Abstract
The E3 ubiquitin ligase membrane-associated ring-CH–type finger 2 (MARCH2) is known to be involved in intracellular vesicular trafficking, but its role in the early secretory pathway between the endoplasmic reticulum (ER) and Golgi compartments is largely unknown. Human ER–Golgi intermediate compartment protein 2 (ERGIC2) and ERGIC3 are orthologs of Erv41 and Erv46 in yeast, proteins that form a heteromeric complex, cycle between the ER and Golgi, and function as cargo receptors in both anterograde and retrograde protein trafficking. Here, we report that MARCH2 directs ubiquitination and subsequent degradation of ERGIC3 and that MARCH2 depletion increases endogenous ERGIC3 levels. We provide evidence that the lysine residues at positions 6 and 8 of ERGIC3 are the major sites of MARCH2-mediated ubiquitination. Of note, MARCH2 did not significantly decrease the levels of an ERGIC3 variant with lysine-to-arginine substitutions at residues 6 and 8. We also show that ERGIC3 binds to itself or to ERGIC2, whereas ERGIC2 is unable to interact with itself. Our results indicate that α1-antitrypsin and haptoglobin are likely to be cargo proteins of ERGIC3. We further observed that α1-antitrypsin and haptoglobin specifically bind to ERGIC3 and that ERGIC3 depletion decreases their secretion. Moreover, MARCH2 reduced secretion of α1-antitrypsin and haptoglobin, and coexpression of the ubiquitination-resistant ERGIC3 variant largely restored their secretion, suggesting that MARCH2-mediated ERGIC3 ubiquitination is the major cause of the decrease in trafficking of ERGIC3-binding secretory proteins. Our findings provide detailed insights into the regulation of the early secretory pathway by MARCH2 and into ERGIC3 function.
Keywords: E3 ubiquitin ligase, protein degradation, intracellular trafficking, ubiquitin, protein secretion, α1-antitrypsin, cargo receptor, endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3), haptoglobin, membrane-associated ring-CH-type finger 2 (MARCH2)
Introduction
Newly synthesized secretory proteins are transported by bulk flow or selectively sorted into COPII2-coated vesicles during ER-to-Golgi transport (1–3). Sorting of soluble cargo proteins requires receptor-mediated transport. Cargo receptors are transmembrane proteins that recognize both cargo proteins in the ER lumen and coat components on the cytoplasmic side during the formation of nascent vesicles (4–6). Cargo receptors cycle between ER and Golgi, binding cargo proteins in one compartment and releasing them in another. The putative human cargo receptor ERGIC3 (also known as hErv46) is a member of a small protein family, which includes ERGIC1 (also called ERGIC32) and ERGIC2 (also named hErv41) (7). Yeast has two homologous family members, ER-derived vesicle protein 41 (Erv41) and Erv46, which form a stable protein complex and are enriched in COPII vesicles (8). Both Erv41 and Erv46 have short tails at N and C termini exposed to the cytosol, whereas their major parts are localized within the lumen (9). The C-terminal cytosolic tails of Erv41 and Erv46 contain export signals required for COPII-dependent transport from the ER (9). The Erv41–Erv46 complex is reported to function as a cargo receptor for the Golgi-resident mannosyltransferase Ktr4 in anterograde protein trafficking (10) and also plays a role in the retrograde transport of a group of escaped ER proteins as a retrograde cargo receptor (11). The recently reported crystal structure of the luminal domain of Erv41 reveals that it is mainly composed of two β-sheets forming an unusual twisted β-sandwich, which is presumed to serve as a cargo-binding site (12). The mammalian counterparts of yeast Erv41 and Erv46 are mainly distributed to the ERGIC and cis-Golgi compartments and cycle between the ER and Golgi (7, 13). Although ERGIC3 is presumed to play a role as a cargo receptor similar to the yeast ortholog Erv46, its cargo proteins have not been identified.
MARCH2 is a member of the membrane-associated RING-CH E3 ubiquitin ligase family (14) and localizes to the ER, Golgi, endosomes, lysosomes, and plasma membrane (14–16). MARCH2 is known to participate in vesicular trafficking between endosomes and the trans-Golgi network as well as the endocytic recycling pathway by binding to syntaxin 6 (15). MARCH2 substrates appear to be membrane proteins and PDZ domain–containing proteins. At its C terminus, MARCH2 has a PDZ-binding motif by which it interacts with and promotes ubiquitination of the cell polarity regulator DLG1, a PDZ domain–containing protein (17). The β-blocker carvedilol induces MARCH2-dependent ubiquitination of β2-adrenergic receptors, leading to its endocytosis and lysosomal trafficking (18). Other known substrates of MARCH2 include cystic fibrosis transmembrane conductance regulator (CFTR) (16, 19) and Dishevelled (20). Although MARCH2 functions in vesicular trafficking, it is not known whether trafficking between the ER and Golgi is regulated by the ligase activity of MARCH2.
In our efforts to identify novel substrates of MARCH2, we have applied a proximity-dependent biotin labeling method recently devised to preferentially label substrates of a specific E3 (21–23). In this study, we identified ERGIC3 as a substrate of MARCH2. We revealed that MARCH2 regulates trafficking of secretory proteins by promoting ubiquitination and subsequent degradation of ERGIC3.
Results
MARCH2 promotes ubiquitination and subsequent degradation of ERGIC3
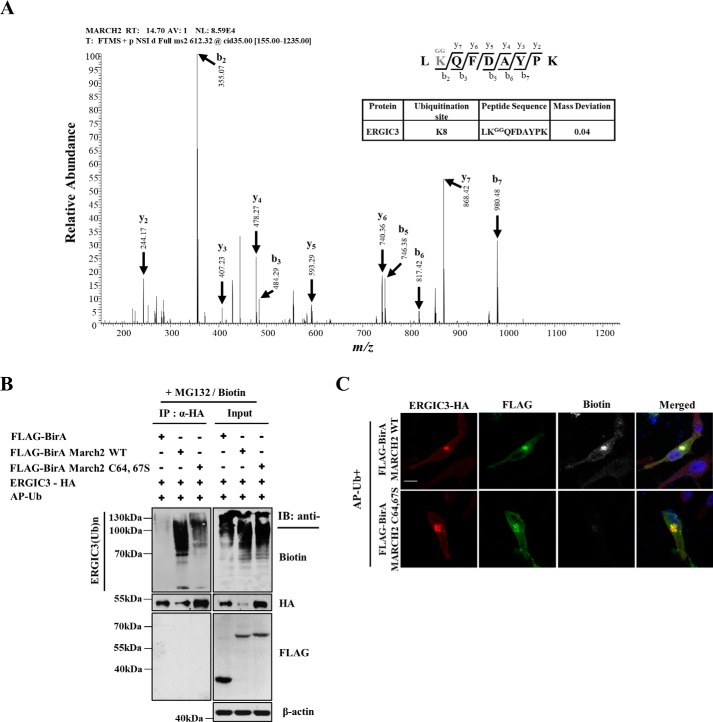
To identify potential MARCH2 substrates, FLAG-BirA-MARCH2 and AP-HA-Ub expression constructs were transfected into HeLa cells. After treating cells with biotin, biotin-labeled proteins were affinity-purified from cell extracts using streptavidin beads and digested with trypsin. Di-Gly antibody–conjugated beads were then used to enrich peptides containing diglycine-modified lysine as a signature of ubiquitination from the tryptic digest. Mass spectrometric analysis of the di-Gly peptide preparation identified a peptide derived from ERGIC3 (Fig. 1A). To directly test whether ERGIC3 is biotin-labeled by FLAG-BirA-MARCH2 and AP-Ub, cells were transfected with ERGIC3-HA, FLAG-BirA-MARCH2, and AP-Ub and biotin-labeled. Immunoprecipitation of ERGIC3-HA and subsequent immunoblotting revealed that ERGIC3 was strongly biotinylated by FLAG-BirA-MARCH2 but much more weakly biotinylated by FLAG-BirA and FLAG-BirA-MARCH2 C64,67S, a catalytically dead MARCH2 variant (Fig. 1B), an observation confirmed by immunofluorescence confocal microscopy (Fig. 1C). These data suggest that ERGIC3 is a candidate substrate of MARCH2.
Figure 1.
