Abstract
Presence of cytotoxic CD8+ T cells (CTL) in tumor microenvironments (TME) is critical for the effectiveness of immune therapies and patients’ outcome, whereas regulatoryT(reg) cells promote cancer progression. Immune adjuvants, including double-stranded (ds)RNAs, which signal via Toll-like receptor-3 (TLR3) and helicase (RIG-I/MDA5) pathways, all induce intratumoral production of CTL-attractants, but also Treg attractants and suppressive factors, raising the question of whether induction of these opposing groups of immune mediators can be separated. Here, we use human tumor explant cultures and cell culture models to show that the (ds) RNA Sendai Virus (SeV), poly-I:C, and rintatolimod (poly-I:C12U) all activate the TLR3 pathway involving TRAF3 and IRF3, and induce IFNα, ISG-60, and CXCL10 to promote CTL chemotaxis to ex vivo-treated tumors. However, in contrast with SeV and poly I:C, rintatolimod did not activate the MAVS/helicase pathway, thus avoiding NFκB− and TNFα-dependent induction of COX2, COX2/PGE2-dependent induction of IDO, IL10, CCL22, and CXCL12, and eliminating Treg attraction. Induction of CTL-attractants by either poly I:C or rintatolimod was further enhanced by exogenous IFNα (enhancer of TLR3 expression), whereas COX2 inhibition enhanced the response to poly-I:C only. Our data identify the helicase/NFκB/TNFα/COX2 axis as the key suppressive pathway of dsRNA signaling in human TME and suggest that selective targeting of TLR3 or elimination of NFκB/TNFα/COX2-driven suppression may allow for selective enhancement of type-1 immunity.
Significance
This study characterizes two different poly-I:C-induced signaling pathways in their induction of immunostimulatory and suppressive factors and suggests improved ways to reprogram the TME to enhance the antitumor efficacy of immunotherapies.
Introduction
The impact of local inflammation on tumor progression is determined by its character. Presence of CD8+ cytotoxic T-cells (CTL) in tumor microenvironments (TME) predicts prolonged survival of patients with multiple types of tumors (1, 2) and effectiveness of different forms of immunotherapies, including PD-1/PD-L1 blockade (3, 4). In contrast, intratumoral accumulation of regulatory T cells (Treg) and myeloid-derived suppressor cells (MDSC) predicts resistance to treatments and accelerated cancer progression (2, 5, 6). Because these different immune subsets are attracted by separate sets of chemokines, significant efforts have been dedicated to selective enhancement of intratumoral production of CTL attracting chemokines (such as CXCL9/MIG, CXCL10/IP10 CXCL11/I-TAC and CCL5/RANTES; ref. 7) while suppressing MDSC- and Treg-attracting chemokines (such as CCL22/MDC or CXCL12/SDF-1; refs. 8, 9).
Toll-like receptor (TLR) ligands have been widely used as adjuvants to enhance systemic immunization and to promote CTL entry into tumor tissues. Ligands for TLR3 (double-stranded RNA; dsRNA; such as poly-I:C and its analogs), TLR4 (LPS and its variant, MPLA), TLR7/8 (imiquimod) or TLR9 have been evaluated extensively in clinical trials (10, 11).
The ability of dsRNA (and other TLR ligands) to simultaneously induce CTL attractants and Treg-attracting chemokines and other tumor-promoting factors (11–13) raises the question of whether these functionally different factors are induced by separate signaling pathways, which can be dissociated to selectively enhance the desirable aspects of inflammation. dsRNA is known to trigger TLR3, which, through TIR domain-containing adaptor TRIF (14, 15), activates TRAF3 (16) and induces nuclear translocation of IRF3, leading to the transcription of type 1 interferons (17, 18). Concomitant activation of TRAF6 by TRIF through RIP-1 leads to nuclear translocation of NFκB and induction of additional pro-inflammatory cytokines such as TNFα (19). In addition to the TLR3/TRIF pathway, dsRNA can also activate the helicases RIG-I and MDA5 present in the cytosol (19, 20). Helicases interact with the mitochondrial anti-viral signaling protein (MAVS) and activate both, type-1 interferon pathway (21) and the NFκB pathway (22, 23). Although NFκB activation can promote the induction of type-1 interferons and the development of type-1 inflammation (24, 25), its intratumoral activation is typically associated with enhanced cancer cell proliferation, resistance to apoptosis, and cancer progression (26, 27).
The induction of NFκB and the associated induction of TNFα in response to either poly-I:C (28) or selective TLR3 ligand rintatolimod (poly-I:C12U; ref. 29), is strongly elevated in mice and other rodent species, compared with humans (28, 29), making mouse and rabbit studies poor models to predict toxicity and other undesirable effects of adjuvants involving different forms of dsRNA (29). Guided by the above considerations, we evaluated the role of different dsRNA-induced signaling pathways in the induction of immunostimulatory and suppressive factors and tested the feasibility of their separation to enhance the pattern of response in complete human TME.
Materials and Methods
Study design and patients
Following informed, written consent, specimens (ovarian tumor, n = 14, see Table 1) were collected during primary debulking surgeries for ovarian cancer under the University of Pittsburgh Institutional Review Board-approved protocol UPCI 07–058 (Prognostic Marker: Acquisition of Blood Samples and Tissue for Research Purposes (Gyn-Onc # 22–096). At least three biological replicates were performed for each experiment with the exact numbers specified in figure legends. All studies have been performed in accordance with the Declaration of Helsinki and other institutional and federal guidelines.
Table 1.
Clinical and pathology data
| Clinicopathological data | Patients (n = 14) |
|---|---|
| n (%) | |
| Age, y | |
| ≤61 | 7 (50) |
| >61 | 7 (50) |
| (range, 36–79) | |
| Stage | |
| I | 8 (57) |
| II | 0 (0) |
| III | 5 (36) |
| IV | 1 (7) |
| Histological grade | |
| Well | 1 (7) |
| Moderate | 2 (14.3) |
| Poor | 9 (64.3) |
| Not reported | 2 |
| Histology | |
| Serous | 7 (50) |
| Endometrioid | 3 (21.4) |
| Mucinous | 2 (14.3) |
| Clear cell | 2 (14.3) |
Tissue explant culture system
The ex vivo whole-tissue culture system, developed in our laboratory at the University of Pittsburgh and which allows us to avoid cell activation during tumor dissociation, has been described in our previous publication (30). In brief, using a 4-mm biopsy punch knife, the cubes of tumor were prepared and placed in antibiotic containing medium for 24 hours, in the absence or presence of the indicated factors and their combinations (Supplementary Fig. S1). The differentially treated tumor issues were harvested for mRNA extraction and culture supernatants were harvested for ELISA and chemotaxis assays (30).
