Abstract
Hypertrophic cardiomyopathy (HCM) is an inherited heart failure condition, mostly found to have genetic abnormalities, and is a leading cause of sudden death in young adults. Whole exome sequencing should be given consideration as a molecular diagnostic tool to identify disease-causing mutation/s. In this study, a HCM family with multiple affected members having history of sudden death were subjected to exome sequencing along with unaffected members. Quality passed variants obtained were filtered for rarity (MAF > 0.5%), evolutionary conservation, pathogenic prediction, and segregation in affected members after removing shared variants present in unaffected members. Only one non-synonymous mutation (p. Glu186Lys or E186K) in exon 6 of P2X7 gene segregated in HCM-affected individuals which was absent in unaffected family members and 100 clinically evaluated controls. The site of the mutation is highly conserved and led to complete loss of function which is in close vicinity to ATP-binding site-forming residues, affecting ATP binding, channel gating, or both. Mutations in candidate genes which were not segregated define clinical heterogeneity within affected members. P2X7 gene is highly expressed in the heart and shows direct interaction with major candidate genes for HCM. Our results reveal a significant putative HCM causative gene, P2X7, for the first time and show that germ-line mutations in P2X7 may cause a defective phenotype, suggesting purinergic receptor involvement in heart failure mediated through arrhythmias which need further investigations to be targeted for therapeutic interventions.
Electronic supplementary material
The online version of this article (10.1007/s11302-019-09660-7) contains supplementary material, which is available to authorized users.
Keywords: HCM, Clinical heterogeneity, Bradycardia, Sudden death, Heart failure
Introduction
Hypertrophic cardiomyopathy (HCM) is a cardiac genetic disease characterized by hypertrophy of the left ventricle and intraventricular septum and in severe cases followed by sudden death. The disease has a prevalence of 1 in 500 globally and is a major cause of sudden cardiac death in young people [10]. A number of genes encoding for sarcomeric and non-sarcomeric proteins are associated with the function of cardiac muscles [9]. It has been estimated that approximately half of the patients with HCM have a germ-line mutation in one of the genes encoding for sarcomeric proteins.
Although most of the mutations associated with HCM are reported in sarcomeric genes but receptor gene expressions are also crucial for the contraction and relaxation of the heart which are regulated by various signaling pathways [4]. Mutations altering these receptor genes may lead to abnormal protein formation, and any disturbances to these receptors functioning may lead to arrhythmias followed by heart failure condition like HCM or even sudden death.
Association of HCM with autosomal dominant mutation makes this disease a clear candidate for the molecular diagnosis through genetic screening. The best possible way to screen the genes in a cost-effective way for the protein-altering mutation is through whole exome sequencing, due to various private mutations being implicated. Whole exome sequencing may not only identify mutations in known candidate genes but also identify the mutations in other unknown genes which may have a potential role in the development of disease.
Materials and methods
A non-consanguineous family with multiple affected individuals was clinically evaluated at the Department of Cardiology, All India Institute of Medical Sciences, New Delhi, India. A total of 9 members in three generations were closely evaluated. Three affected (II-5, II-6, II-8) and three unaffected (I-2, II-3, III-2) individuals as evidenced by 2D echocardiography and ECG examination (Suppl Table 1) were selected for whole exome sequencing. All the age- and sex-matched controls were also subjected to clinical examination by 2D echocardiography and ECG examination. We obtained prior written informed consent as per the guidelines and with approval of the Institutional Ethics Committee of both participating institutes.
DNA was isolated using standard protocols from 3 mL intravenous blood sample. We used SureSelect Human All Exons Kit v5 (Agilent Technologies, USA) for the whole exome capture. Paired-end libraries were prepared following the manufacturer’s protocols (Agilent) after fragmentation (150–200 bp). Approximately 700 ng of the prepared DNA was incubated with biotinylated RNA capture baits to capture coded regions. Then, the captured fragments were sequenced in Illumina HiSeq2500 with 101 bp pair-end reads. We sequenced each of the libraries at over approx. 100 × coverage. All reads were aligned to the reference genome (UCSC Genome Browser hg19). Base quality-score recalibration and local realignment across the gold-standard variant and In-Del list was done, and variants were called. The variant list was then annotated and filtered for all non-synonymous variants, which were predicted deleterious by SIFT and Polyphen2. All the rare variants with a minor allele frequency (MAF) < 0.5% in dbSNP, 1000 genomes, or NHLBI ESP 6500 dataset were excluded. Finally, segregation analysis was performed on these subsets of variants (Suppl Fig. 1).