Identification of MARCH2 substrates by a proximity-directed biotin labeling method. A, the MARCH2-mediated ubiquitinated ERGIC3 was purified using the K-ϵ-GG affinity purification method (see “Experimental procedures”), and the peptides were analyzed by LC-MS/MS. B and C, HeLa cells were cotransfected with AP-Ub and ERGIC3-HA along with FLAG-BirA, FLAG-BirA-MARCH2 WT, or FLAG-BirA-MARCH2 C64,67S for 24 h and then treated with biotin (50 μm) and MG132 (25 μm) for 6 h. The cells were harvested, and the lysates were immunoprecipitated using anti-HA resin. Eluted samples were analyzed by Western blotting (B) and visualized using confocal microscopy (C) with the indicated antibodies. Scale bar, 20 μm. IB, immunoblotting.
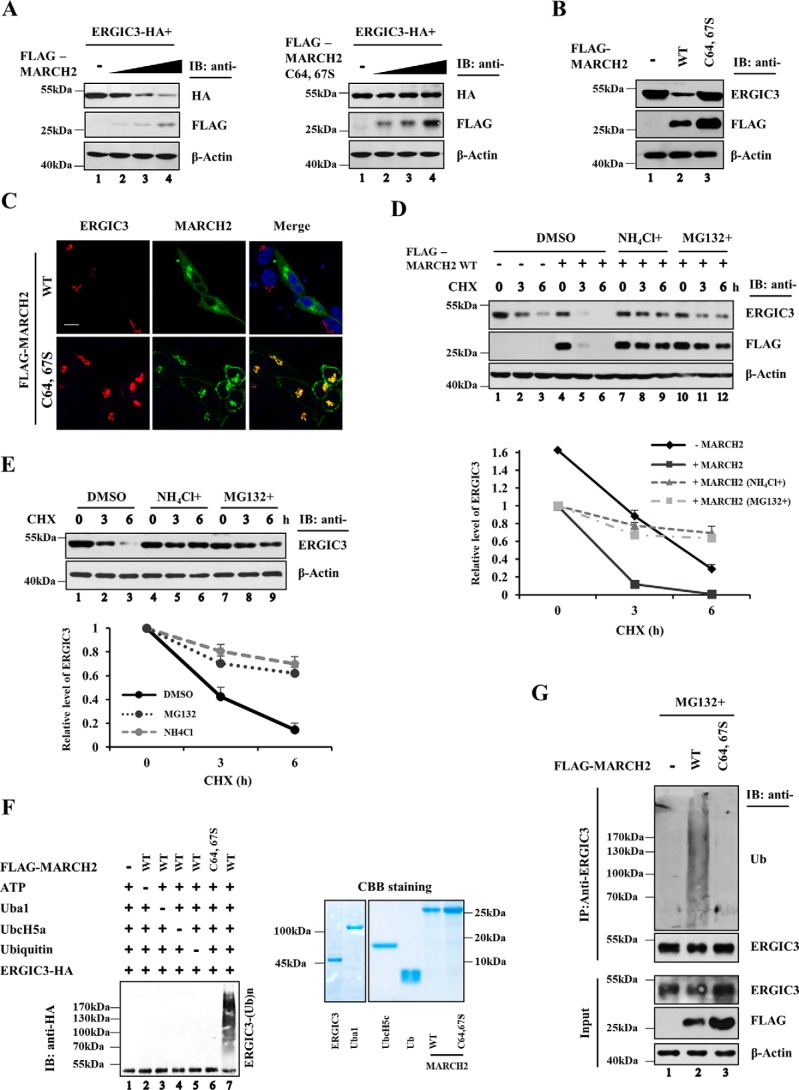
Next, we examined whether MARCH2 overexpression affects ERGIC3 expression by transfecting HeLa cells with FLAG-MARCH2 and ERGIC3-HA expression vectors. Expression of WT MARCH2, but not the ligase-dead MARCH2 C64,67S variant, reduced levels of ERGIC3-HA in a dose-dependent manner (Fig. 2A). Similarly, endogenous ERGIC3 was reduced by expression of WT MARCH2 but not by MARCH2 C64,67S variant (Fig. 2B). Immunofluorescence confocal microscopy confirmed that endogenous ERGIC3 was dramatically reduced only in cells transfected with WT MARCH2 (Fig. 2C). Compared with the WT MARCH2, a significantly greater fraction of the ligase-dead MARCH2 variant was detected in the plasma membrane (Fig. 2C). Because proteasome inhibitor MG132 treatment increases the number of cells with transfected WT MARCH2 localized to the plasma membrane (data not shown), increased expression of the mutant MARCH2 due to the defect in self-ubiquitination may cause more MARCH2 protein to be localized to the plasma membrane. The reduction of ERGIC3 by MARCH2 overexpression was slowed down by treatment with the proteasome inhibitor MG132 or with the lysosome inhibitor NH4Cl in the presence of the translation inhibitor cycloheximide (Fig. 2D). We noted that MARCH2 expression is also affected by proteasome and lysosome inhibitors (Fig. 2D), presumably due to its autoubiquitination and subsequent degradation. In addition, MG132 and NH4Cl increased the stability of endogenous ERGIC3 (Fig. 2E), suggesting that degradation of ERGIC3 is proteasome- and lysosome-dependent.
Figure 2.
MARCH2 promotes the ubiquitination and subsequent degradation of ERGIC3. A, HeLa cells were transiently transfected with plasmid constructs expressing ERGIC3-HA and an increasing amount of FLAG-MARCH2 WT or FLAG-MARCH2 C64,67S. Lysates were analyzed by Western blotting using anti-FLAG, -HA, and -β-actin antibodies. B, cells were transfected with empty vector, FLAG-MARCH2 WT, or FLAG-MARCH2 C64,67S, and then proteins were detected by Western blotting with the indicated antibodies. C, HeLa cells were transfected with FLAG-tagged MARCH2 WT or C64,67S, and cells were immunostained with an immunocytochemistry assay using the indicated antibodies and visualized by confocal microscopy. Scale bar, 20 μm. D, cells were treated with cycloheximide with or without NH4Cl (20 nm) or MG132 (25 μm) for the indicated time in the absence or presence of FLAG-MARCH2 WT. Cells were harvested, and the lysates were analyzed by Western blotting using the indicated antibodies (top). Each protein was quantified using ImageJ and is visualized in the graph (bottom). Error bars show S.D. from the average obtained from three independent experiments. E, HeLa cells were treated with cycloheximide (CHX) alone or with NH4Cl (20 nm) or MG132 (25 μm) for the indicated duration. Cells were harvested, and the lysates were analyzed by Western blotting using the indicated antibodies. The endogenous ERGIC3 expression level was quantified using ImageJ and is visualized in the graph. Error bars represent S.D. from three independent experiments. F, for the in vitro ubiquitination assay of ERGIC3, the purified ERGIC3-HA was incubated with reaction mixtures containing Uba1 (E1), UbcH5c (E2), ubiquitin, ATP, and FLAG-MARCH2 WT or FLAG-MARCH2 C64,67S for 2 h, and the ubiquitination was detected by immunoblotting (IB) with anti-HA antibody (left panel). FLAG-MARCH2 WT, FLAG-MARCH2 C64,67S, and ERGIC3-HA were purified from the transfected HeLa cells with anti-FLAG– and -HA resin–agarose beads. The purified proteins were visualized by Coomassie Brilliant Blue (CBB) staining (right panel). G, HeLa cells were transfected with empty vector, FLAG-MARCH2 WT, or FLAG-MARCH2 C64,67S. Cells were treated with MG132 (25 μm) for 6 h to stabilize ERGIC3. Cells were lysed in 1% SDS and then immunoprecipitated with anti-ERGIC3 antibody and protein G–agarose beads. The ubiquitination of endogenous ERGIC3 was determined by Western blotting using the anti-Ub antibody; other proteins were detected by Western blotting with the indicated antibodies.