Toll-like receptor ligands and modulators of their biologic activity
All TLR-ligands involved in this study were used at the previously established optimal concentrations, which all induced comparable levels of CXCL10 (Supplementary Figs. S2–S4). dsRNA, poly-I:C, a synthetic dsRNA with one strand composed of an inosine polymer and the other-cytidine polymer, known to activate both TLR3 and cytosolic helicases was obtained from (Sigma Aldrich; 20 μg/mL). Rintatolimod (poly-I:C12U), a modified dsRNA through an uracil mismatch at every 12th base position of the C-strand, which is a selective TLR3 ligand with a shorter half-life and lack of helicase-activating function (31, 32) was a gift from Hemispherx Bio and used at 125 μg/mL. Our titration experiments showed that at the concentration of 125 μg/mL of rintatolimod induced the same levels of CXCL10 expression (mRNA and protein levels) as poly-I:C (20 μg/mL) was visible (Supplementary Fig. S2). When indicated, the tissues were treated with IFNα (Life Technologies; 10,000 U/mL) and/or COX½ inhibitor indomethacin (Sigma Aldrich; 50 μmol/L). In some experiments, LPS (Sigma-Aldrich; 250 ng/mL), FSL-1 (Invivogen, 100 ng/mL), imiquimod (Sigma-Aldrich, 10 μg/mL), MPLA(Invivogen; 1 μg/mL) or inhibitory reagents eg. Indomethacin (Sigma-Aldrich, 50 μmol/L), Celecoxib (Biovision; 10 mmol/L), TNFα receptor inhibitor (TNFαRI; R&D Systems, 1 μg/mL) or an inhibitor of the NFκB pathway (BAY, BAY11–7082; Sigma-Aldrich; 10 μmol/L) were used.
Generation of macrophages
PBMCs were isolated from whole-blood samples of healthy donors through lymphocyte separation medium (Ficoll), as described (12, 33). Monocytes were isolated as light fraction of PBMCs through Percoll (GE Healthcare Life Sciences) density gradient centrifugation and cultured for 6 days in 24-well plates in IMDM complete medium, in the presence of GM-CSF (1,000 U/mL). On day 6, the cells were stimulated as indicated above (see: Tissue explant culture system) for 30 minutes, 1, 4, or 24 hours depending on the experiment. After harvesting, the supernatants were used for protein assays and the cells were lysed in RLT buffer for mRNA extraction, used for Western blot analysis or evaluated by immunofluorescence microscopy and image stream, as indicated.
Culture of fibroblasts
Human adult dermal fibroblasts (Thermo Fisher, C0135C) were cultured until confluent status was reached. After harvesting, the cells were reseeded at 2 × 105 cells/well in 24-well plates and treated with IFNα 10,000 U/mL), poly-I:C (20 μg/mL) or rintatolimod (125 μg/mL), LPS (250 ng/mL), FSL-1 (100 ng/mL), Imiquimod (10 μg/mL), MPLA (1 μg/mL) or indomethacin (50 μmol/L) for 24 hours. Supernatants were analyzed for chemokine production and cell lysates were used for mRNA expression analysis.
Generation of CD8+ effector T cells
Naive CD8+ T cells were extracted from whole blood of healthy individuals, as the heavy fraction of PBMCs (via Ficoll), followed by the positive separation of CD8+ T cells using microbeads (Miltenyi). The CD8+ T cells were cocultured with DCs loaded with SEB (1 ng/mL) for 6 days, as described previously (12). On day 6, cells were counted and adjusted to the desired density for chemotaxis assays (12).
TaqMan analysis of immune gene expression
Tumor biopsies or macrophages were taken to extract RNA using the RNeasy Kit (Qiagen). The RNA was used for cDNA synthesis and 25 ng of cDNA was used for mRNA expression analysis by TaqMan on the StepOnePlus system (Applied Biosystems). Commercially-available TaqMan primers (Thermo Fisher Scientific Life Technologies) were purchased to evaluate local expression of the key chemokines involved in the attraction of the effector cells (CCL5 andCXCL10), Treg (CCL22), and MDSCs (CXCL12). Furthermore, expression of EP4, COX2, TNFα, IFNβ, IDO, and IL10 was measured. The expression of each gene was normalized for the HPRT1 housekeeping gene.
Analysis of chemokines in ex vivo tumor explant cultures and cell cultures
Culture supernatants were analyzed by ELISA for the presence of the key chemokines involved in the attraction of the effector cells, Treg and MDSCs as previously described (13). Briefly, ELISA plates were coated with primary antibody at 1μg/mL overnight at room temperature. Next, plates were washed and blocked with 4% BSA in PBS for 3 hours. Samples were loaded (50 μl/well) and after a 1-hour incubation the biotinylated secondary antibody was added (0.5 μg/mL). For detection, Streptavidin-HRP conjugate (Thermo Fisher Scientific Pierce Biotechnology Inc.) was used for 30 minutes. After addition of TMP substrate (Thermo Fisher Scientific Pierce), reaction was stopped with 2% H2SO4 solution. Absorbance, at 450 nm, was measured on Wallace 1420 Victor 2 microplate Reader (PerkinElmer). The antibodies were purchased from R&D Systems.
Chemotaxis assay
Chemotaxis assays were performed in 24 Transwell plates with a 5-μm pore-size polycarbonate filter (#CLS3421; SIGMA). All media and supernatants were brought to room temperature before usage. The lower chamber contained 500 μL of tumor-supernatants and the upper chamber 2 × 105 of either freshly isolated CD4+ T cells or ex vivo-expanded CD8+ T effector cells in 200 μL of IMDM for 1.5 hours at 37°C. The migrated cells in the lower chamber were harvested and stained for either CD8 or CD4 and FoxP3 and measured by flow cytometry (Accuri C6, BD Biosciences; ref. 12). The number of migrated cells per condition was subtracted by the number of cells migrated towards plain media only.
Western blot
Antibody against IκBα was purchased from the Cell Signaling Technology (#9242); the antibody against α-tubulin from the Santa Cruz Biotechnology (Santa Cruz Biotechnology) and ISG60 has been described before (34). Monocyte-derived macrophages were treated as indicated and whole cell lysates were re-suspended in self-made lysis buffer (20 mmol/L HEPES pH 7.4, 1% Triton-X 100, 150 mmol/L NaCl, 1.5 mmol/L MgCl2, 12.5 mmol/L β-glycerophosphate, 2 mmol/L EGTA, 10 mmol/L NaF, 2 mmol/L DTT, 1 mmol/L Na3VO4, 1 mmol/L PMSF plus 1x protease inhibitors). Equal amounts of protein were immunoblotted with IκBα antibody or ISG 60 and a-tubulin antibody. The blots were quantitated by densitometry using the ImageJ software (NIH, Bethesda). The integrated pixel value was determined for each band by multiplying the image intensity by the band area after having subtracted the mean background value.
Image flow analysis
Imaging flow analysis for detection of NFκB nuclear translocation was performed with Amnis Imaging Flow Cytometers (Millipore Sigma) and analyzed with IDEAS Software (Millipore Sigma). Macrophage cultures were left untreated or treated with poly-I:C or rintatolimod at the indicated concentrations and stained with the Amnis NFκB Translocation Kit (ACS10000, Millipore, Sigma) according to the manufacturer’s protocol.
