Abstract
The Musashi family of mRNA translational regulators controls both physiological and pathological stem cell self-renewal primarily by repressing target mRNAs that promote differentiation. In response to differentiation cues, Musashi can switch from a repressor to an activator of target mRNA translation. However, the molecular events that distinguish Musashi-mediated translational activation from repression are not understood. We have previously reported that Musashi function is required for the maturation of Xenopus oocytes and specifically for translational activation of specific dormant maternal mRNAs. Here, we employed MS to identify cellular factors necessary for Musashi-dependent mRNA translational activation. We report that Musashi1 needs to associate with the embryonic poly(A)-binding protein (ePABP) or the canonical somatic cell poly(A)-binding protein PABPC1 for activation of Musashi target mRNA translation. Co-immunoprecipitation studies demonstrated an increased Musashi1 interaction with ePABP during oocyte maturation. Attenuation of endogenous ePABP activity severely compromised Musashi function, preventing downstream signaling and blocking oocyte maturation. Ectopic expression of either ePABP or PABPC1 restored Musashi-dependent mRNA translational activation and maturation of ePABP-attenuated oocytes. Consistent with these Xenopus findings, PABPC1 remained associated with Musashi under conditions of Musashi target mRNA de-repression and translation during mammalian stem cell differentiation. Because association of Musashi1 with poly(A)-binding proteins has previously been implicated only in repression of Musashi target mRNAs, our findings reveal novel context-dependent roles for the interaction of Musashi with poly(A)-binding protein family members in response to extracellular cues that control cell fate.
Keywords: mRNA, oocyte, translation control, polyadenylation, stem cells, cell plasticity, interactome analysis, Musashi, PABP, self-renewal
Introduction
The Musashi family of translational control proteins (Musashi1 and Musashi2) interacts in a sequence-specific manner with target mRNAs and has been shown to be markers of stem and progenitor cell populations in mammalian tissues, where Musashi acts to promote stem cell self-renewal and oppose cell differentiation (1–10). Recent evidence suggests that Musashi may also play a role in controlling plasticity of more differentiated cells (11, 12). Consistent with the physiological control of stem cell maintenance, Musashi1 and Musashi2 have also been implicated pathologically in the promotion of cancer stem cell self-renewal and disease progression (13–24).
The Musashi proteins were originally identified as repressors of target mRNA translation (1, 8, 25, 26). The mechanism of repression has been proposed to involve Musashi1 interaction with PABPC1, the predominant cytoplasmic poly(A)-binding protein (27). Musashi1 was shown to compete with the eIF4G translational initiation factor for the same interaction site within the first two RNA recognition motifs (RRMs)5 of PABPC1. As a consequence of this competition, it was suggested that the Musashi1–PABPC1 interaction prevented recruitment of the large 60S ribosomal subunit and subsequent cap-dependent translation of Musashi1 target mRNAs (27).
Our group was the first to report a critical role for Musashi in promoting activation, rather than repression, of target mRNA translation (28–30). During the transition of oocytes to fertilizable eggs in the frog, Xenopus laevis, gene transcription is suppressed, and all new proteins necessary for oocyte maturation are translated from pre-existing maternal mRNAs (31, 32). Translational activation of these dormant mRNAs involves release of repression and occurs in a sequential manner specified by three evolutionarily conserved mRNA sequence-specific translational regulators: Pumilio, Musashi, and the cytoplasmic polyadenylation element binding (CPEB1) (33, 34). In response to progesterone-triggered maturation of immature oocytes, a signal transduction pathway initiates de-repression of the Pumilio target mRNA, Ringo (35). Newly synthesized Ringo protein stimulates cyclin-dependent kinase (CDK) activity (36–38) that, in turn, initiates phosphorylation of Musashi and translational activation of Musashi target mRNAs, such as the mRNA encoding the MAP kinase kinase kinase, Mos (39). Translation of Mos and subsequent activation of MAP kinase signaling mediate a positive feedback amplification loop that drives robust Musashi activation, as well as activation of cytoplasmic polyadenylation element binding-dependent late class mRNA translation and the all–or–none transition to commit to complete oocyte maturation (29, 30, 34).
Musashi-dependent mRNA translational activation has subsequently been reported in multiple mammalian systems (6, 24, 40–42). The Musashi proteins are thus bifunctional and can switch from mediating repression to promoting activation of target mRNAs in response to appropriate extracellular stimuli (6, 39, 43, 44). We have demonstrated that the switch in Musashi function depends upon the phosphorylation of two conserved serine residues in the C terminus of the protein (39, 43, 44). The exact mechanism by which phosphorylation facilitates de-repression and promotes Musashi's ability to drive translation is unclear, but it may involve altered interaction with, or altered function of, critical co-factors, including PABPC1 (27), the microRNA-binding protein Lin28, which modulates let-7 miRNA biogenesis (45), or the poly(A) polymerase Germline Development 2 (GLD2) (46).
In this study, we employed MS to identify Musashi-specific interacting proteins that are required for Musashi-dependent translational control. We report the identification and characterization of several members of the poly(A)-binding protein family, namely the ePABP, which is functionally required for oocyte maturation and is the most abundant poly(A)-binding protein in immature oocytes (47, 48); PABP4, a newly identified member of the poly(A)-binding protein family that contributes uniquely to aspects of early Xenopus embryo development (49); and PABPC1, although the PABPC1 interaction was restricted to maturing oocytes. Contrary to the model in which interaction with poly(A)-binding protein mediates repression of Musashi target mRNAs, we report that interaction with either ePABP or PABPC1 is necessary to promote Musashi target mRNA translational activation. Interestingly, we observed that Musashi is also associated with PABPC1 in a mammalian cancer cell line and in human stem cells under conditions of Musashi target mRNA translational activation and inhibited stem cell self-renewal. Together, our results indicate that the context-dependent interactions of Musashi with poly(A)-binding proteins confer a differential ability to control target mRNA translation and cell fate.
Results
Musashi interactome is dynamically regulated in response to progesterone stimulation
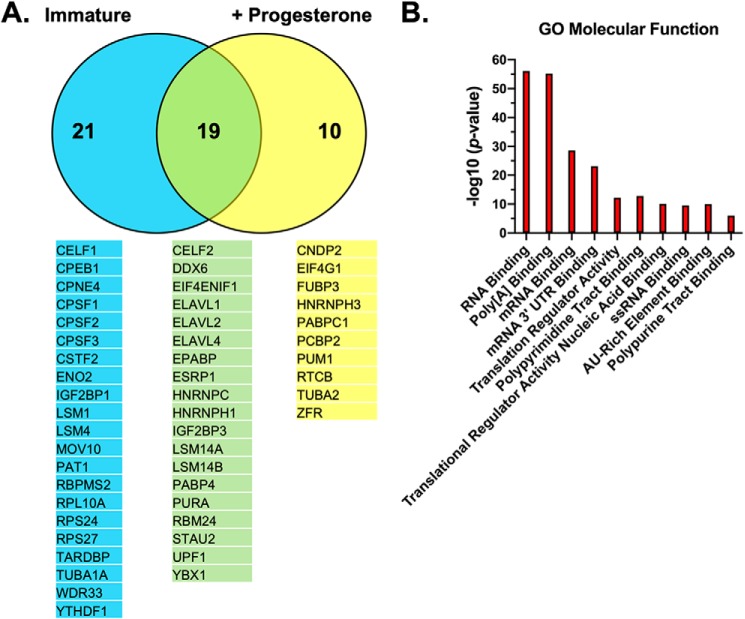
To better understand the molecular components that facilitate Musashi-dependent mRNA translational control, we sought to identify interacting partner proteins in both immature oocytes (where target mRNAs are repressed) and progesterone-stimulated oocytes (where target mRNAs are translationally activated). In three separate experiments, we microinjected immature Xenopus oocytes with RNA encoding either a GST-tagged form of Musashi1 to facilitate recovery of co-associated proteins after partial purification over GSH-Sepharose or the GST moiety alone. Oocytes were incubated overnight to allow expression of the ectopic proteins, and then the GST–Musashi1-injected oocytes or control GST-injected oocytes were each split into two pools, one of which was left untreated (immature) and the other stimulated with progesterone. Following LC-coupled tandem MS, the spectral counts from identified proteins were normalized for protein size, giving us a normalized spectral abundance factor (50), and transformed to log2 with which to compare the relative abundance of each identified protein interacting with either GST–Musashi1 or GST (Table S1). Fifty proteins specifically interacted with the GST–Musashi1 protein but not the GST moiety alone (Fig. 1A). Of these, 40 were found to associate with Musashi1 in immature oocytes (Table S2), and 29 were associated with Musashi1 in progesterone-stimulated oocytes (Table S3). A comparison of the two lists revealed that 19 proteins remain associated with Musashi1 under either experimental condition (and represent a “core” interactome); 21 proteins preferentially associate with Musashi1 only in immature oocytes, and 10 preferentially associate only in maturing oocytes (Table S4). These observations suggest that the Musashi1–mRNA ribonucleoprotein (mRNP) complexes undergo dynamic remodeling in response to progesterone stimulation when Musashi target mRNAs are de-repressed and transition to a state of translational activation. Several known Musashi1 interacting proteins were detected in our analyses, including the cytoplasmic polyadenylation element-binding protein (CPEB1) (51) in immature oocytes and PABPC1 (27) in maturing oocytes. The recovery of PABPC1 only in maturing oocytes likely reflects its reciprocal expression to ePABP, with PABPC1 being present at only low levels in immature oocytes and gradually increasing during maturation and early embryogenesis (48). Interestingly, we detected Musashi1 association with two additional members of the poly(A)-binding protein family (Fig. 1A, the embryonic poly(A)-binding protein ePABP (also called PABPC1L) and PABP4 (also called inducible PABP or PABPC4)) in both immature and maturing oocytes. Gene set enrichment analysis revealed that the 50 identified co-associated proteins are primarily involved in RNA binding and translational regulation (Fig. 1B).
Figure 1.
Mass spectrometry identification of the Musashi1 interactome. A, oocytes were injected with mRNA encoding GST–XMsi1 or the GST moiety alone. The injected oocytes were incubated overnight to express the introduced proteins. Following incubation, one-half of the GST and one-half of the GST–XMsi1-injected oocytes were stimulated to mature with progesterone, and the rest were left untreated (Immature). When 50% of oocytes have reached GVBD, protein lysates were prepared, partially purified over GSH-Sepharose, and subjected to MS. The experiment was repeated on three separate occasions. A total of 50 proteins were identified that specifically interacted with Musashi1. Of these, 21 only interacted significantly with Musashi1 in immature oocytes, 10 only interacted significantly with Musashi1 in progesterone-stimulated oocytes, whereas 19 were common to both conditions. The identified proteins for each category are indicated. B, gene set enrichment analysis was performed on the 50 interacting proteins, and the top 10 significant gene ontology molecular functions are shown graphically.
Musashi associates with ePABP in immature and maturing oocytes
The persistence of ePABP and PABP4 interactions in Musashi mRNP complexes in maturing oocytes and indeed the progesterone-dependent recruitment of PABPC1 were unexpected because Musashi target mRNAs are translationally activated, not repressed, in maturing oocytes. Here, we have sought to determine the role and contribution of ePABP and PABPC1 to Musashi-dependent mRNA translational activation and oocyte maturation.
To confirm the interaction of ePABP with Musashi1 seen by MS, immature oocytes were injected with RNA encoding GST-tagged Xenopus Musashi1 or the GST moiety alone, and the ability of Musashi1 to associate specifically with endogenous ePABP was determined after partial purification over GSH-Sepharose and Western blotting. To ensure that any associations observed were due to protein–protein interactions rather than just co-occupancy of the same mRNA, all GSH pulldown experiments included an RNase1 treatment step. Endogenous ePABP was found to co-associate in an RNA-independent manner with Musashi1 in both immature and progesterone-treated oocytes (Fig. 2A). When compared with levels of ePABP in the input lysates, less than 2% of the cellular ePABP was found in association with Musashi1. No co-association was observed with the GST moiety alone, indicating a specific interaction between Musashi1 and ePABP, verifying our MS findings. Quantitation of ePABP recovery (normalized for the amount of GST–Musashi1 in each pulldown) revealed a 2-fold increase in ePABP association with Musashi1 after progesterone stimulation (Fig. 2B). This increase was observed in progesterone-stimulated oocytes that had not yet completed germinal vesicle (nuclear) breakdown (GVBD(−)), as well as oocytes that had completed GVBD(+). We conclude that increased association of ePABP with Musashi1 is an early event during progesterone-stimulated oocyte maturation, occurring prior to GVBD.
