Abstract
Aims
A better understanding of the expression of cancer/testis antigens (CTAs) in breast cancer might enable the identification of new immunotherapy options, especially for triple‐negative (TN) tumours, which lack expression of the conventional therapeutic targets oestrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2. The aim of this study was to quantify the expression of MAGE‐A and NY‐ESO‐1 CTAs in breast cancer, and relate this to known clinicopathological parameters.
Methods and results
We surveyed MAGE‐A and NY‐ESO‐1 expression in an unselected cohort of 367 breast tumours (of which 65 were TN), with accompanying clinical follow‐up data, by using immunohistochemical analysis of tissue microarrays. Relevant to their potential as vaccine targets in breast cancer, MAGE‐A was expressed in 13% of cases, and NY‐ESO‐1 in 3.8%, with the majority of tumours showing fairly homogeneous staining within individual tissue cores (~85% of cases with staining in >75% of tumour cells). Most NY‐ESO‐1‐positive cases also expressed MAGE‐A (P = 2.06 × 10−9), and both were strongly associated with the TN phenotype (P < 0.0001), with the most proliferative and poorly differentiated cases, in paticular, showing genomic instability. This was characterised by coexpression of c‐Kit and TTK, and overexpression of p53.
Conclusions
MAGE‐A and NY‐ESO‐1 are frequently expressed in TN breast cancer (~47% and 17% of TN cases, respectively), suggesting that targeting them could be feasible in this patient group. Expression is reasonably homogeneous in positive cases, suggesting that immunohistochemical analysis of tissue biopsies would be a reliable companion biomarker.
Keywords: cancer/testis antigens, MAGE‐A, NY‐ESO‐1, TNBC
Introduction
Breast cancer is the most frequently diagnosed cancer in women. It is also a predominant cause of mortality, and the global burden of breast cancer rises every year.1 Approximately 10–20% of breast tumours belong to the triple‐negative (TN) subtype, defined by lack of expression of oestrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), which are companion biomarkers and/or therapeutic targets of hormone and anti‐HER2 therapy. Cytotoxic chemotherapy is the standard‐of‐care therapy, and is very effective in a subset of patients, but responses are not durable in all cases,2 which ultimately relapse with the shortest progression‐free and overall survival of all breast cancers.3 Accordingly, efforts are ongoing to identify predictive markers of chemotherapy efficacy in TN breast cancer (TNBC), as well as alternative treatments for patients who are most likely to develop resistance.
High‐resolution genetic and cellular profiling of TNBCs has revealed subgroups characterised by chromosomal instability (CIN), DNA repair defects, and androgen receptor (AR) signalling.4, 5, 6 The immune microenvironment is also a strong determinant of both the molecular profile and the clinical outcome in TNBC patients, with the poorest prognostic group being characterised by stromal restriction of tumour‐infiltrating lymphocytes (TILs), or a general deficiency of TILs in tumour and stromal compartments.7, 8, 9 Therapeutic strategies retargeting the immune system to cancer cells have proven successful in liquid cancers10, 11, 12, 13 as well as in a few solid tumours.14, 15, 16, 17, 18 This is achieved by manipulating the endogenous immune response19, 20, 21, 22 and/or bypassing it altogether by genetically reprogramming T cells with adoptive transfer of chimeric antigen receptor (CAR) T cells, which are reprogrammed to target tumour antigens.10, 11, 12, 13, 14, 15, 16, 17, 18 Therapeutic cancer vaccines may also be useful as part of regimens aimed at boosting antitumour immunity. Cancer vaccines and CAR T cells can be directed against proteins expressed by many cancers (shared antigens) or neoantigens encoded by mutant transcripts.
Cancer/testis antigens (CTAs) belong to a group of proteins that are expressed in the developing embryo, are restricted to the testis in the adult, and are aberrantly re‐expressed in malignancy, particularly high‐grade and advanced‐stage tumours, including TNBC.23, 24, 25, 26, 27, 28 Members of the melanoma‐associated antigen (MAGE) family and New York Esophageal Squamous Cell Carcinoma‐1 (NY‐ESO‐1) are among the CTAs being actively investigated as cancer immunotherapy targets. They have been shown to evoke spontaneous cytotoxic T‐cell responses in melanoma, oesophageal carcinoma, bladder cancer, and non‐small‐cell lung carcinoma.29, 30 Several studies have evaluated MAGE‐A and NY‐ESO‐1 expression in breast cancer, with variable reports on expression frequency.23, 31, 32, 33, 34 Their therapeutic potential for immunotherapy and the staining homogeneity observed in diverse cohorts led us to investigate the expression of MAGE‐A and NY‐ESO‐1 in our historical cohort of breast cancer patients.
Materials and methods
Tissue Microarrays (TMAs) and Histopathology Review
This study made use of the Queensland Follow‐Up (QFU) cohort, a resource comprising formalin‐fixed paraffin‐embedded breast tumours from patients undergoing surgical resection at the Royal Brisbane Women's Hospital (RBWH) between 1987 and 1994, with accompanying long‐term (up to 30 years) clinical follow‐up information.35, 36, 37, 38, 39 Ethical approval from the Human Research Ethics Committees of the RBWH and the University of Queensland was obtained prior to the commencement of the study. Tumours were sampled in duplicate on TMAs for biomarker studies, and haematoxylin and eosin‐stained whole tissue sections were available for histopathological review.
The review was conducted by an experienced breast pathologist (S.R.L.; parameters included histological subtype, grade, and the presence of lymphovascular invasion and lymphocytic infiltrate; Table 1). We also considered patient age, tumour size, and lymph node (LN) status (whether disease had spread to the LNs at the time of surgery), extracted from diagnostic pathology reports. We selected a range of breast tumour biomarkers implicated in the prognosis and/or pathobiology of breast cancer: (i) hormone receptors, i.e. ER and PR; (ii) Ki67, a marker of proliferation; and (iii) a range of biomarkers implicated in TNBC, including markers of basal/myoepithelial‐like phenotype [epidermal growth factor receptor (EGFR) and the high molecular weight cytokeratins (CKs) CK5/6 and CK14], vimentin (mesenchymal marker), androgen receptor (AR) (can confer luminal‐like intracellular signalling and a luminal‐like phenotype in a proportion of TNBCs with a favourable outcome),40, 41 c‐Kit (associated with primitive, progenitor states42), p53 (overexpression is associated with genomic instability),43 and mitosis‐independent expression of the dual‐specificity protein kinase TTK (implicated in chromosomal instability and poor clinical outcome.35
Table 1.
