SUMMARY
PIWI-interacting RNAs (piRNAs) silence transposons in Drosophila ovaries, ensuring female fertility. Two coupled pathways generate germline piRNAs: the ping-pong cycle, in which the PIWI proteins Aubergine and Ago3 increase the abundance of pre-existing piRNAs, and the phased piRNA pathway, which generates strings of tail-to-head piRNAs, one after another. Proteins acting in the ping-pong cycle localize to nuage, whereas phased piRNA production requires Zucchini, an endonuclease on the mitochondrial surface. Here, we report that Armitage (Armi), an RNA-binding ATPase localized to both nuage and mitochondria, links the ping-pong cycle to the phased piRNA pathway. Mutations that block phased piRNA production deplete Armi from nuage. Armi ATPase mutants cannot support phased piRNA production and inappropriately bind mRNA instead of piRNA precursors. We propose that Armi shuttles between nuage and mitochondria, feeding precursor piRNAs generated by Ago3 cleavage into the Zucchini-dependent production of Aubergine- and Piwi-bound piRNAs on the mitochondrial surface.
eTOC Blurb
In animals, PIWI-interacting RNAs (piRNAs) direct germline transposon silencing. Ge et al. now show that in Drosophila the RNA-binding ATPase protein Armitage uses ATP hydrolysis to selectively bind piRNA precursors, escorting them from the ping-pong piRNA-producing machinery in perinuclear nuage to the phased piRNA-producing pathway on mitochondria.
INTRODUCTION
In animals, PIWI-interacting RNAs (piRNAs) direct PIWI-clade Argonaute proteins to silence germline transposons, ensuring fertility (Huang et al., 2017; Czech and Hannon, 2016). In Drosophila, the cytoplasmic PIWI proteins Aubergine (Aub) and Ago3 increase piRNA abundance via reciprocal cleavage of sense transposon mRNAs and antisense piRNA precursor transcripts, a process termed the ping-pong amplification cycle (Brennecke et al., 2007; Gunawardane et al., 2007). The “initiator” and “responder” piRNAs amplified by the ping-pong cycle go on to direct production of tail-to-head strings of “trailing” piRNAs. Typically, Ago3-catalyzed, piRNA-directed cleavage of piRNA precursor transcripts initiates the production of these phased piRNAs, which are then loaded into Piwi, and, to a lesser extent, Aub. piRNA-bound Piwi, but not unloaded Piwi, can then transit to the nucleus, where it directs Histone 3 lysine 9 trimethylation (H3K9me3) of transposon DNA, silencing transcription by generating heterochromatin (McCue and Slotkin, 2012; Le Thomas et al., 2013; Rozhkov et al., 2013; Yashiro et al., 2018). (Historically, the piRNAs generated by Ago3- or Aub-catalyzed ping-pong cycles were called “secondary” piRNAs, whereas the Piwi-bound piRNAs now known to be phased were termed “primary” piRNAs. Here, we use the terms initiator and responder for ping-pong piRNAs and trailing for phased piRNAs.)
Trailing piRNA production requires Zucchini (Zuc), an endonuclease proposed to simultaneously generate the 3′ end of the preceding immature piRNA (pre-piRNA) and the 5′ end of the pre-piRNA precursor (pre-pre-piRNA) that will produce the next pre-piRNA (Han et al., 2015a; Mohn et al., 2015). Zuc belongs to the phospholipase D superfamily, whose HKD catalytic domain hydrolyzes phosphodiester bonds in phospholipids or nucleic acids (Pane et al., 2007; Selvy et al., 2011). Zuc cleaves single-stranded RNA in vitro, but without the NpU preference expected of an endonuclease generating phased piRNAs (Nishimasu et al., 2012; Ipsaro et al., 2012; Han et al., 2015a; Mohn et al., 2015; Nishida et al., 2018).
Phased piRNA production also requires Armitage (Armi), a member of the Upf1 family of ATP-dependent 5′-to-3′ helicase proteins (Cook et al., 2004; Saito et al., 2010; Haase et al., 2010; Olivieri et al., 2010).
Artificially tethering Armi to a transcript triggers its conversion into piRNAs (Rogers et al., 2017; Pandey et al., 2017). Armi is dispensable for the ping-pong cycle (Malone et al., 2009; Handler et al., 2011). Both recombinant Armi and Mov10l1, the mammalian Armi homolog, use ATP to catalyze 5′-to-3′ RNA duplex unwinding (Vourekas et al., 2015; Pandey et al., 2017), suggesting that, like Upf1, Armi uses ATP to gate its RNA binding. Mov10l1 has also been suggested to remove RNA secondary structures in vivo (Vourekas et al., 2015).
Loss of Zuc, Armi, or other phased-piRNA biogenesis proteins such as Piwi, Gasz (Germ cell protein with Ankyrin repeats, Sterile alpha motif, and leucine Zipper), Minotaur, and Papi, has little effect on ping-pong amplification (Handler et al., 2011; Handler et al., 2013; Baena-Lopez et al., 2013; Vagin et al., 2013; Han et al., 2015a; Hayashi et al., 2016). In contrast, germline phased-piRNA production collapses without the ping-pong machinery, because ping-pong amplification generates the pre-pre-RNAs that feed phased piRNA production (Han et al., 2015a; Mohn et al., 2015). The current model for piRNA biogenesis posits that phased-piRNA production begins when Ago3 cuts a complementary RNA, generating a 5′ monophosphorylated pre-pre-piRNA that can bind Aub. Next, Aub directs an endonuclease—likely Zuc—to cut the pre-pre-piRNA 5′ to the first downstream accessible uridine residue, simultaneously releasing Aub bound to a pre-piRNA and creating a new 5′ monophosphorylated pre-pre-piRNA from which trailing piRNAs can be generated (Mohn et al., 2015; Gainetdinov et al., 2018). Typically, Piwi binds the 5′ end of the new pre-pre-piRNA. Piwi directs Zuc to cut the pre-pre-piRNA 5′ to the next accessible uridine, generating a Piwi-bound, trailing pre-piRNA. The 3′ endonuclease cleavage product can then bind a second Piwi, repeating the process to generate another trailing piRNA and a pre-pre-piRNA. The production of phased piRNAs is believed to be limited by the frequency of ping-pong-directed cleavages: more frequent cleavage yields shorter substrates for phased piRNA production (Mohn et al., 2015).
The initial products of both the ping-pong and phased piRNA pathways are pre-piRNAs bound to PIWI proteins. In flies, the 3′ ends of pre-piRNAs are subsequently trimmed by Nibbler and 2′-O-methylated by the S-adenosyl methionine-dependent methyl transferase, Hen1, completing piRNA biogenesis (Horwich et al., 2007; Saito et al., 2007; Feltzin et al., 2015; Hayashi et al., 2016; Wang et al., 2016). Unlike pre-piRNAs in many animals, fly pre-piRNAs are nearly the same length as mature piRNAs, so that many piRNAs, particularly Piwi-bound piRNAs, are often 2′-O-methylated without 3′ trimming (Hayashi et al., 2016; Gainetdinov et al., 2018).
What couples the ping-pong cycle to the phased piRNA pathway? In Drosophila germline nurse cells, ping-pong piRNAs are thought to be made in perinuclear nuage—electron-dense, non-membrane-bound bodies (Mahowald, 1971; Eddy, 1975)—which contains the piRNA precursor transcripts and proteins required for ping-pong amplification (Liang et al., 1994; Brennecke et al., 2007; Lim and Kai, 2007; Nishida et al., 2009; Patil and Kai, 2010; Zhang et al., 2011; Handler et al., 2011; Zhang et al., 2012a; White-Cooper, 2012; Olivieri et al., 2012; Patil et al., 2014; Mohn et al., 2014; Andress et al., 2016). In contrast, phased piRNAs are likely made on the mitochondrial outer membrane, where phased piRNA biogenesis factors are found (Choi et al., 2006; Saito et al., 2010; Olivieri et al., 2010; Schirle and MacRae, 2012; Handler et al., 2013; Baena-Lopez et al., 2013; Vagin et al., 2013; Huang et al., 2014; Izumi et al., 2016; Nishida et al., 2018). Here, we report that Armi localizes to both nuage and mitochondria and couples the ping-pong cycle to the phased piRNA pathway. Mutations that block phased piRNA production trap Armi on mitochondria. Armi ATPase mutants do not support phased piRNA production and cause Armi to inappropriately bind mRNA instead of piRNA precursors. We propose that Armi shuttles between nuage and mitochondria, feeding precursor piRNAs generated by Ago3 cleavage into the Zucchini-dependent production of Aubergine- and Piwi-bound piRNAs on the mitochondrial surface.
RESULTS
Nuage and Mitochondria are Physically Separate in Nurse Cells
In principle, physical association of nuage and mitochondria could explain the coupling of ping-pong amplification to phased piRNA production. Although nuage often associates with mitochondria (Eddy, 1975; Kloc et al., 2014), particularly in mammals where a nuage-like “cement” resides in the interstices of clustered mitochondria (Eddy, 1974), electron microscopy of Drosophila nurse cells rarely finds nuage apposed to mitochondria (Mahowald, 1970; Dapples and King, 1970; Mahowald, 1971; Liang et al., 1994; Wilsch-Bräuninger et al., 1997; Jaglarz et al., 2011). Quantitative immunofluorescence microscopy further confirms that most mitochondria do not contact nuage (Figure 1A). In stage 3 egg chambers, the 15 germline nurse cell nuclei are well separated, allowing unambiguous detection of cytoplasmic proteins (Figure S1A); immunostaining of these cells readily detected the piRNA biogenesis proteins Aub, a representative ping-pong factor, and Zuc, a representative phased piRNA pathway component. The germline-specific DEAD box protein Vasa served as a marker for nuage, while ATP synthase complex V alpha subunit (ATP5A) identified the inner mitochondrial membrane. In wild-type ovaries, Vasa-containing nuage puncta surround the nurse cell nuclei. Mitochondria are evenly distributed in the cytoplasm, with a tendency to clump near the nucleus (Figure 1A). Quantification of the fluorescent signal detected little Vasa coincident with mitochondria: 18% ± 9% of Vasa signal overlapped with ATP5A, while 88% ± 7% overlapped with Aub (n = 54 images from three egg chambers). Because the wavelength of the emitted light limits the resolution of fluorescence microscopy (λ/[2 × numerical aperture] where λemitted = 525 nm for Alexa Fluor 488), the limit of resolution for our experiments, ~188 nm, is not significantly smaller than the typical nuage puncta (Jaglarz et al., 2011). Consequently, fluorescence microscopy cannot distinguish between the biological finding of few mitochondria associating with nuage and artifacts caused by diffracted light.
Figure 1. Armi localizes to both nuage and mitochondria, physically separate sites of piRNA biogenesis.
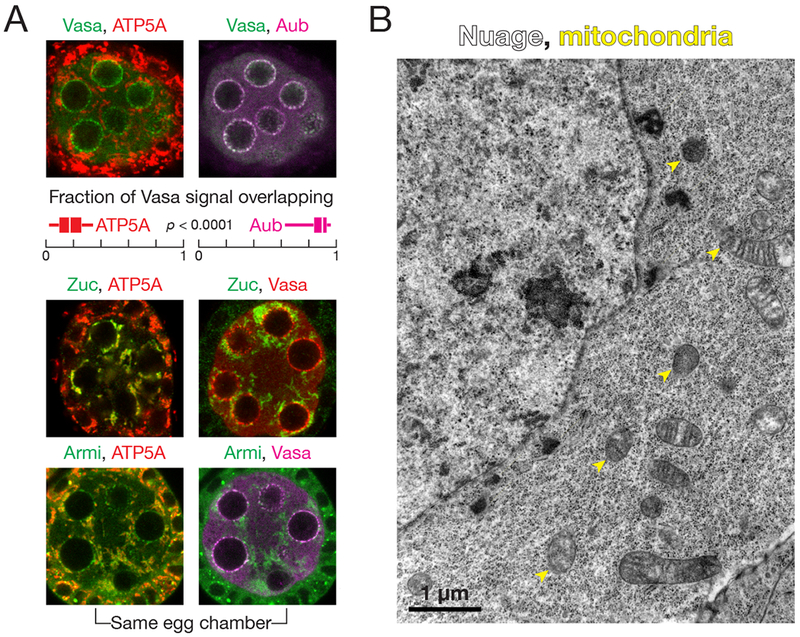
(A) Immunofluorescence detection of Vasa, ATP5A and Aub in wild-type stage 3 egg chambers. Box plots show the fraction of Vasa signal overlapping with ATP5A or Aub. Box comprises the 25th percentile and the 75th percentile of data, with the median indicated; whiskers mark 1.5 × interquartile range. p-value was calculated using the two-tailed Wilcoxon matched-pairs signed rank test. Immunofluorescence of Zuc-3×FLAG or Armi to detect their colocalization with ATP5A or Vasa in wild-type stage 3 egg chambers.
