Abstract
Axon regeneration is a conserved mechanism induced by axon injury that initiates a neuronal response leading to regrowth of the axon. In Caenorhabditis elegans, the initiation of axon regeneration is regulated by the JNK MAP kinase (MAPK) pathway. We have previously identified a number of genes affecting the JNK pathway using an RNAi-based screen. Analysis of these genes, called the svh genes, has shed new light on the regulation of axon regeneration, revealing the involvement of a signaling cascade consisting of a growth factor SVH-1 and its receptor, the tyrosine kinase SVH-2. Here, we characterize the svh-6/tns-1 gene, which is a homolog of mammalian tensin, and show that it is a positive regulator of axon regeneration in motor neurons. We demonstrate that TNS-1 interacts with tyrosine-autophosphorylated SVH-2 and the integrin β subunit PAT-3 via its SH2 and PTB domains, respectively, to promote axon regeneration. These results suggest that TNS-1 acts as an adaptor to link the SVH-2 and integrin signaling pathways.
SIGNIFICANCE STATEMENT The Caenorhabditis elegans JNK MAPK pathway regulates the initiation of axon regeneration. Previously, we showed that a signaling cascade consisting of the HGF-like growth factor SVH-1 and its Met-like receptor tyrosine kinase SVH-2 promotes axon regeneration through activation of the JNK pathway. In this study, we show that the C. elegans tensin, TNS-1, is required for efficient regeneration after axon injury. Phosphorylation of SVH-2 on tyrosine mediates its interaction with the SH2 domain of TNS-1 to positively regulate axon regeneration. Furthermore, TNS-1 interacts via its PTB domain with the integrin β subunit PAT-3. These results suggest that TNS-1 plays a critical role in the regulation of axon regeneration by linking the SVH-2 and integrin signaling pathways.
Keywords: axon regeneration, c-Met, C. elegans, integrin, JNK, tensin
Introduction
Axon regeneration is necessary to restore the nervous system following axon injury, and its success is governed by an interaction between the local extracellular environment and the neuron's intrinsic growth capacity (Rossi et al., 2007). Most invertebrate neurons and mammalian peripheral neurons are able to regenerate, whereas neurons of the mammalian CNS have a limited ability (Case and Tessier-Lavigne, 2005). Successful regeneration depends primarily on neuron-intrinsic regeneration signals, thus these signaling processes are potential targets for regeneration therapies. However, our understanding of these intrinsic signaling pathways is incomplete.
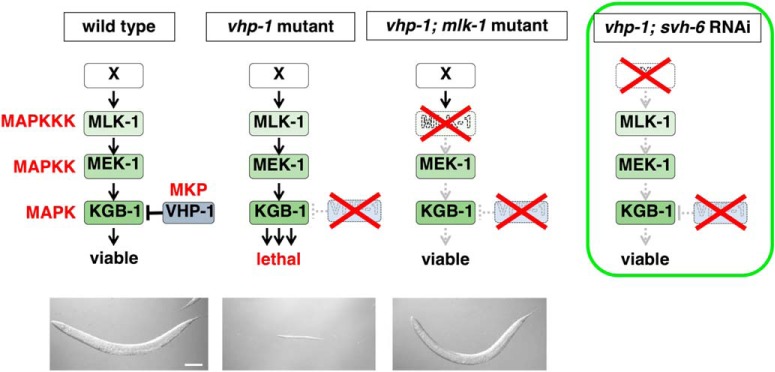
Caenorhabditis elegans is a powerful model system to investigate the mechanisms of axon regeneration following injury (Yanik et al., 2004; Chisholm et al., 2016). Genetic studies in C. elegans have identified a large number of pathways specifically involved in adult axon regeneration (Chen et al., 2011; Nix et al., 2014). It has recently been shown that the C. elegans JNK MAP kinase (MAPK) pathway, consisting of MLK-1 MAPKKK, MEK-1 MAPKK, and KGB-1 JNK, plays a critical role in the initiation of axon regeneration (Nix et al., 2011). The JNK pathway is negatively regulated by VHP-1, a member of the MAPK phosphatase (MKP) family (Camps et al., 2000), which dephosphorylates and thereby inactivates KGB-1 (Mizuno et al., 2004). The vhp-1-null mutation causes developmental arrest at an early larval stage because of hyperactivation of the JNK pathway, and this phenotype is suppressed by mutations in mlk-1, mek-1 or kgb-1 (Fig. 1; Mizuno et al., 2004). We previously identified a number of additional genes functioning in the JNK pathway via a genome-wide RNAi screen for suppressors of vhp-1 lethality (svh genes; Fig. 1; Li et al., 2012; Hisamoto et al., 2018). The svh-1 gene encodes a growth factor-like protein homologous to mammalian HGF. svh-2 encodes a homolog of the mammalian Met, a receptor for HGF. SVH-2 is a receptor tyrosine kinase (RTK) that activates the JNK pathway via tyrosine phosphorylation of the MAPKKK MLK-1 (Li et al., 2012). Thus, the SVH-1–SVH-2 signaling cascade promotes axon regeneration through activation of the JNK pathway (Fig. 2A). The svh-4 gene is identical to ddr-2 and encodes a RTK homologous to the mammalian discoidin domain receptor (DDR), which is activated by collagen (Hisamoto et al., 2016). SVH-4/DDR-2 also modulates the SVH-1–SVH-2–JNK pathway (Fig. 2A). These results suggest that two different RTK networks coordinately regulate axonal regeneration in C. elegans.
Figure 1.
Isolation of svh-6 RNAi. The vhp-1 mutation causes larval arrest because of hyperactivation of the KGB-1 JNK cascade. Downregulation by RNAi of any of the components in the JNK pathway such as a factor that positively regulates the JNK signaling is able to suppress the vhp-1 lethality. Photographs of worms are from previous results (Mizuno et al., 2004). Scale bar, 100 μm.
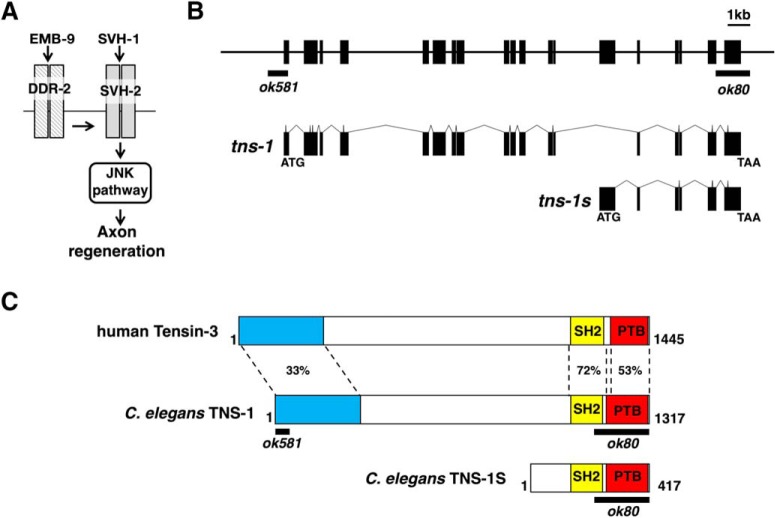
Figure 2.
The tns-1 gene encodes a homolog of Tensin. A, SVH-1–SVH-2 pathway required for axon regeneration in C. elegans. Activation of SVH-2 receptor tyrosine kinase by SVH-1 growth factor activates the JNK pathway. The EMB-9 collagen–DDR-2 pathway modulates the SVH-1–SVH-2 pathway. B, Structures of the tns-1 and tns-1s genes. Boxes indicate exons and lines indicate introns and promoters. The regions deleted in the ok80 and ok581 alleles are indicated by black bars. C, Structure of TNS-1 protein. Schematic diagrams of TNS-1, a short form of TNS-1 (TNS-1S) and human Tensin-3 are shown. The domains shown are the NH2-terminal (blue), SH2 (yellow), and PTB (red) domains. The regions deleted by the ok80 and ok581 alleles are indicated by black bars. Identity (%) in each domain is shown.
The svh-6/tns-1 gene encodes a homolog of mammalian tensin. In this study, we show that TNS-1 regulates axon regeneration by interacting with SVH-2 in a manner dependent on tyrosine-autophosphorylation of SVH-2. This interaction occurs via the TNS-1 SH2 domain, which is essential for axon regeneration. Furthermore, TNS-1 interacts via its PTB domain with the integrin β subunit PAT-3 to positively regulate axon regeneration. These results suggest that TNS-1 plays a positive role in axon regeneration by linking the SVH-2 and integrin signaling pathways.
