteins was measured using A549 cells. Biofilm production was determined by microtiter plate assay. Random amplified polymorphic DNA (RAPD) –PCR was performed for molecular analysis.
Results:
Polymyxin B, piperacillin/tazobactam and meropenem were the most active antibiotics and 9.6% of isolates were ampC overproducers. The prevalence of blaVEB, blaOXA, blaVIM, and blaPER genes were as follow: 22.7%, 3.75%, 6.25% and 3.75%, respectively. A high proportion (83.5%) of isolates was able to produce biofilm. The exoT gene was present in all isolates while exoU was present in about 35% of them. RAPD-PCR revealed 49 patterns among 78 tested isolates in which 34 patterns were detected once.
Conclusion:
Biofilm formation ability and relatively high frequency of exoS may contribute to the persistence of bacteria within lungs of CF patients. Some characteristics of isolates recovered from a single patient after several sampling procedures were similar, while others lacked resemblance.
Keywords: Pseudomonas aeruginosa, Type 3 secretion system, Cystic fibrosis, Cytotoxicity, Biofilm
INTRODUCTION
Pseudomonas aeruginosa, a major opportunistic pathogen causing a diverse range of human infections, is the most frequently isolated pathogen from sputum of patients with cystic fibrosis (CF) lung disease (1). Among multiple pathogenic elements involved in infection process (2), type III secretion system (T3SS) effectors have been well studied and recognized as key for establishing infection (3). Currently, four type III-secreted effector proteins, ExoS, ExoT, ExoU, ExoY, have been recognized in P. aeruginosa (4, 5, 6). Both ExoT and ExoS show ADP-ribosyltransferase and GTPase activating activities. They result in inhibition of actin polymerization, prevent phagocytosis and cell migration, and promote apoptosis (7). ExoU and ExoY have phospholipase and adenylate cyclase activities, respectively (8, 9). Together, these secreted toxins result in disruption of key eukaryotic cellular signaling pathways (10). Furthermore, in clinical studies, the presence of these exoenzymes is associated with a dissatisfactory clinical outcome among patients with P. aeruginosa infection (9). A great deal of evidence has demonstrated that the biofilm formation is responsible, in part, for therapeutic failure and biofilms have been shown to be up to 1000 times more resistant to antibiotics than planktonic, or free-swimming, cells of the same isolate (11). Presence of quorum-sensing molecules in CF sputum, used by P. aeruginosa and other bacteria for cell to cell communication in vitro (12), has been cited as evidence that P. aeruginosa in the CF lung form biofilms (13). On the other hand, due to increased multi-drug resistant isolates and biofilm formation capacity, treatment of infections caused by this bacterium is difficult, so therapeutic options are limited and a careful selection of antibiotic is necessary (14). β-lactam antibiotics are used for the treatment of acute pulmonary exacerbations in CF patients (15). Resistance mechanisms of P. aeruginosa to β-lactam antibiotics are multifactorial, including production of β-lactamase, AmpC over-production, loss of OprD porin and overexpression of efflux-pumps (16).
The distribution of T3SS effector genes amongst clinical isolates of P. aeruginosa remains to be elucidated. Furthermore, the prevalence of effector genes in populations of isolates from different disease sites has not been systematically and thoroughly explored. Hence, in this report, we investigate the distribution of the type III effector proteins-encoding genes and association between cytotoxicity and exo genes in P. aeruginosa strains obtained from cystic fibrosis patients. Other characteristics of isolates such as antibiotic resistant pattern as well as mechanisms of resistance to β-lactam antibiotics, biofilm capacity, and RAPD analysis were also determined.
MATERIALS AND METHODS
Bacterial isolates.
A total of 85 clinical isolates of P. aeruginosa were included in the present study. These isolates were collected from 27 CF patients attending the Children's Medical Center Hospital in Tehran, among which, six patients took part in collection process more than once. The collection process was carried out through April 2010 to June 2011. Primary identification to the species level were done based on colony morphology, pigment production, and Gram staining. All phenotypically different isolates were collected to be observed, though some of them were obtained from one patient. Routine bio-chemical tests used for confirmation were as follow: the ability of oxidase and catalase production, oxidative-fermentative (OF) test, and growth at 42°C on Mueller–Hinton agar (17).
Antibiotic susceptibility test.
The antimicrobial susceptibility profile of isolated P. aeruginoase was determined by using antibiotic containing disks on Mueller Hinton agar according to Clinical and Laboratory Standards Institute (CLSI) guidelines (18). The disks contained the following antibiotics: ceftazidime (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), gentamicin (10 μg), tobramycin (10 μg), piperacillin/tazobactam (100/10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), ticarcillin (75 μg), polymyxin B (300 Unit). Determining of Multidrug resistance (MDR) isolates, as an important and problematic concern, were done based on considering resistance against at least 1 agent in ≥ 3 antimicrobial categories (19).
Detection of bla genes among β-lactam resistant isolates.
