The zinc-finger protein KNUCKLES integrates transcriptional repression with polycomb-mediated silencing of the floral stem cell determinant WUSCHEL.
Abstract
Arabidopsis (Arabidopsis thaliana) floral meristems terminate after the carpel primordia arise. This is achieved through the temporal repression of WUSCHEL (WUS), which is essential for stem cell maintenance. At floral stage 6, WUS is repressed by KNUCKLES (KNU), a repressor directly activated by AGAMOUS. KNU was suggested to repress WUS through histone deacetylation; however, how the changes in the chromatin state of WUS are initiated and maintained to terminate the floral meristem remains elusive. Here, we show that KNU integrates initial transcriptional repression with polycomb-mediated stable silencing of WUS. After KNU is induced, it binds to the WUS promoter and causes eviction of SPLAYED, which is a known activator of WUS and can oppose polycomb repression. KNU also physically interacts with FERTILIZATION-INDEPENDENT ENDOSPERM, a key polycomb repressive complex2 component, and mediates the subsequent deposition of the repressive histone H3 lysine 27 trimethylation for stable silencing of WUS. This multi-step silencing of WUS leads to the termination of floral stem cells, ensuring proper carpel development. Thus, our work describes a detailed mechanism for heritable floral stem cell termination in a precise spatiotemporal manner.
INTRODUCTION
In Arabidopsis (Arabidopsis thaliana), stem cells are confined to a specific domain called the meristem. In the shoot apical meristem (SAM), stem cells are constitutively active and contribute to continuous growth. However, in the floral meristem, stem cells are terminated after initiation of the full complement of floral organs to ensure proper differentiation of the reproductive organs of the flower. The homeobox gene WUSCHEL (WUS), which is expressed in the organizing center (OC) of the SAM and in the floral meristem, plays an important role in the maintenance of the stem cell pool (Mayer et al., 1998). In the SAM, WUS is a direct target of SPLAYED (SYD), an ATP-dependent SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling factor (Kwon et al., 2005), which utilizes the energy of ATP hydrolysis to alter the accessibility of cis-regulatory DNA regions for the transcriptional machinery (Cairns, 2005). WUS is transcriptionally activated by SYD, which is required for the proper maintenance of stem cells (Kwon et al., 2005), and this is accompanied by deposition of the histone trimethylation on lysine 4 of histone H3 (H3K4me3) activation mark (Berger et al., 2011).
WUS is also a target of PRC2-mediated epigenetic repression via histone H3K27me3 (Zhang et al., 2007). H3K27me3 is catalyzed by the polycomb repressive complex2 (PRC2). During reproductive development, the PRC2 complex includes CURLY LEAF (CLF), which is an H3K27 methyltransferase (Goodrich et al., 1997). Another PRC2 component, FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), is a homolog of the Drosophila WD motif-containing protein extra sexcombs (Esc; Ohad et al., 1999). FIE can physically interact with CLF, and knockdown of FIE leads to strong morphological aberrations, suggesting that FIE plays an important role in the control of vegetative and reproductive development (Katz et al., 2004). In addition, EMBRYONIC FLOWER2 (EMF2) can also interact directly with CLF. Mutation of EMF2 results in drastic early flowering phenotypes (Yoshida et al., 2001). TERMINAL FLOWER2 (TFL2)/LIKE HETEROCHROMATIN PROTEIN1 may be a functional component of PRC1 and colocalizes with the repressive mark H3K27me3 throughout the Arabidopsis genome (Turck et al., 2007).
In Arabidopsis, several factors are known to recruit PcG to individual genes. During vernalization, the noncoding RNA COLDAIR recruits PRC2 to the FLOWERING LOCUS C (FLC) locus to silence FLC (Heo and Sung, 2011). Additionally, the transcriptional repressor VIVIPAROUS1/ABI3-LIKE proteins bind FLC cis-elements and recruit PcG for epigenetic silencing (Qüesta et al., 2016; Yuan et al., 2016). During vegetative development, the ASYMMETRIC LEAVES complex can physically interact with PRC2 and recruit it to BREVIPEDICELLUS and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 to stably silence them in differentiating leaves (Lodha et al., 2013). A recent study defined Polycomb response elements and associated transcription factor families that directly recruit PRC2 to developmental genes in Arabidopsis (Xiao et al., 2017). However, it remains unknown how PcG is recruited to specific targets in a precise spatiotemporal manner.
AGAMOUS (AG) directly induces the gene encoding Cys2-His2 (C2H2)-type zinc finger protein KNUCKLES (KNU) to repress WUS in the floral meristem during floral stage 6 (Sun et al., 2009). KNU can physically associate with a repressor complex composed of TOPLESS, HISTONE DEACETYLASE19, and MINI ZINC FINGER2. Within this complex, MINI ZINC FINGER2 binds to the WUS locus and recruits KNU, TOPLESS, and HISTONE DEACETYLASE19, leading to WUS repression through histone deacetylation (Bollier et al., 2018). However, WUS is eventually silenced by H3K27me3-mediated epigenetic memory to terminate the floral meristem (Zhang et al., 2007). Therefore, how the changes in chromatin state of WUS are initiated and how the silenced status of WUS chromatin is maintained to abolish the floral meristem are elusive. Here, we show a multi-step mechanism for WUS silencing. Initial transcriptional repression of WUS is associated with rapid eviction of the chromatin remodeler SYD by KNU, loss of DNA accessibility, and loss of active histone marks on the WUS locus. Subsequently KNU-mediated recruitment of PcG onto the WUS chromatin leads to heritable suppression of floral meristem activities. Thus, our study shows that the repressor protein KNU plays a pivotal role in integrating transcriptional repression with H3K27me3-mediated silencing of WUS in the floral meristem.
RESULTS
Spatial and Temporal Association between KNU and WUS in the Floral Meristem
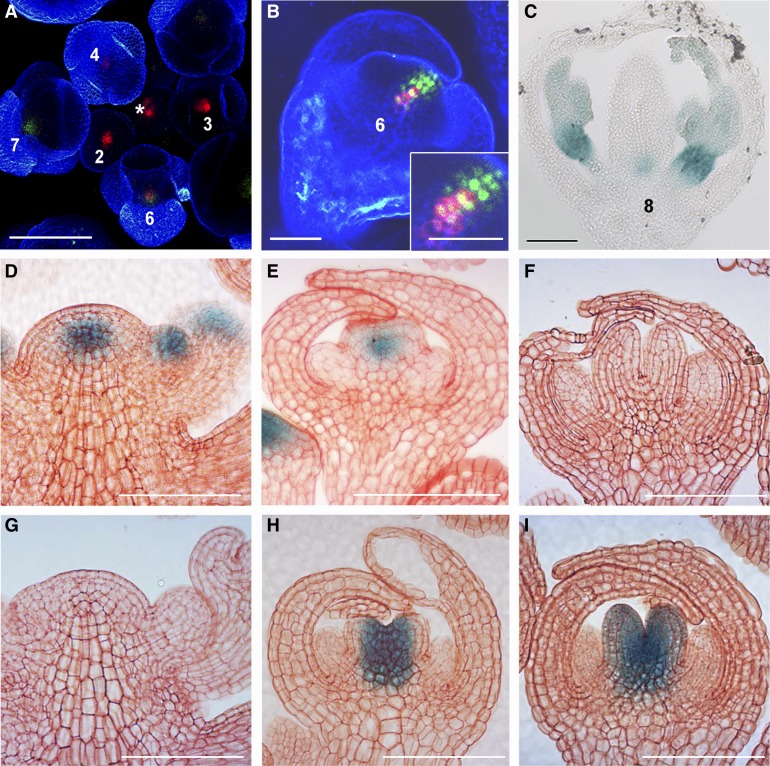
A plant line doubly transgenic for ProKNU:KNU-VENUS (Sun et al., 2014) and ProWUS:green fluorescent protein (GFP)-endoplasmic reticulum (ER; Gordon et al., 2007) was established to examine the spatiotemporal expression patterns of KNU and WUS. Prominent WUS expression was observed in the SAM and in the floral meristems of stages 2 to 6 flower buds (Smyth et al., 1990; Mayer et al., 1998), whereas KNU activity was only detected in flower buds from stage 6 onward (Figures 1A and 1C; Supplemental Figures 1A to 1C). KNU expression was initially detected in the central zone (Figure 1B; Supplemental Figures 1D to 1F), which later became broader, including the top three stem cell layers and OC (Supplemental Figures 1G to 1O). This transient overlap at floral stage 6 in the expression domains of KNU and WUS hints at cell-autonomous repression of WUS by KNU. In the ProWUS:β‑glucuronidase (GUS) line (Bäurle and Laux, 2005), WUS was detected in the SAM and in early stage 6 flower buds, but it was absent from late stage 6 flower buds (Figures 1D to 1F). By contrast, in the ProKNU:KNU-GUS line (Sun et al., 2009), KNU was detected in early stage 6 flower buds in the stem cell niche and in late stage 6 flower buds in developing carpels (Figures 1G to 1I). In floral stage 7, compared with silenced activity of WUS (Supplemental Figure 1P), KNU expression continues at the basal part of developing carpels and starts at the abaxial side of stamen primordia (Supplemental Figure 1Q). KNU expression later converged on the basal central cells of carpels (Figure 1C), which were previously described as silenced late OC cells (Liu et al., 2011), suggesting that KNU may be required in these cells for persistent silencing of WUS.
Figure 1.
KNU and WUS Expression Patterns.
(A) Confocal observation of the doubly transgenic inflorescence for ProWUS:GFP-ER (Gordon et al., 2007) (red) and ProKNU:KNU-VENUS (green), which was confirmed to rescue knu-1 (Sun et al., 2014). *, shoot apical meristem. Numbers, floral stages (Smyth et al., 1990).
(B) Higher magnification of the side view of a stage 6 floral bud of ProWUS:GFP-ER ProKNU:KNU-VENUS. The inset in (B) is the close-up of the central zone, showing WUS and KNU expression in meristematic cells.
(C) ProKNU:KNU-GUS (Sun et al., 2009) staining in a wild-type flower at stage 8. Bars = 50 µm for (A) to (C).
(D) to (I) GUS staining of ProWUS:GUS and ProKNU:KNU-GUS in the wild-type inflorescences, early stage 6 and stage 7 flowers.
(D) to (F) ProWUS:GUS staining in inflorescence (D), early stage 6 flower bud (E), and late stage 6 flower bud (F).
(G) to (I) ProKNU:KNU-GUS staining in inflorescence (G), early stage 6 flower bud (H), and late stage 6 flower bud (I). Bars = 100 μm for (D) to (I).