Materials and Methods
C. elegans strains.
The C. elegans strains used in this study are listed in Table 1. The pat-3(gk804163) mutant was made by outcrossing VC40772 strain (generated by the million mutation project: Thomson et al., 2013) twice with the juIs76 strain and the mutation was verified by DNA sequencing. All strains were maintained on nematode growth medium (NGM) plates and fed with bacteria of the OP50 strain, as described previously (Brenner, 1974). For heat-shock treatment, worms on the NGM dishes were incubated at 37°C for 30 min and then incubated at 20°C for 4 h.
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| KU501 | juIs76 II |
| KU503 | juIs76 II; svh-2(tm737) X |
| KU504 | juIs76 II; mlk-1(km19) V |
| KU1240 | tns-1(ok581) I; juIs76 II |
| KU1241 | tns-1(ok80) I; juIs76 II |
| KU1242 | tns-1(ok80) I; juIs76 II; mlk-1(km19) V |
| KU1243 | tns-1(ok80) I; juIs76 II; kmEx1243 [Punc-25::tns-1] |
| KU1244 | tns-1(ok80) I; juIs76 II; kmEx1244 [Punc-25::tns-1ΔN] |
| KU1245 | tns-1(ok80) I; juIs76 II; kmEx1245 [Punc-25::tns-1(R1071K)] |
| KU1246 | tns-1(ok80) I; juIs76 II; kmEx1246 [Punc-25::tns-1ΔPTB] |
| KU1247 | juIs76 II; mlk-1(km19) V; kmEx1243 [Punc-25::tns-1] |
| KU1248 | tns-1(ok80) I; juIs76 II; kmEx507 [Pmlk-1::mlk-1] |
| KU1249 | tns-1(ok80) I; juIs76 II; kmEx1206 [Punc-25::svh-2] |
| KU1250 | tns-1(ok80) I; juIs76 II; kmEx1202 [Punc-25::ddr-2] |
| KU1251 | wpIs36 I; kmEx1251 [Ptns-1::nls::venus] |
| KU1252 | tns-1(ok80) I; juIs76 II; kmEx1252 [Pmec-7::tns-1] |
| KU1253 | tns-1(ok80) I; juIs76 II; kmEx1253 [Punc-25::tns-1s] (line 1) |
| KU1254 | juIs76 II; kmEx1243 [Punc-25::tns-1] |
| KU1256 | tns-1(ok80) I; juIs76 II; kmEx1256 [Punc-25::hTensin-3] (line 1) |
| KU1257 | tns-1(ok80) I; juIs76 II; svh-2(tm737) X |
| KU1258 | juIs76 II; svh-2(tm737) X; kmEx1243 [Punc-25::tns-1] |
| KU1259 | tns-1(ok80) I; juIs76 II; kmEx468 [Punc-25::max-2] |
| KU1260 | kmEx1260 [Phsp::svh-2::gfp] |
| KU1261 | tns-1(ok80) I; kmEx1260 [Phsp::svh-2::gfp] |
| KU1262 | wpIs36 I; kmEx1262 [Punc-25::tns-1::gfp] |
| KU1263 | tns-1(ok581) I; juIs76 II; ddr-2(ok574) X |
| KU1264 | tns-1(ok581) I; juIs76 max-2(nv162) II |
| KU1265 | juIs76 II; pat-3(gk804163) III |
| KU1266 | juIs76 II; pat-3(gk804163) III; kmEx1253 [Punc-25::pat-3] |
| KU1267 | tns-1(ok80) I; juIs76 II; kmEx1253 [Punc-25::tns-1s] (line 2) |
| KU1268 | tns-1(ok80) I; juIs76 II; kmEx1256 [Punc-25::hTensin-3] (line 2) |
| KU1269 | juIs76 II; maEx247 [Ppat-3::mcherry::H2B::pat-3 3′UTR + Ppat-3::GFP::H2B::pat-3 mutated 3′UTR] |
Isolation of tns-1 and tns-1s cDNAs.
The tns-1 cDNA was amplified by PCR from the pACT C. elegans cDNA library (Sakamoto et al., 2005) using oligonucleotides no. 1: 5′-ACTAGTAATGAAGGATCGAAAAGAAGGTGTACAGGTG-3′ and No.2: 5′-CTCGAGCTTCTTATGACCGATTCTTCTGTGCC-3′. The tns-1s cDNA was also isolated by using oligonucleotides no. 2 and no. 3: 5′-CCTCTACGTCCTCTTTGATTCCATCTTGTATTCCAGATATTTTGT-3′. Both cDNAs were cloned into pCR2.1 TOPO vector (Invitrogen) and verified by sequencing.
Plasmids.
The Ptns-1::nls::venus plasmid was made by amplifying ∼1.5 kb of the tns-1 promoter from the N2 genomic DNA by PCR and inserting it into the pPDnlsVenus vector (Li et al., 2012). Punc-25::tns-1 and Pmec-7::tns-1 were generated by inserting the tns-1 cDNA into the pSC325 and pPD52.102 vectors, respectively. Punc-25::tns-1s was generated by inserting the tns-1s cDNA into the pSC325 vector. Punc-25::tns-1ΔN, Punc-25::tns-1(R1071K), and Punc-25::tns-1ΔPTB were generated by oligonucleotide-directed PCR using Punc-25::tns-1 as a template and were verified by DNA sequencing. Punc-25:: hTensin-3 was generated by inserting the human Tensin-3 cDNA (item #105299, Addgene) into the pSC325 vector. Punc-25::pat-3 was generated by inserting the pat-3 cDNA, which was amplified from the pACT C.elegans cDNA library (Sakamoto et al., 2005) by PCR, into the pSC325 vector, which was verified by DNA sequencing. To construct the Phsp::svh-2::gfp plasmid, a modified svh-2 cDNA that deletes the termination codon was generated by PCR and inserted into the pPD49.78 vector together with the gfp gene from the pPD95.75 vector. Punc-25::tns-1::gfp was generated by inserting a modified tns-1 cDNA lacking the termination codon into the pSC325 vector together with the gfp gene. The T7-TNS-1, T7-TNS-1ΔN, T7-TNS-1ΔN(R1071K), and T7-TNS-1ΔN(ΔPTB) plasmids were constructed by inserting the tns-1, tns-1ΔN, tns-1ΔN(R1071K), and tns-1ΔN(ΔPTB) cDNAs into the pCMVT7 vector, respectively (Kawasaki et al., 1999). To construct GFP-PAT-3-ICD, a 150 bp DNA fragment encoding the COOH-terminal 49 aa of PAT-3 (corresponding to 761–809 aa) was synthesized and subcloned into the pEGFP-C1 vector (Clontech). The pFLAG-Tpr-SVH-2C, pFLAG-Tpr-SVH-2C(Y890F), pFLAG-Tpr-DDR-2C, Pmyo-2::DsRed monomer, and Punc-25::max-2 plasmids were described previously (Li et al., 2012; Hisamoto et al., 2016; Pastuhov et al., 2016).
Transgenic animals.
Transgenic animals were generated according to a basic injection method (Mello et al., 1991). The Punc-25::tns-1 (25 ng/μl), Punc-25::tns-1ΔN (25 ng/μl), Punc-25::tns-1(R1071K) (25 ng/μl), Punc-25::tns-1ΔPTB (25 ng/μl), Ptns-1::nls::venus (25 ng/μl), Pmec-7::tns-1 (25 ng/μl), Punc-25::tns-1s (25 ng/μl), Punc-25::hTensin-3 (25 ng/μl), Punc-25::pat-3 (50 ng/μl), Phsp::svh-2::gfp (45 ng/μl), Punc-25::tns-1::gfp (50 ng/μl), and Pmyo-2::DsRed monomer (25 ng/μl) plasmids were used in kmEx1243 (Punc-25::tns-1 + Pmyo-2::DsRed monomer), kmEx1244 (Punc-25::tns-1ΔN + Pmyo-2::DsRed monomer), kmEx1245 (Punc-25::tns-1(R1071K) + Pmyo-2::DsRed monomer), kmEx1246(Punc-25::tns-1ΔPTB + Pmyo-2::DsRed monomer), kmEx1251 (Ptns-1::nls::venus + Pmyo-2::DsRed monomer), kmEx1252 (Pmec-7::tns-1 + Pmyo-2::DsRed monomer), kmEx1253/kmEx1267 (Punc-25::tns-1s + Pmyo-2::DsRed monomer), kmEx1256/kmEx1268 (Punc-25::hTensin-3 + Pmyo-2::DsRed monomer), kmEx1266 (Punc-25::pat-3 + Pmyo-2::DsRed monomer), kmEx1262 (Punc-25::tns-1::gfp + Pmyo-2::DsRed monomer), and kmEx1260 (Phsp::svh-2::gfp + Pmyo-2::DsRed monomer), respectively. The wpIs36 (Punc-47::mcherry), maEx247 (Ppat-3::mcherry::H2B::pat-3 3′UTR + Ppat-3::gfp::H2B::pat-3 mutated 3′UTR), kmEx468 (Punc-25::max-2), kmEx507 (Pmlk-1::mlk-1), kmEx1206 (Punc-25::svh-2), and kmEx1202 (Punc-25::ddr-2) transgenes were described previously (Li et al., 2012; Burke et al., 2015; Hisamoto et al., 2016; Pastuhov et al., 2016; Shimizu et al., 2018).
