Characterization of the two NSE4 paralogs in Arabidopsis revealed that NSE4A is essential for both genome stability and plant reproduction.
Abstract
The maintenance of genome integrity over cell divisions is critical for plant development and the correct transmission of genetic information to the progeny. A key factor involved in this process is the STRUCTURAL MAINTENANCE OF CHROMOSOME5 (SMC5) and SMC6 (SMC5/6) complex, related to the cohesin and condensin complexes that control sister chromatid alignment and chromosome condensation, respectively. Here, we characterize NON-SMC ELEMENT4 (NSE4) paralogs of the SMC5/6 complex in Arabidopsis (Arabidopsis thaliana). NSE4A is expressed in meristems and accumulates during DNA damage repair. Partial loss-of-function nse4a mutants are viable but hypersensitive to DNA damage induced by zebularine. In addition, nse4a mutants produce abnormal seeds, with noncellularized endosperm and embryos that maximally develop to the heart or torpedo stage. This phenotype resembles the defects in cohesin and condensin mutants and suggests a role for all three SMC complexes in differentiation during seed development. By contrast, NSE4B is expressed in only a few cell types, and loss-of-function mutants do not have any obvious abnormal phenotype. In summary, our study shows that the NSE4A subunit of the SMC5-SMC6 complex is essential for DNA damage repair in somatic tissues and plays a role in plant reproduction.
INTRODUCTION
The eukaryotic nuclear genome is packaged into higher order chromatin structures that are dynamically remodeled during cellular activities (Alabert and Groth, 2012). Key factors establishing and orchestrating chromosome organization are STRUCTURAL MAINTENANCE OF CHROMOSOME (SMC) complexes: cohesin (containing SMC1 and SMC3), condensin (containing SMC2 and SMC4), and the SMC5/6 complex (containing SMC5 and SMC6; reviewed in Hirano, 2006; Jeppsson et al., 2014b; Uhlmann, 2016). The heterodimeric SMC backbone serves as a structural component and a docking platform for additional subunits that vary depending on the complex, thereby enabling a variety of specific assemblies (reviewed in Kegel and Sjögren, 2010; Diaz and Pecinka, 2018). Studies in yeasts and animals showed that cohesin facilitates sister chromatin cohesion, and condensin I and II complexes mediate large-scale chromatin folding and chromosome condensation (reviewed in Hirano, 2012; Uhlmann, 2016). The major activity of the SMC5/6 complex is the maintenance of nuclear genome stability by resolving complex structures and possibly acting as an antagonist of the cohesin complex (reviewed in De Piccoli et al., 2009; Kegel and Sjögren, 2010; Diaz and Pecinka, 2018). The SMC5/6 complex performs many functions, such as the control of unidirectional rDNA replication, neutralizing toxic DNA intermediates during replication, preventing homologous recombination between nonhomologous sequences, and alternative telomere lengthening (Potts and Yu, 2007; Torres-Rosell et al., 2007; Chiolo et al., 2011; Menolfi et al., 2015).
The SMC5/6 complex can be associated with up to six NON-STRUCTURAL ELEMENT (NSE) subunits, which assemble in a combinatorial manner to form three subcomplexes (NSE1-NSE3-NSE4, NSE5-NSE6, and NSE2-SMC5-SMC6) in yeasts (De Piccoli et al., 2009; Duan et al., 2009). Studies in budding yeast, fission yeast, and mammalian cell cultures revealed that the NSE1-NSE3-NSE4 subcomplex binds double-stranded DNA and acts as a binding platform for the heads of SMC5 and SMC6 (Hudson et al., 2011; Palecek and Gruber, 2015; Zabrady et al., 2016. The least evolutionary conserved SMC5/6 complex subunits are NSE5 and NSE6. They interact with the SMC5-SMC6 hinges in budding yeast but with their heads in fission yeast (Pebernard et al., 2006; De Piccoli et al., 2009; Duan et al., 2009). Recently, functional orthologs of NSE5 and NSE6 have been identified in plants and mammals (Yan et al., 2013; Räschle et al., 2015), but their molecular functions remain unclear. NSE2 (also known as METHANE METHYLSULFONATE SENSITIVE21 [MMS21] and HIGH PLOIDY2 [HPY2]) is anchored to SMC5 and has SMALL UBIQUITIN-RELATED MODIFIER E3 ligase activity (Zhao and Blobel, 2005). Many proteins were found to be targets of NSE2 sumoylation, including several SMC5/6 and cohesin subunits, as well as DNA repair proteins in plants, fungi, and animals (Zhao and Blobel, 2005; Pebernard et al., 2006; Potts and Yu, 2007; Huang et al., 2009; Ishida et al., 2009).
Homologs of all SMC5/6 complex subunits were identified in Arabidopsis (Arabidopsis thaliana; Schubert, 2009; Watanabe et al., 2009; Yan et al., 2013; Diaz and Pecinka, 2018). However, our understanding of biological processes controlled by the individual SMC5/6 complex subunits remains limited in plants. Arabidopsis plants mutated in SMC6B (also known as HYPERSENSITIVE TO MMS, IRRADIATION, AND MITOMYCIN C [MMC]) are indistinguishable from the wild type under ambient conditions but are hypersensitive to DNA damaging treatments, show a delayed repair of DNA strand breaks, and have a reduced frequency of homologous recombination (Mengiste et al., 1999; Kozak et al., 2009; Watanabe et al., 2009; Liu et al., 2015). smc6a mutants are viable even under severe DNA damage, but smc6a smc6b double mutation is embryo lethal (Watanabe et al., 2009; Yan et al., 2013), indicating partial functional redundancy. Plants defective in NSE2 are hypersensitive to DNA damage and display a wide range of pleiotropic phenotypes, including leaf and stem malformations, branching defects, reduced meristem size, impaired development of gametes, shortened vegetative phase, and increased drought tolerance (Huang et al., 2009; Ishida et al., 2009; Xu et al., 2013; Zhang et al., 2013; Liu et al., 2014; Yuan et al., 2014; Kwak et al., 2016). SMC5, SMC6, and NSE1, NSE2, NSE3 and NSE4 are evolutionary conserved proteins. In addition, there are two other SMC5/6 complex subunits (collectively named as NSE5 and NSE6) in fungi, animals, and plants, which are presumably functionally conserved but share little sequence similarity (reviewed in Diaz and Pecinka, 2018). In Arabidopsis, both the regulator of systemic acquired resistance SUPPRESSOR OF NPR1-1, INDUCIBLE1 (SNI1) and the ARABIDOPSIS SNI1 ASSOCIATED PROTEIN1 (ASAP1) were found in a complex with SMC5 and SMC6B and were thus proposed as the putative functional orthologs of yeast NSE6 and NSE5, respectively (Yan et al., 2013). Both genes participate in the control of genome stability and suppression of immune hyper-responses, which is a novel and unexpected function of the complex.
The variety of plant phenotypes seen in mutants affecting the SMC5/6 complex suggests that it participates in multiple developmental and cellular pathways possibly linked to stress responses. Currently, the composition of the plant SMC5/6 complex, the roles of individual subunits, and their functional requirement in cellular and developmental processes (besides DNA damage repair) are poorly characterized. In an effort to obtain a more comprehensive functional understanding of the Arabidopsis SMC5/6 complex, we characterized the roles of the NSE4A and NSE4B subunits. We show that NSE4A is involved in repair of zebularine-induced DNA damage in challenged somatic tissues. In addition, NSE4A is essential for reproductive development in Arabidopsis, while the function of NSE4B remains elusive.
RESULTS
The NSE4 Gene Is Duplicated in the Arabidopsis Genome
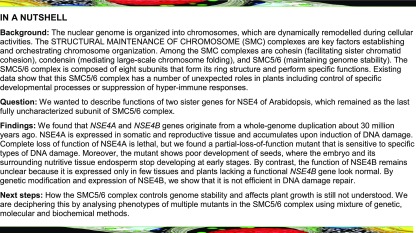
The Arabidopsis genome contains two uncharacterized, putative, NSE4 homologs: NSE4A (At1g51130 encoding a 403 amino acid protein) and NSE4B (At3g20760 encoding a 383 amino acid protein) sharing 65.1% identity at the amino acid level (Figures 1A and 1B). To identify the age of this duplication, we built a NSE4 phylogeny across green plants using the Schizosaccharomyces pombe and Homo sapiens NSE4s as outgroups (Figure 1C; Supplemental Table 1; Supplemental Data Sets 1 to 3). Except forBryophyta and Marchantiophyta, which carry a single NSE4, all other plant genomes contained at least two NSE4 copies. Orthologs of Arabidopsis NSE4A and NSE4B occurred in Arabidopsis lyrata, Capsella rubella, and Eutrema salsugineum. The only exception was Brassica rapa, where both NSE4 copies were derived from NSE4A, while NSE4B was missing. This suggests that the NSE4A and NSE4B originate from the whole-genome duplication event that occurred ∼47 million years ago (MYA) and preceded radiation of the species within Brassicaceae (Kagale et al., 2014). Phylogenetic shadowing of NSE4A and NSE4B promoters revealed that both contain conserved blocks, A1 and B1, respectively, directly upstream of the transcription start site (Supplemental Figure 1; Supplemental Table 2). However, the A1 block was clearly larger and more similar between species, indicating that it may contain key NSE4A cis-regulatory sequences. There was another set of conserved NSE4 paralogs in Poaceae, including Brachypodium distachyon, Hordeum vulgare, and Zea mays (Figure 1C). These paralogs most likely appeared during the Poaceae-specific whole-genome duplication event ∼70 MYA (Paterson et al., 2009). We found a total of six NSE4 copies in rice and four in tomato. Some of these copies were short and grouped with more distantly related species (Figure 1C), raising questions on their origin and functionality. The high frequency of multiple NSE4 copies per genome may indicate rapid NSE4 sub- or neo-functionalization in different plant lineages.
Figure 1.
Basic Characterization of NSE4 Paralogs.
(A) Gene structure of A. thaliana NSE4A and NSE4B with indicated positions of the mutations used in this study. Bars, 100 bp.
(B) Alignment of Arabidopsis NSE4A and NSE4B proteins.
(C) Phylogenetic tree of NSE4 homologs in plants based on the maximum likelihood algorithm (see “Methods”). Fission yeast NSE4/RAD62 and human NSE4 paralogs were used as outgroups. Brassicaceae and Poaceae NSE4 duplications are indicated by the colored squares. Identifiers of the protein sequences used to build the tree are provided as Supplemental Data Set 1.
(D) to (F) Phenotypes of the homozygous wild-type (WT), nse4a-2 (4a-2), nse4a-2 complemented with ProNSE4A:GenomicNSE4A (4a-2 com4A), nse4b-2 (4b-2), and nse4a-2 nse4b-2 (4a-2 4b-2) plants. (D) One-week-old in vitro–grown seedlings. Bar = 10 mm. (E) Three-week-old plants in soil. Bar = 25 mm. (F) Six-week-old mature plants. Bar = 35 mm.