To determine whether MARCH2 directs ERGIC3 degradation by promoting its ubiquitination, we performed in vitro ubiquitination assays using purified components. Robust ubiquitination of ERGIC3 was detected in the presence of ATP, Ub, E1, E2, and MARCH2 (Fig. 2F). Deletion of any component failed to support ubiquitination of ERGIC3. In addition, the MARCH2 C64,67S variant was unable to promote ubiquitination of ERGIC3. We next examined ubiquitination of endogenous ERGIC3 by MARCH2. HeLa cells were transfected with the WT or mutant MARCH2 expression construct. After incubating cells with MG132 for 6 h before harvest, ERGIC3 was immunoprecipitated with an anti-ERGIC3 antibody and tested for the conjugation of Ub by immunoblotting with an anti-Ub antibody. Expression of WT MARCH2, but not the ligase-dead MARCH2 variant, generated a ubiquitination smear of endogenous ERGIC3 (Fig. 2G). Taken together, these results indicate that MARCH2 promotes degradation of ERGIC3 by mediating its ubiquitination.
MARCH2 specifically interacts with ERGIC3
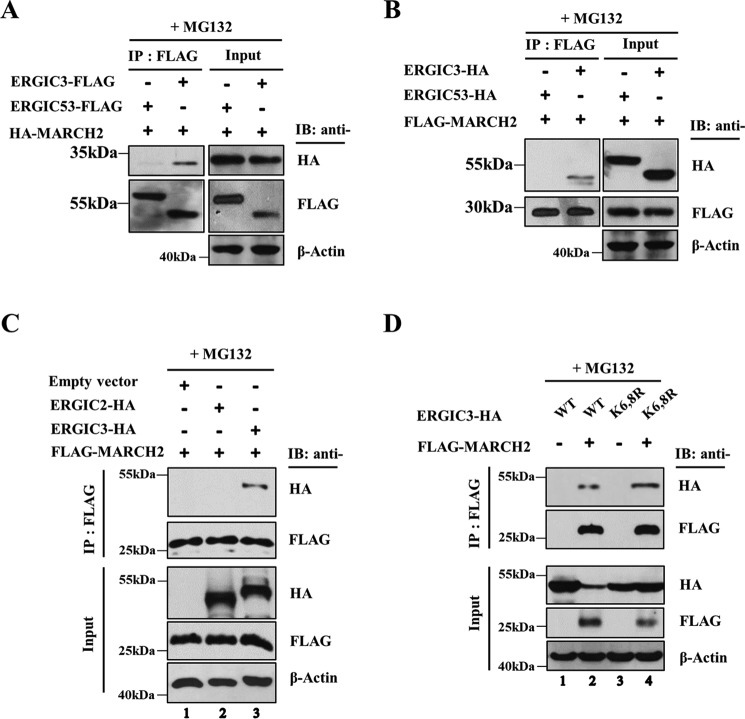
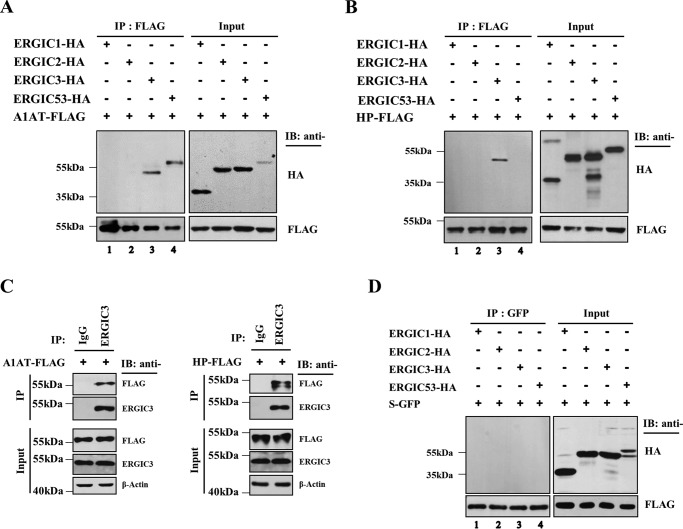
In yeast, Erv46 forms a stable complex with Erv41, and the association of the two proteins stabilizes each other (8). To investigate whether human ERGIC2 and ERGIC3 also form a complex, we analyzed the interaction between ERGIC2 and ERGIC3 by coimmunoprecipitation (co-IP). ERGIC2-FLAG interacted with ERGIC3-HA but not with ERGIC1-HA, ERGIC2-HA, or ERGIC53-HA, another cargo receptor unrelated to ERGIC1–3 (Fig. S1A). In a reciprocal co-IP assay, ERGIC3-FLAG bound to both ERGIC2-HA and ERGIC3-HA efficiently but weakly bound to ERGIC1-HA, whereas it did not interact with ERGIC53-HA (Fig. S1B). This observation suggests that ERGIC3 may form a heteromeric complex with ERGIC2 and a homomeric complex with itself, whereas ERGIC2 is unable to form a homomer.
We next investigated whether depletion of a component of the ERGIC protein complex affected the expression of the binding partner. As shown in Fig. S1C, increasing doses of ERGIC3-specific siRNA decreased not only endogenous ERGIC3 but also ERGIC2-FLAG, suggesting that ERGIC3 was required for the stable expression of ERGIC2. In contrast, whereas the treatment of ERGIC2-specific siRNA efficiently reduced ERGIC2-FLAG, it lowered endogenous ERGIC3 by about 30% and exogenously expressed ERGIC3-FLAG even less (Fig. S1D). Taken together, it seems likely that monomeric ERGIC2 is unstable unless it forms a heteromeric complex with ERGIC3, whereas ERGIC3 is able to form a heteromeric or homomeric complex, which is presumably more stable than the ERGIC3 monomer.
We next examined the interaction between MARCH2 and ERGIC3 by co-IP analysis. MARCH2 coprecipitated with ERGIC3 but not ERGIC53 (Fig. 3A). In the reciprocal experiment, ERGIC3, but not ERGIC53, coprecipitated with MARCH2 (Fig. 3B). In addition, we noted that MARCH2 bound to ERGIC3 but not ERGIC2 (Fig. 3C). Taken together, these results indicated that MARCH2 specifically recognized ERGIC3 as its substrate.
Figure 3.
MARCH2 interacts with ERGIC3. A and B, HeLa cells were transfected either with ERGIC3-FLAG, ERGIC53-FLAG, and HA-MARCH2 (A) or with ERGIC3-HA, ERGIC53-HA, and FLAG-MARCH2 (B). After 24 h, cells were treated with MG132 (25 μm) to stabilize expression of MARCH2 for 6 h. The lysates were immunoprecipitated with FLAG resin and analyzed by Western blotting with the indicated antibodies. C, cells were transfected with ERGIC2-HA or ERGIC3-HA in the presence of FLAG-MARCH2 WT and treated with MG132 (25 μm) for 6 h. The lysates were immunoprecipitated with FLAG resin. Proteins were detected by immunoblotting (IB) using the indicated antibodies. D, HeLa cells were transfected with ERGIC3 WT or ERGIC3 K6,8R in the presence or absence of FLAG-MARCH2. The immunoprecipitated samples from FLAG resin were analyzed by Western blotting with the indicated antibodies.
Endogenous ERGIC3 is increased by siRNA-mediated depletion of MARCH2
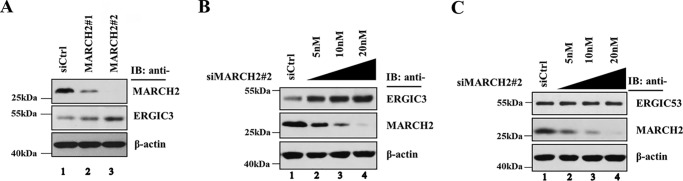
To further explore the effect of MARCH2 on ERGIC3 protein levels, we investigated whether depletion of MARCH2 by siRNA could affect levels of endogenous ERGIC3. As shown in Fig. 4, A and B, endogenous ERGIC3 was increased by siRNA-mediated depletion of MARCH2 in a dosage-dependent fashion. In contrast, MARCH2-specific siRNA had no impact on the levels of endogenous ERGIC53 (Fig. 5C), an observation that suggests that the effect of MARCH2 depletion on ERGIC3 is specific. These data indicate that endogenous MARCH2 is involved in regulating the cellular levels of ERGIC3.
Figure 4.
Endogenous ERGIC3 is increased by siRNA-mediated depletion of MARCH2. A, HeLa cells were treated with control siRNA (siCtrl), siMARCH2#1, or siMARCH2#2 for 48 h. Cells lysates were probed by Western blotting with the indicated antibodies. B and C, HeLa cells were transfected with control siRNA or an increasing amount of siMARCH2#2. Lysates were analyzed by Western blotting using the indicated antibodies. IB, immunoblotting.