The prioritized variant was further validated using Sanger sequencing in all the family members and in well-characterized 100 clinically evaluated controls from the Indian population. For validation and screening of variant c556G>A (p. Glu186Lys) (rs28360451) identified in exon 6 of P2X7, we performed Sanger sequencing of exon 6 of this gene. Specific primers were designed flanking exon 6 of P2X7. The primer sequences were as follows: 5′-CCAAAGACCAAGCCAAGAAAC-3′ (forward) and 5′-CAGAAACCGTGGGAGACAATA-3′ (reverse). We performed standard PCR, and PCR amplicons were checked by gel electrophoresis and then sequenced. DNA sequences were examined using freely available software of Sequence Scanner Software v1.0 (Applied Biosystems). The mutation was confirmed in all three affected members which was absent in all unaffected members and 100 age- and sex-matched clinically evaluated controls (Suppl Table 3 and Suppl Annexure-A).
Phenotypic heterogeneity was addressed by filtering out variants in candidate genes individually. Similar filtering steps were applied as used for the identification of causative mutation except the segregation step. All mutations were checked for quality score and were evaluated for specific functionality of the candidate gene for HCM. A number of mutations carried in different genes were isolated and compared among the affected members to address heterogeneity. Further for the understanding of gene-gene interaction, we analyzed the gene identified in GeneMANIA with candidate genes to reveal any direct or indirect interaction among them. For this, all the candidate genes of HCM and gene identified in the exome sequencing were entered into the software. All interactions (physical, co-expression, co-localization, pathway, and genetic) were summarized and used for further evaluations.
Results
The HCM family had a history of sudden death, and three brothers were affected by the disease while the father died suddenly at the age of 58 years, with no evidence available for the cause of death. Close clinical examination revealed a phenotypic heterogeneity among the three affected members (Suppl Table 1). II-6 and II-5 have early onset of disease (< 30 years) with NYHA II and III respectively. With the symptoms of syncope, pre-syncope, and cardiac arrhythmias, ambulatory ECG (AECG) monitoring during normal routine work showed reduced heart rate (bradycardia) for the patient (II-6) who reported dizziness. Other patients’ AECG did not reveal any arrhythmia episodes. The youngest brother (II-8) with late onset of disease (mid-30s) had severe symptoms, classified under NYHA class III. II-5 died suddenly; however, the cause of death was not confirmed clinically and on pre-cautionary measures, the other two brothers (II-6 & II-8) were implanted with ICD. The mother (I-2) and eldest brother (II-3) were unaffected and the brother’s son (III-2) was also considered for genetic evaluations.