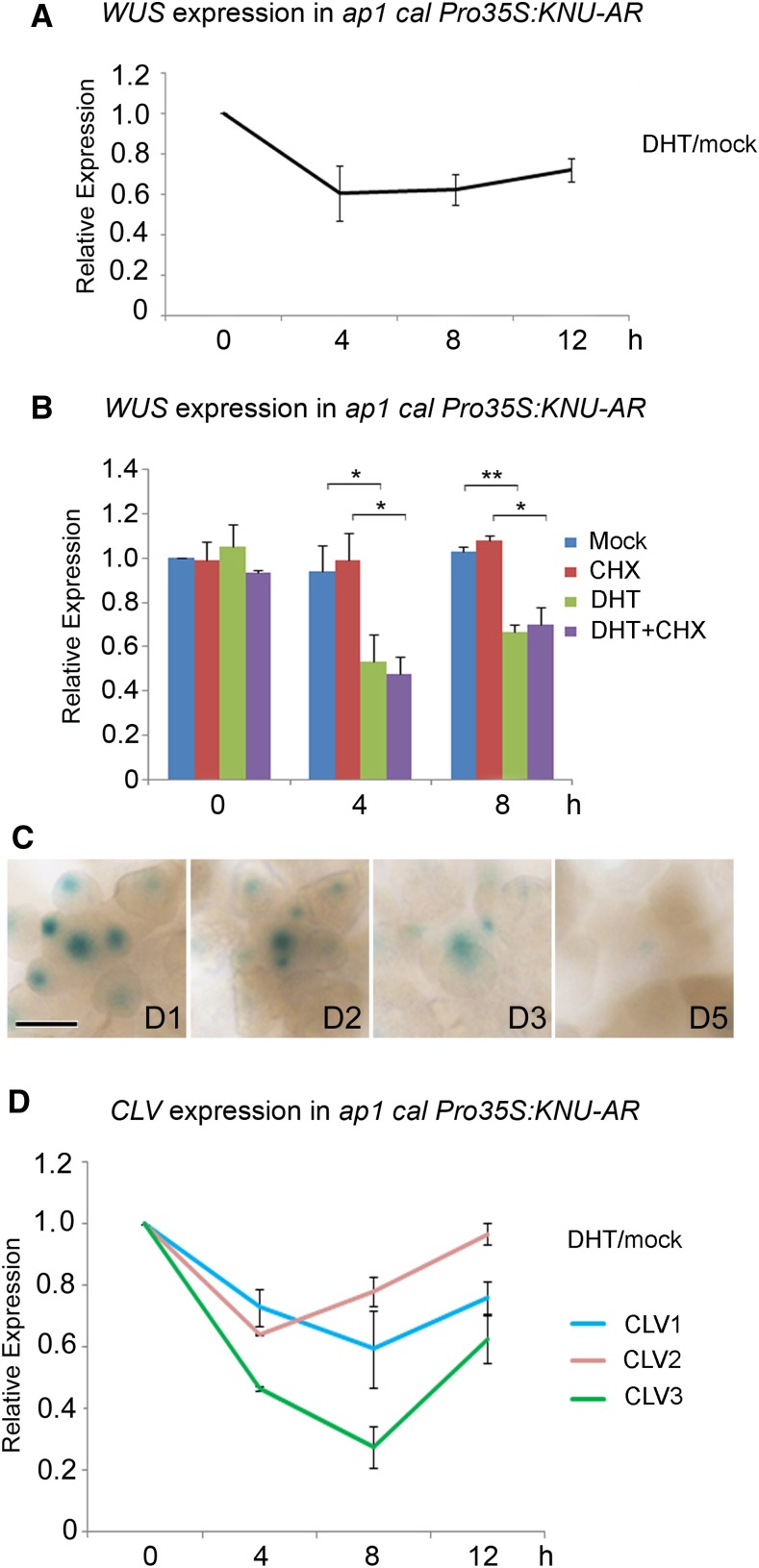
To examine the detailed timing of WUS repression by KNU, a line with inducible KNU activity, apetala1 cauliflower (ap1 cal) Pro35S:KNU-AR, was used for quantitative (q)PCR expression analysis (Figures 2A and 2B). In ap1 cal Pro35S:KNU-AR, the ap1 cal double mutant enriches for meristematic tissues (Alvarez-Buylla et al., 2006), while KNU-AR, a fusion protein between KNU and the steroid hormone ligand binding domain of the androgen receptor (AR), is sufficient to inducibly terminate the floral meristems upon continuous induction of the KNU activity (Sun et al., 2009). A single 5α-androstan-17β-ol-3-one (DHT) treatment lead to a 40% decrease in the WUS mRNA level at 4 h relative to the 0-h time point. At 8 h and 12 h after induction, we observed a trend toward an increase in WUS expression (Figure 2A), which could be explained by a CLAVATA (CLV)-dependent compensation mechanism (Müller et al., 2006). We also treated the ap1 cal Pro35S:KNU-AR plants with cycloheximide (CHX, a protein synthesis inhibitor) alone or with CHX together with DHT and found that WUS repression by KNU is independent of protein synthesis (Figure 2B). In addition, in ProWUS:GUS Pro35S:KNU-AR in response to continuous DHT treatment followed by staining at 1, 2, 3, and 5 d after the start of treatment, WUS expression remains repressed by KNU (Figure 2C). Thus, WUS may be directly repressed by KNU and the repression can be maintained, if KNU is continuously expressed. Because KNU is also expressed in stem cell layers (Figure 1B; Supplemental Figures 1M to 1O), we monitored the expression of CLV genes as well. CLV1, CLV2, and CLV3 expression all show initial reduction by KNU at 4 h after DHT treatment but recovery later (Figure 2D), suggesting CLV genes may also be repressed by KNU.
Figure 2.
Repression of WUS and CLV Genes by KNU.
(A) and (B) WUS expression in ap1 cal Pro35S:KNU-AR after a single DHT treatment, as measured by RT-qPCR. Tip41-like (At4g34270) served as the internal control. Asterisks indicate significant differences between samples treated with different chemicals (*P < 0.05 and **P < 0.01, Student’s t test).
(C) GUS staining in ProWUS:GUS Pro35S:KNU-AR inflorescences from day 1 (D1) to day 5 (D5) after initial DHT treatment (day 0). The ProWUS:GUS Pro35S:KNU-AR inflorescences were treated with 100 nM DHT three times at 1-d intervals. Bar, 100 μm.
(D) Transcript levels of CLV1, CLV2, and CLV3 in ap1 cal Pro35S:KNU-AR inflorescences after a single DHT treatment. Tip41-like (At4g34270) served as the internal control. Error bars in (A), (B), and (D) represent the sd of two biological replicates with three technical replicates each.
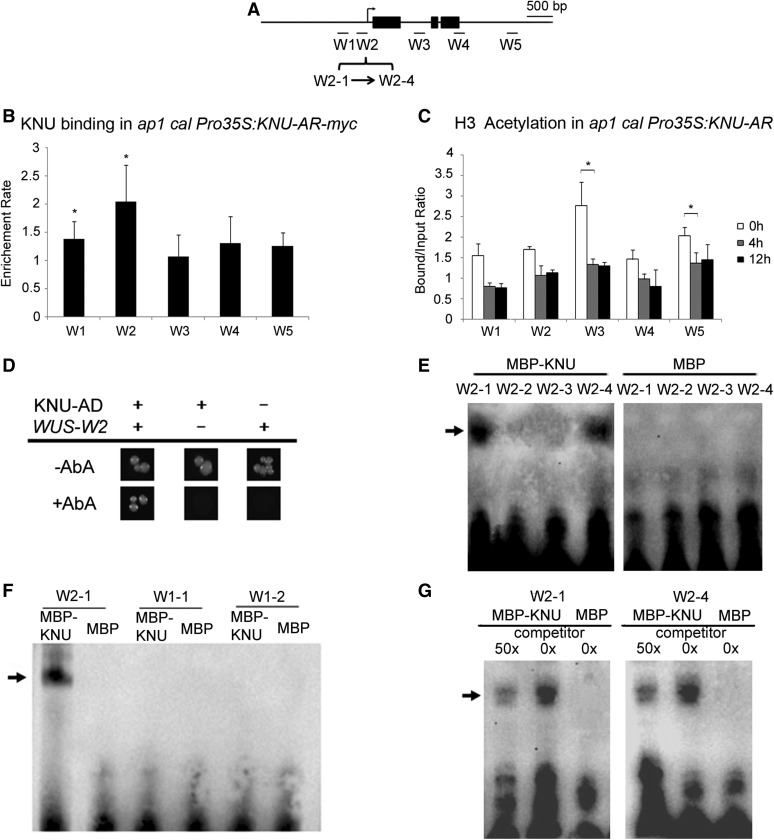
To test whether and in what time course the KNU protein directly binds WUS, we performed chromatin immunoprecipitation (ChIP) in ap1 cal Pro35S:KNU-AR-myc using an antibody against c-Myc. Plants were treated with DHT at 0 and 24 h twice, and inflorescences were harvested at 4 and 48 h, respectively, after the start of initial treatment (Figures 3A and 3B; Supplemental Figures 2A and 2B). An approximate 1.8-fold moderate enrichment of KNU was detected at the WUS promoter in the region from −308 bp to −125 bp upstream of the ATG start codon (primer set W2, Supplemental Table) as early as 4 h (Supplemental Figures 2A and 2B). We also noticed that at 4 h, an approximate 1.6-fold moderate enrichment can be detected within the first intron of WUS (primer set W3, Supplemental Table). At 48 h, we detected stronger (up to twofold) enrichment of KNU binding at the W2 site of the WUS promoter (Figures 3A and 3B). The initial binding at 4 h of KNU at W3 is consistent with a report that KNU is within a histone deacetylation complex associated with the first intron of WUS (Bollier et al., 2018). Because histone acetylation is associated with transcriptional activity (Sawarkar and Paro, 2010), we monitored H3 acetylation level on WUS chromatin upon KNU activation. We found a 40% decrease of H3 acetylation level in the first intron of WUS (primer set W3, Supplemental Table) by 4 h, and no further reduction of H3 acetylation by 12 h (Figures 3A and 3C). This agrees with the recent finding that KNU affects WUS chromatin via H3 deacetylation (Bollier et al., 2018). By contrast, the binding of KNU on the first intron of WUS (W3 region) is not detectable at the 48-h time point (Figures 3A and 3B).
Figure 3.
KNU Binds to the WUS Promoter.
(A) Schematic diagram showing the WUS locus and the promoter region used for ChIP assays, yeast one-hybrid assays, and EMSAs in (B) to (G). Black rectangles represent coding regions. Fragments: W1-1, −648 ∼ −583; W1-2, −582 ∼ −518; W2-1, −308 ∼ −265; W2-2, −264 ∼ −213; W2-3, −212 ∼ −175; W2-4, −174 ∼ −125 bp upstream from the ATG start codon.