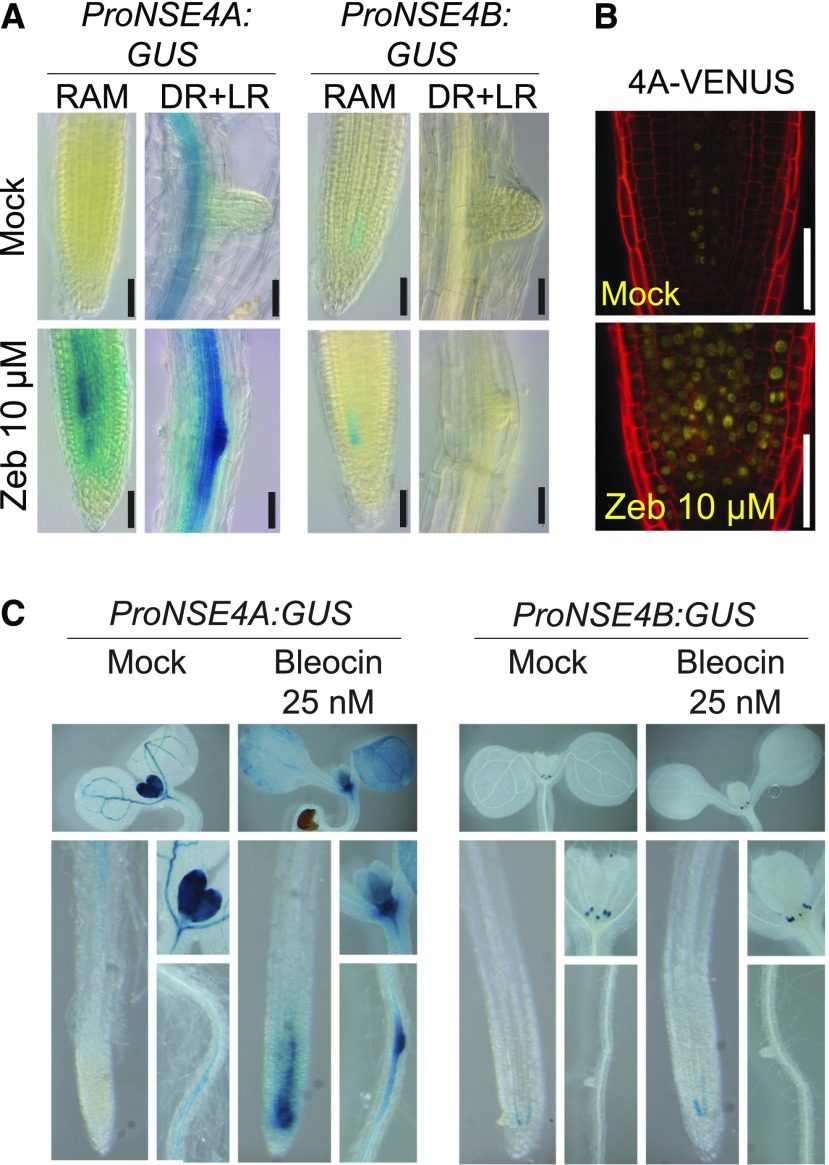
(G) to (I) Analysis of NSE4A and NSE4B promoter activity using the GUS reporter system. (G) One-week-old plants grown as described in (D). Red arrowheads indicate ProNSE4B:GUS signals in the root meristematic zone and leaf stipules (top inset). (H) Fourteen-day-old plants grown in in vitro culture. (I) Flowers at developmental stage (Stg) 10 to 14 (Bowman et al., 1994). Bars = 500 μm.
To assess the role of the NSE4 genes in plant growth and development, we isolated T-DNA insertion mutations in NSE4A and NSE4B (Figure 1A). The nse4a-1 allele carried a T-DNA in the second exon and was lethal as indicated by the absence of homozygous mutants in the progeny of heterozygous parents. However, we recovered viable homozygous nse4a-2 plants carrying a T-DNA insertion in the last exon, 56 amino acids before the stop codon (Supplemental Figure 2). A 3′ rapid amplification of cDNA ends (RACE) technique revealed that the NSE4A transcript in nse4a-2 plants continued into the T-DNA and maintained the reading frame for 201 nucleotides, adding a predicted 67 alien amino acids to the NSE4A protein produced by nse4a-2 mutants. Therefore, nse4a-2 most likely represents a partial loss-of-function allele with a modified C terminus. Juvenile and nonflowering nse4a-2 plants were smaller than the wild type (Figures 1D and 1E) but recovered and were indistinguishable from control plants at flowering (Figure 1F). The nse4b mutant alleles carried T-DNA insertions in the second intron (nse4b-1) and the fifth exon (nse4b-2), respectively. Amplification from cDNA with primer pairs positioned on either side of the T-DNA insertions yielded very low or no products in quantitative PCR, suggesting that both insertions disrupt the NSE4B transcript (Supplemental Figure 3). However, both nse4b-1 and nse4b-2 plants were viable and resembled the wild-type plants (Figures 1D to 1F). Combining the nse4a-2 and nse4b-2 alleles in a homozygous double mutant resulted in a nse4a-2–like phenotype, suggesting that NSE4A and NSE4B do not act redundantly during vegetative development.
To reveal the activity pattern of the NSE4 promoter, we generated stable reporter lines where the NSE4A and NSE4B promoters were fused to the uidA gene encoding β-glucuronidase (GUS; ProNSE4A:GUS and ProNSE4B:GUS). The NSE4A promoter was strongly active in emerging true leaves and weakly active in the vasculature of the cotyledons at 7 d after germination (DAG; Figure 1G). In addition, we observed signals in the stele tissues within the differentiation zone of the root, but there was no ProNSE4A activity in root meristems. At 14 DAG, ProNSE4A was weakly active in all aerial tissues (Figure 1H). Flowers showed ProNSE4A:GUS activity in sepals, the upper half of fully elongated anther filaments, pistils, and anthers (Figure 1I, top). By contrast, ProNSE4B:GUS activity was restricted to the leaf stipules and a small domain in the root apical meristem at 7 DAG (Figure 1G, red arrowheads and insets). This pattern remained unchanged during the entire vegetative phase (Figure 1H). In flowers, ProNSE4B was active in anthers between stages 10 and 12 (Figure 1I). The difference in the expression patterns of NSE4A and NSE4B could be due to the association of the endogenous NSE4B locus with repressive histone H3 Lys-27 trimethylation (Supplemental Figures 4 and 5).
NSE4A Is Expressed in Pollen, Ovules, and Seeds
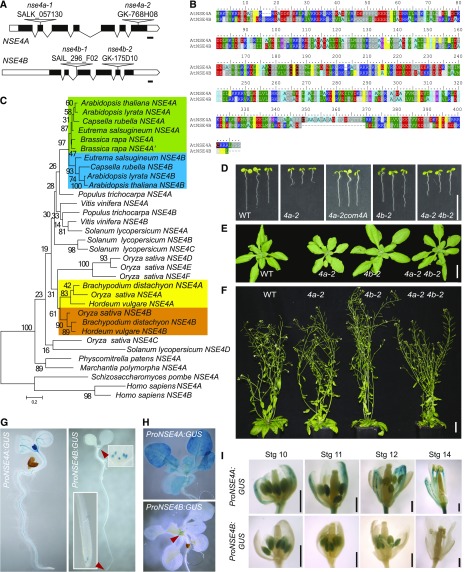
The activity of ProNSE4A and ProNSE4B in flowers prompted us to analyze the reproductive stages in more detail. To get better insight into the expression of the NSE4A protein, we expressed a translational fusion of NSE4A with VENUS (an improved variant of the yellow fluorescent protein; Nagai et al., 2002) under the control of its native promoter (ProNSE4A:NSE4A-VENUS) in the nse4a-2 background. Based on the full complementation of nse4a-2 hypersensitivity to zebularine (Figure 2A), we conclude that the addition of VENUS does not interfere with NSE4A function.
Figure 2.
NSE4 Expression Analysis during Pollen, Ovule, and Seed Development.
(A) Test for functionality of NSE4-VENUS translational fusion line. Wild-type (WT), nse4a-2 (4a-2), and nse4a-2 plants complemented with ProNSE4A:NSE4A:VENUS (4A-VENUS) were germinated and grown on the control and zebularine-containing media for 7 d. Restoration of root growth in 4a-2 NSE4A-VENUS indicates full functionality of the translational fusion protein.
(B) The first two columns show DAPI- and GUS-stained pollen of ProNSE4A:GUS (Pro4A:GUS) reporter line. Stage (Stg) 10 corresponds to the microspore, Stg 11 to bicellular pollen, Stg 12 to tricellular pollen, and Stg 14 to mature pollen from open anthers. The last column shows pollen from the ProNSE4A:NSE4A:VENUS (4A-VENUS) reporter line. Bar = 5 µm.
(C) The ProNSE4B:GUS (Pro4B:GUS) reporter line presented in the same way as in (A). Bar = 5 µm. Stg, stage.
(D) GUS activity of ProNSE4A:GUS (Pro4A:GUS; left) and ProNSE4B:GUS (Pro4B:GUS; right) from ovule primordia to early postfertilization. Stage (Stg) 10, 11, and 12 to 14 show ovule primordia, the nucellus, and developing the embryo sac, respectively. Bars = 50 µm.
(E) ProNSE4A:NSE4A:VENUS (4A-VENUS) signals at the same stages as described in (C). In the ovule primordia of stage (Stg) 11, the megaspore mother cell is almost free of 4A-VENUS signal (arrowheads). However, its expression is greatly increased in the female meiocyte of Stg 11 (arrowhead). Bar = 10 µm.
(F) GUS activity driven by the NSE4A and NSE4B promoters at the indicated hours after pollination (HAP). Reporter lines were pollinated with their own pollen 48 h after emasculation. Bars = 50 µm. e, embryo; ce, chalazal endosperm.
(G) Accumulation of ProNSE4A:NSE4A:VENUS (4A-VENUS) in nuclei of globular-, heart-, torpedo-, and bent cotyledon–stage embryos and syncytial endosperm 72 h after pollination. Left images represent differential interference contrast (DIC), and the right images show the VENUS signal. Bars = 50 µm.
Analysis of the transcription during pollen development revealed strong and weak activity of ProNSE4A and ProNSE4B, respectively (Figures 2B and 2C). The microspores (flower stage 10; Bowman et al., 1994) showed, on average, the strongest signals for both ProNSE4A:GUS and ProNSE4B:GUS, which decreased over subsequent developmental stages. There was practically no transcriptional activity of both genes in mature pollen from open anthers (flower stage 14). At the protein level, NSE4A was present at all pollen stages in the cell lineage leading to the sperm cells, as indicated by VENUS signals in the single nucleus of the unicellular microspore, the generative nucleus of bicellular pollen (flower stage 11), and the two sperm nuclei of tricellular pollen (Figure 2B). No NSE4A-VENUS signal could be observed in the vegetative nucleus.
During ovule development (Figure 2D), we observed ProNSE4A:GUS activity in ovule primordia at flower stage 10, the nucellus at stage 11, and the embryo sac in stages 12 to 14. The transcriptional profile was largely in agreement with NSE4A protein accumulation (Figure 2E). Strong NSE4A-VENUS signals were observed in almost all cells of the nucellus except for the megaspore mother cell, where the fusion protein was barely detectable (Figure 2E, flower stage 10, arrowhead). However, NSE4A-VENUS accumulated strongly in female meiocytes initiating meiotic prophase I (Figure 2E, flower stage 11, arrowhead). The differences between GUS and VENUS signals could be due to different stability of GUS mRNA and/or protein compared with NSE4A-VENUS transcript and/or protein. After pollination, ProNSE4A activity was detected in the embryo and the chalazal endosperm and later (at 96 h after pollination) also in the syncytial endosperm (Figure 2F). This corresponds well with the strong NSE4A-VENUS signals in developing embryos (Figure 2G) and also the prominent localization to the nuclei of the syncytial endosperm (Figure 2G). By contrast, ProNSE4B activity during early ovule development remained largely below detection limit (Figure 2D), and we detected weak activity only in mature embryo sacs, with GUS activity getting stronger after pollination, leading to a clear signal in the early embryo up to the globular stage (Figure 2F).