Association of MAGE‐A and NY‐ESO‐1 expression with histopathological parameters in breast cancer
| MAGE‐A staining: | n | % of cases | P‐value | NY‐ESO‐1 staining: | n | % of cases | P‐valuea | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | MAGE‐A‐positive | MAGE‐A‐negative | Negative | Positive | Total | NY‐ESO‐1‐positive | NY‐ESO‐negative | Negative | Positive | ||||
| Age (years) | Age (years) | ||||||||||||
| >40 | 273 | 237 | 36 | 87 | 13 | NS | >40 | 300 | 287 | 13 | 96 | 4 | NS |
| ≤40 | 35 | 27 | 8 | 77 | 23 | ≤40 | 38 | 38 | 0 | 100 | 0 | ||
| n | 308 | n | 338 | ||||||||||
| Grade | Grade | ||||||||||||
| G1 | 46 | 46 | 0 | 100 | 0 | <0.0001 | G1 | 50 | 50 | 0 | 100 | 0 | 0.023 |
| G2 | 176 | 157 | 19 | 89 | 11 | G2 | 170 | 166 | 4 | 98 | 2 | ||
| G3 | 140 | 106 | 34 | 76 | 24 | G3 | 134 | 124 | 10 | 93 | 7 | ||
| n | 362 | n | 354 | ||||||||||
| Mitotic score | Mitotic score | ||||||||||||
| 1 | 43 | 43 | 0 | 100 | 0 | <0.0001 | 1 | 199 | 196 | 3 | 98 | 2 | 0.0255 |
| 2 | 154 | 137 | 17 | 89 | 11 | 2 | 47 | 44 | 3 | 94 | 6 | ||
| 3 | 126 | 95 | 31 | 75 | 25 | 3 | 107 | 99 | 8 | 93 | 7 | ||
| n | 323 | n | 353 | ||||||||||
| Histological type | Histological type | ||||||||||||
| Ductal NOS | 188 | 161 | 27 | 86 | 14 | NS | Ductal NOS | 207 | 198 | 9 | 96 | 4 | NS |
| Lobular/variants | 37 | 35 | 2 | 95 | 5 | Lobular/variants | 42 | 42 | 0 | 100 | 0 | ||
| Mixed ductolobular | 30 | 26 | 4 | 87 | 13 | Mixed ductolobular | 32 | 31 | 1 | 97 | 3 | ||
| Mixed | 30 | 25 | 5 | 83 | 17 | Mixed ductolobular | 31 | 28 | 3 | 90 | 10 | ||
| Metaplastic | 14 | 9 | 5 | 64 | 36 | Metaplastic | 15 | 15 | 0 | 100 | 0 | ||
| Special types | 24 | 20 | 4 | 83 | 17 | Special types | 27 | 26 | 1 | 96 | 4 | ||
| n | 323 | n | 354 | ||||||||||
| Lymph node status | Lymph node status | ||||||||||||
| Negative | 97 | 84 | 13 | 87 | 13 | NS | Negative | 102 | 100 | 2 | 98 | 2 | NS |
| Positive | 82 | 69 | 13 | 84 | 16 | Positive | 88 | 83 | 5 | 94 | 6 | ||
| n | 179 | n | 190 | ||||||||||
| Tumour size (mm) | Tumour size (mm) | ||||||||||||
| < 20 | 140 | 125 | 15 | 89 | 11 | NS | <20 | 152 | 147 | 5 | 97 | 3 | NS |
| 20–50 | 121 | 98 | 23 | 81 | 19 | 20–50 | 132 | 127 | 5 | 96 | 4 | ||
| >50 | 21 | 18 | 3 | 86 | 14 | >50 | 22 | 20 | 2 | 91 | 9 | ||
| n | 282 | n | 306 | ||||||||||
| Lymphovascular invasion | Lymphovascular invasion | ||||||||||||
| Absent | 236 | 202 | 34 | 86 | 14 | NS | Absent | 262 | 252 | 10 | 96 | 4 | NS |
| Present | 87 | 73 | 14 | 84 | 16 | Present | 92 | 88 | 4 | 96 | 4 | ||
| n | 323 | n | 354 | ||||||||||
| Lymphocytic infiltrate | Lymphocytic infiltrate | ||||||||||||
| Absent | 108 | 105 | 3 | 97 | 3 | <0.0001 | Absent | 123 | 123 | 0 | 100 | 0 | NS |
| Mild | 145 | 117 | 28 | 81 | 19 | Mild | 157 | 150 | 7 | 96 | 4 | ||
| Moderate‐severe | 57 | 42 | 15 | 74 | 26 | Moderate‐severe | 61 | 55 | 6 | 90 | 10 | ||
| n | 310 | n | 341 | ||||||||||
| Central scarring/fibrosis | Central scarring/fibrosis | ||||||||||||
| Absent | 289 | 248 | 41 | 86 | 14 | NS | Absent | 319 | 306 | 13 | 96 | 4 | NS |
| Present | 34 | 27 | 7 | 79 | 21 | Present | 35 | 34 | 1 | 97 | 3 | ||
| n | 323 | n | 354 | ||||||||||
| Tumour border | Tumour border | ||||||||||||
| Infiltrative | 281 | 247 | 34 | 88 | 12 | 0.0023 | infiltrative | 310 | 299 | 11 | 96 | 4 | NS |
| Pushing | 45 | 31 | 14 | 69 | 31 | Pushing | 44 | 41 | 3 | 93 | 7 | ||
| n | 326 | n | 354 | ||||||||||
| HR status | Hormone receptor (HR) status | ||||||||||||
| Negative | 77 | 46 | 31 | 59.7 | 40.3 | <0.0001 | Negative | 68 | 66 | 2 | 97.1 | 2.9 | <0.0001 |
| Positive | 244 | 227 | 17 | 93.0 | 7.0 | Positive | 282 | 270 | 12 | 95.7 | 4.3 | ||
| n | 321 | n | 350 | ||||||||||
| ER status | ER status | ||||||||||||
| Positive | 241 | 225 | 16 | 93 | 7 | <0.0001 | Positive | 270 | 268 | 2 | 99 | 1 | <0.0001 |
| Negative | 83 | 51 | 32 | 61 | 39 | Negative | 85 | 73 | 12 | 86 | 14 | ||
| n | 324 | n | 355 | ||||||||||
| HER2 status (CISH) | HER2 status (CISH) | ||||||||||||
| Negative | 286 | 244 | 42 | 85 | 15 | NS | Negative | 314 | 301 | 13 | 96 | 4 | ns |
| Positive | 30 | 27 | 3 | 90 | 10 | Positive | 31 | 31 | 0 | 100 | 0 | ||