Figure 2.
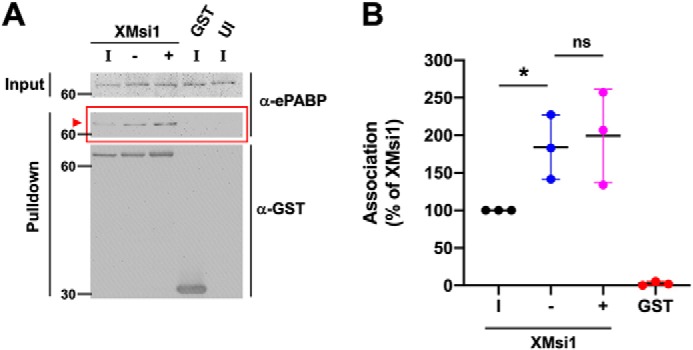
RNA-independent association of ePABP with Musashi1 increases during progesterone-stimulated oocyte maturation. A, oocytes were injected with mRNA encoding GST–XMsi1 or the GST moiety alone. The injected oocytes were incubated overnight to express the introduced proteins. Following incubation, two-thirds of GST–XMsi1-injected oocytes were stimulated to mature with progesterone, and the rest were left untreated (immature). When 50% of the oocytes reached GVBD, lysate was prepared from segregated oocytes that had not (−) or had (+) completed progesterone-stimulated GVBD as well as time-matched immature oocytes (I). Lysates were then subjected to GST-pulldown and treatment with RNase I. Protein co-association was visualized by Western blotting with ePABP (α-ePABP) and GST (α-GST) antibodies. GST–XMsi1 associates with ePABP in an RNA-independent manner (arrowhead). The positions of molecular weight markers are indicated to the left of each panel. B, composite results of three independent experiments reveals increased association of ePABP with Musashi1 following progesterone stimulation. The data were normalized to the levels of ePABP association with GST–XMsi1 in immature oocytes (100% of XMsi1) and for recovered GST-fusion protein levels in each sample. * indicates p < 0.05; ns indicates not significant.
Knockdown of ePABP attenuates Musashi-dependent MAP kinase signaling and blocks oocyte maturation
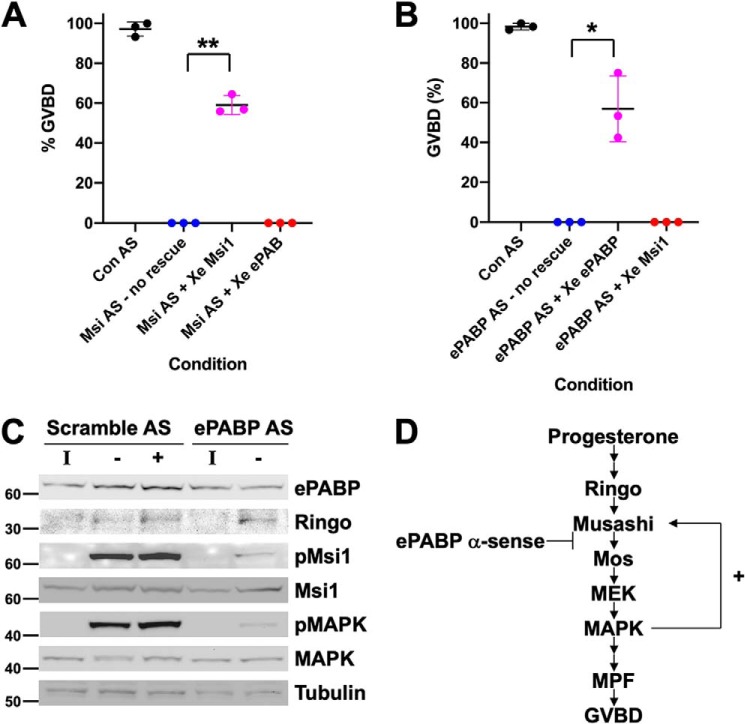
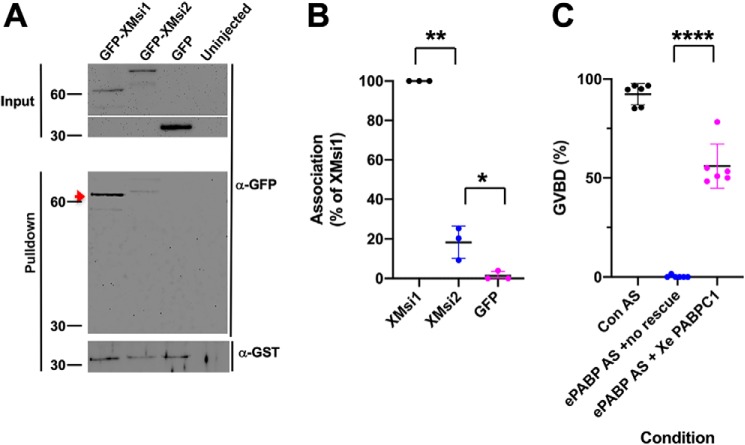
Although it has been proposed that Musashi interaction with PABPC1 correlated with target mRNA repression in a mammalian cell line (27), Musashi has a unique role during the maturation of Xenopus oocytes where it mediates the translational activation of early class mRNAs prior to oocyte GVBD (29, 30, 44, 52, 53). We therefore hypothesized that the interaction of ePABP with Musashi1 may be necessary to promote Musashi target mRNA translation during oocyte maturation. To directly assess the functional role and interdependence of the Musashi1–ePABP interaction, we first injected immature oocytes with Musashi1 and Musashi2 antisense oligonucleotides (Msi AS), which abolish progesterone-dependent maturation (29, 39, 44), and then sought to rescue the deficit with either ectopic Musashi1 or ePABP expression. Although ectopic Musashi1 was able to efficiently rescue maturation, ePABP expression was unable to effect any rescue, even at later time points (Fig. 3A). In a reciprocal experiment, immature oocytes were injected with antisense oligonucleotides that block ePABP translation (49), and the consequences on progesterone-stimulated maturation were assessed. Microinjection of the ePABP antisense oligonucleotides ablated progesterone-stimulated maturation (Fig. 3B), a phenotype indistinguishable from Musashi antisense oligonucleotide injection (Fig. 3A). The block to maturation appeared to be specific, as it could be efficiently rescued by injection of ePABP mRNA containing nucleotide wobble (49) to prevent antisense targeting but maintain correct ePABP amino acid sequence integrity (Fig. 3B) with >90% rescue observed at later time points. By contrast, ectopic expression of Musashi1 was unable to effect any rescue of ePABP AS–injected oocytes. Taken together, these results suggest an interdependence of Musashi1 and ePABP for progesterone-stimulated oocyte maturation.
Figure 3.
Knockdown of ePABP blocks progesterone-stimulated signaling pathways downstream of Musashi that lead to robust MAP kinase activation and oocyte maturation. A, oocytes were injected with antisense oligonucleotides targeting Musashi1/2 (Msi AS) or a scrambled control oligonucleotide (Con AS). Following overnight incubation, the Msi AS-injected oocytes were re-injected with water (no rescue), RNA-encoding GST–XMsi1, or RNA-encoding wobble ePABP and then stimulated to mature with progesterone. Maturation was scored when ∼50% of GST–XMsi1-injected oocytes had reached GVBD. The combined data of three independent experiments is shown. ** indicates p < 0.01. B, oocytes were injected with antisense oligonucleotides targeting ePABP (ePABP AS) or a scrambled control oligonucleotide (Con AS). Following overnight incubation, the ePABP AS-injected oocytes were re-injected with water (no rescue), RNA-encoding wobble ePABP (which is not targeted by ePABP AS), or RNA-encoding GST–XMsi1 and then stimulated to mature with progesterone. Maturation was scored when ∼50% of wobble ePABP-injected oocytes had reached GVBD. The combined data of three independent experiments is shown. * indicates p < 0.05. C, oocytes were co-injected with RNA-encoding GST–Musashi1 (to facilitate analysis of Musashi1 activation status) and either scramble or ePABP antisense oligonucleotides (AS) as indicated, and Western blotting analyses of the indicated components of the oocyte signaling cascade were performed from untreated immature (I) or progesterone-stimulated oocytes. For scramble-injected oocytes, oocytes were segregated at GVBD50 based on whether they had not (−) or had (+) completed GVBD. ePABP antisense-injected oocytes did not mature in response to progesterone and so were harvested when scramble-injected oocytes reached GVBD50. Endogenous ePABP, Ringo, MAP kinase (MAPK), and tubulin (an internal loading control) were analyzed as indicated. The activation status of endogenous MAP kinase was assessed with a phosphorylation (activation)-specific antibody (pMAPK). Ectopically expressed, GST-tagged Musashi1 was detected with GST antibodies (Msi1), and phospho-specific Musashi1 antibodies were used to assess Musashi1 activation status (pMsi1). The positions of molecular weight markers are indicated to the left of each panel. D, schematic showing the early signaling pathway and positive feedback amplification loop leading to full Musashi activation and oocyte maturation GVBD. The observed ePABP antisense effects on progesterone signaling (C) are consistent with a block coincident with Musashi function and attenuated translation of the Musashi target mRNA, Mos (which encodes the primary activator of MAPK signaling in the oocyte, see “Discussion”).
Using protein lysates prepared from oocytes shown in Fig. 3B, the disruption of the oocyte maturation signaling cascade following ePABP antisense injection was analyzed by Western blotting (Fig. 3C). While having little effect on the basal levels of ePABP in immature oocytes, ePABP antisense oligonucleotides completely prevented the progesterone-stimulated increase in ePABP levels in maturing oocytes (Fig. 3C, upper panel). As a control for specificity, scrambled antisense oligonucleotides did not prevent the ePABP increase after progesterone stimulation. Ringo, an atypical CDK partner that is required for initial Musashi1 phosphorylation and activation (39), was translated normally despite ePABP antisense injection (Fig. 3C, Ringo). Consistent with Ringo protein translation and CDK activation, a low level of initial Musashi1-activating phosphorylation was observed in ePABP antisense-injected oocytes (Fig. 3C, pMsi1). However, the robust phosphorylation of Musashi1 seen in control (scrambled antisense-injected) oocytes was blocked by ePABP antisense injection. Furthermore, downstream MAP kinase activation (Fig. 3C, pMAPK) was severely attenuated after ePABP antisense injection, suggesting a block of early class mos mRNA translation, the primary activator of MAP kinase signaling in the oocyte and a known Musashi1 target mRNA. We thus position the effect of the ePABP antisense oligonucleotide block as acting immediately downstream of initial Musashi1 activation (Fig. 3D).
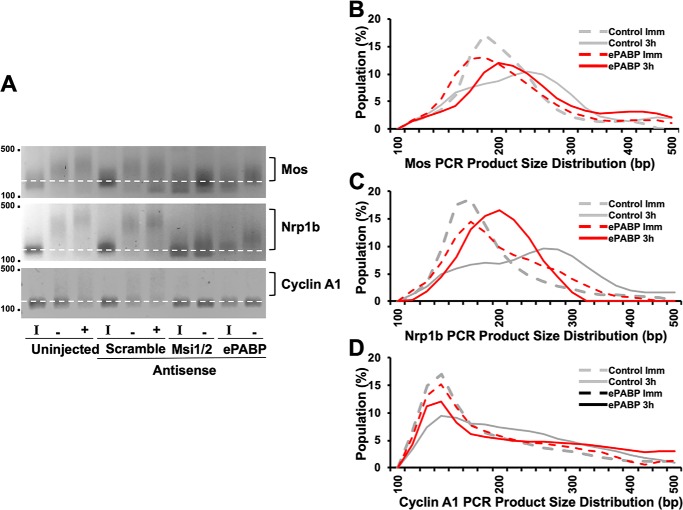
The observed signaling defects in ePABP antisense-injected oocytes suggested that ePABP is required during Musashi-directed early class mRNA translation, particularly for the early mos mRNA. To test this hypothesis directly, we analyzed the polyadenylation status of the Mos and Nrp1b (which encodes Xenopus Musashi1) mRNAs, two established Musashi targets (28, 30, 53). Treatment with ePABP antisense attenuated progesterone-stimulated polyadenylation of both mRNAs compared with control oocytes (Fig. 4, A–C, indicated by reduced PCR product size). This finding indicated that ePABP plays a role in polyadenylation of Musashi-dependent mRNAs, possibly through protection of the newly added poly(A) tail or modulation of the rate of poly(A) addition. We note that the extent of inhibition of polyadenylation was not as great as that seen in Musashi antisense oligonucleotide–injected oocytes. We have previously demonstrated that late class CPE-dependent mRNA translational activation occurs at GVBD and requires prior activation of Musashi-dependent mRNA translation (29, 30, 52, 53). The fact that we do not see CPE-dependent cyclin A1 polyadenylation in either Musashi or ePABP antisense-injected oocytes (Fig. 4, A and D) can thus be explained by the oocytes failing to properly engage the early class (Musashi-dependent) mRNA activation that is required to allow subsequent activation of late class mRNAs and progression to GVBD (54, 55). Taken together, our results indicate that ePABP is necessary for robust Musashi-dependent mRNA polyadenylation and translational activation.
