Abstract
Rationale: The goal was to connect elements of idiopathic pulmonary fibrosis (IPF) pathogenesis, including chronic endoplasmic reticulum stress in respiratory epithelia associated with injury/inflammation and remodeling, distal airway mucus obstruction and honeycomb cyst formation with accumulation of MUC5B (mucin 5B), and associations between IPF risk and polymorphisms in the MUC5B promoter.
Objectives: To test whether the endoplasmic reticulum (ER) stress sensor protein ERN2 (ER-to-nucleus signaling 2) and its downstream effector, the spliced form of XBP1S (X-box–binding protein 1), regulate MUC5B expression and differentially activate the MUC5B promoter variant in respiratory epithelia.
Methods: Primary human airway epithelial (HAE) cells, transgenic mouse models, human IPF lung tissues, and cell lines expressing XBP1S and MUC5B promoters were used to explore relationships between the ERN2/XBP1S pathway and MUC5B. An inhibitor of the pathway, KIRA6, and XBP1 CRISPR-Cas9 were used in HAE cells to explore therapeutic potential.
Measurements and Main Results: ERN2 regulated MUC5B and MUC5AC mRNAs. Downstream XBP1S selectively promoted MUC5B expression in vitro and in distal murine airway epithelia in vivo. XBP1S bound to the proximal region of the MUC5B promoter and differentially upregulated MUC5B expression in the context of the MUC5B promoter rs35705950 variant. High levels of ERN2 and XBP1S were associated with excessive MUC5B mRNAs in distal airways of human IPF lungs. Cytokine-induced MUC5B expression in HAE cells was inhibited by KIRA6 and XBP1 CRISPR-Cas9.
Conclusions: A positive feedback bistable ERN2–XBP1S pathway regulates MUC5B-dominated mucus obstruction in IPF, providing an unfolded protein response–dependent mechanism linking the MUC5B promoter rs35705950 polymorphism with IPF pathogenesis. Inhibiting ERN2-dependent pathways/elements may provide a therapeutic option for IPF.
Keywords: MUC5B, IPF, airway epithelia
At a Glance Commentary
Scientific Knowledge on the Subject
MUC5B mucin-dominated mucus obstruction in distal airways and honeycomb cysts likely contributes to idiopathic pulmonary fibrosis (IPF) pathogenesis. Consistent with this notion, the MUC5B promoter polymorphism (rs35705950, minor allele “T”) is the strongest and most replicated genetic risk factor for IPF.
What This Study Adds to the Field
We identified a novel pathway regulating MUC5B expression that involves activation of a component of the unfolded protein response (i.e., the ERN2 [endoplasmic reticulum to nucleus signaling 2] protein and its downstream target, XBP1S [the spliced form of the X-box–binding protein 1] mRNA, which is translated into a functional transcription factor). XBP1S binds to the MUC5B promoter region and induces expression of MUC5B, but not MUC5AC, in the distal bronchiolar epithelia in vivo and is coexpressed with MUC5B in human IPF lungs with excessive MUC5B accumulation. Importantly, XBP1S differentially activates the MUC5B promoter carrying rs35705950 minor allele “T,” providing a positive feedback mechanism for unfolded protein response–induced differential expression of MUC5B relevant to IPF pathogenesis/risk. Finally, administration of a pathway inhibitor (KIRA6) or deletion of XBP1 by CRISPR/Cas9-mediated lentivirus inhibited cytokine-induced mucin biosynthesis in primary human airway epithelial cells. This study provides mechanistic insights into the pathophysiology of IPF and therapeutic approaches to reduce mucin expression following high-strength, irreversible activation of the unfolded protein response in IPF.
MUC5B is the dominant gel-forming mucin constitutively secreted by respiratory epithelia in health and supports basal mucociliary clearance rates and innate lung defense (1–3). MUC5B-dominated mucus obstruction in distal airways and honeycomb cysts is a common feature in idiopathic pulmonary fibrosis (IPF) (4). The gain-of-function MUC5B promoter variant rs35705950 is the strongest population-wide genetic risk factor associated with the development of IPF, and the risk variant is linked to increased MUC5B expression in both unaffected and IPF subjects (4–7). However, the mechanisms linking abnormal regulation of MUC5B to the development of, and the role of abnormal MUC5B expression/secretion to distal airways disease in, IPF have not been elucidated (6).
Endoplasmic reticulum (ER) stress, particularly in alveolar type 2 epithelial cells (8, 9), contributes to development of IPF in patients that carry SFTPC (surfactant protein C) mutations (10, 11). Interestingly, SFTPC mutations are associated with ectopic expression of MUC5B in alveolar type 2–like cells and increased MUC5B levels are observed in BAL from children with SFTPC mutations (12), suggesting that ER stress in alveolar type 2 or adjacent respiratory epithelial cells is related to increased MUC5B expression. Martino and colleagues (13) demonstrated that the ER stress sensor protein ERN2 (ER-to-nucleus signaling 2), also called IRE1β (inositol-requiring enzyme 1β), is localized to airway mucous cells and required for allergen-induced Muc5b and Muc5ac expression in vivo. Similar to its ubiquitously expressed isoform ERN1 (14), also known as IRE1α (inositol-requiring enzyme 1α), ERN2 contains conserved autophosphorylating kinase domains and distinct endoribonuclease (RNase) domains that share 80% versus 61% homology, respectively (15), with the RNase dominating their functional differences (16). A small molecule KIRA6 has been developed that attenuates ERN1 (IRE1α) RNase activity by inhibiting its kinase domain (17).
Injury and inflammation can trigger kinase-mediated activation of ERN1 RNase activity (18, 19), which splices a 26-nucleotide intron from XBP1 mRNA (20). The spliced XBP1 (XBP1S) mRNA is translated into a transcription factor that regulates genes encoding ER chaperones (21) and elements of the secretory pathway (22), including Muc2 in the gut (23). It is, however, unknown whether ERN2 and XBP1S regulate mucin gene expression in airway epithelia in normal or IPF lungs.
In this study, we tested the hypothesis that a linkage between the unfolded protein response (UPR) and MUC5B expression exists that is mediated by ERN2-triggered XBP1 splicing, which is both sufficient and necessary for MUC5B expression. Furthermore, we hypothesized that XBP1S differentially regulates the MUC5B promoter rs35705950 variant activity, providing a potential mechanism to generate an abnormal UPR-MUC5B positive feedback expression cycle that contributes to IPF pathogenesis. We also explored whether the SPDEF (SAM pointed domain–containing ETS transcription factor), which regulates mucin gene transcription (24, 25), binds to the MUC5B promoter (26), and is expressed in IPF goblet-like cells (27), interacted with this pathway. A series of studies, using the proinflammatory and profibrotic cytokine IL-1β (28) as a tool to induce UPR-dependent MUC5B expression in HAE cells in vitro, recombinant cells expressing XBP1S, and the MUC5B promoters, transgenic mice, and freshly excised normal and IPF lungs, were performed to test these hypotheses.
Methods
Information about methods is available in the online supplement.
Results
ERN2 Colocalizes with and Regulates MUC5B and MUC5AC Expression
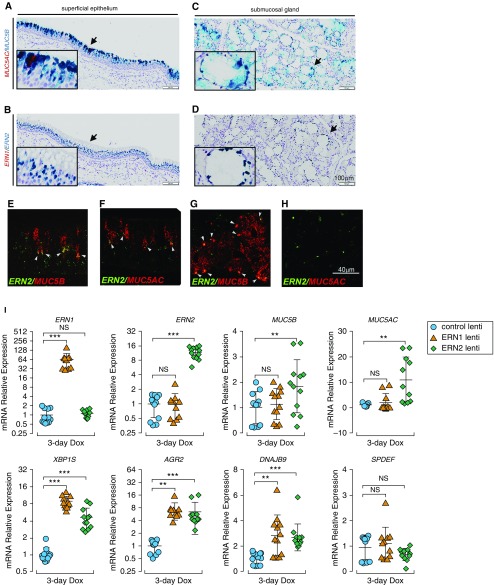
ERN2 regulates mucin gene expression (13), but colocalization of ERN2 expression with individual mucin gene expression in human lung tissues has not been shown. ERN2 mRNA was detected in mucin secretory cells within pseudostratified superficial epithelia by RNAscope assays (Figures 1A and 1B). Robust ERN2 expression was also detected in submucosal glands (SMG) (Figures 1C and 1D). ERN2 mRNA was colocalized with both MUC5B and MUC5AC in superficial epithelium and selectively with MUC5B in SMG (Figures 1E–1H). In contrast, ERN1 mRNA was detected in a wide variety of cell types in superficial airway epithelia and in SMG (see Figures E1A and E1B in the online supplement). The localization pattern of ERN2 suggested a role in regulation of MUC5B and MUC5AC mRNAs.