(B) Transmission electron microscopy image of a wild-type stage 3 egg chamber.
See also Figure S1.
To further test our observation that few mitochondria abut nuage, we used transmission electron microscopy (TEM), a technique that readily distinguishes between nuage, electron-dense, fibrous granules lacking a membrane, and mitochondria, double-membrane-bound organelles with characteristic internal cristae. In TEM images of stage 1 to stage 8 egg chambers, nuage and mitochondria rarely touched: in two independent experiments, none of 31 and two of 191 nuage puncta contacted mitochondria (Figure 1B and S1B). We conclude that Drosophila nurse cell nuage and mitochondria are physically separate, suggesting that one or more piRNA pathway proteins help escort the precursor RNAs generated by the ping-pong cycle from nuage to the mitochondrial surface for processing into phased piRNAs.
Zuc Localizes to Mitochondria, but not Nuage
In cultured ovarian somatic cells, tagged, overexpressed Zuc localizes to mitochondria through its N-terminal mitochondrial localization signal peptide (Saito et al., 2010; Handler et al., 2013). We examined the localization of endogenous Zuc in the germline using a fly strain in which endogenous Zuc bears a C-terminal 3×FLAG tag (Ge et al., 2016). Homozygous flies expressing only Zuc-3×FLAG showed no detectable defects in fertility or piRNA biogenesis, and the tagged Zuc, expressed from its native chromosomal location, was readily detected using anti-FLAG antibody. Essentially all anti-FLAG immunofluorescence coincided with ATP5A; little anti-FLAG staining overlapped with Vasa (Figure 1A). We conclude that within the limits of immunofluorescent detection, Zuc localizes to mitochondria but not nuage.
These experiments cannot exclude the possibility that a small but biologically important population of Zuc is present in nuage and contributes to piRNA biogenesis. To test this possibility, we asked whether Zuc interacts with nuage proteins. We immunoprecipitated 3×FLAG-tagged endogenous Zuc from ovary lysates and identified associated proteins by mass spectrometry. Under native conditions, mass spectrometry failed to detect any known piRNA biogenesis proteins co-immunoprecipitating with Zuc. Because Zuc-interacting proteins may dissociate upon cell lysis, we surveyed a series of membrane-permeable, reversible chemical crosslinkers for their ability to stabilize Zuc protein-protein interactions. Crosslinking has the added benefit of withstanding stringent washes, thus ensuring that any detected interactions occurred in the cell and not after lysis (Mili and Steitz, 2004). The mouse Armi homolog Mov10l1 co-immunoprecipitates with PLD6, the mouse Zuc homolog, when the two proteins are co-expressed in cultured mammalian somatic cells (Vourekas et al., 2015). Therefore, we tested whether Armi co-immunoprecipitated with Zuc using ovaries treated before lysis with the primary amine-to-primary amine crosslinkers paraformaldehyde, disuccinimidyl tartrate, ethylene glycol bis(succinimidyl succinate), or dithiobis(succinimidyl propionate), or the sulfhydryl-to-sulfhydryl crosslinker dithio-bis-maleimidoethane (DTME). Among these, DTME best stabilized the interaction between Zuc and Armi (Figure S1C).
To more comprehensively identify Zuc-associated proteins, we immunoprecipitated 3×FLAG-tagged Zuc from DTME-crosslinked ovaries, and analyzed Zuc-associated proteins by mass spectrometry (Table 1). Among the 39 proteins reproducibly co-immunoprecipitated with Zuc (n = 3), 17 were mitochondrial proteins not known to participate in piRNA biogenesis. An additional four were piRNA pathway proteins known to localize to mitochondria, including the phased piRNA biogenesis proteins Armi, Gasz, and Minotaur (Olivieri et al., 2010; Saito et al., 2010; Handler et al., 2013; Baena-Lopez et al., 2013; Vagin et al., 2013), as well as the mitochondrial outer membrane protein Papi, which is required both to maintain piRNA levels and to trim pre-piRNAs (Honda et al., 2013; Han et al., 2015a; Izumi et al., 2016; Hayashi et al., 2016; Nishida et al., 2018). Sister of Yb (SoYb), a piRNA pathway protein that colocalizes with Armi to cytoplasmic clouds likely to be mitochondria (Handler et al., 2011), also co-purified with Zuc. In all, 21 proteins known to localize to mitochondria or function in phased piRNA production co-purified with Zuc, which strongly suggests that phased piRNAs are produced on the outer surface of mitochondria. In contrast, only a single known component of nuage was enriched in the Zuc immunoprecipitate: Armi.
Table 1.
All data are mean ± S.D. for weighted spectra count (n = 3) and FDR < 0.05 for experimental immunoprecipitate (IP) versus control.
| (A) piRNA biogenesis proteins specifically co-immunoprecipitated with Zuc, Armi or ArmiK729A. See also Table S1. | |||||||
|---|---|---|---|---|---|---|---|
| Protein | Localization | Zuc IP | Control IP for Zuc | Armi IP | ArmiK729A IP | Control IP for Armi | |
| Zucchini (Zuc) | Mitochondria | 31 ± 6 | 0 | – | – | – | |
| Gasz | Mitochondria | 15 ± 5 | 0 | 7 ± 2 | 8.3 ± 0.6 | 0 | |
| Papi | Mitochondria | 11 ± 1 | 0 | – | – | – | |
| Armitage (Armi) | Mitochondria & nuage | 38 ± 3 | 5 ± 4 | 200 ± 90 | 180 ± 80 | 10 ± 20 | |
| Minotaur (Mino) | Mitochondria & ER | 16 ± 4 | 0.7 ± 0.6 | 11 ± 5 | 14 ± 2 | 0.3 ± 0.6 | |
| Sister of Yb (SoYb) | Mitochondria? | 4 ± 4 | 0 | 10 ± 10 | 20 ± 10 | 0 | |
| Vreteno (Vret) | Nuage | – | – | 4 ± 5 | 4 ± 5 | 0 | |
| Shutdown (Shu) | Nuage | – | – | 8 ± 4 | 16 ± 5 | 1 ± 1 | |
| Argonaute 3 (Ago3) | Nuage | – | – | 7 ± 7 | 4 ± 5 | 0.3 ± 0.6 | |
| Spindle E (Spn-E) | Nuage | – | – | 9 ± 9 | 9 ± 9 | 1 ± 2 | |
| Tapas | Nuage | – | – | 8 ± 4 | 8 ± 2 | 2 ± 2 | |
| Aubergine (Aub) | Nuage | – | – | 40 ± 20 | 40 ± 20 | 10 ± 7 | |
| Brother of Yb (BoYb) | Nuage | – | – | 5 ± 6 | 3 ± 4 | 0.3 ± 0.6 | |
| Nibbler (Nbr) | Nuage | – | – | 10 ± 4 | 14 ± 3 | 3.3 ± 0.6 | |
| Qin | Nuage | – | – | 5 ± 7 | 2 ± 4 | 0 | |
| P-element induced wimpy testis (Piwi) | Nucleus | – | – | 16 ± 6 | 20 ± 10 | 9 ± 6 | |
| Vasa (Vas) | Nuage | – | – | 26 ± 6 | 33 ± 2 | 14 ± 2 | |
| Tudor (Tud) | Nuage | – | – | 40 ± 30 | 30 ± 30 | 20 ± 20 | |
| (B) Proteins not known to function in piRNA biogenesis that co-immunoprecipitated with Zuc | |||||||
| Mitochondrial Proteins | Others | ||||||
| Fly name | Human name | Zuc IP | Control IP | Fly name | Human name | Zuc IP | Control IP |
| Miro | Rhot1 | 10 ± 3 | 0 | CG12360 | Trabd | 12 ± 5 | 0 |
| Miga | Miga2 | 11 ± 2 | 1 ± 1 | CG10880 | – | 11 ± 3 | 0.3 ± 0.6 |
| CG9855 | March5 | 5 ± 2 | 0 | nmd | Atad1 | 16 ± 6 | 1 ± 1 |
| Spoon/Yu | Akap1 | 10 ± 4 | 0.7 ± 0.6 | Aldh-III | Aldh3b1 | 9 ± 2 | 0 |
| Marf | Mfn2 | 27 ± 4 | 2 ± 2 | CG7705 | – | 10 ± 3 | 0 |
| Usp30 | Usp30 | 3 ± 4 | 0 | Faf | Faf2 | 7 ± 5 | 0 |
| Men | Me1 | 13 ± 9 | 1 ± 2 | Pex3 | Pex3 | 5 ± 1 | 0.3 ± 0.6 |
| Mtch | Mtch2 | 12 ± 6 | 2 ± 2 | CG2316 | Abcd2 | 5 ± 2 | 0 |
| Ptp61F | Ptpn1 | 5 ± 1 | 0.3 ± 0.6 | Myt1 | PKMYT1 | 4 ± 2 | 0 |
| CG7639 | SAMM50 | 4 ± 3 | 0 | CG1291 | Alg2 | 5 ± 3 | 0.3 ± 0.6 |
| CG1665 | Marc1 | 15 ± 7 | 1.3 ± 0.6 | CG8735 | LNP | 6.3 ± 0.6 | 0.3 ± 0.6 |
| CG7461 | Acadvl | 3 ± 1 | 0 | CG6550 | CDKAL1 | 4 ± 3 | 0 |
| Tom70 | Tomm70a | 28 ± 7 | 5 ± 1 | Sec63 | Sec63 | 7 ± 3 | 1 ± 2 |
| CG3394 | SLC27A1 | 5 ± 4 | 1 ± 1 | CG9853 | Get4 | 5 ± 3 | 0.3 ± 0.6 |
| Pmp70 | Abcd3 | 11 ± 5 | 2 ± 3 | CG6744 | Exd2 | 3 ± 4 | 0 |
| iPLA2-VIA | Pla2g6 | 7 ± 3 | 1.3 ± 0.6 | CG7546 | Bag6 | 10 ± 6 | 2 ± 1 |
| Acsl | Acsl3 | 17 ± 2 | 7 ± 2 | CG6089 | Tmem214 | 13 ± 6 | 2 ± 1 |
Armi-Associated Proteins Suggest Armi Shuttles from Nuage to Mitochondria
For pre-pre-piRNA to be made in the nuage and processed into phased piRNAs on mitochondria, such piRNA precursors need to be delivered from nuage, across the cytoplasm, to the outer face of mitochondria. A likely candidate to serve this role is Armi, a protein found in both compartments (Figure 1; Cook et al., 2004; Handler et al., 2013; Huang et al., 2014; Pandey et al., 2017). To test whether Armi participates in piRNA biogenesis in both nuage and mitochondria, ovaries from flies producing transgenic 3×FLAG-(Myc)6-tagged Armi (FLAG-MYC-Armi) in the germline were crosslinked with DTME before cell lysis, followed by FLAG immunoprecipitation and mass spectrometry. Flies of the same genetic background, but not carrying the transgene served as the control. Of the 95 proteins reproducibly associated with Armi (n = 3), just two (Clueless and Mitochondrial assembly regulatory factor) were mitochondrial proteins without known functions in piRNA biogenesis (Table S1). In contrast, 17 were known piRNA factors (Table 1): the mitochondrial proteins Gasz, Minotaur, SoYb, and Papi; the nuage proteins Vreteno, Shutdown, Ago3, Spindle E (Spn-E), Tapas, Aub, Brother of Yb, Nibbler, Qin, Vasa, and Tudor; and the trailing piRNA-guided protein Piwi. Piwi has been previously shown to interact with Armi (Olivieri et al., 2010; Saito et al., 2010; Haase et al., 2010). We conclude that Armi participates in the piRNA pathway both in nuage and on mitochondria.
In theory, Armi might localize to different subcellular compartments at different stages of egg chamber maturation, or Armi may localize to both nuage and mitochondria within a single nurse cell. To differentiate between these two possibilities, we examined the intracellular location of Armi, ATP5A and Vasa in individual stage 3 egg chambers. Armi colocalized with both ATP5A and Vasa in every nurse cell examined (n = 60) (Figure 1A). Thus, Armi is likely to participate in the delivery of pre-pre-piRNAs from nuage to mitochondria.