Figure 5.
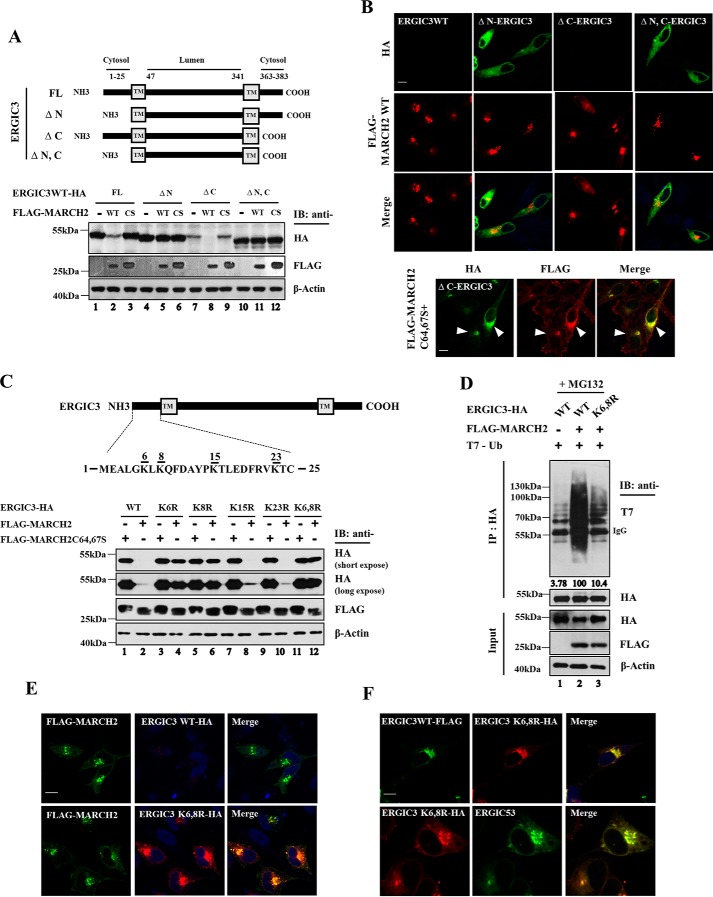
Lysines at positions 6 and 8 of ERGIC3 are required for its ubiquitination by MARCH2. A, a schematic depiction of cytosolic deletion mutants of ERGIC3 (top). Cells were cotransfected with ERGIC3WT-HA, ΔN-ERGIC3-HA, ΔC-ERGIC3-HA, or ΔN,C-ERGIC3-HA and either FLAG-MARCH2 WT or FLAG-MARCH2 C64,67S (bottom). The cell lysates were analyzed by immunoblotting (IB) with the indicated antibodies. B, HeLa cells were transiently cotransfected with ERGIC3 WT-HA, ΔN-ERGIC3-HA, ΔC-ERGIC3-HA, or ΔN,C-ERGIC3-HA along with FLAG-MARCH2 WT. In a separate experiment, cells were cotransfected with ΔC-ERGIC3-HA and FLAG-MARCH2 C64,67S. Cells were immunostained with the indicated antibodies and visualized by confocal microscopy. Colocalization of MARCH2 mutant and ΔC-ERGIC3 is indicated by arrowheads. Scale bar, 20 μm. C, schematic representation of the lysine sites of ERGIC3 (top). HeLa cells were cotransfected with ERGIC3 WT-HA, K6R-HA, K8R-HA, K15R-HA, K23R-HA, or K6,8R-HA and FLAG-MARCH2 C64,67S or WT. The immunoblotting (IB) analysis was performed to characterize the expression profile of ERGIC3 and MARCH2 using the indicated antibodies (bottom). D, HeLa cells were cotransfected with ERGIC3WT-HA or ERGIC3K6,8R-HA and empty vector or FLAG-MARCH2 WT in the presence of T7-Ub. Lysates were immunoprecipitated with anti-HA–agarose beads, and Western blot analysis was performed to determine the ubiquitination of ERGIC3 with the indicated antibodies. E and F, cells were transfected with ERGIC3WT or ERGIC3K6,8R-HA along with FLAG-MARCH2 WT (E) or transfected with ERGIC3K6,8R-HA alone or with ERGIC3WT-FLAG (F). Cells were immunostained with the indicated antibodies and visualized by confocal microscopy. ERGIC53 is a marker for the ERGIC organelle. Scale bar, 20 μm.
Lysines at positions 6 and 8 of ERGIC3 are required for its ubiquitination by MARCH2
ERGIC3 has 25- and 21-amino acid–long cytoplasmic tails at the N and C termini, respectively. To map the ubiquitination sites in ERGIC3, we generated cytoplasmic tail deletion mutants and tested them for MARCH2-mediated degradation. Although the C-terminal tail deletion mutant was decreased by WT MARCH2, mutants with the N-terminal deletion and with deletions at both termini were not affected by MARCH2 expression (Fig. 5A). We employed immunofluorescence confocal microscopy to confirm that expression of the ERGIC3 full-length protein and the C-terminal tail deletion mutant was greatly reduced by coexpressed MARCH2 (Fig. 6B). We noted that the full-length ERGIC3 and the C-terminal deletion mutant were largely colocalized with the ligase-dead MARCH2 variant (Figs. 2B and 5B), whereas mutants with the N-terminal deletion and with deletions at both termini were not colocalized with MARCH2 (Fig. 5B), indicating that the N terminus, but not the C terminus, of ERGIC3 contains information necessary for its proper subcellular localization. Because reduction in expression of the C-terminal deletion mutant by MARCH2 is similar to that of the full-length ERGIC3, these results suggest that the N-terminal cytoplasmic tail contains the major site(s) for MARCH2-directed ubiquitination, an observation that is in agreement with our MS analysis revealing that lysine at position 8 of ERGIC3 is ubiquitinated by BirA-MARCH2 (Fig. 1A).
Figure 6.
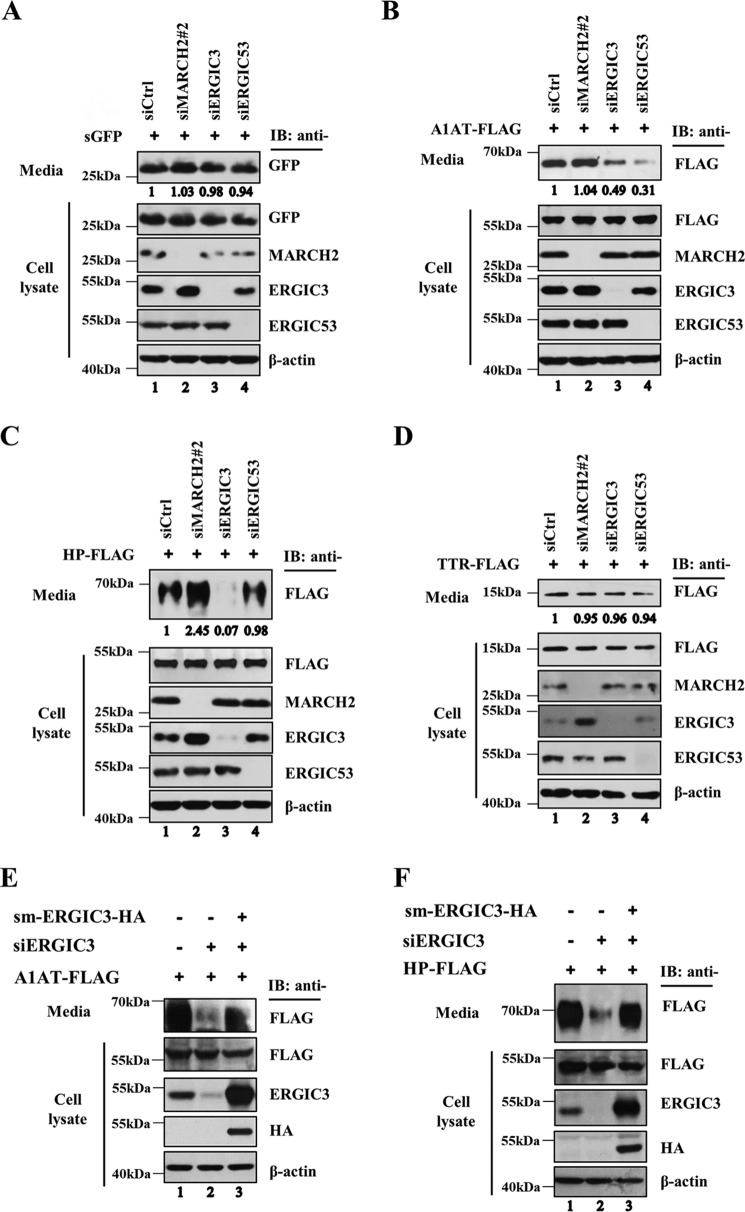
Secretion of α1-antitrypsin and haptoglobin is impaired by ERGIC3 depletion. HeLa cells were treated with negative control siRNA (siCtrl) or siRNAs against MARCH2, ERGIC3, or ERGIC53 for 48 h and then transfected with sGFP (A), α1-antitrypsin (A1AT)-FLAG (B), haptoglobin (HP)-FLAG (C), or transthyretin (TTR) for 24 h. Both the medium and lysates were analyzed by Western blotting using the indicated antibodies. E and F, HeLa cells were treated with negative control siRNA or ERGIC3 siRNA for 48 h and transfected with empty vector or siRNA-resistant mutant ERGIC3-HA (sm-ERGIC3-HA) for 24 h along with α1-antitrypsin-FLAG or haptoglobin-FLAG. Cell lysates and precipitated medium were subjected to SDS-PAGE and immunoblotted (IB) with the indicated antibodies.