Figure 4.
Antisense oligonucleotides targeting ePABP attenuate progesterone-stimulated polyadenylation of Musashi target mRNAs. A, representative experiment analyzing the poly(A) length assay of the Musashi target mRNAs, Mos and Nrp1b (Musashi1). A retardation in migration rate of the PCR products is indicative of poly(A) tail polyadenylation and elongation (52, 53). Polyadenylation (square brackets) of early class Mos and Nrp1b mRNAs, as well as the late class cyclin A1 mRNA, is seen in both uninjected and scrambled control oligonucleotide-injected oocytes treated with progesterone. Oocytes injected with ePABP antisense oligonucleotides show attenuated polyadenylation of the Mos and Nrp1b mRNA populations (indicated by a smaller mode value of PCR product lengths). By contrast, a full block of Mos and Nrp1b polyadenylation is seen in the Msi1/2 antisense oligonucleotide-injected oocytes. The progesterone-stimulated polyadenylation of the late class cyclin A1 mRNA is inhibited in both ePABP and Msi1/2 antisense-treated oocytes, consistent with the observed block to oocyte maturation. I indicates immature oocytes; − indicates progesterone-stimulated oocytes harvested prior to GVBD; + indicates progesterone-stimulated oocytes that had completed GVBD. Because neither Musashi nor ePABP antisense-injected oocytes completed GVBD, samples were harvested when control, scramble antisense-injected oocytes reached GVBD50. The position of DNA size markers are shown to the left of the panels. B–D, graphic representation of the extent of progesterone-stimulated polyadenylation seen in A at the 3-h pre-GVBD time point (−) for Scramble (control) or ePABP AS oocytes. The dashed lines represent the distribution of PCR product size in immature oocytes (Imm); solid lines are the distribution of PCR product size in progesterone-stimulated oocytes prior to GVBD. The gray lines are scrambled, control oligonucleotide-injected oocytes. Red lines represent ePABP antisense-injected oocytes. The peaks indicate the mode of the population of mRNA lengths. The shift of the peak between immature (dashed line) and progesterone-stimulated oocytes (solid line) to a larger size is indicative of mRNA polyadenylation. B, Mos mRNA polyadenylation; C, Nrp1b mRNA polyadenylation; and D, cyclin A1 mRNA polyadenylation.
Deletion mapping reveals two distinct interaction sites for ePABP within the Musashi1 protein
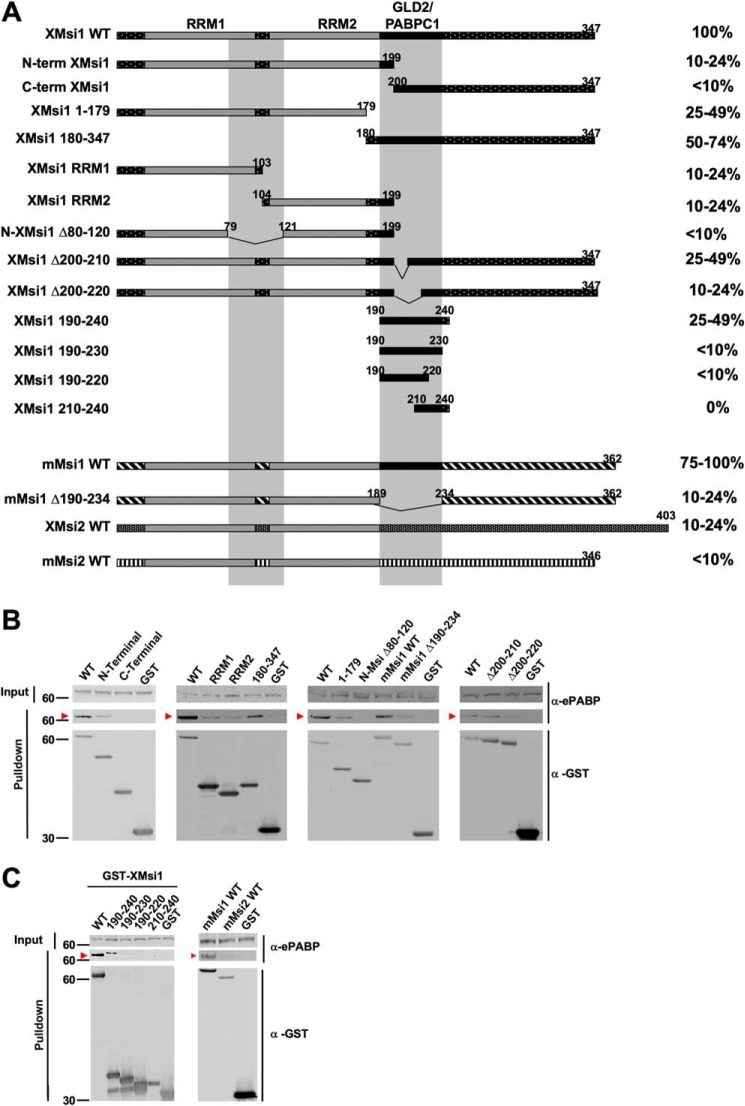
Having established a coincident requirement for both ePABP and Musashi for early mRNA polyadenylation and translational activation, we next sought to map the domain(s) within Musashi1 necessary for ePABP interaction. A series of Xenopus Musashi1 (XMsi1) deletion mutant constructs was generated and tested for interaction with endogenous ePABP (Fig. 5A). Two interaction domains were identified in Musashi1. The first encompasses amino acids 190–240, the previously characterized domain necessary for PABPC1 interaction (27). As can be seen in Fig. 5C (left panel), expression of this domain alone (XMsi 190–240) was sufficient to retain significant interaction with endogenous ePABP, although further truncations within this region either dramatically attenuated ePABP interaction (XMsi1 190–230 and 190–220) or ablated interaction completely (XMsi1 210–240). A second, weaker binding site was identified within the N terminus of Musashi1 spanning the end of the first RNA recognition motif (RRM1) and the start of RRM2 (amino acids 80–120). A comparison of these two interaction domains within Musashi1 revealed no obvious linear amino acid sequence homology.
Figure 5.
Deletion mapping reveals two ePABP interaction domains within amino acids 80–120 and 190–234 of the Musashi1 protein. A, oocytes were injected with mRNA encoding GST-tagged WT or mutant Musashi constructs as indicated, and ePABP interaction was assessed after GST-pulldown and RNase I treatment. For all mapping experiments, injection of the GST moiety alone served as a negative control. Horizontal bars schematically represent the Musashi constructs, and the relative level of ePABP interaction is indicated as a percentage of that observed with WT Xenopus Musashi1 as quantified by densitometry and normalized for the amount of recovered GST fusion protein in each sample. The gray vertical bars mark amino acids 80–120 and 190–234, the deduced ePABP interaction domains. The previously characterized Musashi RNA recognition motifs 1 (RRM1) and 2 (RRM2), as well as GLD2 and PABPC1 interaction domains (GLD2/PABPC1) are indicated. B and C, representative experiments from A showing ePABP co-association (arrowhead) with the indicated Musashi constructs. The positions of molecular weight markers are indicated to the left of each panel.
We observed that while murine Musashi1 was also able to associate with ePABP to a similar degree as observed with Xenopus Musashi1 (Fig. 5B, mMsi1 WT), both the Xenopus Musashi2 (Fig. 7, A and B) and murine Musashi2 proteins (Fig. 5C) exhibited a more than a 4-fold reduced interaction with ePABP. The N-terminal 80–120-amino acid domain is well-conserved between Xenopus Musashi1 and Musashi2 (95% amino acid identity), between Xenopus Musashi1 and murine Musashi1 (100% identity), and between Xenopus Musashi2 and murine Musashi2 (97.5% identity). However, there is considerably more divergence in the C-terminal 190–240-amino acid domain between Xenopus Musashi1 and Xenopus Musashi2 (64% identity) as well as between murine Musashi1 and Musashi2 (64% identity). We propose that the reduced interaction of ePABP with Musashi2 when compared with Musashi1 is primarily due to isoform-specific sequence divergence within the C-terminal 190–240 interaction domain.
Figure 7.

Differential association with ePABP correlates with functional differences between Musashi1 and Musashi2. A, representative GST pulldown experiment to assess association between either GST-tagged Xenopus Musashi1 or Musashi2 and endogenous ePABP. Musashi2 is severely compromised for ePABP binding. Uninjected oocytes (UI) or oocytes injected with RNA encoding the GST moiety alone serve as specificity controls. The positions of molecular weight markers are indicated to the left of each panel. B, graphic summary of three independent experiments assessing association between Musashi1or Musashi2 and ePABP. Association data are normalized to the levels of recovered GST-fusion protein in each sample. Musashi2 is severely compromised for ePABP binding. ** indicates p < 0.01. C, oocytes were injected with antisense oligonucleotides targeting Musashi1 and Musashi2. Following overnight incubation, progesterone-stimulated maturation was measured after injection with mRNA encoding either Xenopus Musashi1 (XMsi1), Musashi2 (XMsi2), a chimeric Musashi2 protein that has the Musashi1 ePABP-binding domain substituted for the Musashi2 ePABP binding domain (Chimera) or water-injected control (No rescue). A summary of three independent experiments is shown graphically. Although Musashi2 is rate-compromised for mediating oocyte maturation relative to similar levels of Musashi1, the chimeric Musahsi2 protein functions as efficiently as Musashi1. * indicates p < 0.05; ** indicates p < 0.01. ns indicates not significant. D, relative protein expression levels of the injected rescue constructs are shown for a representative experiment from C. The positions of molecular weight markers are indicated to the left of each panel.
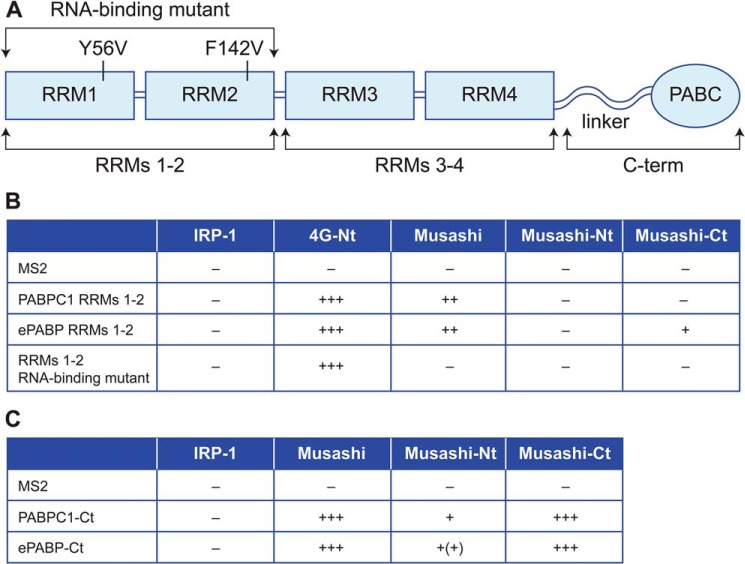
In a reciprocal set of experiments, we sought to map the Musashi1-binding sites within ePABP using yeast two-hybrid analyses. Vertebrate PABPs consist of four RRMs and a C-terminal region containing a variable linker region and a highly-conserved PABC domain (Fig. 6A). As mammalian Musashi1 was reported to repress translation by binding to PABPC1 RRMs 1–2 and disrupting the interaction of PABPC1 with eIF4G (27), we initially focused on this domain. Two RNA-binding proteins, MS2 coat protein (MS2) and iron-regulatory protein (IRP)-1, were utilized as negative controls for specificity, and eIF4G served as a positive control. Surprisingly, neither the N- or C-terminal fragments of Musashi1 interacted with RRMs 1–2 of PABPC1 or ePABP, and although full-length Musashi1 appeared to interact, this was RNA-dependent as point mutations in the RNP motifs of PABPC1 RRMs 1–2 that disrupt RNA binding abrogated this interaction (Fig. 6B). In contrast, the interaction between eIF4G and the RNA-binding mutant form of PABPC1 RRMs 1–2 was maintained (Fig. 6B). PABPC1 or ePABP RRMs 3–4 also failed to interact with either the N- or C-terminal regions of Musashi1 (data not shown). The PABP C-terminal region does not bind RNA but has been characterized as a protein interaction domain. We observed that both full-length Musashi1 and the C-terminal fragment of Musashi1 showed robust interaction with the C terminus of both ePABP and PABPC1, with weaker interactions being observed with the N-terminal region of Musashi1 (Fig. 6C). These yeast two-hybrid mapping experiments are in general agreement with the GST pulldown analyses (Fig. 5, A–C). We conclude that the C-terminal domains of ePABP and PABP1 interact directly with the 190–240 domain (and to a lesser extent the 80–120 domain) of Musashi1.