Genomic DNAs used as template were extracted using DNA Extraction Kit (Accu-Prep® Genomic DNA Extraction Kit, USA Bioneer, Co.) according to the manufacturer's instructions. The blaPER, blaSHV, blaVEB, blaPSE, blaCTX-M, blaOXA, blaOXA-48, blaVIM, blaIMP, blaGIM, blaAIM, blaSPM, blaNDM, blaSIM, blaKPC and blaGES genes were detected by PCR. Amplification of β-lactamases genes was performed using specific primers (16, 20). The amplified PCR products were subjected to electrophoresis in 1.2% agarose gels, stained with ethidium bromide, and visualized in a Gel Doc XR molecular imager (Bio-Rad Laboratories).
Detection of AmpC overproducer among carbapenem resistant isolates.
AmpC overproduction among carbapenem resistant isolates was assayed according to Rodríguez-Martínez et al. method (21). Briefly, the isolates were considered as AmpC over-producers when there was at least a two-fold dilution difference between the MICs (minimal inhibitory concentrations) of IMI (imipenem), MEM meropenem) or CAZ (ceftazidime) and MICs of these antibiotics plus 250 μg COL (cloxacillin).
Sequencing of oprD gene among carbapenem resistant isolates.
PCR amplification of oprD among carbapenem resistant isolates was performed using specific primers (21). Sequences were compared with that of reference P. aeruginosa PAO1.
Detection of genes encoding type-III secretion toxins.
Sequences of specific primers used in amplifying genes encoding type-III secretion toxins, exoT, exoU, exoS, exoY, based on PCR technique as previously described (22, 23).
Cytotoxicity assay.
To determine cytotoxic effect of P. aeruginosa isolates on A549 human lung epithelial cells, isolated bacteria were grown in Luria-Bertani broth. Cultures were centrifuged and resultant pellets were washed three times with sterile PBS. The final bacterial cell pellets were re-suspended in PBS to yield the final concentration of 1.5 × 108 cfu/ml. Human cells suspension were seeded on 96-well flat-bottom tissue culture plates at a density of 1.5 × 105 cells / well with DMEM (Dulbecco's Modified Eagle's medium) and 10% Fetal Bovine Serum (FBS). After an overnight incubation of cell culture at 37°C, wells were rinsed with sterile PBS, followed by the addition of DMEM without phenol red and 1% FBS. Consequently, the diluted bacterial suspension of each P. aeruginoasa isolates were added to the cells and incubated at 37°C for 4 h. PBS without inoculated bacteria was used for negative controls. Eventually, lactate dehydrogenase activity was assayed for quantification of cytotoxicity using the Cytotoxicity Detection Kitplus (Roche, Germany) according to the manufacturer's instructions. The assay was performed in triplicate.
Biofilm formation.
Biofilm formation ability of isolates was determined based on a colorimetric microtiter plates assay as described by Lee et al. (24). In brief, an overnight bacterial culture in Tripticase Soy Broth (TSB) at 37°C was diluted (1:100) by fresh TSB and then inoculated into the sterile flat-bottomed 96-well polystyrene microtiter plates. After an incubation period of 24h at 37°C without shaking, wells were gently washed three times with PBS. 99% methanol was used for biofilm fixation for 15 min, and then the plate was air-dried. Afterwards, Crystal violet 1% (CV) was added for biofilm staining for 20 min. In the next step, unbound CV was removed and the remaining was released by adding 33% acetic acid. Finally, the absorbance of solubilized CV was measured at 590nm using a microtiter plate reader. All the assays were tested in triplicate. Uninoculated medium was considered as control negative sample. The cut-off OD (ODc) was defined as three standard deviations above the mean OD of the negative control. In accordance with the results of microtiter plate tests, Biofilm producers were characterized as follows based on the optical density: non-biofilm producers (OD test <ODc), weak biofilm producers (ODc< OD < 2× ODc), moderate biofilm producers (2× ODc< OD < 4× ODc), and strong biofilm producers (4× ODc< OD).
PCR-RAPD typing of isolates.
The RAPD-PCR technique was used as described by Bulkanov et al. (25). This fingerprinting was carried out by using arbitrary primers, RAPD-208 (5′-AGCGG-GCCAA-3′) and 272 (5′-ACGGCCGACC-3′) (16, 26, 27). This amplification was performed in T-100 Gradient (BioRAD) using PCR Master kit (AMPLIQON, Denmark) according to manufacture guideline, with the following cycling parameters: initial denaturation at 95°C for 5 min followed by 45 cycles of denaturation at 95°C for 1 min, annealing for 1 min at 38.5°C, extension at 72°C for 5 min. The final extension step was continued for another 2 min at 72°C. After visualization of amplified products, genotypic profile of each isolate was determined. BioNumerics software was used in pattern analysis and isolates with ≥85% similarities were considered as indistinguishable (one pattern) types and isolates with <85% similarities were taken as different types. RAPD types were designated numerically. Type-1 was the most frequent pattern among 49 detected patterns.