Immunofluorescence
Macrophages were seeded at a density of 105 cells on rat tail collagen-type 1 coated coverslips. After a period of >6 hours, attached cells were treated with rintatolimod or poly-I:C for 1, 4, and 24 hours. Untreated cultures served as a negative control. As positive control, macrophages were infected with Sendai virus (SeV; 80 HAU/mL) for 18 hours. Following treatments, cells were fixed in 3.33% PFA for 20 minutes at room temperature. After four washes with PBS cells were permeabilized with 0.1% Triton X. Cells, incubated with primary antibodies (MAVS: Bethyl Lab # A300–782A, 1:100; RIP-1 BD # 610459 1:250, TRAF3: Thermo Fisher #PA5–29091, 1:100; IRF3 provided by Dr. S. Sarkar, University of Pittsburgh, 1:500) overnight at 4°C, washed and incubated with a secondary antibody for one hour at room temperature (1:500, Alexa Fluor 488). Cells were then incubated with Hoechst (1:1,000) for 5 minutes and mounted using Prolong gold solution (Thermo Fisher Scientific Invitrogen, # P10144). Imaging was performed using the Carl Zeiss LSM 880 confocal microscope at fixed settings across treatments.
Phospho-CREB activation
Macrophages were differentiated as described above. After treatment with PGE2 (Sigma, 10−6 mol/L) or PGE2 in combination with IFNα, macrophages were harvested and fixed using 4% paraformaldehyde for 15 minutes at room temperature. After washing with PBS, cells were permeabilized by adding ice cold Methanol to a final concentration of 90% for 30 minutes. After two washing steps, blocking was performed with whole mouse IgG (10 μg/mL) for 15 minutes at room temperature. The phospho-CREB antibody (#9198, Cell Signaling Technology) was added and incubated for 1 hour at room temperature. After two washing steps, cells were incubated with fluorochrome conjugated secondary ab (Alexa Fluor 647, #4414, Cell Signaling Technology) for 30 minutes at room temperature in the dark. After another two washing steps, phosphor-CREB activation was measured by flow cytometry (Accuri C6, BD Biosciences).
Mouse studies
Female 6- to 8-week-old C57BL/6 mice were purchased from The Jackson Laboratory and maintained in the Roswell Park Animal Facility, under Institutional Animal Care and Use Committee (IACUC)-approved protocols. Bone marrow was isolated from the femur and tibia of C57BL/6 and cultured in the presence of GM-CSF, as described previously (35). On day 6 to 7, bone marrow–derived macrophages were activated overnight with the indicated concentrations of poly(I:C) or rintatolimod, in the absence or presence of celecoxib, as indicated, before RNA extraction and RT-PCR (TaqMan) analysis, as described previously (36).
Statistical analysis
All data points represent biological replicates. All statistical analyses were conducted using GraphPad Prism 5 software. Multiple treatment conditions of the same tumor sample or culture sample were compared with each other using the nonparametric Wilcoxon matched paired signed-rank. P < 0.05 was considered significant.
Results
Different forms of dsRNA differ in their ability to induce Treg/MDSC-attracting chemokines and suppressive factors in human TME despite being similarly effective in promoting CTL attraction
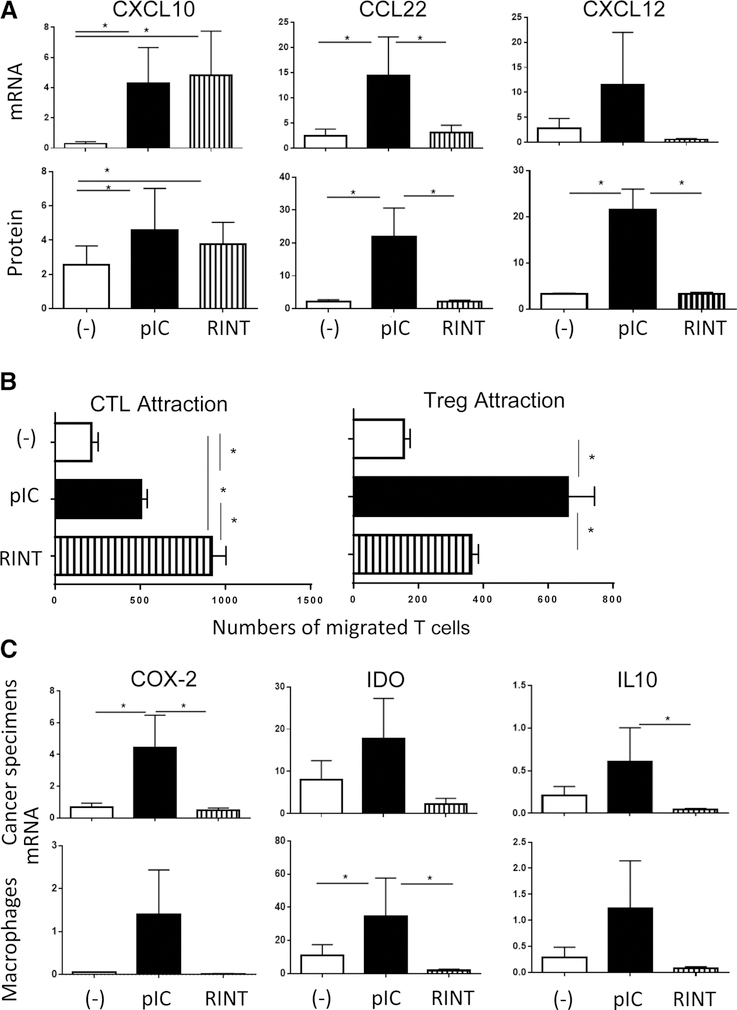
To evaluate the impact of immune adjuvants on complete human microenvironments, we used our previously developed tumor explant model, which includes all elements of TME (Supplementary Fig. S1; ref. 12). As shown in Fig. 1A, fresh biopsies of human ovarian cancer exposed to either poly-I:C or rintatolimod (poly-I:C12U, a selective TLR3 ligand with a shorter systemic half-life and lacking a cytosolic helicase-activating function (31, 32), induced similar levels of the desirable CXCL10. Strikingly, however, only poly-I:C, but not rintatolimod, induced CCL22 or CXCL12 at either mRNA expression level and protein secretion in cancer tissue explants and in macrophage and fibroblast cultures (Fig. 1A; Supplementary Figs. S2 and S3A–S3D), independently on the concentration used (Supplementary Fig. S2). CCL5 production showed an intermediate pattern of regulation, being induced by both forms of dsRNA, but at higher levels by poly-I:C (Supplementary Fig. S3A–S3B).
Figure 1.