Microscopy and laser ablation.
Laser microsurgery for determining axon regeneration was performed as described previously (Li et al., 2012). Standard fluorescent images of transgenic hermaphrodites were observed under a 60× or 100× objective of a Nikon Eclipse E800 fluorescent microscope and photographed with a Hamamatsu ORCA 3CCD or a Zyla-5.5 CCD camera. To quantify axon regrowth, the middle region of the lateral axon of each D-type motor neuron in hermaphrodite was cut by laser. Then the length of each severed axon from the base in the ventral nerve cord to the severed growing end was measured using the ImageJ program (NIH). An Olympus FV-500 and a Zeiss LSM 800 confocal laser microscope system were used to take confocal fluorescent images.
Biochemical experiments using worm extracts.
To extract proteins from worms, the worms collected from NGM dishes were suspended into RIPA buffer [50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 5 mm PMSF, phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich), and protease inhibitor cocktail (Sigma-Aldrich)] and sonicated by Bioraptor UCW-201 (Cosmo Bio) at 4°C. After sonication, the samples were centrifuged at 15,000 × g for 5 min. A small aliquot of the supernatant was boiled with SDS sample buffer and used as a whole lysate. Each remaining supernatant (400 μl) was transferred into a new tube, to which was added 10 μl (bed volume) of Dynabeads Protein G (Invitrogen) coated with anti-GFP (598, rabbit polyclonal, MBL) antibody, and the tube was rotated for 20 min at room temperature. The beads were then washed three times with PBS. The samples were boiled with SDS sample buffer and subjected to immunoblotting as described previously (Li et al., 2012; Hisamoto et al., 2018).
Biochemical experiments using mammalian cells.
For immunoprecipitation, transfected COS-7 cells were lysed in RIPA buffer [50 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulphonyl fluoride, phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich), and protease inhibitor cocktail (Sigma-Aldrich)], followed by centrifugation at 15,000 × g for 12 min. The supernatant was added to 10 μl (bed volume) of Dynabeads protein G (Invitrogen) with anti-T7 (PM022, MBL) and anti-Flag (M2, Sigma-Aldrich) antibodies and rotated for 2 h at 4°C. The beads were then washed three times with ice-cold PBS and subjected to immunoblotting using anti-T7 (T7-Tag, Merck), anti-Flag (PM020, MBL), and anti-GFP (JL-8, Clontech) antibodies.
Statistical analysis.
Statistical analyses were performed as described previously (Pastuhov et al., 2012). In brief, confidence intervals (95%) were calculated by the modified Wald method (https://www.graphpad.com/quickcalcs/confInterval1/) and two-tailed p values were calculated using Fisher's exact test (http://www.graphpad.com/quickcalcs/contingency1). Unpaired t test was executed by using t test calculator (http://www.graphpad.com/quickcalcs/ttest1).
Homology search, phylogenetic analysis, identification of domains, and alignments of amino acids.
Homology search, identification of conserved domains and alignments of amino acids were executed by the NCBI BLAST, NCBI CD-search, and Genetyx-Mac programs, respectively.
Results
SVH-6/TNS-1 is required for efficient axon regeneration
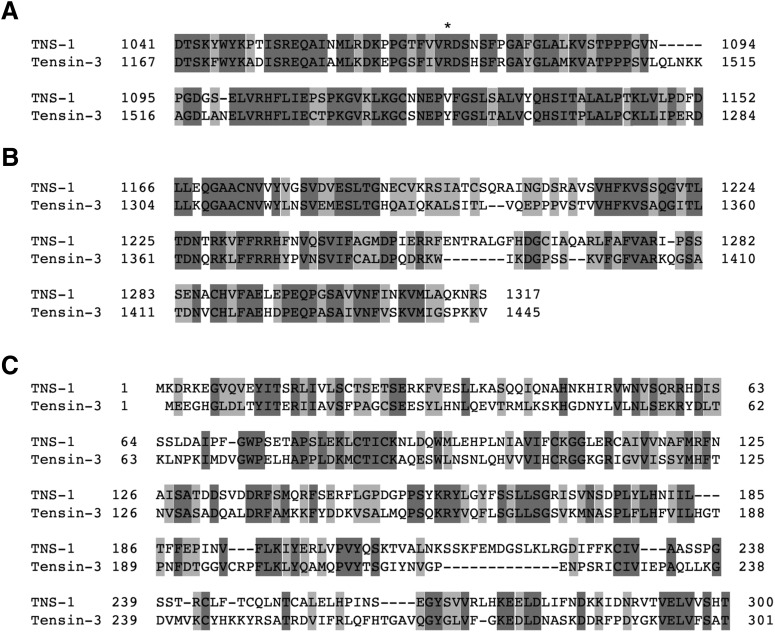
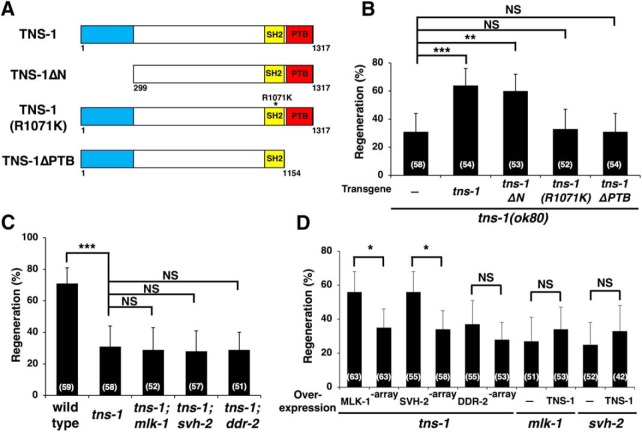
One of the svh genes identified in our previous RNAi screen using vhp-1 mutant animals (Li et al., 2012) was svh-6/tns-1 (hitherto tns-1; Fig. 1), the product of which belongs to the Tensin family (Fig. 2B,C; Lo, 2004). In mammals, there are four members in this family: Tensin-1, Tensin-2, Tensin-3, and Tensin-4 [also called COOH-terminal Tensin-like protein (Cten); Lo, 2004]. TNS-1 contains a Src homology (SH) 2 and a phosphotyrosine-binding (PTB) domain in its COOH-terminal region, which display strong homologies to mammalian Tensin-3 (Figs. 2C, 3A,B). This tandem SH2–PTB domain is unique to the Tensin family (Lo, 2004, 2017). In addition, TNS-1 and Tensin-3 show a weak homology in their NH2-terminal regions (Figs. 2C, 3C). Analysis of C. elegans cDNAs revealed the existence of another shorter transcript, designated tns-1s that has a distinct NH2-terminal domain (Fig. 2B,C).
Figure 3.
Conserved domains between TNS-1 and human Tensin-3. Amino acid alignments of the SH2 (A), PTB (B), and NH2-terminal (C) domains are shown. Identical and similar residues are highlighted with dark and pale gray shadings, respectively. An asterisk indicates an Arg residue essential for binding to phosphotyrosine (A).