RESULTS
Table 1 shows the antibiotic resistance patterns of P. aeruginosa isolates. As shown in Table 1, polymyxin B followed by piperacillin/tazobactam and meropenem were the most active antibiotic against the panel of isolates. Ceftazidime was effective on about 50% of the isolates. Less than half of the isolates were determined as MDR organisms.
Table 1.
The antimicrobial susceptibility patterns of P. aeruginosa isolates
| Antibiotics | N (%) of isolates (n=85) | ||
|---|---|---|---|
| S | I | R | |
| Ceftazidime | 29 (34.11) | 12 (14.11) | 44 (51.76) |
| Aztreonam | 42 (49.41) | 17 (20) | 26 (30.59) |
| Imipenem | 71 (83.52) | 0 | 14 (16.47) |
| Meropenem | 77 (90.58) | 0 | 8 (9.41) |
| Amikacin | 69 (81.17) | 4 (4.7) | 12 (14.12) |
| Gentamicin | 53 (62.35) | 2 (2.35) | 30 (35.29) |
| Tobramycin | 56 (65.88) | 1 (1.17) | 28 (32.94) |
| Piperacillin/Tazobactam | 77 (90.58) | 0 | 8 (9.41) |
| Ciprofloxacin | 57 (67.05) | 8 (9.41) | 20 (23.5) |
| Levofloxacin | 42 (49.41) | 15 (17.64) | 28 (32.9) |
| Ticarcillin | 52 (61.17) | 5 (5.88) | 28 (32.94) |
| Polymyxin B | 85 (100) | 0 | 0 |
S, susceptible; I, intermediate; R, resistant
The prevalence of blaVEB, blaOXA, blaVIM, and blaPER genes were as follow: 22.7%, 3.75%, 6.25%, and 3.75% respectively. All isolates were negative for blaSHV, blaPSE, blaCTX-M, blaOXA-48, blaIMP, blaGIM, blaAIM, blaSPM, blaNDM, blaSIM, blaKPC and blaGES genes. Coexistence of blaVEB and blaPER was observed in 2 isolates.
Also, among 14 carbapenem (imipenem and meropenem) resistant isolates, 5 (35.7%) were positive for blaVIM. In all carbapenem resistant isolates oprD was inactivated by mutations. 12 out of 14 carbapenem resistant isolates, mechanism of oprD inactivation resulted from 1–18 bp deletions and substitution mutations which leads to internal stop codon and frame-shift mutations and in two isolates oprD was not detected by PCR (Table 2). 8 out of 14 carbapenem resistant isolates (57.1%) were AmpC overproducers and showed decrease in MICs for ceftazidime, imipenem and meropenem in presence of cloxacillin (Table 2).
Table 2.
Characteristics of carbapenem resistant isolates
| Isolates | MIC (μg/ml) | AmpC Overproduction | blaVIM | oprD Mutation | |||||
|---|---|---|---|---|---|---|---|---|---|
| CAZ* | CAZ-COL | IMI | IMI-COL | MEM | MEM-COL | ||||
| P1-1 | 32 | 32 | 32 | 16 | 1 | 1 | − | − | 1.N.D† |
| P1-2 | 8 | 2 | 16 | 16 | 16 | 16 | + | − | 1.N.D |
| P1-3 | 8 | 8 | 16 | 16 | 16 | 16 | − | + | 6.N.D |
| P1-4 | 16 | 16 | 8 | 8 | 1 | 1 | − | − | 12.N.D |
| P1-7 | 256 | 256 | 256 | 256 | 128 | 128 | − | − | Not-D‡ |
| P1-8 | 2 | 2 | 16 | 8 | 8 | 8 | − | + | 18.N.D |
| P1-9 | 256 | 256 | 512 | 256 | 256 | 256 | − | − | Not-D |
| P1-10 | 256 | 256 | 32 | 16 | 8 | 1 | + | + | 5.N.D |
| P3-2 | 32 | 32 | 32 | 16 | 8 | 1 | + | − | 4.N.D |
| P3-5 | 2 | 2 | 16 | 1 | 1 | 1 | + | − | 11.N.D |
| P14-1 | 32 | 16 | 32 | 8 | 8 | 1 | + | + | 1.N.D |
| P14-2 | 2 | 2 | 8 | 1 | 1 | 1 | + | + | 7.N.D |
| P17-9 | 2 | 2 | 8 | 1 | 1 | 1 | + | − | 1.N.D |
| P17-10 | 2 | 2 | 8 | 1 | 1 | 1 | + | − | 6.N.D |
CAZ, Ceftazidime; CAZ-COL, Ceftazidime-Cloxacillin; IMI, Imipenem; IMI-COL, Imipenem-Cloxacillin; MEM, Meropenem; MEM-COL, Meropenem - Cloxacillin
Nucleotide Deletion,
Not Determined
Different characteristics of P. aeruginosa isolates including type III secretion-toxin encoding gene patterns, biofilm formation and cytotoxicity effect are shown in Table 3. A high proportion of isolates were able to produce biofilm (83.5%) in which, 20%, 37.64% and 25.88% were determined as weak, moderate, and strong biofilm producers, respectively.
