Abstract
Background
The aim of this study was to clarify the potential function of LINC00922 in regulating the progression of lung cancer and its underling mechanism.
Material/Methods
Relative levels of LINC00922 in lung cancer tissues and cell lines was determined by quantitative polymerase chain reaction. Correlation between LINC00922 levels and pathological indexes of lung cancer patients was analyzed through the chi-square test. Subsequently, regulatory effects of LINC00922 on the proliferative, migratory, and invasive capacities of PC9 and A549 cells were evaluated. Western blot was conducted to determine the role of LINC00922 in mediating protein levels of CXCR4, E-cadherin, and vimentin. Through dual-luciferase reporter gene assay and functional experiments, the potential function of LINC00922/miRNA-204/CXCR4 regulatory loop in mediating the progression of lung cancer was explored.
Results
LINC00922 was highly expressed in lung cancer and correlated to the poor prognosis of lung cancer patients. Overexpression of LINC00922 accelerated PC9 and A549 cells to proliferate, migrate, and invade. CXCR4 was upregulated in lung cancer tissues and cells, which promoted lung cancer cells to migrate and invade. LINC00922 regulated the level of CXCR4 and directly bound to miRNA-204/CXCR4. LINC00922 mediated the cellular behaviors of lung cancer cells via targeting the miRNA-204/CXCR4 axis.
Conclusions
LINC00922 was upregulated in lung cancer, and accelerated lung cancer cells to proliferate, migrate, and invade via targeting the miRNA-204/CXCR4 axis.
MeSH Keywords: Lung Neoplasms; Receptors, CXCR4; RNA, Long Noncoding
Background
Lung cancer is a malignancy worldwide, which seriously threatens human health owing to its high morbidity and mortality [1]. With the development of medical technology, the treatment of lung cancer has made great strides. Nevertheless, the average survival rate for lung cancer patients is 20% and the 5-year survival rate is lower than 15% since most of lung cancer patients are in the middle or advanced stage at diagnosis [2,3]. Currently, individualized therapy based on target genes at the cellular level are being studied, posing a promising application to improve the outcomes of lung cancer patients [4,5]. Therefore, it is of significance to fully elucidate the molecular mechanism of lung cancer.
Long non-coding RNA (lncRNA) mainly exists in nucleus or cytoplasm with a length greater than 200 nucleotides, and does not have protein-encoding functions [6–8]. LncRNA has diverse functions in physiological activities [9–12]. Recent studies have found the crucial functions of non-coding RNA (ncRNA) in regulating the progression of various types of cancers, including lung cancer [13–16]. For example, GAS5 is downregulated in non-small cancer lung cancer (NSCLC) and associated with tumor size and stage [17]. High level of HOTAIR is closely correlated to the prognosis of NSCLC patients, which could mediate the invasion and migration of NSCLC [18]. Overexpression of ANRIL stimulates the metastasis of lung cancer cells, and is related to the poor prognosis of the disease [19]. There are abundant unexplored lncRNAs related to lung cancer. As a newly discovered one, the role of LINC00922 in lung cancer remains unclear.
In 2011, Pandolfi et al. [20] proposed the ceRNA (competing endogenous RNA) hypothesis. They suggested that mRNA, pseudogene transcript and lncRNA sharing the common MRE (miRNA response element) could mediate target gene expression through competitively bind to the same miRNA. The ceRNA hypothesis illustrated a novel regulatory network and provided a creative direction in investigating cellular function and molecular mechanism. Previous studies have already reported that several lncRNAs could bind to target miRNAs, thus regulating cellular behaviors of tumor cells [21–24]. For example, MALATI upregulates SLUG by absorbing miRNA-204, thus accelerating the progression of lung adenocarcinoma [25]. NEAT1 aggravates the malignant progression of NSCLC through sponging miR-377-3p to upregulate E2F2 [26].
In this study, expression pattern of LINC00922 in lung cancer was firstly examined. Moreover, the regulatory effects of LINC00922/miRNA-204/CXCR4 regulatory loop on mediating the progression of lung cancer were further explored. Our study aims to provide novel directions and therapeutic targets for lung cancer.
Material and Methods
Data collection
The expression profiles of lung cancer were downloaded from The Cancer Genome Atlas (TCGA). The dysregulated lncRNAs were obtained by edgeR package. Lung cancer tissues (n=52) and normal lung tissues (n=52) were surgically resected and immediately preserved in liquid nitrogen. Clinical data of 52 lung cancer patients were collected, including age, gender, tumor size, tumor node metastasis (TNM) stage, and the statue of lymph node metastasis. Lung cancer cells A549, PC9, and H460 were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) and 1% penicillin-streptomycin in a 37°C, 5% CO2 incubator. Medium was replaced every 1 to 2 days. Cell passage was conducted until 80% of confluence using trypsin containing 0.05% SDS (sodium dodecyl sulfate) at a ratio of 1: 3. This study was approved by the Ethics Committee of the First Medical Center of Chinese PLA General Hospital. Signed written informed consents were obtained from all participants before the study.
Cell transfection
Cells were transfected with shRNA-LINC00922, vector-LINC00922, shRNA-CXCR4, vector-CXCR4, shRNA-NC, vector-NC, miRNA-204 mimic, miRNA-204 inhibitor, or miRNA-204-NC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Fresh medium was replaced at 6 hours. Transfected cells for 24 hours were harvested for the following experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified using the Nanodrop Spectrophotometer (IMPLEN GmbH, Munich, Germany). The extracted RNA was subjected to reverse transcription to cDNA using the PrimeScript RT reagent Kit (TaKaRa, Otsu, Japan). Finally, complementary deoxyribose nucleic acid (cDNA) was amplified using the SYBR Primix Ex Taq II kit (TaKaRa, Otsu, Japan) on the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Each experiment was repeated in triplicate.