Poly-I:C and rintatolimod both induce CXCL10 and promote CTL attraction to cancer tissues, but differ in their ability to promote Treg attraction and production of suppressive factors. A, The effects of poly-I:C (pIC) and rintatolimod (RINT) on the individual chemokines’ gene expression (relative to HPRT1) and protein secretion (ng/mL) in ex vivo-cultured ovarian cancer explants (n = 8 patients). Note the comparable induction of CXCL10 by both TLR3 ligands, but selective induction of CCL22 and CXCL12 by poly-I:C. B, poly-I:C–treated tumor explants show selectively reduced attraction of CTLs and elevated Treg attraction, compared with the rintatolimod-treated tumors. Data from ex vivo migration assays using either DC-sensitized CD8+ CTLs (n = 9; ref. 50) or CD4+ T cells (Treg identified in post-migration fraction as CD4+/FoxP3+ cells; n = 5) in the upper chamber and differentially treated supernatants of cancer specimens (n = supernatants from tumors of 9 patients for CTL migration and n = 5 patients for Treg migration) in the lower chamber. C, Induction of COX2− and COX2−dependent suppressive factors (9, 33, 40) by the two forms of dsRNA in ovarian cancer specimen (n = 6 patients) and in monocyte-derived macrophages (n = 7) is a selective feature of poly-I:C, but not rintatolimod. Results (mRNA levels, normalized for HPRT) are mean ± SEM. *, P < 0.05 (Wilcoxon signed-rank test).
These differences were reflected by enhanced ability of rintatolimod to promote selective attraction of CTLs, with concomitant reduction in Treg attraction, compared with poly-I:C (Fig. 1B).
Analysis of cancer-associated suppressive factors COX2 (rate-limiting mediator of PGE2 synthesis), IDO and IL10, further showed the selective ability of, poly-I:C, but not rintatolimod, to induce tumor-associated suppressive factors in the tumor biopsies and macrophage cultures (Fig. 1C). The dual character of poly-I:C-driven inflammation was shared by multiple additional TLR ligands, which all induced COX2, IDO, and CCL22, in addition to CXCL10 (Supplementary Fig. S4).
Activation of NFκB- versus type-1 IFN pathway reflects the respective activation of the helicase- versus TLR3-mediated dsRNA recognition
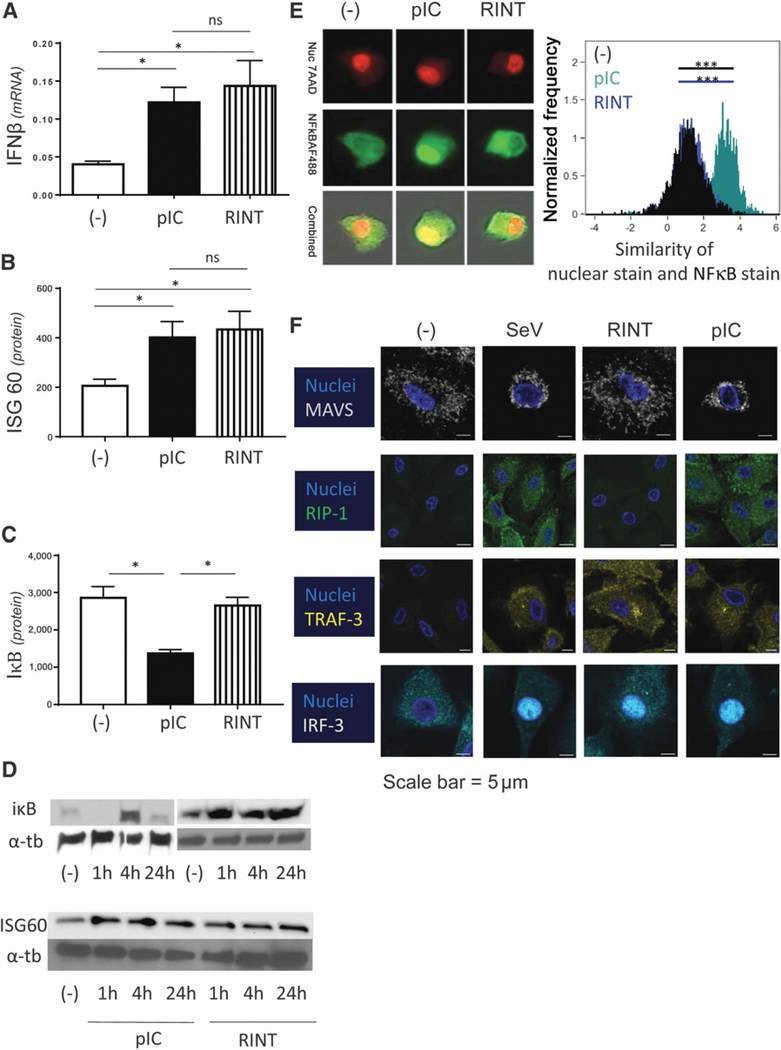
To investigate the mechanisms underlying different patterns of responses to poly-I:C versus rintatolimod, we compared the induction of IFNβ and ISG60 (representatives of type-1 interferon pathway) versus NFκB (as a mediator of theTRAF6- and helicase-mediated pathways of dsRNA recognition).
As shown in Fig. 2A–D, poly-I:C and rintatolimod were both equally potent inducers of IFNβ and ISG60 in macrophage cultures, suggesting their similar ability to activate IRF3. This was in sharp contrast to the activation of the NFκB system, which was driven exclusively by poly-I:C, but not rintatolimod as shown by its inability to degrade IκB, the blocker of nuclear translocation and activation of NFκB (37).
Figure 2.
Poly-I:C and rintatolimod both activate the IFN pathway of dsRNA recognition, but only poly-I:C activates the helicase/MAVS- and RIPK-1–dependent pathways of dsRNA recognition and activates NFκB. A and B, Poly-I:C (pIC) and rintatolimod (RINT) promote similar induction of IFNβ and ISG60 in human macrophages. A. IFNβ mRNA levels (n = 4). B, Similar induction of ISG60 protein (n = 4). Results are presented as the mean values ± SEM of four Western blots measured by densitometry. C, IκB degradation is only visible by poly-I:C treatment (n = 3). IκB degradation measured after 1 hour treatment. Results are presented as the mean values ± SEM of three Western blots measured by densitometry. D, Representative Western blot results of ISG60 and IκB analysis; α-tubulin (a-tb) served as a positive control. E, Unique induction of NFκB nuclear translocation in response to poly-I:C, but not rintatolimod (Amnis image flow). Human macrophages were treated with poly-I:C or rintatolimod, as indicated. Nuclear translocation of NFκB (and cytoplasmic presence of NFκB) was evaluated after 30–40 minutes. Colocalization of nuclear stain and NFκB stain was analyzed using the similarity score. A high similarity score between nuclear stain and NFκB stain (high colocalization of both stains) is seen in the poly-I:C-treated group, whereas the untreated and rintatolimod-treated cells show low similarity scores (the nuclear stain and the NFκB stain do not overlap), demonstrating lack of nuclear NFκB translocation. F, Different patterns of activation of helicase- and TLR3-related signaling pathways by different forms of dsRNA. Human cultured macrophages were treated with rintatolimod or poly-I:C or were infected with SeV (80 HAU/ml) for 20 hours. Note the selective activation of MAVS (helicase-pathway) and RIP-1 (TLR3 adapter responsible for TLR3-dependent activation of NFκB) after treatment with poly-I:C and SeV but not rintatolimod. In contrast, TRAF3, TLR3-dependent activator of type-1 IFN pathway, is similarly activated by rintatolimod, poly-I:C, and SeV, but not in untreated cells. Similarly, nuclear translocation of IRF3, the type-1 IFN pathway-induced transcription factor, is observed after activation with poly-I:C, rintatolimod or SeV but not in the control cells (n = 3). Results are shown as mean ± SEM. *, P < 0.05 (Wilcoxon signed-rank test).