Table 3.
Some characteristics of P. aeruginosa isolates
| Genes | Biofilm density* | % of cytotoxicity | N. of isolates | |||||
|---|---|---|---|---|---|---|---|---|
| exoS | exoY | exoU | exoT | Range | 50%† | 90% | ||
| + | + | + | + | 16 M | 3.41–68.94 | 25.76 | 53.41 | 16 |
| + | + | − | + | 7 W, 16 M, 10 S, 6 N | 0–81.06 | 17.63 | 39.39 | 39 |
| − | + | + | + | 5 S,3 W | 0–81.06 | 16.167 | 24.62 | 8 |
| − | − | − | + | 8 W | 0–21 | 14.77 | 15.91 | 8 |
| − | − | + | + | 4 W | 36.74–81.06 | 37.88 | 50.74 | 4 |
| − | + | − | + | 2 W | 24.38–22.54 | - | - | 2 |
| + | − | + | + | 2 S | 23.11–39.77 | - | - | 2 |
| + | − | - | + | 5 S, 1 N | 6.12–61.36 | 7.2 | 61.36 | 6 |
W, weak; M, moderate; S, strong; N, negative
The cytotoxicity 50: 50% of the isolates shows below this cytotoxicity value and the same principal for the cytotoxicity 90.
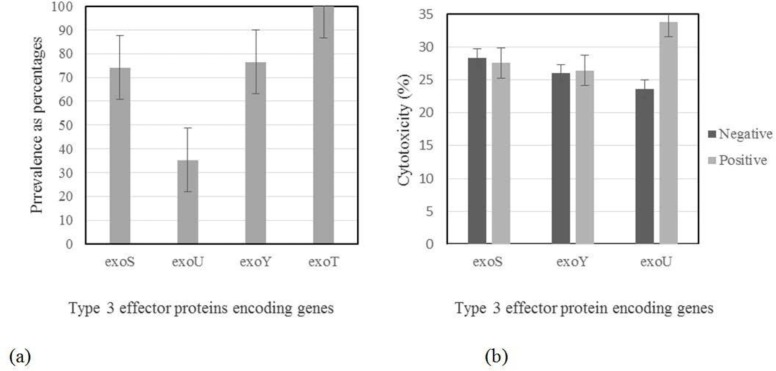
As shown in Fig 1a, exoT was present in all isolates. Most isolates harbored exoS and exoY, while exoU was present in about 35% of isolates. In vitro cytotoxicity against A549 human lung epithelial cells was observed in strains carrying type III secretory genes. The correlation between the presence of exo genes and cytotoxicity is demonstrated in Fig. 1b. It revealed that levels of cytotoxicity were found to be significantly affected by presence of exoU gene.
Fig. 1.
(a) Prevalence (as percentages) of the exoS, b: exoU, c: exoY, d: exoT genes among isolates. Error bars represent standard errors. (b) Correlation of exo genes existence with cytotoxicity of A549 cells. Error bars represent standard errors. Because of observing exoT in all isolates, data related to this gene is not presented in this figure.
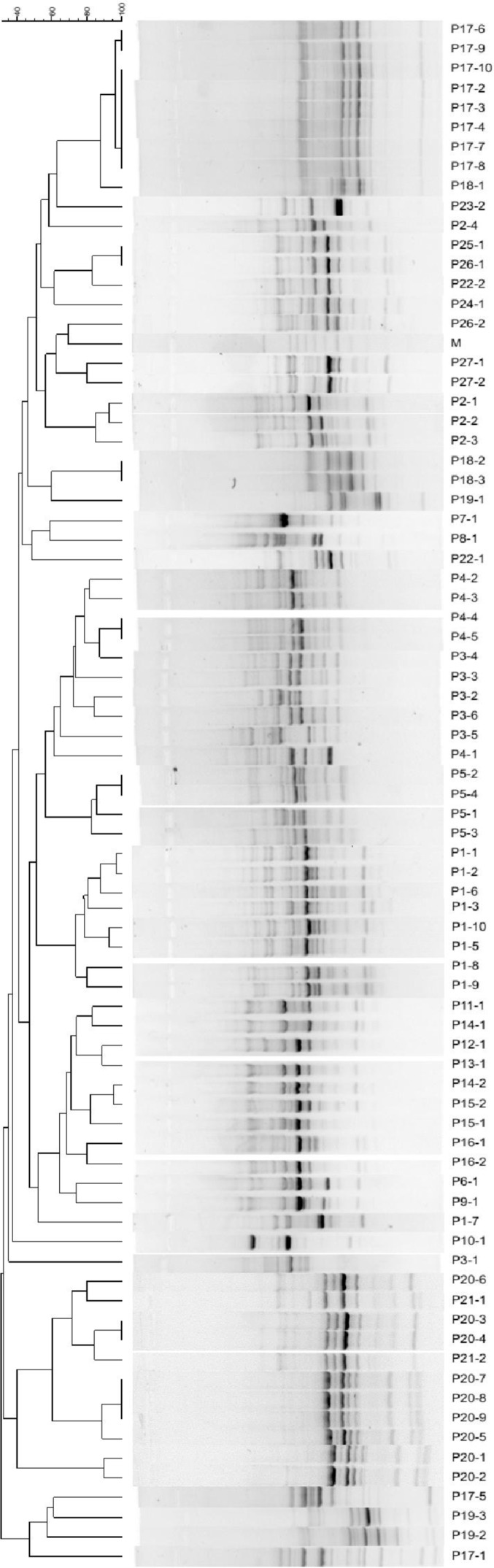
Results based on RAPD-PCR, as a quick method to characterize the heterogeneity in the group by using a cut off value of 85% as the threshold, revealed 49 patterns among 78 tested isolates in which 34 patterns were detected once, while the remaining 15 patterns were repeatedly observed. RAPD-PCR generated dendrogram of 78 P. aeruginosa isolates from 27 different patients is illustrated in Fig. 2.