Western blot
Proteins were extracted from cells or tissues and loaded for electrophoresis. After transferring on a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA), it was blocked in 5% skim milk for 2 hours. Subsequently, the membrane was incubated with primary antibodies at 4°C overnight and secondary antibodies for 2 hours on the other day. Bands were exposed by electrochemiluminescence (ECL) and analyzed by Image Software (NIH, Bethesda, MD, USA).
Dual-luciferase reporter gene assay
LINC00922/CXCR4-Wt and LINC00922/CXCR4-Mut were constructed based on the predicted binding sequences to miRNA-204. Cells were co-transfected with LINC00922/CXCR4-Wt or LINC00922/CXCR4-Mut and miRNA-204-NC/miRNA-204 mimic for 48 hours. After cell lysis, relative luciferase activity was determined. β-galactosidase was used as the internal reference.
Cell Counting Kit-8 (CCK-8)
Cells were seeded in a 96-well plate at a density of 2×104/mL. At the appointed time points, 10 μL of Cell Counting Kit-8 (CCK-8) solution (Dojindo, Kumamoto, Japan) was applied per well. After incubation for 2 hours, the recorded absorbance at 450 nm using a microplate reader was used for plotting the viability curve.
Colony formation assay
Cells were seeded in the 6-well plate with 500 cells per well. Each group had 3 replicate wells. After 2-week incubation, cells were subjected to fixation in 4% paraformaldehyde and 0.1% violet crystal staining for 20 minutes. Finally, colonies were captured for counting under a microscope.
Transwell assay
Transfected cells were subjected to starvation in serum-free medium for 3 hours, and re-suspended in medium containing 1% FBS. 5. X×104 cells were seeded in the upper side of Matrigel-coated Transwell chamber (8 μm, Millipore, Billerica, MA, USA). In the bottom side, 600 μL of medium containing 10% FBS was applied. After 48 hours of incubation, invasive cells in the bottom side were subjected to fixation in 70% ethanol at 4°C for 30 minutes, crystal violet staining for 20 minutes and cell counting using a microscope. Penetrating cells were counted in 5 randomly selected fields per sample. Transwell migration assay was conducted in the same procedures except for Matrigel pre-coating.
Statistical analyses
Statistical Product and Service Solutions (SPSS) 17.0 was used for statistical analysis. Data were represented as mean ±SD (standard deviation). The t-test was used for analyzing intergroup differences. Comparison between groups was done using one-way ANOVA test followed by post hoc test (least significant difference). Correlation between LINC00922 levels and pathological indexes of lung cancer patients was analyzed through the chi-square test. Kaplan-Meier and receiver operating characteristic (ROC) curves were introduced for survival analysis and diagnostic evaluation. P<0.05 indicated the significant difference.
Results
LINC00922 was upregulated in lung cancer and predicted a poor prognosis
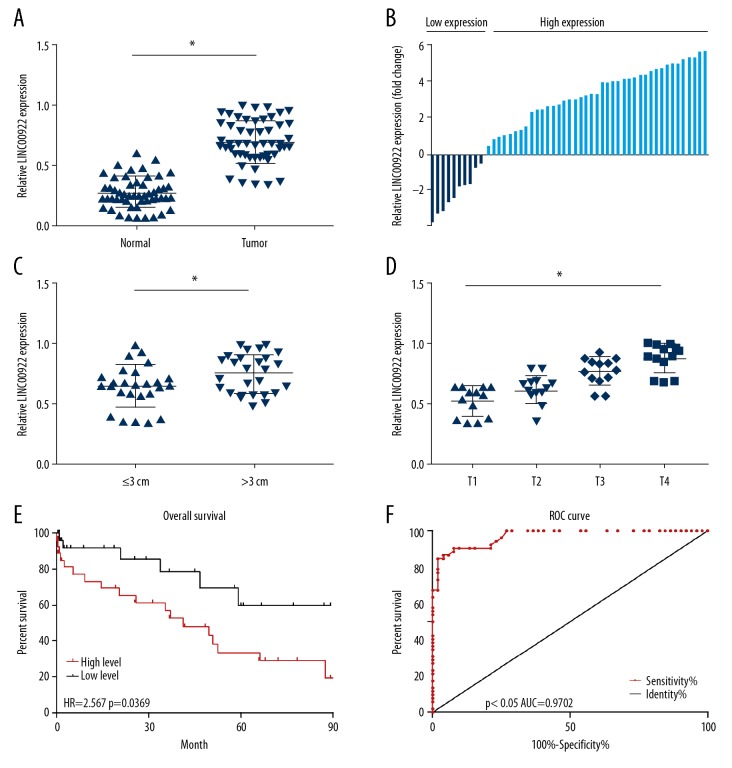
LINC00922 level was higher in lung cancer tissues than normal tissues in the downloaded TCGA datasets analyzed by edgeR package (Figure 1). Expression pattern of LINC00922 in 52 cases of lung cancer tissues and normal lung tissues was determined by qRT-PCR. LING00922 was upregulated in lung cancer tissues as well (Figure 2A). Moreover, 52 lung cancer patients were divided into high-expression and low-expression group based on their LINC00922 level (Figure 2B). As Table 1 depicted, LINC00922 level was correlated to tumor size, TNM stage and lymph node metastasis of lung cancer. In particular, LINC00922 level was higher in lung cancer with tumor size >3 cm compared with those ≤3 cm (Figure 2C). Lung cancer patients in T2, T3, and T4 stage had higher level of LINC00922 relative to those in T1 stage (Figure 2D). Furthermore, follow-up data of enrolled patients were collected for survival analysis. Kaplan-Meier curve revealed a worse overall survival in lung cancer patients with high-level LINC00922 (Figure 2E). ROC curves illustrated the diagnostic potential of LINC00922 in lung cancer with a relatively high sensitivity (AUC=0.9702, Figure 2F). The aforementioned data illustrated that LINC00922 could be a prognostic marker for lung cancer.