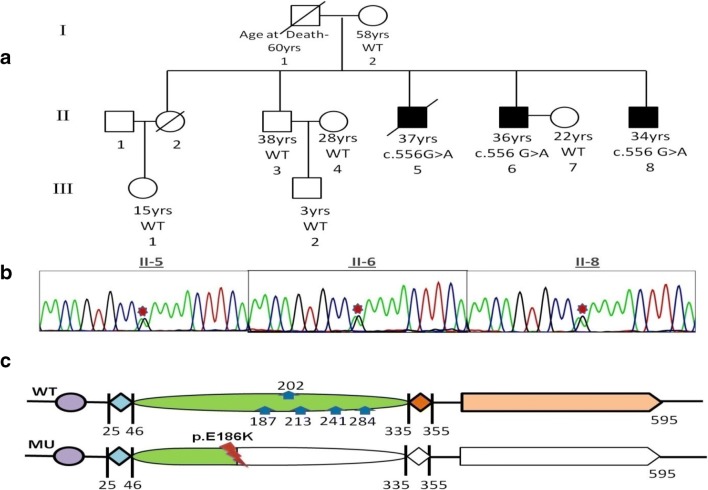
After the analysis of exome data, only one single heterozygous variant c556G>A (p. Glu186Lys) (rs28360451) in exon 6 of P2X7 gene segregated in all three affected members and was absent in unaffected members (Fig. 1a, c). This mutation was previously reported in a study related to affective mood disorder, but the patient was not evaluated for a heart condition [13]. This gene had not been previously reported in HCM patients. Further, we evaluated the allele frequency of the variation in the Exome Aggregation Consortium (ExAC) database, hosting over 61,486 exome sequences. The variant has a MAF of 0.00042 (7/16,512 chromosomes) in South Asian population and a MAF of 0.00011 (7/66,734 chromosomes) in European (non-Finnish) population. In controls, not a single mutated allele (A allele) was found, which suggests a complete absence of this mutation in healthy individuals. All the control samples were found carrying wild type genotype (GG). Absence of mutation (c556G>A in P2X7) in healthy controls and segregation of this mutation in affected members show rarity and deleterious nature of this mutation. A strong association between this mutation and HCM was supported by the fact that all the affected members had heterozygous condition (GA genotype), but the unaffected family members and 100 clinically evaluated controls all were found to be carrying wild type allele (G allele) at this locus. This suggests mutation was completely absent in the 100 clinically evaluated in-house controls as well which indicates that the mutation had a very low frequency in the Indian population. All the sequences of family members and controls were submitted to NCBI through Bankit (Accession Number- MK910589 - MK910696).
Fig. 1.
a Pedigree of the familial HCM in which affected are marked dark. b Sanger sequencing chromatogram of the P2X7 germline mutation (c.556G>A). c The plot of the resulting amino acid change in the P2X7 domain near N-linked glycosylation site 187 (blue arrows) shows loss of function (inactivation of protein) for mutant (MU) compared with wild type (WT)
To address the heterogeneity among the brothers having common causative mutation, we evaluated 24 HCM candidate genes individually for the affected members using all the filters as used earlier except the segregation step. The total numbers of such mutations in these candidate genes are 10, 12, and 16 for affected members II-6 (NYHA II), II-5 (NYHA III), and II-8 (NYHA III) respectively (Suppl Table 2). These mutations in candidate genes which were not segregated with the disease but present in patients may add to the effect of a causative mutation.
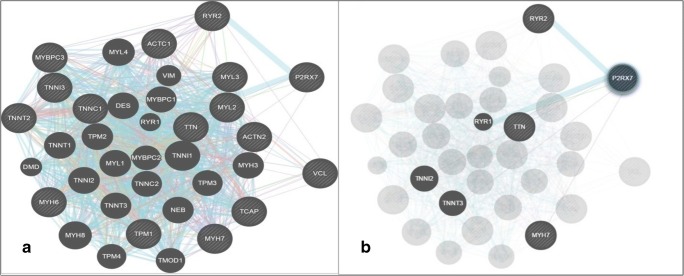
Further investigations on P2X7 gene shows that it had direct or indirect genetic interactions with candidate genes of HCM especially sarcomeric genes. The network involving physical, co-localization, co-expression, similar pathway, and genetic interactions specifies that P2X7 gene interacts with the majority of candidate genes (Fig. 2a.). When we isolated the P2X7 gene for any direct interactions, we found that there is inter-connection between MYH7, TNNI2, TTN, RYR2, and TNNT3 genes which are widely responsible for the hypertrophic cardiomyopathy (Fig. 2b). These interactions suggest that any disturbance in purinergic signaling may directly or indirectly affect these sarcomeric proteins leading to the malfunctioning of the heart system.