Figure 1.
ERN2 is expressed with MUC5B and MUC5AC in mucin-secreting cells in vivo and promotes MUC5B and MUC5AC expression in human airway epithelium in vitro. (A–H) Expression of ERN2, MUC5B, and MUC5AC mRNA was detected using RNAscope assays in pseudostratified superficial epithelium (A, B, E, and F) and submucosal glands (C, D, G, and H) of proximal cartilaginous airways of normal adult human lung. Expression of MUC5AC with MUC5B (A and C) and ERN1 with ERN2 (B and D) was detected by RNAscope duplex chromogenic assays. Insets show high-power view of arrow-pointed areas. Expression of ERN2 with MUC5B (E and G) and ERN2 with MUC5AC (F and H) mRNAs was detected by RNAscope multiplex dual-fluorescent assays in human cartilaginous airway epithelia. Coexpression of red and green signals in the same cell is highlighted by white arrowheads (E–G). Micrographs in A–H are representative of n = 3 lung tissue from donors without preexisting pulmonary diseases. Scale bars: A–D, 100 μm; E–H, 40 μm. (I) Doxycycline (Dox)-induced expression of ERN1 or ERN2 in primary human airway epithelia. Lentiviral stably infected human airway epithelial cells were cultured under acute lung injury conditions for 5 days before 3-day Dox administration (250 ng/ml) in the basolateral acute lung injury media. mRNA expression of ERN1 and ERN2; mucin-related genes MUC5B, MUC5AC, AGR2, and SPDEF; and unfolded protein response genes XBP1S and DNAJB9 was determined by SYBR-Green quantitative (q) RT-PCR or Taqman assays. Graphs represent means ± SD of n = 3–4 human airway epithelial cultures obtained from three non–cystic fibrosis/nonsmoker donors, and data were analyzed with two-way ANOVA followed by Dunnett’s test. **P < 0.01 and ***P < 0.001 compared with control subjects. NS = not significant.
To test if ERN2 regulates MUC5B/MUC5AC expression, primary normal HAE cells were infected with doxycycline (Dox)-inducible lentiviruses expressing ERN1, ERN2, or a control lentivirus (see Figures E1C and E1D). On Dox administration, only ERN2 induced MUC5B and MUC5AC mRNA expression, although both ERN1 and ERN2 induced mRNA expression of XBP1S and its known target genes AGR2 (13) and DNAJB9 (29) (Figure 1I). Neither ERN1 nor ERN2 regulated SPDEF mRNA (Figure 1I). These results identified a specific role of ERN2, independent of ERN1 or SPDEF, in promoting mucin gene expression.
IL-1β Triggers UPR and Mucin Gene Expression in Airway Epithelia
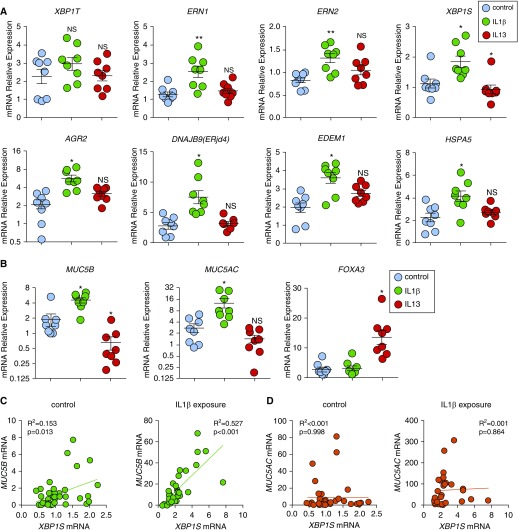
To evaluate induction of the ERN2–XBP1S cascade in a mucus overproduction disease context, primary HAE cells were exposed for 24 hours to recombinant human IL-1β (a proinflammatory/profibrotic cytokine associated with fibrosis in mouse [30] and human IPF [31]). IL-1β effects were compared with the TH2 cytokine IL-13 (Figure 2). IL-13 is a key contributor to asthma pathogenesis and MUC5AC expression (32) and has also been implicated in the fibrotic component of IPF (33). Total XBP1 (XBP1T) was not different among the treatment groups. However, IL-1β induced mRNA expression of ERN1, ERN2, XBP1S, and the XBP1S-regulated UPR genes AGR2, DNAJB9, EDEM1 (34), and HSPA5 (35) within 24 hours (Figure 2A). In contrast, IL-13 did not induce UPR gene expression within 24 hours (Figure 2A). IL-1β exposure over 24 hours also upregulated MUC5B and MUC5AC mRNAs, but not the TH2-dependent MUC5AC regulator, FOXA3 mRNA (Figure 2B). A similar profile was observed when normal HAE cells were exposed to another profibrotic cytokine present in IPF lung, recombinant human IL-17A cytokine (30, 36) for 24 hours (see Figure E2 in the online supplement). In contrast, IL-13 inhibited mRNA expression of both XBP1S and MUC5B, but induced FOXA3 (37, 38). MUC5AC mRNA was not increased following acute IL-13 exposure for 24 hours (Figures 2A and 2B). Thus, acute induction of MUC5B and XBP1S mRNA expression was linked to IL-1β– and IL-17A–induced, but not IL-13–induced, UPR signaling in primary HAE cells.
Figure 2.
IL-1β induces unfolded protein response and XBP1S that is associated with MUC5B expression in human airway epithelial (HAE) cells. (A) Unfolded protein response genes were acutely induced after IL-1β exposure. HAE cells were cultured under acute lung injury conditions for 4 weeks to allow full differentiation before exposure with IL-1β and IL-13. Both IL-1β and IL-13 were added in basolateral acute lung injury media at 10 ng/ml for 24 hours. Expression of unfolded protein response genes XBP1T (total XBP1), ERN1, ERN2, XBP1S, AGR2, DNAJB9, EDEM1, and HSPA5 was quantified by SYBR-green qRT-PCR or Taqman assays. (B) Expression of mucin genes MUC5B and MUC5AC and goblet cell transcription factor FOXA3 was quantified by Taqman assay. Data are presented as mean ± SE of n = 1 culture of HAE cells from eight non–cystic fibrosis, nonsmoker donors, analyzed with paired one-way ANOVA. *P < 0.05 and **P < 0.01 compared with control subjects. HAE cells treated with IL-1β are labeled with green, IL-13 are labeled with red, and control subjects are labeled with blue. (C and D) mRNA expression of XBP1S versus MUC5B (C) and XBP1S versus MUC5AC (D) in HAE cells that were treated basolaterally with or without IL-1β 10 ng/ml (IL-1β exposure or control) for 5 days was analyzed by linear regression. The R2 and P values were compared. Normal HAE cells obtained from n = 40 non–cystic fibrosis donor lungs and n = 33 non–cystic fibrosis donor lungs were tested for control and IL-1β exposure, respectively. NS = not significant.
Consistent with these findings, XBP1S mRNA expression was weakly correlated with MUC5B mRNA at baseline, but this correlation became stronger after 5-day IL-1β exposure (R2 = 0.153, P = 0.013 and R2 = 0.527, P < 0.001 for baseline and IL-1β, respectively) (Figure 2C). In contrast, XBP1S expression was not correlated with MUC5AC mRNA (R2 < 0.001, P = 0.998 and R2 = 0.001, P = 0.864 at baseline and after IL-1β exposure, respectively) (Figure 2D). The strong positive correlation of expression between XBP1S and MUC5B mRNAs after IL-1β, coupled with the association of the XBP1S regulator ERN2 with MUC5B expression in superficial epithelia and SMG (Figure 1), led us to test whether XBP1S regulated MUC5B protein and mRNA expression.
XBP1S Differentially Regulates MUC5B Expression In Vitro
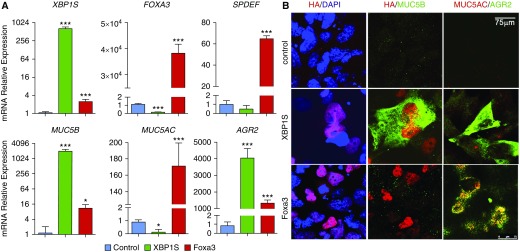
To test whether XBP1S regulates the secretory mucins MUC5B and MUC5AC mRNA expression, the human bronchial epithelial cell line BEAS2B, which expresses low levels of endogenous MUC5B and MUC5AC mRNAs, was transduced with a Dox-inducible lentivirus expressing HA-tagged XBP1S. In parallel, we expressed HA-tagged Foxa3 in BEAS2B cells (see Figures E3A and E3B). Heterologous expression of XBP1S strongly induced MUC5B and AGR2 mRNAs (Figure 3A). Interestingly, XBP1S decreased MUC5AC while having no effect on SPDEF mRNA expression. In contrast, overexpression of Foxa3 strongly induced MUC5AC, SPDEF, and AGR2 mRNAs (Figure 3A). mRNA expression of XBP1S regulated targets DNAJB9, EDEM1, and HSPA5 (29) was also strongly induced by transduced XBP1S lentivirus (see Figure E3C). MUC5B, MUC5AC, and AGR2 protein expression correlated with their mRNA changes in the cells transduced with XBP1S and Foxa3 lentiviruses (Figure 3B). MUC5B protein was not detected in the Foxa3 overexpressing cells by immunofluorescent staining, although MUC5B mRNA was modestly induced (Figures 3A and 3B).