(B) ChIP assay using ap1 cal 35S:KNU-AR-3myc inflorescences harvested 48 h after DHT treatment. Nuclear protein complexes were immunoprecipitated with anti–c-Myc agarose beads, and the enriched DNA was used for qPCR analysis. The y axis shows relative enrichment using anti-HA agarose beads as a control. Mu-like transposons served as a negative control locus and were set to 1. Asterisks indicate significant differences between the control locus (MU) and different primer sets on WUS (*P < 0.05, Student’s t test).
(C) ChIP assay using ap1 cal Pro35:KNU-AR inflorescences harvested at 0, 4, and 12 h after a single 100 nM DHT treatment. The y axis shows the calibrated relative ratio of bound DNAs to input DNAs after IP. Mu-like transposons served as a negative control locus and were set to 1. Error bars represent the sd of three (B) and two (C) biological replicates with three technical replicates each. Asterisks indicate significant differences between different time points at certain primers sets of WUS (*P < 0.05, Student’s t test).
(D) Yeast one-hybrid assays demonstrate that KNU associates with the WUS promoter W2 region. AbA, Aureobasidin A.
(E) to (G) EMSAs confirm that KNU binds to the W2-1 and W2-4 fragments. The arrow indicates the DNA–protein complex. Nonlabeled oligonucleotides were used as competitors. MBP was used as a negative control.
To verify the binding of KNU to the WUS promoter, we performed yeast one-hybrid assays. The result confirmed direct binding of KNU to the W2 region of the WUS proximal promoter (Figures 3A and 3D). The direct binding of KNU was further examined by electrophoretic mobility shift assays (EMSAs). Four fragments for the W2 region (W2-1 to W2-4) were biotin labeled and incubated with maltose binding protein (MBP)–tagged KNU protein. We also used two fragments for the W1 region (W1-1 and W1-2) as controls. W2-1 and W2-4 produced clear band shifts (Figures 3A and 3E), while W1-1 and W1-2 showed no band shifts (Figures 3A and 3F). Moreover, the addition of excess unlabeled competitor probes effectively reduced the amount of shifted bands, demonstrating direct binding of KNU (Figure 3G). To identify the KNU bound cis-element, we first used 5′ upstream sequences of WUS to carry out phylogenetic shadowing with eight Brassicaceae species (O’Malley et al., 2016; Yamaguchi et al., 2018). Three conserved regulatory modules (CRMs) were identified and defined as CRM1, CRM2, and CRM3 (Supplemental Figure 3A). Because the W2 region is within the CRM3, CRM3 was further analyzed and we noticed there is a conserved core aatc motif (Supplemental Figure 3B), which is also found in both W2-1 and W2-4 fragments. Mutating aatc to gggg in W2-1– and W2-4–unlabeled competitor probes cannot reduce the amount of shifted bands (Supplemental Figure 3C), indicating that the aatc motif could be the putative KNU binding cis-element.
Polycomb-Mediated Epigenetic Silencing of WUS Is Delayed Relative to WUS Repression
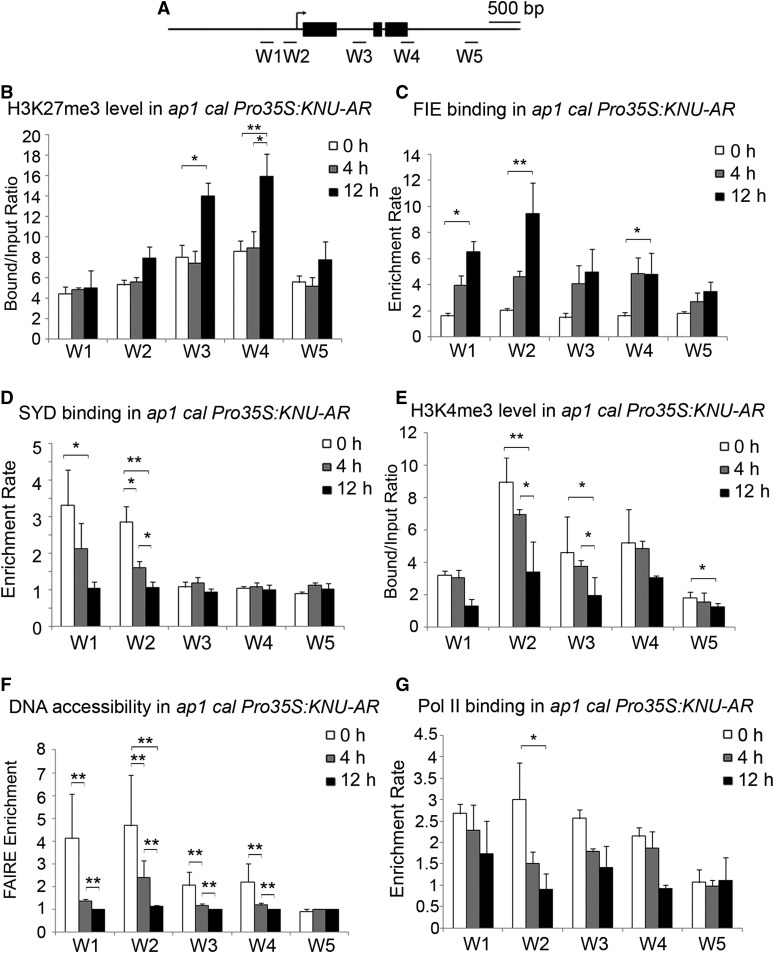
To determine whether the transcriptional repression of WUS by KNU is associated with PcG-mediated silencing of the WUS locus, the H3K27me3 level on WUS was examined at different time points after the DHT treatment in ap1 cal Pro35S:KNU-AR inflorescences. An increase in the H3K27me3 level at the WUS locus was observed at 8 and 12 h, but not at 4 h, after DHT treatment relative to that at the 0-h time point (Figures 4A and 4B; Supplemental Figures 4A and 4B). By contrast, WUS transcript was repressed by KNU within 4 h in the same inducible line (Figures 2A and 2B). Thus, transcriptional repression of WUS by KNU precedes deposition of the repressive mark H3K27me3, which may be required for heritable silencing of WUS at the chromatin level. We also examined dynamic FIE enrichment on WUS chromatin using ap1 cal Pro35S:KNU-AR ProFIE:FIE-VENUS, a line generated by crossing ap1 cal Pro35S:KNU-AR with ProFIE:FIE-VENUS (Sun et al., 2014). An increase in FIE enrichment at the WUS locus was observed at 4 h and 12 h (Figures 4A and 4C). These results suggest that H3K27me3 may be deposited in a KNU-dependent manner and that there are a few to several hours of time lag between PRC2 binding and H3K27me3 deposition.
Figure 4.
WUS Chromatin Dynamics after KNU Activation.
(A) Schematic diagram showing the WUS locus and the promoter region used for ChIP assays in (B) to (G).
(B) H3K27me3 analysis by ChIP assays using ap1 cal Pro35:KNU-AR inflorescences harvested at 0, 4, and 12 h after a single 100 nM DHT treatment.
(C) ChIP assay for FIE enrichment using ProFIE:FIE-VENUS ap1 cal Pro35S:KNU-AR inflorescences harvested at 0, 4, and 12 h. The y axis shows the calibrated relative ratio of bound DNAs to input DNAs after IP.
(D) SYD binding analysis. Inflorescences were harvested at 0, 4, and 12 h. Error bars represent the se of three biological replicates.
(E) H3K4me3 analysis by ChIP assays using ap1 cal Pro35:KNU-AR inflorescences harvested at 0, 4, and 12 h after a single 100 nM DHT treatment.
(F) DNA accessibility at the WUS locus in the context of chromatin assayed by FAIRE upon KNU activation in ap1 cal Pro35S:KNU-AR. The Ta3 retrotransposon (At1g37110) was used as the negative control locus and was set to 1.
(G) ChIP assay for Pol II binding using ap1 cal Pro35S:KNU-AR inflorescences harvested at 0, 4, and 12 h. The y axis shows the calibrated relative ratio of bound DNAs to input DNAs after IP. Mu-like transposons (MU) served as a negative control locus for (B) to (E) and (G), and the relative bound/input ratios or relative enrichment rates on MU were set to 1. Error bars represent the sd of three (see [B] and [E]) and two (see [C], [F], and [G]) biological replicates with three technical replicates each. Asterisks indicate significant differences between different time points at certain primer sets of WUS (*P < 0.05 and **P < 0.01, Student’s t test).
The proximal promoter region (primer set W2, Supplemental Table 1), where KNU binds to the WUS locus, overlaps partially with a region reported to be occupied by SYD, an activator of WUS (Kwon et al., 2005). Thus, we next tested whether KNU competitively inhibits SYD binding to WUS by ChIP assay using a SYD-specific antibody (Kwon et al., 2005). SYD was enriched at the WUS proximal promoter (covering regions including both W1 and W2 primer sets, Supplemental Table) at 0 h. However, SYD binding became weaker (∼40% decrease in occupancy at the W2 region, P < 0.05) at 4 h and even weaker at 12 h (∼30% of the initial occupancy at W2, P < 0.01; Figures 4A and 4D). This suggests that binding of KNU leads to the eviction of SYD from the WUS promoter within 4 h. The eviction of SYD preceded the deposition of H3K27me3 (Figures 4B and 4D; Supplemental Figures 4A and 4B), suggesting that KNU triggers transcriptional repression at least partially by acting antagonistically with SYD.
By opening the WUS locus, SYD may contribute to higher DNA accessibility (Cairns, 2005) and deposition of H3K4me3 (Wu et al., 2012), a mark associated with actively transcribed chromatin (Sawarkar and Paro, 2010). To comprehensively understand the epigenetic events on the WUS chromatin upon KNU activation, the dynamic level of H3K4me3 and DNA accessibility at WUS was examined. At 0 h, in ap1 cal Pro35S:KNU-AR inflorescences, high H3K4me3 levels were detected, which peaked at the proximal promoter of WUS (primer set W2, Supplemental Table). KNU activation resulted in a slight decrease of the H3K4me3 level at 4 h (∼20% decrease at W2) and a further reduction by 12 h (∼50% of initial level at W2; Figures 4A and 4E). To assess the dynamic accessibility of the WUS locus upon KNU activation, we used formaldehyde-assisted isolation of regulatory elements (FAIRE), a method that enriches accessible (nucleosome-depleted) genomic DNA from crosslinked chromatin (Simon et al., 2012). FAIRE revealed an ∼50% decrease of the accessibility at the WUS locus, especially at the KNU-bound region (primer set W2, P < 0.01; Supplemental Table), within 4 h after KNU induction. Twelve hours after KNU induction, accessibility to the WUS locus decreased even further, to 24% of the initial levels (P < 0.01; Figures 4A and 4F). To check whether WUS locus accessibility disrupts RNA polymerase II (Pol II) function, we performed a ChIP assay to examine Pol II enrichment at the WUS locus. We noticed an ∼50% decrease of Pol II enrichment on the proximal promoter of WUS (primer set W2, Supplemental Table) at 4 h and an ∼70% decrease at 12 h (Figures 4A and 4G).