Figure 1.
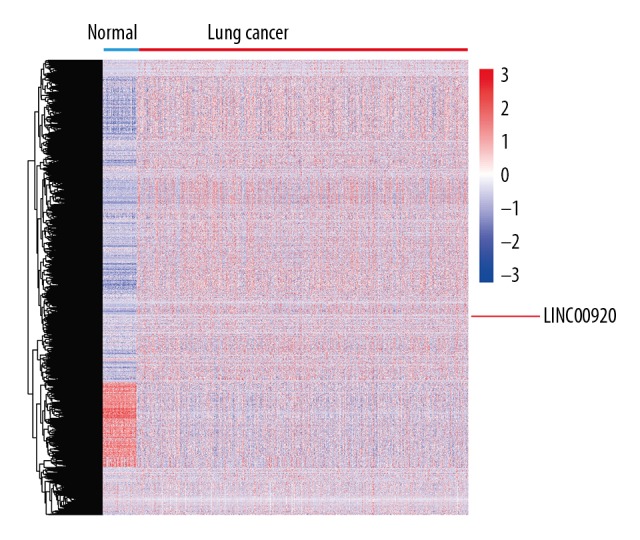
LINC00922 was upregulated in lung cancer in TCGA datasets.
Figure 2.
LINC00922 was upregulated in lung cancer and predicted a poor prognosis. (A) Expression level of LINC00922 in 52 cases of lung cancer tissues and normal lung tissues determined by quantitative real-time polymerase chain reaction. (B) 52 lung cancer patients were divided into high-expression and low-expression group based on their LINC00922 level. (C) LINC00922 level in lung cancer with tumor size >3 cm and those ≤3 cm. (D) LINC00922 level in lung cancer patients in T1, T2, T3, and T4 stage. (E) Kaplan-Meier curve introduced based on LINC00922 level in lung cancer patients for survival analysis. (F) Receiver operating characteristic (ROC) curve introduced for the sensitivity and identity of LINC00922 in lung cancer diagnosis.
Table 1.
Correlation between LINC00922 level and pathological indexes of lung cancer patients (n=52).
| Clinicopathologic features | Number of cases | LINC00922 expression | P-value | |
|---|---|---|---|---|
| Low (n=26) | High (n=26) | |||
| Age (years) | 0.7814 | |||
| ≤56 | 27 | 14 | 13 | |
| >56 | 25 | 12 | 13 | |
| Gender | 0.7775 | |||
| Male | 21 | 11 | 10 | |
| Female | 31 | 13 | 16 | |
| Tumor size | 0.0120 | |||
| ≤3 cm | 29 | 19 | 10 | |
| >3 cm | 23 | 7 | 16 | |
| TNM stage | 0.0125 | |||
| I~II | 25 | 17 | 8 | |
| III~IV | 27 | 9 | 18 | |
| Lymph node metastasis | 0.0261 | |||
| Absent | 24 | 16 | 8 | |
| Present | 28 | 10 | 18 | |
Overexpression of LINC00922 enhanced the proliferative ability of lung cancer cells
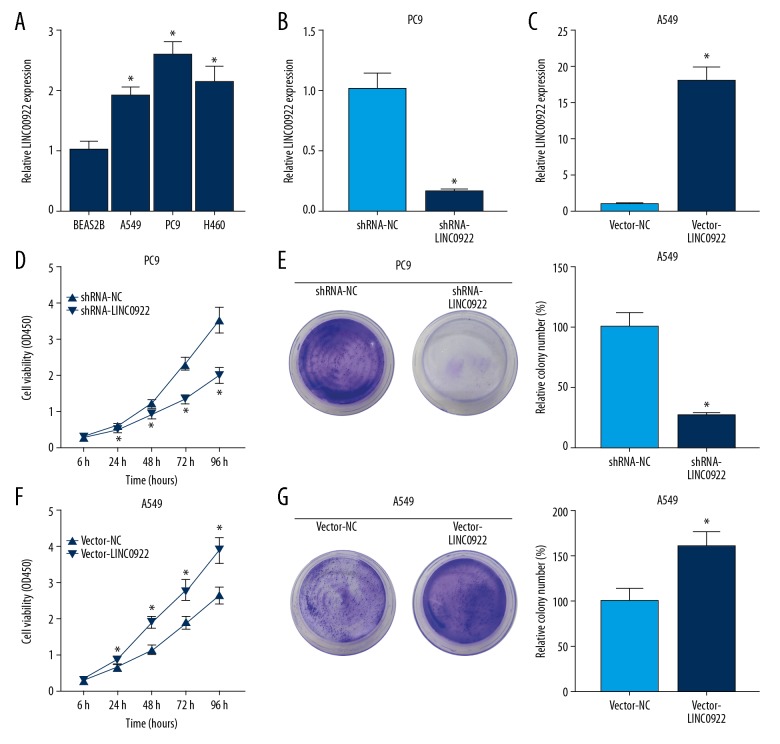
Compared with pulmonary epithelial cells line BEAS2B, LINC00922 was upregulated in lung cancer cell lines A549, PC9, and H460 (Figure 3A). Among the 3 lung cancer cell lines, PC9 and A549 cells expressed the highest and lowest level of LINC00922, respectively. Subsequently, transfection efficacy of shRNA-LINC00922 in PC9 cells and vector-LINC00922 in A549 cells was verified by qRT-PCR (Figure 3B, 3C). CCK-8 and colony formation assay were conducted to evaluate the regulatory effect of LINC00922 on the proliferative capacity of lung cancer. Knockdown of LINC00922 in PC9 cells markedly decreased viability and clonality (Figure 3D, 3E). Conversely, overexpression of LINC00922 in A549 cells elevated viability and clonality (Figure 3F, 3G). These results revealed that LINC00922 overexpression promoted the proliferative capacity of lung cancer cells.