Figure 3.
XBP1S regulates MUC5B expression in vitro. (A) BEAS2B cells were stably transduced with doxycycline-inducible control– (blue bars), XBP1S- (green bars), or Foxa3- (red bars) expressing lentiviruses. Three days after doxycycline administration, mRNA expression of XBP1S, FOXA3, MUC5B, MUC5AC, AGR2, and SPDEF was determined. Graphs represent mean ± SD with n = 3 independent cultures from each stably infected cell line, and data were analyzed with one-way ANOVA followed by Dunnett’s test. (B) Immunofluorescent staining of BEAS2B cells expressing control, XBP1S, and Foxa3 (top, middle, and bottom rows, respectively). Expression of HA-tag that was fused to the N terminus of XBP1S and Foxa3 proteins is shown in the left column with nuclei counterstained with DAPI. Dual staining of HA and MUC5B is shown in the middle column, whereas MUC5AC and AGR2 are shown in the right column. Scale bar, 75 μm. *P < 0.05 and ***P < 0.001 compared to controls.
XBP1S Regulates Muc5b Expression in the Distal Airways In Vivo
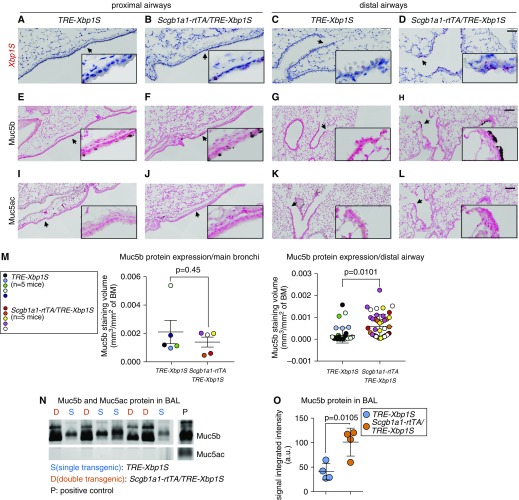
We then tested whether the regulation of MUC5B expression by XBP1S observed in our in vitro studies could be detected in vivo. Transgenic mice (Scgb1a1-rtTA[line2] [39]/TRE-Xbp1S [40]) were generated in which Xbp1S was conditionally expressed in the club cells lining the conducting airways (39) after administration of Dox (see Figures E4A–E4C). In these transgenic mice, Xbp1S mRNA was continuously induced in airway epithelium lining both proximal and distal airways by chronic (4-wk) Dox administration and compared with control littermates (TRE-Xbp1S) (Figures 4A–4D). Conditional expression of Xbp1S did not alter Muc5b expression in proximal airway epithelium, as shown by immunohistochemical staining with a Muc5b antibody (Figures 4E and 4F). However, in the distal airways, chronic Xbp1S expression induced the formation of Muc5b-expressing epithelial cells (Figures 4G and 4H; see Figure E4D), without evidence of overt pulmonary inflammation (see Figure E4E). Muc5ac expression was not detected in proximal or distal airway epithelia of either genotype (Figures 4I–4L). Muc5b protein expression in the proximal and distal airways in Xbp1S conditional expression mice was quantitated by morphometric analysis of Muc5b immunohistochemical staining volume density, which confirmed a selective increase in Muc5b protein expression in the distal airways (Figure 4M). The levels of secreted Muc5b, but not Muc5ac, protein in BAL were also significantly increased in Xbp1S transgenic as compared with control mice (Figures 4N and 4O). Consistent with the in vitro findings, overexpression of Xbp1S did not affect Spdef expression, but did increase expression of mucin and UPR-related genes in the whole lung mRNA (e.g., Clca1 [41] and Agr2) (see Figures E4F–E4H).
Figure 4.
Xbp1S induces Muc5b expression in distal murine airways in vivo. (A–D) Conditional overexpression of Xbp1S mRNA (red) in the epithelium lining from the proximal (A and B) and distal (C and D) airways in Scgb1a1-rtTA/TRE-Xbp1S transgenic mice was detected by RNA in situ hybridization (BaseScope assay) following doxycycline food provision for 4 weeks. Doxycycline-treated littermate TRE-Xbp1S mice served as control animals. (E–H) Cells producing Muc5b protein were detected by immunohistochemical staining with Muc5b antibodies in the epithelial cells resident from the proximal to distal airways. (I–L) Muc5ac protein expression was detected by immunohistochemical staining with Muc5ac antibodies. Images are representative of n = 3–4 of each genotype. Scale bars: A–D, 50 μm; E–L, 100 μm. Insets show higher power views of arrow-pointed areas. (M) Quantification of Muc5b protein expressed in mainstem bronchi versus distal airways. All distal airways with luminal diameters <150 μm (i.e., terminal airways), regardless of staining status, were analyzed for morphometric analyses of Muc5b immunohistochemical staining volume density in left lobes of mouse lungs (n = 8 ± 1.0 and n = 9 ± 1.8 distal airways/mouse, mean ± SD, for control and Xbp1S overexpression mice, respectively). Each colored dot represents measurement of a single airway, and each color indicates measurements obtained from a single mouse (n = 5 mice of each genotype were analyzed). Data represent mean ± SD of staining volume density/distal airway in either genotype, and difference of means between the two groups (denoted by P values) were analyzed by linear mixed-effects model with mouse identification number as random intercept variable. (N) Secretion of Muc5b and Muc5ac protein in BAL was detected by agarose Western blot, and (O) Muc5b protein was semiquantified by densitometry. BM = basement membrane.
XBP1S and SPDEF Are Associated with MUC5B in the Distal Airway Epithelia of Subjects with IPF
XBP1S and SPDEF mRNAs were associated with MUC5AC and MUC5B protein expression in the superficial epithelia lining normal human cartilaginous airways (see Figure E5A). Furthermore, the SMG of normal human and mice, which selectively express MUC5B protein, also expressed XBP1S and SPDEF mRNAs (see Figures E5A and E5B). The XBP1S/MUC5B expression pattern matched that of ERN2 as shown in Figures 1A–1H.