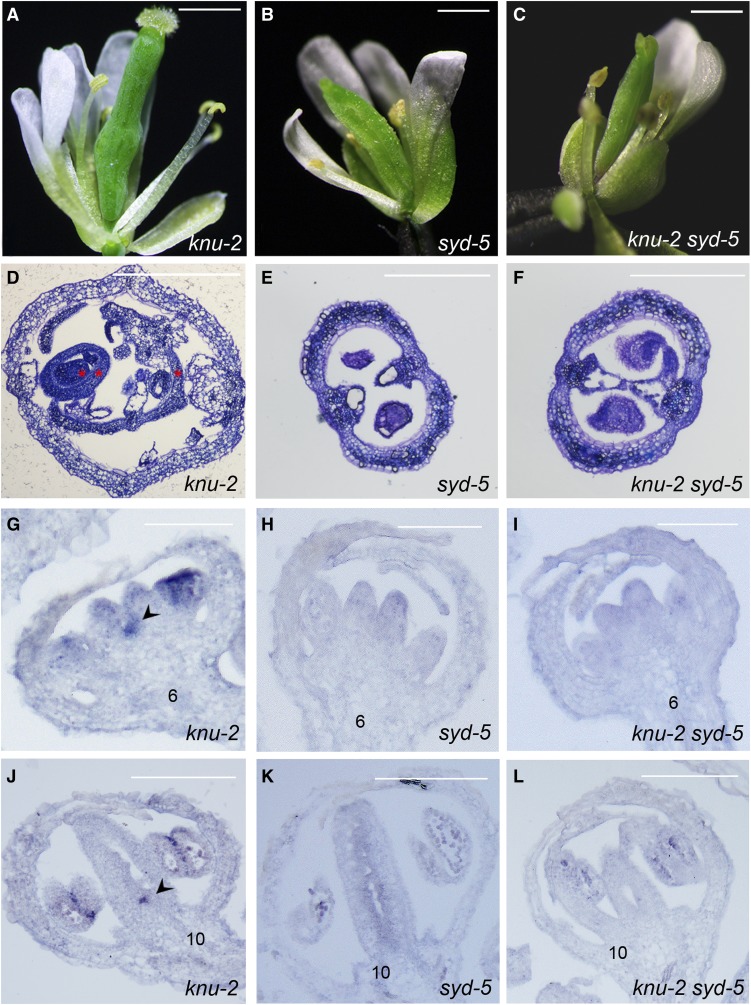
To assess the genetic interaction between KNU and SYD, we crossed a null mutant knu-2 (Sun et al., 2009) with a null allele of syd-5 (Bezhani et al., 2007). The double mutant flowers show a similar phenotype as syd-5 flowers, with defective stigma, while the bulged gynoecia harboring ectopic carpels normally observed in knu-2 flowers were not found in knu-2 syd-5 flowers (Figures 5A to 5F). We also checked WUS expression by in situ hybridization in knu-2, syd-5, and knu-2 syd-5 mutant flowers (Figures 5G to 5L). Consistent with our previous report, prolonged WUS expression was detected in stage 6 and stage 10 flowers of the knu-2 mutant (Figures 5G and 5J; Sun et al., 2009). However, prolonged WUS expression was not observed in stage 6 and stage 10 buds of syd-5 or knu-2 syd-5 mutant flowers (Figures 5H, 5I, 5K, and 5L). These data indicate that SYD function is required for the prolonged WUS activity in knu-2 flowers.
Figure 5.
Genetic Interaction between knu-2 and syd-5.
(A) to (C) Mutation of syd-5 rescues the knu-2 meristem indeterminate phenotype (30 flowers each from knu-2, syd-5, and knu-2 syd-5 were observed, respectively). Flowers at stage 15 of knu-2 (A) have bulged pistils, while the pistils of syd-5 (B) and knu-2 syd-5 (C) flowers at stage 15 were not bulged.
(D) to (F) Cross sections of knu-2 (D), syd-5 (E), and knu-2 syd-5 (F) siliques stained with 0.1% toluidine blue in 0.02% sodium carbonate solution. Within the knu-2 mutant silique, there are three reiterative ectopic carpels, as shown by red asterisks. Bar, 1 mm for (A) to (C).
(G) to (L) WUS expression is prolonged at stage 6 (G) and stage 10 (J) in knu-2 flower buds, but not in stage 6 and stage 10 flower buds of syd-5 (see [H] and [K]) and knu-2 syd-5 (see [I] and [L]). Arrowheads show ectopic WUS expression in meristematic cells. Bar = 100 μm for (G) to (L).
Silencing of WUS Requires KNU-Dependent PcG Recruitment
Next, to examine whether WUS chromatin changes are KNU-dependent during flower development, we examined WUS chromatin status using ap1 cal Pro35S:AP1-GR inflorescences (GR is the steroid-binding domain of the rat glucocorticoid receptor; Wellmer et al., 2006). To avoid the noise generated by WUS expression in stamen primordia after floral stage 7 (Sanders et al., 1999; Deyhle et al., 2007), tissues were harvested on days 0 and 4, roughly corresponding to the synchronized development of flower buds at approximately stages 1 to 2 and stages 6 to 7, respectively (Supplemental Figure 5A; Wellmer et al., 2006).
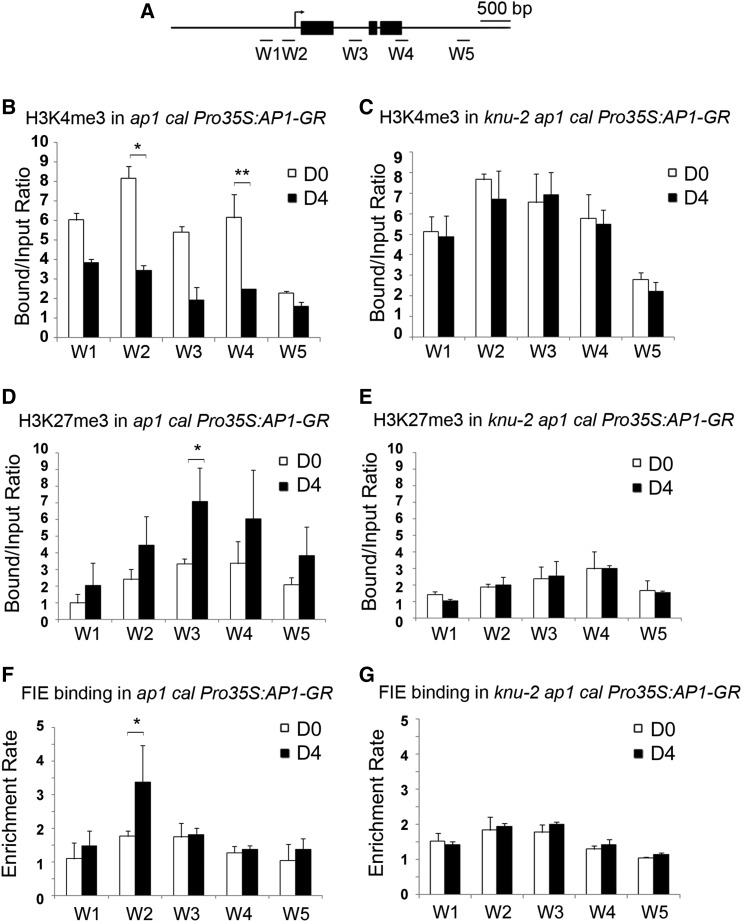
We first assessed H3K4me3 and H3 acetylation levels on WUS chromatin on day 0 and day 4. The ChIP results demonstrate general decreases of H3K4me3 and H3 acetylation levels throughout the entire WUS locus on day 4 (as indicated by primer sets W1 to W5 (Figures 6A and 6B; Supplemental Figures 5B and 5C; Supplemental Table). To test whether the decrease of H3K4me3 and H3 acetylation on WUS is dependent on KNU activity, the knu-2 mutation was introduced into ap1 cal Pro35S:AP1-GR. In the knu-2 ap1 cal Pro35S:AP1-GR inflorescences, we noticed the levels of H3K4me3 and H3 acetylation remains largely unchanged between day 0 and day 4 (Figure 6C; Supplemental Figure 5D). These results suggest that KNU activity is required for the removal of active histone marks of H3K4me3 and H3 acetylation on the WUS locus during flower development.
Figure 6.
Change of WUS Chromatin Status Is KNU Dependent.
(A) Schematic diagram showing the WUS locus used for ChIP assays in (B) to (G).
(B) to (G) The ap1 cal Pro35S:AP1-GR (see [B], [D], and [F]) and knu-2 ap1 cal Pro35S:AP1-GR (see [C], [E], and [G]) inflorescences 0 and 4 d after a single treatment with DEX were sampled.
(B) and (C) H3K4me3 analysis.
(D) and (E) H3K27me3 analysis. The y axis shows the calibrated relative ratio of bound DNAs to input DNAs after IP.
(F) and (G) ChIP assays for FIE binding. The y axis shows the relative enrichment using IgG as a control. Mu-like transposons served as a negative control locus, and the relative bound/input ratios or relative enrichment rates on MU were set to 1. Error bars represent the sd of three (see [B], [D], and [F]) and two (see [C], [E], and [G]) biological replicates with three technical replicates each. Asterisks indicate significant differences between day 0 (D0) and day 4 (D4) at certain primers sets of WUS (*P < 0.05 and **P < 0.01, Student’s t test).
To further investigate dynamic WUS chromatin status change including SYD binding, the DNA accessibility change, and H3K27me3 level during flower development, we also performed ChIP assays using day 0 and day 4 inflorescences of ap1 cal Pro35S:AP1-GR treated with dexamethasone (DEX). SYD enrichment decreases on the WUS promoter (including both W1 and W2 regions) by day 4 compared with day 0 (Supplemental Figure 5E). Similarly, DNA accessibility of the WUS locus also decreases to ∼40% on W2 by day 4 compared with day 0 (Supplemental Figure 5G). By contrast, in knu-2 ap1 cal Pro35S:AP1-GR, no obvious decreases were detected for SYD enrichment and WUS DNA accessibility (Supplemental Figures 5F and 5H). For H3K27me3 assays, the ChIP results showed background levels of H3K27me3 at the WUS locus at day 0, which could be due to the contribution of nonmeristematic cells, and an increase in the H3K27me3 levels throughout the entire WUS locus on day 4 (Figure 6D). The enrichment of H3K27me3 was strongest in the first intron (primer set W3, Supplemental Table), and on day 4 the repressive mark level was nearly twice that observed on day 0 (P < 0.05, Figure 6D). These results indicate that WUS repression at floral stage 6∼7 is associated with the deposition of H3K27me3 on the WUS locus. While in knu-2 ap1 cal1 Pro35S:AP1-GR, only a background level of the H3K27me3 repressive mark was observed at day 0 and day 4 on the WUS locus in the knu-2 ap1 cal Pro35S:AP1-GR plants (Figure 6E). Hence, deposition of the repressive mark H3K27me3 on WUS requires KNU activity.