Figure 3.
Overexpression of LINC00922 enhanced the proliferative ability of lung cancer cells. (A) Expression level of LINC00922 in pulmonary epithelial cells line BEAS2B and lung cancer cell lines A549, PC9, and H460 determined by quantitative real-time polymerase chain reaction (qRT-PCR). (B) Transfection efficacy of shRNA-LINC00922 in PC9 cells determined by qRT-PCR. (C) Transfection efficacy of vector-LINC00922 in A549 cells determined by qRT-PCR. (D) Cell Counting Kit-8 (CCK-8) assay revealed viability in PC9 cells transfected with shRNA-NC or shRNA-LINC00922 at 6, 24, 48, 72, and 96 hours. (E) Colony formation assay revealed the relative colony number in PC9 cells transfected with shRNA-NC or shRNA-LINC00922. (F) CCK-8 assay revealed viability in A549 cells transfected with vector-NC or vector-LINC00922 at 6, 24, 48, 72, and 96 hours. (G) Colony formation assay revealed the relative colony number in A549 cells transfected with vector-NC or vector-LINC00922.
Overexpression of LINC00922 accelerated lung cancer cells to invade and migrate
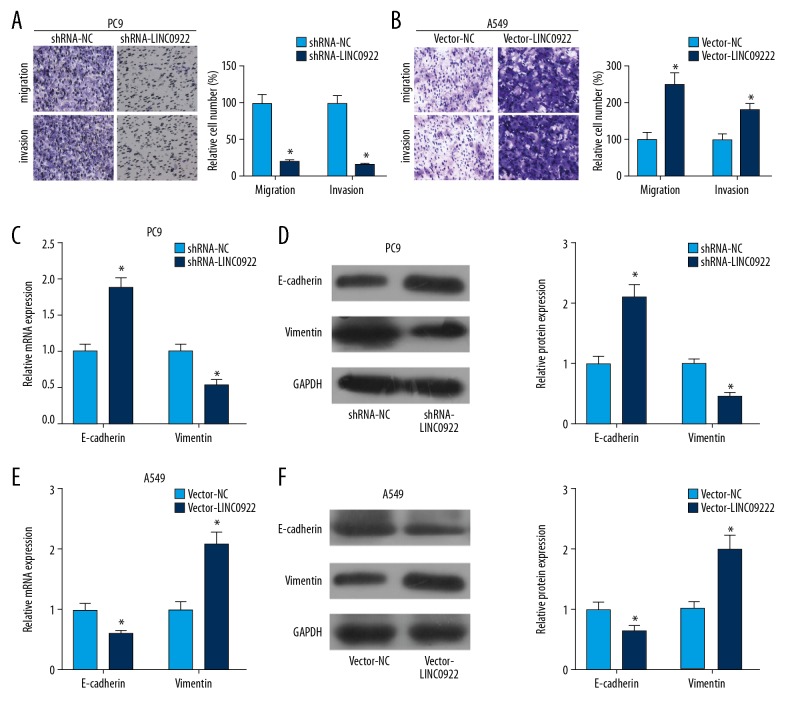
Transwell assay was conducted to evaluate invasive and migratory capacities of lung cancer cells influenced by LINC00922. Transfection of shRNA-LINC00922 attenuated invasive and migratory capacities of PC9 cells (Figure 4A). On the contrary, transfection of vector-LINC00922 in A549 cells enhanced migratory and invasive cell numbers (Figure 4B). It is well known that epithelial-mesenchymal transition (EMT) participates in the migration and invasion of tumor cells. Here, we determined expression levels of EMT-related genes in lung cancer cells influenced by LINC00922. Both mRNA and protein levels of E-cadherin were upregulated, while vimentin was downregulated in PC9 cells transfected with shRNA-LINC00922 (Figure 4C, 4D). Transfection of vector-LINC00922 downregulated E-cadherin and upregulated vimentin in A549 cells (Figure 4E, 4F). It is believed that LINC00922 overexpression enhanced migratory and invasive capacities of lung cancer, and accelerated EMT progression.
Figure 4.
Overexpression of LINC00922 accelerated lung cancer cells to invade and migrate. (A) Transwell assay revealed the invasive and migratory cell number in PC9 cells transfected with shRNA-NC or shRNA-LINC00922. (B) Transwell assay revealed the invasive and migratory cell number in A549 cells transfected with vector-NC or vector-LINC00922. (C) Relative mRNA levels of E-cadherin and vimentin in PC9 cells transfected with shRNA-NC or shRNA-LINC00922. (D) Western blot analyses of E-cadherin and vimentin in PC9 cells transfected with shRNA-NC or shRNA-LINC00922. (E) Relative mRNA levels of E-cadherin and vimentin in A549 cells transfected with vector-NC or vector-LINC00922. (F) Western blot analyses of E-cadherin and vimentin in A549 cells transfected with vector-NC or vector-LINC00922.