We first examined the effects of tns-1 loss-of-function mutations on axon regeneration. The ok80 allele deletes the COOH-terminal SH2 and PTB domains in both the long and short forms of TNS-1 (Fig. 2B,C). The tns-1(ok581) mutation deletes the first 2 exons including the first Met, which affects only the long form (Fig. 2B,C). We assayed the regrowth of laser-severed axons in GABA-releasing D-type motor neurons (Fig. 4A; Yanik et al., 2004). In wild-type animals at the young adult stage, regeneration of axons severed by laser was initiated within 24 h (Fig. 4A,B; Table 2). In contrast, we observed a reduced frequency of axon regeneration in the tns-1(ok80) and tns-1(ok581) mutants (Fig. 4A,B; Table 2). Although ∼40% of axons in tns-1(ok80) mutants regenerated, the lengths of the regrown axons (mean ± SEM; 63.6 ± 1.9 μm, n = 20, p = 0.25) were similar to those found in wild-type animals (mean ± SEM; 67.0 ± 2.1 μm, n = 20). These results indicate that the long form of TNS-1, but not the short form, is involved in axon regeneration following laser axotomy. We also examined whether the short isoform TNS-1S might have a function in axon regeneration. We observed that expression of the tns-1s cDNA from the unc-25 promoter did not rescue the ok80 defect (Fig. 4B; Table 2). Therefore, we focused only on the long form of TNS-1 in our subsequent experiments. Overexpression of tns-1 from the unc-25 promoter had no effect on axon regeneration in wild-type animals (Fig. 4B; Table 2).
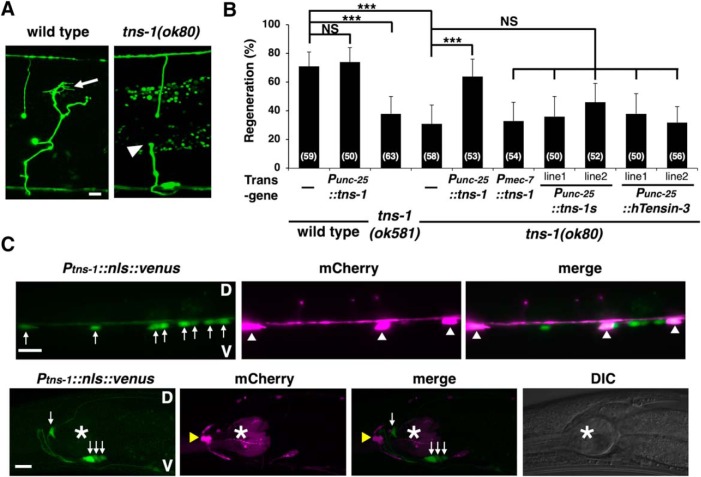
Figure 4.
TNS-1 is required for efficient axon regeneration. A, Representative D-type motor neurons in wild-type and tns-1 mutant animals 24 h after laser surgery. In wild-type animals, a severed axon has regenerated a growth cone (arrow). In tns-1 mutants, the proximal ends of axons failed to regenerate (arrowhead). Scale bar, 10 μm. B, Percentages of axons that initiated regeneration 24 h after laser surgery. Error bars indicate 95% confidence interval (CI). ***p < 0.001 as determined by Fisher's exact test. NS, Not significant. C, Expression of the Ptns-1::nls::venus gene. Fluorescent images and DIC of animals carrying Ptns-1::nls::venus and Punc-47::mcherry are shown. D-type motor neurons are visualized by mCherry under control of the unc-47 promoter. White arrows, white arrowheads, yellow arrowheads, and asterisks indicate cells expressing Ptns-1::nls::venus, D-type motor neurons, RME neuron, and pharynx, respectively. The fluorescent signal in the pharynx is from the injection marker Pmyo-2::DsRed monomer. V, Ventral side; D, dorsal side. Scale bars, 10 μm.
Table 2.
Raw data of genotype tested for axotomy
| Strain | Genotype | n | Regenerated | % | p | Vs |
|---|---|---|---|---|---|---|
| KU501 | Wild-type | 59 | 42 | 71 | — | — |
| KU1240 | tns-1(ok581) | 63 | 24 | 38 | 0.0003 | KU501 |
| KU1241 | tns-1(ok80) | 58 | 18 | 31 | <0.0001 | KU501 |
| KU1253 | tns-1(ok80) + Punc-25::tns-1s (line 1) | 50 | 18 | 36 | 0.68 | KU1241 |
| KU1267 | tns-1(ok80) + Punc-25::tns-1s (line 2) | 52 | 24 | 46 | 0.12 | KU1241 |
| KU1254 | Punc-25::tns-1 | 50 | 37 | 74 | 0.83 | KU501 |
| KU1243 | tns-1(ok80) + Punc-25::tns-1 | 53 | 34 | 64 | 0.0006 | KU1241 |
| KU1252 | tns-1(ok80) + Pmec-7::tns-1 | 54 | 18 | 33 | 0.84 | KU1241 |
| KU1256 | tns-1(ok80) + Punc-25::hTensin-3 (line 1) | 50 | 19 | 38 | 0.54 | KU1241 |
| KU1268 | tns-1(ok80) + Punc-25::hTensin-3 (line 2) | 56 | 18 | 32 | 1.0 | KU1241 |
| KU1244 | tns-1(ok80) + Punc-25::tns-1ΔN | 53 | 32 | 60 | 0.0023 | KU1241 |
| KU1245 | tns-1(ok80) + Punc-25::tns-1(R1071K) | 52 | 17 | 33 | 1.0 | KU1241 |
| KU1246 | tns-1(ok80) + Punc-25::tns-1ΔPTB | 54 | 17 | 31 | 1.0 | KU1241 |
| KU1242 | tns-1(ok80); mlk-1(km19) | 52 | 15 | 29 | 0.84 | KU1241 |
| KU1257 | tns-1(ok80); svh-2(tm737) | 57 | 16 | 28 | 0.84 | KU1241 |
| KU1263 | tns-1(ok80); ddr-2(ok574) | 51 | 15 | 29 | 1.0 | KU1241 |
| KU1264 | tns-1(ok80); max-2(nv162) | 50 | 17 | 34 | 0.84 | KU1241 |
| KU1248 | tns-1(ok80) + Pmlk-1::mlk-1 | 63 | 34 | 54 | 0.017 | KU1241 |
| KU1248* | tns-1(ok80) (-array) | 63 | 22 | 35 | 0.048 | KU1248 |
| KU1249 | tns-1(ok80) + Punc-25::svh-2 | 55 | 31 | 56 | 0.0081 | KU1241 |
| KU1249* | tns-1(ok80) (-array) | 58 | 20 | 34 | 0.024 | KU1249 |
| KU1250 | tns-1(ok80) + Punc-25::ddr-2 | 55 | 18 | 33 | 1.0 | KU1241 |
| KU1250* | tns-1(ok80) (-array) | 53 | 15 | 28 | 0.68 | KU1250 |
| KU1259 | tns-1(ok80) + Punc-25::max-2 | 50 | 31 | 62 | 0.0018 | KU1241 |
| KU1259* | tns-1(ok80) (-array) | 57 | 23 | 40 | 0.033 | KU1259 |
| KU504 | mlk-1(km19) | 51 | 14 | 27 | <0.0001 | KU501 |
| KU1247 | mlk-1(km19) + Punc-25::tns-1 | 53 | 18 | 34 | 0.53 | KU504 |
| KU503 | svh-2(tm737) | 52 | 13 | 25 | <0.0001 | KU501 |
| KU1258 | svh-2(tm737) + Punc-25::tns-1 | 42 | 14 | 33 | 0.49 | KU503 |
| KU1265 | pat-3(gk804163) | 71 | 23 | 32 | <0.0001 | KU501 |
| KU1266 | pat-3(gk804163) + Punc-25::pat-3 | 50 | 31 | 62 | 0.0016 | KU1265 |
Asterisks indicate the non-transgenic sibling controls.
Next, we tested whether TNS-1 can act cell-autonomously in axon regeneration. The tns-1 cDNA was expressed in tns-1(ok80) mutant animals using the unc-25 or mec-7 promoter. Expression of tns-1 in D-type motor neurons by the unc-25 promoter, but not in sensory neurons by the mec-7 promoter, rescued the axon regeneration defect of D-type motor neurons observed in tns-1(ok80) mutants (Fig. 4B; Table 2), thus demonstrating that TNS-1 can function cell autonomously in damaged D-type motor neurons. We next tested whether the mammalian Tensin-3 could substitute for C. elegans TNS-1 in axon regeneration, but found that expression of human Tensin-3 from the unc-25 promoter failed to rescue the tns-1 defect in axon regeneration (Fig. 4B; Table 2). Notably, the effect of tns-1 is injury-specific, because the tns-1(ok80) or tns-1(ok581) mutation otherwise has no obvious effect on nerve development.