Since the PRC2 complex is responsible for the deposition of H3K27me3 on WUS, the transgenic lines ap1 cal ProFIE:FIE-VENUS Pro35S:AP1-GR and ap1 cal ProEMF2:EMF2-VENUS Pro35S:AP1-GR (Wellmer et al., 2006; Sun et al., 2014) were used to examine the change in binding of PRC2 at the WUS locus. On day 0, only background levels of FIE binding were detected at WUS, whereas on day 4 there was a distinct enrichment of FIE (up to twofold) at the WUS proximal promoter region (P < 0.05, Figure 6F; primer set W2, Supplemental Table 1). Similarly, approximately a twofold enrichment of EMF2 was detected in the same region (P < 0.05; primer set W2, Supplemental Table) on day 4 compared with the background level on day 0 (Supplemental Figures 5B and 5I). In the knu-2 mutant background, enrichment of both FIE and EMF2 were detected at background levels at the WUS locus on days 0 and 4 (Figure 6G; Supplemental Figure 5J). These data show that recruitment of PcG to WUS is dependent on KNU activity during flower development.
PcG Activity Is Required for the Stable Silencing of WUS
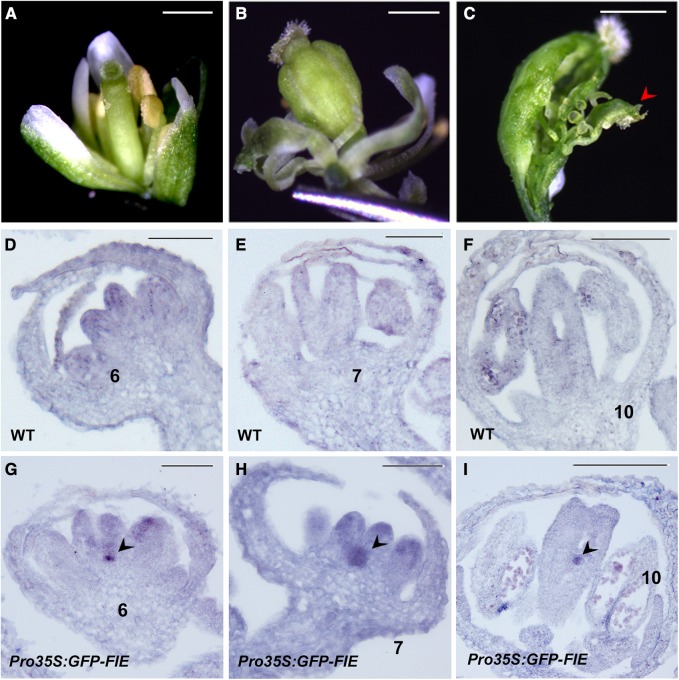
To determine whether WUS expression during flower development is affected in PcG mutants, the floral phenotypes were examined in the following mutants: tfl2-1, emf2-1, clf-28, fie-11/+ (fie-11 homozygous is embryonically lethal and cannot germinate), and the transgenic cosuppression line Pro35S:GFP-FIE, with a mostly silenced FIE activity (Katz et al., 2004). Generally, mutants of each PcG component show distinct floral phenotypes. The tfl2-1 mutant displays fused terminal flowers with two carpels in each gynoecium (Larsson et al., 1998). The emf2-1 flowers produce two fused infertile carpels surrounded by six small sessile leaves (Yoshida et al., 2001; Sung et al., 2003). The flower phenotypes of the clf-28 and fie-11/+ mutants are indistinguishable from the wild type (Guitton et al., 2004; Doyle and Amasino, 2009; Deng et al., 2013). None of these mutants displayed an indeterminate floral phenotype, possibly due to heterozygosity (for fie-11/+) or due to the activity of redundantly acting PcG factors. By contrast, unlike the wild-type flowers with two fused carpels, more than half (57 of 100 plants) of the Pro35S:GFP-FIE cosuppression plants produced multiple fused carpels with ectopic carpel-like tissue inside (Figures 7A to 7C). In the wild-type flower buds, WUS mRNA was mostly undetectable from stage 6 onward (Figures 7D to 7F; Lenhard et al., 2001; Lohmann et al., 2001). On the contrary, WUS persisted in the stage 6 and stage 7 carpel primordia and even in carpels of stage 10 flowers of the FIE cosuppression line (Figures 7G to 7I). The ectopic WUS in FIE-silenced flowers provides genetic evidence that PcG-mediated H3K27me3 plays an essential role in stable silencing of WUS from stage 6 onward, which is required for termination of the floral stem cells.
Figure 7.
PcG Activity Is Required for the Stable Silencing of WUS.
(A) The wild-type flower.
(B) and (C) Indeterminate phenotypes of flowers of the Pro35S:GFP-FIE cosuppression line showing multiple carpels (B) and an internal ectopic carpelloid organ (red arrowhead) within the primary carpels (C).
(D) to (F) Through in situ hybridization, expression of WUS was not detected in stages 6 (D), 7 (E), and 10 (F) wild-type flower buds. WT, wild type.
(G) to (I) Prolonged WUS expression in FIE-silenced line Pro35S:GFP-FIE flower buds at stages 6 (G), 7 (H), and 10 (I). Black arrowheads show ectopic WUS expression in meristematic cells. Bars = 1 mm in (A) to (C); 50 µm in (D) to (I).
We also tested the effect of ectopic KNU expression in the polycomb mutant background, and generated tfl2-1 Pro35S:KNU-AR and clf-28 Pro35S:KNU-AR (Figures 8A to 8F). After KNU activation with continuous DHT treatment, the treated plants showed a similar floral phenotype as tfl2-1 and clf-28 flowers, respectively (Figures 8B, 8C, 8E, and 8F). This is in stark contrast with the premature termination of floral meristems observed in Pro35S:KNU-AR continuously treated with DHT (Figures 8A and D; Sun et al., 2009). These results demonstrate that repression of WUS by KNU requires PcG activity. By contrast, loss of clf or tfl2 activity in the knu mutant background revealed no or only a slight increase in indeterminacy (Supplemental Figures 6A to 6F). We further monitored time-course WUS expression in clf-28 Pro35S:KNU-AR upon DHT treatment. WUS is repressed to ∼0.6-fold at 4 h upon KNU activation but recovered to ∼1-fold at 8 h and later time points (Supplemental Figure 6G). In tfl2-1 Pro35S:KNU-AR with DHT treatment, WUS expression showed a similar trend as in clf-28 Pro35S:KNU-AR (Supplemental Figure 6H). These results suggest that in clf-28 or tfl2-1 mutant backgrounds, WUS is initially repressed by KNU, but the repression is not stable without PcG activity. In addition, we found that KNU can still bind to the WUS promoter in the clf-28 null mutant background (Supplemental Figures 6I and 6J), suggesting that PRC2 is not involved in the recruitment of KNU. Thus, KNU-dependent recruitment of PRC2 is necessary for stable silencing of WUS.
Figure 8.
Repression of WUS by KNU Requires PcG Activity.
(A) to (F) Flowers of Pro35S:KNU-AR (see [A] and [D]), tfl2-1 Pro35S:KNU-AR (see [B] and [E]), and clf-28 Pro35S:KNU-AR (see [C] and [F]) after mock (see [A], [B], and [C]) or continuous DHT treatments (once a day for five continuous days; see [D] to [F]). After continuous induction of KNU, carpels were lost in the wild-type background (D), but not in the tfl2-1 or clf-28 mutant backgrounds (see [E] and [F]). Bar, 1 mm.
KNU Recruits PcG to WUS through Physical Interaction with FIE
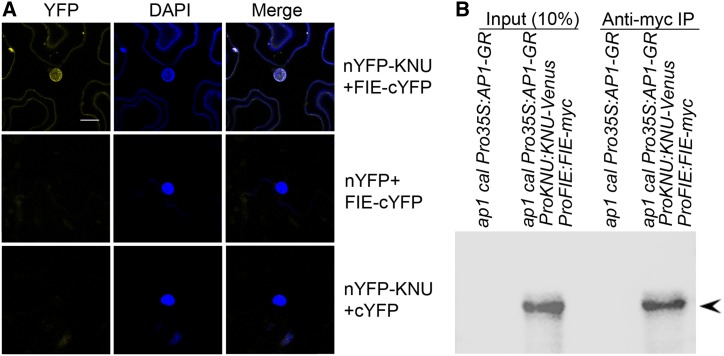
To examine the molecular basis of KNU-dependent PcG recruitment, we tested for a physical interaction between KNU and PcG factors, including FIE, CLF, SWINGER, EMF2, and TFL2 by yeast two-hybrid analysis. The use of the KNU full-length coding sequence as bait revealed an interaction between KNU and the WD repeat domain of FIE. Truncation of either the reported N-terminal C2H2 zinc finger domain or the C-terminal EAR-like motif of KNU (Payne et al., 2004) negated the interaction (Supplemental Figures 7A and 7B). To verify the yeast result, bimolecular fluorescence complementation (BiFC) analysis was performed in tobacco (Nicotiana tabacum) leaves (Ohad et al., 2007). BiFC revealed an in vivo interaction between KNU and FIE in the nucleus (Figure 9A). By contrast, another C2H2 zinc finger protein, ZFP11, does not show in vivo interaction with FIE in the nucleus of tobacco leaves (Supplemental Figure 7C). Furthermore, coimmunoprecipitation (co-IP) analysis of nuclear extracts from stage 6 flower buds of ap1 cal Pro35S:AP1-GR ProKNU:KNU-VENUS ProFIE:FIE-myc confirmed the in vivo interaction of KNU and FIE in Arabidopsis (Figure 9B; Supplemental Figures 7D and 7E). Overall, these results provide evidence that KNU physically interacts with FIE in the nucleus and may recruit PcG.
Figure 9.
KNU Physically Interacts with FIE.
(A) BiFC analysis of the interaction between KNU and FIE. DAPI, fluorescence of 4′,6-diamidino-2-phenylindole; Merge, merged images for yellow fluorescent protein (YFP) and DAPI. KNU and FIE were fused to nYFP and cYFP to generate nYFP-KNU and FIE-cYFP, respectively. Vectors containing only nYFP or cYFP were used as controls. nYFP, N-terminal version of YFP; cYFP, C-terminal version of YFP. Bars = 20 µm.