The N-terminal cytoplasmic tail of ERGIC3 contains four lysine residues. We substituted these lysine residues with arginine and monitored their degradation by MARCH2. Substitution mutants K6R and K8R, but not K15R and K23R mutants, were significantly stabilized against MARCH2-mediated degradation compared with WT ERGIC3 (Fig. 5C), suggesting that Lys-6 and Lys-8 are involved in MARCH2-directed ubiquitination of ERGIC3. To generate a more stable version of ERGIC3 against MARCH2, both lysines at position 6 and 8 were substituted with arginines. As shown in Fig. 6C, expression of the mutant K6,8R was hardly affected by MARCH2. This observation was confirmed by analysis using immunofluorescence confocal microscopy (Fig. 5E). To investigate the possibility that the stability of the mutant K6,8R against MARCH2 is due to the impairment of MARCH2 binding to the mutant, we examined the interaction of MARCH2 and the double-point mutant K6,8R. Co-IP analysis revealed that MARCH2 bound to both the WT and the mutant ERGIC3 (Fig. 3D), indicating that lysine residues of ERGIC3 at positions 6 and 8 are not involved in MARCH2 interaction. In addition, immunofluorescence confocal microscopy revealed that the WT and the mutant ERGIC3 had identical distribution patterns in the cell (Fig. 5F), indicating that mutations at Lys-6 and Lys-8 of ERGIC3 did not disturb its intracellular localization. Using ubiquitination assays and subsequent quantification by scanning the blot image, we noted that the ubiquitination of the double-point mutant K6,8R by MARCH2 was reduced to about 10% of that of the WT (Fig. 5D). Taken together, these results indicate that Lys-6 and Lys-8 of ERGIC3 are required for its ubiquitination by MARCH2.
Secretion of α1-antitrypsin and haptoglobin is impaired by ERGIC3 depletion
Because the Erv41–Erv46 complex in yeast is known to function as a cargo receptor in anterograde and retrograde protein trafficking, we examined the possibility that ERGIC3 possess a similar function in human cells by monitoring protein secretion following ERGIC3 depletion. GFP, a foreign protein not normally produced in human cells, does not require a specific cargo receptor and instead uses a bulk flow mode of intracellular trafficking when expressed as a secretory protein (1). For use as a marker for protein trafficking by a bulk flow process, we generated a secretory version of GFP (sGFP) by fusing the signal peptide of α1-antitrypsin to GFP. The secretion of sGFP was monitored by analyzing the levels of sGFP in the culture medium. In accordance with the expectation that the secretion of sGFP is governed by the bulk flow mechanism, the secretion of sGFP was not affected by siRNA-mediated depletion of MARCH2, ERGIC3, or ERGIC53 (Fig. 6A). We next examined the effect of MARCH2, ERGIC3, or ERGIC53 depletion on three well-known secretory proteins (i.e. α1-antitrypsin, haptoglobin, and transthyretin). ERGIC53-specific siRNA treatment lowered secreted α1-antitrypsin (Fig. 6B, lane 4), confirming that ERGIC53 functions as a cargo receptor of α1-antitrypsin (24). Depletion of ERGIC3 by siRNA was also found to lower secreted α1-antitrypsin (Fig. 6B, lane 3), suggesting that ERGIC3 plays a similar role in secretion of α1-antitrypsin. MARCH2 depletion did not significantly affect the secretion of α1-antitrypsin, although it increased ERGIC3 (Fig. 6, A–D). Secretion of haptoglobin was decreased by depletion of ERGIC3 but not by that of ERGIC53 (Fig. 6C), revealing the specificity of cargo receptors in cargo selection. Interestingly, MARCH2 depletion, which increased ERGIC3, resulted in elevation of secreted haptoglobin (Fig. 6C). These data suggested that ERGIC3 may be a limiting factor regulating intracellular trafficking and secretion of haptoglobin. In contrast to α1-antitrypsin and haptoglobin, transthyretin secretion was not significantly affected by depletion of MARCH2, ERGIC3, or ERGIC53 (Fig. 6D), again showing the specificity in cargo selection.
Because ERGIC2 forms a complex with ERGIC3, we examined the effect of ERGIC2 knockdown on secretion of haptoglobin. As shown in Fig. S2, although ERGIC2 knockdown lowered haptoglobin secretion, the effect of ERGIC2 depletion on haptoglobin secretion was much smaller than that of ERGIC3 depletion. Because ERGIC3 is able to interact not only with ERGIC2 but with ERGIC3 itself, it is tempting to speculate that the ERGIC3 homomeric complex functions as a cargo receptor for haptoglobin trafficking as the ERGIC2–ERGIC3 complex does.
To further confirm that siRNA-mediated knockdown of ERGIC3 lowered secretion of α1-antitrypsin and haptoglobin due to the suppression of ERGIC3 expression, we complemented ERGIC3 knockdown cells with the siRNA-resistant ERGIC3 transgene with silent missense mutations. Expression of ERGIC3 almost completely restored secretion of α1-antitrypsin (Fig. 6E) and haptoglobin (Fig. 6F) in ERGIC3-knockdown cells. Taken together, these results suggest that ERGIC3 is involved in trafficking of a specific subset of secretory proteins such as α1-antitrypsin and haptoglobin.
ERGIC3 interacts with α1-antitrypsin and haptoglobin
To further explore the role of ERGIC3 in trafficking of α1-antitrypsin and haptoglobin, we investigated whether ERGIC3 could interact with α1-antitrypsin and haptoglobin. Cells were transfected with FLAG-tagged α1-antitrypsin together with ERGIC1-HA, ERGIC2-HA, ERGIC3-HA, or ERGIC53-HA. Immunoprecipitation of intracellular α1-antitrypsin-FLAG and subsequent immunoblotting using anti-HA antibodies revealed that α1-antitrypsin specifically bound to ERGIC3 but not ERGIC1 or ERGIC2 (Fig. 7A). Importantly, α1-antitrypsin also bound to ERGIC53, a known cargo receptor of α1-antitrypsin (Fig. 7A, lane 4). We carried out a similar coimmunoprecipitation analysis with haptoglobin. As shown in Fig. 7B, intracellular haptoglobin specifically interacted with ERGIC3 but not ERGIC1, ERGIC2, or ERGIC53. In addition, we found that endogenous ERGIC3 interacted with α1-antitrypsin-FLAG and haptoglobin-FLAG in co-IP analysis using anti-ERGIC3 antibody (Fig. 7C). In contrast, sGFP did not associate with ERGIC1–3 or ERGIC53 (Fig. 7D). These data strengthen our conclusion that ERGIC3 functions as a cargo receptor of α1-antitrypsin and haptoglobin.