Figure 6.
Yeast two-hybrid analyses reveal a direct interaction between Musashi1 and the C terminus of ePABP and PABPC1. A, schematic representation of the structure of vertebrate PABPs and the specific constructs used for the interaction analyses. B, yeast two-hybrid analysis using transcription activator domain fusions with full-length Musashi, or Musashi N terminus (−Nt, amino acids 1–198), or C terminus (−Ct, amino acids 199–347) with DNA-binding domain fusions of X. laevis PABP1 and ePABP RRMs 1–2 or an RNA-binding defective version of PABPC1 RRMs 1–2. C, yeast two-hybrid analysis using DNA-binding domain fusions of PABP1 and ePABP C-terminal regions (−Ct) and the indicated Musashi transcription activator domain fusion constructs. IRP-1 and eIF4G N terminus (4G-Nt) represent negative and positive controls. The relative strength of tested interactions (assessed by colony growth and LexA-dependent β-gal expression) are represented by + symbols. The data in B and C are derived from three independent experiments.
Differential ePABP interaction correlates with functional differences between Musashi1 and Musashi2
Our interaction mapping of ePABP-binding sites within Musashi1 and Musashi2 revealed that ePABP binding to Musashi2 was significantly reduced compared with Musashi1 (Figs. 5 and 7, A and B). To assess whether the differential interaction with ePABP reflected any functional differences between Musashi1 and Musashi2, we assayed the ability of Musashi1 and Musashi2 to rescue oocytes that had been previously injected with Musashi antisense oligonucleotides. Given sufficient time, Musashi2 was eventually able to rescue maturation to the same extent as Musashi1. However, when assessed at the time when Musashi1 had effected maturation of 50% of the oocytes, Musashi2 was compromised in rescue ability (Fig. 7C). This rescue deficit was a consequence of the reduced rate at which Musashi2 was able to mediate progesterone-stimulated maturation because Musashi2 was expressed to similar levels as the Musashi1 protein (Fig. 7D). We reasoned that the stronger interaction of Musashi1 with ePABP was necessary for the rapid endogenous mRNA translational activation and consequently determined the rate of maturation. In theory, an enhancement of ePABP interaction with Musashi2 would allow Musashi2 to function at the same rate as Musashi1 in the antisense rescue assay. To test this idea, we generated a chimeric Musashi2 protein where amino acids 190–240 of Musashi2 were substituted with amino acids 190–240 from Musashi1, creating a full-length Musashi2 protein encoding the Musashi1 ePABP interaction domain. The resulting chimeric Musashi2 protein showed a robust gain of function in our antisense rescue assay, and it was functionally indistinguishable from the WT Musashi1 protein (Fig. 7C, Chimera). We propose that the relative ePABP interaction strength may contribute to the difference between the functional capacities of Musashi1 and Musashi2 protein isoforms to exert timely translational activation during Xenopus oocyte maturation.
Our MS also detected interaction of PABPC1 with Musashi1, although this was restricted to maturing oocytes (Fig. 1A). We thus sought to confirm this interaction and determine whether PABPC1 also interacted differentially with Musashi1 and Musashi2. We co-expressed GST-tagged Xenopus PABPC1 with either GFP-tagged Musashi1 or Musashi2 and assessed their ability to co-associate in an RNA-independent manner. Consistent with the ePABP interaction results, PABPC1 was found to preferentially interact with Musashi1, and to a lesser extent with Musashi2 (Fig. 8, A and B). We next determined whether the PABPC1 isoform could function to promote progesterone-dependent Musashi target mRNA translation and maturation of oocytes. We expressed GST-tagged Xenopus PABPC1 in ePABP antisense oligonucleotide-injected oocytes and assessed the ability of PABPC1 to exert rescue of progesterone-dependent maturation. Importantly, PABPC1 was able to efficiently rescue progesterone-stimulated oocyte maturation (Fig. 8C) with >90% rescue observed at later time points. At least in the context of the maturing oocyte, PABPC1 and ePABP appear to be functionally redundant as ectopic PABPC1 rescued ePABP antisense-treated oocytes at the same rate and to the same extent as ectopically expressed ePABP. We conclude that PABPC1, like ePABP, associates with Musashi and facilitates Musashi-dependent mRNA translational activation.
Figure 8.
PABPC1 can functionally compensate for loss of ePABP during oocyte maturation. A, oocytes were co-injected with mRNA-encoding GST-tagged Xenopus PABPC1 and GFP-tagged Xenopus Musashi1 (GFP-XMsi1), Xenopus Musashi2 (GFP-XMsi2), or the GFP moiety alone. Following overnight incubation, the oocytes were lysed and subjected to GST pulldown, and PABPC1 co-association was assessed by GFP Western blotting. A strong interaction with Musashi1 is seen (arrowhead), but a greatly reduced interaction with Musashi2 is evident even after correction for recovered GST–PABPC1 in the samples. PABPC1 did not interact with the GFP moiety alone. B, ability of Musashi1 or Musashi2 to interact with PABPC1 was assessed in three independent experiments, and the data are summarized graphically. Association data were normalized to the levels of recovered GST–PABPC1 in each sample. * indicates p < 0.05; ** indicates p < 0.01. C, oocytes were injected with antisense oligonucleotides targeting ePABP. Following overnight incubation, the oocytes were reinjected with either water (No rescue) or PABPC1 mRNA (PABPC1 rescue) and then stimulated with progesterone, and the extent maturation was scored (% GVBD). The graph shows the combined data from six independent experiments. ****, p < 0.0001.
PABPC1 is associated with Musashi in mammalian cells under conditions that promote Musashi target mRNA translation
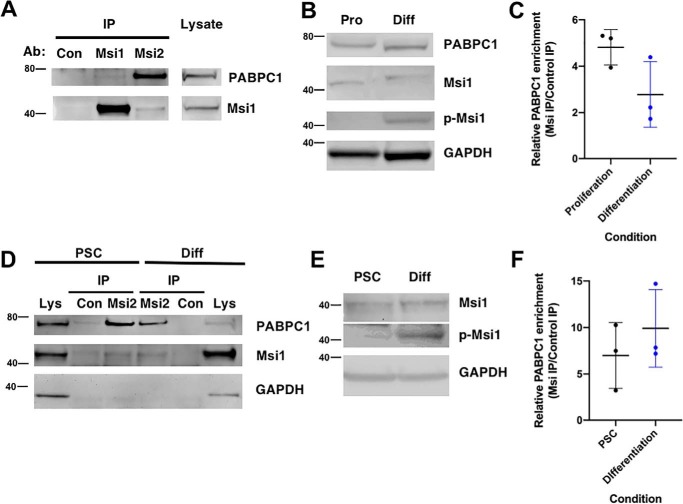
We next sought to determine whether endogenous Musashi associates with PABPC1 in mammalian cells under conditions of Musashi target mRNA de-repression and translation. We have previously reported that differentiation of primary embryonic rat neuronal stem/progenitor cells or human SH-SY5Y neuroblastoma cells results in the de-repression and translation of Musashi target mRNAs (6, 43). When proliferating cells are switched to media to promote differentiation, mammalian Musashi is subject to activating phosphorylation on the sites conserved with the Xenopus Musashi1 protein, resulting in translation of target mRNAs (43, 44). An assessment of Musashi1-activating phosphorylation thus serves as an indicator of de-repression and translation of target mRNAs.
We utilized the human SH-SY5Y neuroblastoma cell line and a human-induced pluripotent stem cell line (iPSCs) to assess the interaction of PABPC1 under proliferating conditions where Musashi directs repression or under conditions where Musashi1 is phosphorylated to allow de-repression and translation of target mRNAs through differentiation of SH-SY5Y cells or iPSCs along a neural progenitor lineage. Two antibodies were found to immunoprecipitate human Musashi1. One antibody (Abcam 52865) efficiently immunoprecipitated endogenous Musashi1 and weakly retained association of PABPC1, whereas a second antibody (Abcam 114107) immunoprecipitated both Musashi1 and Musashi2 and more effectively preserved association of PABPC1 (Fig. 9A). We utilized the Abcam 114107 antibody for all subsequent immunoprecipitations. When SH-SH5Y cells were cultured to promote differentiation, the differentiating SH-SY5Y cells exhibited a significant increase in Musashi1-activating phosphorylation (Fig. 9B, p-Msi1) without significant change in overall levels of Musashi1 (Fig. 9B, Msi1). Notably, over three independent experiments, no statistically significant changes in the levels of Musashi–PABPC1 interaction were observed when Musashi1 was immunoprecipitated from either proliferating or from differentiating SH-SY5Y cells (Fig. 9C). Similarly, the Abcam 114107 antibody immunoprecipitated Musashi1 and co-associated PABPC1 from either proliferating human iPSCs or iPSCs differentiated to adherent neural progenitor cells (Fig. 9D). When the iPSCs were differentiated to neural progenitor cells, a significant increase in activating Musashi1 phosphorylation was observed (Fig. 9E). Over three independent experiments, immunoprecipitation of endogenous Musashi1 revealed no significant changes in the level of co-associated PABPC1 in proliferating iPSCs (PSC) or in differentiated cell populations (Fig. 9F). We conclude that PABPC1 interaction persists with Musashi1 under conditions of target mRNA de-repression and translation. These findings are consistent with PABPC1 interacting with Musashi1 to facilitate translation of Musashi–target mRNAs in differentiating cell contexts.
Figure 9.
PABPC1 is associated with Musashi in mammalian cells under conditions of target mRNA translation. A, representative Western blotting showing rabbit IgG (Con) or Musashi1 immunoprecipitation using two different antibodies (Msi1, Abcam 52865; Msi2, Abcam 114107) from the lysate of proliferating SH-SY5Y cells. Recovery of Musashi1 and co-associated PABPC1 is shown along with levels of the proteins in starting lysate (1/10th volume of that used for immunoprecipitation). The positions of molecular weight markers are indicated to the left of each panel. B, activating phosphorylation of Musashi1 (p-Msi1) is observed in the lysate of differentiated (Diff) but not proliferating (Pro) SH-SY5Y cells. C, composite results of three independent experiments reveals that PABPC1 is present in immunoprecipitated Musashi1 protein complexes from both proliferating and differentiated SH-SY5Y cells. No statistically significant difference in Musashi1/PABPC1 co-association was observed between proliferating and differentiated SH-SY5Y cells. D, representative Western blotting showing rabbit IgG (Con) or Musashi1 immunoprecipitation using Msi2 (Abcam 114107) antibodies from lysate of proliferating iPSCs (PSC) or differentiated (Diff) iPSCs. Recovery of Musashi1 and co-associated PABPC1 is shown along with levels of the proteins in their respective starting lysate (1/10th volume of that used for immunoprecipitation). E, activating phosphorylation of Musashi1 (p-Msi1) is significantly increased in the lysate of differentiated (Diff) rather than proliferating iPSCs. F, composite results of three independent experiments reveal PABPC1 is present in immunoprecipitated Musashi1 complexes from both proliferating and from differentiated iPSCs. No statistically significant difference in Musashi1/PABPC1 co-association was observed between proliferating and differentiated iPSCs.
Discussion
In this study, we demonstrate that Musashi1 interacts with several members of the poly(A)-binding protein family, including ePABP and PABPC1, to promote translational activation of target mRNAs. As such, our data necessitate a fundamental revision of the model that describes a repressive-only function for the Musashi interaction with poly(A)-binding proteins. Our mapping data indicate that Musashi1 possesses two domains for interaction with ePABP, whereas in reciprocal experiments Musashi1 was shown to interact with the C-terminal domain of both ePABP and PABPC1. Our data further indicate that differential interaction of Musashi1 or Musashi2 with poly(A)-binding proteins may underlie differences in their functional properties.