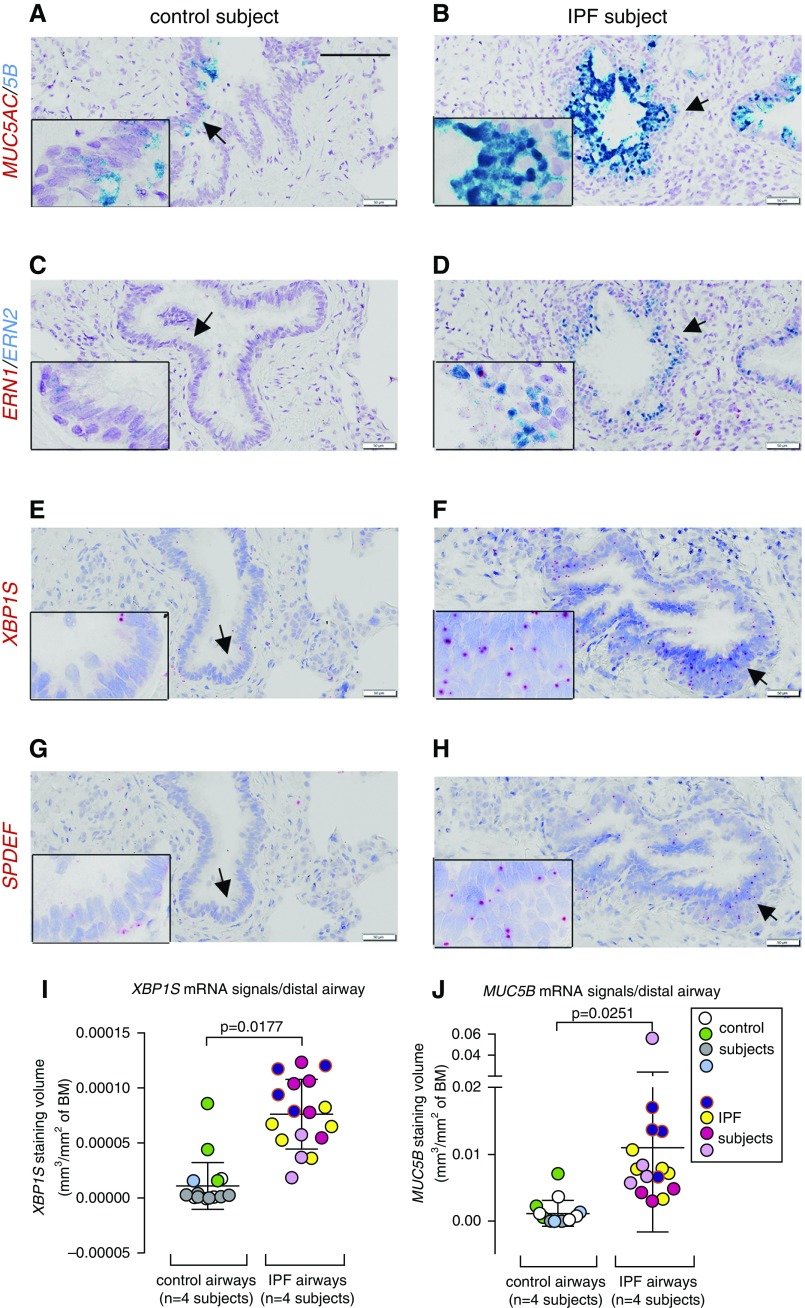
Distal airways (i.e., noncartilaginous airways) are the most affected region by MUC5B-dominated mucus obstruction in IPF, and they are also the sites exhibiting increased Muc5b expression in Xbp1S transgenic mice. Because of the absence of antibodies that reliably detect expression of XBP1S and SPDEF by immunohistochemical staining, we applied RNAscope and Basescope to identify these genes. IPF distal airways from human subjects exhibited increased MUC5B expression associated with increased ERN2, XBP1S, and SPDEF expression compared with control subjects (Figure 5; see Figures E6A–E6L). Significantly increased XBP1S and MUC5B mRNA signal volume density in the distal airways of IPF versus control subjects was confirmed by morphometric analyses (Figures 5I and 5J). Epithelial cells within honeycomb cysts in IPF heterogeneously expressed MUC5B, SPDEF, ERN2, and XBP1S (see Figures E6M–E6P vs. E6Q–E6T).
Figure 5.
Expression of MUC5AC/MUC5B, ERN1/ERN2, XBP1S, and SPDEF mRNAs in the distal airways of human subjects without idiopathic pulmonary fibrosis (IPF) and with IPF. (A–H) mRNA expression of MUC5AC/MUC5B (A and B) and ERN1/ERN2 (C and D) was detected by RNAscope Duplex assays, and XBP1S (E and F) and SPDEF (G and H) mRNAs were detected by BaseScope assays in distal airways (<0.5 mm) of control subjects (n = 4) and patients with IPF (n = 8). Scale bar, 100 μm. Insets show higher power views of arrow-pointed areas. (I and J) Quantification of XBP1S (I) and MUC5B (J) mRNA signals in the distal human airways (including all the airways with luminal diameter <500 μm) (i.e., terminal airways, regardless of staining status) was determined by morphometric analysis of staining volume density detected by Basescope and RNAscope, respectively (n = 4 of control subjects and n = 4 of IPF subjects). Analysis for XBP1S mRNA signal quantification was performed with n = 4.5 ± 1.9 and n = 4.3 ± 1 distal airways/subject, mean ± SD, for control and IPF, respectively. For MUC5B mRNA signal quantification, analysis was performed with n = 3.75 ± 1.3 and n = 4 ± 0.8 distal airways/subject, means ± SD, for control and IPF, respectively. Each colored dot represents measurement of one airway, and each color indicates measurements obtained from the same subject. Data represent mean ± SD of signal volume density/distal airway in either group, and the differences of means between the two groups (denoted by P values) were analyzed by linear mixed-effects model with subject identification number as random intercept variable. BM = basement membrane.
XBP1S Activates the MUC5B Promoter and Differentially Regulates MUC5B Expression in the Context of the rs35705950 Variant
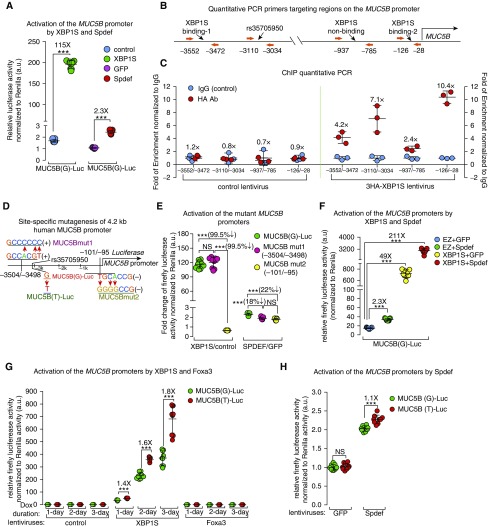
To further characterize MUC5B transcriptional regulation, MUC5B promoter–XBP1S interactions were directly tested. Relevant to IPF pathogenesis, we speculated that XBP1S might differentially regulate the rs37505950 polymorphic locus on the MUC5B promoter. SPDEF binds to its consensus binding motif on the MUC5B promoter in a region (−3.4 to −2.7 kb) that contains the variant locus (−3.1 kb) (26), and SPDEF mRNA is expressed in IPF epithelia (27), suggesting it might also regulate the MUC5B promoter and its variant in IPF. Accordingly, we compared XBP1S and SPDEF in the regulation of MUC5B promoter activity, using a 4.2-kb human MUC5B promoter (see Figure E7A) cloned into the episomal luciferase reporter pREP4-Luc (42) to produce MUC5B(G)-Luc (carrying the major “G” allele at rs35705950 locus). XBP1S strongly activated the MUC5B promoter compared with Spdef (115- vs. 2.3-fold induction, respectively) (Figure 6A). We then evaluated the conservation of the cis-elements on the MUC5B promoter among approximately 100 vertebrate species, and found two highly conserved regions: surrounding the rs35705950 variant locus, and in the proximal region of the transcription start site. Two predicted (in silico) XBP1S binding motifs (GCCACGT) (43, 44) were found in the −3.5-kb and −0.1-kb loci of the MUC5B promoter, with the latter positioned in the conserved proximal region of the MUC5B promoter (see Figures E7B–E7E).
Figure 6.
XBP1S differentially regulates MUC5B promoter variant activity. (A) MUC5B (G)-Luc carrying “G” allele at the rs35705950 locus was transfected into BEAS2B cells transduced with XBP1S versus control, and Spdef versus GFP lentiviruses, followed by doxycycline administration for 24 hours before luciferase activity measurement. (B) Regions evaluated in chromatin immunoprecipitation (ChIP)-qPCR studies with the 3HA-tagged XBP1S were selected with respect to the two predicted XBP1S consensus binding sites, the rs35705950 locus, and a negative control region (nonconserved, predicted nonbinding) on the MUC5B promoter. (C) ChIP was performed in the BEAS2B cells transduced with control (left) or 3HA-XBP1S (right) lentiviruses using an HA antibody or control IgG to pull down XBP1S-bound chromatin. qPCR was performed to measure the enrichment of chromatin fragments that cover the four selected regions. Fold change of the chromatin enrichment pulled down by the HA antibody versus control IgG was compared between mean values of two groups and labeled on top of the data from each group. Graphs present means ± SD of n = 3 independent experiments. (D) Two putative conserved XBP1S binding motifs on MUC5B promoter were mutated to generate MUC5Bmut1 and MUC5Bmut2. The “G” allele was mutated to “T” at locus of rs35705950 to generate MUC5B(T)-Luc. (E) MUC5Bmut1, MUC5Bmut2, or MUC5B(G)-Luc were transfected in BEAS2B cells expressing XBP1S and its control, Spdef and GFP. Fold changes of promoter activity induced by XBP1S versus control, and Spdef versus GFP, were compared among three luciferase vectors at 24 hours. (F) BEAS2B cells were infected with lentiviruses expressing 1) control and GFP, 2) control and Spdef, 3) XBP1S and GFP, and 4) XBP1S and Spdef. Luciferase activity was determined at 24 hours after doxycycline administration. (G) Equal amounts of MUC5B(G)-Luc or MUC5B(T)-Luc plasmids were transfected in BEAS2B cells stably infected with control, XBP1S, and Foxa3 lentiviruses. Luciferase activity was determined at 1, 2, and 3 days after doxycycline administration. (H) Equal quantities of MUC5B(G)-Luc and MUC5B(T)-Luc luciferase reporter vectors were transfected into BEAS2B cells stably expressing GFP and Spdef. Cells were collected 3 days after transfection for luciferase assays. Firefly luciferase activity was normalized to TK-Renilla (cotransfected with firefly luciferase constructs) by the dual luciferase assay; graphs present means ± SD of n = 9 independent experiments, analyzed with two-tailed, unpaired t test (A, G, and H) and one-way ANOVA-Tukey test (E and F). ***P < 0.001. NS = not significant.