(B) In vivo interaction between KNU and FIE shown by co-IP. Nuclear extracts from stage 6 flower buds of ap1 cal Pro35S:AP1-GR and ap1 cal Pro35S:AP1-GR ProKNU:KNU-VENUS ProFIE:FIE-myc were incubated with anti–c-Myc agarose beads. The co-IPed KNU fusion protein (arrowhead) was detected by anti-GFP antibody.
DISCUSSION
KNU Mediates Multi-Step Silencing of WUS in the Floral Meristem
Here, we show that the C2H2 zinc finger protein KNU binding site (W2) on the WUS promoter overlaps with the region occupied by the SWI/SNF chromatin remodeling factor SYD, which is required for the maintenance of WUS transcription (Kwon et al., 2005). Our ChIP assay showed that SYD binds to both W1 and W2 in vivo, and in our model, SYD binding is prevented by KNU. We also performed an EMSA experiment to test KNU binding to the W1 region of the WUS promoter. The result suggests that KNU does not bind to W1 (Figure 3F), at least in vitro. The W1 and W2 sites are less than 200 bp apart and SYD is a component of a protein complex (Bezhani et al., 2007); hence, the KNU and SYD binding sites likely partially overlap. Our findings also agree with the reported SYD binding region (Kwon et al., 2005). Besides, W3 also contains one aatc sequence. KNU may initially bind to the core sequence in W3 to deacetylate the WUS locus (Bollier et al., 2018).
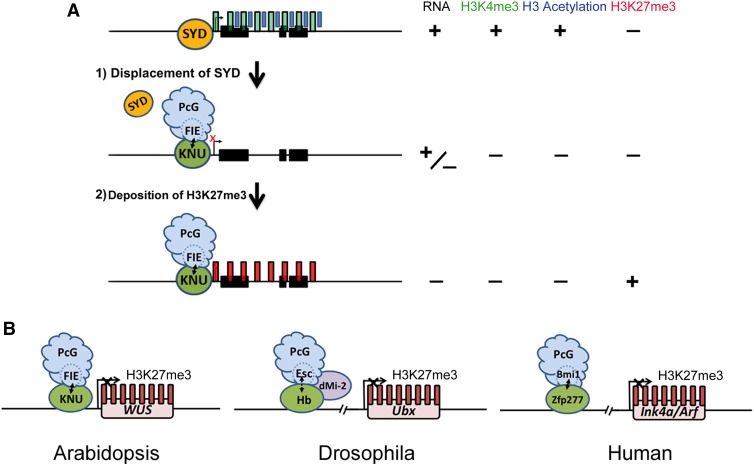
Our genetic analyses confirmed that meristem indeterminacy in knu is dependent on SYD activity (Figures 5A to 5F). KNU induction causes the rapid eviction of the SYD protein (Figure 4D). We also show that recruitment of PcG on the WUS locus is KNU dependent and that PcG activity is necessary for stable silencing of WUS by KNU (Figures 6F and 6G; Figure 8; Supplemental Figures 5I and 5 J). KNU physically interacts with the key PRC2 component FIE and recruits PRC2 to the WUS promoter to silence WUS via H3K27me3 (Figures 4B, 4C, 6F, 6G, 9A, and9B). WUS repression was initiated prior to H3K27me3 accumulation (Figures 2A and 4B; Supplemental Figures 4A and 4B). Therefore, the termination of floral stem cells may occur in sequential steps: initial transcriptional repression of WUS associated with rapid eviction of active chromatin remodelers, loss of DNA accessibility, loss of active histone marks, and subsequent PcG-mediated silencing of the WUS chromatin (Figure 10A). These multi-steps are integrated by KNU, which plays an important role in the programmed termination of the floral stem cells.
Figure 10.
Models.
(A) Working model on how KNU represses WUS. Before stage 6, when the active histone marks H3K4me3 and H3 acetylation on WUS are detected, the active transcription of WUS is maintained by the SWI/SNF ATPase SYD. During the early floral stage 6, the KNU protein, which is induced by AG, may associate with PRC2 and the KNU-PRC2 complex directly binds to the WUS promoter by competing with SYD. That leads to eviction of SYD from the WUS promoter and a decrease in the active marks H3K4me3 and H3 acetylation. Both the repressor activity of KNU (mediated by histone deacetylation) and the eviction of SYD simultaneously lead to transcriptional repression of WUS. Subsequently, the KNU-PRC2 complex deposits the repressive mark H3K27me3 onto WUS, which is maintained by continuous expression of KNU after stage 6.
(B) A conserved mechanism for the recruitment of PcG to target genes by C2H2 zinc finger proteins in plants and animals. In Arabidopsis, the C2H2 zinc finger protein KNU binds to the WUS promoter and recruits PcG to WUS via direct interaction with FIE, a homolog of Esc of Drosophila. In Drosophila, the C2H2 zinc finger protein Hunchback (Hb), which binds the Ultabithorax (Ubx) promoter, together with dMi-2 recruits PcG to UBX via interaction with Esc. In human cells, the C2H2 zinc finger protein Zfp277, which binds the Ink4a/Arf locus, directly interacts with Bmi1, a PRC1 factor for PcG recruitment to Ink4a/Arf. (For simplicity, gene size and structures are not to scale.)
In human cells, nuclear hormone receptors require SWI/SNF chromatin remodeling factors at each successive round of transcription initiation at the promoter in vivo through maintaining open chromatin structures and active H3K4me3 marks (McNally et al., 2000; Nagaich et al., 2004). KNU induction also caused rapid reduction of DNA accessibility and H3K4me3 marks (Figures 4E and 4F). Furthermore, we show that H3 acetylation on the WUS locus decreased after KNU activation (Figure 3C). This is consistent with a recent finding that KNU recruits a histone deacetylase–containing complex to the WUS locus (Bollier et al., 2018). In vitro, PcG and SWI/SNF factors have mutually exclusive activities (Bar-Ziv et al., 2016). Thus, KNU causes H3 deacetylation, H3K4 demethylation, and SYD dissociation simultaneously, which would be prerequisite steps prior to H3K27me3-mediated epigenetic silencing of the WUS locus. One possibility is that the KNU-PcG complex forms first and acts together to remove SYD and then trigger the H3K27me3 deposition on WUS. Deposition of H3K27me3 on WUS may require reduced transcription, which was also implicated at the FLC locus, where H3K27me3 deposition on the gene body occurs only after transcription is decreased (Buzas et al., 2011). In animal cells, reduced transcriptional activity and histone deacetylation also occur prior to H3K27me3 accumulation on gene body (Kehle et al., 1998). Persistent silencing of WUS beyond floral stage 6 may still require KNU, as KNU-GUS activity can still be observed in the late OC cells (Figure 1C; Liu et al., 2011). In those cells, the silenced status of WUS chromatin may be stably maintained by KNU through the continuous recruitment of PcG as well as histone deacetylase and through the prevention of SYD binding.
Our early work has shown that KNU, a repressor activated by AG, plays a critical role in the termination of WUS (Sun et al., 2009). Hereby in this study, we provide a precise mechanism toward understanding the nature of events in WUS repression during floral meristem termination. Although studies show that WUS may be mildly but directly repressed by AG from floral stage 3 onward through chromatin looping as an early event (Liu et al., 2011; Guo et al., 2018), our work unveils the final concrete events at floral stage 6 for WUS termination, especially the sequential epigenetic changes on WUS chromatin. Furthermore, our work not only agrees with a recent report that WUS could be initially repressed through histone deacetylation (Bollier et al., 2018) but also introduces a player, SYD, into the complex regulatory network, whose displacement from the WUS promoter is a critical initial step in repression of WUS expression. Furthermore, stable silencing of WUS is achieved by the recruitment of PcG and following deposition of H3K27me3. Thus, our study demonstrates the final details for stem cell termination with high resolution in plants during floral organ differentiation.
Continuous Activity of KNU Is Required for WUS Silencing
We noticed that KNU binds the WUS proximal promoter region (W2) for PRC2 recruitment (Figure 3; Supplemental Figures 2A and 2B), while H3K27me3 peaks on the first intron (W3) and 3′ untranslated region (W4) of WUS (Figures 4A and 4B; Supplemental Figures 4A and 4B). This suggests that recruitment of PRC2 and deposition of H3K27me3 are not always at the same location. For instance, FIE binds to the ∼1-kb upstream promoter region of the KNU transcriptional start site (Sun et al., 2014), while H3K27me3 can only be detected on KNU coding sequences (CDS; Sun et al., 2009). Another example is that the AS1-AS2 complex recruits PRC2 at promoter regions of BREVIPEDICELLUS and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2, while H3K27me3 peaks at transcribed regions or at the CDS of the two genes (Lodha et al., 2013).
For H3K27me3-mediated silencing of WUS, we noticed only continuous KNU activity (continuous DHT treatments on ap1 cal Pro35S:KNU-AR) resulted in stable repression of WUS (Figure 2C), while single induction of KNU activity (single DHT treatment on ap1 cal Pro35S:KNU-AR) led to a transient decrease of WUS mRNA due to transcriptional repression (Figure 2A). We also observed a similar transient repression in PcG mutant backgrounds (Supplemental Figures 6G and 6H). Epigenetic silencing memory may not be successfully established by transient KNU activity since H3K27me3 deposition happens later than transcriptional repression. After WUS expression is initially repressed by transient KNU activity, a compensatory mechanism for WUS recovery by a CLV3-dependent mechanism may exist (Figures 2A and 2D; Müller et al., 2006). Furthermore, we show that CLV3 is repressed by KNU within 4 h (Figure 2D). These may hint at a tight repressive mechanism to terminate stem cell activity within a narrow time window. After WUS is fully silenced in the presumptive floral meristems, WUS starts to be induced in emerging ovule and stamen primordia. How this spatial and temporal specific reactivation of WUS is regulated is an interesting topic for further study.
Conserved Mechanism for PcG Recruitment to Target Genes via C2H2 Zinc Finger Proteins
Here, we show that in Arabidopsis, the C2H2 zinc finger protein KNU recruits PcG to WUS via physical interaction with the WD motif-containing protein FIE. There is also another C2H2 zinc finger protein SUPERMAN, which directly interacts with CLF, a histone methyltransferase and key component of PRC2, and negatively regulates the expression of auxin biosynthesis genes YUCCA1/4 (Xu et al., 2018). In Drosophila early embryogenesis, the C2H2 zinc finger transcription factor Hunchback, which forms a complex with the FIE putative ortholog Esc, recruits PcG to homeotic Hox genes. This maintains the repression of Hox genes in cells outside the Hox expression domain throughout development (Kehle et al., 1998). In human cells, the C2H2 zinc finger protein Zfp277 binds to the Ink4a/Arf tumor suppressor locus and directly recruits the PRC1 factor Bmi1 for stable silencing of the Ink4a/Arf (Negishi et al., 2010). Hence, PcG recruitment to target genes via C2H2 zinc finger family proteins could be conserved not only among different plant species (Xiao et al., 2017) but also among different kingdoms (Figure 10B). In Drosophila, quiescent stem cells can be activated in adult tissues for homeostasis or repair upon tissue damage (Otsuki and Brand, 2018). After damaged tissue has regenerated, the stem cell activity should be repressed. However, the mechanism underlying the precise temporal control of stem cell activity during regeneration is largely unknown both in plants and in animals (Takemura and Nakato, 2017). It would be interesting to establish whether epigenetic-mediated multi-step termination of key stemness gene(s), as we show here in flower development, is conserved in animals, and to further assess whether C2H2 zinc finger proteins have a central role in this multi-step silencing of their target loci in both the plant and animal kingdoms.
