Abstract
Purpose
The activity of the cationic antimicrobial peptide WLBU2 was evaluated against planktonic cells and biofilms of multi-drug resistant (MDR) Acinetobacter baumannii and Klebsiella pneumoniae, alone and in combination with classical antimicrobial agents.
Methods
Control American Type Culture Collection (ATCC) strains and MDR clinical isolates of A. baumannii and K. pneumoniae were utilized. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of WLBU2 alone and in combination with antimicrobials were determined by classical methods. The Calgary biofilm device was used to determine the minimum biofilm eradication concentration (MBEC). The MTT assay was used to determine the cytotoxicity of agents on eukaryotic cells. The electrophoretic mobility shift assay was used to evaluate the ability of WLBU2 to bind bacterial DNA.
Results
The WLBU2 MIC and MBC values were identical indicating bactericidal activity. The MIC/MBC values ranged from 1.5625 to 12.5 µM. At these concentrations, Vero cells and human skin fibroblasts were viable. The MBEC of WLBU2 ranged from 25 to 200 µM. A significant loss of eukaryotic cell viability was observed at the MBEC range. The combination of sub-inhibitory concentrations of WLBU2 with amoxicillin-clavulanate or ciprofloxacin for K. pneumoniae, and with tobramycin or imipenem for A. baumannii, demonstrated synergism, leading to a significant decrease in MIC and MBEC values for some isolates and ATCC strains. However, all combinations were associated with considerable loss in eukaryotic cells’ viability. WLBU2 did not demonstrate the ability to bind bacterial plasmid DNA.
Conclusion
WLBU2 in combination with antimicrobials holds promise in eradication of MDR pathogens.
Keywords: antimicrobial peptide, synergy, combination therapy, biofilm, multi-drug resistance, bacteria
Introduction
Bacterial resistance to conventional antimicrobials is increasing year after year and is a global health emergency.1 Biofilm formation is a common mechanism to overcome the activity of antimicrobials and the host immune response. Due to treatment difficulties, biofilm-associated infections have led to significant rates of morbidity and mortality among the community and the health-care settings.2,3 Hence, developing safe alternate therapies with diverse mechanisms of action research is urgently needed.4–6
Antimicrobial peptides (AMPs) are immune effector molecules that are being considered as potential alternatives for conventional antimicrobial agents. AMPs demonstrate multiple mechanisms of action, including formation of transmembrane pores, which lead to lysis of microorganisms, the deterioration of bacterial viability by interfering with cell wall biosynthesis,7,8 the disruption of biochemical processes, and enhancement and activation of the immune response. Some AMPs are effective against biofilm and multi-drug resistant (MDR) bacteria, with rapid killing kinetics.9 Hence, AMPs have advantages over antimicrobial agents, as the bacteria are less likely to produce and transfer resistance genes against the peptides.7
Most AMPs are cationic peptides with an amphipathic structure that selectively targets bacterial membranes via electrostatic forces.10 WLBU2 is a 24-residue engineered cationic amphipathic peptide (eCAP) that consists of only three types of amino acids: tryptophan, valine, and arginine. The WLBU2 sequence was rationally designed by amino acid substitutions from precursor peptides to have an ideal amphipathic helix conformation to maximize antimicrobial properties, while minimizing epithelial cell cytotoxicity.11–13
Acinetobacter baumannii is a significant Gram-negative MDR pathogen associated with urinary tract infections, pulmonary infections, wound infection, and infections of other tissues and organs. A. baumannii has the ability to form biofilms, leading to increased virulence.14,15 Klebsiella pneumoniae is a Gram-negative, encapsulated, opportunistic pathogen associated with pneumonia, meningitis, urinary tract infections, sepsis, and surgical wound site infections. K. pneumoniae is the main cause of health care-associated Klebsiella infections, mostly involving the respiratory and urinary tracts.16
This study provides insights on the activity of WLBU2 against planktonic and biofilm-producing MDR A. baumannii and K. pneumoniae, and in combination with classical antimicrobial agents. This study also examined the potential toxicity of WLBU2-antimicrobials combinations on human skin fibroblast cells and Vero cells, and the ability of WLBU2 to bind bacterial DNA. The findings pave the way for future investigations that can potentially culminate in the development of treatments for difficult to treat and biofilm-associated infections.
Materials and methods
The study was approved by Jordan University of Science and Technology (JUST) research committee. Requirement for approval by the institutional review board of JUST was waived as the study did not involve the study of human subjects, human data or tissue, or animals.
Bacterial strains
Forty-three K. pneumoniae (n=24) and A. baumannii (n=19) clinical isolates, which were previously isolated and stored at −80°C from four major hospitals in Jordan were utilized in this study. All isolates had an MDR phenotype (resistance to at least three antimicrobial agent classes; data not shown). Six A. baumannii and K. pneumoniae control strains obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) were also included (Table 1).
Table 1.
ATCC strains utilized in the study
| Species | ATCC number | Characteristics |
|---|---|---|
| Acinetobacter baumannii | BAA-1605 | Resistance to ceftazidime, gentamicin, ticarcillin, piperacillin, aztreonam, cefepime, ciprofloxacin, imipenem, and meropenem. Susceptible to amikacin and tobramycin |
| 19606 | Quality control strain | |
| Klebsiella pneumoniae | BAA-2146 | Quality control strain |
| BAA-1705 | Carbapenem-resistant (imipenem and ertapenem) | |
| BAA-1706 | Quality control strain | |
| 700603 | Control for extended beta-lactamase production | |
| Quality control strain. |
Abbreviation: ATCC, American Type Culture Collection.
WLBU2
WLBU2 is a peptide consisting of 24-amino acid residues. The amino acid sequence is (RRWVRRVRRWVRRVVRVVRRWVRR). The molecular weight is 3398 g/mol (~3400 g/mol). WLBU2 was synthesized by GL Biochem (Minhang Qu, Shanghai Shi, People's Republic of China). Synthesis was based on the solid phase method and standard Fmoc chemistry. The confirmation and purification of the synthesized peptide was performed by mass spectrometry and HPLC. The purity was 95.71%. WLBU2 concentration conversion from µM to µg/mL can be done by multiplying the value in µM by (3.4).