Fig. 2.
RAPD-PCR generated dendrogram for 78 P. aeruginosa isolates of 27 different patients.
Properties of the isolates obtained from some of the patients who have been taking part in sampling for several times, during a year, are demonstrated in Table 4.
Table 4.
Detailed properties of isolates obtained from patients who took part in samplings for several times
| Patients | Attending dates | Isolates | Gene pattern of T3SS* | Cytotoxicity (%) | Biofilm density† | Antibiotic resistant pattern‡ | RAPD Type§ |
|---|---|---|---|---|---|---|---|
| P1 | 2010/7/6 | P1-1 | S+U+ | 15.2 | - | ATM, IMI, GM, TM, CIP, LE, TC | 8 |
| P1-2 | S−U+ | 24.4 | W | ATM, IMI, MEM, CIP, LE, TC | 8 | ||
| 2010/9/29 | P1-3 | S+U− | 8.8 | M | CAZ, ATM, IMI, MEM, GM, TM, LE, CPM | 33 | |
| P1-4 | S+U− | 7.2 | W | CAZ, ATM, IMI, AK, GM, TM,TC | 37 | ||
| P1-5 | S+U− | 7.0 | W | CAZ, ATM, AK, GM, TM, TC | 9 | ||
| P1-6 | S+U− | 17.9 | M | CAZ, ATM, GM, TM | 8 | ||
| 2010/11/1 | P1-7 | S+U− | 81.1 | W | CAZ, ATM, IMI, MEM, GM, TM, CIP, TC | 40 | |
| P1-8 | S+U− | 11.8 | M | ATM, IMI, MEM, GM, TM, CIP, TC | 22 | ||
| 2011/4/4 | P1-9 | S+U+ | 3.5 | W | CAZ, ATM, IMI, MEM, AK, GM, TM, PTZ, CIP, TC | 23 | |
| P1-10 | S+U− | 73.2 | W | CAZ, ATM, IMI, MEM, GM, TM, CIP, LE, TC | 9 | ||
| P14 | 2011/1/3 | P14-1 | S+U− | 29.6 | N | CAZ, IMI | 12 |
| 2011/3/21 | P14-2 | S+U+ | 28.5 | W | ATM, IMI, MEM | 12 | |
| P5 | 2010/7/29 | P5-1 | S+U− | 23.2 | W | CAZ, ATM, PTZ | 7 |
| 2010/9/17 | P5-2 | S+U+ | 14.1 | W | CAZ | 7 | |
| P5-3 | S−U+ | 81.1 | M | CAZ, ATM, GM, TM, CIP, TC | 51 | ||
| 2010/10/25 | P5-4 | S+U− | 20.9 | S | CAZ | 7 | |
| P3 | 2010/4/12 | P3-1 | S+U+ | 37.9 | W | GM, LE | 28 |
| 2010/9/28 | P3-2 | S+U− | 26.3 | M | CAZ, IMI, MEM, LE | 6 | |
| 2011/14/16 | P3-5 | S−U+ | 5.0 | W | IMI, LE | 6 | |
| P3-6 | S−U+ | 16.0 | - | ATM, LE | 50 | ||
| P17 | 2010/7/20 | P17-1 | S−U+ | 37.9 | - | CAZ, GM, TM, TC | 38 |
| P17-2 | S+U− | 17.7 | S | CAZ, GM, TM, TC | 1 | ||
| P17-3 | S+U− | 15.6 | S | CAZ, GM, TM, TC | 1 | ||
| P17-4 | S+U− | 32.2 | S | CAZ, GM, TM, TC | 1 | ||
| 2010/5/26 | P17-5 | S+U− | 9.5 | S | ATM, GM, TM, CIP, TC | 44 | |
| 2010/10/5 | P17-6 | S+U− | - | M | GM, LE, TC | 1 | |
| 2011/6/11 | P17-8 | S+U− | 17.7 | S | GM, TM, LE, TC | 1 | |
| P17-9 | S+U− | 11.8 | S | ATM, IMI, GM, TM, CIP, TC | 1 | ||
| P17-10 | S+U− | 10.3 | S | ATM, IMI, GM, TM, CIP, LE, TC | 1 | ||
| P20 | 2010/7/9 | P20-1 | S−U+ | 22.7 | M | LE | 15 |
| P20-2 | S+U+ | 57.2 | S | CAZ, LE | 15 | ||
| P20-3 | S−U+ | 16.7 | M | LE | 13 | ||
| 2010/9/8 | P20-4 | S+U− | 39.4 | S | CAZ, TC | 13 | |
| P20-5 | S+U+ | 26.2 | S | CAZ | 14 | ||
| P20-6 | S+U− | 17.8 | W | CAZ | 35 | ||
| 2011/4/19 | P20-7 | S−U+ | 16.7 | M | CAZ | 14 | |
| P20-8 | S−U+ | 50.8 | S | CAZ | 14 | ||
| P20-9 | S−U+ | 24.62 | W | CAZ, ATM, PTZ, LE, TC | 14 |
Type 3 secretion system,
W, weak; M, moderate; S, strong; N, negative
AK, Amikacin; ATM, Aztreonam; CTZ, Ceftazidime; CIP, Ciprofloxacin; GM, Gentamicin; IMI, Imipenem; LE, Levofloxacin; MEM, Meropenem; PTZ, Piperacillin/ Tazobactam; PB, Polymyxin B; TC, Ticarcillin; TM, Tobramycin.