| n | 316 | n | 345 | ||||||||||
| Ki67 | Ki67 | ||||||||||||
| Negative | 192 | 177 | 15 | 92 | 8 | <0.0001 | Negative | 192 | 188 | 4 | 98 | 2 | 0.0386 |
| Positive | 124 | 93 | 31 | 75 | 25 | Positive | 124 | 115 | 9 | 93 | 7 | ||
| n | 316 | n | 316 | ||||||||||
| Basal marker: CK14, CK5, EGFR | Basal marker: CK14, CK5. EGFR | ||||||||||||
| Negative | 216 | 199 | 17 | 92 | 8 | <0.0001 | Negative | 236 | 235 | 1 | 100 | 0 | <0.0001 |
| Positive | 87 | 58 | 29 | 67 | 33 | Positive | 91 | 78 | 13 | 86 | 14 | ||
| n | 303 | n | 327 | ||||||||||
| Vimentin | Vimentin | ||||||||||||
| Negative | 284 | 253 | 31 | 89 | 11 | <0.0001 | Negative | 316 | 308 | 8 | 97 | 3 | 0.0016 |
| Positive | 38 | 21 | 17 | 55 | 45 | Positive | 38 | 32 | 6 | 84 | 16 | ||
| n | 322 | n | 354 | ||||||||||
| Androgen receptor | Androgen receptor | ||||||||||||
| Negative | 35 | 19 | 16 | 54 | 46 | <0.0001 | Negative | 38 | 30 | 8 | 79 | 21 | <0.0001 |
| Positive | 278 | 248 | 30 | 89 | 11 | Positive | 305 | 300 | 5 | 98 | 2 | ||
| n | 313 | n | 343 | ||||||||||
CISH, chromogenic in‐situ hybridisation; CK, cytokeratin; EGFR, epidermal growth factor receptor; ER, oestrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; MAGE, melanoma‐associated antigen; NOS, not otherwise specified; NS, not significant; NY‐ESO‐1, New York Esophageal Squamous Cell Carcinoma‐1. st.
Chi‐square test or Fisher's exact test.
Immunohistochemical (IHC) Analysis
Analysis of the CTAs were performed by staining the QFU TMA slides with antibodies against MAGE‐A (Santa Cruz Biotechnology, Santa Cruz, CA, USA; sc‐20034, 1:500 dilution), NY‐ESO‐1 (Santa Cruz Biotechnology; sc‐53869, 1:30 dilution), p53 (Dako, Santa Clara, CA, USA; M7001, 1:150 dilution), c‐Kit (Dako; A4502, 1:1000 dilution), TTK (Abcam, Cambridge, UK; ab11108, 1:100 dilution), HER2 (Dako; A0485, 1:200 dilution), ER (Novocastra, Newcastle upon Tyne, UK; NCL‐L‐ER‐6F11, 1:100 dilution), PR (Novocastra; NCL‐L‐PGR‐312, 1:200 dilution), vimentin (Dako; M0725, 1:400 dilution), AR (Dako; M3562, 1:50 dilution) and Ki67 (Dako; M724001‐2, 1:200 dilution) with the MACH 1 Universal HRP‐Polymer (Biocare Medical; (Pacheco, CA, USA; Cat. no. M1U539 G, L10) or Vectastain Universal ABC (Vector Laboratories, Burlingame, CA, USA) kit. Slides were scanned with the Aperio ScanScope T2 digital system (Buffalo Grove, IL, USA), and core images were then segmented into individual images for scoring (Aperio Spectrum TMA module).
Samples were assessed in a blinded manner by two observers (A.P. and P.K.dC). For MAGE‐A and NY‐ESO‐1, scoring was based purely on intensity, owing to its homogeneity of staining in the tissue cores. A score in a range of 0–3 (0 = negative; 1 = weak; 2 = moderate; 3 = strong) was assigned for each cellular compartment—cytoplasm and nucleus. However, the majority of the cases (65%) expressed CTAs homogeneously throughout the tumour compartment of duplicate cores, and there were no obvious subcellular expression patterns. Hence, cytoplasmic and nuclear component scoring were not considered separately for further analysis. Tumour cell percentage staining was subsequently stratified as either >75% or <75%, as this adequately described the positive cases. IHC analysis was performed for the markers, the results for some of which have been published previously.35, 36, 38, 39 The cut‐off value for HER2 positivity was based on a silver in‐situ hybridisation (SISH) score of >6 and an IHC score of 3+ if SISH was unsuccessful. Once HER2 positivity had been determined, ER status and PR status were examined; staining of ≥1% of the tumour cell nuclei was considered to be positive. After scoring for ER positivity if the sample was negative for both ER/PR and HER2, it was assigned to the TN subtype. For distinction between TN, basal‐like, and non‐basal, if ≥1% of tumour cells were positive for either EGFR or CK14 or CK5/6, the tumour was considered to be TN basal‐like. However, with relevance to the clinical context, TN/basal‐like is not a clinically defined subtype, and we therefore assigned any tumours that fell into the TN category to the TN group.