LINC00922 upregulated CXCR4 in lung cancer
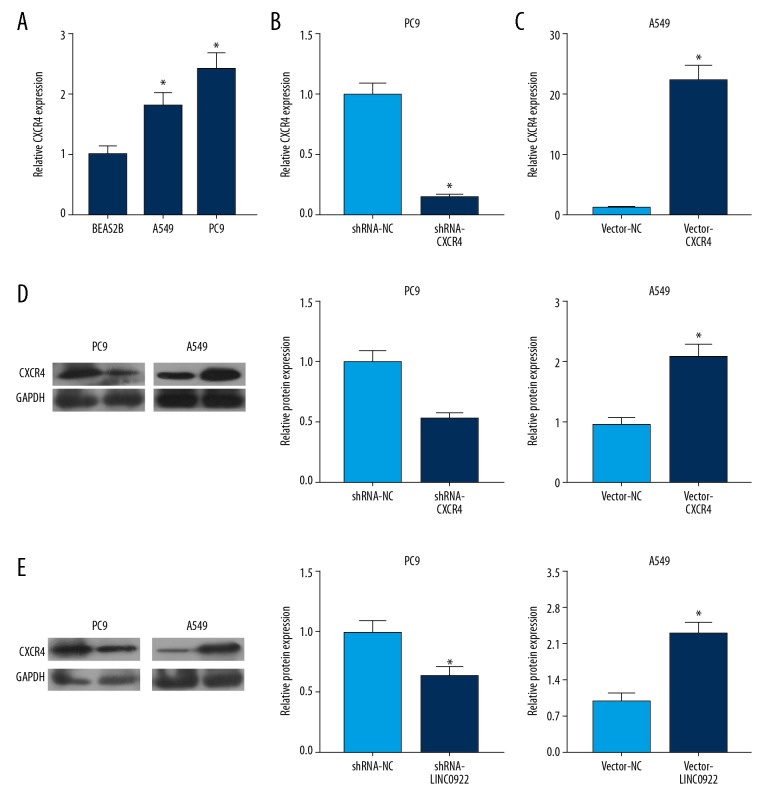
Considering the carcinogenic role of LINC00922 in lung cancer, it is speculated that LINC00922 might exert its function by mediating CXCR4 level. As qRT-PCR data revealed, CXCR4 was upregulated in lung cancer cells (Figure 5A). To further clarify the role of CXCR4 in the progression of lung cancer, shRNA-CXCR4 and vector-CXCR4 were constructed. Transfection of shRNA-CXCR4 downregulated mRNA and protein levels of CXCR4 in PC9 cells (Figure 5B, 5D, right panel). Meanwhile, transfection of vector-CXCR4 markedly upregulated its level in A549 cells (Figure 5C, 5D, left panel). Furthermore, it is demonstrated that knockdown of LINC00922 downregulated CXCR4 level, and LINC00922 overexpression upregulated CXCR4 level in lung cancer cells (Figure 5E). Therefore, LINC00922 positively regulated CXCR4 expression in lung cancer.
Figure 5.
(A–E) LINC00922 upregulated CXCR4 in lung cancer. (A) Transwell assay revealed the invasive and migratory cell number in PC9 cells transfected with shRNA-NC or shRNA-CXCR4. (B) Transwell assay revealed the invasive and migratory cell number in A549 cells transfected with vector-NC or vector-CXCR4. (C) Western blot analyses of E-cadherin and vimentin in PC9 cells transfected with shRNA-NC or shRNA-CXCR4. (D) Western blot analyses of E-cadherin and vimentin in A549 cells transfected with vector-NC or vector-CXCR4.
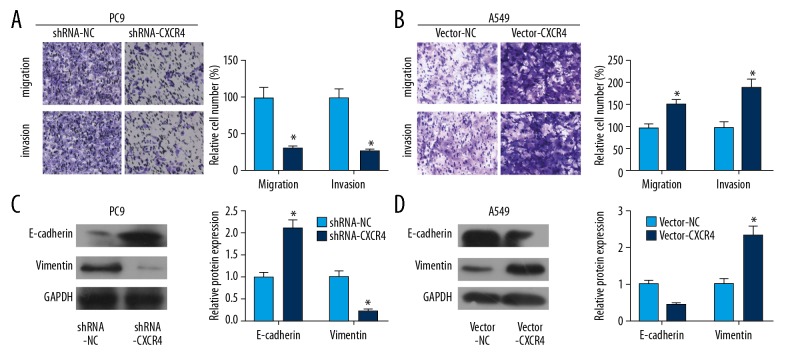
Overexpression of CXCR4 accelerated lung cancer cells to invade and migrate
Through Transwell assay, we found that transfection of shRNA-CXCR4 in PC9 cells decreased migratory and invasive cell numbers, and transfection of vector-CXCR4 in A549 cells obtained the opposite trends (Figure 6A, 6B). Furthermore, overexpression of CXCR4 upregulated E-cadherin and downregulated vimentin in lung cancer cells (Figure 6C). Knockdown of CXCR4 showed the opposite trends at protein levels of E-cadherin and vimentin (Figure 6D). CXCR4 was believed to be an oncogene in lung cancer cells.
Figure 6.
CXCR4 facilitated the invasive and migrated ability of lung cancer. (A) Knockdown of CXCR4 inhibited the invasive and migrated ability of PC9 cell line. (B) Overexpression of CXCR4 facilitated the invasive and migrated ability of A549 cell line. (C, D) The invasive and migrated markers were dysregulated while knocking down or overexpression of CXCR4.