Numerical types are obtained as a result of RAPD PCR method.
DISCUSSION
As antibiotic resistant pattern shows, less than half of the present isolates were resistant to all tested antibiotics, except cefatazidime. The difference in antibiotic resistant pattern between the obtained results in the present and other studies (28) may be due to either the different common colones present in a certain region or different antibiotic treatment pattern. The frequency of MDR isolates in the present study was considerably less than the previous investigation done by our team on isolates from wound infections of burn patients (about 92%) (23). Results obtained by Rao et al. (28) even showed less frequency of MDR isolates from CF patients.
As shown in Table 2, AmpC overproduction and oprD mutations were associated with resistance to imipenem and meropenem. Furthermore, AmpC overproduction was associated with resistance to ceftazidime. In the present investigation, results revealed that the association between OprD gene deficiency and resistance to imipenem and meropenem is similar to other literatures. Mutations in oprD gene reportedly lead to increased MIC to imipenem (16).
To date, presence of different β-lactamases genes (TEM, SHV, CTXM, PER, VEB, GES and IBC families) were reported among P. aeruginosa isolates (16, 20). In the present study, VEB type of β-lactamases was the most predominant ESBLs. The results of this study demonstrated blaVIM as the most common MBLs (Metallo-β-lactamases) found in carbapenem-resistant bacteria, including carbapenem-resistant P. aeruginosa in non CF patients (16, 20).
It was shown that type-III secretion system is an important virulence mechanism and cytotoxins encoded by exoS, exoT, exoU and exoY genes may be important contributors to the dissemination of the organism from the site of infection, bacterial evasion of the host immune response and inhibition of DNA synthesis leading to host cell death (29). The majority of P. aeruginosa strains carry exoT and exoY genes; however, the presence of exoS and exoU differ noticeably between the isolates and appear to be mutually exclusive (29). As the previous study done by our team on P. aeruginosa isolated from burn patients (23), exoT was the most prevalent cytotoxin gene found in isolates of CF patient and was reported in all isolates (100%), though exoY was not as frequent as exoT (about 79%). In contrast to our previous results that showed exoS as the least prevalent gene (29%) (23), present study revealed that exoU has less frequency than other cytotoxin genes (36.58%). Data showed that the least prevalence of exoU in CF patients, as a chronic disease, is in agreement with results reported by Feltman et al. (22), which may be due to the straight correlation between exoU and acute infections. So, the high prevalence of exoU gene in burn isolates, as was reported before is predictable. Relatively high frequency of isolates harboring exoS (about 80%) may be related to hypothesis presented by Feltman et al. (22). They suggested that production of exoS may provide P. aeruginosa isolates with an advantage in colonizing or persisting in CF lung. Furthermore, exoS may be genetically linked to a second factor that is important in the pathogenesis of P. aeruginosa pulmonary colonization and infection in CF patients. Taken together, our findings suggest that at least some of the genes encoding type III secretion effector proteins are present in all P. aeruginosa isolates, though the prevalence of them was different. Variation observed in the prevalence of exo genes may be the consequence of clinical specimen used for collecting isolates.
In the present study, results related to in vitro cytotoxic effect of strains carrying type III secretion proteins concur with the previous studies, in which the type III secretion system is reported to be the primary contributor to the virulence of P. aeruginosa and may lead to the increased morbidity and mortality in patients and animals (23).