Antimicrobial agents
Four antimicrobials (Cayman Chemicals, Ann Arbor, MI, USA) were used for susceptibility tests. Imipenem and tobramycin were utilized for A. baumannii experiments, while amoxicillin/clavulanate and ciprofloxacin were utilized for K. pneumoniae experiments. The antimicrobial agent choices were based on the recommendations by the Clinical and Laboratories Standards Institute (CLSI) antimicrobial susceptibility testing standards, 26th edition (2016).
Cell lines
Eukaryotic cell viability experiments utilized Vero cells (African green monkey kidney epithelial cells; ATCC CCL-81) and human skin fibroblast cells (ATCC PCS-201–012). Both cell lines were obtained from the ATCC.
Quantitative biofilm formation assay
The 96-well tissue culture plate method was utilized for quantitative evaluation of biofilm formation as described previously with modifications.17 Briefly, the clinical isolates and ATCC strains were subcultured from frozen glycerol stock onto nutrient agar to obtain pure-well isolated colonies. Next, two to three colonies were inoculated in 10 mL of trypticase soy broth with 1% glucose. The broth cultures were incubated at 37°C for 24 hrs and were diluted 1:100 with fresh medium. Individual wells of a sterile 96-well plate were filled with 200 µL of the diluted cultures. Negative control wells contained sterile broth media. The microtiter plate was incubated for 24 hrs at 37°C. The contents of each well were discarded by inversion and gentle tapping on absorbent paper towels, and the wells were washed with 200 μL of PBS (pH 7.2) three to four times to remove free-floating planktonic bacteria. Next, 200 μL of 0.1% crystal violet solution in water were added per well for 30 mins at room temperature to stain the biofilms. The plate was rinsed with deionized water to remove excess stain. Finally, 200 μL of absolute ethanol were added to each well to solubilize the stain. Absorbance at 575 nm (OD575) was measured for each well to obtain quantitative data on biofilm formation, using an Epoch ELISA plate reader (BioTek, Winooski, VT, USA).
The test was carried out in duplicates and the average was calculated for each bacterial strain. The cutoff optical density (ODc) for biofilm formation was defined as three standard deviations above the mean OD of the negative control. Optical density data were interpreted according to Table 2.
Table 2.
Interpretation of optical density data for detection of biofilm formation
| Average OD value | Interpretation |
|---|---|
| OD≤ODc | No biofilm formation |
| ODc<OD≤2xODc | Weak biofilm formation |
| 2xODc<OD≤4xODc | Moderate biofilm formation |
| 4xODc<OD | Strong biofilm formation |
Notes: All OD values were measured at 575 nm; ODc, average OD of negative control +3x standard deviation of the negative controls.18
Abbreviation: ODc, optical density cutoff value.
Minimum inhibitory and bactericidal concentrations of WLBU2 against planktonic bacteria
To determine the lowest concentration of WLBU2 required to inhibit visible growth of the microorganisms, WLBU2 was dissolved in PBS (pH=7.3) to achieve 100 µM or 340 µg/mL. The peptide was serially diluted twofold from 100 to 1.563 µM or 340 to 5.313 µg/mL. The clinical isolates and ATCC strains were cultured in sterile Muller-Hinton broth (MHB) (Oxoid, Basingstoke, Hampshire, UK), incubated aerobically overnight, and diluted with fresh MHB media to reach 106 CFU/mL. Next, 50 μL of the respective peptide concentrations and 50 µL of bacterial suspension were added to wells of sterile 96-well polypropylene microtiter plates. Plates were incubated at 37°C for 20 hrs in a humidified incubator. The minimum inhibitory concentration (MIC) of the peptide capable of inhibiting visible bacterial growth was quantitatively determined by measuring OD600 for each well using an Epoch ELISA plate reader (BioTek). All MIC determinations were made in triplicate. Sterile MHB (Oxoid) was used as negative control. Wells having bacteria alone without the peptide served as positive controls.19
The minimum bactericidal concentration (MBC) of WLBU2 needed to kill ≥99.9% of bacteria was determined by inoculating 10 μL from the wells demonstrating the MIC concentration or higher, on Muller-Hinton agar (Oxoid) (incubated for 24 hrs at 37°C) to count viable cells. The lowest concentration that led to ≥99.9% decrease in CFUs/mL was considered the MBC.
MIC of antimicrobial agents
The minimum concentration capable of inhibiting visible bacterial growth was determined for each antimicrobial agent. A specific weigh for each antimicrobial agent was dissolved in PBS (pH =7.3) to yield stock concentrations. The stock concentrations corresponded to the upper limits of MIC values as indicated by CLSI antimicrobial susceptibility testing guidelines (2016). Bacterial density was fixed to approximately 106 CFU/mL. Next, 50 μL of the bacteria were added to wells having 50 μL of each antimicrobial agent concentration (twofold serial dilution) in 96-well plates, and the plates were incubated aerobically overnight at 37°C. The MIC values were determined by measuring OD600 using an Epoch ELISA plate reader (BioTek).
Synergism between WLBU2 and antimicrobial agents against planktonic bacteria
Briefly, 25 μL of the WLBU2 at its sub-MIC level for each respective isolate (ie, concentration=25% of MIC) was added to a 50 μL inoculum of 105 CFU/mL planktonic bacteria, in respective wells of a 96 well microtiter plates, in triplicates. Next, serial dilutions of the antimicrobial agents at 25 μL volumes were added to the wells, and the plates were incubated at 37°C for 24 hrs. Absorbance was measured as an indicator of growth inhibition (OD600) using an Epoch ELISA plate reader (BioTek).20 The CLSI, 2016 concentration values for resistance, intermediate susceptibility, and susceptibility, respectively, in μg/mL were 128, 64, and 32 for amoxicillin-clavulanate, 16, 8, and 4 for ciprofloxacin, 64, 32, and 16 for tobramycin, and 32, 16, and 8 for imipenem.