Fig. 2.
a Interaction networks (physical, co-expression, co-localization, pathway, and genetic) of the P2X7 gene with other HCM candidate genes identified for risk stratification with sarcomeric genes. bP2X7 direct network interaction with genes associated with HCM
Discussion
P2X7 or P2X7 (MIM 602566; Ref. Seq. accession number NC_000012.12) encodes for purinergic receptor P2X, ligand-gated ion channel 7 which is an ionotropic cell surface ATP receptor and involved in cell permeabilization [11]. P2X7 has been shown with significant associations with many other diseases related to the heart, brain, spinal cord, kidney etc. through different signaling pathways [6]. Assessment of the expression levels in Human Protein Atlas revealed that P2X7 is well expressed in cardiac myocytes. Studies show that the P2X7 mutation effects on two main functional properties, namely ATP-activated channel and ATP-induced dye uptake pore formation. Alterations in P2X7 gene is the causal factor of bone disorders, infectious disease, malignancies, and inflammatory and cardiovascular disorders [15].
In a study, HEK293 cells expressed with mutant receptor (E186K) for P2X7 gene were completely non-responsive to stimulations as compared with wild type (WT) receptor. For further understanding, an experiment shows no apparent dye uptake in cells expressing E186K as compared with WT. These effects showed that E186K led to complete loss of function [13]. Glu186 is located towards the C terminal end of α2 helix and is in close vicinity to ATP-binding site-forming residues (Gln187)(Fig. 1c) suggesting E186K may affect ATP binding, channel gating, or both leading to loss of function [13]. Lenertz et al. [8] reported role of the glycosylation in receptor function and found that residue N187 is critical for receptor trafficking which also supports that E186K affects the receptor function more prominently.
Glu186Lys mutation is extremely rare in the general population; according to the dataset, ESP6500, the MAF of the mutant allele in all ethnic groups, is not detected. As there is no previous constructive exome database for Indian population, we could not estimate allele frequency in the Indian population. In order to exclude the potential confounding factor due to the effect of ethnicity on the association, we studied age- and sex-matched controls (200 chromosomes) recruited from the same geographic region as patients which shows a complete absence of this conserved mutation. We also queried the Exome Aggregation Consortium (ExAC) database, a total 61,486 exome sequences depository which contains allele frequency data generated by a wide variety of large scale projects. It was found that, in the South Asian population, the c.556G>A (p. Glu186Lys) variant had a MAF of 0.00042 in 7/16,512 chromosomes. This is a little higher than the frequency observed in the European (non-Finnish) population (MAF = 0.00011) with 7/66,734 chromosomes. However, the ExAC dataset contains data from thousands of disease samples; therefore, the estimated frequency of this mutation is biased and not ideal for comparison with our HCM subjects but these frequency data suggest the involvement of this mutation in disease expression.
Involvement of P2X7 in heart function is not very well understood. As it is an ATP-gated cation selective channel, the mechanism by which ATP works are likely to be mediated through activation of different receptors of P2X family. The P2X7 expression in the heart was confirmed in different studies. In the sinoatrial node and atria of the adult heart, the presence of the P2X7 receptor has already been detected through immunohistochemistry, qPCR, and in situ hybridization. Within the P2 receptor family, P2X7 is the only member associated with NOD-like receptor (NLR) P3 inflammasome [11]. In a study, at the protein level, induced vs genetic P2X7 downmodulation had an opposite effect; induced had a decrease in NLRP3 protein expression whereas genetic was associated with a striking increase in NLRP3 expression [5]. Activation of the NLRP3 inflammasome in CNTg mice promotes myocardial inflammation and systolic dysfunction through the production of pro-inflammatory IL-1β leading to heart failure condition [2]. This could be one of the possible pathways through which genetically downmodulation of P2X7 leads to heart failure. Taken together, these initial studies suggest that the use of IL-1β or NLRP3 inhibition is a promising therapy for the treatment of this heart failure condition.
Other evidence supporting our finding has come from quite disparate studies. A study [17] which computationally prioritized cardiac channel genes associated with cardiomyopathy using machine learning approaches prioritized 26 genes, of which P2X7 is a prominent candidate. A similar kind of study identified P2X7 as an ion channel gene related to cardiomyopathy using a novel decision forest strategy [18]. An independent study also showed that Caveloin(-/-) mice showed the clinical phenotype of cardiomyopathy and associated over-expression of P2X7, the molecular physiology of which has not been established [1].