High frequency of biofilm producers in the present study (more than 90%), as in other report, suggests that P. aeruginosa is able to form biofilms within the lungs of CF patients and this mode of growth likely facilitates persistence of the bacterium within this niche (30) and biofilm development in CF airway is one of the most striking features of P. aeruginosa adaptation to airways of patients with CF (31). However, CF isolates can be highly variable with respect to biofilm density. In the current study, the prevalence rate of strong biofilm producers was about 25%, whereas Perez et al. (32) could not find such a biofilm state among CF patients. No correlation was observed between biofilm formation and cytotoxicity.
Because of the presence of problems related to phenotypic variations of bacterial isolates throughout the period of infection in CF patients (33) and the difficulty of ascertaining the reinfection of patients or difficulty in determining whether the patient is infected by different strains over a period of time, or whether the isolated strains differ according to the respiratory tract site where the sample was taken (34), molecular typing was selected as a more sensitive and specific method in this study. RAPD typing of the 78 P. aeruginosa isolates gave 49 different RAPD fingerprints, 34 of which contained only 1 strain. The dominant RAPD type amongst the remaining 15 fingerprints contained 9 strains. Presence of most isolates harboring unique RAPD patterns may be due to the acquisition of these isolates from environmental sources where horizontal gene transfer occurs and may result in acquisition of new genes.
As shown in Table 4, our results revealed that more than one P. aeruginosa strains can be isolated from a CF patient in one time attendance for sampling, though a given strain was not recovered from a given patient after a period of time. In other words, isolation of different strains with different properties from one patient while attending for several times revealed the possibility of both phenotypic and genotypic alterations in causative strains. Furthermore, the similarity between some of the genotypic aspects of different strains, such as the same RAPD pattern in isolates 1 and 2 in patient 20 (P20) was observed, though another genotypic pattern related to exo genes was different. On the other hand, despite the presence of analogous exo genes and RAPD patterns in isolates P1-5 and P1-10, which were recovered from the same patient, their cytotoxic capability were drastically different. Moreover, recovery of an isolate with a similar RAPD pattern, 7, from patient 5, was observed in each round of sampling, though isolation of another different pattern, 51, from the mentioned patient, was restricted to a defined round of sampling. Interestingly, cytotoxicity of isolates with pattern 7 from patient 5, which were recovered constantly, was low and the last isolated strain with this pattern has a strong biofilm intensity. Failure of the treatment in CF patients may be due to the strong ability of causative P. aeruginosa to form biofilm. On the other hand, isolated strain with pattern 51, recovered once, from patient 5 was significantly more cytotoxic than other isolates with pattern 7. However, treatment and management of patient 5 was not acceptable and victorious. Such findings revealed that isolation of stable strains present in the whole duration of the disease along with new strains showing new different RAPD patterns in CF patients may be due to the acquisition of the latter strains from hospitals where they took part in sampling. Undoubtedly, antibiotic therapy against such new and transient strains may result in its elimination from patients, while stable strains with strong ability to form biofilm do not respond to such treatment and can be isolated for several times. However, it is well documented that in the initial stages of infection, when the bacteria are mainly planktonic and not yet persistent, their eradication may be easier and so it reveals the importance of early identification of infection (35).
CONCLUSION
To our knowledge, the compelling role of biofilm formation capacity and cytotoxicity effect of isolates in bacterial invasion process validate probable and powerful therapeutic strategies other than conventional and traditional antibiotic treatment. Furthermore, relatively high frequency of antibiotic resistance among isolated P. aeruginosa may persuade the pharmaceutical industry to introduce new antibiotics to solve the resistance problem.
ACKNOWLEDGEMENTS
This research has been supported by Tehran University of Medical Sciences & Health Services Grant No. 97-01-30-38032.
REFERENCES
- 1.Keating D, Crowe MJ, Kennedy B, Salmon A, Britton D, Gallagher CG, et al. Molecular detection of an atypical, highly resistant, clonal Pseudomonas aeruginosa isolate in cystic fibrosis patients. J Cyst Fibros 2013;12:141–146. [DOI] [PubMed] [Google Scholar]
- 2.Valadbeigi H, Tabatabaei RR, Malek A, Sekawi Z, Raftari M, Parvaneh K, et al. Genomic diversity and virulence genes among clinical isolates of Pseudomonas aeruginosa. Clin Lab 2014;60:363–367. [DOI] [PubMed] [Google Scholar]
- 3.Engel J, Balachandran P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 2009;12:61–66. [DOI] [PubMed] [Google Scholar]
- 4.Iglewski BH, Sadoff J, Bjorn MJ, Maxwell ES. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci U S A 1978;75:3211–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yahr TL, Goranson J, Frank DW. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol 1996; 22:991–1003. [DOI] [PubMed] [Google Scholar]