Figure 7.
ERGIC3 interacts with α1-antitrypsin and haptoglobin. HeLa cells were transfected with ERGIC1-HA, ERGIC2-HA, ERGIC3-HA, or ERGIC53-HA along with α1-antitrypsin (A1AT)-FLAG (A) or haptoglobin (HP)-FLAG (B). The lysates were immunoprecipitated with anti-FLAG resin. Eluted samples from anti-FLAG resin were subjected to SDS-PAGE and detected by immunoblotting (IB) with the indicated antibodies. C, HeLa cells were transfected with α1-antitrypsin-FLAG or haptoglobin-FLAG for 24 h. Cell lysates were immunoprecipitated using anti-ERGIC3 antibody with protein G–agarose beads in 1% NP-40 buffer, and the precipitates were analyzed by Western blotting using the indicated antibodies. D, HeLa cells were cotransfected with sGFP (S-GFP) and ERGIC1-HA, ERGIC2-HA, ERGIC3-HA, or ERGIC53-HA. Lysates were immunoprecipitated using anti-GFP and then incubated with protein G–agarose beads. Eluted samples were analyzed by immunoblotting using the indicated antibodies.
MARCH2 overexpression leads to reduced secretion of α1-antitrypsin and haptoglobin, which is rescued by the ubiquitination-resistant version of ERGIC3
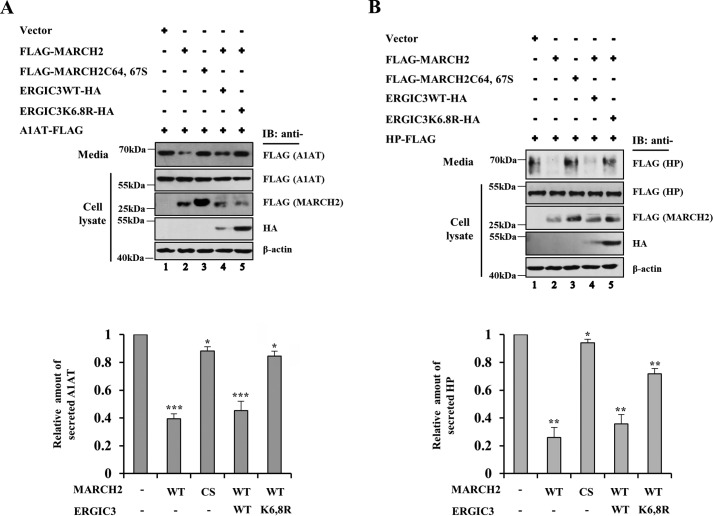
Because MARCH2 promotes ubiquitination and subsequent degradation of ERGIC3 and because ERGIC3 functions as a cargo receptor of α1-antitrypsin and haptoglobin, we examined whether MARCH2 affected secretion of α1-antitrypsin and haptoglobin. As shown in Fig. 8A, secreted α1-antitrypsin was reduced by WT MARCH2 but not by the ligase-dead MARCH2 variant, whereas levels of the intracellular α1-antitrypsin were not significantly affected. Furthermore, coexpression of the ubiquitination-resistant ERGIC3 variant, but not the WT ERGIC3, significantly restored secretion of α1-antitrypsin (Fig. 8A, lanes 4 and 5), suggesting that MARCH2-mediated ubiquitination of ERGIC3 is the major cause of the observed reduction in α1-antitrypsin secretion by MARCH2. Haptoglobin secretion was similarly affected by MARCH2 and ERGIC3 (Fig. 8B). Taken together, these results indicate that MARCH2 negatively regulates ERGIC3, which in turn reduces secretion of its cargos such as α1-antitrypsin and haptoglobin.
Figure 8.
MARCH2 expression leads to reduced secretion of α1-antitrypsin and haptoglobin, which is rescued by the ubiquitination-resistant version of ERGIC3. A and B, HeLa cells were cotransfected with the indicated constructs and α1-antitrypsin (A1AT)-FLAG (A) or haptoglobin (HP)-FLAG (B). Both the precipitated medium and lysates were analyzed by immunoblotting (IB) using the indicated antibodies. Quantification of secreted proteins (α1-antitrypsin and haptoglobin) was carried out from three-independent experiments, and the relative amount is shown as the mean ± S.D. (error bars).*, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Discussion
MARCH2 is involved in vesicular trafficking (e.g. endosome-to-Golgi network) and the endocytic recycling or degradation pathway. However, the potential involvement of MARCH2 in regulation of the early secretory pathway between the ER and Golgi compartments has not been characterized. In this study, we provide novel insight into the MARCH2-mediated regulation of protein secretion. Our data reveal that MARCH2 directs ubiquitination of ERGIC3 to modulate trafficking and secretion of ERGIC3-dependent secretory proteins.
In yeast, Erv41 and Erv46 appear to form an obligatory heteromeric complex. We have shown that ERGIC2 and ERGIC3 also interact with each other and that ERGIC3, but not ERGIC2, undergoes homomeric interaction (Fig. S1). In conjunction with interdependence of ERGIC protein stability (Fig. S1), our data support the notion that ERGIC3 forms a heteromeric complex with ERGIC2, similar to the yeast proteins Erv41 and Erv46, as well as a homomeric complex by itself.
We found that MARCH2 specifically interacts with and ubiquitinates ERGIC3 (Figs. 2 and 3), leading to its degradation. MARCH2-mediated ubiquitination of ERGIC3 was confirmed in several experimental contexts. Ubiquitination sites of ERGIC3 were identified by MS and deletion/point-mutation analyses (Figs. 1 and 5). The ubiquitination-resistant version of ERGIC3 attenuated MARCH2-dependent inhibition of ERGIC3-binding cargo protein secretion (Fig. 8). Thus, this study reveals that the early secretory pathway can be regulated by ubiquitin-dependent degradation of a cargo receptor.
Ktr4, a Golgi-resident protein, is the only known cargo of the Erv41-Erv46 complex in ER-to-Golgi anterograde transport (10); however, its cargo receptor–binding domain and sorting signal for ER exit have not been identified. With little known about cargo proteins of the ERGIC2–ERGIC3 complex in human cells, we examined hepatic secretory proteins as potential candidate cargos because secretion of α1-antitrypsin is known to require cargo receptor–mediated exit from the ER. Our data provide evidence suggesting that the ERGIC2–ERGIC3 complex and the ERGIC3 homomeric complex are likely to function as cargo receptors for α1-antitrypsin and haptoglobin. Depletion of ERGIC3 reduced secretion of α1-antitrypsin and haptoglobin (Fig. 6). ERGIC3 binds to α1-antitrypsin and haptoglobin under neutral pH conditions (Fig. 7). Depletion of ERGIC3 does not appear to cause a general secretion defect because secretion of transthyretin was not affected. As reported previously (24), ERGIC53 interacts with α1-antitrypsin, and its depletion reduced secretion of α1-antitrypsin (Figs. 6B and 7B). Thus, α1-antitrypsin appears to use at least two cargo receptors for anterograde transport from the ER. We have observed the binding of ERGIC3 to α1-antitrypsin and haptoglobin but have failed to detect their interaction with ERGIC2. Similarly, Ktr4 binds to Erv46, but not Erv41, in yeast. This result suggests that ERGIC3 (and its yeast ortholog Erv46) may be the major cargo-recognition subunit of the receptor and supports the notion that that the ERGIC3 homomeric complex may be fully functional as a cargo receptor. Structural analysis of the Erv41 luminal domain revealed that it contains an unusual β-sandwich with limited similarity to other known β-sandwich domains (12). Erv46 and ERGIC3 are expected to have a β-sandwich similar to that of Erv41; however, they contain large insertions and more cysteine residues. Further analysis of the structural differences in the cargo receptor subunits may provide clues to the cargo-binding domain and cargo selectivity.
In addition to its role in the anterograde trafficking, the Erv41–Erv46 complex is reported to function as a cargo receptor in retrieving escaped ER-resident proteins through retrograde trafficking to the ER. These ER proteins include Gls1 and Fpr2, components of folding machinery in the ER (11). Assuming that the ERGIC2–ERGIC3 complex plays a role similar to that of the yeast complex in retrograde trafficking, we cannot exclude the possibility that inefficient folding of proteins in the ER due to reduced folding machinery may contribute to the decreased secretion of α1-antitrypsin and haptoglobin by ERGIC3 depletion.