In summary, these results confirmed NSE4A to be a nuclear protein, as expected for a DNA repair factor, and revealed a dynamic expression pattern of NSE4A during sporogenesis, gametogenesis, embryogenesis, and endosperm development. The high levels of NSE4A during meiosis and in the proliferating fertilization products may be linked with its DNA repair function, for example, during meiotic crossing-over or to ensure genome integrity during the fast mitoses in embryo and endosperm.
NSE4A Plays a Role in Seed Development
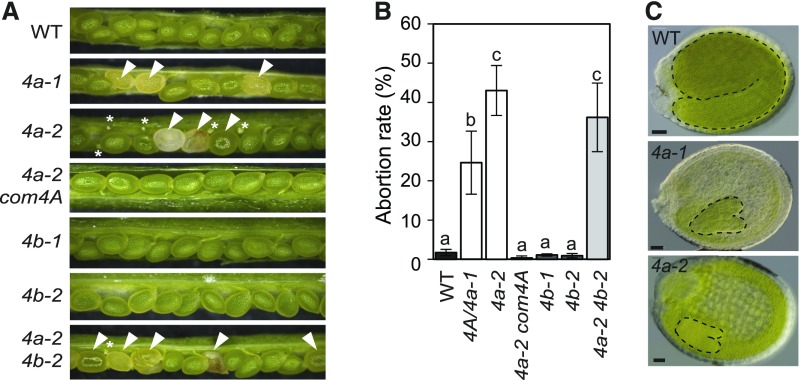
Prompted by NSE4 expression in seeds, we analyzed fertility of nse4a and nse4b mutants 2 weeks after pollination (Figures 3A and 3B). In contrast to the wild-type plants, siliques from nse4a-1/NSE4A heterozygotes produced 28.8% abnormal seeds (pale seeds representing delayed embryos and/or aborted seeds; n = 1402, Figures 3A and 3B). Fertility was even more impaired in homozygous nse4a-2 plants, with approximately one-half (53.4%) of the seeds developing normally, 22% showing early aborted ovules, and 24.6% showing abnormally large seeds with a glossy surface and liquid endosperm (n = 1008). Clearing of abnormal nse4a-1 and nse4a-2 seeds revealed that the embryos were arrested at the heart or heart-to-torpedo transition stages, respectively (Figure 3C; Supplemental Figure 6A). A NSE4A genomic construct could fully rescue the nse4a-2 mutant seed phenotype (up to 96.5% normal seeds, n = 949), confirming that embryo unviability is a consequence of the loss of NSE4A function (Figures 3A and 3B). To test whether the increased frequency of abnormal seeds in nse4a-1 heterozygous plants (28.8% observed versus expected 25%) is due to preferential transmission of the mutant allele or a partial gametophytic maternal effect, nse4a-1/NSE4A heterozygous plants were self-pollinated and reciprocally crossed to the wild-type plants. The frequency of late aborted seeds resulting from these crosses was scored (Supplemental Figure 6B). Reciprocal crosses resulted in 0.6 to 2.0% late aborted seeds, indistinguishable from the wild-type control, while self-pollinated nse4a-1/NSE4A heterozygous plants produced 23.9% late aborted seeds. These results indicate that nse4a-1 is a zygotic embryo-lethal mutation. By contrast, and in agreement with the NSE4B expression pattern, nse4b-1 and nse4b-2 single mutants were fully fertile, while the nse4a-2 nse4b-2 double mutant showed a similar phenotype as the nse4a-2 single mutant (Figures 3A and 3B). Hence, NSE4A is required for normal seed development, while NSE4B is dispensable.
Figure 3.
NSE4A Is Necessary for Seed Development.
(A) Seed phenotypes in the wild-type (WT), heterozygous self-pollinated NSE4A/nse4a-1 (4a-1), homozygous nse4a-2 (4a-2), homozygous nse4a-2 complemented with genomic NSE4A locus (4a-2 com4A), nse4b-1 (4b-1), nse4b-2 (4b-2), and homozygous 4a-2 4b-2 double mutant plants. Abnormally developing seeds are indicated by white arrowheads. Nondeveloping ovules are indicated by white asterisks.
(B) Quantification of aborted seeds in the genotypes listed in (A). Error bars indicate sd between means of three biological replicates. Each replicate was represented by one plant from which 140 to 300 seeds were analyzed. All plants were grown at the same time. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05). WT, wild type.
(C) Equally old cleared wild-type (WT), pale self-pollinated NSE4A/nse4a-1 (4a-1), and large nse4a-2 (4a-2) seeds. Additional nse4-2 seeds are shown in Supplemental Figure 6A. Embryos were outlined by black dashed lines for easier visibility. Bars = 50 µm.
NSE4A Is Involved in Somatic DNA Damage Repair
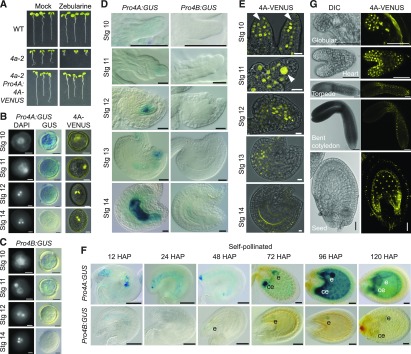
Next, we tested which of the Arabidopsis NSE4 paralogs is involved in DNA damage repair. First, we scored for the transcriptional response of NSE4A and NSE4B to drug treatment using the promoter-GUS reporter lines (Figure 4). No induction was observed for ProNSE4B:GUS upon treatment with DNA damaging agents including zebularine (10 µM), which (similarly to the related drug 5-azacytidine; reviewed in Stingele and Jentsch, 2015; Tretyakova et al., 2015) generates enzymatic DNA–protein crosslinks by covalently trapping DNA Methyltransferase 1 class enzymes, and bleocin (25 nM), which causes DNA strand breaks (Figures 4A and 4C). By contrast, ProNSE4A became active throughout the entire meristematic zone and in the emerging lateral roots (Figures 4A and 4C), indicating that NSE4A is activated by different types of DNA damage. This transcriptional activation was accompanied by protein accumulation as indicated by NSE4A-VENUS signals within a larger area of the root apical meristem of stressed reporter plants (Figure 4B).
Figure 4.
NSE4A Is Induced Upon DNA Damage Stimulus.
(A) Transcriptional response of the ProNSE4A and ProNSE4B promoters after 7 d of treatment with 10 µM zebularine (Zeb) in the root apical meristem (RAM) and differentiated root (DR) section with emerging lateral roots (LR). Scale bars = 50 µm.
(B) nse4a-2 ProNSE4A:NSE4A:VENUS (4A-VENUS) accumulation in the RAM under control conditions and with 10 µM zebularine (Zeb). Error bars = 50 µm.
(C) Transcriptional response of the ProNSE4A and ProNSE4B promoters to 25 nM bleocin treatment. Each composite image shows (from top to down and from left to right) the following: cotyledons and the first pair of true leaves, main root apical meristem, detail of the first pair of true leaves, and differentiated root zone.
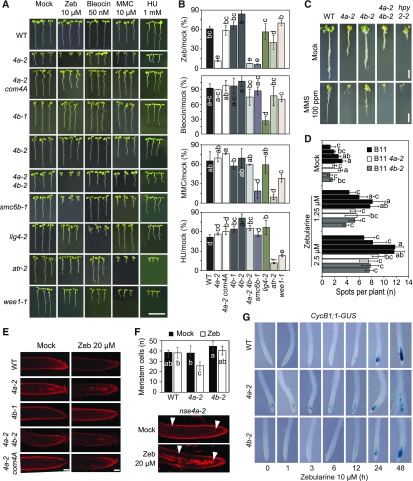
Subsequently, we assessed the functional contribution of the NSE4 genes to plant survival upon drug-induced DNA damage. To this aim, we monitored the growth of the wild-type, nse4a-2 single mutant, nse4a-2 complemented with NSE4A genomic construct (ProNSE4A:NSE4A:TerNSE4A), nse4b (both alleles), and nse4a-2 nse4b-2 double mutant plants on media containing 10 µM zebularine, 50 nM bleocin, 10 µM MMC, or 1 mM hydroxyurea (HU; Figures 5A and 5B; Supplemental Figure 7). In a separate assay, we applied the DNA alkylating agent methyl methanesulfonate (MMS; Fig. 5C), which caused poor growth of the Arabidopsis smc6b-3 (mim-1) mutant (Mengiste et al., 1999). As positive controls, we used the drug-sensitive ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED (ATR) signaling kinase mutant (atr-2), the DNA LIGASE4 mutant (lig4-2), WEE1 KINASE HOMOLOG mutant (wee1-1), and mutants in the two SMC5/6 complex subunits, SMC6B (smc6b-1) and HPY2 (hpy2-2; De Schutter et al., 2007; Ishida et al., 2009; Yuan et al., 2014; Liu et al., 2015). The nse4b-1 and nse4b-2 single mutants were not hypersensitive to any of the applied genotoxic treatments (Figures 5A to 5C). The nse4a-2 single and nse4a-2 nse4b-2 double mutants were indistinguishable from the wild type under MMC, bleocin, and HU stress, but they were strongly hypersensitive to zebularine and MMS (Figures 5A to 5C). By contrast, smc6b-1 was also hypersensitive to MMC treatment, which could be due to the fact that nse4a-2 is only a partial loss-of-function allele. To test for effect on homologous recombination (HR) rates, we generated nse4a-2 nse4b-2 double mutants carrying the reporter N1DC1 No. 11 (B11) with 566 bp overlap of GUS recombination substrate in direct orientation (Puchta et al., 1995). The plants were grown for 10 d in media containing low amounts of zebularine (1.25 and 2.5 µM) to avoid lethality. We used multiple independent lines of each analyzed genotype, which showed a zebularine dose-dependent increase in HR rate, but no significant differences between the wild-type, nse4a-2, and nse4b-2 lines (Figure 5D). This result differs from those published for hpy2 and smc6 mutants, which showed reduced HR rates (Mengiste et al., 1999; Watanabe et al., 2009; Yuan et al., 2014). On the one hand, this may suggest that NSE4 proteins are not controlling single strand annealing type of HR in Arabidopsis. On the other hand, these results should be interpreted with caution because nse4a-2 is not a null allele and nse4b mutants are not sensitive to DNA damage treatments.
Figure 5.
NSE4A Is Involved in Somatic DNA Damage Repair.
(A) Sensitivity to genotoxic stress. The wild-type (WT), nse4a-2 (4a-2), nse4b-1 (4b-1), nse4b-2 (4b-2), nse4a-2 nse4b-2 (4a-2 4b-2), nse4a-2 complemented with genomic NSE4A locus (4a-2 com4A), smc6b-1, lig4-2, atr-2, and wee1-1 plants were germinated and maintained for 1 week on 10 µM zebularine (Zeb), 50 nM bleocin, 10 µM MMC, or 1 mM HU. Bar = 10 mm.