Minimum biofilm eradication concentration (MBEC) of WLBU2 against biofilm-forming bacteria
The Calgary biofilm device (Innovotech, Edmonton, Alberta, Canada) method is a reliable assay of biofilm formation. In this study, the Calgary biofilm device was used to determine WLBU2’s MBEC against ATCC strains and several representative biofilm-forming clinical isolates. Briefly, 150 µL of bacterial suspensions having a density of 105 CFU/mL were transferred into wells of a 96-well MBEC plate. Next, the plate’s lid having the pegs was placed, and the plate was put on a platform shaker incubator set at 110 rpm and 37ºC for 20 hrs, to allow for biofilm formation on lid pegs. Negative control wells had 150 µL of sterile MHB (Oxoid, ). Following incubation, the lid pegs were washed three times with sterile PBS to remove non-adherent cells.
Next, a challenge plate was prepared by transferring 200 µL of each WLBU2 concentration (twofold serial dilutions made in PBS) to respective wells of the plate. Next, lid pegs were placed, and the plate was incubated at 37ºC for 18 hrs. The lid pegs were subsequently placed into a recovery plate, containing fresh MHB (Oxoid) and the plate was sonicated for 30 mins in a water bath. Finally, the plate was incubated with the lid pegs at 37ºC for 20 hrs. To determine MBEC values, growth in each well was determined by measuring OD600 using an Epoch ELISA plate reader (BioTek). The MBEC value for each strain corresponded with the lowest WLBU2 concentration resulting in growth inhibition. All samples were run in triplicates.
Antimicrobial agents’ MBEC in the presence of WLBU2 at sub-MBEC
The experiment was performed as described for the MBEC assay above with the following changes: the challenge plate was prepared by adding 25 μL of WLBU2 at its sub-MBEC level for each respective isolate (ie, concentration=25% of MBEC) in respective wells of 96-well microtiter plates. Next, 25 μL of serial dilutions of the antimicrobial agents were added, and the plates were incubated at 37°C for 24 hrs. The MBEC values for the antimicrobial agents corresponded with the lowest concentration that resulted in growth inhibition. All samples were run in triplicates.
Cell viability assay
The MTT assay was used to assess potential reduction in cell viability after treatment of eukaryotic cells with WLBU2 alone and in combination with antimicrobial agents. Briefly, Vero cells and human skin fibroblasts were seeded in a flat-bottomed 96-well plate at 5000 cells (in 100 μL) per well, and incubated at 37ºC for 24 hrs under 5% CO2. Next, medium was removed, and 100 μL of fresh media containing different concentrations of the WLBU2 alone or in combination with the antimicrobial agents at the indicated CLSI resistance cutoff values were adder per respective wells. The plates were incubated at 37ºC for 24 hrs, and medium was replaced with fresh medium containing 30 μL (2.5 mg/mL) MTT solution (Sigma-Aldrich, St. Louis, MO, USA). The plate was incubated for 4–6 hrs at 37°C, under 5% CO2. After medium/MTT was removed, 100 μL DMSO were added to dissolve the formazan crystals. Cell survival rates were calculated by measuring absorbance at 540 nm using an Epoch ELISA plate reader (BioTek). Medium without treatment was used for the positive control wells. All tests were run in triplicates.
The true relative value of viable cells was calculated using the equation: (Sample A570-Background A650)/(Control A570-Background A650)×100.
DNA binding assay
The ability of the WLBU2 to bind bacterial plasmid DNA was investigated using the electrophoretic mobility shift assay. In brief, various concentrations of WLBU2 (range: 200–1.563 µM or 680–5.313 µg/mL) were incubated with 250 ng of bacterial plasmid DNA (pUC19; 2686 base pairs, NEB, USA) in 30 μL binding buffer (10 mM Tris-HCl and 1 mM EDTA buffer, pH 8.0) at room temperature for 30 mins. After incubation, 20 μL of the mixture were analyzed by electrophoretic separation on 1.5% agarose gel. Each gel also included 7 μL of 1 Kb DNA ladder (NEB, USA) per well. DNA migration was visualized by ethidium bromide staining followed by UV transillumination.4
Data analysis
Data analysis was performed using Microsoft Excel version 2016 (Microsoft, Redmond, Washington, USA) and GraphPad Prism version 6.05 (GraphPad Software, San Diego, CA, USA). All values represent means of multiple runs.
Results
Quantitative biofilm detection using 96-well plate assay
Four K. pneumoniae clinical isolates (ID# 6, 12, 18, and 20) were moderate biofilm producers, while the remaining 20 isolates and all 19 A. baumannii clinical isolates were strong biofilm producers. K. pneumoniae ATCC strains (BAA-2146, BAA-1706 and 700603) were strong biofilm producers, while BAA-1705 showed moderated biofilm production. A. baumannii ATCC strains 19606 showed moderate biofilm production, while BAA-1605 showed strong biofilm production.
MIC and MBC for WLBU2 against planktonic bacteria
MIC and MBC values were identical for each respective isolate and control strain (Table 3). MIC/MBC values for WLBU2 ranged from 3.125 to 12.500 µM or 10.625 to 42.500 µg/mL for K. pneumoniae clinical isolates (mean=7.943 µM or 27.006 µg/mL), and 1.563 to 12.500 µM or 5.313 to 42.500 µg/mL for A. baumannii clinical isolates (mean=7.484 µM or 25.446 µg/mL).
Table 3.
MIC and MBC results of WLBU2 against study isolates
| K. pneumoniae ID# | A. baumannii ID# | MIC/MBC value (µM; [µg/mL]) |
|---|---|---|
| – | 36 | 1.563 [5.313] |
| 2, 4, 8, 16, 23 | 32, 38, 41 | 3.125 [10.625] |
| ATCC BAA-2146, ATCC BAA-1705, ATCC BAA-1706 | ATCC BAA-1605 | |
| ATCC 19606 | ||
| 1, 5–7, 10, 12, 17, 18, 20, 21 | 26, 28, 29, 31, 34, 35, 37, 39, 42 | 6.250 [21.250] |
| ATCC 700603 | ||
| 3, 9, 11, 13–15, 19, 22, 24 | 25, 27, 30, 33, 40, 43 | 12.500 [42.500] |
Note: MICs indicated represent values for each respective clinical isolate or control strain.
Abbreviations: ID#, identification number; MIC, minimum inhibitory concentration; MBC, minimum bactericidal concentration.
MIC of antimicrobial agents
Table 4 summarizes the MIC values of the study isolates and control strains against antimicrobial agents.