To test whether XBP1S physically interacted with the MUC5B promoter, we performed ChIP-qPCR, using the HA antibody detecting the 3HA-Tag fused at the N terminus of XBP1S protein (see Figures E8A and E8B). The region (−126/−28, containing the −0.1 kb XBP1S consensus binding motif) proximal to the MUC5B transcription start site showed highly enriched binding by XBP1S compared with that of the distal region (−3552/−3472, containing the −3.5-kb putative binding sites) (10.4-fold vs. 4.2-fold of HA/IgG ChIP-qPCR). Although there is no XBP1S consensus binding motif predicted adjacent to the rs35705950 locus, XBP1S ChIP-qPCR detected enrichment of chromatin fragments (−3110/−3034) in this region compared with a nonconserved, XBP1S-negative binding region (−937/−785) (7.1-fold vs. 2.4-fold of HA/IgG ChIP-qPRC, respectively) (Figures 6B and 6C). Likewise, the promoter region of AGR2 (−540/−398), a mucin chaperon protein (45) and a known XBP1S-regulated gene (13), was also bound to XBP1S (see Figure E8C).
To further evaluate these findings, we mutated the −3.5-kb and −0.1-kb consensus binding sites, denoted as MUC5Bmut1 and MUC5Bmut2, respectively (Figures 6D; see Figure E8D). Compared with control subjects, XBP1S-induced luciferase activity was not affected in MUC5Bmut1, but almost completely lost in MUC5Bmut2 (reduced by 99.5%), whereas Spdef-induced activation was marginally reduced in both mutants (Figure 6E). Thus, the proximal XBP1S binding motif located on the −0.1-kb of cis-element was critical for XBP1S-induced MUC5B promoter activation, likely through direct protein–DNA interaction, whereas the distal −3.5-kb motif was dispensable. Neither site seemed to be critically important for Spdef-mediated activation alone (Figure 6E).
Because SPDEF regulates MUC5B expression and physically interacts with MUC5B promoter region that contains rs35705950 locus (26), and the ChIP-qPCR showed the evidence of XBP1S interaction with the rs35705950-containing chromatin fragments, we tested whether there was a synergy between XBP1S and SPDEF to activate the MUC5B promoter. Spdef synergistically enhanced XBP1S activation of the MUC5B promoter (three to four times higher than XBP1S alone on MUC5B[G]-Luc activity) (Figure 6F). In contrast, XBP1S marginally activated the MUC5AC promoter and inhibited SPDEF-induced MUC5AC promoter activation (see Figure E8E).
To evaluate potential links between XBP1S and MUC5B expression in the context of IPF, we tested whether the human MUC5B promoter variant rs35705950 affected regulation by XBP1S, Spdef, Foxa3, or by the combination of XBP1S and Spdef. Induction of MUC5B(T)-Luc (carrying the minor risk “T” allele) (see Figure E8F) by XBP1S was greater than MUC5B(G)-Luc at all times tested (80% higher after 3-d induction) (Figure 6G). In contrast, there was little difference between the two genotypes in regulation by Foxa3 or Spdef (Figures 6G and 6H). Spdef increased XBP1S-mediated activation of both MUC5B(T)-Luc and MUC5B(G)-Luc promoters (see Figure E8G). However, because this synergy was proportional for each genotype, this interaction did not enhance the XBP1S differential activation of MUC5B(T)-Luc versus MUC5B(G)-Luc. Thus, our data emphasize an XBP1S-dependent, differential upregulation of the gain-of-function rs35705950 IPF-risk variant as a key pathway in the regulation of MUC5B in IPF.
Loss of XBP1 Activity Inhibits MUC5B Expression Basally and after IL-1β Exposure
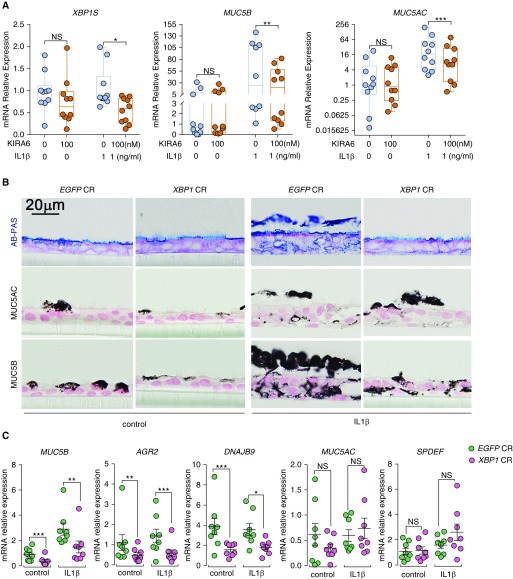
Because the ERN2/XBP1S cascade may be a therapeutic target for mucus obstruction dominated by MUC5B in IPF, the use of a pharmacologic inhibitor and CRISPR/Cas9-mediated DNA-based approaches to interdict this regulatory pathway were explored. Because KIRA6 is known for its attenuation effect of ERN1 RNase activity, we tested whether KIRA6 also decreases ERN2 RNase activity in human bronchial epithelial cells. BEAS2B cells expressing ERN1 or ERN2 were transfected with a XBP1 splicing-luciferase reporter (pCAX-F-XBP1ΔDBD-Luc) (46), followed by KIRA6 administration. KIRA6 moderately inhibited both ERN1- and ERN2-activated XBP1 mRNA splicing (see Figures E9A and E9B). Second, administration of KIRA6 (100 nM) to IL-1β-exposed, but not unchallenged, HAE cells was sufficient to inhibit IL-1β–induced XBP1 splicing and reduce MUC5AC and MUC5B mRNA expression without causing cytotoxicity (Figure 7A; see Figure E9C).
Figure 7.
KIRA6 and XBP1 CRISPR inhibit IL-1β–induced MUC5B expression in primary human airway epithelial (HAE) cells. (A) KIRA6 partially inhibits XBP1S, MUC5B, and MUC5AC. Primary HAE cells were cultured under acute lung injury (ALI) condition for 4 weeks to allow full differentiation before IL-1β exposure. At 24 hours after IL-1β (1 ng/ml in basolateral ALI media) exposure, KIRA6 was added in ALI media of the HAE cells preexposed with IL-1β at the final concentration of 100 nM. Expression of XBP1S, MUC5B, and MUC5AC mRNAs was determined 72 hours after KIRA6 treatment by SYBR green qRT-PCR or Taqman assays. Data represent n = 1 HAE cell culture from 10 independent non–cystic fibrosis/nonsmoker donors. Data were analyzed with two-tailed, ratio paired Student’s t test. (B) XBP1 CRISPR inhibited MUC5B expression. Primary HAE cells were stably infected with lentiviruses expressing control CRISPR (EGFP CR) or XBP1 CRISPR (XBP1 CR) and cultured under ALI for 4 weeks to allow full differentiation before IL-1β exposure (1 ng/ml). Five days after exposure with and without IL-1β, HAE cells were collected for histologic analysis, including alcian blue/periodic acid–Schiff (AB-PAS), MUC5AC, and MUC5B immunohistochemical staining. Micrographs are representatives of lenti-CRISPR-infected HAE cells obtained from three non–cystic fibrosis, nonsmoker donors. (C) Expression of MUC5B, AGR2, DNAJB9, MUC5AC, and SPDEF mRNAs in XBP1 CRISPR-targeted cells was analyzed with Taqman assays. Graphs represent mean ± SE with n = 1 HAE cell culture from eight different non–cystic fibrosis, nonsmoker donors. Data were analyzed with two-tailed, paired Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001. NS = not significant.
We next tested whether genetic deletion of XBP1S may also be therapeutic. A guide RNA was designed to target the human XBP1 gene, the precursor of XBP1S. An XBP1 CRISPR-Cas9 (XBP1 CR) lentivirus produced mutagenesis of approximately 85% of the XBP1 alleles compared with a control CRISPR (EGFP CR) (see Figure E10) in normal HAE cells, and XBP1CR inhibited MUC5B mRNA and protein expression and AGR2 and DNAJB9 mRNAs at baseline and after exposure to IL-1β (Figures 7B and 7C). In contrast, MUC5AC mRNA/protein and SPDEF mRNA were not affected by the XBP1 CRISPR (Figure 7C), suggesting XBP1S selectively regulates MUC5B expression in a SPDEF-independent manner.