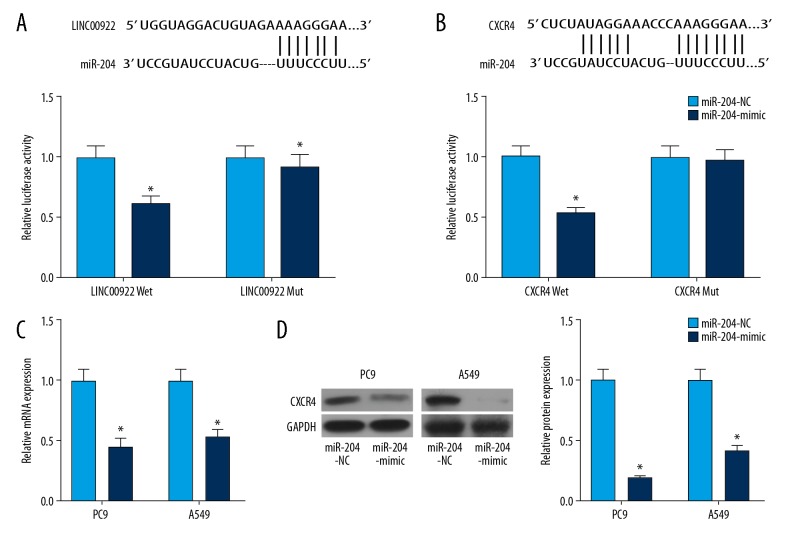
MiRNA-204 bound to LINC00922 and CXCR4
MiRNA-204 was identified to bind to the 3′UTR sequences of LINC00922. Overexpression of miRNA-204 quenched the luciferase activity of LINC00922 Mut, indicating the binding between miRNA-204 and LINC00922 (Figure 7A). Similarly, miRNA-204 could bind to CXCR4 as dual-luciferase reporter gene assay confirmed (Figure 7B). After verifying the transfection efficacy of miRNA-204 mimic, transfection of miRNA-204 mimic remarkably downregulated CXCR4 level (Figure 7C, 7D).
Figure 7.
MiRNA-204 bound to LINC00922 and CXCR4. (A) Binding sequences of LINC00922 and miRNA-204 (upper panel). Relative luciferase activity in cells co-transfected with LINC00922-Wt/LINC00922-Mut and miRNA-204-NC/miRNA-204 mimic (bottom panel). (B) Binding sequences of CXCR4 and miRNA-204 (upper panel). Relative luciferase activity in cells co-transfected with CXCR4-Wt/CXCR4-Mut and miRNA-204-NC/miRNA-204 mimic (bottom panel). (C) Transfection efficacy of miRNA-204 mimic in PC9 and A549 cells determined by quantitative real-time polymerase chain reaction. (D) Western blot analysis of CXCR4 in PC9 and A549 cells transfected with miRNA-204-NC or miRNA-204 mimic.
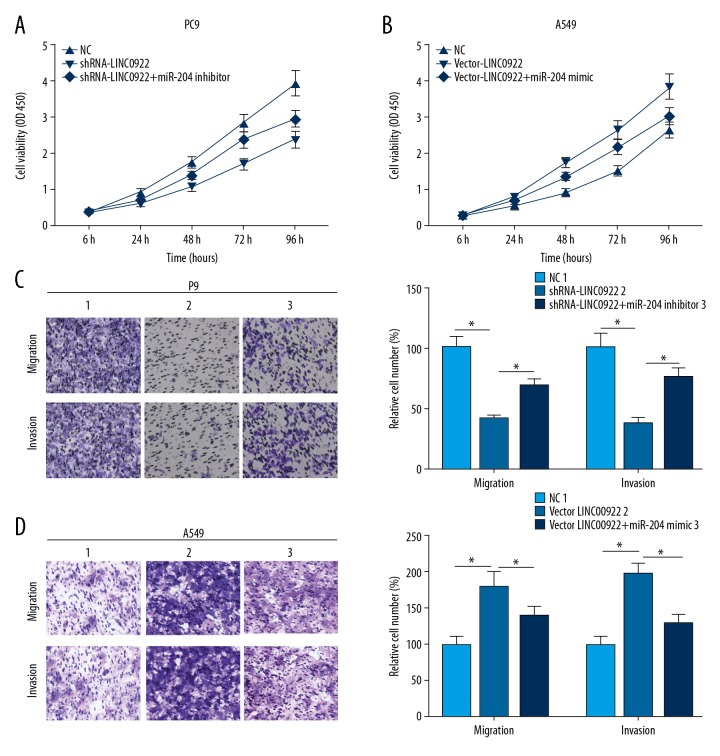
MiRNA-204 reversed the promotive effects of LINC00922 on invasive and migratory capacities of lung cancer cells
LINC00922 could be a ceRNA to competitively bind to miRNA-204, thus mediating the cellular behaviors of lung cancer cells. Here, LINC00922 knockdown inhibited viability of PC9 cells, which was partially reversed after miRNA-204 knockdown (Figure 8A). In A549 cells transfected with vector-LINC00922, the viability markedly increased, which was reversed by transfection of miRNA-204 mimic (Figure 8B). Identically, inhibited migratory and invasive capacities of PC9 cells with LINC00922 knockdown were partially reversed by miRNA-204 knockdown (Figure 8C). Similarly, the increased migration and invasion in A549 cells overexpressing LINC00922 were reversed by overexpression of miRNA-204 (Figure 8D).
Figure 8.