Once HER2 status and ER status had been determined, Ki67 expression was scored high if staining was observed in 10% of the tumour cell nuclei, and low if it was observed in <10% of the tumour cell nuclei. Biomarkers such as TTK and c‐Kit were scored purely on intensity, owing to the homogeneity of staining in the tissue cores. A score in a range of 0–3 (0 = negative; 1 = weak; 2 = moderate; 3 = strong) was assigned. Scoring for p53, vimentin and AR was based on an IHC score, which was derived by multiplying the intensity and percentage of the tumour cell staining. It was further stratified into a binary scoring system, i.e. 0–1 (<60 = 0/negative and >60 = 1/positive for p53; >0 = 1/positive for AR and vimentin).
Statistical analysis was performed with prism (v7). Associations between MAGE‐A, NY‐ESO‐1 and clinicopathological parameters were evaluated with the chi‐square test and Fisher's exact test. Relationships with breast cancer‐specific survival were investigated with Kaplan–Meier analysis, with log‐rank tests being used to assess significance. P‐values of <0.05 were considered to be significant.
Results
Cohort Demographics and Clinicopathology
Among the 367 cases, the median patient age at diagnosis was 59 years, the median follow‐up was 5.2 years, and the median follow‐up of patients who were alive was 21.5 years. These cases were collected between 1987 and 1994. Most of the cases were grade 2 (49%), and LN status was available for 56% of the cohort (27% LN‐positive; 29% LN‐negative) (Figure 1A). Invasive ductal carcinoma (IDC) was the major histological subtype (58%), followed by lobular variants (12%), mixed histologies (ductolobular, 9%; others collectively, 9%), metaplastic (5%), and other special types (collectively, 7%; Figure 1A). IHC staining showed that 75% of the cohort were ER‐positive, and SISH analysis identified ERBB2 amplification in 10% of cases.
Figure 1.
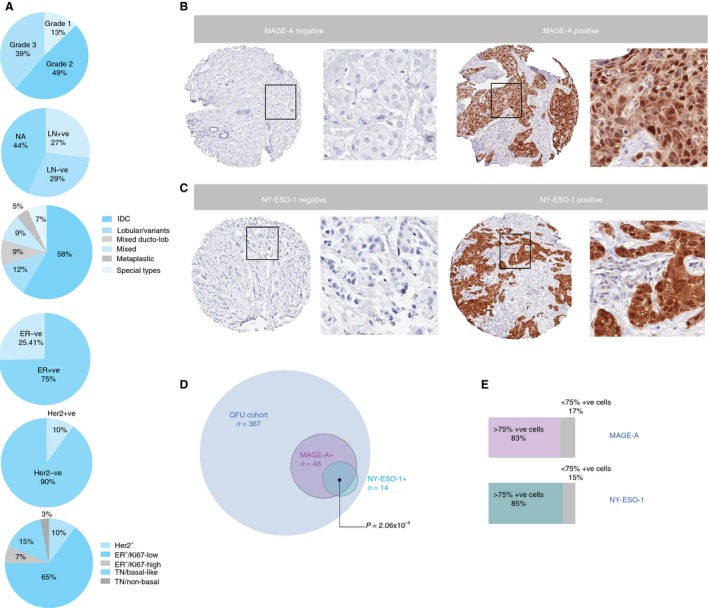
A, Distribution of the Queensland Follow‐Up patient population according to: grade, lymph node involvement, histological subtype, oestrogen receptor positivity, HER2 status, and various prognostic subtypes. B,C, Representative images of MAGE‐A and NY‐ESO‐1 staining on breast tumour tissue microarray cores, shown at low and high magnification. D, Concomitant expression of MAGE‐A and NY‐ESO‐1. E, Proportion of MAGE‐A‐positive and NY‐ESO‐1‐positive tumours expressing the respective antigens in >75% of cells within duplicate cores.
Cases were also categorised according to their expression of a panel of prognostic biomarkers: ER, PR, HER2, Ki67, and expression of EGFR and/or high molecular weight cytokeratins (CK5/6 and CK14), which are associated with a basal‐like phenotype. According to this scheme,35 the majority of the cohort was categorised as ER‐positive/Ki67‐low (65%), followed by TN/basal‐like (15%), HER2‐positive (10%), ER‐positive/Ki67‐high (7%) and TN/non‐basal (3%) (Figure 1A).
Expression of MAGE‐A and NY‐ESO‐1 in Invasive Breast Cancer
MAGE‐A and NY‐ESO‐1 showed homogeneous staining, with positivity in both cytoplasmic and nuclear tumour cell compartments in 13% (n = 48) and 3.8% (n = 14) of cases, respectively (Figure 1B–D). CTA staining was relatively homogeneous, with >75% of tumour cells being stained in the majority of the MAGE‐A‐positive (83%) and NY‐ESO‐1‐positive (85%) cases (Figure 1E). This criterion was employed as a threshold to note homogeneity, and not as a cut‐off for positivity. Interestingly, 12 of the 14 NY‐ESO‐1‐positive cases coexpressed MAGE‐A (Figure 1D; P = 2.06 × 10−9). Analysis of clinicopathological parameters showed that expression of both CTAs was associated with Ki67 expression and grade (driven mostly by the mitotic score component; Table 1). This was underpinned by strong enrichment of expression in TN tumours (86% of which were TN/basal‐like for MAGE‐A, and all of which were TN/basal‐like for NY‐ESO‐1 (Figure 2Ai,ii). Taking advantage of data generated as part of previous QFU cohort studies,35, 36, 37, 38, 39 we further examined the phenotypic features of MAGE‐A‐expressing and NY‐ESO‐1‐expressing TNBCs, and found positive associations with c‐Kit, p53, and mitosis‐independent TTK expression (Figure 2B). The CTAs (particularly NY‐ESO‐1) were also associated with expression of vimentin (a marker of mesenchymal differentiation), and were inversely associated with AR expression (Table 1), although these associations did not reach statistical significance within the TN group (not shown).