We have previously shown that site-specific phosphorylation is required to facilitate de-repression and to promote Musashi target mRNA translation, both in the oocyte system and mammalian cells (39, 43, 44). In the oocyte, the initial “trigger” phosphorylation of these activating sites occurs via early Ringo/CDK-mediated phosphorylation (39). Robust Musashi1 phosphorylation was shown to require feedback phosphorylation by MAP kinase signaling, a downstream effector of the Musashi target mRNA encoding Mos, a MAP kinase signaling activator (39). We found that robust activating phosphorylation of Musashi1 is blocked in ePABP antisense-injected oocytes (Fig. 3C), although synthesis of Ringo protein occurs normally in ePABP antisense-injected oocytes and the initial Ringo/CDK-mediated trigger phosphorylation of Musashi1 (39) seems unperturbed. The failure to achieve full Musashi1 activation may be explained by the compromised polyadenylation of the mos mRNA (Fig. 4) and the consequent failure to attain robust activation of MAP kinase and the MAP kinase–dependent feedback amplification of Musashi1 phosphorylation (Fig. 3D) (39). We conclude that the loss of ePABP interaction with Musashi resulted in attenuation of translation of Musashi target mRNAs, including the mRNA encoding Mos, and that this compromised downstream MAP kinase signaling.
Our MS data suggest that the Musashi interactome undergoes dynamic regulation in response to progesterone stimulation. Of note, the differential enrichment of two proteins, CPEB1 and PABPC1, exemplify the remodeling of Musashi mRNP complexes. We have previously reported that the interaction of Musashi1 with CPEB1 is specific, RNase1-insensitive (i.e. it does not simply occur via mRNA co-occupancy), and occurs via an indirect association (51). Here, we see the interaction of Musashi1 with CPEB1 is restricted to immature oocytes. The CPEB1 population undergoes significant degradation in response to progesterone stimulation (56), likely explaining the lack of a significant Musashi1 co-association in maturing oocytes. A failure to detect PABPC1 in the immature oocyte data set while observing association in maturing oocytes likely reflects the low abundance of this protein isoform in the immature oocyte and the subsequent progesterone-dependent accumulation of PABPC1 in the maturing oocyte (48).
It is noteworthy that approximately one-third (16/50) of the proteins detected as Musashi1 partner proteins in the oocyte are have been previously identified as partner proteins of the human Musashi2 protein (57). Moreover, over half (9/16) belong to the core Musashi1 interactome that is shared between immature and maturing oocytes (Table S4). We speculate that these proteins may play a conserved role in controlling the assembly and/or function of Musashi mRNP complexes on target mRNAs.
Consistent with repression of Musashi target mRNAs in immature oocytes, a number of the co-associating proteins are known to contribute to translational repression (Fig. 1A), including DDX6, LSM14A/B, PAT1, and EIF4ENIF1 (58–61). Although association with PAT1 decreases in maturing oocytes, the other proteins remain associated, and it will be interesting in future studies to determine how their activity is modified within Musashi mRNP complexes to allow progesterone-dependent target mRNA translation.
The increased interaction (∼2-fold) of ePABP with Musashi1 after progesterone stimulation (Fig. 2) also supports the idea that Musashi interactions with partner proteins are dynamically regulated in response to extracellular cues. The mechanism underlying differential ePABP co-association is unclear, but several nonexclusive possibilities may be considered. First, the increased ePABP interaction may represent ePABP recruitment to a previously unbound Musashi1 after progesterone stimulation. We note that Musashi1 protein increases in response to progesterone potentially providing a reservoir of available targets for ePABP interaction (28). Second, the two sites of interaction on Musashi1 may allow engagement of a second ePABP to a Musashi1 protein already bound to ePABP. Third, the increase may represent multimerization of ePABP bound to Musashi1.
We report a strong interaction of ePABP with Musashi C-terminal amino acids 190–240 as well as weaker interaction with Musashi N-terminal amino acids 80–120 (Fig. 5). Interestingly, three proteins have now been demonstrated to interact with the C-terminal amino acids 190–240 region of Musashi1: PABPC1 (27); GLD2 (46); and ePABP (this study). GLD2 interacts strongly with amino acids 190–220 of the Musashi1 protein (46), although this same region interacted poorly with ePABP (Fig. 5C). By contrast, ePABP interacted strongly with amino acids 190–240, interacted weakly with amino acids 190–230, and failed to interact with amino acids 210–240, indicating that ePABP has a larger footprint of interaction across this region of the Musashi1 protein (Fig. 5C). Whether GLD2 and either ePABP or PABPC1 can interact simultaneously to the same Musashi1 protein remains to be determined. Although antisense knockdown experiments indicate that all three proteins promote Musashi-dependent translational activation, their relative contributions will require generation of point mutations that selectively attenuate their interactions within the 190–240 region of Musashi1. With regard to the poly(A)-binding proteins, our data indicate an attenuation of Musashi target mRNA polyadenylation in ePABP-depleted oocytes (Fig. 4). These findings support a role for ePABP in augmenting the ability of GLD2 to promote progesterone-dependent polyadenylation and/or protecting newly extended poly(A) tails to facilitate Musashi target mRNA translation.
Employing yeast two-hybrids assays, Musashi1 was shown to interact directly with the C-terminal domains of ePABP and PABPC1 (Fig. 6). Contrary to a previous report (27), no RNA-independent interaction of Musashi1 with RRMs 1 and 2 of either ePABP or PABPC1 was observed. One possible explanation for the discrepancy between these findings is that the prior study employed co-immunoprecipitation in the presence of RNase A (a pyrimidine-specific endoribonuclease), which may not have fully degraded mRNAs in the sample, thus allowing recovery of Musashi1 and PABP due to mRNA transcript co-occupancy rather than protein–protein interaction. Indeed, in the yeast two-hybrid assay, an interaction with full-length Musashi1 and RRMs 1 and 2 was seen, but this was abolished when an RNA-binding mutant form of the PABPC1 RRM 1 and 2 domain was employed. Of note, the RNA-binding mutant form of the PABPC1 RRM 1 and 2 domain was still able to interact with eIF4G (Fig. 6). Because Musashi1 interacts with the C-terminal domain of PABPs and not the eIF4G-interacting RRMs 1 and 2, our data do not support a direct competition between Musashi1 and eIF4G for the same site of interaction on PABPs as proposed (27). Nonetheless, it is possible that Musashi1 interaction with the C-terminal domain of PABPC1 or ePABP mediates an indirect attenuation of eIF4G binding through a conformation change.
Irrespective of the precise mechanism of repression, our findings add to the current understanding of Musashi-dependent translational control and extend the existing model in several ways. First, Musashi1 interaction with PABPs is not simply limited to exerting repression. Musashi1 remains associated with ePABP and PABPC1 under conditions of Musashi target mRNA de-repression and translational activation (Figs. 1, 2, and 9). Thus, the regulatory switch from mRNA repression to translational activation does not require dissociation of PABPs, but rather stimulus-dependent modification of Musashi activity in the presence of retained PABP association. Second, association of PABPs with Musashi is required to promote target mRNA translational activation in response to appropriate differentiation cues. Like GLD2, the other characterized co-factor activator (46), PABPs remain constitutively associated with Musashi1 irrespective of whether target mRNAs are repressed or translated. Thus, the activity of the regulatory complex must be controlled in a stimulus-dependent manner. Although this switch in regulatory function involves site-specific phosphorylation of the Musashi1 and Musashi2 proteins, it may also require additional modification of co-associated factors, perhaps including the PABPs (62, 63), to facilitate formation of a regulatory complex capable of stimulating de-repression and translation of Musashi target mRNAs.
Musashi1 and Musashi2 have been shown to act redundantly in several physiological systems, including neural stem cell self-renewal and Xenopus oocyte maturation, as well as pathologically in propagation of colon cancer stem cell function (23, 30, 44, 64). However, Musashi1 and Musashi2 have been reported to have distinct functions in pancreatic cells (65) and a differential ability to support Zika virus replication (42). We show here that Musashi1 and Musashi2 have overlapping but nonidentical functions in the oocyte, as Musashi2 functions less effectively than the Musashi1 isoform in promoting maturation. Consistent with the idea of nonidentical functions, several reports have demonstrated isoform-specific interaction with partner proteins. Musashi1, but not Musashi2, can interact with Lin28 (45) suggesting that regulation of miRNA biosynthesis may be a unique property of Musashi1. Similarly, mammalian GLD2 can interact with mammalian Musashi1 but not mammalian Musashi2, although the functional consequence of this altered GLD2 interaction has yet to be established (46). To add to this list of isoform-specific differences, we show here that ePABP and PABPC1 have dramatically reduced interaction with Musashi2 compared with Musashi1 and that this reduced interaction correlates with diminished capacity to promote cell cycle progression during oocyte maturation. Indeed, domain-swap experiments revealed that substitution of Musashi2 amino acids 190–240 for the corresponding Musashi1 sequence recapitulated full activity to the chimeric Musashi2 protein (Fig. 7). Our data support a model where differential interactions with partner proteins underlie functional distinctions between the Musashi1 and Musashi2 isoforms.
In summary, we provide evidence for a dynamic interaction of Musashi with members of the poly(A)-binding protein family and demonstrate that poly(A)-binding protein interaction with Musashi is necessary to promote target mRNA translation, in addition to mediating repression. These findings extend earlier models of Musashi-dependent translational control and provide new insights into the regulation of co-associated factor interactions as Musashi switches from a repressor to an activator of translation. Our results also highlight an important Musashi isoform-specific difference in partner protein interactions that can modulate the ability of Musashi1 and Musashi2 to control the translational output of target mRNAs.
Experimental procedures
Oocyte culture and microinjections
Dumont stage VI immature X. laevis oocytes were isolated and cultured as described previously (66). Oocytes were microinjected using a Nanoject II Auto-Nanoliter Injector (Drummond Scientific). mRNA for oocyte injection was made by linearization of the plasmid and in vitro transcription using SP6 (Promega) or T7 (Invitrogen) RNA polymerase as appropriate. Oocytes were stimulated to mature with 2 μg/ml progesterone. The appearance of a white spot on the animal pole was used to score the rate of oocyte maturation as it indicates germinal vesicle (nuclear) breakdown. Where indicated, progesterone-stimulated oocytes were segregated when 50% of the oocytes completed GVBD (GVBD50) into those that had not (−) or had (+) completed GVBD. In the event of ambiguous morphology, oocytes were fixed for 10 min in ice-cold 10% TCA and dissected for the presence or absence of a germinal vesicle. Animal protocols were approved by the UAMS Institutional Animal Care and Use committee, in accordance with Federal regulations.
Oocyte lysis and sample preparation
For co-association experiments, oocytes were lysed in 10 μl/oocyte of ice-cold Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 2 mm EDTA, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm PMSF, 1× protease inhibitor (Thermo Fisher Scientific)). Yolk and cell debris were removed by centrifugation at 12,000+ rpm for 10 min in a refrigerated tabletop centrifuge at 4 °C. For each sample, half-oocyte equivalents of lysate were prepared in NuPAGE sample loading buffer and electrophoresed through a 10% NuPAGE gel (Invitrogen). Where required, a portion of the lysate was transferred immediately following lysis to STAT-60 (Tel-Test, Inc.) for RNA extraction using the manufacturer's protocol followed by a subsequent purification by precipitation in 4 m LiCl at −80 °C for 30 min and centrifugation at 12,000 rpm for 10 min in a refrigerated tabletop centrifuge.
Pulldown and RNase treatment
Oocytes were injected with 57.5 ng of each in vitro transcribed mRNA and incubated for 16 h at 18 °C. Lysates were prepared as described above. 300 μl of oocyte lysate was added to 450 μl of ice-cold Nonidet P-40 lysis buffer and incubated with 50 μl of 50% GSH-Sepharose–conjugated bead slurry (GE Healthcare) at 4 °C for 6 h with gentle rotation. Beads were then gently pelleted by centrifugation at 500 rpm for 5 min; the supernatant was removed and replaced with 500 μl of fresh Nonidet P-40 lysis buffer, and the beads were inverted to mix, and the buffer was then removed. This process was repeated three times at 4 °C. On the third wash, 200 units of RNase1 (Ambion) was added and incubated for 15 min at 37 °C. Following final centrifugation, all Nonidet P-40 was removed, and 50 μl of NuPAGE sample loading buffer (Invitrogen) was added. Beads were incubated for 10 min at 70 °C and then crushed by centrifugation at 12,000 rpm for 10 min. Finally, 45 μl of the sample was loaded per each lane of a 10% NuPAGE gel (Invitrogen) and electrophoresed.