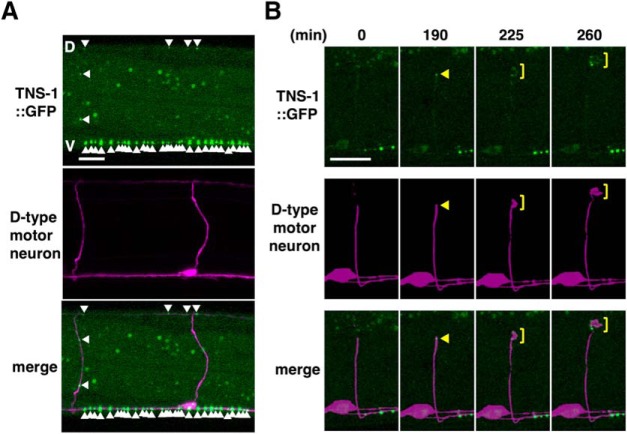
We investigated the expression pattern of tns-1 using a reporter construct consisting of the tns-1 promoter, a nuclear localization signal and the fluorescent protein VENUS (Ptns-1::nls::venus). Consistent with the cell-autonomous function of TNS-1 in axon regeneration, we found that the tns-1 gene is expressed in ventral motor neurons, including the Dorsal D-type (DD) and Ventral D-type (VD), as well as a subset of cells in heads (Fig. 4C). We next examined the intracellular localization of TNS-1 in D-type motor neurons using the Punc-25::tns-1::gfp gene. We observed that TNS-1::GFP was distributed in a punctate pattern along the ventral and dorsal nerve cords in D-type motor neurons (Fig. 5A). Following laser ablation of the axons, TNS-1::GFP accumulated at the severed ends and the growth cones (Fig. 5B).
Figure 5.
Localization of TNS-1::GFP in D-type motor neurons. A, Fluorescent images of animals carrying Punc-25::tns-1::gfp and Punc-47::mcherry are shown. D-type motor neurons are visualized by mCherry under control of the unc-47 promoter. White arrowheads indicate punctate localization of TNS-1::GFP in D-type motor axons. V, Ventral nerve cord; D, dorsal nerve cord. Scale bar, 10 μm. B, Fluorescent images of severed axons in animals carrying Punc-25::tns-1::gfp and Punc-47::mcherry are shown. Each image was taken at the indicated times (minutes) after laser surgery. Yellow arrowheads and yellow square brackets indicate the tip of the proximal injured axon and growth cones, respectively. Scale bar, 10 μm.
To examine whether the SH2 and PTB domains in TNS-1 are essential for axon regeneration, we generated two mutants, one that replaces Arg-1071 in the SH2 domain with Lys (R1071K) and the other that deletes amino acids 1155–1317, which contain the PTB domain (ΔPTB; Fig. 6A). Neither expression of TNS-1(R1071K) nor TNS-1(ΔPTB) from the unc-25 promoter was able to rescue the tns-1(ok80) mutant phenotype (Fig. 6B; Table 2). These results indicate that both the SH2 and PTB domains are required for TNS-1 function in axon regeneration.
Figure 6.
The involvement of TNS-1 in the JNK pathway in C. elegans. A, Domains in TNS-1. Structures of TNS-1 are shown. B–D, Percentages of axons that initiated regeneration 24 h after laser surgery are shown. Error bars indicate 95% confidence interval (CI). *p < 0.05, **p < 0.01, ***p < 0.001 as determined by Fisher's exact test. NS, Not significant.
TNS-1 functions in the JNK pathway regulating axon regeneration
As our RNAi screen for svh genes was intended to identify genes involved in the JNK pathway (Li et al., 2012; Hisamoto et al., 2018), we next investigated the relationship between TNS-1 and the JNK pathway in the regulation of axon regeneration. MLK-1 functions as a MAPKKK in the JNK pathway (Mizuno et al., 2004). When we generated tns-1(ok80); mlk-1(km19) double-mutants, we found that the phenotype of the double-mutant was indistinguishable from that of either single mutant (Fig. 6C; Table 2). This indicates that TNS-1 acts in the same pathway with MLK-1, and thus may function in the JNK pathway.
We next asked at what point TNS-1 acts in the JNK pathway during axon regeneration. Overexpression of mlk-1 suppressed the regeneration defect observed in tns-1(ok80) mutants (Fig. 6D; Table 2). In contrast, overexpression of tns-1 failed to suppress the mlk-1 defect (Fig. 6D; Table 2). These results suggest that TNS-1 functions upstream of MLK-1 in the JNK pathway. Activation of the JNK cascade following axonal injury is mediated by SVH-2, a homolog of the RTK Met, and its ligand SVH-1, a HGF-like growth factor. Activated SVH-2 in turn tyrosine phosphorylates MLK-1 (Li et al., 2012). In addition, DDR-2, another RTK that contains a discoidin domain, further modulates the SVH-1–SVH-2 pathway (Fig. 2A; Hisamoto et al., 2016). We confirmed that TNS-1 functions in the same pathway as SVH-2 and DDR-2, because the phenotype of tns-1(ok80); svh-2(tm737) or tns-1(ok80); ddr-2(ok574) double-mutants was indistinguishable from that of the tns-1(ok80) single mutant (Fig. 6C; Table 2). Because TNS-1 functions upstream of MLK-1, TNS-1 might act further downstream on SVH-2 or DDR-2. We thus examined whether overexpression of svh-2 or ddr-2 might reverse the defect in axon regeneration observed in tns-1 mutants. We found that overexpression of svh-2, but not of ddr-2, suppressed the regeneration defect in tns-1(ok80) mutants (Fig. 6D; Table 2). Furthermore, overexpression of tns-1 was unable to suppress the svh-2 defect (Fig. 6D; Table 2). Overexpression of mlk-1, svh-2, and ddr-2 does not increase the frequency of axon regeneration in a wild-type background (Li et al., 2012; Hisamoto et al., 2016). These results suggest that TNS-1 functions downstream of DDR-2 and may act on SVH-2 in the regulation of axon regeneration.
TNS-1 associates with tyrosine-autophosphorylated SVH-2 via the SH2 domain
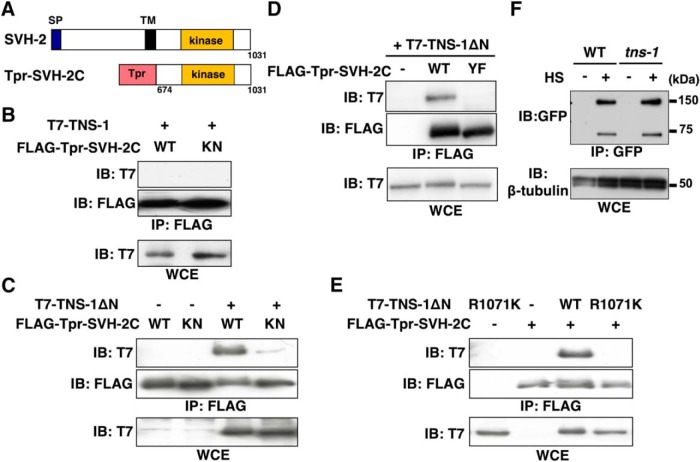
Because the TNS-1 protein contains an SH2 domain (Fig. 2C), we examined whether TNS-1 might associate with SVH-2 following SVH-2 autophosphorylation on tyrosine. To generate a constitutively active SVH-2, we fused the cytoplasmic region of SVH-2 (amino acids 674–1031) directly to the NH2-terminus of a dimerization leucine zipper motif [translocated promoter region (Tpr); Fig. 7A; Li et al., 2012]. The Tpr leucine zipper forces dimer formation and results in ligand-independent, constitutive activation of the Tpr-SVH-2C protein (Li et al., 2012). However, we failed to detect an interaction between T7-tagged, full-length TNS-1 and FLAG-tagged Tpr-SVH-2C coexpressed in mammalian COS-7 cells (Fig. 7B).
Figure 7.
Relationship between TNS-1 and SVH-2. A, Schematic diagrams of SVH-2 are shown. SP, Signal peptides; TM, transmembrane domain. B–E, COS-7 cells were transfected with plasmids encoding FLAG-Tpr-SVH-2C (WT), FLAG-Tpr-SVH-2C(K767R) (KN), FLAG-Tpr-SVH-2C(Y890F) (YF), T7-TNS-1, T7-TNS-1ΔN, and T7-TNS-1ΔN(R1071K), as indicated. Whole-cell extracts (WCE) and immunoprecipitated complexes obtained with anti-FLAG antibody (IP: FLAG) were analyzed by immunoblotting (IB). F, Effects of the tns-1 mutation on SVH-2 protein levels. Wild-type and tns-1(ok80) mutant animals carrying Phsp::svh-2::gfp were subjected to heat shock (+HS) at 37°C for 30 min and incubated at 20°C for additional 4 h. Animal lysates were prepared and immunoprecipitated with anti-GFP antibody. The SVH-2::GFP immunoprecipitate was blotted with anti-GFP antibody. β-tubulin was used for the loading control.