Figure 2.
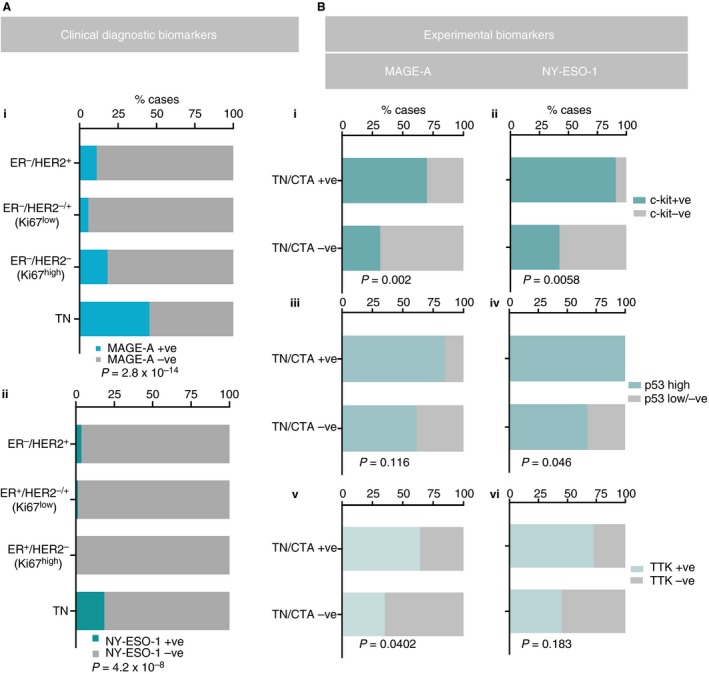
A,B, Chi‐square or Fisher's exact test analysis of associations between expression of MAGE‐A/NY‐ESO‐1 and clinical diagnostic or experimental biomarkers (c‐Kit, p53, and TTK1).
Expression of MAGE‐A and NY‐ESO‐1 in Breast Cancer is Not Associated With Survival
The median overall survival time of all cases in this present study was 13.6 years. According to Kaplan–Meier survival analysis, MAGE‐A‐positive cases within the whole cohort showed a 40% decrease in breast cancer‐specific survival (BCSS) at 25 years (Figure 3A), and MAGE‐A‐positive cases within the TN group showed a 35% decrease in BCSS at 5 years (Figure 3C), which was not significantly different from the MAGE‐A‐negative cases. NY‐ESO‐1‐positive cases showed a trend towards a poorer outcome (Figure 3B), but there was no difference after accounting for TN status (Figure 3D).
Figure 3.
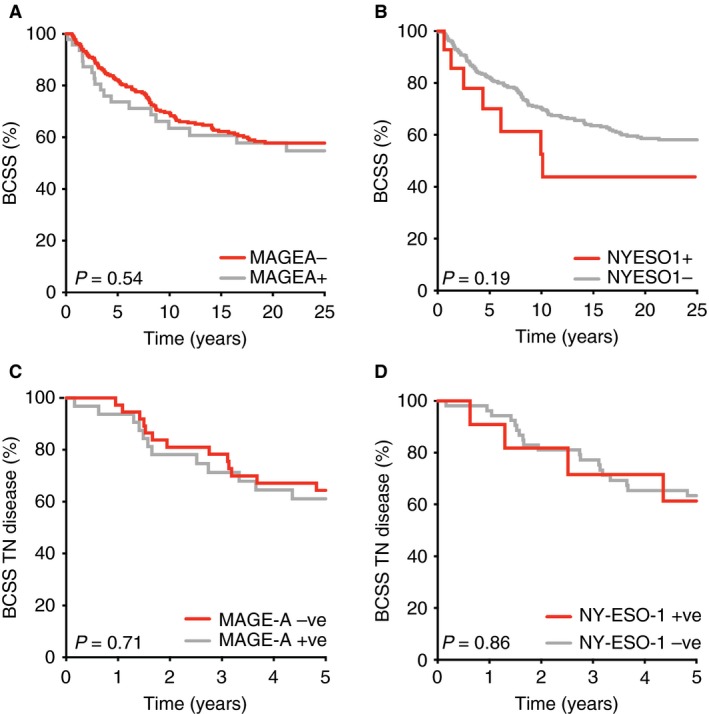
Kaplan–Meier survival analysis of the whole Queensland Follow‐Up cohort (A,B) or triple‐negative breast cancer (TNBC) cases (C,D) according to MAGE‐A and NY‐ESO‐1 expression. For TNBC, the analysis focuses on the first 5 years after diagnosis, which is the period that is most determinant of long‐term outcome. [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
We investigated the expression of MAGE‐A and NY‐ESO‐1 in invasive breast cancer, and found prevalent expression within the TN group. Previous reports have shown variable expression of MAGE‐A and NY‐ESO‐1, ranging between 17% and 74%, and between 2% and 40%, respectively,31, 44 and the expression levels of both were higher in TNBC,31, 44, 45, 46, 47, 48 which corroborates our findings. Taking advantage of the large pre‐existing dataset on experimental biomarkers in the QFU cohort, we found that the CTAs are expressed most prevalently in the TN tumours showing salient features of poor differentiation/primitive phenotype, proliferation, and genomic instability. This was indicated by coexpression of c‐Kit and TTK, and overexpression of p53. In addition, we found that these CTAs were expressed concomitantly and in the majority of the cells within a tumour, suggesting that they have favourable features as cancer vaccine targets. Our study strengthens the rationale for targeting CTAs to broaden the therapeutic options for TNBC.