From all evidences, mutation (E186K) in P2X7 gene leading to the loss of function of this receptor could be the possible causative variant leading to heart failure in patients but the clinical heterogeneity among patients in the same family carrying the same mutation was addressed by other variations present in candidate genes. II-6 with the least number of mutations had mild severity (NYHA class II) than the other two having more mutations and classified under NYHA III. Delayed onsets of disease in II-8 can be explained with three mutations in the MYBPC3 gene. Previous studies [12] had reported that patients with MYBPC3 mutation had late onset but with 13 other mutations in candidate genes other than MYBPC3, II-6 had more severe symptoms after onset. II-5, who died suddenly, had two mutations each in TPM1 and RYR2. These two genes (TPM1 and RYR2) had been reported to be associated with sudden death [7, 16]. P2X7 as an ion-channel for the calcium signaling and responsible for the arrhythmias (reduced heart rate) [14], possibly which in combination with mutations in TPM1 and RYR2, makes it more fatal leading to sudden death.
In terms of gene-gene interaction of P2X7 gene with other sarcomeric genes, it is quite evident from the analysis that this gene interacts directly with the MYH7 and TTN. MYH7 and TTN, in most of cases, are found to be mutated in HCM patients, so any disturbances in their genetic product may lead to abnormal functioning of the heart. This suggests that ATP-ligated ion-channel P2X7 gene had a common mechanistic pathway with sarcomeric genes causing hypertrophy and leading to HCM.
If we summarize all the evidences, a single mutation in P2X7 gene segregates in all three patients and gene-gene interaction with other sarcomeric genes makes this gene a candidate for HCM. The family had a history of sudden death, and all three patients had common symptoms such as arrhythmias. P2X7 gene which is highly expressed in heart and is one of the candidate genes for cardiac arrhythmias shows a possible mechanistic insight for the disease progression leading to sudden death. NLPR3 inflammasome could be the possible mechanistic pathway target for this P2X7genetic abnormality. A recent review on the role of P2X7 as emerging therapeutic target states P2X7 receptor inhibition mediated through downregulation of NLRP3 decreases arrhythmia after myocardial infarction and prolongs cardiac survival [3]. The present study also implies on targeting P2X7/NLRP3 as a possible therapeutic target for the cardiomyopathy patients having a putative mutation in the P2X7 gene.
Conclusion
To the best of our knowledge, our exome study is the first to report the association of P2X7 gene in familial HCM in the Indian population which indicates the involvement of purinergic signaling in cardiomyocytes. Clinical heterogeneity can be understood by consideration of mutations in related genes as per disease condition. Our data warrant further research into deleterious mutations of the P2X7 gene for exploration and the feasibility of developing targeted therapies against HCM in individuals with P2X7 variants by compensating the loss of function effect caused by these variants.
Accession numbers
The accession numbers for the whole exome sequences reported in this paper are SRR1980932, SRR1980950, SRR1980978, SRR1980979, SRR1980980, and SRR1980981. The accession numbers for targeted sequences (Sanger sequencing) of exon 6 of the P2X7 gene reported in this paper are MK910589 - MK910696.
Electronic supplementary material
(DOCX 5778 kb)
Acknowledgments
The authors acknowledge CSIR-Institute of Genomics and Integrative Biology, New Delhi, for exome sequencing facility and patients and their family for participating in the study.
Funding
This work was supported by Department of Biotechnology (DBT), Government of India, India (BT/PR 5767/MED/12/563/2012) and ICMR Emeritus Medical Scientist Fellowship (ICMR/74/1/2016-Pers,EMS) to VRR.