Mammalian cell culture and immunoprecipitation of endogenous Musashi
The SH-SY5Y human neuroblastoma cell line (ATCC) was cultured in DMEM/F-12 medium with 10% fetal bovine serum (Thermo Fisher Scientific), and human-induced pluripotent stem cells (catalog no. KYOU-DXR0109B, ATCC) were cultured in Essential 8 Medium (Thermo Fisher Scientific), on vitronectin-coated dishes (Thermo Fisher Scientific) with 5% CO2, as per the supplier's protocol. Neuronal differentiation was induced in SH-SY5Y cells through 1 week of culture in retinoic acid-containing medium (DMEM/F-12 with 1× B27 with vitamin A supplement (Thermo Fisher Scientific), further supplemented with 1 μm retinoic acid (Sigma)). Differentiation to neural stem cells was induced in human iPSCs through 1 week of culture in PSC Neural Induction Medium on vitronectin-coated dishes (Thermo Fisher Scientific). Neural progenitor cells exhibited greater than 10-fold loss of pluripotency gene expression (Nanog) and gain of neural progenitor gene expression (sox1), as per the supplier's protocol (Thermo Fisher Scientific). Cell lysate was prepared after rinsing the cells twice with cold DPBS (Thermo Fisher Scientific) followed by lysis with Nonidet P-40 lysis buffer (250–500 μl per 10-cm dish) and clearing through centrifugation. Musashi1 or Musashi1/2 was immunoprecipitated from cleared cell lysate (100–250 mg) with 10 μg of antibody (anti-Musashi1 catalog no. ab52865 (Abcam) or anti-Musashi2 catalog no. ab114107 (which also immunoprecipitates Musashi1, Abcam)) or normal rabbit IgG (control, catalog no. 12-370, Millipore) and 20 μl of protein A–agarose (catalog no. ab193254, Abcam) with overnight incubation at 4 °C with rocking. Immunocomplexes were washed twice with lysis buffer and dissociated at 100 °C, 10 min, with NuPAGE gel loading buffer (Thermo Fisher Scientific) with added 10 mm DTT and separated by electrophoresis (NuPAGE, Thermo Fisher Scientific).
Western blotting
Following electrophoresis, NuPAGE gels were transferred to a 0.2-μm pore-size nitrocellulose membrane (Protran; Midwest Scientific). The membrane was blocked with 5% nonfat dried milk or 1% BSA in TBST (20 mm Tris, pH 7.5, 150 mm NaCl, 0.05% Tween 20) for 60 min at room temperature or overnight at 4 °C. Following incubation with primary antibody, filters were washed three times for 10 min in TBST, incubated with horseradish peroxidase-conjugated secondary antibody or protein A HRP-conjugate (catalog no. 12291, Cell Signaling Technologies), and then washed three times for 10 min in TBST. Blots were developed using enhanced chemiluminescence in a Fluorchem 8000 Advanced Imager (Alpha Innotech Corp.). Western blots were quantified using Fluorchem FC2 software (Alpha Innotech Corp.)
Antibodies
The Xenopus ePABP antibody (67) was used at 1:5000 dilution. Antisera for phosphorylation-specific Musashi1 S322 (39) was used at 1:1000 dilution. The Xenopus Ringo antibody was a generous gift from Dr. Angel Nebreda and used at 1:1000 dilution. Additional antibodies used for immunodetection were as follows: anti-glyceraldehyde-3-phosphate dehydrogenase (1:5000, catalog no. AM4300, Ambion/Life Technologies, Inc.); anti-GST (1:5000, catalog no. SC-138, Santa Cruz Biotechnology); anti-p44/42 MAPK (ERK1/2) (1:1000, catalog no. 4695, Cell Signaling Technologies); anti-phospho-p44/42 MAPK (ERK1/2) (1:1000, catalog no. 4370, Cell Signaling Technologies); anti-Musashi (1:500, catalog no. 2154, Cell Signaling Technologies); anti-Musashi1 (1:1000, catalog no. ab21628, Abcam); anti-Musashi2 (1:000, catalog no. ab50829, Abcam); anti-PABP1 (1:000, catalog no. 4992, Cell Signaling Technologies); and anti-tubulin (1:5000, catalog no. ab7291, Abcam). All working antibody preparations were made in TBST + 0.5% nonfat milk or 0.1% BSA.
Polyadenylation assays
cDNAs for polyadenylation assays were synthesized using RNA ligation-coupled PCR as described (52). The increase in PCR product length is specifically due to extension of the poly(A) tail (28, 52, 53). The same reverse primer P1′ was used for all reactions and has the sequence 5′-GCTTCAGATCAAGGTGACCTTTTT-3′. The Mos forward primer has the sequence 5′-GCAAGGATATGAAAAAAAGATTTC-3′. The Nrp1b (Xenopus Musashi1) primer has the sequence 5′-CAATACTGCAATGTACAATGTACTGC-3′. The cyclin A1 primer has the sequence 5′-CATTGAACTGCTTCATTTTCCCAG-3′. Deducting the size of the mRNA-specific 3′ UTR sequence prior to the site of poly(A) addition and the size of the ligated P1 DNA oligonucleotide from the mode of the PCR product size in immature oocytes, we deduce the mos mRNA has an ∼60-residue adenylate tail; the Nrp1b mRNA has an ∼60-residue adenylate tail, and the cyclin A1 mRNA has an ∼30-residue adenylate tail in control immature oocytes. For Mos and Nrp1b, the poly(A) tails are extended by ∼60 and 70 adenylate residues, respectively, after 3 h of progesterone stimulation (see Fig. 4, B–D).
Antisense oligodeoxynucleotide injections and rescue
Antisense oligodeoxynucleotide 5′-CGGCTCCGGTTGCATTCATGTTTG-3′ was used to target endogenous ePABP mRNA (49). Antisense oligodeoxynucleotides targeting Xenopus Musashi1 and Musashi2 have been described previously (29). Control oocytes were injected with randomized oligonucleotide with the sequence 5′-TAGAGAAGATAATCGTCATCTTA-3′ (36). A total of 100 ng of antisense oligonucleotides was injected for each condition, and oocytes were incubated at 18 °C for 16 h. For Musashi rescue assays, Musashi1/2 antisense-injected oocytes were subsequently injected with 23 ng of RNA encoding WT Musashi1, Musashi2, or the chimeric Musashi2 protein that contained the Musashi1 ePABP-binding domain. The oocytes were then incubated for 1 h at room temperature to allow expression of the protein, then stimulated to mature with progesterone. For rescue of ePABP antisense-injected oocytes, oocytes were co-injected with 57.5 ng of wobble Xenopus ePABP mRNA, which is resistant to the antisense oligonucleotides (49) or Xenopus PABPC1 mRNA, which is not targeted by the ePABP oligonucleotides. Following overnight incubation, oocytes were stimulated with progesterone, and the extent of GVBD scored.
Liquid chromatography–coupled tandem mass spectrometry
We used MS to identify Musashi1-associated protein complexes. 240 oocytes were each injected with 115 ng of mRNA encoding GST–XMsi1 or GST alone. We estimate by Western blot analysis that this results in a 5–10-fold overexpression of GST–XMsi1 to the endogenous Musashi1 protein. Following overnight incubation, the oocytes from each injection were split into two separate pools and either left untreated or stimulated with progesterone. All oocytes were collected when progesterone-stimulated samples had reached GVBD50 and lysed in ice-cold Tween 20 buffer (0.1% Tween 20, 20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 2 mm EDTA, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm PMSF, 1× protease inhibitor (Thermo Fisher Scientific)). A GSH pulldown for each condition was then performed. 600 μl of oocyte lysate was added to 250 μl of 50% GSH-Sepharose–conjugated bead slurry (GE Healthcare) at 4 °C for 6 h with gentle rotation. Beads were then gently pelleted by centrifugation at 500 rpm for 5 min; the supernatant was removed and replaced with 1 ml of fresh Tween 20 lysis buffer. This process was repeated three times at 4 °C, and following the final centrifugation, all lysis buffer was removed, and 50 μl of LDS sample loading buffer (Invitrogen) was added and incubated overnight at 4 °C for 6 h with gentle rotation. Beads were incubated for 10 min at 70 °C and then crushed by centrifugation at 12,000 rpm for 10 min. Finally, 45 μl of the sample was loaded to each lane of a 10% NuPAGE gel (Invitrogen) and electrophoresed. Following electrophoresis, the gel was fixed by a 5-min incubation with 10% acetic acid and 16% methanol at room temperature. Following incubation, fixative was replaced with deionized water and microwaved for 45 s. The water was poured off and replaced with GelCode Blue (Pierce) and microwaved again for 30 s to begin staining. The gel was then gently rocked for 30 min at room temperature. Finally, the GelCode Blue was replaced with 1% acetic acid and gently rocked for 1 h. Protein gel bands were then excised and subjected to in-gel trypsin digestion. Gel slices were destained in 50% methanol (Thermo Fisher Scientific), 100 mm ammonium bicarbonate (Sigma), followed by reduction in 10 mm tris(2-carboxyethyl)phosphine (Pierce) and alkylation in 50 mm iodoacetamide (Sigma). Gel slices were then dehydrated in acetonitrile (Fisher), followed by addition of 100 ng of porcine sequencing grade-modified trypsin (Promega) in 100 mm ammonium bicarbonate (Sigma) and incubation at 37 °C for 12–16 h. Peptide products were then acidified in 0.1% formic acid (Pierce). Tryptic peptides were separated by reverse phase Jupiter Proteo resin (Phenomenex) on a 100 × 0.075-mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 40-min gradient from 97:3 to 35:65 buffer A/B ratio. (Buffer A = 0.1% formic acid, 0.5% acetonitrile; buffer B = 0.1% formic acid, 75% acetonitrile.) Eluted peptides were ionized by electrospray (1.9 kV) followed by MS/MS analysis using collision-induced dissociation on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). MS data were acquired using the FTMS analyzer in profile mode at a resolution of 60,000 over a range of 375 to 1500 m/z. MS/MS data were acquired for the top 15 peaks from each MS scan using the ion trap analyzer in centroid mode and normal mass range with a normalized collision energy of 35.0. Proteins were identified by searching against the UniprotKB database restricted to Homo sapiens (177,579 entries) using MaxQuant (version 1.5.3.8, Max Planck Institute) with a parent ion tolerance of 3 ppm and a fragment ion tolerance of 0.5 Da, fixed modification of carbamidomethyl on C, and variable modifications, including oxidation on M, and acetyl on the peptide N terminus. Scaffold (Proteome Software) was used to verify MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established with less than 1.0% false discovery by the Scaffold Local FDR algorithm and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (68). The pulldown and MS experiments were repeated in triplicate, and a p value representing the t test significance of an interaction with GST–Musashi1 versus the GST moiety alone after correction of assigned spectral hits for molecular weight of the target protein (log2 normalized spectral abundance factor (50)) was derived for each identified protein. Fifty proteins were found to specifically interact with Musashi1 (Table S1). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (69) with the dataset identifier PXD013585 and 10.6019/PXD013585. Gene set enrichment analysis (70) was used to query the molecular signatures database (MSigDB) for the top 10 gene ontology molecular functions.
Yeast two-hybrid assays (Y2H)
Y2H analysis was performed in strain L40 as described in Ref. 71, with the strength of interaction being scored by the extent of blue color apparent within a fixed time.
Statistical analysis
All quantitated data are presented as the mean ± S.E. Statistical significance was assessed by one-way analysis of variance followed by the Bonferroni post hoc test or by Student's t test when only two groups were compared. A probability of p < 0.05 was adopted for statistical significance.
Plasmids and plasmid construction
The derivation of a number of constructs used in this study has been previously described. These include the following: GST–XMsi1 (30); GST-XMsi2, GST-mMsi1, GST-mMsi2, GST-mMsi1 Δ190–234, GST-N-term XMsi1, GST-C-term XMsi1, GST–XMsi1 Δ200–210, GST–XMsi1 Δ200–220, GST–XMsi1 190–240, GST–XMsi1 190–230, GST–XMsi1 190–220, GST–XMsi1 210–240, and pXen GFP (46); XePABP (wobble) and XPABPC1 (49); LexA-MS2, PAB 1–2, PAB-Rd (RRMs 1 and 2 RNA-binding mutant), PAB 3–4, PAB-Ct, and pACT-IRP and pACT-4GNt (71); and BTM e1–2, BTM e3–4, and BTM-eCt (67). Plasmid constructs generated exclusively for this study are detailed in Table 1.