Mutations in p53 can result in a stable non‐functional protein that accumulates in the nucleus, and that gives rise to an IHC phenotype mimicking overexpression;43, 49 thus, p53 overexpression in cancer is a surrogate for functional abrogation. CIN and aneuploidy are interrelated and are crucial hallmarks of cancer, which is partly contributed to by inadequacy of p53.50, 51, 52, 53, 54, 55 This promotes aberrant DNA damage repair and augments mutagenesis.56 Through association with p53 coexpression, our findings imply that NY‐ESO‐1‐positive tumours reflect these chromosomal abnormalities, and have a proliferative advantage. However, we observed no association between MAGE‐A and p53, which, perhaps, could be attributed to MAGE‐A inhibiting its function;57 one possible suggested mechanism is blocking of the interaction between p53 and chromatin, thus making p53 unable to regulate tumour cell proliferation and apoptosis.57, 58 Furthermore, we found a strong association between MAGE‐A expression and mitosis‐independent expression of the spindle assembly checkpoint protein TTK, which is crucial for chromosomal alignment and centrosome duplication, and is a marker for CIN in TNBC.35, 59, 60
Re‐expression of CTAs in cancer is thought to give cancer cells stem cell‐like properties.61 Interestingly, we found striking coexpression of both CTAs with the mammary luminal progenitor marker c‐Kit (Figure 2Bi,ii), and enrichment with vimentin (Table 1), suggesting that these tumours are in relatively primitive states of differentiation.62 Basal‐like breast cancers often show luminal progenitor‐like phenotypes, and may even originate from this cell type in the premalignant breast.42 Therefore, this striking coexpression could be part of the association with the basal‐like phenotype (Table 1). This strong correlation might also indicate a programmed state of dedifferentiation involving multiple stem cell markers, whereby a demethylation programme drives the expression of these CTAs, as is evident in ovarian and colon cancer.63, 64
If cancer vaccine or CAR T‐cell therapies are to be implemented for treatment in certain cases, efficacy would be determined, in part, by antigen abundance and heterogeneity. An extensive IHC analysis of the expression of eight CTAs in 454 IDCs revealed frequent coexpression in ER‐positive as compared with ER‐negative tumours,47 consistent with our findings on MAGE‐A and NY‐ESO‐1 coexpression. This raises the possibility that therapies simultaneously targeting multiple CTAs may elicit more efficient antitumour responses than single‐antigen approaches. In terms of expression heterogeneity, we found that the majority of cases expressed MAGE‐A and NY‐ESO‐1 in >75% of tumour cells within duplicate cores, which is also a potentially favourable feature in terms of therapeutic use. To maximise the efficacy of vaccination‐based strategies, it would be informative to analyse the distribution of multiple ‘actionable’ CTAs within individual tumours in a larger cohort, in order to identify combinations that could achieve the greatest breadth of coverage.
Immunotherapy is an attractive strategy to target TNBC, especially in patients with minimal residual disease after neoadjuvant chemotherapy, because of their statistically poor prognosis.46, 48 Analysing MAGE‐A and NY‐ESO‐1 expression in post‐neoadjuvant chemotherapy surgical samples may therefore aid in recognising patients who are suitable for vaccination strategies. Interestingly, adoptive T‐cell therapy targeting MAGE‐A3 has shown promise in the metastatic setting,65 and another trial is ongoing (Identifier no. NCT02111850).66 We hypothesise that CTAs may be expressed more frequently in the chemoresistant cells selected after neoadjuvant therapy, owing to their association with features that promote clonal selection (primitive phenotype, high proliferation, and CIN). Features of high proliferation, evasion of apoptosis, and a primitive/stem‐like state, which is considered to be an epithelial–mesenchymal transition state, perhaps confer chemoresistance to these cells, as is evident in multiple myeloma.67
Finally, our study was performed on a small number of TN tumours, so our findings are worth following up in a larger cohort. Nonetheless, given the promising benefits of immunotherapy for this group of patients with currently limited interventional strategies, it provides the rationale for targeting CTAs in TNBC.
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
A. Raghavendra, J. M. Saunus and S. R. Lakhani designed the study. P. Kalita‐de Croft, A. Raghavendra, A. C. Vargas and C. E. Smart performed the research. P. Kalita‐de Croft analysed the data and wrote the manuscript. J. M. Saunus, P.T. Simpson and S. R. Lakhani contributed to writing and editing of the manuscript.
Acknowledgements
We are thankful to Mrs Lynne Reid for technical assistance. We also acknowledge the Brisbane Breast Bank, which facilitated QFU cohort assembly, TMA construction, and cohort characterisation. This work was funded by the Australian National Health and Medical Research Council (NHMRC program grant, APP1017028), a Ludwig training fellowship to A. C. Vargas, and an Australian Government Endeavour Award to A. Raghavendra.
Raghavendra A, Kalita‐de Croft P, Vargas A C, Smart C E, Simpson P T, Saunus J M & Lakhani S R (2018) Histopathology 73, 68–80. 10.1111/his.13498 Expression of MAGE‐A and NY‐ESO‐1 cancer/testis antigens is enriched in triple‐negative invasive breast cancers
A.R. and P.K.C. contributed equally to this work.