METHODS
Plant Materials and Chemical Treatments
Except for tfl2-1, emf2-1, and the Pro35S:GFP-FIE cosuppression line (Colombia background), all plants were of the Landsberg erecta background and all were grown at 22°C under continuous light conditions. The spectrum contains equal levels of blue light (430 to 460 nm) and red light (630 to 660 nm). The light intensity was ∼100 μmol m−2 s−1 at the soil surface level. Plants were photographed using a stereomicroscope (Carl Zeiss Micro-Imaging). Scanning electron microscopy images were acquired using an electron microscope (JSM-6360LV, JEOL).
DEX (D4902, Sigma-Aldrich), DHT (A8380, Sigma-Aldrich), and CHX (01,810, Sigma-Aldrich) treatments were performed by inverting the plants and submerging the inflorescences for 1 min in a solution containing either 1 µM DEX (for Pro35S:AP1-GR) or 100 nM DHT (for Pro35S:KNU-AR) or 10 µM CHX (for Pro35S:KNU-AR) together with 0.015% (v/v) Silwet L-77. The time of initial DEX or DHT treatment was defined as day 0 or 0 h. For the continuous treatment, the inflorescences were submerged more than 2 min in the solution repeatedly at 1-d intervals.
RNA Extraction and Expression Analyses
Total RNA was isolated from inflorescences using the RNeasy Plant Mini Kit (Qiagen). RT was performed using the ThermoScript III RT-PCR system (Invitrogen). Quantitative real-time PCR assays were performed in triplicate using the 7900HT fast real-time PCR system (Applied Biosystems) and the KAPA SYBR FAST ABI Prism qPCR Kit (KAPA Biosystems). Expression assays were performed using RNAs from different batches of independently prepared plant inflorescences (biological replicates using different plants), and each was run in triplicate (technical replicates). Tip41-like (At4g34270) served as the internal reference gene (Czechowski et al., 2005).
Phylogenetic Shadowing and cis-Element Identification
Phylogenetic shadowing was performed as described previously (Yamaguchi et al., 2018). The 5′ intergenic sequence of the Arabidopsis (Arabidopsis thaliana) WUS promoter was obtained from the The Arabidopsis Information Resource website. Using the Arabidopsis WUS promoter sequence as a query, similar sequences from eight Brassicaceae were obtained through National Center for Biotechnology Information BLASTn. The resulting eight promoter sequences were aligned by mVISTA (http://genome.lbl.gov/vista/mvista/submit.shtml). Three CRMs were identified and defined as CRM1, CRM2, and CRM3. Because KNU bound to the CRM3 region, CRM3 was characterized in detail by ClustalW (https://www.genome.jp/tools-bin/clustalw) and a conserved and putative aatc KNU cis-element was identified.
In Situ Hybridization
Nonradioactive in situ hybridizations were performed as described previously (Carles et al., 2005). To produce a WUS-specific antisense probe, a pMHwus16 clone carrying WUS cDNA was used as a template for in vitro transcription.
GUS Staining
GUS staining was performed as described previously (Ito et al., 2003) and observed on paraffin sections or plastic sections (Yamaguchi et al., 2018). Photographs were taken using a stereomicroscope (Carl Zeiss Micro-Imaging).
Vector Construction and Plant Transformation
ProFIE:FIE-myc was prepared as follows. First, the pENTR-D (Invitrogen)–based ProFIE:FIE-VENUS-myc (Sun et al., 2014) was digested with SfoI to release the VENUS fragment. Subsequently, the digested vector was self-ligated to generate ProFIE:FIE-myc. Finally, the pENTR-D–based ProFIE:FIE-myc was recombined into pKGW (Invitrogen) using LR-recombinase (Invitrogen). ProFIE:FIE-myc was introduced into ap1 cal Pro35S:AP1-GR ProKNU:KNU-VENUS. The transformation was performed using the Agrobacterium tumefaciens-mediated floral dipping method (Clough and Bent, 1998).
ChIP Assays
ChIP experiments were performed as described previously (Ito et al., 1997), with slight modifications. Inflorescences were ground in liquid nitrogen and postfixed with 1% (w/v) formaldehyde for 10 min. Chromatin was isolated and solubilized by sonication to generate DNA fragments with an average length of 400 bp. After incubation with salmon sperm DNA/protein-A agarose beads (Millipore), the solubilized chromatin was incubated overnight with anti-H3K27me3 antibody (07-449, Millipore), anti-H3K4me3 antibody (07-473, Millipore), or anti–acetyl-H3 antibody (06-599, Millipore; 1:200 dilution used for all histone modification ChIP experiments), or the solution containing sonicated DNA fragments was incubated overnight with anti–c-Myc-Agarose beads (A7470; Sigma-Aldrich; 1:20 dilution used) or anti–hemagglutinin (HA)-Agarose beads as a control (A2095, Sigma-Aldrich; 1:20 dilution used, for KNU ChIP). The relative enrichment for KNU on the WUS locus was taken as the ratio between Myc and HA. For Pol II ChIP Pol II antibody (ab817, Abcam; 1:200 dilution used) was used, and for FIE ChIP and EMF2 ChIP GFP-Trap beads (gta-20, Chromotek; 1:20 dilution used) were used. DNA fragments were recovered from the purified DNA–protein complexes and then used for enrichment tests by real-time PCR analysis in triplicates. The primary ratio between the bound DNA after IP and the input DNA before IP was calculated for all the representative primer sets spanning the WUS genomic region, and the ratios were plotted to show the relative changes in the levels of the H3K27me3 mark, H3K4me3 mark, and H3 acetylation. For SYD ChIP, ChIP was performed as described previously (Wu et al., 2015), with minor modifications. The SYD antibody was used as in a previous publication (Kwon et al., 2005). For the SYD ChIP result in the supplemental data (Supplemental Figures 5E and 5F), due to a lack of SYD antibody, we generated a tagging line of knu-2/+ ap1 cal Pro35S:AP1-GR ProSYD:SYD-GFP and used GFP-Trap beads (gta-20, Chromotek; 1:20 dilution used). ChIP sample was normalized to the related input sample. The percent input data were converted to enrichment rate and plotted to facilitate comparison of time-course occupancy levels. For all ChIP experiments, Mu-like transposons (Supplemental Table) served as a negative control locus and the relative enrichment rate was set to 1. ChIP assays were performed using inflorescences from different batches of independently prepared plant materials (biological replicates with using different plants), each run in triplicate (technical replicates).
Formaldehyde-Assisted Isolation of Regulatory Elements
FAIRE was performed as described previously (Wu et al., 2015), with minor modifications. Inflorescence tissues (0.5 g) were harvested after 1-min treatment in a solution containing 100 nM DHT at 22°C. Tissues were crosslinked with 1% (w/v) formaldehyde under vacuum for 10 min, replaced by 125 mM Gly in buffer 1 (Omidbakhshfard et al., 2014) for 5 min at room temperature, and rinsed with ice-cold water three times. Chromatin was isolated by grinding in buffer 1, filtered through four layers of miracloth, washed with buffers (Omidbakhshfard et al., 2014), and sonicated to produce DNA fragment shorter than 500 bp. One volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to extract the chromatin, and samples were mixed for 2 min by vortex. After ethanol precipitation, DNA was purified again using a QIAquick DNA Purification Kit (Qiagen). DNA was determined for crosslinked and noncrosslinked FAIRE samples with a Light Cycler 480 (Roche) and Light Cycler 480 release 1.5.1.62 SP software (Roche) using FastStart DNA essential DNA Green Master (Roche). The Ta3 retrotransposon (At1g37110, Supplemental Table; Johnson et al., 2002) was used as the negative control locus for FAIRE experiments. FAIRE experiments were repeated twice, and the combined data are shown.
Yeast One-Hybrid Assays
Yeast one-hybrid assays were performed as described previously (Sparkes et al., 2006). WUS-W2 (−308 to −125 bp upstream of the ATG start codon) fragment was cloned into the pAbAi vector digested with SmaI and XhoI, creating WUS-W2-AbAi. WUS-W2-AbAi was linearized by digestion with BstBI prior to transformation of the yeast strain Y1H Gold. The full-length cDNA of KNU was isolated and cloned into the pGADT7 activation domain (AD) vector, creating the pAD-KNU plasmid. The pAD-KNU vector was subsequently transformed into the yeast strain containing the WUS-W2-AbAi constructs. Activation of the yeast was observed after 3 d on selection plates (synthetic dextrose/−Ura) containing 200 ng mL−1 aureobasidin A.
Yeast Two-Hybrid Assay
To construct the vectors for yeast two-hybrid assays, the full-length and truncated versions of KNU as well as the WD repeat domain of FIE were amplified and cloned into pGADT7 or pGBKT7 (Clontech). The yeast two-hybrid assay was performed using the Yeastmaker Yeast Transformation System 2 (Clontech) according to the manufacturer’s instructions.
BiFC Assay
The full-length coding sequences of KNU and FIE were cloned into pGreen vectors and transformed into Agrobacterium. The Agrobacteria were co-infiltrated into tobacco (Nicotiana benthamiana) leaves of 3-week-old plants as described previously (Sparkes et al., 2006).
Co-IP Experiment
The flower buds at stage 6 from ap1 cal Pro35S:AP1-GR and ap1 cal Pro35S:AP1-GR ProKNU:KNU-VENUS ProFIE:FIE-myc were ground with a mortar and pestle in liquid nitrogen, and proteins were prepared in IP buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% (v/v) Nonidet P-40, 5% (v/v) glycerol, 1 mM dithiobis, and 1× Complete Protease Inhibitor Cocktail) and then incubated with anti–c-Myc agarose beads (Sigma-Aldrich) for 4 h at 4°C on a rotator. After incubation, the beads were washed four times with ice-cold Dulbecco’s PBS buffer and then eluted by boiling in SDS sample buffer. Samples were separated on 10% (w/v) SDS-PAGE gels, transferred to a nitrocellulose membrane, and then probed with anti-GFP antibody (Cell Signaling Technology). Detection was according to the procedures in the Clarity Western ECL Substrate Kit (Bio-Rad). Images showing immunoblot results were taken using the Chemi DOC XRS+ Imaging System (Bio-Rad). Co-IP experiments were repeated twice, and representative data are shown.