Discussion
IPF is a rapidly progressive and typically fatal lung disease of unknown etiology. Although therapies that slow disease progression are available, these treatments do not restore function and their long-term effects on mortality are uncertain (47). Along with the classic features of myofibroblast differentiation, matrix deposition, and fibroblast proliferation, the abnormal abundance of MUC5B in the distal IPF lung is now widely recognized. After the strong genetic link between the MUC5B promoter variant rs35705950 and IPF risk was identified (4), the role of this variant in IPF has been the focus of research to understand the mechanisms triggering excessive MUC5B production and whether targeting this pathway could be a therapeutic option for IPF.
The transcription factors SPDEF (26), nuclear factor-κB (48, 49), and FOXA2 (7, 50) bind to both MUC5B and MUC5AC promoters; regulate their gene expression; and, hence, lack the specificity needed to differentially regulate these two mucins. The activation of XBP1S in airway epithelial cells in in vitro and in vivo models (Figures 3 and 4) describes at least one mucin selective regulatory mechanism. Specifically, XBP1S selectively induced the mRNA and promoter activities of MUC5B, but not MUC5AC, in vitro (Figures 3 and 6). The coexpression of XBP1S and MUC5B in airway epithelia of the human lung is consistent with a regulatory relationship between these two genes (Figure 5; see Figures E5 and E6). Finally, the selectivity of XBP1S for MUC5B expression was observed in vivo in a mouse model where selective induction of Muc5b, but not Muc5ac, resulted from Xbp1S overexpression driven by the Scgb1a1 promoter in mice. The mouse studies also raised the possibility that Xbp1S-mediated regulation of Muc5b expression dominates in distal, not proximal, airways (Figure 4). This notion is reinforced by previous studies in which the same promoter driving overexpression of Spdef (24, 25) or Foxa3 (51) induced goblet cell metaplasia in proximal but not distal airways. XBP1S-mediated increase of MUC5B expression associated with the gain-of-function of the MUC5B promoter variant in human distal airways is consistent with describing abnormal peripheral airways mucus accumulation in human IPF lungs (6, 52), and in a mouse model of Muc5b overexpression in the distal lung (53). Thus, the XBP1S regulation of MUC5B may provide a mechanism for selective regulation of MUC5B and a spatial (distal vs. proximal) pattern key to IPF pathogenesis.
The MUC5B promoter variant rs35705950 is a gain-of-function, expression quantitative trait locus polymorphism. Changing the “G” to a “T” increases MUC5B promoter activity in A549 cells (52). Although the SPDEF (26) and FOXA2 (7) DNA binding regions overlap with rs35705950 locus in the MUC5B promoter, no data are available as to their differential regulation of the activities of the MUC5B wild-type versus variant promoters. In contrast, we show that XBP1S differentially regulated MUC5B promoter variant activity. Promoter activity was proportionally increased when SPDEF was coexpressed with XBP1S for each variant. These data provide a mechanism linking ER stress–induced activation of the UPR transcription factor XBP1S and the mucous cell transcription factor SPDEF to net MUC5B promoter activity (Figure 6). Expression of SPDEF in IPF respiratory epithelia has been previously reported (27), and we detected expression of both XBP1S and SPDEF with MUC5B in distal airway epithelia in IPF (Figures 5E–5J). This finding is consistent with roles for XBP1S and, to a lesser degree, SPDEF in regulating the MUC5B promoter and its variant activities in IPF in vivo and in vitro.
Our episomal luciferase promoter and ChIP qPCR data indicated that the proximal −0.1-kb predicted XBP1S binding site, but not the distal −3.5-kb predicted site, dominates XBP1S binding and regulation of the MUC5B promoter activity. These data are also consistent with the presence of highly conserved cis regulatory elements in the proximal region of the MUC5B promoter among approximately 100 vertebrate species (see Figures E7B and E7E). Despite the fact that XBP1S differentially regulates rs35705950 promoter activity, and binds to the chromatin fragment surrounding this variant locus (Figures 6C and 6G), there is no predicted XBP1S binding motif in this region. We speculate that this differential regulation conferred by this site to MUC5B regulation may be mediated through cis regulatory protein–protein interactions. For example, interactions between XBP1S and SPDEF, the latter which physically interacts with its binding motif in this region (26), may contribute to the rs35705950-dependent differential regulation of MUC5B transcription. XBP1S and SPDEF protein–protein interaction studies, and XBP1S whole genome-wide ChIP-seq, are needed to address this conjecture. Furthermore, studies are also needed to elucidate whether DNA methylation and/or FOXA2, transcription factor that interacts with a consensus binding motif adjacent to rs35705950 locus (7), play roles in differential XBP1S-mediated regulation of MUC5B wild-type versus variant promoter activities.
Upstream of XBP1S are two ER stress sensors, ERN1 and ERN2. Different from its role in the gut, where it suppresses the mucin gene Muc2 (54), ERN2 increased MUC5AC and MUC5B mRNA expression in normal HAE cells (Figure 1I). Furthermore, ERN2 expression correlated with MUC5B mRNA expression in mucin-secretory cells in the proximal airway superficial epithelia and mucous cells in SMG (Figures 1E and 1G; see Figure E5B), suggesting a role in regulating mucin production in health. Although both ERN1 and ERN2 increased XBP1S, only the ERN2–XBP1S UPR pathway promoted mucin production. The mechanism underlying this selectivity for ERN2 mucin regulatory activity is unclear. One plausible explanation is that overexpression of ERN1 triggers an inhibitory pathway that blocks XBP1S-induced activation of MUC5B, perhaps reflecting a regulated IRE1-dependent decay of mRNA activity (55). An equally plausible explanation is that a separate ERN2-specific factor is required to promote MUC5B after XBP1S induction. Interestingly, mRNA expression of ERN2, but not ERN1, was induced in IPF/usual interstitial pneumonia compared with control lung tissues (1.75-fold; P = 8 × 10−7) (56), suggesting that ERN2 has a unique role in development of pulmonary fibrosis. Our detection of ERN2 mRNA in distal airways in IPF is consistent with this notion (Figures 5C and 5D; see Figure E6).
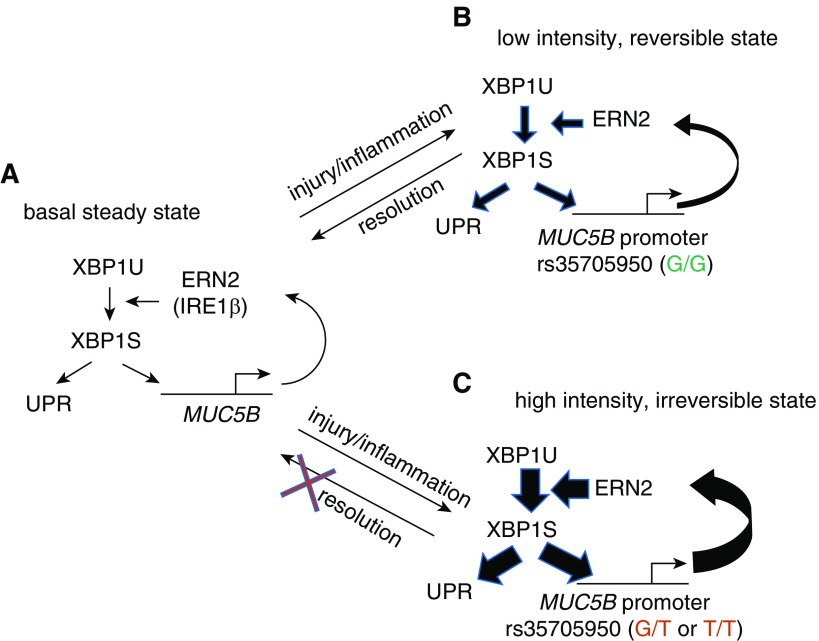
Our data support the model depicted in Figure 8 of ERN2/XBP1S-mediated regulation of MUC5B and its promoter variant in normal and IPF distal airway epithelia. In normal distal airway secretory cells, we postulate that a steady-state balance of MUC5B transcription/synthesis is maintained by a cycle of basal MUC5B transcription, production of MUC5B in the ER, which triggers ERN2 activation and XBP1S formation (Figure 8A). Note that a positive feedback relationship characterizes each component of this MUC5B regulatory pathway (i.e., it is a positive feedback system). Importantly, the positive feedback systems can exhibit both a low stimulus strength, low-intensity, and reversible state; and a high stimulus strength, high-intensity, and irreversible state (i.e., these systems are “bistable”) (57). In response to insults that produce injury and/or inflammation, which accelerate MUC5B transcription, ER stress is induced, ERN2 activated, and spliced XBP1 increases UPR gene and MUC5B transcription rates (Figure 8B). The signal strength of this response is relatively small because of the modest effect of XBP1S on regulation of MUC5B transcription in the context of the G/G genotype. Importantly, a “small” response is reversible on removal of the injury/cytokine stimulus. However, the presence of the MUC5B promoter minor variant (carrying “G/T” or “T/T” allele) amplifies XBP1S-induced MUC5B transcription. This amplification produces a high stimulus strength, reflecting a high rate of MUC5B synthesis that may produce an irreversible positive feedback state (Figure 8C). This state alone may be sufficient to trigger cellular responses that produce impaired host defense and accelerated cell senescence and/or damage. In addition, the persistence of this response may sensitize these “high-intensity” cells to second hits with other agents (e.g., oxidant stress), which further accentuate ER stress and abnormal cellular responses.