(B) Quantitative data for (A) calculated as the relative root length under drug versus control conditions. Error bars represent sd between means of three biological replicates. The replicates were grown on separate screening plates, and each contained at least 25 plants. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05). WT, wild type.
(C) Sensitivity to MMS. Representative phenotypes of the wild-type (WT), 4a-2, 4b-2, 4a-2 4b-2 double mutant, and hpy2-2 plants grown for 1 week in control liquid medium and then for 3 weeks in control and 100 ppm MMS-containing media. Bar = 10 mm.
(D) Analysis of DNA damage repair by homologous recombination using B11 reporter line in the wild-type (WT), 4a-2, and 4b-2 backgrounds. Identically colored columns represent individual lines obtained from segregating hybrid populations. Error bars represent mean of three biological replicates, each with at least 30 plants. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05).
(E) Cell death assay. PI-stained roots from living Arabidopsis seedlings treated without (Mock) and with 20 µM zebularine (Zeb) for 24 h. WT, wild type.
(F) Meristem size estimation. Plants from (E) were used to estimate the number of cells within the root apical meristem (indicated by white arrowheads). Error bars in graph indicate sd among primary roots from 5 to 12 analyzed plants per each genotype. All plants were grown at the same time. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05). WT, wild type; Zeb, zebularine.
(G) G2/M cell cycle progression in nse4a-2 and nse4b-2 analyzed by ProCycB1;1:CycB1;1:GUS (CycB1;1-GUS) after exposure to 10 µM zebularine for the indicated number of hours.
Inhibition of root growth in response to DNA damage is frequently accompanied by increased cell death. Therefore, we monitored the amount of dead cells using the propidium iodide (PI) uptake assay in control and 20 µM zebularine-treated plants (Figure 5E). While there were no or few dead cells in the wild-type and nse4b-2 plants, nse4a-2 single and nse4a-2 nse4b-2 double mutant plants showed a drastic increase upon zebularine treatment. The drug sensitivity phenotype (growth and cell death) of nse4a-2 to zebularine is directly due to the loss of NSE4A activity as shown by complementation using an NSE4A genomic construct (Figures 5A, 5B, and 5E). We noticed that the root meristem was partially disorganized in zebularine-treated nse4a-2 plants. Therefore, we estimated the meristem size by counting the number of cells in the cortex layer between the quiescent center and the differentiation zone (Figure 5F). The wild-type and nse4b-2 roots contained 38 to 45 cells, and this number did not change significantly after 24 h of 20 µM zebularine treatment (analysis of variance, post hoc Duncan’s test, P > 0.05). By contrast, nse4a-2 showed a significant 31% reduction to 26 cells upon zebularine treatment. To test the effect of the mutation on cell cycle regulation, we introduced a G2/Mitosis DNA damage reporter, which utilizes a translational fusion between CyclinB1;1 and GUS (Colón-Carmona et al., 1999), into nse4a-2 and nse4b-2 mutant backgrounds. The chimeric protein accumulates specifically in the G2 phase of cycling cells and is destroyed at the onset of mitosis, resulting in a loss of the signal. Double homozygous lines were exposed to 10 µM zebularine for up to 48 h, and the domain of GUS expression was monitored (Figure 5G). The nse4a-2 roots showed an increased number of GUS-positive cells already at 0 h, indicating a prolonged G2 phase. After 48 h of treatment, meristems of nse4a-2 plants were damaged, as indicated by an abnormal root morphology and root hairs emerging close to the root tips. The response in nse4b-2 and the wild type was slower, less severe, and similar between the two (Figure 5G).
Collectively, these results demonstrate that NSE4A responds to genotoxic stress, is likely involved in DNA repair of zebularine-induced DNA–protein crosslinks, and is required to promote cell division in response to this genotoxic drug, possibly to actively propagate cells after repair.
Loss of NSE4A Function Causes Upregulation of DNA Damage Repair and Immune Response Genes
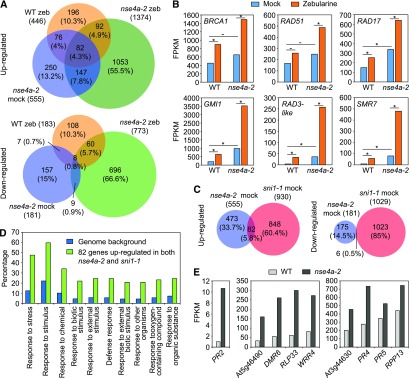
We analyzed the effect of the nse4a-2 mutation on gene expression by RNA sequencing using dissected shoot apices from the 10-d-old wild-type and nse4a-2 plants treated without (mock) and with 20 μM zebularine for 24 h (Figure 6; Supplemental Data Set 4). In mock-treated nse4a-2, we identified 555 significantly upregulated genes and 181 significantly downregulated genes relative to the mock-treated wild type (Figure 6A; DESeq, adjusted P < 0.05; the same parameters apply to the whole section). In zebularine-treated wild-type plants, we found 446 significantly upregulated genes and 183 significantly downregulated genes, that is, many more than we identified in a previous study (Liu et al., 2015). This difference is most likely due to the treatment in liquid media, allowing for a more intense uptake of zebularine compared with the previously used solid media. Zebularine treatment of nse4a-2 plants had the strongest effect, leading to upregulation of 1374 genes and downregulation of 773 genes compared with mock-treated nse4a-2 control plants. Upregulated genes included several prominent DNA damage repair markers (Figure 6B). These data suggest that the SMC5-SMC6 complex is not required for transcriptional upregulation of DNA damage repair genes, but loss of its functionality triggers a more intense DNA damage response (Figure 6B).
Figure 6.
Transcriptome Analysis of nse4a-2 Plants.
(A) Venn diagrams of genes significantly (DESeq, adjusted P < 0.05) up- and downregulated in dissected shoot apices of the 20 µM zebularine (zeb)–treated wild-type (WT zeb/WT mock), mock-treated nse4a-2 (nse4a-2 mock/WT mock), and 20 µM zeb-treated nse4a-2 (nse4a-2 zeb/nse4a-2 mock) plants. The data are based on two RNA sequencing replicates.
(B) mRNA abundance of DNA damage repair marker genes expressed as fragments per kilobase per million of reads (FPKM) based on data shown in (A). Asterisks and dashes indicate statistically significant and nonsignificant, respectively, differences between groups indicated by horizontal bar in DESeq (adjusted P-value < 0.05). WT, wild type; BRCA1, BREAST CANCER SUSCEPTIBILITY1; RAD51, RADIATION SENSITIVE51; RAD17, RADIATION SENSITIVE17; GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED1; RAD3-like,RADIATION SENSITIVE3-like, At1g20750; SMR7, SIAMESE-RELATED7.
(C) Venn diagrams of significantly up- and downregulated genes in nse4a-2 (see [A]) and sni1-1 (sni1-1 mock/wild type (WT) mock; ATH1 expression microarrays, adjusted P < 0.05) plants.
(D) Gene ontology (GO) term analysis of 82 genes significantly upregulated in both nse4a-2 and sni1-1 (see [C]) using agriGO v2.0. Top 10 GO term categories are shown as input relative to Arabidopsis genomic background/reference. The full list of significant GO terms is available in Supplemental Table 3.
(E) Examples of significantly (DESeq, adjusted P-value < 0.05) upregulated defense-related genes in dissected shoot apices of mock-treated nse4a-2 plants. DMR6, DOWNY MILDEW RESISTANT6; RLP33, RECEPTOR LIKE PROTEIN33; WRR4, WHITE RUST RESISTANCE4; RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13.
Previous microarray-based expression analysis of sni1-1 suggested a link between function of the SMC5/6 complex and immune responses (Mosher et al., 2006). Comparison of the transcriptomes from nse4a-2 and sni1-1 mutants revealed 82 (5.8%) commonly upregulated and 6 (0.5%) commonly downregulated genes (Figure 6C; Supplemental Data Set 5). The upregulated genes were mainly associated with stress responses, defense responses to (biotic) stimuli, and responses to other organisms (Figure 6D; Supplemental Table 3), which was described for SNI1 (Mosher et al., 2006; Yan et al., 2013) but is new information for NSE4A. The upregulated genes in nse4a-2 plants included PATHOGENESIS-RELATED GENE2 (PR2; also known as BETA-1,3-GLUCANASE2), PR4, PR5, and several TOLL/INTERLEUKIN-1 RECEPTOR-NUCLEOTICDE BINDING SIGNAL-LEUCINE RICH REPEAT genes (At5g46490, WHITE RUST RESISTANCE4, At3g44630; Figure 6E; Supplemental Data Set 5). This indicates that mutations affecting the SMC5/6 complex cause constitutive expression of immune response genes and lead to activation of other DNA damage repair pathways, most likely due to accumulation of spontaneous DNA damage.
NSE4A and NSE4B Interact with the Same SMC5/6 Complex Subunits
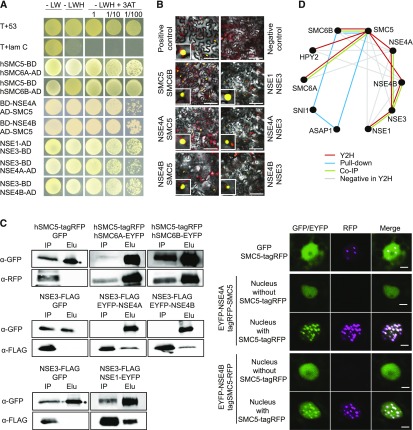
In plants, the architecture of SMC5/6 complex remains unknown. Based on fungal and animal models, we assume that NSE4 may act as a central subunit interacting with SMC5 and SMC6, and possibly several other NSEs (Duan et al., 2009; Hudson et al., 2011). To test whether this hypothesis holds true for both NSE4 paralogs, we performed yeast two-hybrid (Y2H) assays. The assay conditions were optimized using the positive (T+53) and the negative (T+lam C) controls, and we suppressed protein auto-activation by adjusting the 3-amino-1,2,4-triazole (3-AT) concentrations (Figure 7A; Supplemental Figure 8; Supplemental Table 4). As a control, we confirmed the interaction of SMC6A and SMC6B hinges with the SMC5 hinge (Figure 7A). Subsequently, we tested for interactions of full-length SMC5 or SMC6 with NSE4A and NSE4B. While the interaction between both NSE4 paralogs and SMC5 was positive (Figure 7A), we did not observe yeast growth when testing interactions with SMC6A and SMC6B. This remained true even after switching the tag positions (N- and C-terminal positions) and extensive optimization (Supplemental Figure 8). Within the NSE1-NSE3-NSE4 subcomplex, we measured positive interactions of both NSE4 paralogs with NSE3 and confirmed (Li et al., 2017) the interaction of NSE1 with NSE3 (Figure 7A). However, we did not detect interactions between NSE4A or NSE4B and NSE1. To validate the interactions identified by Y2H, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana and analyzed signals using confocal microscopy (Figure 7B). In all cases, the signals were localized to the nucleus and confirmed that both NSE4A and NSE4B are able to interact with SMC5 and NSE3. Moreover, we tested protein–protein interactions using coimmunoprecipitation (co-IP) assays in N. benthamiana and validated (1) the interactions of the SMC5 hinge with the hinges of SMC6A and SMC6B, (2) the interaction of NSE3 with NSE4A and NSE4B, and (3) the interaction of NSE3 with NSE1 (Figure 7C, left; Supplemental Figure 9). We could not evaluate the interactions of NSE4A and NSE4B with the full-length SMC5 protein using co-IPs because, despite extensive optimization, SMC5 did not reach detectable levels following transfection in N. benthamiana leaves as assayed by protein gel blotting. However, the presence of tagRED FLUORESCENT PROTEIN (tagRFP)-SMC5 modified the nuclear distribution of both NSE4A-ENHANCED YELLOW FLUORESCENT PROTEIN (EYFP) and NSE4B-EYFP from a dispersed to a speckled pattern (Figure 7C, right).