Because both MARCH2 and cargo proteins interact with ERGIC3 but not ERGIC2, a domain unique to ERGIC3 is expected to be involved in their binding. It is tempting to speculate that MARCH2 and cargo proteins compete with each other for binding to ERGIC3, thus making cargo-free ERGIC3 a preferential substrate of MARCH2.
The activity of ER in protein synthesis and export responds dynamically to various extracellular and intracellular stimuli; however, how the early secretory pathway is regulated by cargo receptors remains poorly understood. Further analyses will be required to understand the precise mechanism by which alteration in activity of cargo receptors regulates COPII-dependent protein trafficking.
Experimental procedures
Plasmids
Complementary DNAs (cDNAs) encoding human ERGIC1, ERGIC2, ERGIC3, ERGIC53, α1-antitrypsin, haptoglobin, and MARCH2 were purchased from DNASU Plasmid Repository (Tempe, AZ). Plasmids expressing epitope-tagged proteins were constructed by subcloning each PCR-amplified cDNA (primers listed in Table S1) into pcDNA3.1 (Invitrogen), pYR (25), or BirA expression vectors (21, 23). The construct for sGFP was generated by inserting α1-antitrypsin signal-peptide sequences into the pEGFP-N1 vector (Clontech).
Site-direct mutagenesis
The Lys-6 and Lys-8 residues of ERGIC3 and the Cys-64 and Cys-67 residues of the MARCH2 RING domain were altered to arginine and serine, respectively, using site-directed mutagenesis. To generate the siRNA-resistant ERGIC3 transgene, silent missense mutations were introduced by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol and the primers listed in Table S1. The accuracy of all the constructs was confirmed by sequencing.
RNAi
The siRNAs against MARCH2, ERGIC2, ERGIC3, and ERGIC53 were generated by Bioneer Korea, and transfections were accomplished using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. siRNA oligonucleotide sequences were as follow: siMARCH2#1, 5′-CCGUGCAUAAGAGCUGUCUGGAGAA-3′ and 5′-UUCUCCAGACAGCUCUUAUGCACGG-3′, sense and antisense, respectively; siMARCH2#2, 5′-1GCGACAUGGUGUGUUUCCUGUUCAU-3′ and 5′-AUGAACAGGAAACACACCAUGUCGC-3′, sense and antisense, respectively; siERGIC2, 5′-GCCAUGAAGUGUCAAUAUG-3′ and 5′-CATATTGACACTTCATGGC-3′, sense and antisense, respectively; siERGIC3, 5′-GUCCAUGACUUGCAGAGCUUUGG-3′ and 5′-CCAAAGCUCUGCAAGUCAUGGAC-3′, sense and antisense, respectively; siERGIC53, 5′-GGACAGAAUCGUAUUCAUC-3′ and 5′-GAUGAAUACGAUUCUGUCC-3′, sense and antisense, respectively.
Cell culture, transfection, and reagents
HeLa cells were cultured in DMEM (WELGENE) supplemented with 10% fetal bovine serum (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2. Cells were transiently transfected using Lipofectamine 3000, Lipofectamine RNAiMAX, or polyethylenimine (Sigma). MG132 was purchased from Cayman Chemical Co.(Ann Arbor, MI, USA), and biotin and cycloheximide were from Sigma.
Immunoprecipitation and Western blot analysis
Cells were transiently transfected with plasmids as indicated in the figure legends and lysed using RIPA buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% NP-40, 5 mm EDTA, 1% sodium deoxycholate, 0.1% SDS, 1% aprotinin, 0.1 mm Na3VO4, 50 mm NaF) 24 h post-transfection. Lysates were centrifuged at 12,000 × g at 4 °C for 15 min. The supernatants were diluted using RIPA buffer and then incubated with anti-FLAG M2–agarose (Sigma) or anti-HA resin (Sigma) overnight at 4 °C. After incubation, the samples were washed with RIPA buffer and eluted with 2× SDS sample buffer. To characterize protein expression levels, protein samples were separated by SDS-PAGE, transferred to nitrocellulose membranes (Pall Corp., Pensacola, FL), and visualized by Western blotting using the following antibodies: anti-HA from Roche Applied Science (catalog number 12CA5; 1:2,000), anti-FLAG from Sigma (catalog number F3165; 1:10,000), anti-T7 from Novagen (catalog number 69522; 1:10,000), anti-biotin from Sigma (catalog number B7653; 1:10,000), anti-ERGIC3 from Abcam (catalog number ab129179; 1:50,000), anti-MARCH2 from Abcam (catalog number ab76397; 1:1,000), anti-ERGIC53 from Santa Cruz Biotechnology (catalog number sc-365158; 1:1,000), and anti-β-actin from Sigma (catalog number A2228, 1:10,000).
Affinity purification of biotinylated proteins
To purify biotinylated proteins using BirA-MARCH2, HeLa cells were transfected with FLAG-BirA-MARCH2– and AP-HA-Ub–encoding constructs for 24 h and then treated with biotin for 4 h. The biotinylated-ubiquitinated proteins were purified as described previously (21, 23).
Protein digestion and affinity purification of ubiquitinated peptides
The biotinylated-ubiquitinated proteins immobilized on beads were reduced, alkylated, trypsinized into peptides, and purified using the ubiquitin branch motif (K-ϵ-GG) affinity purification method as described previously (21, 23).
Mass spectrometry
K-ϵ-GG affinity–purified peptides were detected and quantitated by LC-MS/MS on an LTQ Orbitrap Velos (Thermo Fisher Scientific Inc.) as described previously (21, 23).
Ubiquitination assay
HeLa cells were cotransfected with a T7-ubiquitin–encoding construct along with the expression constructs as indicated in the figure legends. After 24 h, cells were treated with MG132 (25 μm) for 6 h and then lysed using RIPA buffer. Cell lysates were boiled at 90 °C for 10 min and centrifuged at 10,000 × g for 15 min at 4 °C. Lysates were then diluted with 1% NP-40 lysis buffer (50 mm Tris-Cl, 150 mm NaCl, pH 7.4, 1% NP-40, 0.2 mm PMSF). For immunoprecipitation of HA-tagged ERGIC3, the lysates were incubated with anti-HA–agarose beads overnight at 4 °C. Samples were washed with 1% NP-40 lysis buffer three times and eluted in 2× SDS sample buffer. The samples were analyzed with Western blotting using antibodies as indicated in the figure legends.
Immunofluorescence
The immunofluorescence assay was performed as described previously (21). The following antibodies were used for proteins as indicated in the figure legends: anti-ERGIC3 (catalog number ab129179; 1:600), anti-ERGIC53 from Santa Cruz Biotechnology (catalog number sc-365158; 1:100), anti-FLAG from Sigma (catalog number F3165; 1:200), and anti-HA from Roche Applied Science (clone 12; 1:200).
In vitro ubiquitination assay
Purified ERGIC3 WT-HA was incubated with or without 100 ng of HA-Uba1, 250 ng of an E2 (UbcH5c; E2-627; Boston Biochem), 1 μg of ubiquitin (sigma), and 5 mm ATP in the absence or presence of the purified FLAG-MARCH2 WT or FLAG-MARCH2 C64,67S in reaction buffer (40 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 1 mm DTT) for 2 h. After the ubiquitination reaction, the samples were boiled in 2× SDS sample buffer and analyzed by immunoblotting with anti-HA antibody.
In vivo ubiquitination assay
To analyze the ubiquitination of endogenous ERGIC3, HeLa cells were transfected with empty vector, FLAG-MARCH2 WT, or FLAG-MARCH2 C64,67S. After 24 h, cells were treated with MG132 (25 μm) for 6 h and then lysed using RIPA buffer containing 1% (w/v) SDS and 10 mm N-ethylmaleimide. The lysates were boiled at 90 °C for 10 min, centrifuged at 10,000 × g for 15 min at 4 °C, and then diluted with 1% NP-40 lysis buffer. To immunoprecipitate the endogenous ERGIC3, cell lysates were incubated with 500 ng of anti-ERGIC3 antibody and protein G–agarose beads (Invitrogen) overnight at 4 °C. Samples were washed with 1% NP-40 lysis buffer three times, eluted in 2× SDS sample buffer, and analyzed by Western blotting using antibodies as indicated in the figure legends.