As shown in Fig. 2D, degradation of IκB was evident after 1 hour of stimulation by poly-I:C, with its re-synthesis and re-appearance observed at 4 hours and return to baseline levels at 24 hours. As expected, no degradation by rintatolimod is apparent at any time point. Image flow analysis demonstrated that nuclear translocation of NFκB is selectively induced by poly-I:C, but not by rintatolimod (Fig. 2E; the similarity score reflects colocalization between 7-AAD nuclear stain and NFκB stain). In contrast to the untreated cells and rintatolimod-treated cells, which showed no colocalization of NFκB and the nuclear marker, the poly-I:C-treated cells showed strong colocalization between nuclear stain and NFκB stain.
To directly evaluate the impact of both forms of dsRNA on two upstream NFκB–activating pathways of dsRNA recognition, RIP-1 (TLR3-dependent pathway) and MAVS (cytoplasmic helicase-dependent pathway), we performed immunofluorescence analysis of the upstream regulatory proteins, MAVS and RIP-1. As shown in Fig. 2F, poly-I:C treatment activated both MAVS and RIP-I, similar to the SeV, whereas no activation of either MAVS or RIP-I was observed after rintatolimod exposure. In sharp contrast, both poly-I:C and rintatolimod promoted similar induction of TRAF3 and IRF3 nuclear translocation, the key mediators of the TLR3/TRAF pathway of type-1 IFN induction.
Positive feedback between TNFα to NFκB is involved in the dsRNA-triggered TNFα and COX2 activation
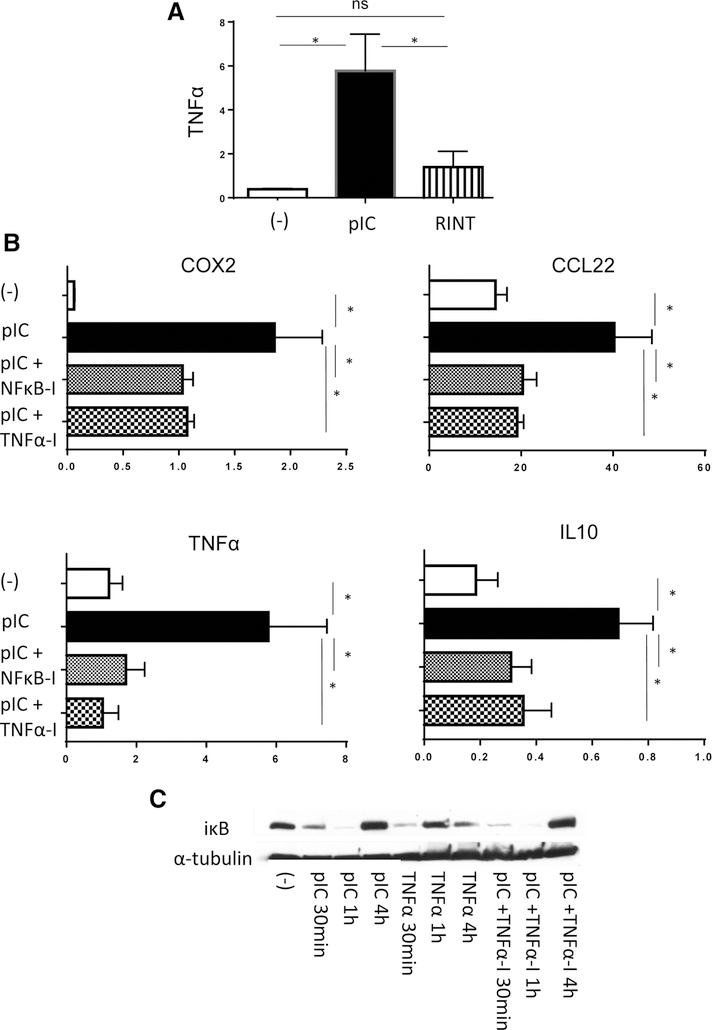
TNFα is a major pro-inflammatory cytokine, which both activates NFκB and is induced itself as a result of NFκB activation (38). TNFα is also a potent inducer of COX2 (38), the key mediator of PGE2 synthesis (39), which coordinates intratumoral production of multiple other suppressive factors and Treg/MDSC attractants (9, 33, 39, 40). To investigate the sequence of events and the role of the interplay between TNFα, NFκB and COX2 during the induction of antitumor-and tumor-promoting aspects of dsRNA signaling, we investigated the regulation of TNFα and NFκB by poly-I:C or rintatolimod, and the impact of their blockade upon the induction of each other and COX2.
As shown in Fig. 3A, poly-I:C-treated macrophages showed significantly higher levels of TNFα expression compared with untreated-or rintatolimod-treated cells (which showed similar levels of TNFα). To test whether endogenous TNFα and activated NFκB are needed for poly-I:C-driven COX2 induction we performed inhibitory assays using BAY11–7082, an NFκB inhibitor and soluble TNFαRI, as a competitive TNFα receptor antagonist. Suppression of COX2 was observed when combining poly-I:C with either one of these inhibitors, indicating that both NFκB and TNFα are involved in COX2 induction (Fig. 3B).
Figure 3.
Effective COX2 induction depends on the positive feedback between poly-I:C–induced NFκB activation and endogenous TNFα. A, Effective TNFα induction in human macrophages exposed to poly-I:C (pIC) but not rintatolimod (RINT; n = 6). B, Poly-I:C–induced expression of TNFα and COX2 (as well as COX2–dependent suppressive factors CCL22 and IL10) is suppressed by NFκB inhibitor BAY11–7082 (NFκB-I) or TNF receptor inhibitor TNFαRI (TNFα-I) in poly-I:C–stimulated human macrophages (n = 6; mRNA levels, normalized for HPRT). C, Poly-I:C triggers IκB degradation (30 minutes and 1 hours) in macrophages, whereas the TNFα-induced degradation is faster and is compensated already at 1 hour. Note that the IκB degradation triggered by Poly-I:C cannot be prevented by TNRαRI. Results are mean ± SEM. *, P < 0.05 (Wilcoxon signed-rank test).
Interestingly, blocking either TNFα or NFκB, each suppressed COX2, CCL22, IL10 and TNFα expression (Fig. 3B), indicating that dsRNA-triggered TNFα induction involves NFκB and the existence of a poly-I:C-NFκB–TNFα-dependent feed forward loop. In support of the direct TNFα-independent ability of poly-I:C to activate NFκB within minutes of dsRNA exposure, IκB degradation occurred within 30 minutes of either poly-I:C or TNFα treatment, and could not be prevented by TNFα blockade (Fig. 3C), although such blockade clearly inhibited the poly-I:C-induced expression of COX2, as well as IL10 and CCL22, again indicating the existence of a feed forward loop (Fig. 3B). Cumulatively, these data indicate that dsRNA triggers initial NFκB activity in a TNFα-independent fashion, but that NFκB-dependent TNFα induction is critically needed for the optimal COX2 induction and COX2–dependent induction of suppressive factors and Treg and MDSC attractants (Fig. 3; Supplementary Fig. S3).