Table 4.
Study isolates’ MIC values against antimicrobial agents
| Species | Antimicrobial agent | Isolate ID# (MIC value) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| K. pneumoniae | Amoxicillin-clavulanate | – | – | 8, 12, ATCC 700603 (32 μg/mL) |
| 1–7, 9–11, 13–24 (>32 μg/mL) | ||||
| Ciprofloxacin | 12, ATCC 700603 (1 μg/mL) | – | 7, ATCC BAA-1706 (8 μg/mL) | |
| 1–6, 8–11, 13–24 (>8 μg/mL) | ||||
| A. baumannii | Tobramycin | ATCC 19606 (4 μg/mL) | ATCC BAA-1605 (MIC =8 μg/mL) | 36, 43 (16 μg/mL) |
| 25–35, 37–42 (>16 μg/mL) | ||||
| Imipenem | ATCC 19606 (2 μg/mL) | – | 26, 30, ATCC BAA-1605 (8 μg/mL) 25, 27–29. 31–43 (>8 μg/mL) |
|
Note: MICs indicated represent values for each respective clinical isolate or control strain.
Abbreviations: ID#, identification number; MIC, minimum inhibitory concentration.
Synergistic activity of WLBU2 at 25% MIC with antimicrobial agents against planktonic bacteria
Bacterial isolates that demonstrated intermediately susceptible or resistant MIC values against antimicrobial agents were utilized for potential synergistic activity testing between WLBU2 at its sub-inhibitory concentration (25% of WLBU2 MIC for each respective isolate), with antimicrobial agents at different concentrations. Results are indicated in Table 5.
Table 5.
Result of synergistic testing between sub-inhibitory WLBU2 concentrations and antimicrobial agents for planktonic cells
| Species | Antimicrobial agent | Isolate ID# | MIC of antimicrobial agent alone (μg/mL) | Synergism test | ||
|---|---|---|---|---|---|---|
| Concentration of WLBU2 used for synergism test (µM; [μg/mL]) | Observed antimicrobial agent MIC (μg/mL) in presence of WLBU2 | Synergism observed (interpretation) | ||||
| K. pneumoniae | Amoxicillin-clavulanate | 8 | 32 | 0.781 [2.655] | >32 | No |
| 12 | 32 | 1.563 [5.314] | 8 | Yes (susceptible) | ||
| ATCC BAA-2146 | >32 | 0.781 [2.655] | >32 | No | ||
| ATCC BAA-1705 | >32 | 0.781 [2.655] | 16 | Yes (intermediate) | ||
| ATCC BAA-1706 | >32 | 0.781 [2.655] | >32 | No | ||
| ATCC 700603 | 32 | 1.563 [5.314] | >32 | No | ||
| Ciprofloxacin | 7 | 8 | 1.563 [5.314] | 2 | Yes (intermediate) | |
| BAA-2146 | >4 | 0.781 [2.655] | >4 | No | ||
| BAA-1705 | >4 | 0.781 [2.655] | >4 | No | ||
| BAA-1706 | 8 | 0.781 [2.655] | 4 | Yes (resistant) | ||
| A. baumannii | Tobramycin | 36 | 16 | 0.391 [1.329] | >16 | No |
| 43 | 16 | 0.781 [2.655] | >16 | No | ||
| ATCC BAA-1605 | 8 | 0.781 [2.655] | >16 | No | ||
| Imipenem | 26 | 8 | 1.563 [5.314] | 8 | No | |
| 30 | 8 | 0.781 [2.655] | 2 | Yes (susceptible) | ||
| ATCC BAA-1605 | 8 | 0.781 [2.655] | 4 | Yes (intermediate) | ||
Note: Concentration of WLBU2 used for synergism test corresponded with 25% of MIC values for each respective clinical isolate or control strain.
Abbreviations: ID#, identification number; MIC, minimum inhibitory concentration.
Overall, synergism was observed for K. pneumoniae between WLBU2 and amoxicillin-clavulanate in two instances of six, and for ciprofloxacin in two instances of four. Synergism was observed for A. baumannii between WLBU2 and tobramycin in none of the three instances, and for imipenem in two instances of three.
MBEC
MBEC testing was done for WLBU2 alone, followed by synergy testing between WLBU2 at 25% of MBEC values combined with antimicrobial agents at different concentrations. The tests were done for all ATCC strains and for representative clinical isolates that demonstrated synergism between WLBU2 (at sub-MIC values for planktonic cells) and the antimicrobial agents. Results are indicated in Table 6.
Table 6.
MBEC results of WLBU2 alone, and of antimicrobial agents in combination with sub-MBEC WLBU2 concentrations
| Species | Antimicrobial agent | Isolate ID# | MBEC of WLBU2 alone (µM; [μg/mL]) | Synergism test | ||
|---|---|---|---|---|---|---|
| Concentration of WLBU2 used for synergism test (µM; [μg/mL]) | Observed antimicrobial agent MBEC(μg/mL) in presence of WLBU2 | Synergism observed (interpretation) | ||||
| K. pneumoniae | Amoxicillin-clavulanate | 12 | 100 [340] | 25 [85] | 8 | Yes (susceptible) |
| ATCC BAA-2146 | >200 [>680] | 50 [170] | 8 | Yes (susceptible) | ||
| ATCC BAA-1705 | 25 [85] | 8.125 [27.625] | 8 | Yes (susceptible) | ||
| ATCC BAA-1706 | 100 [340] | 25 [85] | 8 | Yes (susceptible) | ||
| ATCC 700603 | >200 [>680] | 50 [170] | 8 | Yes (susceptible) | ||
| Ciprofloxacin | 7 | 50 [170] | 12.500 [42.500] | 1 | Yes (susceptible) | |
| BAA-2146 | >200 [>680] | 50 [170] | 1 | Yes (susceptible) | ||
| BAA-1705 | 25 [85] | 8.125 [27.625] | 4 | No | ||
| BAA-1706 | 100 [340] | 25 [85] | 1 | Yes (susceptible) | ||
| ATCC 700603 | >200 [>680] | 50 [170] | 1 | Yes (susceptible) | ||
| A. baumannii | Tobramycin | ATCC BAA-1605 | 50 [170] | 12.500 [42.500] | 8 | Yes (intermediate) |
| ATCC 19606 | 50 [170] | 12.500 [42.500] | 8 | Yes (intermediate) | ||
| Imipenem | 30 | 100 [340] | 25 [85] | 2 | Yes (susceptible) | |
| ATCC BAA-1605 | 50 [170] | 12.500 [42.500] | 2 | Yes (susceptible) | ||
| ATCC 19606 | 50 [170] | 12.500 [42.500] | 2 | Yes (susceptible) | ||
Note: Concentration of WLBU2 used for synergism test corresponded with 25% of MBEC values for each respective clinical isolate or control strain.