Analysis of secreted proteins
Cells were transiently transfected with the secreted protein expression plasmids. After 24 h, cells were washed twice with phosphate-buffered saline (PBS) and incubated in serum-free medium (DMEM) for 12 h. The conditioned medium was collected by centrifugation at 2,000 × g for 5 min and concentrated using trichloroacetic acid (TCA; 25%) at 4 °C for 30 min. The precipitated proteins were washed with cold acetone and dried at 50 °C for 5 min. Dried samples were dissolved in 100 μl of SDS sample buffer. Aliquots (25 μl) of protein samples were used for Western blot analysis as described above.
Statistical analysis
Statistical analysis was performed with all the data produced from three independent experiments, and data are expressed as mean ± S.D. Statistical significance was determined using Student's t test. All p values <0.05 were considered significant.
Author contributions
W. Y. data curation; W. Y. formal analysis; W. Y. and E.-B. C. investigation; W. Y. methodology; W. Y. writing-original draft; S. K. and J.-B. Y. conceptualization; S. K. and J.-B. Y. supervision; S. K. and J.-B. Y. writing-review and editing; J.-B. Y. funding acquisition.
Supplementary Material
This work was supported by the National Research Foundation of Korea (NRF) Grants NRF-2019R1A2C1010777 and NRF-2018R1A2B6003455 funded by the Korea government (Ministry of Science and ICT (MSIT)) and the Brain Korea 21 (BK21) PLUS Program (to J.-B. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD013601.
- COPII
- coat protein complex II
- MARCH2
- membrane-associated ring-CH–type finger 2
- ER
- endoplasmic reticulum
- ERGIC
- ER–Golgi intermediate compartment protein
- Erv
- ER-derived vesicle protein
- HA
- hemagglutinin
- Ub
- ubiquitin
- AP
- acceptor protein
- C64,67S
- C64S,C67S
- co-IP
- coimmunoprecipitation
- K6,8R
- K6R,K8R
- sGFP
- secretory version of GFP
- DMEM
- Dulbecco's modified Eagle's medium
- RIPA
- radioimmune precipitation assay
- NP-40
- Nonidet P-40
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Barlowe C., and Helenius A. (2016) Cargo capture and bulk flow in the early secretory pathway. Annu. Rev. Cell Dev. Biol. 32, 197–222 10.1146/annurev-cellbio-111315-125016 [DOI] [PubMed] [Google Scholar]
- 2. Zanetti G., Pahuja K. B., Studer S., Shim S., and Schekman R. (2011) COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14, 20–28 10.1038/ncb2390 [DOI] [PubMed] [Google Scholar]
- 3. Gomez-Navarro N., and Miller E. (2016) Protein sorting at the ER-Golgi interface. J. Cell Biol. 215, 769–778 10.1083/jcb.201610031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dancourt J., and Barlowe C. (2010) Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79, 777–802 10.1146/annurev-biochem-061608-091319 [DOI] [PubMed] [Google Scholar]
- 5. Geva Y., and Schuldiner M. (2014) The back and forth of cargo exit from the endoplasmic reticulum. Curr. Biol. 24, R130–R136 10.1016/j.cub.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y., Sirkis D. W., and Schekman R. (2014) Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol. 30, 169–206 10.1146/annurev-cellbio-100913-013012 [DOI] [PubMed] [Google Scholar]
- 7. Breuza L., Halbeisen R., Jenö P., Otte S., Barlowe C., Hong W., and Hauri H. P. (2004) Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279, 47242–47253 10.1074/jbc.M406644200 [DOI] [PubMed] [Google Scholar]
- 8. Otte S., Belden W. J., Heidtman M., Liu J., Jensen O. N., and Barlowe C. (2001) Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J. Cell Biol. 152, 503–518 10.1083/jcb.152.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Otte S., and Barlowe C. (2002) The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 21, 6095–6104 10.1093/emboj/cdf598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noda Y., Hara T., Ishii M., and Yoda K. (2014) Distinct adaptor proteins assist exit of Kre2-family proteins from the yeast ER. Biol. Open 3, 209–224 10.1242/bio.20146312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shibuya A., Margulis N., Christiano R., Walther T. C., and Barlowe C. (2015) The Erv41-Erv46 complex serves as a retrograde receptor to retrieve escaped ER proteins. J. Cell Biol. 208, 197–209 10.1083/jcb.201408024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biterova E. I., Svärd M., Possner D. D., and Guy J. E. (2013) The crystal structure of the lumenal domain of Erv41p, a protein involved in transport between the endoplasmic reticulum and Golgi apparatus. J. Mol. Biol. 425, 2208–2218 10.1016/j.jmb.2013.03.024 [DOI] [PubMed] [Google Scholar]
- 13. Orci L., Ravazzola M., Mack G. J., Barlowe C., and Otte S. (2003) Mammalian Erv46 localizes to the endoplasmic reticulum-Golgi intermediate compartment and to cis-Golgi cisternae. Proc. Natl. Acad. Sci. U.S.A. 100, 4586–4591 10.1073/pnas.0730885100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., and Früh K. (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78, 1109–1120 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura N., Fukuda H., Kato A., and Hirose S. (2005) MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol. Biol. Cell 16, 1696–1710 10.1091/mbc.e04-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng J., and Guggino W. (2013) Ubiquitination and degradation of CFTR by the E3 ubiquitin ligase MARCH2 through its association with adaptor proteins CAL and STX6. PLoS One 8, e68001 10.1371/journal.pone.0068001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao Z., Huett A., Kuballa P., Giallourakis C., and Xavier R. J. (2008) DLG1 is an anchor for the E3 ligase MARCH2 at sites of cell-cell contact. Cell. Signal. 20, 73–82 10.1016/j.cellsig.2007.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han S. O., Xiao K., Kim J., Wu J. H., Wisler J. W., Nakamura N., Freedman N. J., and Shenoy S. K. (2012) MARCH2 promotes endocytosis and lysosomal sorting of carvedilol-bound β2-adrenergic receptors. J. Cell Biol. 199, 817–830 10.1083/jcb.201208192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia D., Qu L., Li G., Hongdu B., Xu C., Lin X., Lou Y., He Q., Ma D., and Chen Y. (2016) MARCH2 regulates autophagy by promoting CFTR ubiquitination and degradation and PIK3CA-AKT-MTOR signaling. Autophagy 12, 1614–1630 10.1080/15548627.2016.1192752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee H., Cheong S. M., Han W., Koo Y., Jo S. B., Cho G. S., Yang J. S., Kim S., and Han J. K. (2018) Head formation requires Dishevelled degradation that is mediated by March2 in concert with Dapper1. Development 145, dev143107 10.1242/dev.143107 [DOI] [PubMed] [Google Scholar]
- 21. Deshar R., Moon S., Yoo W., Cho E. B., Yoon S. K., and Yoon J. B. (2016) RNF167 targets Arl8B for degradation to regulate lysosome positioning and endocytic trafficking. FEBS J. 283, 4583–4599 10.1111/febs.13947 [DOI] [PubMed] [Google Scholar]
- 22. Cho E. B., Yoo W., Yoon S. K., and Yoon J. B. (2018) β-Dystroglycan is regulated by a balance between WWP1-mediated degradation and protection from WWP1 by dystrophin and utrophin. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2199–2213 10.1016/j.bbadis.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 23. Deshar R., Yoo W., Cho E. B., Kim S., and Yoon J. B. (2019) RNF8 mediates NONO degradation following UV-induced DNA damage to properly terminate ATR-CHK1 checkpoint signaling. Nucleic Acids Res. 47, 762–778 10.1093/nar/gky1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nyfeler B., Reiterer V., Wendeler M. W., Stefan E., Zhang B., Michnick S. W., and Hauri H. P. (2008) Identification of ERGIC-53 as an intracellular transport receptor of α1-antitrypsin. J. Cell Biol. 180, 705–712 10.1083/jcb.200709100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Min K. W., Hwang J. W., Lee J. S., Park Y., Tamura T. A., and Yoon J. B. (2003) TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278, 15905–15910 10.1074/jbc.M213070200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.