Blockers of PGE2 synthesis and signaling selectively eliminate undesirable aspects of poly-I:C-induced tissue response
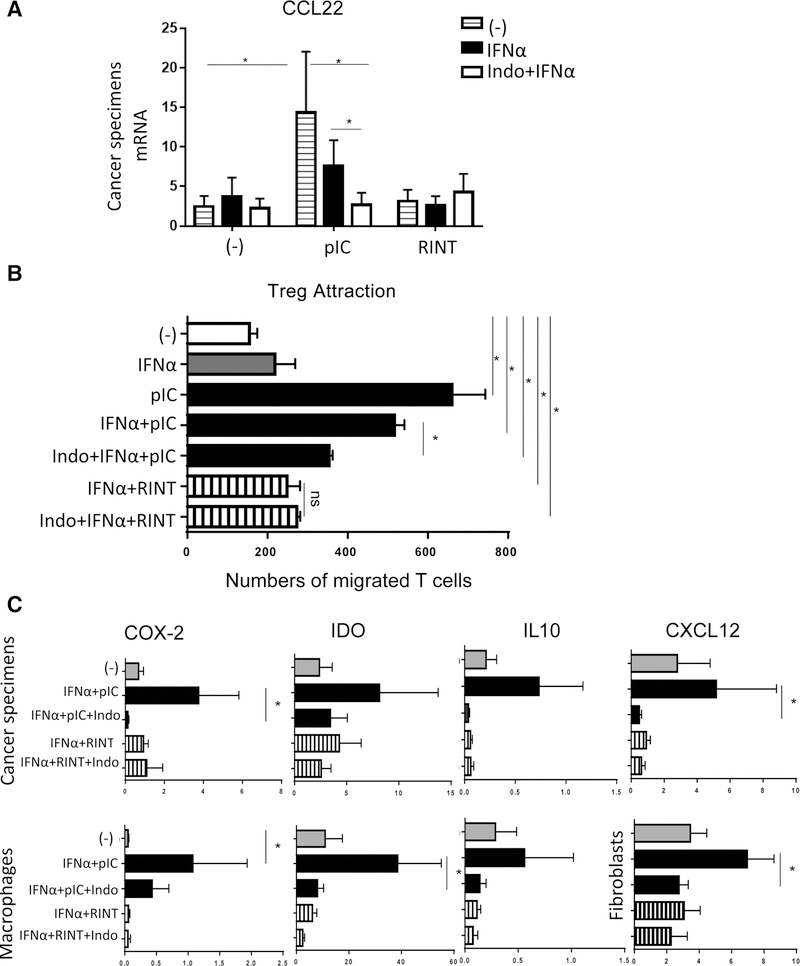
Because exogenous IFNα enhances the expression ofTLR3 (41, 42), but, paradoxically, suppresses the poly-I:C–driven induction of CCL22 (12, 13, 30), we tested whether it can affect the differences between the suppressive aspects of poly-I:C and rintatolimod-driven inflammation in human ovarian cancer TME. As shown in Fig. 4A, the addition of IFNα, partially eliminated the induction of CCL22 by poly-I:C, but did not abrogate the differences between the two forms of dsRNA. Because IFNα suppressed the expression of EP4, the main receptor-mediating suppressive effects of PGE2 (9, 33, 39, 40), blocking the PGE2-induced CREB phosphorylation (Supplementary Fig. S5A–S5B), and we previously observed that COX2-driven PGE2 production orchestrates the production of the MDSC/Treg attractant CXCL12 and multiple other suppressive factors in OvCa TME (9, 33, 40), we tested whether the overall differences in the patterns of tumor–tissue responses to poly-IC versus rintatolimod can be abolished by the addition of a COX½ inhibitor, indomethacin. Indeed, the combination of IFNα and indomethacin reduced the levels of poly-I:C-induced CCL22 induction down to the baseline (and to the levels observed with rintatolimod; Fig. 4A), with similar effects being observed when combining indomethacin with poly-I:C versus rintatolimod, in the absence of IFNα (Supplementary Fig. S3D).
Figure 4.
Combination of IFNα and indomethacin counteracts the induction of suppressive factors by poly-I:C. A, Undesirable CCL22 induction by poly-I:C (pIC) is partially suppressed by IFNα (which suppresses EP4; see Supplementary Fig. S5A) and abrogated by the combination of IFNα and indomethacin, down to the baseline levels observed with rintatolimod (RINT). Data from tumor samples from n = 8 patients. B, IFNα alone and the combination of IFNα and indomethacin suppress the ability of poly-I:C to promote Treg migration to the treated tumors (n = 5). C, Combination of IFNα and indomethacin (Indo) abrogates the induction of COX2– and COX2–dependent suppressive factors IDO, IL10 in poly-I:C–treated cancer specimen (n = 6), and macrophage cultures (n = 7). Because CXCL12 is not produced by human macrophages, human fibroblast cultures were used to evaluate CXCL12 induction (n = 5). Results (mRNA levels normalized for HPRT) are mean ± SEM. *, P < 0.05 (Wilcoxon signed-rank test).
These observations were further supported by the functional studies, showing that PGE2 suppression reduced the levels of Treg attraction to poly-I:C-treated tumors, abrogating the functional differences between these two variants of dsRNA (Fig. 4B).
In accordance with the mobilization of the suppressive COX2/PGE2 system as the central differentiating element between the adjuvant activity of different forms of dsRNA, the poly-I:C–dependent induction of COX2, IL10, IDO, and CXCL12 induction in cancer specimens, macrophage cultures and fibroblast cultures, was suppressed by addition of IFNα and indomethacin, down to the baseline levels observed with rintatolimod (Fig. 4C). COX2 dependence of these effects was confirmed by the observations that both, indomethacin and celecoxib (selective COX2 inhibitor) suppressed the undesirable effects of poly-I:C signaling (Supplementary Fig. S5C).
Discussion
The ability of dsRNA and other TLR ligands to promote the induction and effector phases of cytotoxic type-1 responses, mediated by CTLs, Th1 and NK cells, is critical to their ability to act as “danger signals” alerting the immune system to infections and facilitating the clearance of the relevant pathogens, and has been used in the development of immune stimulants in cancer immunotherapy.
Unexpectedly, our current data demonstrate that whereas two forms of dsRNA, poly-I:C and a selective TLR3 ligand, rintatolimod, are both similarly effective in inducing the intratumoral production of type-1 interferon and type-1 interferon-dependent CTL attractant CXCL10, only poly-I:C promoted the expression of the MDSC/Treg attractants CXCL12 and CCL22 and amplified the production of multiple tumor-associated suppressive factors, in human isolated macrophages and fibroblasts and with ex vivo-treated explants of whole ovarian cancer tumor tissues. Selective ability of poly-I:C to activate the helicase pathway of dsRNA recognition led to NFκB–dependent induction of TNFα and COX2 and the resulting mobilization of Treg/MDSC-attracting chemokines and suppressive factors, while limiting the effectiveness of poly-I:C as inducer of CXCL10 and CTL attraction. Because multiple other TLRs (which signal through MyD88) are known to activate the NFκB/TNFα pathway (43), and all induce COX2 and additional suppressive factors (Supplementary Fig. S4), our current data suggest that selective targeting of these factors (rather than NFκB) can be achieved without inhibiting the IFN pathway and may be used to enhance the selectivity and antitumor potency of multiple TLR–based adjuvants.