- 6.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, a novel adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci U S A 1998;95:13899–13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbieri JT, Sun J. Pseudomonas aeruginosa ExoS and ExoT. Rev Physiol Biochem Pharmacol 2004;152:79–92. [DOI] [PubMed] [Google Scholar]
- 8.Ochoa CD, Alexeyev M, Pastukh V, Balczon R, Stevens T. Pseudomonas aeruginosa exotoxin Y is a promiscuous cyclase that increases endothelial tau phosphorylation and permeability. J Biol Chem 2012;287:25407–25418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Mol Microbiol 2004;53:1279–1290. [DOI] [PubMed] [Google Scholar]
- 10.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2000;2:1051–1060. [DOI] [PubMed] [Google Scholar]
- 11.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999;284:1318–1322. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 2001;35:439–468. [DOI] [PubMed] [Google Scholar]
- 13.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000;407:762–764. [DOI] [PubMed] [Google Scholar]
- 14.Hengzhuang W, Ciofu O, Yang L, Wu H, Song Z, Oliver A, et al. High β-lactamase levels change the pharmacodynamics of β-lactam antibiotics in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2013;57:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Döring G, Flume P, Heijerman H, Elborn JS. Treatment of lung infection in patients with cystic fibrosis: Current and future strategies. J Cyst Fibros 2012;11:461–479. [DOI] [PubMed] [Google Scholar]
- 16.Kalantar-Neyestanaki D, Mirsalehian A, Rezagholizadeh F, Jabalameli F, Taherikalani M, Emaneini M. Determination of extended spectrum beta-lactamases, metallo-β-lactamases and AmpC-β-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns 2014;40:1556–1561. [DOI] [PubMed] [Google Scholar]
- 17.Mahon CR, Lehman DC, Manuselis G. Text book of diagnostic microbiology. 3rd edn Philadelphia, PA, USA: 2007; p:564–586. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. M100-S25. 2016;35: 52–54. [Google Scholar]
- 19.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18:268–281. [DOI] [PubMed] [Google Scholar]
- 20.Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns 2010;36:70–74. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009;53:4783–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feltman H, Schulert G, Khan S, Jain M, Peterson M, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 2001;147:2659–2669. [DOI] [PubMed] [Google Scholar]
- 23.Jabalameli F, Mirsalehian A, Khoramian B, Aligholi M, Khoramrooz SS, Asadollahi P, et al. Evaluation of biofilm production and characterization of genes encoding type III secretion system among Pseudomonas aeruginosa isolated from burn patients. Burns 2012;38:1192–1197. [DOI] [PubMed] [Google Scholar]
- 24.Lee B, Schjerling CK, Kirkby N, Hoffmann N, Borup R, Molin S, et al. Mucoid Pseudomonas aeruginosa isolates maintain the biofilm formation capacity and the gene expression profiles during the chronic lung infection of CF patients. APMIS 2011;119:263–274. [DOI] [PubMed] [Google Scholar]
- 25.Bukanov N, Nathan Ravi V, Miller D, Srivastava K, Berg DE. Pseudomonas aeruginosa corneal ulcer isolates distinguished using the arbitrarily primed PCR DNA fingerprinting method. Curr Eye Res 1994;13:783–790. [DOI] [PubMed] [Google Scholar]
- 26.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol 1996;34:1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruimy R, Genauzeau E, Barnabe C, Beaulieu A, Tibayrenc M, Andremont A. Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun 2001;69:584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao P, McCaughan J, McCalmont M, Goldsmith CE, Hall V, Millar BC, et al. Comparison of antibiotic susceptibility patterns in Pseudomonas aeruginosa isolated from adult patients with cystic fibrosis (CF) with invasive Pseudomonas aeruginosa from non-CF patients. J Cyst Fibros 2012;11:349–352. [DOI] [PubMed] [Google Scholar]
- 29.Lee VT, Smith RS, Tümmler B, Lory S. Activities of Pseudomonas aeruginosa effectors secreted by the Type III secretion system in vitro and during infection. Infect Immun 2005;73: 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 2004;70:6188–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folkesson A, Jelsbak L, Yang L, Krogh Johansen H, Ciofu O, Høiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 2012;10:841–851. [DOI] [PubMed] [Google Scholar]
- 32.Perez LRR, Costa MCN, Freitas ALP, Barth AL. Evaluation of biofilm production by Pseudomonas aeruginosa isolates recovered from Cystic fibrosis and non-cystic fibrosis patients. Braz J Microbiol 2011;42:476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horrevortz AM, Borst J, Puyk RJT, de Ridder R, Dzoljic-Danilovic G, Degener JE, et al. Ecology of Pseudomonas aeruginosa in patients with cystic fibrosis. J Med Microbiol 1990;31:119–124. [DOI] [PubMed] [Google Scholar]
- 34.Ortiz-Herrera M, Gerónimo-Gallegos A, Cuevas-Schacht F, Pérez-Fernández L, Coria-Jiménez R. [RAPD-PCR characterization of Pseudomonas aeruginosa strains obtained from cystic fibrosis patients]. Salud Publica Mex 2004;46:149–157. [DOI] [PubMed] [Google Scholar]
- 35.Cohen-Cymberknoh M, Gilead N, Gartner S, Rovira S, Blau H, Mussaffi H, et al. Eradication failure of newly acquired Pseudomonas aeruginosa isolates in cystic fibrosis. J Cyst Fibros 2016;15:776–782. [DOI] [PubMed] [Google Scholar]