Figure 7.
Analysis of Protein–Protein Interactions.
(A) Y2H assays. T+53, positive control and T+lam C, negative control. Domain position before/after the gene name indicates N- or C-terminal fusions, respectively. Autoactivation controls, negatively tested combinations, and used 3-AT concentrations are provided in Supplemental Figure 8 and Supplemental Table 4. -LW, without leucine and tryptophan; -LWH, without leucine, tryptophan and histidine; h, hinge domain, BD, binding domain, AD, activation domain.
(B) BiFC validation of interactions indicated by Y2H. Insets show nuclei with positive signals. Bars = 50 µm.
(C) co-IP and colocalization assays. Right panel displays co-IP analysis. Whole blots are shown in Supplemental Figure 9. Right panel shows changes in EYFP-NSE4A and EYFP-NSE4B localization after addition of SMC5-tagRFP. Elu, elution (proteins collected by green fluorescent protein trapping); GFP, GREEN FLUORESCENT PROTEIN trapping; RFP, RED FLUORESCENT PROTEIN; IP, input (total protein extract); h, hinge domain.
(D) Model of protein–protein interactions within Arabidopsis SMC5/6 complex based on Y2H and BiFC (red lines), pull-down (Yan et al., 2013), and co-IP (green lines) experiments. Negatively tested combinations in Y2H are indicated by gray lines. Interaction between HPY2 and SMC5 was published previously (Xu et al., 2013).
In summary, the results from Y2H, BiFC, and co-IP assays together with published data allow us to conclude that individual Arabidopsis SMC5/6 complex subunits interact and that SMC5 recruits NSE4A and NSE4B into speckled domains in the nucleus (Figure 7C). Based on these experiments, we developed a model for interactions between SMC5/6 complex subunits in Arabidopsis (Figure 7D).
The NSE4B Protein Can Partially Substitute NSE4A Protein Functions
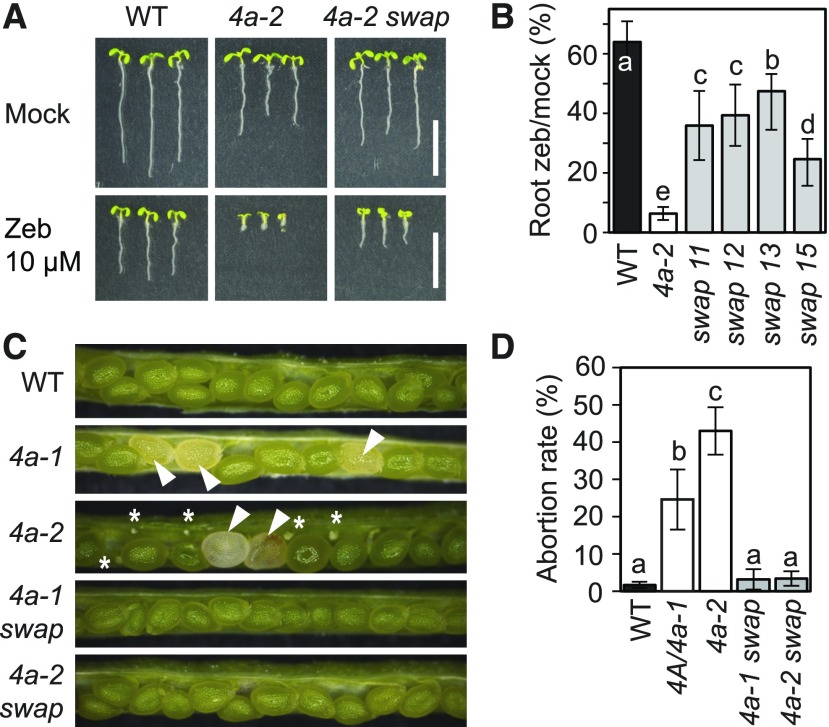
The NSE4A and NSE4B paralogs show little overlap in their expression patterns and loss-of-function phenotypes. To test whether NSE4A and NSE4B also diverged functionally, we developed a promoter swap construct consisting of the NSE4B genomic coding sequence (CDS) under the control of the NSE4A promoter (ProNSE4A:GenomicNSE4B:TerNSE4B). This construct was transformed into homozygous nse4a-2 plants, and individuals heterozygous or homozygous for the promoter swap construct were selected in the T2 generation and tested for zebularine sensitivity in the T3 generation. While the control nse4a-2 plants were strongly hypersensitive, several independent promoter swap lines showed rescue, albeit incomplete, of the drug sensitivity phenotype, with average roots length being intermediate between those of nse4a-2 and the wild-type plants (Figures 8A and 8B).
Figure 8.
Analysis of NSE4B Functions.
(A) Zebularine (Zeb) hypersensitivity assay. Wild-type (WT), nse4a-2 (4a-2), and nse4a-2 complemented with ProNSE4A:GenNSE4B:TerNSE4B (4a-2 swap) line 13 were germinated and kept on control and 10 µM Zeb-containing media for 1 week. Bar = 10 mm.
(B) Quantitative data for root length of zebularine (zeb)–treated versus control plants as described in (A). Lines 11, 12, 13, and 15 represent independent promoter swap transgenic lines. Error bars indicate sd between the means from two biological replicates. Each replicate consisted from at least 20 plants per line grown on separate screening plates at different times. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05). WT, wild type.
(C) Analysis of seed development phenotypes in the wild type (WT), heterozygous NSE4A/nse4a-1 (4a-1), and 4a-2. The two bottom pictures show homozygous nse4a-1 and nse4a-2 containing homozygous promoter swap line 13 (4a-1 swap and 4a-2 swap). White arrowheads indicate aberrantly developing seeds and asterisks aborted ovules.
(D) Quantification of abortion rates in the genotypes described in (C). Error bars indicate sd between means of three biological replicates (plants), each with at least 300 scored seeds. Values marked with the same letter do not differ according to Duncan’s multiple range test (P ≤ 0.05). WT, wild type.
In addition, the broader expression domain of NSE4B in the promoter swap lines was able to rescue the seed abortion phenotype of nse4a-2 (Figures 8C and 8D). Furthermore, NSE4B expression in the nse4a-1 background allowed the recovery of homozygous nse4a-1 plants (24% viable nse4a-1/nse4a-1 plants in the progeny of a NSE4A/nse4a-1;ProNSE4A:GenomicNSE4B:TerNSE4B segregating parent; n = 92, Supplemental Tables 5 and 6).
Taken together, these results demonstrate that NSE4A and NSE4B have similar biochemical activities that are fully exchangeable during seed development but only partially in DNA damage responses.
DISCUSSION
The SMC5/6 complex plays a crucial role in the maintenance of genome stability in eukaryotes (De Piccoli et al., 2009; Kegel and Sjögren, 2010; Jeppsson et al., 2014b; Diaz and Pecinka, 2018). Some of its subunits remain poorly characterized in plants, including the two NSE4 homologs. Here, we demonstrate that NSE4A is involved in preserving genome stability and controls seed development. NSE4B is barely active during normal development and nonresponsive to drug-induced genotoxic stress.
NSE4A Is an Essential Gene in Arabidopsis
The NSE4 paralogs of Arabidopsis originate from the whole-genome duplication event (α) that occurred ∼47 MYA in Brassicaceae (Kagale et al., 2014). Surprisingly, there were at least two NSE4A copies in all vascular plants analyzed, with the highest number of six copies inOryza sativa. The NSE4 amplifications are family specific and much more frequent than duplications of any other SMC5/6 complex members in plant genomes (reviewed in Diaz and Pecinka, 2018). Our data from Arabidopsis and published data from humans (Hudson et al., 2011) suggest that at least some of these duplicated copies differ in their expression domains. We found that both NSE4A and NSE4B can interact with the core subunits SMC5 and NSE3, but not with NSE1, with the latter two representing members of the NSE1-NSE3-NSE4 subcomplex (Palecek and Gruber, 2015). However, in spite of extensive optimization, we did not detect interactions of the NSE4 proteins with SMC6B. This interaction is very likely to exist in Arabidopsis but seems particularly difficult to confirm as indicated by previous studies in Saccharomyces cerevisiae and S. pombe (Palecek et al., 2006; Duan et al., 2009; J. Palecek, personal communication). This is possibly caused by a steric hindrance due to the specific conformation of SMC6 and NSE4 proteins or the absence of an activating and/or stabilizing component.
A strong nse4a mutation was homozygous lethal, and self-pollinated heterozygotes showed 28.8% seed abortion. This resembles the phenotypes of smc5, nse1, nse3, and asap1 mutants and the sm6a smc6b double mutant, which show embryonic or cotyledon-stage seedling death in Arabidopsis (Watanabe et al., 2009; Xu et al., 2013; Yan et al., 2013; Li et al., 2017). However, we also found a hypomorphic nse4a-2 allele, which likely produces a protein with a modified C terminus. This allele alleviates the problem of homozygous lethality encountered in the loss-of-function allele nse4a-1, thereby enabling the analysis of NSE4A functions during plant development and genotoxic stress. Its phenotypes partially resemble those of HPY2 and SNI1 mutants, which survive but are strongly affected in development and fertility (Li et al., 1999; Huang et al., 2009; Ishida et al., 2009).
NSE4A Is Involved in Sporogenesis, Gametogenesis, and Seed Development
We observed prominent and dynamic expression of NSE4A during Arabidopsis reproductive development. In the male gametophyte, NSE4A was expressed in the generative cell lineage but absent in the vegetative cell. This is consistent with the observation that the sperm nucleus is rich in the components of active chromatin control, while the vegetative nucleus has lost multiple repressive chromatin modifications and will no longer divide (Schoft et al., 2009; Slotkin et al., 2009; Abdelsamad and Pecinka, 2014). However, the function of NSE4A in pollen development remains unknown. Possibly, NSE4A secures a faster or more accurate response, which is not detected under laboratory conditions, upon environmental challenges affecting genome integrity in the germline.
NSE4A is also broadly expressed in ovule primordia, with a notable accumulation in the female meiocyte. Thus, besides its role in male meiosis (Liu et al., 2014), the SMC5/6 complex may play a role during female meiosis, possibly in the process of DNA replication, meiotic recombination, or DNA damage repair. During embryo sac development and early seed development, NSE4A was expressed in synergids and the central cell and later in the embryo and the syncytial and chalazal endosperm. NSE4A expression at these stages may be interpreted as a functional requirement for genome integrity safeguarding processes, which involve DNA repair as a consequence of the challenges posed by rapid DNA replication and chromatin dynamics in these tissues (Baroux et al., 2007; Baroux and Autran, 2015). Genome integrity is necessary to ensure the proper differentiation and functioning of the progeny and to avoid the propagation of genetic mutations. In addition, but not exclusively, the high levels of NSE4A in the syncytial endosperm may play a role in the detoxification of endogenously occurring replication-derived toxic DNA structures. DNA replication produces a high frequency of inter-twining between nascent chromatids, DNA supercoils, and X-shaped toxic DNA replication intermediates, which all require (to different extents) SMC5/6 functions for resolution (Jeppsson et al., 2014a; Menolfi et al., 2015; reviewed in Diaz and Pecinka, 2018).