MiRNA-204 reversed the promotive effects of LINC00922 on invasive and migratory capacities of lung cancer cells. PC9 cells were transfected with NC, shRNA-LINC00922 or shRNA-LINC00922+miRNA-204 inhibitor, respectively. (A) Cell Counting Kit-8 (CCK-8) assay revealed viability in PC9 cells at 6, 24, 48, 72, and 96 hours. (B) CCK-8 assay revealed viability in A549 cells at 6, 24, 48, 72, and 96 hours. (C) Transwell assay revealed the invasive and migratory cell number in PC9 cells. A549 cells were transfected with NC, vector-LINC00922 or vector-LINC00922+miRNA-204 mimic, respectively. (D) Transwell assay revealed the invasive and migratory cell number in A549 cells.
Discussion
Lung cancer ranks first place in tumor death throughout the world, with a high mortality and mobility, and it is reported that approximately 2 2000 lung cancer cases are diagnosed yearly [27,28]. The prognosis of lung cancer is relatively poor, and the overall 5-year survival rate is only 16.6% according to previous studies [28]. Because most lung cancer patients are diagnosed at an advanced stage, and the metastatic loci and drug-resistance are easily developed at the early stage of treatment, the clinical outcomes of lung cancer remain poor [29]. Similar to other human malignant cancers, the imbalance of oncogenes and tumor-suppressor genes is found during the tumorigenesis, development, and progression of lung cancer [30].
RNA sequencing (RNA-seq) is a recently developed sequencing technology that recognizes unmapped genes, unrecognized non-coding RNAs, and splice variants [31–33]. RNA-seq provides a novel tool for clinical diagnosis and treatment of lung cancer [31–33]. Multiple miRNAs and lncRNAs have been proven to be closely related to the diagnosis and prognosis of lung cancer [34,35]. With the contribution of next-generation sequencing, transcriptome sequencing of a large number of tumor samples and survival analysis, various tumor hallmarks have emerged [36]. In this study, LINC00922 was upregulated in lung cancer and closely related to poor prognosis. It is speculated that LINC00922 served as an ontogenetic role in lung cancer. Overexpression of LINC00922 accelerated lung cancer cells to proliferate, migrate, and invade. Furthermore, LINC00922 had a certain impact on EMT, manifesting as upregulated E-cadherin and downregulated vimentin after overexpression of LINC00922.
CXCR4, a chemokine receptor, and its only agonist, CXCL12, have been reported to regulate the progression of solid tumors, including prostate cancer [37], breast cancer [38], colon cancer [39], and lung cancer [40,41]. CXCL16 has with many biological functions. Huang et al. [42] suggested that CXCL16 is highly expressed in various tumors such as breast cancer, pancreatic cancer, and prostatic cancer. Ke et al. [43] showed that CXCL16 is highly expressed in NSCLC patients, and also influences survival time. Moreover, silencing of CXCL16 not only suppresses proliferation of A549 and PC-9 cells, but also decreases their invasive ability [44]. In this study, CXCR4 was upregulated in lung cancer tissues. Its level was closely related to a poor prognosis of lung cancer patients. CXCR4 was upregulated in lung cancer cells as well. Overexpression of CXCR4 accelerated the proliferation and metastasis of lung cancer cells. EMT-related genes were found to be regulated by CXCR4 in lung cancer. Meanwhile, we further demonstrated that LINC00922 bound to CXCR4 and positively regulated its level in lung cancer cells. The specific mechanism of CXCR4 to promote lung cancer progression needs further investigations.
LncRNAs are reported to serve as ceRNAs to sponge miRNA, thus downregulating miRNA expressions [45]. For example, UCA1 upregulates the oncogene ERBB4 in lung cancer via binding to miR-193-3p [46]. LncRNA NORAD promotes cell proliferation and glycolysis in NSCLC by acting as a sponge for miR-136-5p [47]. To clarify whether LINC00922 served as a ceRNA to sponge a certain miRNA and further mediated CXCR4, dual-luciferase reporter gene assay was conducted to verify the binding between miRNA-204 to LINC00922 and CXCR4. It showed that LINC00922 upregulated CXCR4 by competitively binding to miRNA-204. A previous study had already confirmed the inhibitory effect of miRNA-204 on the progression of NSCLC by mediating SIX1 level [48]. Here, our results illustrated that silencing of miRNA-204 could reverse the attenuated proliferative, migratory and invasive abilities in lung cancer cells with LINC00922 knockdown. Conversely, overexpression of miRNA-204 reversed the promotive effects of overexpressed LINC00922 on the cellular behaviors of lung cancer. The aforementioned results indicated that miRNA-204 could reverse the promotive role of LINC00922 in mediating the malignant behaviors of lung cancer cells, presenting as a tumor-suppressor gene.
Conclusions
LINC00922 was upregulated in lung cancer, and accelerated lung cancer cells to proliferate, migrate, and invade via the miRNA-204/CXCR4 axis as a ceRNA.