Table 1.
Plasmid construction
The technique, strategy, and where appropriate primer sequences used to generate the plasmid constructs utilized in this study are indicated
For PCR fragment generation, template was subjected to PCR amplification using the indicated primers. Resulting PCR fragments were then purified using agarose gel electrophoresis followed by clean up using a QIAquick Gel Extraction Kit (Qiagen). Next, the fragments and destination vector were digested using the indicated restriction enzymes and again purified and cleaned up using gel electrophoresis and the QIAquick kit. The fragment and vector were then ligated using the T7 Quick Ligase (New England Biolabs). Finally, the ligated fragment/vector was used to transform competent DH5-α E. coli. For PCR-directed mutagenic deletion, template was subjected to PCR amplification of the entire plasmid. Primer sequence “looped out” the desired sequence for deletion. For restriction fragment subcloning, template DNA and destination vector were separately digested with the indicated restriction enzymes and the desired fragments isolated and purified using agarose gel electrophoresis followed by clean up using a QIAquick gel extraction kit (Qiagen). The fragment and vector were then ligated using the T7 Quick Ligase (New England Biolabs). Finally, the ligated fragment/vector was used to transform competent DH5-α E. coli.
| Construct | Methodology |
|---|---|
| GST-XMsi1 1–179 | (+)5′-GCGCGATCGATGGCGACAGAAGCGCCCCAG-3′ |
| (−)5′-CGCGCTCGAGCTAAACCATTTTATTGTTGAT-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 | |
| Vector: pXen1 | |
| Restriction enzymes: 5′ClaI 3′XhoI | |
| GST-XMsi1 180–347 | (+)5′-GCGCGATCGATGGCGTGTAAGAAGGCCCAGCC-3′ |
| (−)5′-CGCGCGTCTAGATCAGTGGTAGCCGTTGGTAAAAGC-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 | |
| Vector: pXen1 | |
| Restriction enzymes: 5′ClaI 3′XbaI | |
| GST-XMsi1 RRM1 | (+)5′-GCGCGATCGATGGCGACAGAAGCGCCCCAG-3′ |
| (−)5′-GCGCGTCTAGACTACTTGGGTTGAGCTCTACGAGGAAA-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 N-terminal | |
| Vector: pXenI | |
| Restriction enzymes: 5′ClaI 3′XbaI | |
| GST-XMsi1 RRM2 | (+)5′-GCGCGATCGATGGTAACACGGACAAAGAAGATT-3′ |
| (−)5′-GCGTCTAGATCGGCCTCTCACAGACCCTGTTG-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 N-terminal | |
| Vector: pXen1 | |
| Restriction enzymes: 5′ClaI 3′XbaI | |
| GST-N-terminal XMsi1 Δ80–120 | (+)5′-GGAGTGGACAAAGTTTTGGCTACAGTTGAAGATGTGAAAC-3′ |
| (−)5′-GGTTCACATCTTCAACTGTAGCCAAAACTTTGTCCACTCC-3′ | |
| Technique: PCR-directed mutagenic deletion | |
| Template: GST-XMsi1 N-terminal | |
| Vector: pXen1 | |
| pACT-XMsi1 | (+)5′-GCGCCATGGAGACAGAAGCGCCCCAGCC-3′ |
| (−)5′-GCGGAATTCTCAGTGGTAGCCGTTGGTAAAAGC-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 | |
| Vector: pACT2 | |
| Restriction enzymes: 5′NcoI 3′EcoRI | |
| pACT-XMsi1-Nt (1–198) | (+)5′-GCGCCATGGAGACAGAAGCGCCCCAGCC-3′ |
| (−)5′-GCGGAATTCTCAGCCTCTCACAGACCCTGTTGGTGAC-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 | |
| Vector: pACT2 | |
| Restriction enzymes: 5′NcoI 3′EcoRI | |
| pACT-XMsi1-Ct (199–347) | (+)5′-GCGCCATGGAGCGATCTCGGGTCATGCTATATGG-3′ |
| (−)5′-GCGGAATTCTCAGTGGTAGCCGTTGGTAAAAGC-3′ | |
| Technique: PCR fragment generation | |
| Template: GST-XMsi1 | |
| Vector: pACT2 | |
| Restriction enzymes: 5′NcoI 3′EcoRI | |
| GST-XMsi2 Chimera | (A) 5′-GCGCATCGATGGAGGCAGATGGGAGC-3′ |
| (B) 5′-CACAGACCCTGTTGGTGACATGACTTCCTTAGGCTGTGC-3′ | |
| (C) 5′-GCACAGCCTAAGGAAGTCATGTCACCAACAGGGTCTGTG-3′ | |
| (D) 5′-AAAACCTGGAAACTGATAGCTATATCCAGGTGCAATGCC-3′ | |
| (E) 5′-ATTGCACCTGGATATAGCTATCAGTTTCCAGGTTTTCC-3′ | |
| (F) 5′-GCGCCTCGAGTCAATGGTATCCATTTG-3′ | |
| Primers A and B were combined with pXen-XMsi2 to PCR the N-terminal 189 amino acids of XMsi2. Primers E and F were combined with pXen-XMsi2 to PCR the C-terminal 163 amino acids. Primers C and D were combined with pXen-XMsi1 to PCR the ePABP-binding domain (amino acids 190–240). The products from these three PCRs were then combined along with primers A and F to create the chimeric PCR product. This product was then digested with 3′-ClaI and 5′-XhoI then ligated into pXen1. | |
| GST-XPABPC1 | (+)5′-GCGCATCGATATGAATCCCAGTGCTCCCAGC-3′ |
| (−)5′-GCGCCTCGAGTTAAGCAGTTGGCACTCCAGTTGCA-3′ | |
| Technique: PCR fragment generation | |
| Template: pET XPABPC1-FLAG (49) | |
| Vector: pXen1 | |
| Restriction enzymes: 5′ClaI 3′XhoI | |
| GFP XMsi1 | Technique: restriction fragment subclone |
| Template: pXen XMsi1 (30) | |
| Vector: pXen GFP (46) | |
| Restriction enzymes: 5′ClaI 3′XbaI | |
| GFP XMsi2 | Technique: Restriction fragment subclone |
| Template: pXen XMsi2 (46) | |
| Vector: pXen GFP (46) | |
| Restriction enzymes: 5′ClaI 3′XhoI |
Author contributions
C. E. C., M. C. M., S. D. B., S. G. M., W. A. R., N. K. G., A. J. T., and A. M. M. formal analysis; C. E. C., S. D. B., L. L. H., and N. K. G. validation; C. E. C., M. C. M., S. D. B., L. L. H., S. G. M., W. A. R., N. K. G., and A. J. T. investigation; C. E. C., L. L. H., and S. G. M. methodology; C. E. C., M. C. M., and A. M. M. writing-original draft; M. C. M., N. K. G., G. V. C., A. J. T., and A. M. M. conceptualization; M. C. M., G. V. C., and A. M. M. funding acquisition; S. D. B. data curation; S. D. B., N. K. G., G. V. C., A. J. T., and A. M. M. writing-review and editing; N. K. G., A. J. T., and A. M. M. supervision; A. M. M. project administration.
Supplementary Material
Acknowledgments
The University of Arkansas for Medical Sciences (UAMS)Translational Research Institute was supported by National Institutes of Health NCRR and National Center for Advancing Translational Sciences Grants UL1TR000039 and KL2TR000063. The University of Arkansas for Medical Sciences Center for Translational Neuroscience was supported by National Institutes of Health NIGMS Grant P30 GM110702. We acknowledge the UAMS Proteomics Facility for mass spectrometric support. A. M. M. acknowledges training received through the advanced course in Proteomics/Bioinformatics held at the Welcome Genome Campus/European Bioinformatics Institute, Cambridge, United Kingdom.
This work was supported by a Sturgis Diabetes Research Pilot Award, the Arkansas Breast Cancer Research Program, the Arkansas BioSciences Institute, intramural funding from the University of Arkansas for Medical Sciences (UAMS) College of Medicine Research Council, National Institutes of Health IDeA Program Award P30 GM110702 NIGMS Pilot Grant, National Institutes of Health Grant RO1 HD93461 (to A. M. M., M. C. M., and G. V. C.), and the UAMS Medical Research Endowment. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession nos. PXD013585 and 10.6019/PXD013585.
- RRM
- RNA recognition motif
- Msi AS
- Musashi1/2 antisense oligonucleotide
- GVBD
- germinal vesicle breakdown
- GST
- glutathione S-transferase
- ePABP
- embryonic poly(A)-binding protein
- MAP
- mitogen-activated protein
- MAPK
- mitogen-activated protein kinase
- PABP
- poly(A)-binding protein
- PMSF
- phenylmethylsulfonyl fluoride
- mRNP
- mRNA ribonucleoprotein
- iPSC
- induced pluripotent stem cell line
- Y2H
- yeast two-hybrid
- CDK
- cyclin-dependent kinase
- CPE
- cytoplasmic polyadenylation element
- DMEM
- Dulbecco's modified Eagle's medium.