Figure 8.
A “bistable” model of ERN2/XBP1S-mediated regulation of MUC5B and its promoter variant in distal airway epithelia of idiopathic pulmonary fibrosis. (A) Basal MUC5B transcription produces MUC5B mRNAs that are translated into MUC5B protein at the endoplasmic reticulum (ER). The MUC5B protein synthetic load is sensed by ERN2 (and perhaps also by ERN1), which leads to splicing of XBP1U (unspliced XBP1) to XBP1S. XBP1S regulates basal level expression of MUC5B and unfolded protein response (UPR) genes. Note, each step reflects a positive regulatory interaction. (B) In a rs35705950 (G/G) distal airway mucin-secreting cell, inflammatory cytokines or other types of injury/stimuli stimulate MUC5B expression. An increased MUC5B mRNA load fuels increased MUC5B synthesis in ER, increased ERN2 activity, and XBP1S formation, and positive feedback by XBP1S on MUC5B transcriptional activity. The strength of this interaction is relatively weak, reflecting the “low-intensity state” of the XBP1S–MUC5B (G/G) promoter interaction. On removal of the stress-causing stimuli, the cell can revert to a low MUC5B basal secretion, low UPR activation state. (C) In a MUC5B rs35705950 (G/T or T/T) promoter distal airway mucus-secreting cell, stress signaling triggers overloaded MUC5B mRNA and protein synthesis, which in turn leads to ERN2 hyperactivation and increased XBP1S formation. Note, the regulatory interaction in the MUC5B promoter between XBP1S and rs35705950 locus is amplified by the presence of G/T or T/T at that site. This positive feedback reflects a “high-intensity state” of interaction, which produces a high-intensity signal response that imposes significant ER stress and may be irreversible. This state is sufficient to trigger an abnormal response to impair innate host defense and contribute to cell senescence. A second hit (e.g., oxidative stress) may signal these already “high-intensity” cells to accentuate ER stress–induced injury further, resulting in a profibrotic microenvironment that promotes fibroblast proliferation and myofibroblast differentiation (not shown in the illustration).
One approach to turn off this positive feedback (“vicious”) cycle is to inhibit the ERN2/XBP1S stimulation of MUC5B transcription. KIRA6 is a type II kinase inhibitor that stabilizes an inactive ATP-binding site conformation in ERN1, and dose-dependently inhibits ERN1 kinase and RNase activities and XBP1 splicing in vitro and in vivo (17). We showed that KIRA6 not only inhibits ERN1-, but also ERN2-induced XBP1 splicing in BEAS2B cells (see Figure E9B). Because Ern2 is required for both Muc5b and Muc5ac production in vivo (13), we speculated that following IL-1β exposure, KIRA6 not only would inhibit the kinase and RNase activities of ERN1, but likely that of ERN2, because of their conserved kinase domain homology. Consistent with this notion, KIRA6 inhibited IL-1β-stimulated increases in XBP1 splicing and MUC5AC and MUC5B mRNA expression levels in HAE cells (Figure 7A). Suppression of IL-1β–induced MUC5B expression likely reflected inhibition of ERN2-mediated XBP1 splicing, whereas the reduced MUC5AC expression was likely mediated by direct attenuation of ERN2 activity (Figure 1I). In these studies, IL-1β was used to generate a cell model that mimicked the phenotypes of IPF airway cells in terms of ER stress and mucin overproduction. Future studies of KIRA6 efficacy will require studies in focused models of IPF pathogenesis.
Targeting the XBP1 gene directly might also be a therapeutic option for suppressing excessive MUC5B in IPF. Both loss of function of XBP1 by CRISPR/Cas9-mediated deletion of XBP1 and gain of function by overexpression of Xbp1S selectively regulated MUC5B, without affecting MUC5AC expression (Figures 4, 7B, and 7C). Whereas MUC5B is the dominant mucin expressed in the distal airways of IPF lung, MUC5AC is also expressed (see Figures E6A and E6E). Our data suggest that if modulation of MUC5B alone is therapeutically preferred, targeting XBP1S may be an ideal approach. If reducing MUC5AC is also needed, targeting SPDEF (58) and/or ERN2 may be preferable.
In conclusion, we have demonstrated a novel molecular mechanism for positive regulation of MUC5B transcription by XBP1S in airway epithelium in both health and disease. XBP1 splicing can be triggered by inflammatory cytokines or other types of stimuli that trigger ER stress (e.g., injury) (59), mediated by ERN2 in mucin secretory cells (Figure 8). A positive feedback cycle, particularly in the context of the rs35705950 allele, may provide a mechanism to generate distal airway cells chronically expressing a maladaptive UPR response. The specificity of the ERN2–XBP1S pathway to increase MUC5B expression, and its ability to regulate MUC5B promoter activity in a rs35705950-specific manner, provides a rationale for designing therapeutic approaches to prevent and treat MUC5B-dominated mucus hyperproduction linked to UPR activation in pulmonary diseases, such as IPF.
Acknowledgments
Acknowledgment
The authors thank the laboratory of Jeffrey Whitsett at Cincinnati Children’s Hospital Medical Center for donating the Scgb1a1-rtTA (line 2) transgenic mice necessary for in vivo study and providing plasmids, antibodies, and other reagents for performing biochemistry assays in this study; the members of Marsico Lung Institute Tissue Procurement and Cell Culture Core for providing HAE cells and lung tissue; Dr. Gianni Carraro of Pulmonary Research, Cedars Sinai Medical Center, Los Angeles, California, for helping in idiopathic pulmonary fibrosis tissue procurement and processing; the members of the Marsico Lung Institute Molecular Biology Core, Kristy A. Terrell for assistance with mouse genotyping, and Lisa C. Morton for contributing to molecular biology aspects of this work; Dr. Arunava Ghosh for assistance in lactate dehydrogenase measurement; Histological Core, Kim Burns for tissue processing and performing hematoxylin-eosin, alcian blue/periodic acid–Schiff staining; Dr. Michael Chua for assistance with microscopy; and Eric C. Roe for editorial assistance.
Footnotes
This work was supported by the NHLBI P01 HL110873, P50 HL060280, P50 HL107168, R01 HL080396, P50 HL084934, UH2/UH3 HL123645 (R.C.B.), and P01 HL108808; National Institute of Diabetes and Digestive and Kidney Diseases P30 DK065988; and Cystic Fibrosis Foundation BOUCHE15R0 (R.C.B.) and R026-CR11 (W.K.O’N.).