Loss of Phased piRNA Production Depletes Armi from the Nuage
If Armi delivers pre-pre-piRNAs from nuage to mitochondria, but plays no other role in piRNA amplification, then loss of Armi should block production of phased piRNAs without perturbing ping-pong. Consistent with this prediction, removing Armi genetically or by germline RNAi disrupts piRNA phasing but not ping-pong piRNA amplification (Handler et al., 2011; Han et al., 2015a). Loss of Zuc similarly disrupts piRNA phasing but not ping-pong (Han et al., 2015a).
Recycling of Armi from mitochondria appears to require phased piRNA production. First, compared to wild type, less Armi was associated with nuage and more with mitochondria in zucH169Y mutant ovaries, which produce appropriately localized but catalytically inactive Zuc (Figures 2A, S2 and S3). Armi was similarly depleted from nuage but not mitochondria in minotaurz3-5967 mutant ovaries (Figures 2A and S4). These data suggest that recycling of Armi from mitochondria back to nuage requires phased piRNA production. In support of the idea that wild-type Armi distribution requires both piRNA amplification and phased piRNA synthesis, inactivating the ping-pong pathway by mutating the catalytic triad of Ago3 from DDH to AAH traps Armi in nuage and increases the amount of Armi associated with Ago3 (Huang et al., 2014).
Figure 2. Phased piRNA production promotes nuage localization of Armi.
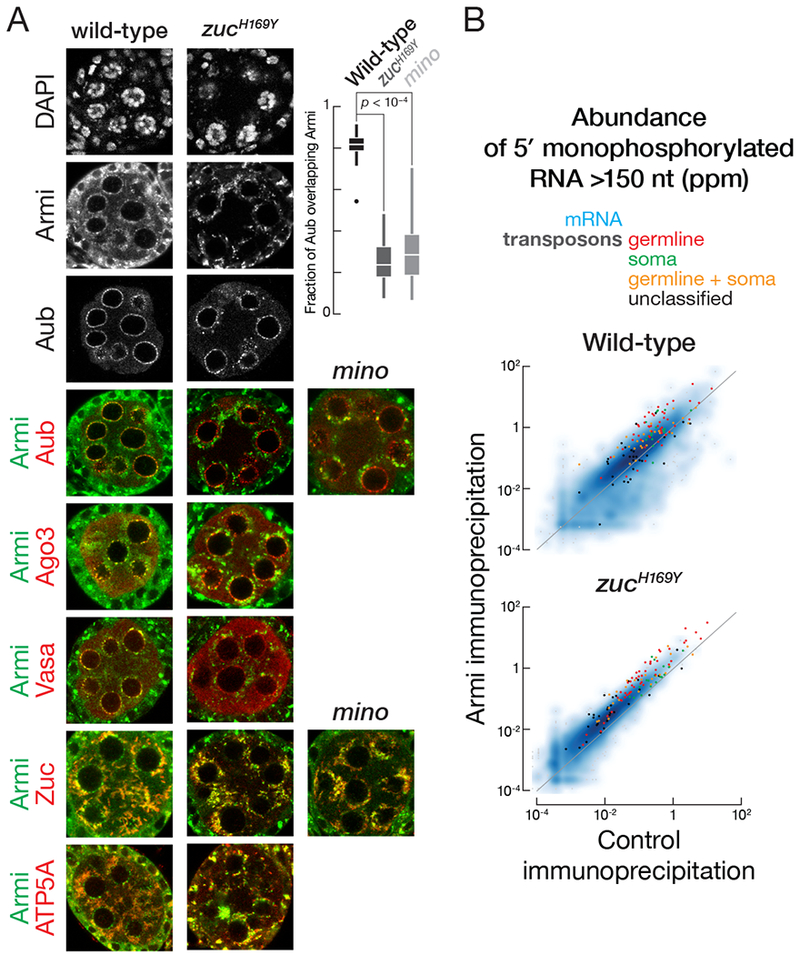
(A) Immunofluorescence co-detection of Armi and Aub, Ago3, Vasa, Zuc-3×FLAG or ATP5A in wild-type or zucH169Y stage 3 egg chambers or of Armi and Aub or Zuc-3×FLAG in minotaurz3-5967 stage 3 egg chambers. Box plots show the fraction of Aub signal overlapping with Armi in wild-type, zucH169Y or minotaurz3-5967 stage 3 egg chambers. Box comprises the 25th percentile and the 75th percentile of data, with the median indicated; whiskers mark 1.5 × interquartile range. p-value was calculated using the two-tailed Mann Whitney test.
(B) Scatter plots of the abundance of transposon- or gene-mapping 5′ monophosphorylated RNA >150 nt co-immunoprecipitated with Armi or control in wild-type or zucH169Y mutant ovaries.
Armi Binds Pre-pre-piRNAs During Ping-pong and Phased piRNA Biogenesis
If Armi delivers pre-pre-piRNAs from nuage to mitochondria, then Armi should bind pre-pre-piRNAs. To test this prediction, we immunoprecipitated FLAG-Myc-Armi from the germline and sequenced associated RNAs >150 nt long and bearing a 5′ monophosphate, the terminal structure found on pre-pre-piRNAs (Han et al., 2015a; Gainetdinov et al., 2018). Ovaries were crosslinked with paraformaldehyde before lysis to ensure detection of only protein-RNA interactions present in vivo. Ovaries without the FLAG-Myc-Armi transgene served as a negative control. Because FLAG-Myc-Armi was expressed only in the germline, sequencing data was normalized to the background of reads mapping uniquely to the somatic follicle cell-specific piRNA cluster flamenco. Immunoprecipitated Armi was specifically associated with transposon mapping RNAs with the characteristics of pre-pre-piRNAs (Figure 2B). First, a higher fraction of transposon-derived reads was antisense in the Armi immunoprecipitates than in the control (61% vs. 45%), and, second, the Armi-associated RNA favored a 5′ uridine (Figure 3A).
Figure 3. Armi interacts with pre-pre-piRNA during the nuage and mitochondrial phases of piRNA production.
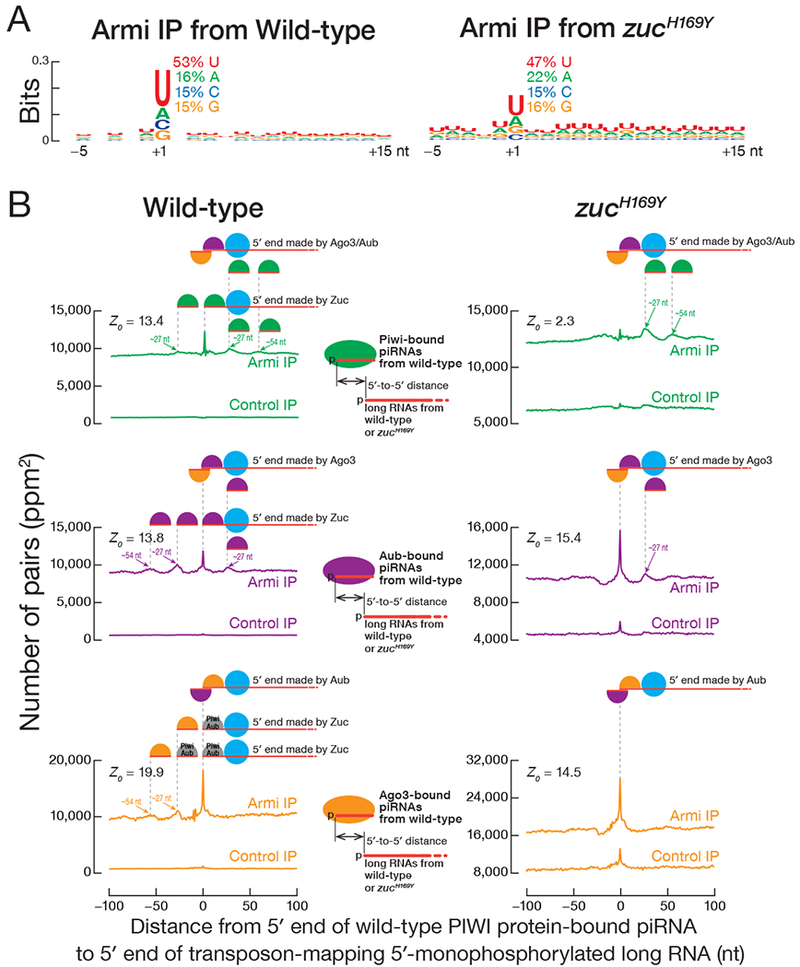
(A) Genomic nucleotide bias around the 5′ ends (nt position 1) of 5′ monophosphorylated antisense transposon RNA >150 nt co-immunoprecipitated with Armi. Each RNA 5′ end was weighted equally, ignoring abundance.
(B) Distance on the same genomic strand from 5′ ends of PIWI protein-bound piRNAs to 5′ ends of 5′ monophosphorylated long RNAs co-immunoprecipitated with Armi or control in wild-type or zucH169Y mutant ovaries.
The current model for Drosophila germline piRNA biogenesis posits that in the nuage, an initiator piRNA directs Ago3 (and to a lesser extent, Aub) to cleave a piRNA precursor transcript, generating a 5′ monophosphorylated end that binds Aub. This Aub-bound pre-pre-piRNA is then transferred to the mitochondrial surface to be cleaved by Zuc. Zuc cleavage generates an Aub-bound responder pre-piRNA from the 5′ end of the pre-pre-piRNA, while the rest of the pre-pre-piRNA is processed into a chain of trailing piRNAs loaded into Piwi and, to a smaller extent, Aub. The hypothesis that Armi transports the Aub-bound pre-pre-piRNA from nuage to mitochondria predicts that Armi interacts with two types of transposon-derived 5′ monophosphorylated RNA: (1) RNAs whose 5′ ends are established by Ago3 or Aub cleavage, i.e., ping-pong piRNA precursors; and (2) RNAs whose 5′ ends are made by Zuc and that generate Piwi- or Aub-bound trailing piRNAs.
Our data suggest that Armi binds pre-pre-piRNAs produced both in the ping-pong and phased piRNA biogenesis pathways. First, we compared the sequences of mature piRNAs bound to Piwi (Han et al., 2015a) with the sequences of transposon-mapping, 5′ monophosphorylated RNAs—i.e., putative pre-pre-piRNAs—co-immunoprecipitated with Armi. Consistent with Armi binding pre-pre-piRNAs in the phased piRNA pathway, the 5′ ends of RNAs associated with Armi often corresponded to a Piwi-bound piRNA (Z0 = 13.4). That is, Piwi-bound, phased piRNAs typically arise from the 5′ ends of Armi-bound pre-pre-piRNAs (Figure 3B, top left panel). Second, the 5′ ends of RNAs associated with Armi also frequently corresponded to an Aub- or Ago3-bound piRNA (Han et al., 2015a, Z0 = 13.8 middle panel, left, and Z0 = 19.9, bottom panel, left in Figure 3B), indicating that Armi associates with the immediate precursors of responder piRNAs in the ping-pong piRNA pathway.
The current model also predicts that in the absence of Zuc, only pre-pre-piRNAs arising from ping-pong slicing are produced. That is, loss of Zuc should restrict pre-pre-piRNA generation to the precursors whose 5′ ends are generated by Ago3 or Aub cleavage. Compared to wild-type Armi-associated long RNA, fewer 5′ ends of the Armi-associated RNAs from zucH169Y ovaries corresponded to Piwi-bound piRNAs from wild-type (Figures 3B, compare the two top panels). Instead, the 5′ ends of the Armi-associated RNAs from zucH169Y corresponded to Aub- or Ago3-bound piRNAs found in wild-type (Figure 3B, middle- and bottom-right panels). Because trailing piRNAs are not produced in zucH169Y mutants (Han et al., 2015a; Mohn et al., 2015), the 5′ ends of the Piwi- or Aub-bound, but not Ago3-bound piRNAs from wild-type ovaries map 27 nt or 54 nt (one or two pre-piRNA lengths) downstream from the 5′ ends of the pre-pre-piRNAs associated with Armi in zucH169Y mutant (Figure 3B). In fact, pre-pre-piRNAs whose 5′ ends correspond to an Aub-bound responder piRNA accumulate in zucH169Y. That is, the pre-pre-piRNAs whose 5′ ends correspond to Piwi- or Aub-bound trailing piRNAs are not generated without Zuc function. We conclude that in addition to binding pre-pre-piRNAs that fuel phased piRNA production, Armi also binds pre-pre-piRNAs typically generated by Ago3 guided by an initiator piRNA—the piRNA pathway intermediates that must be delivered from the nuage to the mitochondria.