Abbreviations: ID#, identification number; MBEC, minimum biofilm eradication concentration.
The ability of WLBU2 alone to eradicate biofilms was assessed against K. pneumoniae clinical isolates 7 and 12, and A. baumannii clinical isolate 30. The measured MBEC values were 100 µM (340 µg/mL), 50 µM (170 µg/mL), and 100 µM (340 µg/mL), respectively. MBEC values ranged from 25 to >200 µM or 85 to >680 µg/mL for K. pneumoniae ATCC strains and were 50 µM or 170 µg/mL, for each of the two A. baumannii ATCC strains (Table 6).
MBEC values were also determined for amoxicillin-clavulanate, ciprofloxacin, tobramycin, and imipenem, when combined with WLBU2 (at 25% MBEC). WLBU2 demonstrated synergism against K. pneumoniae in four of four instances when combined with amoxicillin-clavulanate, and in three of four instances when combined with ciprofloxacin. WLBU2 demonstrated synergism against A. baumannii in two of two instances when combined with tobramycin, and in three of three instances when combined with imipenem.
Cytotoxicity tests
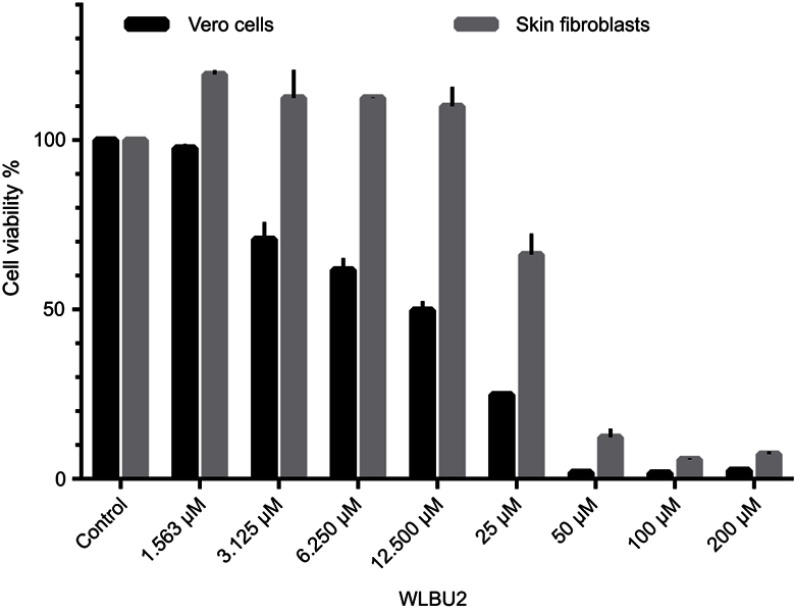
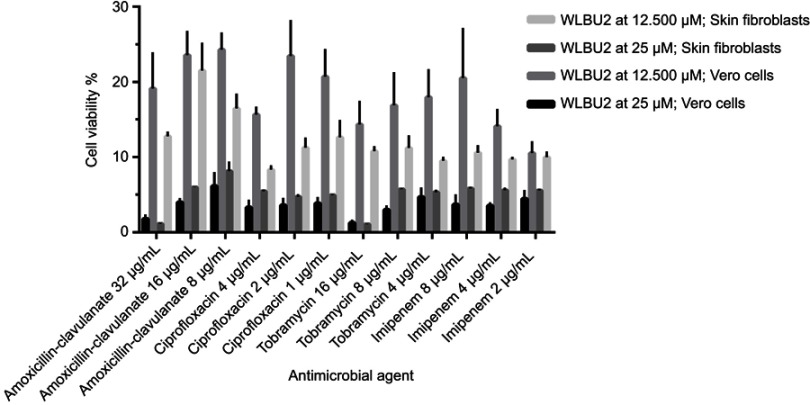
Vero cells and human skin fibroblast were used in MTT assays for viability testing of WLBU2 alone (at 1.563–200 µM or 5.313–680 µg/mL) (Figure 1), and of WLBU2 at 25 µM (85 µg/mL) or 12.500 µM (42.5 µg/mL) combined with antimicrobial agents at different concentrations (Figure 2).
Figure 1.
Viability of Vero cells and human skin fibroblasts exposed to WLBU2. Vero cells or human skin fibroblasts were incubated with different concentrations of WLBU2 or media alone (control) for 24 hrs. Cell viability was determined using the MTT assay. Experiments were run in triplicates. Bars represent means±standard error of the mean. WLBU2 concentrations in µM correspond to 5.313, 10.625, 21.250, 42.500, 85, 170, 340, 680 µg/mL, respectively.
Figure 2.
Viability of Vero cells and skin fibroblasts exposed to WLBU2 combined with antimicrobial agents. Vero cells or human skin fibroblasts were incubated with WLBU2 at 12.500 µM (42.5 µg/mL) or 25 µM (85 µg/mL) combined with antimicrobial agents at the indicated concentrations for 24 hrs. Cell viability was determined using the MTT assay. Experiments were run in triplicates. Bars represent means±standard error of the mean.
Vero cells and human skin fibroblasts were viable at WLBU2 concentrations ≤3.125 µM (10.625 µg/mL) and ≤12.500 µM (42 µg/mL), respectively (Figure 1). Both cell types showed markedly reduced cell viability when WLBU2 was tested in combination with antimicrobial agents at all tested concentrations (Figure 2).