Author Contributions: G.C. designed and performed experiments, analyzed results, and wrote the manuscript. L.S., K.O., T.K., R.C.G., M.B.M., A.A., J.M.L., E.A.H.-R., A.S.V., and A.L.-B. performed experiments and reviewed the manuscript. C.M.P.R., S.H.R., and B.R.S. prepared and provided reagents, cells, tissue specimens, interpreted results, reviewed the manuscript and provided comments and conducted discussions. Y.D. and P.E.S. generated and provided transgenic mice and antibody. H.D. performed statistical analysis. W.K.O’N. and R.C.B. conceived strategies, supervised the project, interpreted results, and wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201810-1972OC on April 11, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livraghi-Butrico A, Grubb BR, Wilkinson KJ, Volmer AS, Burns KA, Evans CM, et al. Contribution of mucus concentration and secreted mucins Muc5ac and Muc5b to the pathogenesis of muco-obstructive lung disease. Mucosal Immunol. 2017;10:395–407. doi: 10.1038/mi.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, et al. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am J Respir Crit Care Med. 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans CM, Fingerlin TE, Schwarz MI, Lynch D, Kurche J, Warg L, et al. Idiopathic pulmonary fibrosis: a genetic disease that involves mucociliary dysfunction of the peripheral airways. Physiol Rev. 2016;96:1567–1591. doi: 10.1152/physrev.00004.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helling BA, Gerber AN, Kadiyala V, Sasse SK, Pedersen BS, Sparks L, et al. Regulation of MUC5B expression in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2017;57:91–99. doi: 10.1165/rcmb.2017-0046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;302:L721–L729. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korfei M, Ruppert C, Mahavadi P, Henneke I, Markart P, Koch M, et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:838–846. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson WE, Crossno PF, Polosukhin VV, Roldan J, Cheng DS, Lane KB, et al. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: association with altered surfactant protein processing and herpesvirus infection. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1119–L1126. doi: 10.1152/ajplung.00382.2007. [DOI] [PubMed] [Google Scholar]
- 11.Lawson WE, Grant SW, Ambrosini V, Womble KE, Dawson EP, Lane KB, et al. Genetic mutations in surfactant protein C are a rare cause of sporadic cases of IPF. Thorax. 2004;59:977–980. doi: 10.1136/thx.2004.026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liptzin DR, Watson AM, Murphy E, Kroehl ME, Dishop MK, Galambos C, et al. MUC5B expression and location in surfactant protein C mutations in children. Pediatr Pulmonol. 2015;50:1270–1276. doi: 10.1002/ppul.23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martino MB, Jones L, Brighton B, Ehre C, Abdulah L, Davis CW, et al. The ER stress transducer IRE1β is required for airway epithelial mucin production. Mucosal Immunol. 2013;6:639–654. doi: 10.1038/mi.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyoshi K, Katayama T, Imaizumi K, Taniguchi M, Mori Y, Hitomi J, et al. Characterization of mouse Ire1 alpha: cloning, mRNA localization in the brain and functional analysis in a neural cell line. Brain Res Mol Brain Res. 2000;85:68–76. doi: 10.1016/s0169-328x(00)00243-6. [DOI] [PubMed] [Google Scholar]
- 15.Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 16.Imagawa Y, Hosoda A, Sasaka S, Tsuru A, Kohno K. RNase domains determine the functional difference between IRE1alpha and IRE1beta. FEBS Lett. 2008;582:656–660. doi: 10.1016/j.febslet.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, et al. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prischi F, Nowak PR, Carrara M, Ali MM. Phosphoregulation of Ire1 RNase splicing activity. Nat Commun. 2014;5:3554. doi: 10.1038/ncomms4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 22.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, et al. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest. 2009;119:2914–2924. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, et al. SPDEF regulates goblet cell hyperplasia in the airway epithelium. J Clin Invest. 2007;117:978–988. doi: 10.1172/JCI29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo M, Tomoshige K, Meister M, Muley T, Fukazawa T, Tsuchiya T, et al. Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Mol Med. 2017;9:462–481. doi: 10.15252/emmm.201606711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J Clin Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson MS, Madala SK, Ramalingam TR, Gochuico BR, Rosas IO, Cheever AW, et al. Bleomycin and IL-1beta-mediated pulmonary fibrosis is IL-17A dependent. J Exp Med. 2010;207:535–552. doi: 10.1084/jem.20092121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter AG, Perkins GD, Chavda A, Sapey E, Harper L, Thickett DR. Neutrophil chemotaxis in granulomatosis with polyangiitis (Wegener’s) and idiopathic pulmonary fibrosis. Eur Respir J. 2011;38:1081–1088. doi: 10.1183/09031936.00161910. [DOI] [PubMed] [Google Scholar]
- 32.Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, Paquet AC, et al. IL-13 and epidermal growth factor receptor have critical but distinct roles in epithelial cell mucin production. Am J Respir Cell Mol Biol. 2007;36:244–253. doi: 10.1165/rcmb.2006-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray LA, Zhang H, Oak SR, Coelho AL, Herath A, Flaherty KR, et al. Targeting interleukin-13 with tralokinumab attenuates lung fibrosis and epithelial damage in a humanized SCID idiopathic pulmonary fibrosis model. Am J Respir Cell Mol Biol. 2014;50:985–994. doi: 10.1165/rcmb.2013-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 35.Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci USA. 2011;108:8885–8890. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi S, Li Z, Yang HZ, Liu H, Wang JP, Ma YG, et al. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Immunol. 2011;187:3003–3014. doi: 10.4049/jimmunol.1004081. [DOI] [PubMed] [Google Scholar]
- 37.Park SW, Verhaeghe C, Nguyenvu LT, Barbeau R, Eisley CJ, Nakagami Y, et al. Distinct roles of FOXA2 and FOXA3 in allergic airway disease and asthma. Am J Respir Crit Care Med. 2009;180:603–610. doi: 10.1164/rccm.200811-1768OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajavelu P, Chen G, Xu Y, Kitzmiller JA, Korfhagen TR, Whitsett JA. Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J Clin Invest. 2015;125:2021–2031. doi: 10.1172/JCI79422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perl AK, Zhang L, Whitsett JA. Conditional expression of genes in the respiratory epithelium in transgenic mice: cautionary notes and toward building a better mouse trap. Am J Respir Cell Mol Biol. 2009;40:1–3. doi: 10.1165/rcmb.2008-0011ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123:455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long AJ, Sypek JP, Askew R, Fish SC, Mason LE, Williams CM, et al. Gob-5 contributes to goblet cell hyperplasia and modulates pulmonary tissue inflammation. Am J Respir Cell Mol Biol. 2006;35:357–365. doi: 10.1165/rcmb.2005-0451OC. [DOI] [PubMed] [Google Scholar]
- 42.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 43.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder BW, Verhaeghe C, Park SW, Nguyenvu LT, Huang X, Zhen G, et al. AGR2 is induced in asthma and promotes allergen-induced mucin overproduction. Am J Respir Cell Mol Biol. 2012;47:178–185. doi: 10.1165/rcmb.2011-0421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwawaki T, Akai R. Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem Biophys Res Commun. 2006;350:709–715. doi: 10.1016/j.bbrc.2006.09.100. [DOI] [PubMed] [Google Scholar]
- 47.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 48.Fujisawa T, Chang MM, Velichko S, Thai P, Hung LY, Huang F, et al. NF-κB mediates IL-1β- and IL-17A-induced MUC5B expression in airway epithelial cells. Am J Respir Cell Mol Biol. 2011;45:246–252. doi: 10.1165/rcmb.2009-0313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujisawa T, Velichko S, Thai P, Hung LY, Huang F, Wu R. Regulation of airway MUC5AC expression by IL-1beta and IL-17A; the NF-kappaB paradigm. J Immunol. 2009;183:6236–6243. doi: 10.4049/jimmunol.0900614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young HW, Williams OW, Chandra D, Bellinghausen LK, Pérez G, Suárez A, et al. Central role of Muc5ac expression in mucous metaplasia and its regulation by conserved 5′ elements. Am J Respir Cell Mol Biol. 2007;37:273–290. doi: 10.1165/rcmb.2005-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen G, Korfhagen TR, Karp CL, Impey S, Xu Y, Randell SH, et al. Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am J Respir Crit Care Med. 2014;189:301–313. doi: 10.1164/rccm.201306-1181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakano Y, Yang IV, Walts AD, Watson AM, Helling BA, Fletcher AA, et al. MUC5B promoter variant rs35705950 affects MUC5B expression in the distal airways in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;193:464–466. doi: 10.1164/rccm.201509-1872LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hancock LA, Hennessy CE, Solomon GM, Dobrinskikh E, Estrella A, Hara N, et al. Muc5b overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat Commun. 2018;9:5363. doi: 10.1038/s41467-018-07768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuru A, Fujimoto N, Takahashi S, Saito M, Nakamura D, Iwano M, et al. Negative feedback by IRE1β optimizes mucin production in goblet cells. Proc Natl Acad Sci USA. 2013;110:2864–2869. doi: 10.1073/pnas.1212484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang IV, Coldren CD, Leach SM, Seibold MA, Murphy E, Lin J, et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax. 2013;68:1114–1121. doi: 10.1136/thoraxjnl-2012-202943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 58.Chen G, Volmer AS, Wilkinson KJ, Deng Y, Jones LC, Yu D, et al. Role of Spdef in the regulation of Muc5b expression in the airways of naive and mucoobstructed mice. Am J Respir Cell Mol Biol. 2018;59:383–396. doi: 10.1165/rcmb.2017-0127OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen AC, Burr L, McGuckin MA. Oxidative and endoplasmic reticulum stress in respiratory disease. Clin Transl Immunology. 2018;7:e1019. doi: 10.1002/cti2.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]