We next considered the possibility that the NH2-terminal region in TNS-1 might be an inhibitory domain, preventing interactions by forming a closed conformation. We tested this using a construct that deleted the NH2-terminal 298 aa of TNS-1 and tagged this with T7 (TNS-1ΔN; Figs. 3C, 6A). Immunoprecipitation analysis now showed that TNS-1ΔN associated with Tpr-SVH-2C (Fig. 7C). If this interaction is of functional significance, the NH2-terminal domain of TNS-1 should be dispensable for axon regeneration. Indeed, the Punc-25::tns-1ΔN transgene was able to rescue the tns-1 defect in axon regeneration (Fig. 6B; Table 2). Furthermore, we found that a catalytically inactive Tpr-SVH-2C(K767R) mutant failed to associate with TNS-1ΔN (Fig. 7C), suggesting that this interaction depends on the tyrosine-autophosphorylation of Tpr-SVH-2C. We have previously shown that the Tyr-890 autophosphorylation site is essential for the function of SVH-2 in axon regeneration (Li et al., 2012). We found that the mutant Tpr-SVH-2C(Y890F) also did not associate with TNS-1ΔN (Fig. 7D). We next tested whether the SH2 domain in TNS-1 is required for its interaction with Tpr-SVH-2C. We found that TNS-1ΔN(R1071K), a mutated form defective in the SH2 domain (Fig. 6A), was unable to interact with Tpr-SVH-2C (Fig. 7E), suggesting that the COOH-terminal SH2 domain is important for the interaction with Tpr-SVH-2C.
In mammalian cells, it is known that the Met tyrosine kinase receptor by itself not stably expressed, but that Tensin-4 interacts with and stabilizes Met by inhibiting its endocytosis and subsequent lysosomal degradation (Muharram et al., 2014). This raised the possibility that the C. elegans Met-like receptor SVH-2 would be also stabilized by TNS-1 in a manner similar to mammalian Met. To test this possibility, we expressed SVH-2::GFP fusion proteins from the heat shock promoter in wild-type and tns-1(ok80) mutant animals. After heat-shock treatment of the animals for 30 min and incubation for additional 4 h, animal lysates were prepared, immunoprecipitated, and immunoblotted with anti-GFP antibody. In wild-type animals, we detected the full-length SVH-2::GFP and its processed form lacking the NH2-terminal extracellular domain (Fig. 7F). We found that the tns-1 mutation had no effect on SVH-2::GFP expression levels (Fig. 7F). These results suggest that TNS-1 does not affect the stabilization of SVH-2 proteins.
TNS-1 acts as an adaptor to link the SVH-2 and integrin signaling pathways
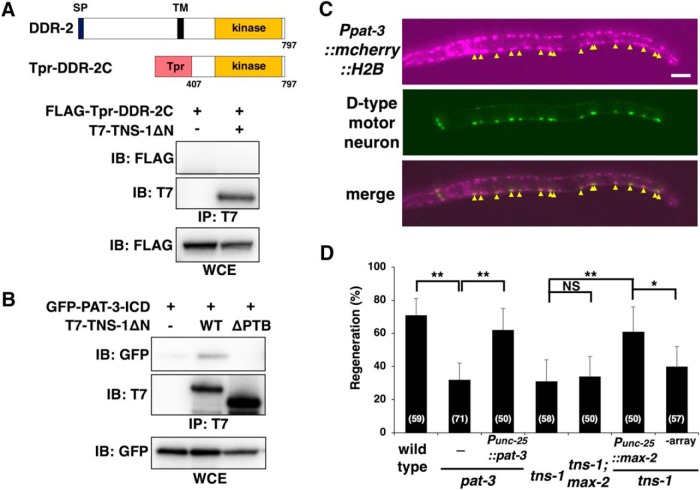
How does TNS-1 regulate SVH-2–JNK signaling in axon regeneration? We have previously demonstrated that upon injury, the DDR-2 and SVH-2 receptor tyrosine kinases coordinately regulate axon regeneration upstream of the JNK pathway and their interaction is mediated by the scaffold protein SHC-1 (Hisamoto et al., 2016). Because TNS-1 and SHC-1 contain SH2 and PTB domains, we examined whether TNS-1 also interacts with DDR-2. We coexpressed FLAG-Tpr-DDR-2C containing the cytoplasmic domain (amino acids 407–797) of DDR-2 (Fig. 8A; Hisamoto et al., 2016) with T7-TNS-1ΔN in COS-7 cells. However, TNS-1ΔN was unable to associate with FLAG-Tpr-DDR-2C (Fig. 8A).
Figure 8.
Relationship between TNS-1 and PAT-3. A, Interaction of TNS-1 with DDR-2. COS-7 cells were transfected with plasmids encoding FLAG-Tpr-DDR-2C and T7-TNS-1ΔN, as indicated. Whole-cell extracts (WCE) and immunoprecipitated complexes obtained with anti-T7 antibody (IP: T7) were analyzed by immunoblotting (IB). Schematic diagrams of DDR-2 are shown in the top part. SP, Signal peptides; TM, transmembrane domain. B, Interaction of TNS-1 with PAT-3. COS-7 cells were transfected with plasmids encoding GFP-PAT-3-ICD, T7-TNS-1ΔN (WT) and T7-TNS-1ΔN(ΔPTB) (ΔPTB), as indicated. Whole-cell extracts (WCE) and immunoprecipitated complexes obtained with anti-T7 antibody (IP: T7) were analyzed by immunoblotting (IB). C, Expression of the Ppat-3::mcherry::H2B::pat-3 3′UTR gene. Fluorescent images of animals carrying Ppat-3::mcherry::H2B::pat-3 3′UTR and Punc-25::gfp are shown. D-type motor neurons are visualized by GFP under control of the unc-25 promoter. Yellow arrowheads indicate D-type motor neurons. Anterior is the left. Scale bar, 20 μm. D, Percentages of axons that initiated regeneration 24 h after laser surgery are shown. Error bars indicate 95% confidence interval (CI). *p < 0.05, **p < 0.01 as determined by Fisher's exact test. NS, Not significant.
Muharram et al. (2014) recently demonstrated that mammalian Tensin-4 forms complexes with Met and integrin β1. C. elegans contains one gene, pat-3, encoding the integrin β subunit (Gettner et al., 1995). Therefore, we examined whether TNS-1 associates with PAT-3. When T7-TNS-1ΔN was coexpressed with the GFP-tagged intracellular domain of PAT-3 (GFP-PAT-3-ICD) in COS-7 cells, immunoprecipitation analysis revealed that TNS-1ΔN associated with PAT-3-ICD (Fig. 8B). The interaction between mammalian Tensin-4 and integrin β1 is known to occur through the Tensin-4 PTB domain (Calderwood et al., 2003; Katz et al., 2007). We next examined whether the TNS-1 PTB domain is required for binding to PAT-3-ICD. Deletion of the PTB domain abolished the ability of TNS-1ΔN to associate with PAT-3-ICD (Fig. 8B), suggesting that the PTB domain is important for the interaction of TNS-1 with PAT-3.
It is known that PAT-3 functions in D-type motor neurons to regulate commissural axon navigation (Poinat et al., 2002). To confirm that the pat-3 gene is expressed in D-type motor neurons, we used a reporter construct consisting of the pat-3 promoter and the mCherry::H2B fluorescent protein (Ppat-3::mcherry::H2B::pat-3 3′UTR). We found that the pat-3 gene is expressed in D-type motor neurons (Fig. 8 C). Furthermore, we found that the pat-3(gk804163) mutant was defective in axon regeneration (Fig. 8D; Table 2). The pat-3(gk804163) mutant substitutes a leucine in place of the proline residue in the NPXY motif, Asn-Pro (790)-Ile-Tyr, which is involved in the interaction with the PTB-containing protein (Calderwood et al., 2003). To exclude the possibility that the pat-3 phenotype might have a contribution from background mutations, we confirmed that the expression of pat-3 in D neurons from the unc-25 promoter was able to rescue the pat-3 defect in axon regeneration (Fig. 8D; Table 2). This result is also consistent with the expression of pat-3 in D neurons.