References
- 1. Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol. Biomarkers Prev. 2017; 26; 444–457. [DOI] [PubMed] [Google Scholar]
- 2. Cameron D, Brown J, Dent R et al Adjuvant bevacizumab‐containing therapy in triple‐negative breast cancer (BEATRICE): primary results of a randomised, phase 3 trial. Lancet Oncol. 2013; 14; 933–942. [DOI] [PubMed] [Google Scholar]
- 3. Costa RLB, Gradishar WJ. Triple‐negative breast cancer: current practice and future directions. J. Oncol. Pract. 2017; 13; 301–303. [DOI] [PubMed] [Google Scholar]
- 4. Dannenfelser R, Nome M, Tahiri A et al Data‐driven analysis of immune infiltrate in a large cohort of breast cancer and its association with disease progression, ER activity, and genomic complexity. Oncotarget 2017; 8; 57121–57133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burstein MD, Tsimelzon A, Poage GM et al Comprehensive genomic analysis identifies novel subtypes and targets of triple‐negative breast cancer. Clin. Cancer Res. 2015; 21; 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lehmann BD, Bauer JA, Chen X et al Identification of human triple‐negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011; 121; 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loi S, Michiels S, Salgado R et al Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann. Oncol. 2014; 25; 1544–1550. [DOI] [PubMed] [Google Scholar]
- 8. Loi S, Sirtaine N, Piette F et al Prognostic and predictive value of tumor‐infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node‐positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin‐based chemotherapy: BIG 02‐98. J. Clin. Oncol. 2013; 31; 860–867. [DOI] [PubMed] [Google Scholar]
- 9. Adams S, Gray RJ, Demaria S et al Prognostic value of tumor‐infiltrating lymphocytes in triple‐negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014; 32; 2959–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davila ML, Riviere I, Wang X et al Efficacy and toxicity management of 19‐28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014; 6; ra25224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grupp SA, Kalos M, Barrett D et al Chimeric antigen receptor‐modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013; 368; 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maude SL, Frey N, Shaw PA et al Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014; 371; 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor‐modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011; 365; 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le DT, Uram JN, Wang H et al PD‐1 Blockade in tumors with mismatch‐repair deficiency. N. Engl. J. Med. 2015; 372; 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herbst RS, Baas P, Kim DW et al Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): a randomised controlled trial. Lancet 2016; 387; 1540–1550. [DOI] [PubMed] [Google Scholar]
- 16. Robert C, Thomas L, Bondarenko I et al Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 2011; 364; 2517–2526. [DOI] [PubMed] [Google Scholar]
- 17. Hodi FS, O'Day SJ, McDermott DF et al Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010; 363; 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brahmer J, Reckamp KL, Baas P et al Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N. Engl. J. Med. 2015; 373; 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dushyanthen S, Teo ZL, Caramia F et al Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat. Commun. 2017; 8; 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loi S, Dushyanthen S, Beavis PA et al RAS/MAPK activation is associated with reduced tumor‐infiltrating lymphocytes in triple‐negative breast cancer: therapeutic cooperation between MEK and PD‐1/PD‐L1 immune checkpoint inhibitors. Clin. Cancer Res. 2016; 22; 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dua I, Tan AR. Immunotherapy for triple‐negative breast cancer: a focus on immune checkpoint inhibitors. Am. J. Hematol. Oncol. 2017; 13; 20–27. [Google Scholar]
- 22. Stagg J, Allard B. Immunotherapeutic approaches in triple‐negative breast cancer: latest research and clinical prospects. Ther. Adv. Med. Oncol. 2013; 5; 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grigoriadis A, Caballero OL, Hoek KS et al CT‐X antigen expression in human breast cancer. Proc. Natl Acad. Sci. USA 2009; 106; 13493–13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gure AO, Chua R, Williamson B et al Cancer‐testis genes are coordinately expressed and are markers of poor outcome in non‐small cell lung cancer. Clin. Cancer Res. 2005; 11; 8055–8062. [DOI] [PubMed] [Google Scholar]
- 25. Velazquez EF, Jungbluth AA, Yancovitz M et al Expression of the cancer/testis antigen NY‐ESO‐1 in primary and metastatic malignant melanoma (MM)—correlation with prognostic factors. Cancer Immun. 2007; 7; 11. [PMC free article] [PubMed] [Google Scholar]
- 26. Napoletano C, Bellati F, Tarquini E et al MAGE‐A and NY‐ESO‐1 expression in cervical cancer: prognostic factors and effects of chemotherapy. Am. J. Obstet. Gynecol. 2008; 198; 99e1–99e7. [DOI] [PubMed] [Google Scholar]
- 27. Andrade VC, Vettore AL, Felix RS et al Prognostic impact of cancer/testis antigen expression in advanced stage multiple myeloma patients. Cancer Immun. 2008; 8; 2. [PMC free article] [PubMed] [Google Scholar]
- 28. Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002; 188; 22–32. [DOI] [PubMed] [Google Scholar]
- 29. Jager E, Stockert E, Zidianakis Z et al Humoral immune responses of cancer patients against ‘Cancer‐Testis’ antigen NY‐ESO‐1: correlation with clinical events. Int. J. Cancer 1999; 84; 506–510. [DOI] [PubMed] [Google Scholar]
- 30. van der Bruggen P, Traversari C, Chomez P et al A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991; 254; 1643–1647. [DOI] [PubMed] [Google Scholar]
- 31. Bandic D, Juretic A, Sarcevic B et al Expression and possible prognostic role of MAGE‐A4, NY‐ESO‐1, and HER‐2 antigens in women with relapsing invasive ductal breast cancer: retrospective immunohistochemical study. Croat. Med. J. 2006; 47; 32–41. [PMC free article] [PubMed] [Google Scholar]
- 32. Mischo A, Kubuschok B, Ertan K et al Prospective study on the expression of cancer testis genes and antibody responses in 100 consecutive patients with primary breast cancer. Int. J. Cancer 2006; 118; 696–703. [DOI] [PubMed] [Google Scholar]
- 33. Sugita Y, Wada H, Fujita S et al NY‐ESO‐1 expression and immunogenicity in malignant and benign breast tumors. Cancer Res. 2004; 64; 2199–2204. [DOI] [PubMed] [Google Scholar]