While SMC5/6 complex null mutations lead to early seed abortion, the hypomorphic nse4a-2 mutant produced large glossy seeds with liquid endosperm, which turned brown at later stages and aborted. Seed phenotypes similar to nse4a-1 or nse4a-2 were reported for nse1, nse3, and mms21/hpy2 mutants (Liu et al., 2014; Li et al., 2017). Studies in S. cerevisiae revealed that the SMC5/6 complex is loaded by the Sister chromatid cohesion protein 1 subunit of the cohesin complex to specific sites during DNA replication (Jeppsson et al., 2014a). This could explain the similarity of SMC5/6 complex and cohesin mutant seed phenotypes and indicates that both complexes cooperate during seed development. This may be supported by the identification of cohesin, and also condensin, mutants in a screen focusing on aberrant seed development (Liu et al., 2002; Tzafrir et al., 2002) and underlines the importance of maintaining genome stability during seed development (reviewed in Diaz and Pecinka, 2017).
NSE4A, but Not NSE4B, Is Required for Resistance to Genotoxic Stress
The functions of the SMC5/6 complex are widely associated with the maintenance of genome stability (Kegel and Sjögren, 2010; Wu and Yu, 2012; Jeppsson et al., 2014b); however, it was not clear which of the Arabidopsis NSE4 paralogs confers this function. We observed activation of NSE4A, but not NSE4B, in response to genotoxic treatments with drugs inducing various types of DNA damage. In addition, the viable and phenotypically almost wild-type nse4a-2 plants were hypersensitive to the cytidine analog zebularine and the alkylating agent MMS, but not to other treatments. Lack of sensitivity to bleocin, MMC, and HU could be caused by the fact that the mutation we analyzed is not a complete loss-of-function allele and/or that such damages can be processed by SMC5/6-independent pathways. We have previously shown that smc6b mutants are hypersensitive to zebularine-induced damage (Liu et al., 2015). This suggests that the SMC5/6 complex is essential for detoxification from complex toxic structures, such as zebularine-induced DNA damage. DNA repair in response to zebularine treatment is mediated both by ATAXIA TELANGIECTASIA-MUTATED and ATR kinases (Liu et al., 2015), which are known to phosphorylate proteins at Ser followed by Gln or Thr followed by Gln motifs (Awasthi et al., 2015). NSE4A contains two adjacent Thr-Gln motifs at amino acids 361 to 365 (TQDTQ), which makes it a good candidate for a direct target of phosphorylation by ATM and/or ATR.
Recent studies from nonplant models suggest that the SMC5/6 complex acts as an ATP-dependent intermolecular linker, which helps resolving toxic DNA structures at late-replicating sites and also prevents recombination between nonhomologous sequences (Chiolo et al., 2011; Kanno et al., 2015; Menolfi et al., 2015). In Arabidopsis, the SMC5/6 complex promotes the association of sister chromatids and is required for normal levels of homologous recombination (Mengiste et al., 1999; Hanin et al., 2000; Watanabe et al., 2009; Yuan et al., 2014). In addition to its role in somatic DNA damage repair, there is emerging evidence that the SMC5-SMC6 complex also plays a role in immune responses (Yan et al., 2013) and meiosis (Yuan et al., 2014). Our data indirectly support a meiotic role of NSE4A as it strongly accumulates in female meiocytes. However, the exact molecular mechanism of genome maintenance by the SMC5/6 complex remains unknown.
NSE4B and NSE4A Have Primarily Diversified Transcriptionally, and NSE4B Is Not Responsive to DNA Damage
In Arabidopsis, the functions of NSE4B are less clear than those of NSE4A. NSE4B single mutants are morphologically indistinguishable from the wild type and do not worsen the phenotype of a weak nse4a mutant. We found that NSE4B is silenced throughout most of development, except for a small domain in the root apical meristem, leaf stipules, and the embryo up to the globular stage. Based on the results of in silico analyses, which revealed an extensive coverage of the NSE4B locus by histone H3 Lys-27 trimethylation, we hypothesize that NSE4B is controlled by the Polycomb Repressive Complex 2 (reviewed in Mozgova and Hennig, 2015). To explore NSE4B’s function in the nonsilenced state, we swapped its promoter with that of NSE4A and tested whether NSE4B expressed in the pattern of NSE4A can complement the nse4a phenotypes. The seed abortion phenotype was fully complemented, but we found only a partial rescue under DNA damaging conditions. This points to the dual function of the SMC5/6 complex described in budding yeast (Menolfi et al., 2015): a DNA damage-independent function during DNA replication and a DNA damage-dependent function in DNA repair. Both NSE4A and NSE4B seem capable of performing the first function, while DNA damage repair can be done only by NSE4A in Arabidopsis.
METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) wild type and mutants were in the Col background: nse4a-1 (SALK_057130), nse4a-2 (GK-768H08), nse4b-1 (SAIL_296_F02), nse4b-2 (GK-175D10), smc6b-1 (SALK_ SALK_101968C), hpy2-2 (SAIL_77_G06), atr-2 (SALK_032841C), wee1-1 (GK-270E05), and lig4-2 (SALK_044027C). We also used a cyclin-GUS line containing the ProCYCB1;1:CYCB1;1:GUS construct (Colón-Carmona et al., 1999) and the B11 line containing an intramolecular type of HR substrate (Puchta et al., 1995). For promoter reporter constructs, regions 18,943,545 to 18,941,640 and 7,260,588 to 7,258,919 bp upstream of the NSE4A and NSE4B transcription start sites, respectively, were PCR amplified, cloned into pDONOR221, and recombined into the binary Gateway vector pGWB553 containing the uidA gene encoding GUS. The final plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and then into Arabidopsis Col using the floral dip method (Zhang et al., 2006). T1 generation seeds were screened on one half Murashige and Skoog (MS) plates containing 25 µg/L hygromycin B (Duchefa Biochemie), and resistant plants were transferred to soil. T2 populations with ∼75% resistant seedlings, indicating single locus T-DNA insertions, were considered for further analyses. For promoter swap experiments, the NSE4A promoter and genomic region of NSE4B were PCR amplified and cloned into the pGWB550 vector by MultiSite Cloning Gateway (Thermo Fisher Scientific). The construct was transformed into the nse4a-2 background using the floral dip method. To construct the NSE4A-fluorescent protein translational fusion, the NSE4A promoter, CDS, terminator, VENUS N-terminal tag, and a BASTA resistance cassette were cloned using Gibson assembly (New England Biolabs) into pGGA000, pGGC000, pGGE000, pGGB000, and pGGF000, respectively, to generate entry clones. The Greengate cloning reaction was performed as described previously (Lampropoulos et al., 2013), and the multi entry cassette was assembled into the pAGM4723 backbone. nse4a-2 mutant plants were transformed with this construct using the floral dip method. For nse4a-2 complementation analysis, the NSE4A promoter and genomic region of NSE4A were PCR amplified and cloned into the pGWB550 vector by MultiSite Cloning Gateway (Thermo Fisher Scientific). Plant transformation and screening of transformants were performed exactly as for the promoter swap experiment. Plants were emasculated ∼48 h prior to pollination in crossing experiments.
Phylogenetic Analysis and Shadowing
NSE4 protein sequences were retrieved from the National Center for Biotechnology Information and Phytozome (Supplemental Table 1). The protein alignment was performed using the MUSCLE algorithm (Edgar, 2004), and the resulting alignment was submitted to Gblocks (Castresana, 2000). Curation and selection of aligned blocks were performed in Gblocks using less stringent parameters. Bootstrap probabilities for each node were calculated with 100 replicates. Original sequences, alignments, and blocks are provided as Supplemental Data Sets 1, 2, and 3, respectively.
Promoter sequences from all analyzed species were retrieved from Phytozome (Supplemental Table 2). Promoter regions of NSE4A and NSE4B were submitted individually to mVISTA (Frazer et al., 2004), and sequence conservation was calculated using LAGAN program (Brudno et al., 2003). The Arabidopsis sequences were used as references for pairwise comparisons (Supplemental Figure 2).
Plant Growth Conditions and Drug Treatments
For genotyping, crossing, and seed production plants were grown in 7 × 7-cm pots filled with peat bog in a climatic chamber under controlled long-day conditions (at 16 h with an ∼200 μmol m−2 s−1 light intensity and 21°C during day; 8 h at 19°C during night) with standard 70% humidity.
For in vitro experiments, sterilized seeds were evenly spread on sterile one half Murashige and Skoog (MS) medium with or without zebularine (Sigma-Aldrich), MMC (Duchefa Biochemie), bleocin (Calbiochem), and HU (Sigma-Aldrich) in concentrations specified in the text and grown at 16 h with 150 μmol m−2 s−1 light:8 h dark at 21°C. Seven-day-old plants were used for root length measurements. For MMS experiment, sterilized seeds were grown in one half MS medium for 5 d and then transferred to liquid one half MS medium with and without 100 ppm MMS, and grown for 26 d. Roots from 20 to 25 seedlings per genotype were straightened, and in total three replicates were performed. For RNA sequencing, seeds were germinated on drug-free on half MS solid medium, and 9-d-old plants were carefully transferred to liquid one half MS medium with or without 20 μM zebularine. After 24 h, plants were washed with drug-free liquid one half MS medium; their leaves, hypocotyl, and roots were removed; and shoot apices were flash-frozen in liquid nitrogen for later use.
Nucleic Acid Isolation, cDNA Synthesis, and PCR
For DNA isolation, leaf material of plants at the rosette stage was harvested, and DNA was isolated using the DNeasy Plant Mini kit (QIAGEN), following the manufacturer’s instructions. For RNA isolation, floral buds were collected, shock-frozen in liquid nitrogen, and kept at −80°C until use. Total RNA isolation was performed with QIAzol (QIAGEN), and the RNA integrity was assessed by formaldehyde agarose gel electrophoresis. cDNA synthesis was performed from 1 µg of total RNA as starting material, using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) with oligo(dT) primers according to manufacturer’s instructions. Primers used in this study are provided in Supplemental Tables 7 and 8. For 3′ RACE PCR, we performed in total four nested PCR reactions using the primer combinations listed in Supplemental Table 8. The first PCR was performed using a1/100 (v/v) dilution of cDNA synthesized from the nse4a-2 mutant. Afterwards, the PCR product was gel purified and used for the subsequent nested PCR reaction. This step was repeated until the fourth reaction. PCR product obtained from the fourth reaction was cloned into the pJET1.2 vector and sequenced.