We have previously shown that integrin activates MAX-2 MAP4K through the CED-10 GTPase and that the activated MAX-2 phosphorylates MLK-1 MAPKKK, which leads to activation of the JNK pathway (Fig. 9; Pastuhov et al., 2016). The above results raised the possibility that TNS-1 links the SVH-2 and integrin signaling pathways by forming a complex of these three proteins to activate the JNK pathway in axon regeneration (Fig. 9). To test this possibility, we examined the genetic interaction between tns-1 and max-2. We found that the max-2(nv162) mutation did not enhance the axon regeneration defect observed in tns-1(ok80) mutants (Fig. 8D; Table 2). This result confirms that TNS-1 functions in the same pathway as MAX-2. Furthermore, overexpression of max-2 by the unc-25 promoter rescued the axon regeneration defect of tns-1(ok80) mutants (Fig. 8D; Table 2). These results suggest that TNS-1 links the SVH-2–MLK-1 pathway with the integrin–MAX-2–MLK-1 pathway (Fig. 9).
Figure 9.
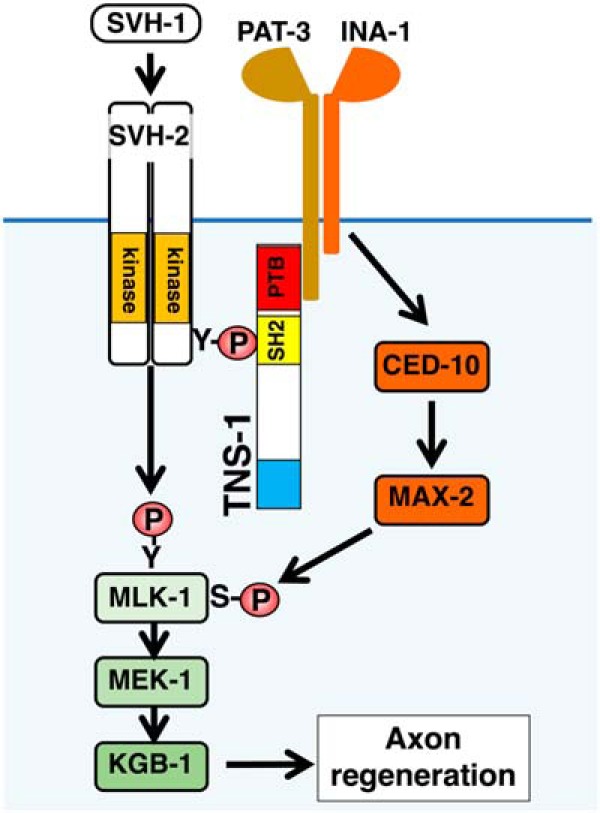
Relationship between SVH-2–MLK-1 and integrin–MAX-2–MLK-1 pathways in axon regeneration. PAT-3 makes a complex with INA-1, which activates MAX-2 through CED-10. Activated MAX-2 acts as a MAP4K for MLK-1 MAPKKK.
Discussion
A number of studies have shown that axon regeneration in C. elegans is regulated by the JNK MAPK pathway (Nix et al., 2011). This pathway is antagonized by the action of VHP-1, a MAPK phosphatase (Mizuno et al., 2004). The importance of this negative regulation of MAPK signaling is underscored by the fact that vhp-1 mutants are arrested in their development at an early larval stage (Mizuno et al., 2004). Mutations that inactivate various components in the JNK pathway can suppress the vhp-1 developmental arrest, indicating that the vhp-1 phenotype is caused by hyperactivation of the JNK pathway. This phenotype has made it possible to isolate regulators of the JNK pathway by genome-wide RNAi screens, which led to the identification of the svh genes (Li et al., 2012). Examination of various svh genes, svh-1, -2, -3, -4, -8, -13, and -15, have revealed them to also function as regulators of axon regeneration, thereby underscoring the importance of the JNK pathway in this process (Li et al., 2012, 2015; Pastuhov et al., 2012; Hisamoto et al., 2014, 2016, 2018; Shimizu et al., 2018). SVH-1 is homologous in sequence to mammalian HGF, whereas SVH-2 is homologous to the mammalian receptor for HGF, Met. This suggests that SVH-1 and SVH-2 may also function as a ligand-receptor pair in axon regeneration (Li et al., 2012; Hisamoto et al., 2014). In mammals, exogenous expression of HGF also promotes axon regeneration in many axotomized neurons (Kitamura et al., 2007; Esaki et al., 2011), suggesting that the HGF–Met signaling cascade is important for axon regeneration.
In this study, we undertook a functional characterization of the C. elegans ortholog of tensin, SVH-6/TNS-1, a protein implicated in SVH-2 signaling in axon regeneration. Four members of the Tensin family are found in mammals: Tensin-1, Tensin-2, Tensin-3, and Tensin-4/Cten, whereas C. elegans has only one known example. Mammalian Tensin, specifically Tensin-1, was first identified as a component of focal adhesions involving actin filaments (Wilkins et al., 1986). Tensin-1 has three distinct regions involved in binding to actin, two of which are located in the NH2-terminal region (Lo et al., 1994). In addition to its ability to bind actin, the NH2-terminal region of Tensin-1 is responsible for its localization to focal adhesions (Chen and Lo, 2003). Recently, it was shown the C. elegans M01E11.7, corresponding to TNS-1, is localized in the dense bodies, M-line, and cell attachment sites in muscle (Meissner et al., 2011). This suggests that TNS-1 is also a component of focal adhesions in C. elegans muscle. Thus, it is likely that TNS-1 has two different roles in C. elegans, one as a component of focal adhesions in muscles and second as a regulator of axon regeneration in neurons. The NH2-terminal domain of TNS-1 is dispensable for its function in axon regeneration and plays an inhibitory role in its interaction with SVH-2. These results suggest that under normal conditions, TNS-1 function is inhibited by the NH2-terminal region. Axon injury may relieve this inhibition, resulting in the association of TNS-1 with SVH-2, which, in turn, leads to the activation of the JNK pathway promoting axon regeneration.
The presence of SH2 and PTB domains in tandem is unique to tensin (Lo, 2004, 2017). Because SH2 domains typically mediate binding to phosphotyrosine, this domain may be essential for Tensin function. Mammalian tensins use the SH2 domain to recruit various tyrosine-phosphorylated signaling proteins, such as Met and EGFR (Lo, 2017). Recently, Muharram et al. (2014) found that mammalian Tensin-4 interacts with tyrosine-autophosphorylated Met via the SH2 domain and stabilizes Met protein by inhibiting its endocytosis. We demonstrate that the C. elegans TNS-1 SH2 domain is required for its interaction with tyrosine-autophosphorylated SVH-2 Met-like receptor, and is also required for its function in axon regeneration. However, our analysis of TNS-1 protein levels in tns-1 mutants shows that it is not involved in the stabilization of SVH-2.
How does TNS-1 regulate the SVH-2–JNK signaling pathway in axon regeneration? We show that the PTB domain of TNS-1 is essential for its function in axon regeneration. Mammalian tensin's PTB domain binds to β integrin cytoplasmic tails (Calderwood et al., 2003). We demonstrate that the C. elegans TNS-1 PTB domain is required for its association with the cytoplasmic region of the integrin β subunit PAT-3. We have recently shown that the integrin pathway also promotes axon regeneration through activation of the JNK cascade (Pastuhov et al., 2016; Hisamoto et al., 2018). In this pathway, the integrin α subunit INA-1 transduces a signal to the guanine nucleotide exchange factor complex, CED-2/CrkII, CED-5/DOCK180, and CED-12/ELMO, which activates the GTPase CED-10/Rac. CED-10, when bound by GTP, interacts with and activates the Ste20-related protein kinase MAX-2, which phosphorylates and activates MLK-1 (Pastuhov et al., 2016). These results suggest that TNS-1 brings PAT-3 in close proximity to SVH-2 by associating with both proteins and then SVH-2 and MAX-2 phosphorylate MLK-1 at tyrosine and serine residues, respectively (Fig. 9). One explanation as to why overexpression of svh-2 suppresses the tns-1 defect in axon regeneration is that overexpressed SVH-2 is in close proximity to components of the integrin–MAX-2–MLK-1 pathway. Similarly, when MAX-2 is overexpressed, it can access the SVH-2–MLK-1 pathway even in the absence of TNS-1. Thus, it is likely that TNS-1 links Met-like SVH-2 and integrin–MAX-2 with MLK-1 MAPKKK signaling. Our study describes a novel mechanism involving the TNS-1 adaptor protein in the SVH-2–JNK signaling pathway to regulate axon regeneration after neuronal injury.
Footnotes
This work was supported by Grants from the Ministry of Education, Culture and Science of Japan (N.H., S.I.P., H.H., and K.M.), and the Project for Elucidating and Controlling Mechanisms of Aging and Longevity from Japan Agency for Medical Research and Development, AMED, under Grant JP18gm5010001 (N.H.). We thank Caenorhabditis Genetic Center and C. elegans Knockout Consortium for materials.