Compliance with ethical standards
We obtained prior written informed consent as per the guidelines and with approval of the Institutional Ethics Committee of both participating institutes.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barth K, Pfleger C, Linge A, Sim J, Surprenant A, Steinbronn N, Strasser R, Kasper M. Increased P2X7R expression in atrial cardiomyocytes of caveolin-1 deficient mice. Histochem Cell Biol. 2010;134:31–38. doi: 10.1007/s00418-010-0716-8. [DOI] [PubMed] [Google Scholar]
- 2.Bracey NA, Beck PL, Muruve DA, Hirota SA, Guo J, Jabagi H, Wright JR, Jr, et al. The Nlrp3 inflammasome promotes myocardial dysfunction in structural cardiomyopathy through interleukin-1β. Exp Physiol. 2013;98(2):462–472. doi: 10.1113/expphysiol.2012.068338. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, He L, Li L, Chen L. The P2X7 purinergic receptor: an emerging therapeutic target in cardiovascular diseases. Clin Chim Acta. 2018;479:196–207. doi: 10.1016/j.cca.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 4.Erlinge D, Burnstock G. P2 receptors in cardiovascular regulation and disease. Purinerg Signal. 2008;4(1):1–20. doi: 10.1007/s11302-007-9078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franceschini A, Capece M, Chiozzi P, Falzoni S, Sanz JM, Sarti AC, Bonora M, Pinton P, Di Virgilio F. The P2X7 receptor directly interacts with the NLRP3 inflammasome scaffold protein. FASEB J. 2015;29(6):2450–2461. doi: 10.1096/fj.14-268714. [DOI] [PubMed] [Google Scholar]
- 6.Górecki DC. P2X7 purinoceptor as a therapeutic target in muscular dystrophies. Curr Opin Pharmacol. 2019;47:40–45. doi: 10.1016/j.coph.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Ingles J, Semsarian C. Sudden cardiac death in the young: a clinical genetic approach. Intern Med J. 2007;37:32–37. doi: 10.1111/j.1445-5994.2006.01241.x. [DOI] [PubMed] [Google Scholar]
- 8.Lenertz LY, Wang Z, Guadarrama A, Hill LM, Gavala ML, Bertics PJ. Mutation of putative N-linked glycosylation sites on the human nucleotide receptor P2X7 reveals a key residue important for receptor function. Biochemistry. 2010;49:4611–4619. doi: 10.1021/bi902083n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maron B. Hypertrophic cardiomyopathy. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 10.Maron B. Hypertrophic cardiomyopathy: an important global disease. Am J Med. 2004;116:63–65. doi: 10.1016/j.amjmed.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Martinez CM. P2X7 receptor in cardiovascular disease: the heart side. Clin Exp Pharmacol Physiol. 2019;46:513–526. doi: 10.1111/1440-1681.13079. [DOI] [PubMed] [Google Scholar]
- 12.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, Kristinsson A, Roberts R, Sole M, Maron BJ, Seidman JG, Seidman CE. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 13.Roger S, Mei Z, Baldwin J, Dong L, Bradley H, Baldwin SA, Surprenant A, Jiang LH. Single nucleotide polymorphisms that were identified in affective mood disorders affect ATP-activated P2X7 receptor functions. J Psychiatr Res. 2010;44:347–355. doi: 10.1016/j.jpsychires.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Skaper SD, Debetto P, Giusti P. P2X7 receptors in neurological and cardiovascular disorders. Cardiovasc Psychiatry Neurol. 2009;861324:1–13. doi: 10.1155/2009/861324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sluyter R, Stokes L. Significance of P2X7 receptor variants to human health and disease. Recent Pat DNA Gene Seq. 2011;5:41–54. doi: 10.2174/187221511794839219. [DOI] [PubMed] [Google Scholar]
- 16.VanDriest SL, Will ML, Atkins DL, Ackerman MJ. A novel TPM1 mutation in a family with hypertrophic cardiomyopathy and sudden cardiac death in childhood. Am J Cardiol. 2000;90:1123–1127. doi: 10.1016/S0002-9149(02)02780-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Shao D, Zhang S, Wang Y. Prioritization of candidate disease genes by enlarging the seed set and fusing information of the network topology and gene expression. Mol BioSyst. 2014;10:1400–1408. doi: 10.1039/C3MB70588A. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Wang G, Feng J, Zhang L, Li J. Identifying ion channel genes related to cardiomyopathy using a novel decision forest strategy. Mol BioSyst. 2014;10(9):2407–2414. doi: 10.1039/C4MB00193A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 5778 kb)