Armi ATPase Activity Enables Correct Substrate Selection
The Armi helicase domain belongs to the Upf1-like Superfamily 1. Modeling the Armi helicase core predicts that its structure is highly similar to human Upf1 (sequence similarity: 45%; coverage of the alignment: 90%), especially the residues at the ATP-binding pocket, which are completely conserved (Figure 4A). Upf1 is a core factor in nonsense-mediated mRNA decay (NMD), and mutating lysine 498 to alanine in helicase motif I of human Upf1 disrupts ATP- but not RNA-binding; Upf1K498A loses its selectivity for mRNAs with premature stop codons and instead binds RNA promiscuously (Weng et al., 1996; Weng et al., 1998; Lee et al., 2015).
Figure 4. ArmiK729A ATPase mutant fails to support phased piRNA production.
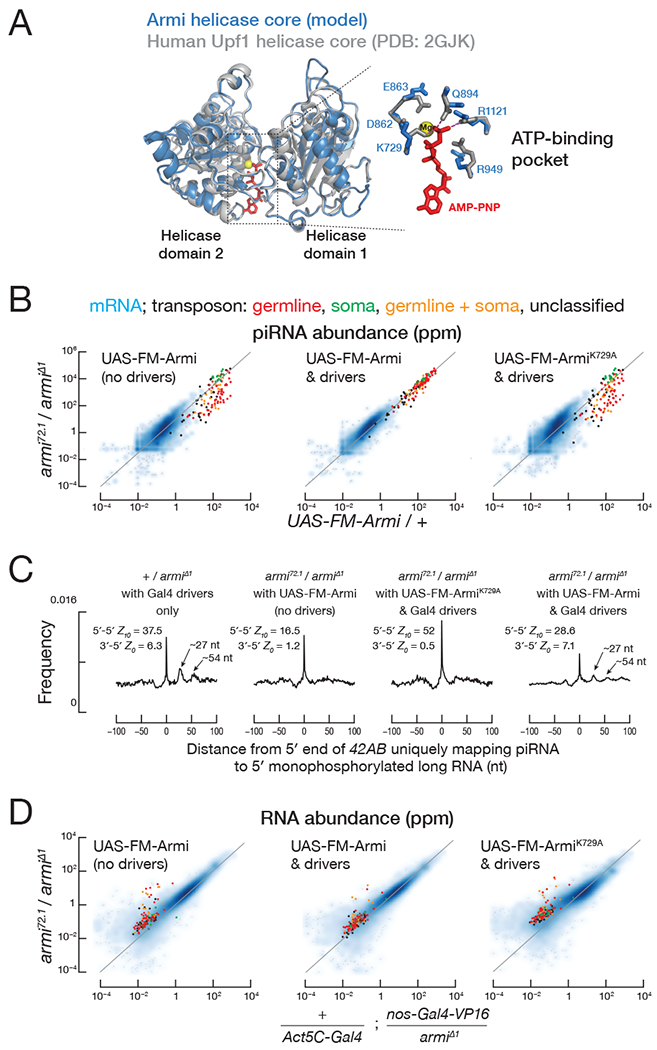
(A) Predicted Armi helicase core superimposed on the human Upf1 helicase core (Cheng et al., 2007). The enlarged view of the Armi ATP-binding pocket shows amino acids surrounding the adenosine-5′-(β,γ-imido)triphosphate (“ANP”) and a magnesium ion present in the Upf1 structure.
(B) Scatter plot of the abundance of transposon- or gene-mapping piRNAs from the indicated genotype.
(C) Distance on the same genomic strand from 5′ ends of piRNAs uniquely mapping to 42AB to the 5′ ends of 5′ monophosphorylated RNA >150 nt from the indicated genotype. Z-scores indicate the significance of the 5′-to-5′ distance = 10 nt on opposite genomic strands (ping-pong) and the 3′-to-5′ distance = 1 nt on the same genomic strand (phasing) for piRNAs.
(D) Scatter plot of the abundance of transposon- or gene-mapping RNA >150 nt or piRNAs from the indicated genotype.
See also Figure S5.
The corresponding amino acid substitution in Armi, K729A, does not support piRNA biogenesis: unlike the transgene expressing wild-type Armi, transgenic ArmiK729A failed to rescue piRNA production in armi72.1/Δ1 ovaries, which lack germline Armi. Compared to the wild-type transgene, the abundance of piRNAs from germline-specific transposons fell nearly tenfold in ArmiK729A ovaries (median = 11%, n = 47), a level statistically indistinguishable from armi72.1/Δ1 alone (median = 9%; Wilcoxon matched-pairs signed rank test p = 0.07; Figure 4B). Like armi72.1/Δ1, ArmiK729A ovaries produced mainly ping-pong piRNAs; in contrast, armi72.1/Δ1 rescued with a wild-type Armi transgene made both ping-pong and phased piRNAs (Figure 4C). Unlike wild-type transgenic Armi, the ArmiK729A transgene failed to support germline transposon silencing (Figure 4D), and the flies showed defects in both egg laying and hatching (Figures S5A). The defect does not reflect a dearth of ArmiK729A: the abundance of the transgenic wild-type and K729A mutant proteins were comparable (Figure S5B). Neither increasing nor decreasing Armi abundance relative to the endogenous level had a measurable effect on piRNA levels, demonstrating that Armi abundance does not limit piRNA production (Figure S5C).
To test whether ArmiK729A interacts with piRNA precursors, we sequenced the 5′ monophosphorylated RNA co-immunoprecipitated with transgenic ArmiK729A in the germline of wild-type ovaries. Compared to transgenic wild-type Armi, ArmiK729A bound 18-fold more mRNA degradation products, as judged by the ArmiK729A median enrichment over wild-type Armi (Figure 5A). Both ArmiK729A and wild-type Armi bound germline transposon-derived pre-pre-piRNAs (Figures 2B and 5A). These data suggest that, like Upf1K498A, ArmiK729A loses substrate selectivity when it cannot bind ATP.
Figure 5. Armi ATPase activity enables correct substrate selection.
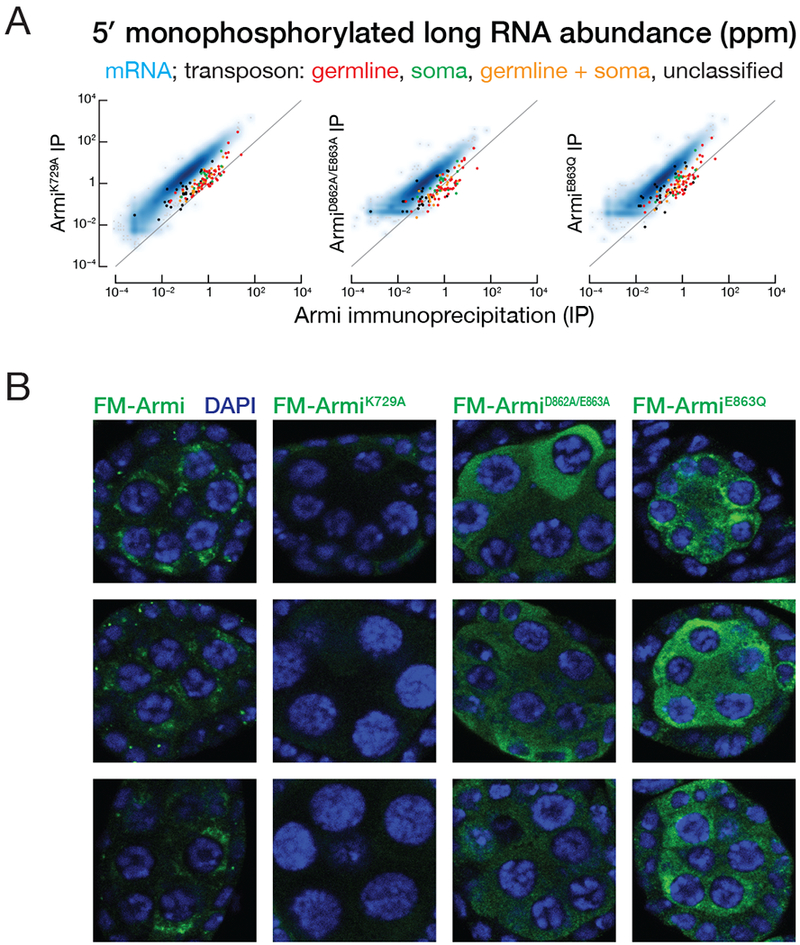
(A) Scatter plot of the abundance of transposon- or gene-mapping 5′ monophosphorylated RNA >150 nt co-immunoprecipitated with transgenic FLAG-Myc-ArmiK729A, FLAG-Myc-ArmiDE862,863AA, FLAG-Myc-ArmiE863Q versus FLAG-Myc-Armi.
(B) Immunofluorescence detection using anti-FLAG antibody of transgenic FLAG-Myc-Armi, FLAG-Myc-ArmiK729A, FLAG-Myc-ArmiDE862,863AA or FLAG-Myc-ArmiE863Q in armi mutant germ cells from stage 3 egg chambers.
See also Figure S6.
Artificially tethering Armi to a transcript triggers piRNA production (Rogers et al., 2017; Pandey et al., 2017). Yet the mRNA degradation products associated with ArmiK729A do not fuel an increase in mRNA-derived piRNAs (Figure S5C). Why do the mRNA fragments associated with ArmiK729A fail to enter the phased piRNA pathway? Perhaps ATP-binding or hydrolysis by Armi is required for additional steps in phased piRNA biogenesis. Alternatively, mRNA degradation products may be associated with proteins that prevent their delivery to mitochondria.
Another Upf1 mutant, Upf1D637A/E638A, still binds ATP but binds RNA promiscuously (Lee et al., 2015). Similarly, mutant Armi bearing the corresponding amino acid substitution, D862A/E863A, inappropriately bound mRNA degradation products (Figure 5A). Finally, mutation of the DEAD motif (E339Q) of the DEAD-box protein Vasa slows release of the products of ATP hydrolysis—ADP and inorganic phosphate—and traps Vasa on RNA (Xiol et al., 2014). The analogous amino acid substitution in Armi, E863Q, also increased the association of Armi with mRNA degradation products (Figure 5A). We conclude that ATP binding and hydrolysis, as well as subsequent release of the resulting ADP and inorganic phosphate, are required for Armi to discriminate between pre-pre-piRNAs intended to produce transposon-silencing piRNAs and mRNA degradation products that are normally excluded from the piRNA pathway.
Unlike the punctate perinuclear localization of transgenic wild-type Armi, ArmiK729A was dispersed throughout the cytoplasm, distributing the immunofluorescence signal over a larger area (Figure 5B, Figure S6). Yet ArmiK729A remains associated with the same set of piRNA pathway proteins that co-immunoprecipitate with wild-type Armi (Table 1), including the nuage proteins Vreteno, Shutdown, Ago3, Spn-E, Tapas, Aub, Brother of Yb, Nibbler, Qin, and Vasa, and the mitochondrial proteins Gasz, Minotaur, SoYb and Papi. These data suggest that RNA-protein rather than protein-protein interactions with Armi determine its subcellular localization. Supporting this view, both ArmiD862A/E863A and ArmiE863Q are partially dispersed in the cytoplasm (Figure 5B). We hypothesize that the loss of pre-pre-piRNA selectivity leads to Armi inappropriately binding mRNA degradation products, in turn causing it to disperse throughout the cytoplasm.
DISCUSSION
Armi Moves between Nuage and Mitochondria
Our data suggest that Armi links the nuage and mitochondrial phases of piRNA biogenesis by associating with piRNA precursors and protein factors in both compartments. We propose that pre-pre-piRNAs are generated in the nuage by initiator piRNA-directed cleavage, bound by Aub, then accompanied by Armi as they transit to the mitochondria, where Zuc generates both responder ping-pong and trailing phased pre-piRNAs. We cannot currently determine whether transfer of Armi-bound pre-pre-piRNAs is an active process or simply the consequence of passive diffusion. In either case, the interactions of Armi with mitochondrial proteins likely serve to anchor Armi-associated pre-pre-piRNAs to the mitochondrial surface when it arrives at the organelle. A functional piRNA pathway is required for release of Armi from nuage or mitochondria: catalytically inactive Ago3, a mutation that blocks ping-pong cleavage, traps Armi in nuage (Huang et al., 2014), while zucH169Y or minotaurz3-5967, which cannot generate phased piRNAs, traps Armi on mitochondria. Finally, in Armi mutants that promiscuously bind mRNA, Armi disperses throughout the cytoplasm.