Although only poly-I:C, but not rintatolimod, required COX2 inhibition for its optimal effectiveness, both forms of dsRNA benefited from their combination with IFNα, consistent with the ability of IFNα to enhance the expression of TLR3 (41, 42), in addition to suppressing the key receptor mediating suppressive effects of PGE2, EP4. Interestingly, not only CXCL10, but particularly CCL5, which is NFκB and type-1 IFN dependent and may contribute to CTL attraction (12, 44), benefitted from the combination of rintatolimod with exogenous IFNα (Supplementary Fig. S3C), raising the possibility that IFNα may facilitate the ability of TLR3 to induce partial, helicase-independent, NFκB activation.
The mechanism of the interplay between type-1 IFNs and PGE2 in the regulation of the inflammatory responses to dsRNA and other TLR-Ls is a subject of our continued analyses. Previous reports demonstrated that IFN enhances the expression of TLR3 (41, 42), whereas PGE2 blocks the induction of endogenous type-1 IFNs (45). Our preliminary data indicate that IFNα blocks the expression of the PGE2 receptor EP4, and the PGE2-induced phospho-CREB activation (Supplementary Fig. S5A–S5B). These observations suggest that in the absence of exogenous IFNα, PGE2, which is present at significant levels in OvCa TME at baseline (9, 33) and further amplified by the poly-I:C-driven activation of the NFκB/COX2/PGE2 system (the current data), limits the levels of endogenous IFNβ, making its induction suboptimal, and favoring the suppressive aspects of inflammation.
Interestingly, we observed that TNFα, which similarly effective as poly-I:C in inducing NFκB and that endogenous TNFα production, was needed for the optimal mobilization of the COX2/PGE2 pathway in poly-I:C-activated cells. These observations and the ability of both NFκB-blockade and TNFα blockade to suppress COX2 induction indicate the scenario where the early poly-I:C–driven NFκB activation is needed for the initial induction of endogenous TNFα, leading to the establishment of a prolonged feed forward loop between poly-I:C, NFκB, and TNFα and the resulting induction of COX2, representing the negative component of poly-I:C signaling (Supplementary Fig. S6). Such feed forward loop is consistent with lack of complete abrogation of the poly-I:C–induced IκB degradation by TNFαRI, since TNFαRI blocks only the TNFα-related part of the loop.
Previous reports have shown that while poly-I:C activates both TLR3 and cytoplasmic helicases (MDA5 and RIG-1), leading to activation of MAVS and NFκB (46, 47), rintatolimod selectively activates only the TLR3 pathway (31,32). Our current data show that the optimal induction of type-1 IFNs, type-1 IFN-dependent CXCL10 and CTL attraction, requires neither the cytoplasmic helicase pathway nor the TLR3- induced RIP-1–mediated NFκB pathway, demonstrating a separation of the type-1–inducing versus suppressive pathways of signaling of dsRNA.
Although NFκB activation can amplify type-1 inflammation (26, 27, 48), the dominant aspects of its activation in TMEs are enhanced cancer cell proliferation, resistance to spontaneous and treatment-induced apoptosis, as well as tumor-associated neo-angiogenesis and metastatic spread (26, 27). The possibility to induce type-1 inflammation in cancer tissues without concomitant activation of NFκB and the resulting induction of COX2, one of the orchestrators of tumor-promoting inflammatory response, may facilitate the development of new treatments avoiding multiple levels of the negative aspects of tumor-associated inflammation.
Our follow-up work evaluates the impact of different forms of dsRNA (and other TLR-Ls, which all, with the exception of rintatolimod, promoted substantial induction of COX2, IL10 and CCL22 in human cells; see Supplementary Fig. S4) upon the accumulation of immune cells in mouse tumor tissues and antitumor effectiveness of immune checkpoint blockade, cancer vaccines and adoptive T-cell therapies. Because COX2 has been recently shown to direct mouse TMEs toward preferential production of tumor-promoting chemokines and cytokines, limiting the effectiveness of immune therapies (49), we expect that COX2 blockade will be uniformly advantageous to the antitumor impact of TLR ligands in mouse models.
However, murine cells do not distinguish between different forms of dsRNA with regard to NFκB activation and TNFα induction (28, 29) and produce TNFα also in response to rintatolimod injection, resulting in its enhanced toxicity in rodents compared with primates (29). Because the differential induction of COX2 and CCL22 in the human cells responding to poly-IC versus rintatolimod results from the differential NFκB activation and TNFα induction (current results), lack of such differences in rodent cells may complicate the use of mouse models to evaluate the TLR3-selective and nonselective dsRNAs with regard to Treg attraction. Indeed, our preliminary mouse data indicate that mouse macrophages behave similarly to human cells with regard to poly-IC- or rintatolimod-induced CXCL10 production, but, in sharp in contrast to human macrophages and whole tumor explants from patients with cancer, they elevate CCL22 production both in response to poly-I:C and to rintatolimod, in a COX2–independent manner (Supplementary Fig. S7A–S7C).
In summary, our current data demonstrate the molecular separation of the immunostimulatory and suppressive pathways of dsRNA recognition in human cancer tissues and the feasibility of their separate modulation to improve the selectivity of inflammation induced by dsRNA adjuvants. We show that the TLR3-driven activation of type-1 IFN pathway and intratumoral induction of CTL attractants by dsRNA, can proceed in the absence of the helicase/MAVS or TLR3/RIPK1-driven activation of the NFκB/TNFα pathway and the resulting induction of COX2– and COX2–dependent suppressive factors and Treg/MDSC-attracting chemokines. Our data help to understand the interplay between the immunostimulatory and suppressive functions of immune adjuvants and facilitate the development of improved therapies.
Supplementary Material
Acknowledgments
The authors thank Dr. William M. Mitchell (Vanderbilt University) for critical comments and discussions. This work has been supported by the NIH/NCI grants P01CA132714 (to P. Kalinski, R.P. Edwards, and D.L. Bartlett), P50CA159981 (to R.P. Edwards, P. Kalinski, and F. Modugno) and P30CA047904 (all authors), the NIH/NIAID grant AI118896 (to S. Sarkar) and by the Deutsche Forschungsgemeinschaft (research fellowship #TH 2172/1–1 to M.-N. Theodoraki) and the Rustum Family Foundation (to P. Kalinski).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclaimer
This work involves commercially available reagents and materials, with the exception of rintatolimod, which was obtained under an MTA from Hemispherx, Bio and the tumor samples from patients with ovarian cancer.