- 34. Theurillat JP, Ingold F, Frei C et al NY‐ESO‐1 protein expression in primary breast carcinoma and metastases: correlation with CD8+ T‐cell and CD79a+ plasmacytic/B‐cell infiltration. Int. J. Cancer 2007; 120; 2411–2417. [DOI] [PubMed] [Google Scholar]
- 35. Al‐Ejeh F, Simpson PT, Saunus JM et al Meta‐analysis of the global gene expression profile of triple‐negative breast cancer identifies genes for the prognostication and treatment of aggressive breast cancer. Oncogenesis 2014; 3; e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Junankar S, Baker LA, Roden DL et al ID4 controls mammary stem cells and marks breast cancers with a stem cell‐like phenotype. Nat. Commun. 2015; 6; 6548. [DOI] [PubMed] [Google Scholar]
- 37. Field S, Uyttenhove C, Stroobant V et al Novel highly specific anti‐periostin antibodies uncover the functional importance of the fascilin 1‐1 domain and highlight preferential expression of periostin in aggressive breast cancer. Int. J. Cancer 2016; 138; 1959–1970. [DOI] [PubMed] [Google Scholar]
- 38. Burgess JT, Bolderson E, Saunus JM et al SASH1 mediates sensitivity of breast cancer cells to chloropyramine and is associated with prognosis in breast cancer. Oncotarget 2016; 7; 72807–72818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernandez‐Perez S, Cabrera E, Salido E et al DUB3 and USP7 de‐ubiquitinating enzymes control replication inhibitor Geminin: molecular characterization and associations with breast cancer. Oncogene 2017; 36; 4802–4809. [DOI] [PubMed] [Google Scholar]
- 40. Aleskandarany MA, Abduljabbar R, Ashankyty I et al Prognostic significance of androgen receptor expression in invasive breast cancer: transcriptomic and protein expression analysis. Breast Cancer Res. Treat. 2016; 159; 215–227. [DOI] [PubMed] [Google Scholar]
- 41. Farmer P, Bonnefoi H, Becette V et al Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005; 24; 4660–4671. [DOI] [PubMed] [Google Scholar]
- 42. Lim E, Vaillant F, Wu D et al Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009; 15; 907–913. [DOI] [PubMed] [Google Scholar]
- 43. Vousden KH, Lane DP. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007; 8; 275–283. [DOI] [PubMed] [Google Scholar]
- 44. Badovinac Crnjevic T, Spagnoli G, Juretic A, Jakic‐Razumovic J, Podolski P, Saric N. High expression of MAGE‐A10 cancer‐testis antigen in triple‐negative breast cancer. Med. Oncol. 2012; 29; 1586–1591. [DOI] [PubMed] [Google Scholar]
- 45. Wang H, Sang M, Geng C, Liu F, Gu L, Shan B. MAGE‐A is frequently expressed in triple negative breast cancer and associated with epithelial–mesenchymal transition. Neoplasma 2016; 63; 44–56. [DOI] [PubMed] [Google Scholar]
- 46. Curigliano G, Viale G, Ghioni M et al Cancer‐testis antigen expression in triple‐negative breast cancer. Ann. Oncol. 2011; 22; 98–103. [DOI] [PubMed] [Google Scholar]
- 47. Chen YT, Ross DS, Chiu R et al Multiple cancer/testis antigens are preferentially expressed in hormone‐receptor negative and high‐grade breast cancers. PLoS One 2011; 6; e17876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ademuyiwa FO, Bshara W, Attwood K et al NY‐ESO‐1 cancer testis antigen demonstrates high immunogenicity in triple negative breast cancer. PLoS One 2012; 7; e38783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sjogren S, Inganas M, Norberg T et al The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J. Natl Cancer Inst. 1996; 88; 173–182. [DOI] [PubMed] [Google Scholar]
- 50. Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature 1997; 386; 623–627. [DOI] [PubMed] [Google Scholar]
- 51. Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998; 396; 643–649. [DOI] [PubMed] [Google Scholar]
- 52. Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest. 2012; 122; 1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Foijer F, Xie SZ, Simon JE et al Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy‐induced stress. Proc. Natl Acad. Sci. USA 2014; 111; 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Manning AL, Benes C, Dyson NJ. Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation. Oncogene 2014; 33; 2487–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 2012; 24; 809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011; 333; 1895–1898. [DOI] [PubMed] [Google Scholar]
- 57. Glazer CA, Smith IM, Bhan S et al The role of MAGEA2 in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 2011; 137; 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marcar L, Ihrig B, Hourihan J et al MAGE‐A cancer/testis antigens inhibit MDM2 ubiquitylation function and promote increased levels of MDM4. PLoS One 2015; 10; e0127713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maire V, Baldeyron C, Richardson M et al TTK/hMPS1 is an attractive therapeutic target for triple‐negative breast cancer. PLoS One 2013; 8; e63712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006; 38; 1043–1048. [DOI] [PubMed] [Google Scholar]
- 61. Yang P, Huo Z, Liao H, Zhou Q. Cancer/testis antigens trigger epithelial–mesenchymal transition and genesis of cancer stem‐like cells. Curr. Pharm. Des. 2015; 21; 1292–1300. [DOI] [PubMed] [Google Scholar]
- 62. Kijima T, Maulik G, Ma PC et al Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c‐Kit in small cell lung cancer cells. Cancer Res. 2002; 62; 6304–6311. [PubMed] [Google Scholar]
- 63. Kim R, Kulkarni P, Hannenhalli S. Derepression of cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation. BMC Cancer 2013; 13; 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siebenkas C, Chiappinelli KB, Guzzetta AA et al Inhibiting DNA methylation activates cancer testis antigens and expression of the antigen processing and presentation machinery in colon and ovarian cancer cells. PLoS One 2017; 12; e0179501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. MAGE‐A3‐targeted autologous CD4+ T cells may target metastatic cancer. Cancer Discov. 2017. [Google Scholar]
- 66. Lu Y‐C, Parker L, Lu T et al A phase I study of an HLA‐DPB1*0401‐restricted T cell receptor targeting MAGE‐A3 for patients with metastatic cancers: 30th Annual Meeting and Associated Programs of the Society for Immunotherapy of Cancer 4/11/15; National Harbor, MD, USA. J. Immunother. Cancer 2015; 3; 158. [Google Scholar]
- 67. Cho HJ, Mei A, Fukui J et al MAGE‐a mediate resistance to chemotherapy in multiple myeloma through regulation of Bcl‐2 proteins. Blood 2016; 128; 3277. [Google Scholar]