We envision that in nuage, Armi binds to 5′ monophosphorylated pre-pre-piRNAs generated by Ago3-catalyzed cleavage of piRNA precursors during ping-pong amplification (Figure 6). The resulting ternary complex of Armi, Aub, and pre-pre-piRNA is then released into the cytoplasm. When the complex encounters the surface of a mitochondrion, it can be retained by protein-protein interactions, such as Armi binding to Gasz or other mitochondrial proteins (Handler et al., 2013). On the mitochondrial outer membrane, Zuc and other piRNA pathway proteins act to release an Aub-bound responder pre-piRNA and to convert the rest of the pre-pre-piRNA into trailing piRNAs bound to Piwi. In this model, the complete conversion of a pre-pre-piRNA into pre-piRNAs releases Armi to the cytoplasm, allowing it to recycle to the nuage by diffusing through the cytosol and then once again stably binding an Aub-bound pre-pre-piRNA in nuage.
Figure 6. A model for the role of Armi in piRNA biogenesis.
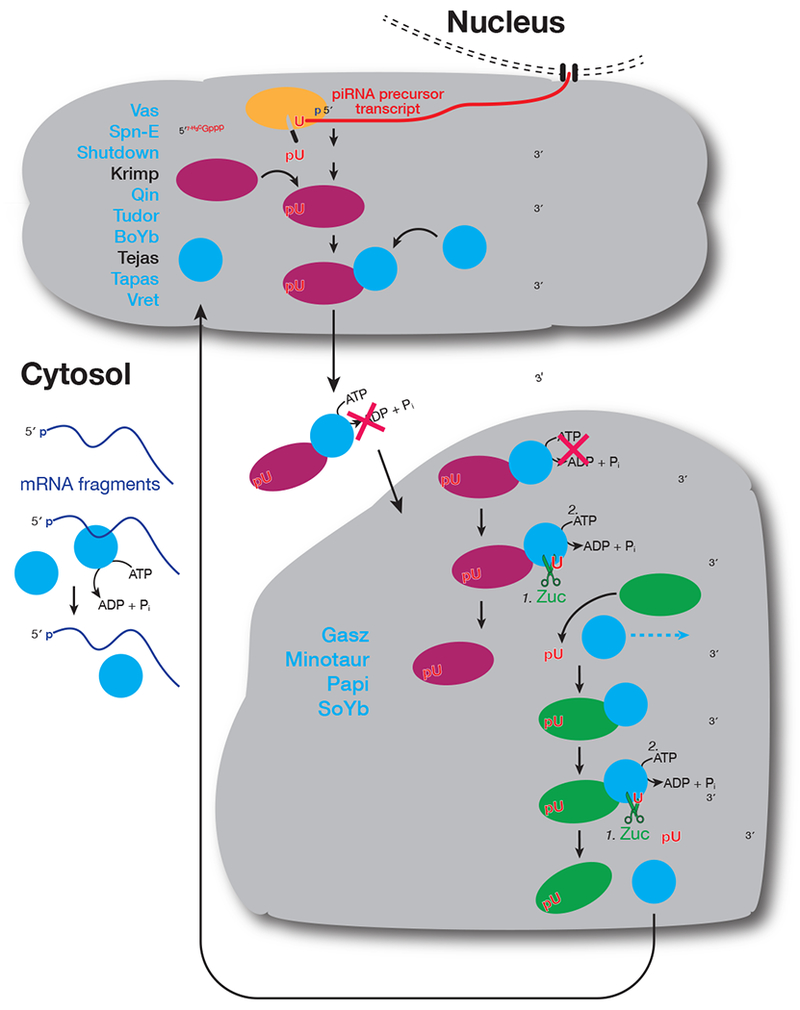
In addition to the three PIWI proteins and Zuc, other piRNA factors interacting with Armi are shown in blue.
Our model predicts that Aub should be detected on mitochondria (Figure 6). However, we did not detect Aub on mitochondria by immunofluorescence in either wild-type or zucH169Y mutants (Figure 2A, Figure S2 and S3); Aub was also not recovered crosslinked to immunopurified Zuc (Table 1A), suggesting that if a subpopulation of Aub is present on mitochondria, it is below the detection limit of immunofluorescence or immunoprecipitation. A potential explanation for our failure to detect mitochondrial Aub is that the rate-determining step for Zuc liberating an Aub-bound pre-piRNA is the speed of the encounter of Zuc and an Aub-bound pre-pre-piRNA. Similarly, in zucH169Y mutants, the Aub-responder pre-piRNA complex may still be released by Ago3-mediated cleavage, severing the RNA connection linking Armi with Aub.
The movement of piRNA proteins between nuage and cytoplasm has been reported for Vasa, Tudor, Aub, Tejas, Spn-E and Ago3 (Xiol et al., 2014; Webster et al., 2015; Andress et al., 2016). Except for Tejas, all of these proteins associate with Armi (Table 1A), suggesting that they accompany Armi when it moves from the nuage into the cytoplasm. Interestingly, none of these five proteins co-immunoprecipitated with Zuc, suggesting that they dissociate from the Armi-pre-pre-piRNA complex in the cytoplasm or after reaching the mitochondrial outer membrane but before Zuc engages Armi (Figure 6). In fly germline nurse cells, nuage and mitochondria are physically separate, a property that facilitated our study of Armi re-localization between the two compartments. However, in some animal germ cells, such as those of mice and frogs, nuage and mitochondria are directly apposed (Eddy, 1975). We anticipate that Armi plays a similar role in these species, escorting pre-pre-piRNA generated in nuage to the mitochondria for conversion into piRNAs. However, without the clear separation of nuage and mitochondria of flies, visualizing Armi dynamics will likely be challenging.
Uncharacterized proteins associate with Zuc or Armi
At least 127 proteins interact with Zuc or Armi, including 32 uncharacterized proteins (Tables 1 and S1), some of which may correspond to piRNA pathway proteins whose functions overlap with those of known piRNA factors, causing them to be missed by previous screens. For example, Armi interacts with the Tudor domain-containing protein CG9925, which is predicted to be one of the two fly orthologs of mammalian TDRD1 (Table S1). Zuc interacts with the Tudor domain-containing proteins Spoon and Papi, a pair predicted to have similar functions (Handler et al., 2011). Individually, depleting the paralogous proteins CG9925 or CG9684 or the paralogs Spoon or Papi by RNAi does not de-repress transposons, suggesting redundant roles within each pair (Handler et al., 2011). Loss of Papi in cultured Bombyx mori (silk moth) BmN4 cells decreases piRNA levels (Izumi et al., 2016; Nishida et al., 2018), but papi mutant flies and Tdrkh (the mammalian Papi homolog) mutant mice show little change in piRNA abundance (Han et al., 2015a; Saxe et al., 2013). The B. mori and mouse genomes both encode Papi paralogs similar to fly Spoon (BGIBMGA006841 and Akap1). The finding that both Spoon and Papi interact with Zuc suggests that Spoon may compensate for loss of Papi in Drosophila.
Armi ATPase Confers Substrate Discrimination in piRNA Biogenesis
The promiscuous mRNA binding of Armi ATPase mutants (Figure 5A) calls to mind the aberrant binding of Upf1 ATPase mutants to mRNA that are not NMD substrates (Lee et al., 2015). The Upf1 ATPase activity is proposed to be rapidly activated on non-target mRNAs, promoting dissociation; on NMD targets the ATPase is temporarily inhibited allowing Upf1 to dwell longer (Lee et al., 2015). ATP or ADP decreases the association between the Upf1 helicase domain and single-stranded RNA (Cheng et al., 2007).
We envision (Figure 6) that in the cytoplasm, the Armi ATPase allows it to rapidly dissociate from mRNA; in nuage, Armi ATP-binding or ATPase activity is likely inhibited by interactions with proteins such as Aub, which stabilize Armi binding to pre-pre-piRNAs and cause Armi to accumulate in nuage. After an Armi- and Aub-bound pre-pre-piRNA arrives at the mitochondrial surface, Zuc cleavage liberates an Aub-bound pre-piRNA from the 5′ end of the pre-pre-piRNA, activating the Armi ATPase and allowing Armi to translocate 5′-to-3′ along the pre-pre-piRNA. Armi translocation would allow the newly shortened pre-pre-piRNA to bind a new Piwi or Aub at its 5′ end. Binding of Piwi or Aub would then restore inhibition of the Armi ATPase until Zuc makes the next cleavage to liberate a trailing piRNA. Because Aub and Piwi are not freely available in the cytosol, the ATPase activity of de-localized Armi is not inhibited, enabling rapid dissociation of Armi from incorrect RNA substrates, conferring substrate selectivity.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Phillip D. Zamore (phillip.zamore@umassmed.edu), or by completing the request form at www.zamorelab.umassmed.edu/reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Stocks
Transgenic 3×FLAG-6xMyc-Armi:
cDNA corresponding to armi mRNA isoform A (NM_001014556; 3558 bp) was cloned into pENTR-D-TOPO (Invitrogen); K729A, DE862AA and E863Q mutations were generated by site-directed mutagenesis. Wild-type and mutant sequences were subcloned into the modified Gateway vector pPFM-attB, which includes UASp sites, a 3×FLAG-6×Myc N-terminal tag, and an attB site for site-specific integration by the PhiC31 integrase-mediated transgenesis system. The pPFM-attB-armi plasmid was injected into attP40 flies carrying the attP landing site at cytological band 25C7 in chromosome 2L. Rainbow Transgenic Flies, Inc. (Camarillo, CA, USA) performed injections and established transgenic lines UAS-FM-Armi, UAS-FM-ArmiK729A, UAS-FM-ArmiDE862AA or UAS-FM-ArmiE863Q.
Rescue of armi germline null mutants:
Armi function is needed in somatic follicle cells for ovary development, and armi null flies (armiΔ1) develop rudimentary ovaries (Olivieri et al., 2010). armi72.1, an incomplete excision of a P-element inserted into the armi 5′-untranslated region (UTR), produces somatic but not germline Armi (Olivieri et al., 2010). armiΔ1 removes the armi, CycJ, and CG14971 coding sequences. We therefore used trans-heterozygous armi72.1/armiΔ1 as the armi germline null mutant background. To test whether the UAS-FM-Armi transgene rescues armi72.1/armiΔ1, armi72.1 was recombined with the third-chromosome, germline-specific Gal4 driver P{GAL4:: VP16-nos.UTR}CG6325[MVD1] (Bloomington #4937) to yield armi72.1, nos-Gal4-VP16. A second-chromosome Gal4 driver, P{Act5C-GAL4}25FO1 (Bloomington #4414), which is expressed throughout the ovary, was also included to increase UAS-FM-Armi expression. Rescue flies had the genotype: w1118; UAS-FM-Armi/Act5C-Gal4; armi72.1, nos-Gal4-VP16/armiΔ1 (Armi rescue) or w1118; UAS-FM-ArmiK729A/Act5C-Gal4; armi72.1, nos-Gal4-VP16/armiΔ1 (ArmiK729A rescue).
Germline-specific overexpression of transgenic Armi:
UAS-FM-Armi was overexpressed using the third-chromosome, germline-specific Gal4 driver P{GAL4::VP16-nos.UTR}CG6325[MVD1]. Tpo overexpress UAS-FM-Armi in zucH169Y background, UAS-FM-Armi was recombined with zucSG63, a point mutant allele changing histidine 169 to tyrosine (henceforth zucH169Y), then crossed in trans to zucHM27, a null allele (Pane et al., 2007). The same third-chromosome germline-specific Gal4 driver P{GAL4::VP16-nos.UTR}CG6325[MVD1] was used to express UAS-FM-Armi in a zucH169Y background.
Other stocks:
Wild-type and the H169Y mutant endogenous Zuc, tagged with 3×FLAG strains have been described previously (Ge et al., 2016). minotaurz3-5967 (Vagin et al., 2013) was crossed in trans to Df(3R)ED6280 (Bloomington #29667) to obtain minotaurz3-5967 mutants.
METHOD DETAILS
Female Fertility
Female fertility was tested as described in (Li et al., 2009a) except five two-day-old female virgins were mated at 25°C to three Oregon R males in a small cage (70 mm tall) on a 60 mm diameter grape juice agar plate dabbed with yeast paste (Lesaffre Red Star bakers active dry yeast). The plate was removed from the cage after 24 h, and the number of eggs laid counted. The plate was further incubated at 25°C for 24 h, then the number of eggs hatched were counted. Plates were replaced and scored daily for 8 days.