References
- 1.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66. [DOI] [PubMed] [Google Scholar]
- 2.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A 2005;102:18538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti–PD-Ll antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res 2012;72:2162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musha H, Ohtani H, Mizoi T, Kinouchi M, Nakayama T, Shiiba K, et al. Selective infiltration of CCR5(+)CXCR3(+) T lymphocytes in human colorectal carcinoma. Int J Cancer 2005;116:949–56. [DOI] [PubMed] [Google Scholar]
- 8.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10: 942–9. [DOI] [PubMed] [Google Scholar]
- 9.Obermajer N, Muthuswamy R, Odunsi K, Edwards RP, Kalinski P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res 2011;71:7463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iribarren K, Bloy N, Buque A, Cremer I, Eggermont A, Fridman WH, et al. Trial Watch: Immunostimulation with Toll-like receptor agonists in cancer therapy. Oncoimmunology 2016;5:e1088631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridnour LA, Cheng RY, Switzer CH, Heinecke JL, Ambs S, Glynn S, et al. Molecular pathways: toll-like receptors in the tumor microenvironment–poor prognosis or new therapeutic opportunity. Clin Cancer Res 2013;19:1340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muthuswamy R, Berk E, Junecko BF, Zeh HJ, Zureikat AH, Normolle D, et al. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res 2012;72:3735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muthuswamy R, Wang L, Pitteroff J, Gingrich JR, Kalinski P. Combination of IFNalpha and poly-I:C reprograms bladder cancer microenvironment for enhanced CTL attraction. J Immunother Cancer 2015;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003;301:640–3. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, et al. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat Immunol 2003;4:1223–9. [DOI] [PubMed] [Google Scholar]
- 16.Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 2006;439:204–7. [DOI] [PubMed] [Google Scholar]
- 17.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol 2005;17:1367–78. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 2000;13:539–48. [DOI] [PubMed] [Google Scholar]
- 19.Akira S TLR signaling. Curr Top Microbiol Immunol 2006;311:1–16. [DOI] [PubMed] [Google Scholar]
- 20.Lopez CB, Moltedo B, Alexopoulou L, Bonifaz L, Flavell RA, Moran TM. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J Immunol 2004;173:6882–9. [DOI] [PubMed] [Google Scholar]
- 21.Saha SK, Pietras EM, He JQ, Kang JR, Liu S-Y, Oganesyan G, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. Embo j 2006;25:3257–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, Kawai T, Kumar H, Sato S, Yonehara S, Akira S. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J Immunol 2006;176:4520–4. [DOI] [PubMed] [Google Scholar]
- 23.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, et al. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol 2007;8:592–600. [DOI] [PubMed] [Google Scholar]
- 24.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 2008;134:85–96. [DOI] [PubMed] [Google Scholar]
- 25.Balachandran S, Beg AA. Defining emerging roles for NF-kappaB in antivirus responses: revisiting the interferon-beta enhanceosome paradigm. PLoS Pathog 2011;7:e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005;5: 749–59. [DOI] [PubMed] [Google Scholar]
- 27.Van Waes C Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res 2007;13:1076–82. [DOI] [PubMed] [Google Scholar]
- 28.Lundberg AM, Drexler SK, Monaco C, Williams LM, Sacre SM, Feldmann M, et al. Key differences in TLR3/poly I:C signaling and cytokine induction by human primary cells: a phenomenon absent from murine cell systems. Blood 2007;110:3245–52. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell WM, Nicodemus CF, Carter WA, Horvath JC, Strayer DR. Discordant biological and toxicological species responses to TLR3 activation. Am J Pathol 2014;184:1062–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muthuswamy R, Corman JM, Dahl K, Chatta GS, Kalinski P. Functional reprogramming of human prostate cancer to promote local attraction of effector CD8+ T cells. Prostate 2016;76:1095–105. [DOI] [PubMed] [Google Scholar]
- 31.Gowen BB, Wong MH, Jung KH, Sanders AB, Mitchell WM, Alexopoulou L, et al. TLR3 is essential for the induction of protective immunity against Punta Toro Virus infection by the double-stranded RNA (dsRNA), poly(I:C12U), but not Poly(I:C): differential recognition of synthetic dsRNA molecules. J Immunol 2007;178:5200–8. [DOI] [PubMed] [Google Scholar]
- 32.Trumpfheller C, Caskey M, Nchinda G, Longhi MP, Mizenina O, Huang Y, et al. The microbial mimic poly IC induces durable and protective CD4+ T cell immunitytogether with a dendritic cell targeted vaccine. Proc Natl Acad Sci U S A 2008;105:2574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obermajer N, Muthuswamy R, Lesnock J, Edwards RP, Kalinski P. Positive feedback between PGE2 and COX2 redirects the differentiation of human dendritic cells toward stable myeloid-derived suppressor cells. Blood 2011;118:5498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 2014;40:936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Son YI, Egawa S, Tatsumi T, Redlinger RE Jr, Kalinski P, Kanto T. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J Immunol Methods 2002;262:145–57. [DOI] [PubMed] [Google Scholar]
- 36.Obermajer N,Urban J, Wieckowski E, Muthuswamy R, Ravindranathan R, Bartlett DL, et al. Promoting the accumulation of tumor-specific T cells in tumor tissues by dendritic cell vaccines and chemokine-modulating agents. Nat Protoc 2018;13:335–57. [DOI] [PubMed] [Google Scholar]
- 37.Baeuerle PA. IkappaB-NF-kappaB structures: at the interface of inflammationcontrol. Cell 1998;95:729–31. [DOI] [PubMed] [Google Scholar]
- 38.Ruddle NH.Tumor necrosis factor (TNF-alpha) and lymphotoxin (TNF-beta). Curr Opin Immunol 1992;4:327–32. [DOI] [PubMed] [Google Scholar]
- 39.Kalinski P Regulation of immune responses by prostaglandin E2. J Immunol 2012;188:21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong JL, Obermajer N, Odunsi K, Edwards RP, Kalinski P. Synergistic COX2 induction by IFNgamma and TNFalpha self-limits type-1 immunity in the human tumor microenvironment. Cancer Immunol Res 2016; 4:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun 2001;2:349–55. [DOI] [PubMed] [Google Scholar]
- 42.Tissari J, Siren J, Meri S, Julkunen I, Matikainen S. IFN-alpha enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J Immunol 2005; 174:4289–94. [DOI] [PubMed] [Google Scholar]
- 43.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 2007;13:460–9. [DOI] [PubMed] [Google Scholar]
- 44.Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappaB and IFN-regulatory factor transcription factors. J Immunol 2000;164: 5352–61. [DOI] [PubMed] [Google Scholar]
- 45.Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 2014;40:554–68. [DOI] [PubMed] [Google Scholar]
- 46.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 2006;103:8459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, et al. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 2005;433:887–92. [DOI] [PubMed] [Google Scholar]
- 48.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004;118:285–96. [DOI] [PubMed] [Google Scholar]
- 49.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, et al. Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 2015;162:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watchmaker PB, Berk E, Muthuswamy R, Mailliard RB, Urban JA, Kirkwood JM, et al. Independent regulation of chemokine responsiveness and cytolytic function versus CD8+ T cell expansion by dendritic cells. J Immunol 2010;184:591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.