DNA binding assay
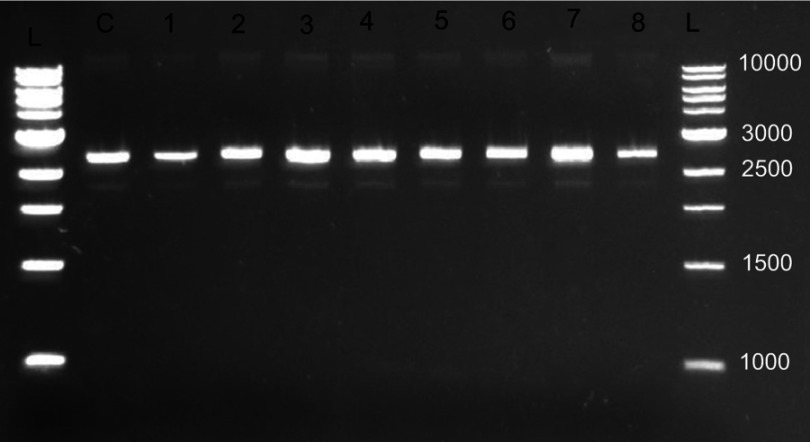
WLBU2 did not demonstrate DNA binding ability when tested with pUC19 plasmid DNA (Figure 3).
Figure 3.
Representative gel electrophoresis result for WLBU2 DNA binding assay. DNA binding assay was utilized to determine the ability of WLBU2 to bind bacterial plasmid DNA (pUC19 plasmid, 2686 base pairs); L: 1 KB DNA ladder (values are in base pairs); C: DNA alone (8.33 ng/µL); from 1 to 8 twofold diluted WLBU2 (200–1.563 µM or 680–5.313 µg/mL) combined with fixed DNA concentration (8.33 ng/µL); Electrophoresis was done at 140 V for 45 mins on 1% agarose; DNA was visualized under a UV transilluminator; no band shifts occurred for lanes 1–8 indicating no binding between WLBU2 and DNA.
Discussion
WLBU2 is one of the newly synthesized cAMPs, that displayed promising activity against Gram-positive and Gram-negative bacterial pathogens, MDR pathogens, and bacterial biofilms.21,22
The current study utilized MDR clinical isolates of A. baumannii and K. pneumoniae. The clinical isolates were from a variety of sources, including the urinary tract, blood, synovial fluid, bronchial washings, sputum, and wound material (data not shown). Several ATCC strains were included as controls. The findings of this study may pave the way for alternate therapies for bacterial pathogens that are highly resistant to classical antimicrobial agents and that produce hard to treat biofilm-associated infections.
The clinical isolates and the ATCC strains were first tested for the ability to produce biofilms using the reliable 96-well tissue culture plate method using crystal violet staining. All A. baumannii and 88.33% of K. pneumoniae clinical isolates, and most ATCC strains demonstrated strong biofilm formation. Only 16.78% of K. pneumoniae clinical isolates and one ATCC strain from each species demonstrated moderate biofilm production. Other methods folr evaluation of biofilm formation include the Congo red agar (CRA) method. However, CRA is not a reliable biofilm assay, as it was reported to have little sensitivity, specificity, and accuracy.23,24
The MIC of WLBU2 was determined against planktonic A. baumannii and K. pneumoniae clinical isolates and the ATCC strains utilizing a concentration range of 100–1.563 µM or 340–5.313 µg/mL. This range was selected based on previous reports on WLBU2 that indicated MIC values well below 100 µM or 340 µg/mL.11,12
Mean MIC values for WLBU2 were 7.943 µM (27.006 µg/mL) and 7.484 µM (25.446 µg/mL) for K. pneumoniae and A. baumannii clinical isolates, respectively. Importantly, all individual MBC and MIC values were identical, indicating that WLBU2 is bactericidal. This is compatible with previous reports indicating that engineered cationic peptides (eCAPs) have fast killing activity on bacterial cells25 For comparison, WLBU2 showed MICs of 1.5–3.2 µM against XDR A. baumannii, 2.9–4.7 µM against XDR K. pneumoniae, and 9.3 µM against K. pneumoniae KP2 strain.12 These results are comparable to the study’s findings.
Another study investigated the activity of two eCAPs; WLBU2 and WR12, along with colistin and LL37, against 142 clinical isolates. The clinical isolates were Gram-positive methicillin-resistant S. aureus, vancomycin-resistant enterococci, and Gram-negative Enterobacteriaceae, including K. pneumoniae, Enterobacter aerogenes, E. cloacae, E. coli, and A. baumannii. The mean MICs of eCAPs were ≤10 µM against both Gram-negative and Gram-positive bacteria.22
Amoxicillin-clavulanate and ciprofloxacin were used for MIC testing against MDR K. pneumoniae. Tobramycin and imipenem were used for MIC testing against A. baumannii. The antimicrobial agents were chosen based on recommendations indicated in the “Performance Standards for Antimicrobial Susceptibility Testing”, CLSI-2016, 26th edition. Most of the clinical isolates and ATCC strains were resistant to the antimicrobial agents.
The synergism test between WLBU2 and the four antimicrobial agents against planktonic bacteria was done by adding WLBU2 at its sub-inhibitory MIC for each respective isolate, with antimicrobial agents at different concentrations. Most clinical isolates selected for testing had MICs at the resistance cutoff values. Synergism was observed in several instances. Synergism between AMPs and antimicrobial agents has been described previously. A study from 2014 investigated the activity of four chimeric cationic peptides against 19 MDR A. baumannii isolates. The peptides showed MIC values between 3.125 and 12.5 µM against all isolates. The isolates were resistant to all tested antimicrobial agents (ampicillin, cefotaxime, ciprofloxacin, erythromycin, tobramycin, and polymyxin) except polymyxin. The four peptides showed synergy against one or two isolates when combined with cefotaxime, ciprofloxacin, or erythromycin.26
The ability of WLBU2 to eradicate biofilms was investigated using the MBEC assay, the standard method to evaluate agents’ ability to eradicate bacterial biofilms. All K. pneumoniae and A. baumannii ATCC strains were subjected to the MBEC assay using WLBU2 alone and in combination with antimicrobial agents. Representative clinical isolates in planktonic form that demonstrated synergism between WLBU2 and antimicrobial agents were also utilized for MBEC testing. When tested alone, WLBU2 demonstrated high MBEC values. Hence, we investigated whether combining antimicrobial agents with WLBU2 at 25% MBEC would lead to biofilm eradication. Indeed, the combinations led to biofilm eradication in several instances.