Footnotes
Source of support: This study was supported by National Natural Science Foundation (81573026)
Conflict of interest
None.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Rusthoven CG, Kavanagh BD, Karam SD. Improved survival with stereotactic ablative radiotherapy (SABR) over lobectomy for early stage non-small cell lung cancer (NSCLC): Addressing the fallout of disruptive randomized data. Ann Transl Med. 2015;3:149. doi: 10.3978/j.issn.2305-5839.2015.06.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–99. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 4.Kris MG, Johnson BE, Berry LD. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–88. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: A new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 10.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–35. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–33. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–83. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghidini M, Braconi C. Non-coding RNAs in primary liver cancer. Front Med (Lausanne) 2015;2:36. doi: 10.3389/fmed.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 18.Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin L, Gu ZT, Chen WH, Cao KJ. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol. 2015;10:14. doi: 10.1186/s13000-015-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language? Cell. 2011;146:353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou M, Wang X, Shi H, et al. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H, He Y, Lin L, et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37:1683–91. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 23.Jiao C, Song Z, Chen J, et al. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36:2960–66. doi: 10.3892/or.2016.5121. [DOI] [PubMed] [Google Scholar]
- 24.Gao XH, Li J, Liu Y, et al. ZNF148 modulates TOP2A expression and cell proliferation via ceRNA regulatory mechanism in colorectal cancer. Medicine (Baltimore) 2017;96:e5845. doi: 10.1097/MD.0000000000005845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099–107. [PMC free article] [PubMed] [Google Scholar]
- 26.Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soo RA, Lim SM, Syn NL, et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: Current controversies and future directions. Lung Cancer. 2018;115:12–20. doi: 10.1016/j.lungcan.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 29.Ma XP, Zhang W, Wu BQ, et al. Correlations between mRNA levels of centrosomal protein 55 (CEP55) and clinical features of patients with lung cancer. Med Sci Monit. 2018;24:3093–97. doi: 10.12659/MSM.907266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu M, Wang M, Jiang Y, et al. Gambogic acid induces apoptosis of non-small cell lung cancer (NSCLC) cells by suppressing notch signaling. Med Sci Monit. 2018;24:7146–51. doi: 10.12659/MSM.912563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du X, Qi F, Lu S, et al. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of miR-99b and miR-192. Biomed Pharmacother. 2018;101:656–62. doi: 10.1016/j.biopha.2018.02.113. [DOI] [PubMed] [Google Scholar]
- 32.Zhan S, Wang C, Yin F. MicroRNA-29c inhibits proliferation and promotes apoptosis in non-small cell lung cancer cells by targeting VEGFA. Mol Med Rep. 2018;17:6705–10. doi: 10.3892/mmr.2018.8678. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Q, Zang Q, Jiang ZM. Enhanced expression of non-coding miR 92a expression is implicated in the development of lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:1028–34. doi: 10.26355/eurrev_201802_14385. [DOI] [PubMed] [Google Scholar]
- 34.Ye ZH, Wen DY, Cai XY, et al. The protective value of miR-204-5p for prognosis and its potential gene network in various malignancies: A comprehensive exploration based on RNA-seq high-throughput data and bioinformatics. Oncotarget. 2017;8:104960–80. doi: 10.18632/oncotarget.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audoux J, Philippe N, Chikhi R, et al. DE-kupl exhaustive capture of biological variation in RNA-seq data through k-mer decomposition. Genome Biol. 2017;18:243. doi: 10.1186/s13059-017-1372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha S, Yang CD, Hong HC, et al. Integrated microRNA-mRNA analysis reveals miR-204 inhibits cell proliferation in gastric cancer by targeting CKS1B, CXCL1 and GPRC5A. Int J Mol Sci. 2017;19:87. doi: 10.3390/ijms19010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Molecular mechanisms of bone metastasis. Cancer Genomics Proteomics. 2016;13:1–12. [PubMed] [Google Scholar]
- 38.Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Des Devel Ther. 2015;9:4953–64. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroda K, Fukuda T, Krstic-Demonacos M, et al. MiR-663a regulates growth of colon cancer cells, after administration of antimicrobial peptides, by targeting CXCR4-p21 pathway. BMC Cancer. 2017;17:33. doi: 10.1186/s12885-016-3003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao H, Guo L, Zhao H, et al. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget. 2015;6:5022–40. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirbe AC, Morgan EA, Weilbaecher KN. The CXCR4/SDF-1 chemokine axis: A potential therapeutic target for bone metastases? Curr Pharm Des. 2010;16:1284–90. doi: 10.2174/138161210791034012. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Zhang J, Cui ZM, et al. Expression of the CXCL12/CXCR4 and CXCL16/CXCR6 axes in cervical intraepithelial neoplasia and cervical cancer. Chin J Cancer. 2013;32(5):289–96. doi: 10.5732/cjc.012.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ke C, Ren Y, Lv L, et al. Association between CXCL16/CXCR6 expression and the clinicopathological features of patients with non-small cell lung cancer. Oncol Lett. 2017;13(6):4661–68. doi: 10.3892/ol.2017.6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang K, Liu Y, Eer D, et al. High CXC chemokine ligand 16 (CXCL16) expression promotes proliferation and metastasis of lung cancer via regulating the NF-κB Pathway. Med Sci Monit. 2018;24:405–11. doi: 10.12659/MSM.906230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yonesaka K, Tamura K, Kurata T, et al. Small interfering RNA targeting survivin sensitizes lung cancer cell with mutant p53 to adriamycin. Int J Cancer. 2006;118:812–20. doi: 10.1002/ijc.21350. [DOI] [PubMed] [Google Scholar]
- 46.Gao W, Weng T, Wang L, et al. Long non coding RNA NORAD promotes cell proliferation and glycolysis in non small cell lung cancer by acting as a sponge for miR 136 5p. Mol Med Rep. 2019;19(6):5397–405. doi: 10.3892/mmr.2019.10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crnkovic-Mertens I, Hoppe-Seyler F, Butz K. Induction of apoptosis in tumor cells by siRNA-mediated silencing of the livin/ML-IAP/KIAP gene. Oncogene. 2003;22:8330–36. doi: 10.1038/sj.onc.1206973. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, Zhu Y, Ma T, et al. MiR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett. 2014;588:3703–12. doi: 10.1016/j.febslet.2014.08.016. [DOI] [PubMed] [Google Scholar]