References
- 1. Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., and Okano H. (2001) The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900 10.1128/MCB.21.12.3888-3900.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagata T., Kanno R., Kurihara Y., Uesugi S., Imai T., Sakakibara S., Okano H., and Katahira M. (1999) Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 287, 315–330 10.1006/jmbi.1999.2596 [DOI] [PubMed] [Google Scholar]
- 3. Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., and Katahira M. (2012) Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 40, 3218–3231 10.1093/nar/gkr1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Andrés-Aguayo L., Varas F., and Graf T. (2012) Musashi 2 in hematopoiesis. Curr. Opin. Hematol. 19, 268–272 10.1097/MOH.0b013e328353c778 [DOI] [PubMed] [Google Scholar]
- 5. Horisawa K., Imai T., Okano H., and Yanagawa H. (2010) The Musashi family RNA-binding proteins in stem cells. Biomol. Concepts 1, 59–66 10.1515/bmc.2010.005 [DOI] [PubMed] [Google Scholar]
- 6. MacNicol M. C., Cragle C. E., and MacNicol A. M. (2011) Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle 10, 39–44 10.4161/cc.10.1.14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sutherland J. M., Siddall N. A., Hime G. R., and McLaughlin E. A. (2015) RNA binding proteins in spermatogenesis: an in depth focus on the Musashi family. Asian J. Androl. 17, 529–536 10.4103/1008-682X.151397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okano H., Kawahara H., Toriya M., Nakao K., Shibata S., and Imai T. (2005) Function of RNA-binding protein Musashi1 in stem cells. Exp. Cell Res. 306, 349–356 10.1016/j.yexcr.2005.02.021 [DOI] [PubMed] [Google Scholar]
- 9. MacNicol A. M., Wilczynska A., and MacNicol M. C. (2008) Function and regulation of the mammalian Musashi mRNA translational regulator. Biochem. Soc. Trans. 36, 528–530 10.1042/BST0360528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fox R. G., Park F. D., Koechlein C. S., Kritzik M., and Reya T. (2015) Musashi signaling in stem cells and cancer. Annu. Rev. Cell Dev. Biol. 31, 249–267 10.1146/annurev-cellbio-100814-125446 [DOI] [PubMed] [Google Scholar]
- 11. Odle A. K., Akhter N., Syed M. M., Allensworth-James M. L., Benes H., Melgar Castillo A. I., MacNicol M. C., MacNicol A. M., and Childs G. V. (2017) Leptin regulation of gonadotrope gonadotropin-releasing hormone receptors as a metabolic checkpoint and gateway to reproductive competence. Front. Endocrinol. 8, 367 10.3389/fendo.2017.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odle A. K., Beneš H., Melgar Castillo A., Akhter N., Syed M., Haney A., Allensworth-James M., Hardy L., Winter B., Manoharan R., Syed R., MacNicol M. C., MacNicol A. M., and Childs G. V. (2018) Association of Gnrhr mRNA with the stem cell determinant Musashi: a mechanism for leptin-mediated modulation of GnRHR expression. Endocrinology 159, 883–894 10.1210/en.2017-00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith A. R., Marquez R. T., Tsao W. C., Pathak S., Roy A., Ping J., Wilkerson B., Lan L., Meng W., Neufeld K. L., Sun X. F., and Xu L. (2015) Tumor suppressive microRNA-137 negatively regulates Musashi1 and colorectal cancer progression. Oncotarget 6, 12558–12573 10.18632/oncotarget.3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hemmati H. D., Nakano I., Lazareff J. A., Masterman-Smith M., Geschwind D. H., Bronner-Fraser M., and Kornblum H. I. (2003) Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 15178–15183 10.1073/pnas.2036535100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito T., Kwon H. Y., Zimdahl B., Congdon K. L., Blum J., Lento W. E., Zhao C., Lagoo A., Gerrard G., Foroni L., Goldman J., Goh H., Kim S. H., Kim D. W., Chuah C., et al. (2010) Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768 10.1038/nature09171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanemura Y., Mori K., Sakakibara S., Fujikawa H., Hayashi H., Nakano A., Matsumoto T., Tamura K., Imai T., Ohnishi T., Fushiki S., Nakamura Y., Yamasaki M., Okano H., and Arita N. (2001) Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation 68, 141–152 10.1046/j.1432-0436.2001.680208.x [DOI] [PubMed] [Google Scholar]
- 17. Kharas M. G., Lengner C. J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., Gozo M., Einhorn W., Lane S. W., Scholl C., Fröhling S., et al. (2010) Musashi2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16, 903–908 10.1038/nm.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oskarsson T., Acharyya S., Zhang X. H., Vanharanta S., Tavazoie S. F., Morris P. G., Downey R. J., Manova-Todorova K., Brogi E., and Massagué J. (2011) Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 17, 867–874 10.1038/nm.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sureban S. M., May R., George R. J., Dieckgraefe B. K., McLeod H. L., Ramalingam S., Bishnupuri K. S., Natarajan G., Anant S., and Houchen C. W. (2008) Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology 134, 1448–1458 10.1053/j.gastro.2008.02.057 [DOI] [PubMed] [Google Scholar]
- 20. Toda M., Iizuka Y., Yu W., Imai T., Ikeda E., Yoshida K., Kawase T., Kawakami Y., Okano H., and Uyemura K. (2001) Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia 34, 1–7 10.1002/glia.1034 [DOI] [PubMed] [Google Scholar]
- 21. Wang X. Y., Penalva L. O., Yuan H., Linnoila R. I., Lu J., Okano H., and Glazer R. I. (2010) Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 9, 221 10.1186/1476-4598-9-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X. Y., Yu H., Linnoila R. I., Li L., Li D., Mo B., Okano H., Penalva L. O., and Glazer R. I. (2013) Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget 4, 739–750 10.18632/oncotarget.1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li N., Yousefi M., Nakauka-Ddamba A., Li F., Vandivier L., Parada K., Woo D. H., Wang S., Naqvi A. S., Rao S., Tobias J., Cedeno R. J., Minuesa G., Y K., Barlowe T. S., et al. (2015) The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep. 13, 2440–2455 10.1016/j.celrep.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwon H. Y., Bajaj J., Ito T., Blevins A., Konuma T., Weeks J., Lytle N. K., Koechlein C. S., Rizzieri D., Chuah C., Oehler V. G., Sasik R., Hardiman G., and Reya T. (2015) Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell 17, 152–164 10.1016/j.stem.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Battelli C., Nikopoulos G. N., Mitchell J. G., and Verdi J. M. (2006) The RNA-binding protein Musashi1 regulates neural development through the translational repression of p21(WAF-1). Mol. Cell. Neurosci. 31, 85–96 10.1016/j.mcn.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 26. Okabe M., Imai T., Kurusu M., Hiromi Y., and Okano H. (2001) Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature 411, 94–98 10.1038/35075094 [DOI] [PubMed] [Google Scholar]
- 27. Kawahara H., Imai T., Imataka H., Tsujimoto M., Matsumoto K., and Okano H. (2008) Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 181, 639–653 10.1083/jcb.200708004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arumugam K., Macnicol M. C., and Macnicol A. (2012) Autoregulation of Musashi1 mRNA translation during Xenopus oocyte maturation. Mol. Reprod. Dev. 79, 553–563 10.1002/mrd.22060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arumugam K., Wang Y., Hardy L. L., MacNicol M. C., and MacNicol A. M. (2010) Enforcing temporal control of maternal mRNA translation during oocyte cell cycle progression. EMBO J. 29, 387–397 10.1038/emboj.2009.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Charlesworth A., Wilczynska A., Thampi P., Cox L. L., and MacNicol A. M. (2006) Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 25, 2792–2801 10.1038/sj.emboj.7601159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wickens M., Goodwin E. B., Kimble J., Strictland S., and Hentze M. W. (2000) in Translational Control of Gene Expression (Sonenberg N., Hershey J., and Mathews M. B., eds), pp. 295–370, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Lasko P. (2009) Translational control during early development. Progr. Mol. Biol. Transl. Sci. 90, 211–254 10.1016/S1877-1173(09)90006-0 [DOI] [PubMed] [Google Scholar]
- 33. Cragle C. E., and MacNicol A. M. (2014) in Xenopus Development (Kloc M. and Kubiak J. Z, eds), pp. 38–59, Wiley-Blackwell, NJ [Google Scholar]
- 34. MacNicol M. C., and MacNicol A. M. (2010) Developmental timing of mRNA translation -integration of distinct regulatory elements. Mol. Reprod. Dev. 77, 662–669 10.1002/mrd.21191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Padmanabhan K., and Richter J. D. (2006) Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 20, 199–209 10.1101/gad.1383106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferby I., Blazquez M., Palmer A., Eritja R., and Nebreda A. R. (1999) A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 13, 2177–2189 10.1101/gad.13.16.2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karaiskou A., Perez L. H., Ferby I., Ozon R., Jessus C., and Nebreda A. R. (2001) Differential regulation of Cdc2 and Cdk2 by RINGO and cyclins. J. Biol. Chem. 276, 36028–36034 10.1074/jbc.M104722200 [DOI] [PubMed] [Google Scholar]
- 38. Lenormand J. L., Dellinger R. W., Knudsen K. E., Subramani S., and Donoghue D. J. (1999) Speedy: a novel cell cycle regulator of the G2/M transition. EMBO J. 18, 1869–1877 10.1093/emboj/18.7.1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arumugam K., MacNicol M. C., Wang Y., Cragle C. E., Tackett A. J., Hardy L. L., and MacNicol A. M. (2012) Ringo/CDK and MAP kinase regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J. Biol. Chem. 287, 10639–10649 10.1074/jbc.M111.300681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuwako K., Kakumoto K., Imai T., Igarashi M., Hamakubo T., Sakakibara S., Tessier-Lavigne M., Okano H. J., and Okano H. (2010) Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron 67, 407–421 10.1016/j.neuron.2010.07.005 [DOI] [PubMed] [Google Scholar]
- 41. Park S. M., Deering R. P., Lu Y., Tivnan P., Lianoglou S., Al-Shahrour F., Ebert B. L., Hacohen N., Leslie C., Daley G. Q., Lengner C. J., and Kharas M. G. (2014) Musashi2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 211, 71–87 10.1084/jem.20130736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chavali P. L., Stojic L., Meredith L. W., Joseph N., Nahorski M. S., Sanford T. J., Sweeney T. R., Krishna B. A., Hosmillo M., Firth A. E., Bayliss R., Marcelis C. L., Lindsay S., Goodfellow I., Woods C. G., and Gergely F. (2017) Neurodevelopmental protein Musashi1 interacts with the Zika genome and promotes viral replication. Science 357, 83–88 10.1126/science.aam9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacNicol A. M., Hardy L. L., Spencer H. J., and MacNicol M. C. (2015) Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev. Biol. 15, 15 10.1186/s12861-015-0064-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. MacNicol M. C., Cragle C. E., McDaniel F. K., Hardy L. L., Wang Y., Arumugam K., Rahmatallah Y., Glazko G. V., Wilczynska A., Childs G. V., Zhou D., and MacNicol A. M. (2017) Evasion of regulatory phosphorylation by an alternatively spliced isoform of Musashi2. Sci. Rep. 7, 11503 10.1038/s41598-017-11917-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kawahara H., Okada Y., Imai T., Iwanami A., Mischel P. S., and Okano H. (2011) Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 286, 16121–16130 10.1074/jbc.M110.199166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cragle C., and MacNicol A. M. (2014) Musashi-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germline development-2. J. Biol. Chem. 289, 14239–14251 10.1074/jbc.M114.548271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim J. H., and Richter J. D. (2007) RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 21, 2571–2579 10.1101/gad.1593007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Voeltz G. K., Ongkasuwan J., Standart N., and Steitz J. A. (2001) A novel embryonic poly(A)-binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 15, 774–788 10.1101/gad.872201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gorgoni B., Richardson W. A., Burgess H. M., Anderson R. C., Wilkie G. S., Gautier P., Martins J. P., Brook M., Sheets M. D., and Gray N. K. (2011) Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 108, 7844–7849 10.1073/pnas.1017664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., and Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 10.1021/pr060161n [DOI] [PubMed] [Google Scholar]
- 51. MacNicol M. C., Cragle C. E., Arumugam K., Fosso B., Pesole G., and MacNicol A. M. (2015) Functional integration of mRNA translational control programs. Biomolecules 5, 1580–1599 10.3390/biom5031580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Charlesworth A., Cox L. L., and MacNicol A. M. (2004) Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 279, 17650–17659 10.1074/jbc.M313837200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Charlesworth A., Ridge J. A., King L. A., MacNicol M. C., and MacNicol A. M. (2002) A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 21, 2798–2806 10.1093/emboj/21.11.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Moor C. H., and Richter J. D. (1997) The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 17, 6419–6426 10.1128/MCB.17.11.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sheets M. D., Fox C. A., Hunt T., Vande Woude G., and Wickens M. (1994) The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 8, 926–938 10.1101/gad.8.8.926 [DOI] [PubMed] [Google Scholar]
- 56. Reverte C. G., Ahearn M. D., and Hake L. E. (2001) CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev. Biol. 231, 447–458 10.1006/dbio.2001.0153 [DOI] [PubMed] [Google Scholar]
- 57. Vu L. P., Prieto C., Amin E. M., Chhangawala S., Krivtsov A., Calvo-Vidal M. N., Chou T., Chow A., Minuesa G., Park S. M., Barlowe T. S., Taggart J., Tivnan P., Deering R. P., Chu L. P., et al. (2017) Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat. Genet. 49, 866–875 10.1038/ng.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Minshall N., Thom G., and Standart N. (2001) A conserved role of a DEAD box helicase in mRNA masking. RNA 7, 1728–1742 10.1017/S135583820101158X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minshall N., Reiter M. H., Weil D., and Standart N. (2007) CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 282, 37389–37401 10.1074/jbc.M704629200 [DOI] [PubMed] [Google Scholar]
- 60. Tanaka K. J., Ogawa K., Takagi M., Imamoto N., Matsumoto K., and Tsujimoto M. (2006) RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 281, 40096–40106 10.1074/jbc.M609059200 [DOI] [PubMed] [Google Scholar]
- 61. Nakamura Y., Tanaka K. J., Miyauchi M., Huang L., Tsujimoto M., and Matsumoto K. (2010) Translational repression by the oocyte-specific protein P100 in Xenopus. Dev. Biol. 344, 272–283 10.1016/j.ydbio.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 62. Brook M., McCracken L., Reddington J. P., Lu Z. L., Morrice N. A., and Gray N. K. (2012) The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem. J. 441, 803–812 10.1042/BJ20111474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Friend K., Brook M., Bezirci F. B., Sheets M. D., Gray N. K., and Seli E. (2012) Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem. J. 445, 93–100 10.1042/BJ20120304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sakakibara S., Nakamura Y., Yoshida T., Shibata S., Koike M., Takano H., Ueda S., Uchiyama Y., Noda T., and Okano H. (2002) RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. U.S.A. 99, 15194–15199 10.1073/pnas.232087499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Szabat M., Kalynyak T. B., Lim G. E., Chu K. Y., Yang Y. H., Asadi A., Gage B. K., Ao Z., Warnock G. L., Piret J. M., Kieffer T. J., and Johnson J. D. (2011) Musashi expression in beta-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2, e232 10.1038/cddis.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Machaca K., and Haun S. (2002) Induction of maturation-promoting factor during Xenopus oocyte maturation uncouples Ca2+ store depletion from store-operated Ca2+ entry. J. Cell Biol. 156, 75–85 10.1083/jcb.200110059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilkie G. S., Gautier P., Lawson D., and Gray N. K. (2005) Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol. Cell. Biol. 25, 2060–2071 10.1128/MCB.25.5.2060-2071.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 69. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gray N. K., Coller J. M., Dickson K. S., and Wickens M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19, 4723–4733 10.1093/emboj/19.17.4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.