Ovary Isolation and Crosslinking
Flies were grown at 25°C. Unless otherwise noted, 0–3-day-old female flies were fed yeast paste for two days before ovary dissection. Ovaries (30–50 mg) were dissected into dissection buffer (5 mM HEPES, pH 7.2, 128 mM NaCl, 2 mM KCl, 4 mM MgCl2, 1.8 mM CaCl2, 35.5 mM sucrose) and transferred to 1.7 ml microfuge tubes on ice. Next, dissection buffer was replaced with 1 ml of 2 mM dithio-bis-maleimidoethane (DTME, Thermo Fisher #22335), 5 mM disuccinimidyl tartrate (DST, Thermo Fisher #20589), 5 mM ethylene glycol bis-succinimidyl succinate (EGS, Thermo Fisher #21565) or 5 mM dithio-bis-succinimidyl propionate (DSP, Thermo Fisher #PI22586) in dissection buffer or 0.2% (w/v) methanol-free paraformaldehyde (PFA, Thermo Fisher #PI28908) in 0.1 M sodium phosphate, pH 7.3. Crosslinking was performed for 10 min (PFA), 15 min (DTME) or 30 min (DST, EGS, DSP), then the crosslinking solution removed and ovaries washed three times for 5 min with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, (1×TBS) at room temperature. The ovaries were frozen in liquid nitrogen and stored at −80°C. PFA crosslinking was reversed for protein an alysis by heating at 95°C for 30 min and for RNA experiments by heating at 65°C for 2 h. DTME crosslinking was reversed by heating at 37°C for 30 min in the presence of 10 mM dithiothreitol (DTT); DST crosslinking was reversed by incubating at room temperature for 30 min in the presence of 15 mM sodium periodate; EGS crosslinking was reversed by heating at 37°C for 3 h in the presence of 1 M hydroxylamine-HCl; DSP crosslinking was reversed by heating at 95°C for 5 min in the presence of 100 mM DTT.
Immunofluorescence
Intact ovaries were fixed in 4% methanol-free paraformaldehyde (w/v) in 0.1 M sodium phosphate, pH 7.3, for 10 min, rotating at room temperature, then washed for 5, 10, and 15 min at room temperature in phosphate buffer supplemented with 0.1% (w/v) Triton X-100 (PBT). After removing the wash buffer, 100 μl phosphate buffer was added and ovaries separated into ovarioles by repetitive pipetting using a P200 pipette with a tip cut to enlarge the orifice. Disrupted ovarioles were transferred to 0.2 mL tubes and incubated rotating at 4°C overnight with primary antibodies in PBT supplemented with 5% (v/v) normal donkey serum (Sigma #D9663). Primary antibodies were diluted 1:1000 for rabbit polyclonal anti-Armi C-terminal peptide (gift of William Theurkauf; Cook et al., 2004), 1:200 for mouse clone 2F8A9 monoclonal anti-Armi N-terminal peptide (1.3 mg/ml; gift of Mikiko Siomi; Saito et al., 2010), 1:500 for mouse monoclonal clone M2 anti-FLAG (Sigma), 1:200 for mouse monoclonal anti-ATP5A (Abcam 15H4C4), 1:2000 for rabbit polyclonal anti-Aub (MA514, 2.4 mg/mL, (Li et al., 2009a), 1:2000 for mouse monoclonal anti-Ago3 (gift from Julius Brennecke; Senti et al., 2015), 1:50 rat monoclonal anti-Vasa (Developmental Studies Hybridoma Bank).
The next day, ovarioles were washed three times with PBT for 5, 10, and 15 min at room temperature. Secondary antibodies diluted in PBT were added and the ovarioles incubated in the dark, rotating at room temperature for 2 h. All secondary antibodies are from Thermo Fisher, produced in donkey or goat, against mouse, rabbit or rat IgG (H+L), highly cross-adsorbed against close species, and conjugated to Alexa Fluor 488 or Alexa Fluor 594 (for three color experiments), or Alexa Fluor 488, Alexa Fluor 546 or Alexa Fluor 633 (for four color experiments).
Ovarioles were next washed twice in PBT for 10 min each at room temperature, then incubated with 0.5 μg/mL 4′,6-diamidino-2-phenylindole diluted in 0.3 M NaCl, 0.03 M sodium citrate for 15 min at room temperature, and washed again with PBT for 10 min. Wash buffer was removed and a drop of VECTASHIELD Mounting Medium (Vector Laboratories #H-1000) added. After gentle pipetting to mix, the ovarioles in mounting medium were transferred to a glass slide using a P200 pipette with a cut tip and covered with a 0.13–0.17 mm thick cover slip (VWR #48393106). The cover slip was gently pressed with a Kimwipe to absorb extra mounting medium, then sealed with nail polish. Images were captured using a Leica TCS SP5 II laser scanning confocal microscope using a 63× HCX PL APO CS oil immersion objective (NA = 1.4) and 1 μM-thick z-stacks.
Transmission Electron Microscopy
Fly ovaries were quickly dissected in dissecting solution and transferred to 1.7 ml microfuge tubes on ice. Dissecting solution was removed and ovaries fixed in 2.5% glutaraldehyde (w/v) in 0.1 M sodium cacodylate buffer, pH 7.2, overnight at 4°C. Samples were processed and analyzed at the University of Massachusetts Medical School Electron Microscopy Core Facility using standard procedures. Briefly, intact ovaries were rinsed three times in the fixation buffer and post-fixed with 1% osmium tetroxide (w/v) for 1 h at room temperature. Samples were then washed three times with water for 10 min each and dehydrated using a graded series of 10%, 30%, 50%, 70%, 85%, and 95% ethanol, finishing with three changes in 100% ethanol. Samples were then infiltrated first with two changes 100% propylene oxide and then with a mixture of 50% propylene oxide/50%SPI-Pon 812 resin. The sample was incubated in seven successive changes of fresh 100% SPI-Pon 812 resin over 2 days, then polymerized at 68°C in flat molds. The samples were then reoriente d for horizontal sections of the center of individual ovaries. Thin sections (~70 nm) were placed on gold support grids, and stained with lead citrate and uranyl acetate to increase contrast. Sections were examined using a CM10 with 80 KV accelerating voltage, and images captured using a Gatan TEM CCD camera.
Immunoprecipitation
Zuc immunoprecipitation:
freshly dissected zucWT-3×FLAG ovaries were crosslinked with DTME and kept on ice. For each mg of the ovary pellet, 4 μl ice-cold lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% IGEPAL CA-630 (Sigma), 1% Empigen BB (w/v, Sigma), 0.1% SDS (w/v), 0.5 mM DTT) containing protease inhibitor cocktail (1 mM AEBSF (4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride [EMD Millipore #101500], 0.3 μM Aprotinin [Bio Basic Inc #AD0153], 20 μM Bestatin [Sigma Aldrich #B8385], 10 μM E-64 [(1S,2S)-2-(((S)-1-((4-Guanidinobutyl)amino)-4-methyl-1-oxopentan-2-yl)carbamoyl)cyclopropanecarboxylic acid; VWR #97063], and 10 μM Leupeptin [Fisher Scientific #108975]. Ovaries were homogenized with a motorized plastic pestle (Fisher Scientific #12141364) for 30 strokes on ice. The tube containing ovary lysate was then submerged in an ice-water bath and sonicated (Branson Digital Sonifier Model 450) at 40% amplitude for 2 min total sonication (8 cycles of 15 s sonication followed by 1 min rest). Lysate was centrifuged at 13,000 × g for 10 min at 4°C to remove insoluble material and the supernatan t retained. Mouse monoclonal clone M2 anti-FLAG antibody (clone M2, Sigma) or mouse anti-GFP antibody (clone GF28R, Invitrogen, as negative control) was added at 6 μg antibody per 1 ml of lysate and incubated rotating 2 h at 4°C, before transferring the supernatant to a new tube containing protein G paramagnetic Dynabeads (1/10 volume of the lysate before removing the buffer) washed three times with lysis buffer. The tube was further rotated for 1 h at 4°C. Protein G beads were collected usin g a magnetic stand, washed at room temperature for 1 min successively with a volume equal to that of the original lysate of wash buffer (50 mM Tris, pH 7.4, 1 mM EDTA, 0.05% v/v NP-40, and 0.1% w/v SDS) containing 150 mM, 300 mM, 500 mM, 750 mM and 150 mM NaCl. Beads were then washed with 0.05% v/v NP-40 in water. Finally, beads were eluted by incubating in elution buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.05% NP-40, 0.1% SDS, 1 mM DTT) containing 0.5 μg/μl (175 μM) 3×FLAG peptide (Sigma) for 10 min at room temperature with occasional mixing to keep beads suspended.
Armi immunoprecipitation:
Ovaries with germline-specific overexpression of Flag-Myc-tagged Armi were dissected and immunoprecipitated as described for Zuc, except that the cleared lysate was diluted by adding four volumes 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5 mM DTT, and protease inhibitor cocktail. For each 1 ml diluted lysate, 6 μg mouse clone M2 monoclonal anti-FLAG antibody (Sigma) was added. An amount of Protein G Dynabead suspension equal to 1/10 volume of the diluted lysate was used.
Western Blotting
For each mg tissue, ovaries were homogenized with a motorized plastic pestle (Thermo Fisher #12141364) in 4 μl ice-cold lysis buffer (100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate, 1 mM DTT) containing protease inhibitor cocktail. Lysate was centrifuged at 13,000 × g for 10 min at 4°C and an equal volume of 2× denaturing loading buffer (100 mM Tris-HCl, pH 6.8, 4% (w/v) SDS, 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol, 200 mM DTT) was added to the supernatant and heated to 95°C for 5 min.
The lysate was resolved by electrophoresis through a 4–20% gradient polyacrylamide gel (Bio-Rad Laboratories #5671085). Proteins were transferred to a 0.45 μm pore nitrocellulose membrane (Amersham #GE10600002) in a Mini Trans-blot tank at 15 V overnight. The membrane was blocked in Blocking Buffer (Rockland Immunochemicals #MB-070) at 4°C for 5 h or overnigh t, then incubated overnight at 4°C in with primary antibodies diluted in Blocking Buffer. Primary antibodies were diluted 1:2000 for mouse clone 2F819 monoclonal anti-Armi N-terminal peptide (1.3 mg/ml; gift from Mikiko Siomi; Saito et al., 2010), 1:500 for goat anti-Armi C-terminal peptide (Santa Cruz dD-17), 1:10,000 for mouse clone M2 monoclonal anti-FLAG (Sigma), and 1:3000 for rabbit anti-Dicer-2 (Abcam ab4732).
The membrane was washed three times, 5 min each, with TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% v/v Tween 20) at room temperature, incubated for 1 h at room temperature with secondary antibodies conjugated to IRDye 680RD or 800CW (LICOR Biosciences) diluted 1:20,000 in TBST, and then washed five times, 5 min each in TBST at room temperature. All secondary antibody steps were conducted in the dark. Signal was detected using the Odyssey Infrared Imaging System.