RNA Sequencing and Microarray Analysis
RNA for RNA sequencing was isolated using RNeasy Plant Mini kit (QIAGEN) with additional on-column DNase I digestion according to manufacturer’s instructions. RNA sequencing was performed with two biological replicates per experimental point. The libraries were prepared from 1 μg of total RNA with RNA integrity number >7.8 (Bioanalyzer, Agilent) using TruSeq type RNA kit (Illumina) at the Cologne Genome Centre and sequenced as 100-bp single-end reads on a HiSeq2500 instrument (Illumina). Reads were trimmed and quality filtered with FAST-X tools (http://hannonlab.cshl.edu/fastx_toolkit/). This yielded an average of 18.5 million high-quality reads per library. The reads were mapped to the TAIR10 Arabidopsis reference genome using Tophat2 (Kim et al., 2013) with default settings. The coverage of individual genes was retrieved with the Qualimap from the set of uniquely mapped reads and significance (adjusted P-value < 0.05) of mRNA level changes estimated with the DESeq package (Anders and Huber, 2010). Publicly available sni1-1 Affymetrix Arabidopsis ATH1 GeneChip array data (Nottingham Arabidopsis Science Centre experiment ID 389, slides 20561 to 20566; Gene Expression Omnibus Series: GSE6827; Mosher et al., 2006) were analyzed using rma protocol with Bioconductor in R. Venn diagrams were drawn using BioVenn online tool (http://www.biovenn.nl/).
GUS Histochemical Staining
The staining protocol was adapted according to different tissues. Vegetative tissues were stained as described previously (Liu et al., 2015). Inflorescences were dissected under an MZ16FA stereomicroscope (Leica Microsystems), fixed for 30 min in ice-cold 4% (v/v) formaldehyde in 1× PBS buffer, washed three times for 5 min each in 1× PBS, and infiltrated with GUS staining solution (Stangeland and Salehian, 2002) under vacuum. After 10 to 15 min, the vacuum was released and samples were incubated at 37°C for 3 d, followed by overnight clearing in 70% (v/v) ethanol. Subsequently, inflorescences were rinsed with water and mounted in Petri dishes containing agarose and water. For staining of ovules and young seeds, developing siliques were first opened and fixed in 90% (v/v) cold acetone at −20°C for 45 min. Afterwards, they were rinsed three times with 100 mM phosphate buffer, transferred to GUS staining solution, vacuum infiltrated for 5 min, and stained at 37°C for 48 h. After staining, pistils and siliques were quickly rinsed with phosphate buffer and mounted in 8:2:1 chloral hydrate solution. In order to avoid loss of signal when we observed weak GUS staining, we performed a less severe clearing. We dissected pistils and immediately transferred them to GUS solution. Staining of ovules was performed as described previously (Vielle-Calzada et al., 2000). After clearing, mounted ovules where immediately imaged using a microscope (Zeiss). For GUS and 4′,6-diamidino-2-phenylindole (DAPI) costaining of pollen grains, flowers were opened and fixed in cold 3:1 ethanol:acetic acid (v/v) for 30 min. Afterwards, they were rinsed three times with phosphate buffer, infiltrated with GUS staining solution for 10 to 15 min, and stained for 48 h at 37°C in dark. Next, GUS-stained anthers were dissected, rinsed with phosphate buffer, transferred to a microscopic slide, further dissected with a needle in DAPI solution (0.4 μg/mL DAPI, 0.1 M sodium phosphate buffer, pH 7, 0.1% (v/v) Triton X-100, and 1 mM EDTA), covered with a cover slip, and then used for microscopy.
Hoyer’s Clearing
Clearing of seeds was performed as described by Liu and Meinke (1998).
Cell Cycle Arrest
The double homozygous nse4a-2 ProCYCB1;1:CYCB1;1:GUS and nse4b-2 ProCYCB1;1:CYCB1;1:GUS plants were grown for 5 d in liquid one half MS medium; transferred to liquid one half MS supplemented with 10 µM zebularine for 0, 1, 3, 6, 12, 24, or 48 h; GUS stained overnight; cleared in 70% (v/v) ethanol; and imaged using an MZ16FA stereomicroscope (Leica Microsystems).
Confocal Microscopy
For cell death analysis, seeds from transgenic lines were grown on vertically positioned plates with one half MS medium for 4 d and then transferred for 1 d to liquid one half MS medium with 20 µM zebularine. Seedlings were stained with 10 μg mL−1 PI solution (Sigma-Aldrich) for 3 min, followed by a rinsing step with sterilized water, and were placed on slides in a drop of water and then evaluated using an LSM700 laser scanning confocal microscope (Zeiss). For subcellular localization of NSE4A-VENUS in roots, transgenic lines expressing ProNSE4A:VENUS:NSE4A:TerNSE4A were grown for 5 d in either solid one half MS or one half MS supplemented with 10 µM zebularine. Afterwards, seedlings were stained with PI, and imaged with a TCS SP8 confocal microscope (Leica Microsystems). For imaging of ovules, pistils were quickly dissected in a drop of water, and ovules from different stages were mounted on a slide with a drop of water and placed on ice. After few minutes, preparations were observed using a TCS SP8 confocal microscope (Leica Microsystems)
Y2H Assay and BiFC
The full-length CDSs of Arabidopsis SMC5, SMC6A, SMC6B, NSE1, and NSE3 were PCR amplified from cDNA. SMC5, SMC6A, and SMC6B were cloned via restriction digest (Supplemental Table 7) into the vector pGADT7 (Clontech), while NSE1 and NSE3 were cloned into the gateway compatible vector pGADT-GW (Lu et al., 2010) to produce a protein fusion with the GAL4 DNA activation domain (AD) in N-terminal orientation. In order to produce a protein fusion with the GAL4 DNA binding domain (BD), the SMC5, NSE4A, and NSE4B PCR fragments were cloned via restriction (Supplemental Table 7) digest into pGBKT7 and NSE1 and NSE3 were cloned via gateway into pGBKT7-GW (Lu et al., 2010). In order to avoid negative results due to interference of BD or AD domain with possible interactors, all genes were cloned into both C-terminal pGBKCg and pGADCg Y2H vectors, to produce C-terminally tagged GAL4 AD and BD fusion proteins, respectively, with exception of NSE4B, which was only cloned into the pGADCg vector. The hinge and fragments of coils of SMC5 (corresponding to amino acids 415 to 699), SMC6A (amino acids 367 to 670), and SMC6B (amino acids 358 to 691) were cloned into the pGBKCg and pGADCg vectors to test for interaction with the core subunits. The GAL4-based interaction was tested in the yeast strain AH109 (Clontech). Cotransformed yeast strains were selected on synthetic defined/–Leu/–Trp medium. Protein–protein interactions were tested using stringent (synthetic defined/–Leu/–Trp/–His) selection medium supplemented with defined concentrations of 3-AT (Supplemental Table 4). The interaction between pGADT7-T and pBKT7-53 was used as the positive control and that between pGADT7-T and pBKT7-LamC was used as the negative control. For BiFC, we used the same CDSs as for the Y2H experiments. The SMC5, SMC5 (hinge), and NSE3 sequences were cloned into pBATL-nYFP, and NSE4A, NSE4B, NSE1, and SMC6B hinge sequences were cloned into pBaTL-cYFP. Both plasmids produce C-terminal fusion proteins. Nicotiana benthamiana leaves were transformed for transient expression as described previously (Tian et al., 2011). YFP fluorescence was observed using an LSM 700 confocal microscope (Zeiss).
Coimmunoprecipitation and Localization Assays
Here, we used the same entry clones as for Y2H and BiFC assays. Hinge SMC5 was cloned into pGWB560, to produce a C-terminal fusion with tagRFP protein. Hinge SMC6A, hinge SMC6B, NSE1 CDS, NSE4A CDS, and NSE4B CDS were cloned into pGWB541 to produce a C-terminal tagged EYFP proteins. NSE3 CDS was cloned into pGWB611 to produce a C-terminal FLAG fusion protein. To test interactions of SMC5 with NSE4s, full-length SMC5 CDS was cloned into pGWB561 to produce an N-terminal tagRFP fusion, while both NSE4A and NSE4B CDS were cloned into pGWB542 to produce N- and C-terminal tagged EYFP fusion proteins. As a negative control we used pSY1, containing GFP CDS driven by 35S promoter. Afterwards, the expression clones were transformed into A. tumefaciens strain GV3101.
Fluorescent or epitope tag–conjugated proteins were transiently expressed in N. benthamiana leaves by Agrobacterium-mediated infiltration. Leaves were harvested at 4 or 5 d after inoculation, and immunoprecipitation was performed with a µMACS GFP isolation kit (Miltenyi Biotec). Approximately 1 to 2 g of plant material was homogenized in threefold volume of µMACS lysis buffer containing protease inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich), and then the lysate was filtered through two layers of miracloth. Afterwards, the lysate was mixed with anti-GFP antibody-conjugated magnetic beads and was incubated at 4°C for 60 min with gentle rotation. The GFP-conjugated proteins were purified using a magnetic column according to the manufacturer’s instructions. The immunoprecipitated proteins were analyzed by protein gel blotting using an anti-GFP antibody at 1/1000 (v/v; ab290, Abcam), an anti-tagRFP antibody at 1/500 (v/v; R10367, Thermo Fisher Scientific), or an anti-FLAG antibody at 1/5000 (v/v; 3022-100, BioVision) as primary antibodies and horseradish peroxidase–conjugated anti-mouse IgG antibody at 1/15000 (v/v; W402, Promega) or horseradish peroxidase–conjugated anti-rabbit IgG antibody at 1/15000 (v/v; MB4458, MBL) as secondary antibodies. The chemiluminescences from target proteins of each antibody were visualized with ImmunoStar LD (Wako) on Fusion Pulse system (Vilber Lourmat).
For the localization analysis of GFP, EYFP-NSE4A, and EYFP-NSE4B proteins simultaneously expressed with tagRFP-SMC5. Five days after inoculation, leaves were observed under an inverted FV1200 laser confocal microscope equipped with a GaAsP detector (Olympus) with an excitation wavelength with 473 nm for GFP/EYFP and 559 nm for tagRFP.
Statistical Analysis
The values were examined by one-way analysis of variance and post hoc comparison by Duncan’s multiple range test (P ≤ 0.05). Statistical analyses except for RNA sequencing and microarray analysis were performed using STATISTICA 13 software (StatSoft). Fisher’s test was used to calculate the adjusted P-value (q-value) in RNA sequencing and microarray analysis. Raw data and detailed results of the statistical analyses are provided in Supplemental Data Set 6.
Accession Numbers
The following gene names and symbols are associated with this publication: ASAP1 (AT2G28130), ATR (AT5G40820), LIG4 (AT5G57160), HPY2 (AT3G15150), NSE1 (AT5G21140), NSE3 (AT1G34770), NSE4A (AT1G51130), NSE4B (AT3G20760), SMC5 (AT5G15920), SMC6A (AT5G07660), SMC6B (AT5G61460), SNI1 (AT4G18470), WEE1 (AT1G02970). RNA sequencing reads are deposited in Gene Expression Omnibus as the study number GSE113310.
Supplemental Data
Supplemental Figure 1. Phylogenetic shadowing of NSE4A and NSE4B promoters.
Supplemental Figure 2. Characterization of NSE4A mutation in nse4a-2.
Supplemental Figure 3. Characterization of nse4b-1 and nse4b-2 mutations.
Supplemental Figure 4. Chromatin environment of the NSE4A genomic region.
Supplemental Figure 5. Chromatin environment of the NSE4B genomic region.
Supplemental Figure 6. Seed phenotypes of NSE4A mutants.
Supplemental Figure 7. Mutant sensitivity to hydroxyurea (HU).
Supplemental Figure 8. Y2H assays (autoactivation and negative results).
Supplemental Figure 9. Whole blots from coimmunoprecipitation (co-IP) assays.
Supplemental Table 1. Protein sequences used in the phylogenetic analysis.
Supplemental Table 2. Promoter sequences used for the phylogenetic shadowing.