Overall, an increase in WLBU2 MBEC levels was observed compared to planktonic MIC values against K. pneumoniae and A. baumannii. Similarly, two- to fourfold higher MBEC levels of WLBU2 were observed for the treatment of P. aeruginosa abiotic and biotic biofilms compared to the MIC values against planktonic cells.27 Furthermore, it was reported by others that ciprofloxacin, erythromycin, and tobramycin alone were not able to reduce or inhibit biofilm formation by A. baumannii. However, when the agents were combined with chimeric cationic peptides, a reduction in biofilm formation was observed,26 which is similar to what was observed for WLBU2.
A study has indicated that NK-18 (an eCAP) has the ability to not only associate with the bacterial plasma membrane, but also to internalize and target bacteria DNA. The study also reported that another membrane-active AMP, magainin 2, could not bind plasmid DNA even at 128 μg/mL.4 A study of two synthetic short peptides with potent activity against S. aureus, namely, RRIKA and RR, demonstrated that both peptides were able to bind plasmid DNA and delay its electrophoretic migration in agarose, especially at higher peptide concentrations.5 The ability of WLBU2 to bind DNA as a potential mechanism for antimicrobial activity has not been previously investigated. WLBU2 at up to the tested 200 µM (680 µg/mL) level was not able to retard DNA mobility. Hence, WLBU2 mediates its activity via mechanisms other than binding of DNA.
MTT assay was used for measuring the viability of Vero cells and human skin fibroblasts. Vero cells (African green monkey kidney epithelial cells) were utilized to assess toxicity should WLBU2 alone or in combination with antimicrobial agents be used in vivo. Human skin fibroblasts were utilized to assess toxicity should WLBU2 alone or in combination with antimicrobial agents be used for topical applications on human skin. The International Organization for Standardization (ISO) guidelines for cytotoxicity testing (ISO 10993–5:2009) state that an agent is cytotoxic if cell viability falls below 70%.28,29 WLBU2 effects on cell viability were tested at a wide range of concentrations. For both cell types, cell viability was reduced to ≤12% at high WLBU2 concentrations; 200–50 µM or 680–170 µg/mL. However, these concentrations corresponded with effective biofilm eradication, suggesting that WLBU2 will lead to considerable cell toxicity if used at MBEC values.
Vero cells and human skin fibroblasts viability percentages were >70% at WLBU2 concentrations ≤3.125 µM (10.625 µg/mL) and ≤12.500 µM (42.5 µg/mL), respectively. Fortuitously, these values were similar to WLBU2’s MIC values against planktonic cells. Hence, WLBU2 could be used at this range to eradicate planktonic cells without significant host cell toxicity.
Since cell viability was decreased at WLBU2 concentrations ≥50 µM (170 µg/mL), and since WLBU2 at sub-MBEC values (ie, <50 µM or 170 µg/mL) demonstrated synergistic activity in eradicating biofilms of several isolates when combined with antimicrobial agents, we investigated the viability of Vero and human skin fibroblasts when treated with WLBU2 at 25 and 12.500 µM (85and 42.5 µg/mL) combined with antimicrobial agents at different concentrations. Surprisingly, a significant decrease in cell viability was observed for all tested combinations. For comparison, a study of WLBU2’s potential toxicity on J774 macrophages demonstrated reduced viability at 3–100 µM for 1 hr, with 50% of the cells being lost at a concentration of 25 μM.11
Despite the loss of cell viability when WLBU2 was combined with the antimicrobial agents, combination results showed excellent and substantial synergistic effect on some bacterial isolates and ATCC strains. This suggests that the combination could be useful for in vitro disinfection of planktonic bacteria and associated-biofilms, rather than in vivo application.
A study on eCAPs indicated that, WLBU2 had no cytotoxicity at concentration of ≤20 µM for 6 hrs, using both hemolytic assay and MTT assay against peripheral blood mononuclear cells (PBMCs). The study also investigated antibacterial activity under acidic conditions for several eCAPs. WLBU2 displayed no significant difference in activity at acidic pH compared with neural pH against P. aeruginosa and MRAS.12 The viability results of WLBU2 against human skin fibroblasts in the current study are consistent with those reported on PBMCs, although in our protocol the exposure duration was 24 hrs instead of the 6 hrs used for PBMCs, which likely leads to an overestimation of potential toxicity.
The reasonably good selective toxicity of WLBU2 against bacteria coupled with low toxicity against eukaryotic cells may be explained by eCAPs’ ability to form weak hydrophobic interactions with eukaryotic membranes due to the presence of cholesterol, and the formation of strong electrostatic interactions with the negatively charged bacterial membranes.30
Conclusion
The MIC values of WLBU2 ranged from 12.500 to 3.125 µM (42.5to 10.625 µg/mL) against the clinical isolates and ATCC strains. High WLBU2 concentrations were needed to eradicate bacterial biofilms. However, at these concentrations, viability of Vero cells and human skin fibroblasts was reduced to <50%. In contrast, at MIC values for planktonic cells, acceptable viability was observed (≥70%).
The combinations between WLBU2 sub-inhibitory concentrations and antimicrobial agents demonstrated synergism in several instances, suggesting the ability of using WLBU2 at lower MBEC levels to eradicate biofilms upon combination with antimicrobial agents. Nonetheless, eukaryotic cell toxicity of WLBU2 combined with antimicrobial agents was excessively high, potentially limiting future in vivo applications. Finally, WLBU2 did not demonstrate the ability to bind bacterial DNA.
Additional assays and cell types should be utilized to investigate potential cytotoxicity by WLBU2.
Based on the WLBU2’s potent killing activity of MDR A. baumannii and K. pneumoniae, in vivo animal studies of WLBU2 pharmacokinetics and potential utility to treat bacterial infections, including those by intracellular bacteria might be useful. Similar investigations could be carried out for Gram-positive bacteria. The anti-bacterial mechanisms of WLBU2 alone and combined with antimicrobial agents should be elucidated, for instance, the assessment of transmembrane pores formation by scanning electron microscopy, and effects on bacterial metabolism. The potential synergism between WLBU2 and other cationic-short AMPs is still unknown and could be the topic for future investigations. The observed high eukaryotic toxicity between WLBU2 and the antimicrobial agents even at low concentrations merit future investigations, especially as potential agents in the treatment of human tumors.