Mass spectrometry
FLAG immunoprecipitation was eluted as described above, and DTME was reverse-crosslinked. LC/MS/MS digestion and analysis were carried out by the University of Massachusetts Proteomics Core: the eluted immunoprecipitation reaction (for Zuc immunoprecipitation, eluate from ~350 mg of ovary tissue; for Armi immunoprecipitation, from ~40 mg ovary tissue) was denatured in 2× denaturing loading buffer and electrophoresed for 20 min at 100 V through an SDS-polyacrylamide gel to separate proteins from lower molecular weight contaminants. The entire protein region of the gel excised as a single band and subjected to in-gel trypsin digestion after reduction with DTT and alkylation with iodoacetamide. Peptides eluted from the gel were lyophilized and re-suspended in 25 μl 5% acetonitrile, 0.1% TFA. A 3 μl injection was loaded at 4.0 μl/min for 4 min onto a 100 μM I.D. fused-silica pre-column packed with 2 cm of 5 μM (200Å) Magic C18AQ (Bruker-Michrom) run in 5% acetonitrile, 0.1% formic acid using a NanoAcquity UPLC (Waters). Peptides were loaded onto a 75 μm I.D. gravity-pulled analytical column packed with 25 cm of 3 μm Magic C18AQ particles (100 Å), and eluted at 300 nl/min over 60 min using a 5–35% linear gradient of mobile phase B (acetonitrile + 0.1% formic acid) in mobile phase A (water + 0.1% formic acid). Ions were introduced by positive electrospray ionization via liquid junction into a Q Exactive hybrid mass spectrometer (Thermo Fisher). Mass spectra were acquired over m/z 300–1750 at 70,000 resolution (m/z 200) and data-dependent acquisition selected the top 10 most abundant precursor ions for tandem mass spectrometry by HCD fragmentation using an isolation width of 1.6 Da, maximum fill time of 110 ms, AGC target of 106, collision energy of 27, and a resolution of 17,500 (m/z 200). For data analysis, raw data files were peak processed with Proteome Discoverer 2.1 (Thermo Fisher) followed by identification using Mascot Server 2.5 against the Drosophila melanogaster Uniprot FASTA file downloaded 5/2016. Search parameters included Trypsin/P specificity, up to two missed cleavages, fixed carbamidomethyl on cysteine, and the variable modifications of oxidized methionine, pyroglutamic acid for N-terminal glutamine peptides, and N-terminal acetylation of the protein. Assignments were made using a 10 ppm mass tolerance for the precursor and 0.05 Da mass tolerance for the fragments. All non-filtered search results were then loaded into Scaffold Viewer v4.8.4 (Proteome Software, Inc.) with thresholding to a peptide FDR of 1%, for subsequent peptide/protein validation and label free quantitation.
Armi Structure Modeling
The predicted Armi helicase core, amino acids 692 to 1160, was submitted to I-TASSER (Roy et al., 2010). The modeled structure was superimposed on human Upf1 helicase core structure (PDB ID 2GJK) using PyMol v1.3 (Schrodinger, LLC).
Small RNA-seq Libraries
Small RNA libraries were constructed as described (Han et al., 2015a). Briefly, total RNA (50 μg) was purified by 15% urea polyacrylamide gel electrophoresis, selecting for 18–30 nt small RNAs using 18 nt and 30 nt size markers. Half of the purified sRNAs were oxidized with 25 mM NaIO4 to deplete miRNAs and enrich for siRNAs and piRNAs (Li et al., 2009a). To reduce ligation bias, a 3′ adaptor with three random nucleotides at its 5′ end was used (5′-rApp NNN TGG AAT TCT CGG GTG CCA AGG /ddC/-3′). The 3′ adaptor was ligated to RNAs using purified, recombinant truncated, K227Q mutant T4 RNA ligase 2 at 16°C overnight, small RNA precipitated and size selected by comparison to 18 nt and 30 nt size markers ligated to the same 3′ adaptor. To exclude 2S rRNA from sequencing libraries, 10 pmol 2S blocker oligo was added before 5′ adaptor ligation (Wickersheim and Blumenstiel, 2013). 5′ adaptor was ligated using T4 RNA ligase (Life Technologies #AM2141) at 25°C for 2 h, followed by reverse-transcription using AMV reverse transcriptase (New England Biolabs #M0277L) and PCR using AccuPrime Pfx DNA polymerase (Invitrogen #12344-024). PCR products were purified from a 2% agarose gel, and the DNA recovered from the gel with QIAquick gel extraction kit (Qiagen). Length distribution and quality of the libraries were analyzed using a Bioanalyzer 2100 (Agilent). Libraries were quantified using KAPA library quantification kit (Kapa Biosystems), before being sequenced on a NextSeq500 (Illumina) to obtain 75 nt single-end reads.
The sequence of the 3′ adapter, including its 5′ three random nucleotides, was removed from raw reads. Small RNA-seq analysis was performed with piPipes 1.4 (Han et al., 2015b). Briefly, reads were aligned to rRNA and miRNA hairpin sequences using Bowtie 1.0.0 (Langmead et al., 2009). Unaligned reads were mapped to fly genome release dm3 using Bowtie and 23–29 nt piRNAs were retained for analysis. The abundance of piRNAs overlapping genes and transposons were apportioned by the number of times each piRNA aligned in the genome. Division of transposon families into germline, somatic and intermediate groups was according to Wang et al. (2015).
Ping-pong Z10 (5′-to-5′ distance on opposite genomic strands = 10 nt) and phasing Z1 (3′-to-5′ distance on the same genomic strand = 1 nt) scores were calculated as described (Zhang et al., 2011; Han et al., 2015a).
RNA-seq Libraries
RNA-seq libraries were constructed as previously described (Zhang et al., 2012b) with modifications, including the use of unique molecular identifiers to eliminate PCR duplicates (Fu et al., 2018). To deplete ribosomal RNA, RNA was hybridized in 10 μl with a pool of 186 rRNA antisense oligos (0.05 μM/each) in 10 mM Tris-HCl, pH 7.4, 20 mM NaCl, heated to 95°C, cooled at −0.1°C/sec to 22°C, and then incubated at 22°C for 5 min. Thermostable RNase H (10 U, Lucigen #H39500) were added and incubated at 45°C for 30 min in 20 μl containing 50 mM Tris-H Cl, pH 7.4, 100 mM NaCl, and 20 mM mgCl2. RNA was treated with 4 U Turbo DNase (Thermo Fisher #AM2238) in 50 μl at 37°C for 20 min, then purified using RNA C lean & Concentrator-5 (Zymo Research #R1016), to enrich for RNA >150 nt. RNA-seq libraries were sequenced using a NextSeq500 (Illumina) to obtain 75 + 75 nt, paired-end reads.
RNA-seq analysis was performed with piPipes. Briefly, RNAs were first aligned to rRNA sequences using Bowtie2 (v2.2.0; Langmead and Salzberg, 2012) and unaligned reads mapped to fly genome release dm3 using STAR (v2.3.1; (Dobin et al., 2013). The numbers of reads overlapping genes and transposons were apportioned by the number of times each read aligned in the genome. Germline, somatic and intermediate transposon family classification was according to Wang et al. (2015).
5′ monophosphorylated long RNA Sequencing
5′ monophosphorylated long RNA-seq libraries were constructed as described (Han et al., 2015a) with modifications, including the use of unique molecular identifiers to eliminate PCR duplicates (Fu et al., 2018). PFA-crosslinked Armi immunoprecipitation eluate was reversed by adding an equal volume of 200 mM Tris-Cl, pH 7.5, 25 mM EDTA, pH 8.0, 300 mM NaCl, 2% w/v SDS containing 0.4 mg/ml proteinase K and incubating at 50°C for 1 h then 65°C for 2 h, befor e extracting with an equal volume of acid 5:1 phenol:chloroform pH 4.5 (AMRESCO LLC, Solon, OH, USA), and centrifuged at 20,800 × g for 5 min at room temperature. One-tenth volume 3 M sodium acetate and three volumes 100% ethanol were added to the aqueous phase and the RNA precipitated by on ice for 1 h. The precipitate was recovered by centrifugation at 20,800 × g for 15 min at 4° washed with 70% (v/v) ethanol, air dried, and dissolved in water. rRNA depletion and RNA Clean & Concentrator-5 purification were as described for RNA-seq. A 5′-adapter with Unique Molecular Identifier (UMI) comprising an equimolar mix of 5′-GUU CAG AGU UCU ACA GUC CGA CGA UC (N3) CGA (N3) UAC (N3)-3′ and 5′-GUU CAG AGU UCU ACA GUC CGA CGA UC (N3) AUC (N3) AGU (N3)-3′ (Fu et al., 2018) was ligated to 5′-monophosphorylated RNAs using T4 RNA ligase (Ambion) at 25°C for 2 h. The ligation reaction was purified using 1.5× volume of Ampure XP beads suspension. Reverse transcription using Superscript III (Life Technologies) used a primer containing degenerate sequences at its 3′ end (5′-GCA CCC GAG AAT TCC ANN NNN NNN-3′). The reverse transcription reaction was digested with 1 μl RNase H (Ambion) at 37°C for 20 min, and purified using 1.5 × volume of Ampure XP beads. Purified cDNA was amplified by PCR #1, using a pair of primers that anneal to the 5′-(5′-CTA CAC GTT CAG AGT TCT ACA GTC CGA-3′) or to 3′-(5′-GCC TTG GCA CCC GAG AAT TCC A-3′) adapter. The PCR reaction was mixed with 0.7× volume of Ampure beads, and the supernatant was transferred to a new tube containing 0.5× volume of Ampure beads (total 1.2× volume) to purify 200–400 nt PCR products. Subsequently, PCR #2 was carried out using the purified products of PCR #1 and the same barcoded primer set as the small RNA library cloning protocol. PCR #2 was purified with 1.1× volume of Ampure beads. All PCR reactions used Phusion polymerase (NEB). 5′ monophosphorylated long RNA-seq libraries were sequenced using a NextSeq500 (Illumina) to obtain 75 + 75 nt, paired-end reads.
Sequences of unique molecular identifiers (UMIs; NNNCCCNNNCCCNNN) were extracted from the 5′ section of read 1. PCR duplicates were removed with umi_tools 0.5.3 using the “directional” mode (Fu et al., 2018), and reads were then analyzed using piPipes. Briefly, sequences were first aligned to rRNA using Bowtie2. Unaligned reads were then mapped using STAR to fly genome release dm3, and alignments with soft clipping of ends were removed with SAMtools 1.0.0 (Li et al., 2009b). In addition to sequencing depth, libraries were normalized using reads uniquely mapping to flamenco. The abundance of reads overlapping genes and transposons was apportioned by the number of times each read aligned to the genome. Division of transposon families into germline, somatic and intermediate groups was according to Wang et al. (2015).
Distance probability analyses (Han et al., 2015a) were done without considering abundance, and species uniquely mapping to flamenco were used to normalize libraries. To determine the significance of piRNA reads sharing 5′ ends with 5′ monophosphorylated long RNAs, the Z0 score for the peak at 0 was calculated using positions 1–50 as background.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image Quantification
Confocal images were quantified using custom-built scripts in CellProfiler (Kamentsky et al., 2011). To measure the amount of Aub signal overlapping with Armi, primary Armi and Aub objects in separate fluorescent channels were identified using the adaptive Otsu thresholding method. “Threshold correction factor” and “lower bound on threshold” were empirically determined using representative test images. Once optimized, the same object identification settings were applied to all samples. Armi objects were then used as masking objects to mask Aub objects. The amount of signal in masked Aub objects was divided by the amount of signal in total Aub objects for each image. The amount of Vasa signal overlapping with ATP5A or Aub was quantified similarly.
Proteome Quantification
To detect proteins enriched in immunoprecipitation, Fisher’s exact test was performed in Scaffold Viewer with weighted spectra and a threshold of Benjamini-Hochberg multiple test-corrected p < 0.05 from three biological replicates to compare the proteome of target protein immunoprecipitation to that of the control immunoprecipitation.
DATA AND SOFTWARE AVAILABILITY
Sequencing data are available from the National Center for Biotechnology Information Sequence Read Archive using accession number PRJNA495886. Proteomics data are available from Mass spectrometry Interactive Virtual Environment MSV000083634 (doi: 10.25345/C5262Z).