Supplemental Table 3. Gene ontology (GO) term analysis of 82 genes upregulated in nse4a-2 and sni1-1.
Supplemental Table 4. 3-AT concentrations used in yeast-two hybrid experiments.
Supplemental Table 5. Punnett square indicating frequencies of genotypes in F2 generation of self-pollinated F1 hybrid nse4a-1/nse4a-2 T/0.
Supplemental Table 6. Promoter swap rescues nse4a-1 lethality.
Supplemental Table 7. Primers used in this study.
Supplemental Table 8. Primer combinations used in 3′ RACE PCR of NSE4A in nse4a-2.
Supplemental Data Set 1. Protein sequences used to build NSE4 phylogenetic tree (FASTA file).
Supplemental Data Set 2. Alignment from the full length NSE4 protein sequences submitted to Gblocks server (FASTA file).
Supplemental Data Set 3. Gblocks output. Conserved blocks from NSE4 protein alignment (FASTA file).
Supplemental Data Set 4. Transcriptomic study of nse4a-2 under control and zebularine stress conditions.
Supplemental Data Set 5. Comparison of nse4a-2 and sni1-1 induced transcriptional changes.
Supplemental Data Set 6. Background data for and the results of the statistical analyses.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Andreas Finke, Fen Yang, and Jan Palecek for helpful discussions; Peter Doerner for the cyclin reporter construct; and Barbara Eilts, Regina Gentges, and Barbara Piwowarczyk for assistance. This work was supported by funding from the Max Planck Society to A.P., P.P. and P.Y.G.; Deutsche Akademische Austauschdienst Dienst scholarships (A/12/77772 to M.D. and ST21 2015/16 to A.N.); the University of Zurich to U.G.; European Regional Development Fund project "Plants as a tool for sustainable global development" (CZ.02.1.01/0.0/0.0/16_019/0000827 to H.J. and A.P.); the Czech Academy of Sciences Purkyně Fellowship (to A.P.); Czech Science Foundation (19-13848S to A.P.) and the Ministry of Education, Youth and Sports, Czech Republic (LTC18026 to A.P.). The work of Matsunaga lab was supported bythe Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI (grants 15H05955 and 15H05962 to S.M.).
AUTHOR CONTRIBUTIONS
A.P. and M.D. designed the research with help from C.B., T.S., U.G., and S.M. on specific aspects. M.D., P.P., A.N., T.S., H.J., C.B., and P.Y.G. performed the experiments and analyzed the data. A.P. and M.D. wrote the article with contributions from all authors. All authors approved the final version of this article.
Footnotes
Articles can be viewed without a subscription.
References
- Abdelsamad A., Pecinka A. (2014). Pollen-specific activation of Arabidopsis retrogenes is associated with global transcriptional reprogramming. Plant Cell 26: 3299–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C., Groth A. (2012). Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13: 153–167. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi P., Foiani M., Kumar A. (2015). ATM and ATR signaling at a glance. J. Cell Sci. 128: 4255–4262. [DOI] [PubMed] [Google Scholar]
- Baroux C., Autran D. (2015). Chromatin dynamics during cellular differentiation in the female reproductive lineage of flowering plants. Plant J. 83: 160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C., Pecinka A., Fuchs J., Schubert I., Grossniklaus U. (2007). The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization. Plant Cell 19: 1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Mansfield S.G., Modrusan Z., Reiser L., Fischer R.L., Haughn G.W., Feldman K.A., Webb M.C. (1994). Ovules. In Arabidopsis, Bowman J., ed (New York: Springer; ), pp. 297–331. [Google Scholar]
- Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Batzoglou S.; NISC Comparative Sequencing Program (2003). LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S.U., Polyzos A., Costes S.V., Karpen G.H. (2011). Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- De Piccoli G., Torres-Rosell J., Aragón L. (2009). The unnamed complex: What do we know about Smc5-Smc6? Chromosome Res. 17: 251–263. [DOI] [PubMed] [Google Scholar]
- De Schutter K., Joubès J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman T., Kushnir S., Inzé D., De Veylder L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M., Pecinka A. (2017). Seeds as emerging hotspot for maintenance of genome stability. Cytologia (Tokyo) 82: 467–480. [Google Scholar]
- Diaz M., Pecinka A. (2018). Scaffolding for repair: Understanding molecular functions of the SMC5/6 complex. Genes (Basel) 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Yang Y., Chen Y.-H., Arenz J., Rangi G.K., Zhao X., Ye H. (2009). Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5–6 subcomplex and the hinge regions of Smc5 and Smc6. J. Biol. Chem. 284: 8507–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32: W273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin M., Mengiste T., Bogucki A., Paszkowski J. (2000). Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J. 24: 183–189. [DOI] [PubMed] [Google Scholar]
- Hirano T. (2006). At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7: 311–322. [DOI] [PubMed] [Google Scholar]
- Hirano T. (2012). Condensins: Universal organizers of chromosomes with diverse functions. Genes Dev. 26: 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., Shi S., Wang J., Wang Y., Xie Q., Yang C. (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60: 666–678. [DOI] [PubMed] [Google Scholar]
- Hudson J.J.R., Bednarova K., Kozakova L., Liao C., Guerineau M., Colnaghi R., Vidot S., Marek J., Bathula S.R., Lehmann A.R., Palecek J. (2011). Interactions between the Nse3 and Nse4 components of the SMC5-6 complex identify evolutionarily conserved interactions between MAGE and EID Families. PLoS One 6: e17270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K., Carlborg K.K., Nakato R., Berta D.G., Lilienthal I., Kanno T., Lindqvist A., Brink M.C., Dantuma N.P., Katou Y., Shirahige K., Sjögren C. (2014a). The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 10: e1004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K., Kanno T., Shirahige K., Sjögren C. (2014b). The maintenance of chromosome structure: Positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15: 601–614. [DOI] [PubMed] [Google Scholar]
- Kagale S., Robinson S.J., Nixon J., Xiao R., Huebert T., Condie J., Kessler D., Clarke W.E., Edger P.P., Links M.G., Sharpe A.G., Parkin I.A.P. (2014). Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Berta D.G., Sjögren C. (2015). The Smc5/6 complex is an ATP-dependent intermolecular DNA linker. Cell Reports 12: 1471–1482. [DOI] [PubMed] [Google Scholar]
- Kegel A., Sjögren C. (2010). The Smc5/6 complex: More than repair? Cold Spring Harb. Symp. Quant. Biol. 75: 179–187. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak J., West C.E., White C., da Costa-Nunes J.A., Angelis K.J. (2009). Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst.) 8: 413–419. [DOI] [PubMed] [Google Scholar]
- Kwak J.S., Son G.H., Kim S.-I., Song J.T., Seo H.S. (2016). Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes Flowering Locus C through its E3 ligase activity. Front. Plant Sci. 7: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A., Sutikovic Z., Wenzl C., Maegele I., Lohmann J.U., Forner J. (2013). GreenGate---A novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zou W., Jian L., Qian J., Deng Y., Zhao J. (2017). Non-SMC elements 1 and 3 are required for early embryo and seedling development in Arabidopsis. J. Exp. Bot. 68: 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339. [DOI] [PubMed] [Google Scholar]
- Liu C.-M., Meinke D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16: 21–31. [DOI] [PubMed] [Google Scholar]
- Liu C.M., McElver J., Tzafrir I., Joosen R., Wittich P., Patton D., Van Lammeren A.A.M., Meinke D. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29: 405–415. [DOI] [PubMed] [Google Scholar]
- Liu C.-H., Finke A., Díaz M., Rozhon W., Poppenberger B., Baubec T., Pecinka A. (2015). Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell 27: 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Shi S., Zhang S., Xu P., Lai J., Liu Y., Yuan D., Wang Y., Du J., Yang C. (2014). SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol. 14: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W.T., Harada J.J., Rothstein S.J., Cui Y. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 61: 259–270. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Revenkova E., Bechtold N., Paszkowski J. (1999). An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18: 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menolfi D., Delamarre A., Lengronne A., Pasero P., Branzei D. (2015). Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 60: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Durrant W.E., Wang D., Song J., Dong X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I., Hennig L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66: 269–296. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90. [DOI] [PubMed] [Google Scholar]
- Palecek J.J., Gruber S. (2015). Kite proteins: A superfamily of SMC/kleisin partners conserved across bacteria, archaea, and eukaryotes. Structure 23: 2183–2190. [DOI] [PubMed] [Google Scholar]
- Palecek J., Vidot S., Feng M., Doherty A.J., Lehmann A.R. (2006). The Smc5-Smc6 DNA repair complex. bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J. Biol. Chem. 281: 36952–36959. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Pebernard S., Wohlschlegel J., McDonald W.H., Yates J.R. III, Boddy M.N. (2006). The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 26: 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P.R., Yu H. (2007). The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14: 581–590. [DOI] [PubMed] [Google Scholar]
- Puchta H., Swoboda P., Hohn B. (1995). Induction of intrachromosomal homologous recombination in whole plants. Plant J. 7: 203–210. [Google Scholar]
- Räschle M., et al. (2015). DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348: 1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft V.K., Chumak N., Mosiolek M., Slusarz L., Komnenovic V., Brownfield L., Twell D., Kakutani T., Tamaru H. (2009). Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V. (2009). SMC proteins and their multiple functions in higher plants. Cytogenet. Genome Res. 124: 202–214. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B., Salehian Z. (2002). An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol. Biol. Report. 20: 107–114. [Google Scholar]
- Stingele J., Jentsch S. (2015). DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 16: 455–460. [DOI] [PubMed] [Google Scholar]
- Tian G., Lu Q., Zhang L., Kohalmi S.E., Cui Y. (2011). Detection of protein interactions in plant using a gateway compatible bimolecular fluorescence complementation (BiFC) system. J. Vis. Exp. 55: 3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., Lisby M. (2007). The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9: 923–931. [DOI] [PubMed] [Google Scholar]
- Tretyakova N.Y., Groehler A. IV, Ji S. (2015). DNA-protein cross-links: Formation, structural identities, and biological outcomes. Acc. Chem. Res. 48: 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I., McElver J.A., Liu Cm C.M., Yang L.J., Wu J.Q., Martinez A., Patton D.A., Meinke D.W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128: 38–51. [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. (2016). SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17: 399–412. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J.-P., Baskar R., Grossniklaus U. (2000). Delayed activation of the paternal genome during seed development. Nature 404: 91–94. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Pacher M., Dukowic S., Schubert V., Puchta H., Schubert I. (2009). The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yu H. (2012). The Smc complexes in DNA damage response. Cell Biosci. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Yuan D., Liu M., Li C., Liu Y., Zhang S., Yao N., Yang C. (2013). AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 161: 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Wang W., Marqués J., Mohan R., Saleh A., Durrant W.E., Song J., Dong X. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 52: 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., Lai J., Xu P., Zhang S., Zhang J., Li C., Wang Y., Du J., Liu Y., Yang C. (2014). AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Repair (Amst.) 21: 140–147. [DOI] [PubMed] [Google Scholar]
- Zabrady K., Adamus M., Vondrova L., Liao C., Skoupilova H., Novakova M., Jurcisinova L., Alt A., Oliver A.W., Lehmann A.R., Palecek J.J. (2016). Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 44: 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qi Y., Liu M., Yang C. (2013). SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F). J. Integr. Plant Biol. 55: 83–95. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao X., Blobel G. (2005). A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]