Acknowledgments
This work was financially supported by the Deanship of Research of Jordan University of Science and Technology [Grant number 20170221].
Data availability
Raw data are available upon request from the corresponding author.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.World Health Organization. Implementation of the global action plan on antimicrobial resistance. WHO GAP AMR Newsl. 2017;2017(30):1–4 [Google Scholar]
- 2.Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. [DOI] [PubMed] [Google Scholar]
- 3.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236(2):163–173. doi: 10.1016/j.femsle.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 4.Yan J, Wang K, Dang W, et al. Two hits are better than one: membrane-active and DNA binding-related double-action mechanism of NK-18, a novel antimicrobial peptide derived from mammalian NK-lysin. Antimicrob Agents Chemother. 2013;57(1):220–228. doi: 10.1128/AAC.01619-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohamed MF, Hamed MI, Panitch A, Seleem MN. Targeting methicillin-resistant Staphylococcus aureus with short salt-resistant synthetic peptides. Antimicrob Agents Chemother. 2014;58(7):4113–4122. doi: 10.1128/AAC.02578-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalatzis PG, Castillo D, Katharios P, Middelboe M. Bacteriophage interactions with marine pathogenic vibrios: implications for phage therapy. Antibiotics (Basel). 2018;7(1):15. doi: 10.3390/antibiotics7030077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sierra JM, Fuste E, Rabanal F, Vinuesa T, Vinas M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin Biol Ther. 2017;17(6):663–676. doi: 10.1080/14712598.2017.1315402 [DOI] [PubMed] [Google Scholar]
- 8.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7(5):545–594. doi: 10.3390/ph7050545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batoni G, Maisetta G, Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta. 2016;1858(5):1044–1060. doi: 10.1016/j.bbamem.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 10.Chu HL, Yip BS, Chen KH, et al. Novel antimicrobial peptides with high anticancer activity and selectivity. PLoS One. 2015;10(5):e0126390. doi: 10.1371/journal.pone.0126390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelbaqi S. Novel Engineered Cationic Antimicrobial Peptides Have a Broad-spectrum Activity Against: Francisella Tularensis, Burkholderia Pseudomalli and Yersinia Pestis. Pittsburgh: University of Pittsburgh; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Mietzner TA, Montelaro RC. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob Agents Chemother. 2013;57(6):2511–2521. doi: 10.1128/AAC.02218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deslouches B, Phadke SM, Lazarevic V, et al. De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob Agents Chemother. 2005;49(1):316–322. doi: 10.1128/AAC.49.1.316-322.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safari M, Mozaffari Nejad AS, Bahador A, Jafari R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J Biol Sci. 2015;22(4):424–429. doi: 10.1016/j.sjbs.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardbari AM, Arabestani MR, Karami M, Keramat F, Alikhani MY, Bagheri KP. Correlation between ability of biofilm formation with their responsible genes and MDR patterns in clinical and environmental Acinetobacter baumannii isolates. Microb Pathog. 2017;108:122–128. doi: 10.1016/j.micpath.2017.04.039 [DOI] [PubMed] [Google Scholar]
- 16.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Toole GA. Microtiter dish biofilm formation assay. JoVE (journal of Visualized Experiments). 2011;30(47):e2437–e2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lashua LP, Melvin JA, Deslouches B, Pilewski JM, Montelaro RC, Bomberger JM. Engineered cationic antimicrobial peptide (eCAP) prevents Pseudomonas aeruginosa biofilm growth on airway epithelial cells. J Antimicrob Chemother. 2016;71(8):2200–2207. doi: 10.1093/jac/dkw143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wikler MA, Alder J. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, M7. 11th ed. Pennsylvania: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 20.Yan H, Hancock RE. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob Agents Chemother. 2001;45(5):1558–1560. doi: 10.1128/AAC.45.5.1558-1560.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang K, Gitter B, Ruger R, et al. Antimicrobial peptide-modified liposomes for bacteria targeted delivery of temoporfin in photodynamic antimicrobial chemotherapy. Photochem Photobiol Sci. 2011;10(10):1593–1601. doi: 10.1039/c1pp05100h [DOI] [PubMed] [Google Scholar]
- 22.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2015;59(2):1329–1333. doi: 10.1128/AAC.03937-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305–311. [PubMed] [Google Scholar]
- 24.Knobloch JK, Horstkotte MA, Rohde H, Mack D. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med Microbiol Immunol. 2002;191(2):101–106. doi: 10.1007/s00430-002-0124-3 [DOI] [PubMed] [Google Scholar]
- 25.Hancock RE, Falla T, Brown M. Cationic bactericidal peptides In: Advances in Microbial Physiology. Vol. 37 Elsevier, New York; 1995:135–175. [DOI] [PubMed] [Google Scholar]
- 26.Gopal R, Kim YG, Lee JH, et al. Synergistic effects and antibiofilm properties of chimeric peptides against multidrug-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother. 2014;58(3):1622–1629. doi: 10.1128/AAC.02473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Burns JL, Montelaro RC. Engineered cationic antimicrobial peptides (eCAPs) to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother. 2014;59(2):3914–3937. AAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velasco-Ortega E, Jos A, Camean AM, Pato-Mourelo J, Segura-Egea JJ. In vitro evaluation of cytotoxicity and genotoxicity of a commercial titanium alloy for dental implantology. Mutat Res. 2010;702(1):17–23. doi: 10.1016/j.mrgentox.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 29.Haghi AK, Thomas S, Pourhashemi A, Hamrang A, Klodzinska E. Nanomaterials and Nanotechnology for Composites: Design, Simulation and Applications. CRC Press, Florida; 2015. [Google Scholar]
- 30.Biggin PC, Sansom MS. Interactions of alpha-helices with lipid bilayers: a review of simulation studies. Biophys Chem. 1999;76(3):161–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data are available upon request from the corresponding author.