The authors declare no competing financial interests.
References
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SL, Hammell M, Ambros V (2015) Robust distal tip cell pathfinding in the face of temperature stress is ensured by two conserved microRNAs in Caenorhabditis elegans. Genetics 200:1201–1218. 10.1534/genetics.115.179184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Fujioka Y, de Pereda JM, García-Alvarez B, Nakamoto T, Margolis B, McGlade CJ, Liddington RC, Ginsberg MH (2003) Integrin β cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A 100:2272–2277. 10.1073/pnas.262791999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Nichols A, Arkinstall S (2000) Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J 14:6–16. 10.1096/fasebj.14.1.6 [DOI] [PubMed] [Google Scholar]
- Case LC, Tessier-Lavigne M (2005) Regeneration of the adult central nervous system. Curr Biol 15:R749–R753. 10.1016/j.cub.2005.09.008 [DOI] [PubMed] [Google Scholar]
- Chen H, Lo SH (2003) Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem J 370:1039–1045. 10.1042/bj20021308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O'Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD (2011) Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron 71:1043–1057. 10.1016/j.neuron.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Hutter H, Jin Y, Wadsworth WG (2016) The genetics of axon guidance and axon regeneration in Caenorhabditis elegans. Genetics 204:849–882. 10.1534/genetics.115.186262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki S, Kitoh J, Katsumi S, Goshima F, Kimura H, Safwat M, Yamano K, Watanabe N, Nonoguchi N, Nakamura T, Coffin RS, Miyatake SI, Nishiyama Y, Murakami S (2011) Hepatocyte growth factor incorporated into herpes simplex virus vector accelerates facial nerve regeneration after crush injury. Gene Ther 18:1063–1069. 10.1038/gt.2011.57 [DOI] [PubMed] [Google Scholar]
- Gettner SN, Kenyon C, Reichardt LF (1995) Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol 129:1127–1141. 10.1083/jcb.129.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto N, Li C, Yoshida M, Matsumoto K (2014) The C. elegans HGF/plasminogen-like protein SVH-1 has protease-dependent and -independent functions. Cell Rep 9:1628–1634. 10.1016/j.celrep.2014.10.056 [DOI] [PubMed] [Google Scholar]
- Hisamoto N, Nagamori Y, Shimizu T, Pastuhov SI, Matsumoto K (2016) The C. elegans dischoidin domain receptor DDR-2 modulates the Met-like RTK-JNK signaling pathway in axon regeneration. PLoS Genet 12:e1006475. 10.1371/journal.pgen.1006475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto N, Tsuge A, Pastuhov SI, Shimizu T, Hanafusa H, Matsumoto K (2018) Phosphatidylserine exposure mediated by ABC transporter activates the integrin signaling pathway promoting axon regeneration. Nat Commun 9:3099. 10.1038/s41467-018-05478-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, Ben-Chetrit N, Tarcic G, Lindzen M, Avraham R, Liao YC, Trusk P, Lyass A, Rechavi G, Spector NL, Lo SH, et al. (2007) A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat Cell Biol 9:961–969. 10.1038/ncb1622 [DOI] [PubMed] [Google Scholar]
- Kawasaki M, Hisamoto N, Iino Y, Yamamoto M, Ninomiya-Tsuji J, Matsumoto K (1999) A Caenorhabditis elegans JNK signal transduction pathway regulates coordinated movement via type-D GABAergic motor neurons. EMBO J 18:3604–3615. 10.1093/emboj/18.13.3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Iwanami A, Nakamura M, Yamane J, Watanabe K, Suzuki Y, Miyazawa D, Shibata S, Funakoshi H, Miyatake S, Coffin RS, Nakamura T, Toyama Y, Okano H (2007) Hepatocyte growth factor promotes endogenous repair and functional recovery after spinal cord injury. J Neurosci Res 85:2332–2342. 10.1002/jnr.21372 [DOI] [PubMed] [Google Scholar]
- Li C, Hisamoto N, Nix P, Kanao S, Mizuno T, Bastiani M, Matsumoto K (2012) The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat Neurosci 15:551–557. 10.1038/nn.3052 [DOI] [PubMed] [Google Scholar]
- Li C, Hisamoto N, Matsumoto K (2015) Axon regeneration is regulated by Ets-C/EBP transcription complexes generated by activation of the cAMP/Ca2+ signaling pathways. PLoS Genet 11:e1005603. 10.1371/journal.pgen.1005603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SH. (2004) Tensin. Int J Biochem Cell Biol 36:31–34. 10.1016/S1357-2725(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Lo SH. (2017) Tensins. Curr Biol 27:R331–R332. 10.1016/j.cub.2017.02.041 [DOI] [PubMed] [Google Scholar]
- Lo SH, Janmey PA, Hartwig JH, Chen LB (1994) Interactions of tensin with actin and identification of its three distinct actin-binding domains. J Cell Biol 125:1067–1075. 10.1083/jcb.125.5.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, Rogalski T, Viveiros R, Warner A, Plastino L, Lorch A, Granger L, Segalat L, Moerman DG (2011) Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS One 6:e19937. 10.1371/journal.pone.0019937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10:3959–3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Hisamoto N, Terada T, Kondo T, Adachi M, Nishida E, Kim DH, Ausubel FM, Matsumoto K (2004) The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J 23:2226–2234. 10.1038/sj.emboj.7600226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muharram G, Sahgal P, Korpela T, De Franceschi N, Kaukonen R, Clark K, Tulasne D, Carpén O, Ivaska J (2014) Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells. Dev Cell 29:421–436. 10.1016/j.devcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix P, Hisamoto N, Matsumoto K, Bastiani M (2011) Axon regeneration requires co-activation of p38 and JNK MAPK pathways. Proc Natl Acad Sci U S A 108:10738–10743. 10.1073/pnas.1104830108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, Bastiani M (2014) Axon regeneration genes identified by RNAi screening in C. elegans. J Neurosci 34:629–645. 10.1523/JNEUROSCI.3859-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuhov SI, Fujiki K, Nix P, Kanao S, Bastiani M, Matsumoto K, Hisamoto N (2012) Endocannabinoid-Goα signalling inhibits axon regeneration in Caenorhabditis elegans by antagonizing Gqα-PKC-JNK signalling. Nat Commun 3:1136. 10.1038/ncomms2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuhov SI, Fujiki K, Tsuge A, Asai K, Ishikawa S, Hirose K, Matsumoto K, Hisamoto N (2016) The core molecular machinery used for engulfment of apoptotic cells regulates the JNK pathway mediating axon regeneration in Caenorhabditis elegans. J Neurosci 36:9710–9721. 10.1523/JNEUROSCI.0453-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinat P, De Arcangelis A, Sookhareea S, Zhu X, Hedgecock EM, Labouesse M, Georges-Labouesse E (2002) A conserved interaction between β1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr Biol 12:622–631. 10.1016/S0960-9822(02)00764-9 [DOI] [PubMed] [Google Scholar]
- Rossi F, Gianola S, Corvetti L (2007) Regulation of intrinsic neuronal properties for axon growth and regeneration. Prog Neurobiol 81:1–28. 10.1016/j.pneurobio.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Sakamoto R, Byrd DT, Brown HM, Hisamoto N, Matsumoto K, Jin Y (2005) The Caenorhabditis elegans UNC-14 RUN domain protein binds to the kinesin-1 and UNC-16 complex and regulates synaptic vesicle localization. Mol Biol Cell 16:483–496. 10.1091/mbc.e04-07-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Pastuhov SI, Hanafusa H, Matsumoto K, Hisamoto N (2018) The C. elegans BRCA2-ALP/Enigma complex regulates axon regeneration via a Rho GTPase-ROCK-MLC phosphorylation pathway. Cell Rep 24:1880–1889. 10.1016/j.celrep.2018.07.049 [DOI] [PubMed] [Google Scholar]
- Thomson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, Kieffer A, Lau J, Lee N, Miller A, Raymant G, Shen B, Shendure J, Taylor J, Turner EH, Hillier LW, et al. (2013) The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res 23:1749–1762. 10.1101/gr.157651.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JA, Risinger MA, Lin S (1986) Studies on proteins that co-purify with smooth muscle vinculin: identification of immunologically related species in focal adhesions of nonmuscle and Z-lines of muscle cells. J Cell Biol 103:1483–1494. 10.1083/jcb.103.4.1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A (2004) Neurosurgery: functional regeneration after laser axotomy. Nature 432:822. 10.1038/432822a [DOI] [PubMed] [Google Scholar]