Supplementary Material
Supplemental Table S1. Proteins specifically co-immunoprecipitated with wild-type Armi or ArmiK729A in DTME-crosslinked ovaries (FDR < 0.05). Related to Table 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Armi C-terminal peptide | Cook et al., 2004 | |
| Mouse monoclonal anti-Armi N-terminal peptide clone 2F8A9 | Saito et al., 2010 | |
| Mouse monoclonal anti-FLAG clone M2 | Sigma-Aldrich | Cat# F1804 RRID:AB_262044 |
| Mouse monoclonal anti-ATP5A clone 15H4C4 | Abcam | Cat# ab14748 RRID:AB_301447 |
| Rabbit polyclonal anti-Aub (MA514) | Li et al., 2009a | |
| Mouse monoclonal anti-Ago3 | Senti et al., 2015 | |
| Rat monoclonal anti-Vasa | Developmental Studies Hybridoma Bank | Cat# anti-vasa RRID:AB_760351 |
| Goat polyclonal anti-Armi C-terminal peptide (dD-17) | Santa Cruz Biotechnology | Cat# sc-34564 RRID:AB_1134112 |
| Rabbit polyclonal anti-Dicer-2 | Abcam | Cat# ab4732 RRID:AB_449344 |
| IRDye 680RD Donkey anti-mouse | LI-COR Biosciences | Cat# 926-68072 RRID:AB_10953628 |
| IRDye 800CW Goat anti-mouse | LI-COR Biosciences | Cat# 926-32210 RRID:AB_621842 |
| Deposited Data | ||
| Raw sequencing data from D. melanogaster | This paper | SRA: PRJNA495886 |
| Raw proteomics data from D. melanogaster | This paper | MassIVE: doi:10.25345/C5262Z |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w1118; UAS-FM-Armi; + | This paper | |
| D. melanogaster: w1118; UAS-FM-ArmiK729A; + | This paper | |
| D. melanogaster: w1118; UAS-FM-ArmiDE862AA; + | This paper | |
| D. melanogaster: w1118; UAS-FM-ArmiE863Q; + | This paper | |
| D. melanogaster: w1118; +; armi72.1/TM3, Sb | Cook et al., 2004 | BDSC:8544 RRID:BDSC_8544 |
| D. melanogaster: w1118; +; armiΔ1/TM3, Sb | Olivieri et al., 2010 | |
| D. melanogaster: w1118; +; P{GAL4::VP16-nos.UTR}CG6325MVD1 | Bloomington Drosophila Stock Center | BDSC:4937 RRID:BDSC_4937 |
| D. melanogaster: w1118; +; armi72.1, P{GAL4::VP16-nos.UTR}CG6325MVD1 | This paper | |
| D. melanogaster: y1 w*; P{Act5C-GAL4}25FO1/CyO, y+; + | Bloomington Drosophila Stock Center | BDSC:4414 RRID:BDSC_4414 |
| D. melanogaster: w1118; zucSG63/CyO; + | Pane et al., 2007 | |
| D. melanogaster: w1118; zucSG63, UAS-FM-Armi/CyO ; + | This paper | |
| D. melanogaster: w1118; zucHM27/CyO; + | Pane et al., 2007 | |
| D. melanogaster: w1118; zuc-3×FLAG; + | Ge et al., 2016 | |
| D. melanogaster: w1118; zucH169Y-3×FLAG/CyO; + | Ge et al., 2016 | |
| D. melanogaster: w1118; +; minotaurz3-5967/TM3, Sb | Vagin et al., 2013 | |
| D. melanogaster: w1118; +; Df(3R)ED6280, P{3′.RS5+3.3′}ED6280/TM6C, cu1 Sb1 | Bloomington Drosophila Stock Center | 29667 RRID:BDSC_29667 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3×FLAG Peptide | Sigma-Aldrich | Cat# F4799 |
| normal donkey serum | Sigma-Aldrich | Cat# D9663 |
| dithio-bis-maleimidoethane (DTME) | Thermo Fisher Scientific | Cat# 22335 |
| disuccinimidyl tartrate (DST) | Thermo Fisher Scientific | Cat# 20589 |
| ethylene glycol bis-succinimidyl succinate (EGS) | Thermo Fisher Scientific | Cat# 21565 |
| dithio-bis-succinimidyl propionate (DSP) | Thermo Fisher Scientific | Cat# PI22586 |
| methanol-free paraformaldehyde (PFA) | Thermo Fisher Scientific | Cat# PI28908 |
| VECTASHIELD Mounting Medium | Vector Laboratories | Cat# H-1000 RRID:AB_2336789 |
| Software and Algorithms | ||
| CellProfiler | Kamentsky et al., 2011 | |
| I-TASSER | Roy et al., 2010 | https://zhanglab.ccmb.med.umich.edu/I-TASSER |
| Scaffold Viewer | Proteome Software, Inc. | |
| Bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2 |
| STAR | Dobin et al., 2013 | https://code.google.com/archive/p/rna-star/ |
| piPipes | Han et al., 2015b | http://bowhan.github.io/piPipes/ |
| SAMtools | Li et al., 2009b | http://samtools.sourceforge.net |
| BEDtools | Quinlan and Hall, 2010 | https://code.google.com/archive/p/bedtools/ |
| umitools | Fu et al., 2018 | https://github.com/weng-lab/umitools |
Highlights.
The ping-pong and phased piRNA biogenesis pathways are physically separate
Armi couples piRNA biogenesis in nuage to phased piRNA production on mitochondria
Armi binds piRNA precursors during both ping-pong and phased piRNA production
Armi ATPase mutants inappropriately bind mRNA and fail to support piRNA production
ACKNOWLEDGEMENTS
We thank John Leszyk and Michelle Dubuke at the UMass Proteomics Core, Keith Reddig at the UMass Electron Microscopy Core, Christina Baer at the Sanderson Center for Optical Experimentation, and Haiwei Mou, Cha San Koh, and members of the Zamore laboratory for help and discussions. This work was supported by NIH grant R37GM062862 to P.D.Z. and P01HD078253 to Z.W. and P.D.Z. The UMass Electron Microscopy Core Facility is supported by grant S10RR027897 from the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Andress A, Bei Y, Fonslow BR, Giri R, Wu Y, Yates JR, and Carthew RW (2016). Spindle-E cycling between nuage and cytoplasm is controlled by Qin and PIWI proteins. J Cell Biol 213, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez LA, Alexandre C, Mitchell A, Pasakarnis L, and Vincent JP (2013). Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140, 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, and Song H (2007). Structural and functional insights into the human Upf1 helicase core. EMBO J 26, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, and Frohman MA (2006). A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8, 1255–1262. [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, and Theurkauf WE (2004). The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116, 817–829. [DOI] [PubMed] [Google Scholar]
- Czech B, and Hannon GJ (2016). One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci 41, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapples CC, and King RC (1970). The development of the nucleolus of the ovarian nurse cell of Drosophila melanogaster. Z Zellforsch Mikrosk Anat 103, 34–47. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM (1974). Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec 178, 731–757. [DOI] [PubMed] [Google Scholar]
- Eddy EM (1975). Germ plasm and the differentiation of the germ cell line. Int Rev Cytol 43, 229–280. [DOI] [PubMed] [Google Scholar]
- Feltzin VL, Khaladkar M, Abe M, Parisi M, Hendriks GJ, Kim J, and Bonini NM (2015). The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell 14, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu PH, Beane T, Zamore PD, and Weng Z (2018). Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 19, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I, Colpan C, Arif A, Cecchini K, and Zamore PD (2018). A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol Cell 71, 775–790.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge DT, Tipping C, Brodsky MH, and Zamore PD (2016). Rapid Screening for CRISPR-Directed Editing of the Drosophila Genome Using white Coconversion. G3 (Bethesda) 6, 3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, and Siomi MC (2007). A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, and Hannon GJ (2010). Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24, 2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, and Zamore PD (2015a). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Zamore PD, and Weng Z (2015b). pi Pipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics 31, 593–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, and Brennecke J (2013). The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50, 762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, and Brennecke J (2011). A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30, 3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R, Schnabl J, Handler D, Mohn F, Ameres SL, and Brennecke J (2016). Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 539, 588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, Murali R, Mourelatos Z, and Kirino Y (2013). Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 19, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, and Zamore PD (2007). The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Huang H, Li Y, Szulwach KE, Zhang G, Jin P, and Chen D (2014). AGO3 Slicer activity regulates mitochondria-nuage localization of Armitage and piRNA amplification. J Cell Biol 206, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Fejes Tóth K, and Aravin AA (2017). piRNA Biogenesis in Drosophila melanogaster. Trends Genet 33, 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, and Hannon GJ (2012). The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, and Tomari Y (2016). Identification and Functional Analysis of the Pre-piRNA 3′ Trimmer in Silkworms. Cell 164, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglarz MK, Kloc M, Jankowska W, Szymanska B, and Bilinski SM (2011). Nuage morphogenesis becomes more complex: two translocation pathways and two forms of nuage coexist in Drosophila germline syncytia. Cell Tissue Res 344, 169–181. [DOI] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, and Carpenter AE (2011). Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Jedrzejowska I, Tworzydlo W, and Bilinski SM (2014). Balbiani body, nuage and sponge bodies—germ plasm pathway players. Arthropod Struct Dev 43, 341–348. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, and Tóth KF (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Pratt GA, Martinez FJ, Yeo GW, and Lykke-Andersen J (2015). Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol Cell 59, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, and Zamore PD (2009a). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000, G. P. D. P. S. (2009b). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, and Lasko P (1994). Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120, 1201–1211. [DOI] [PubMed] [Google Scholar]
- Lim AK, and Kai T (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A 104, 6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP (1970). Intercellular migration of centrioles in the germarium of Drosophila melanogaster. An Electron Microscopic Study. The Journal of Cell Biology 45, 306–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP (1971). Polar granules of Drosophila. 3. The continuity of polar granules during the life cycle of Drosophila. J Exp Zool 176, 329–343. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, and Hannon GJ (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue AD, and Slotkin RK (2012). Transposable element small RNAs as regulators of gene expression. Trends Genet 28, 616–623. [DOI] [PubMed] [Google Scholar]
- Mili S, and Steitz JA (2004). Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, and Brennecke J (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, and Brennecke J (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364–1379. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, and Siomi MC (2009). Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28, 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Sakakibara K, Iwasaki YW, Yamada H, Murakami R, Murota Y, Kawamura T, Kodama T, Siomi H, and Siomi MC (2018). Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis. Nature 555, 260–264. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi MC, and Nureki 0 (2012). Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491, 284–287. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, and Brennecke J (2012). The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47, 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, and Brennecke J (2010). An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29, 3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Homolka D, Chen KM, Sachidanandam R, Fauvarque MO, and Pillai RS (2017). Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet 13, e1006956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, and SchQpbach T (2007). zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Anand A, Chakrabarti A, and Kai T (2014). The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in the piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol 12, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, and Kai T (2010). Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20, 724–730. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AK, Situ K, Perkins EM, and Toth KF (2017). Zucchini-dependent piRNA processing is triggered by recruitment to the cytoplasmic processing machinery. Genes Dev 31, 1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, and Zhang Y (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, and Hannon GJ (2013). Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, and Siomi MC (2010). Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24, 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, and Siomi MC (2007). Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21, 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, and Lin H (2013). Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J 32, 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, and MacRae IJ (2012). The crystal structure of human Argonaute2. Science 336, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvy PE, Lavieri RR, Lindsley CW, and Brown HA (2011). Phospholipase D: enzymology, functionality, and chemical modulation. Chem Rev 111, 6064–6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, and Brennecke J (2015). piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 29, 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Yu Y, Jankowska A, Luo Y, Wasik KA, Malone CD, Harrison E, Rosebrock A, Wakimoto BT, Fagegaltier D, Muerdter F, and Hannon GJ (2013). Minotaur is critical for primary piRNA biogenesis. RNA 19, 1064–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, and Wang PJ (2015). The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev 29, 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma Z, Niu K, Xiao Y, Wu X, Pan C, Zhao Y, Wang K, Zhang Y, and Liu N (2016). Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3′ ends in Drosophila. Development 143, 530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, and Zamore PD (2015). Slicing and Binding by Ago3 or Aub T rigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol Cell 59, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A, Li S, Hur JK, Wachsmuth M, Bois JS, Perkins EM, Patel DJ, and Aravin AA (2015). Aub and Ago3 Are Recruited to Nuage through Two Mechanisms to Form a Ping-Pong Complex Assembled by Krimper. Mol Cell 59, 564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, and Peltz SW (1996). Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16, 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, and Peltz SW (1998). ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4, 205–214. [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H (2012). Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersheim ML, and Blumenstiel JP (2013). Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. Biotechniques 55, 269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsch-Bräuninger M, Schwarz H, and Nüsslein-Volhard C (1997). A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol 139, 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, Kaksonen M, Cusack S, Ephrussi A, and Pillai RS (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711. [DOI] [PubMed] [Google Scholar]
- Yashiro R, Murota Y, Nishida KM, Yamashiro H, Fujii K, Ogai A, Yamanaka S, Negishi L, Siomi H, and Siomi MC (2018). Piwi Nuclear Localization and Its Regulatory Mechanism in Drosophila Ovarian Somatic Cells. Cell Rep 23, 3647–3657. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, and Theurkauf WE (2012a). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, and Zamore PD (2012b). Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, and Zamore PD (2011). Heterotypic piRNA Ping-Pong requires Qin, a protein with both E3 ligase and Tudor domains. Mol Cell 44, 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Proteins specifically co-immunoprecipitated with wild-type Armi or ArmiK729A in DTME-crosslinked ovaries (FDR < 0.05). Related to Table 1.
Data Availability Statement
Sequencing data are available from the National Center for Biotechnology Information Sequence Read Archive using accession number PRJNA495886. Proteomics data are available from Mass spectrometry Interactive Virtual Environment MSV000083634 (doi: 10.25345/C5262Z).