Confocal Microscopy Imaging
To observe the reporter lines in Arabidopsis inflorescences, the transgenic seeds were sown in soil and the inflorescences were plucked and mounted on slides. The older floral buds were then carefully removed or spaced out to expose the SAM and early-stage floral buds. Dissected inflorescences were incubated with FM4-64 dye (50 µg/mL) for 45 min on slides. Plants were imaged with a Zeiss LSM 510 upright (with motorized stage) confocal microscope using an EC Plan-Neofluar 40×/1.30 Oil differential interference contrast or a Plan-Apochromat 20×/0.8 objective lens. GFP was stimulated with an argon laser at 488 nm at 60 to 70% of its output with emission filtered with a 505 to 530 nm band-pass filter. VENUS was stimulated with an argon laser at 514 nm at 65 to 80% of its output with emission filtered with a 530 to 600 nm band-pass filter. FM4-64 dye emission was filtered with a 585 nm long-pass filter. The z-stack was acquired using a 512 × 512 pixel frame, and the three-dimensional projections of the obtained z-stacks were generated with Zeiss LSM Image Brower version 4 and adjusted with Adobe Photoshop.
Electrophoretic Mobility Shift Assay
The DNA fragment of KNU CDS was inserted into the pMAL-c5G vector digested with NdeI and BamHI, which was expressed in Transetta (DE3) Chemically Competent Cells (TransGen Biotech) to produce MBP-tagged KNU protein. The recombinant fusion protein was purified using Amylose Resin (NEB) following the manufacturer’s instructions. The probes were labeled with biotin and annealed before use. The EMSA kit (Thermo Fisher Scientific) was used for binding reactions and detection of biotin-labeled probe. The nonlabeled probes were used as competitors at 50 times the concentration of labeled probes. For individual probes, the detailed length information is provided in the Figure 3A legend.
Statistical Analyses
Student’s t tests were performed using SPSS version 21 software to determine significance, whenever groups were compared, and are described in the corresponding figures (Figures 2B, 4B to 4G, and 6B to 6G; Supplemental Figures 2B, 4B, and 5C to 5J). Statistical significance was suggested at the indicated P-values. Detailed results of statistical analyses are available in Supplemental File.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: KNU (At5g14010), WUS (At2g17950), AP1 (At1g69120), CAL (At1g26310), SYD (At2g28290), FIE (At3g20740), TFL2 (At5g17690), CLF (At2g23380), CLV1 (At1g75820), CLV2 (At1g65380), CLV3(At1g69970), EMF2 (At5g51230), ZFP11 (At2g42410).
Supplemental Data
Supplemental Figure 1. Confocal observation of the doubly transgenic inflorescence for ProWUS:GFP-ER (red) and ProKNU:KNU-VENUS (green) in stage 6 flower buds, together with KNU and WUS expression patterns in stage 7 flower buds.
Supplemental Figure 2. ChIP assay for KNU binding on WUS.
Supplemental Figure 3. KNU binds conserved core sequence aatc on WUS promoter.
Supplemental Figure 4. H3K27me3 analysis of WUS chromatin upon KNU activation.
Supplemental Figure 5. Change of WUS chromatin status is KNU-dependent.
Supplemental Figure 6. Genetic interaction between KNU with PcG factors, WUS expression in PcG mutants, and KNU binding on WUS in clf-28 mutant background.
Supplemental Figure 7. KNU physically interacts with FIE in yeast and co-immunoprecipitation of KNU and FIE.
Supplemental Table. List of primer sequences used in this study; primer names are followed by their sequences (5′ to 3′).
Supplemental File. Statistical analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yu Hao for providing some constructs for the yeast two-hybrid assay, Haruka Sawada for help with the GUS staining sections, T. Laux for the ProWUS:GUS seeds, and N. Ohad for the Pro35S:GFP-FIE seeds. This work was supported by the Fundamental Research Funds for the Central Universities and National Natural Science Foundation of China (grant 31670308 to B.S.) and was also supported by grants from Temasek Life Sciences Laboratory, the National Research Foundation Singapore (Competitive Research Programme Award NRFCRP001-108), the NAIST Foundation, the Grant-in-Aid for Scientific Research A (grant 15H02405), the Grant-in-Aid for Scientific Research on Innovative Areas (grants 15H01234, 15H01356, 17H05843, and 18H04839), and the Grant-in-Aid for Challenging Exploratory Research (grants 15K14549 and 18K19342 to T.I.).
AUTHOR CONTRIBUTIONS
B.S. and T.I. designed research; B.S., Y.Z., J.C., E.S., N.Y., J.X., L.-S.L., W.-Y.W., and X.G. performed research; D.W. revised the article; and T.I. and B.S. wrote the article.
References
- Alvarez-Buylla E.R., García-Ponce B., Garay-Arroyo A. (2006). Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated Arabidopsis thaliana floral MADS-box genes. J. Exp. Bot. 57: 3099–3107. [DOI] [PubMed] [Google Scholar]
- Bar-Ziv R., Voichek Y., Barkai N. (2016). Chromatin dynamics during DNA replication. Genome Res. 26: 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I., Laux T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N., Dubreucq B., Roudier F., Dubos C., Lepiniec L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23: 4065–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani S., Winter C., Hershman S., Wagner J.D., Kennedy J.F., Kwon C.S., Pfluger J., Su Y., Wagner D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollier N., Sicard A., Leblond J., Latrasse D., Gonzalez N., Gévaudant F., Benhamed M., Raynaud C., Lenhard M., Chevalier C., Hernould M., Delmas F. (2018). At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30: 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas D.M., Robertson M., Finnegan E.J., Helliwell C.A. (2011). Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant J. 65: 872–881. [DOI] [PubMed] [Google Scholar]
- Cairns B.R. (2005). Chromatin remodeling complexes: Strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15: 185–190. [DOI] [PubMed] [Google Scholar]
- Carles C.C., Choffnes-Inada D., Reville K., Lertpiriyapong K., Fletcher J.C. (2005). ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132: 897–911. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Buzas D.M., Ying H., Robertson M., Taylor J., Peacock W.J., Dennis E.S., Helliwell C. (2013). Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics 14: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyhle F., Sarkar A.K., Tucker E.J., Laux T. (2007). WUSCHEL regulates cell differentiation during anther development. Dev. Biol. 302: 154–159. [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Amasino R.M. (2009). A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151: 1688–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548. [DOI] [PubMed] [Google Scholar]
- Guitton A.E., Page D.R., Chambrier P., Lionnet C., Faure J.E., Grossniklaus U., Berger F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981. [DOI] [PubMed] [Google Scholar]
- Guo L., Cao X., Liu Y., Li J., Li Y., Li D., Zhang K., Gao C., Dong A., Liu X. (2018). A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 94: 1083–1097. [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Ito T., Takahashi N., Shimura Y., Okada K. (1997). A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 38: 248–258. [DOI] [PubMed] [Google Scholar]
- Ito T., Sakai H., Meyerowitz E.M. (2003). Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr. Biol. 13: 1524–1530. [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao X., Jacobsen S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367. [DOI] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J.A., Bienz M., Müller J. (1998). dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897–1900. [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Chen C., Wagner D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19: 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A.S., Landberg K., Meeks-Wagner D.R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jürgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814. [DOI] [PubMed] [Google Scholar]
- Liu X., Kim Y.J., Müller R., Yumul R.E., Liu C., Pan Y., Cao X., Goodrich J., Chen X. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M., Marco C.F., Timmermans M.C. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803. [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- McNally J.G., Müller W.G., Walker D., Wolford R., Hager G.L. (2000). The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 287: 1262–1265. [DOI] [PubMed] [Google Scholar]
- Müller R., Borghi L., Kwiatkowska D., Laufs P., Simon R. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich A.K., Walker D.A., Wolford R., Hager G.L. (2004). Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14: 163–174. [DOI] [PubMed] [Google Scholar]
- Negishi M., Saraya A., Mochizuki S., Helin K., Koseki H., Iwama A. (2010). A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One 5: e12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Shichrur K., Yalovsky S. (2007). The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol. 145: 1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Winck F.V., Arvidsson S., Riaño-Pachón D.M., Mueller-Roeber B. (2014). A step-by-step protocol for formaldehyde-assisted isolation of regulatory elements from Arabidopsis thaliana. J. Integr. Plant Biol. 56: 527–538. [DOI] [PubMed] [Google Scholar]
- Otsuki L., Brand A.H. (2018). Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360: 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T., Johnson S.D., Koltunow A.M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749. [DOI] [PubMed] [Google Scholar]
- Qüesta J.I., Song J., Geraldo N., An H., Dean C. (2016). Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353: 485–488. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Sawarkar R., Paro R. (2010). Interpretation of developmental signaling at chromatin: The Polycomb perspective. Dev. Cell 19: 651–661. [DOI] [PubMed] [Google Scholar]
- Simon J.M., Giresi P.G., Davis I.J., Lieb J.D. (2012). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sun B., Xu Y., Ng K.H., Ito T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23: 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Looi L.S., Guo S., He Z., Gan E.S., Huang J., Xu Y., Wee W.Y., Ito T. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- Sung Z.R., Chen L., Moon Y.H., Lertpiriyapong K. (2003). Mechanisms of floral repression in Arabidopsis. Curr. Opin. Plant Biol. 6: 29–35. [DOI] [PubMed] [Google Scholar]
- Takemura M., Nakato H. (2017). Drosophila Sulf1 is required for the termination of intestinal stem cell division during regeneration. J. Cell Sci. 130: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F., Alves-Ferreira M., Dubois A., Riechmann J.L., Meyerowitz E.M. (2006). Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Sang Y., Bezhani S., Yamaguchi N., Han S.K., Li Z., Su Y., Slewinski T.L., Wagner D. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 109: 3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Yamaguchi N., Xiao J., Bargmann B., Estelle M., Sang Y., Wagner D. (2015). Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49: 1546–1552. [DOI] [PubMed] [Google Scholar]
- Xu Y., et al. (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37: e97499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Huang J., Tatsumi Y., Abe M., Sugano S.S., Kojima M., Takebayashi Y., Kiba T., Yokoyama R., Nishitani K., Sakakibara H., Ito T. (2018). Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 9: 5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Yanai Y., Chen L., Kato Y., Hiratsuka J., Miwa T., Sung Z.R., Takahashi S. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Luo X., Li Z., Yang W., Wang Y., Liu R., Du J., He Y. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48: 1527–1534. [DOI] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]