Abstract
The LRRK2/MUC19 gene region constitutes a high-risk genetic locus for the occurrence of both inflammatory bowel diseases (IBDs) and Parkinson’s disease. We show that dendritic cells (DCs) from patients with Crohn’s disease (CD) and lymphoblastoid cell lines derived from patients without CD but bearing a high-risk allele (rs11564258) at this locus as heterozygotes exhibited increased LRRK2 expression in vitro. To investigate the immunological consequences of this increased LRRK2 expression, we conducted studies in transgenic mice over-expressing Lrrk2 and showed that these mice exhibited more severe colitis induced by dextran sodium sulfate (DSS) than did littermate control animals. This increase in colitis severity was associated with lamina propria DCs that showed increased Dectin-1–induced NF-κB activation and proinflammatory cytokine secretion. Colitis severity was driven by LRRK2 activation of NF-κB pathway components including the TAK1 complex and TRAF6. Next, we found that membrane-associated LRRK2 (in association with TAB2) caused inactivation of Beclin-1 and inhibition of autophagy. HCT116 colon epithelial cells lacking Beclin-1 exhibited increased LRRK2 expression compared to wild-type cells, suggesting that inhibition of autophagy potentially could augment LRRK2 proinflammatory signaling. We then showed that LRRK2 inhibitors decreased Dectin-1–induced TNF-α production by mouse DCs and ameliorated DSS-induced colitis, both in control and Lrrk2 transgenic animals. Finally, we demonstrated that LRRK2 inhibitors blocked TNF-α production by cultured DCs from patients with CD. Our findings suggest that normalization of LRRK2 activation could be a therapeutic approach for treating IBD, regardless of whether a LRRK2 risk allele is involved.
INTRODUCTION
The prevailing view of the pathogenesis of major inflammatory bowel diseases (IBDs), such as Crohn’s disease (CD) and ulcerative colitis (UC), is that they are caused by dysregulated and therefore enhanced immune responses to microbial organisms in the gut microbiome that ultimately result in gut inflammation. Major support for this view comes from studies of murine models of gut inflammation that invariably show that germ-free mice do not develop inflammation in the face of immune abnormalities that usually result in spontaneous inflammation such as interleukin-10 (IL-10) deficiency or tumor necrosis factor–α (TNF-α) hyperexpression (1). Important support for this notion comes from the fact that the various single-nucleotide polymorphisms (SNPs) associated with these diseases affect the risk for disease development through their effects on mucosal immune homeostasis. Prime examples of this include (i) the disease polymorphisms associated with the NOD2 (nucleotide binding oligomerization domain containing 2) gene that adversely affect host defense function or an immunoregulatory function that limits excessive innate responses (2) and (ii) the protective polymorphisms associated with the IL-23 receptor that result in decreased proinflammatory T helper 17 cell (TH17) responses (3).
In view of the many risk polymorphisms that have been associated with IBD, further studies of the mechanism by which polymorphisms cause or prevent disease represent a potentially fruitful approach to understanding disease pathogenesis as well as a means of identifying new treatments. Among the risk polymorphisms awaiting, such further study are those nested in the LRRK2/MUC19 gene region. This region can be considered a disease “hotspot” in view of the fact that mutations and SNPs in this gene region are risk factors associated with several major diseases including Parkinson’s disease (PD) (4, 5), IBD (6), and leprosy (7). With respect to IBD, this polymorphism has particular significance because the SNPs in the LRRK2/MUC19 locus have an odds ratio (OR) exceeded only by that of the IL-23R locus among those loci shared by CD and UC. Thus, the elucidation of the contribution of the LRRK2/MUC19 locus to IBD pathogenesis may be particularly important to the understanding of disease pathogenesis.
LRRK2 (leucine-rich repeat kinase 2) is a large protein with several identifiable domains including a ROC (Ras of complex proteins) domain, a COR (C-terminal of Roc) domain, a kinase domain, and an LRR (leucine-rich repeat) domain. This suggests that it can have several functions in different types of cells. Mutations in this gene are a cause of autosomal dominant PD, but precisely how these mutations lead to abnormal neuronal function and the symptoms of PD is not well understood (8). In the gastrointestinal (GI) tract, LRRK2 is present in various antigen-presenting cells, suggesting that it plays some role in regulating the cytokines that mediate inflammation. Here, we analyzed LRRK2 function in a mouse model of colitis and in dendritic cells (DCs) from patients with CD and established that LRRK2 has an important role in Dectin-1–mediated innate immune responses.
RESULTS
LRRK2 expression in DCs from CD patients
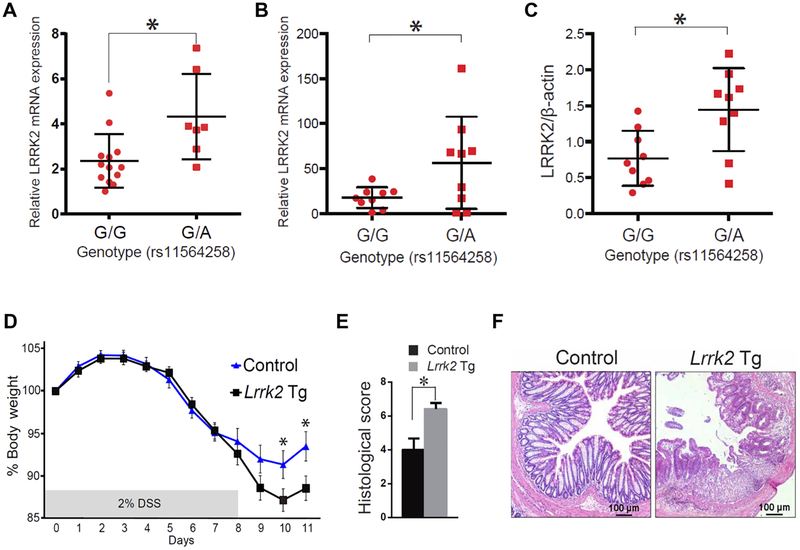
To evaluate how a LRRK2 risk allele may affect LRRK2 expression, we focused on the risk allele at rs1156258 located downstream of LRRK2 and associated with one of the highest risks for IBD among the alleles in the LRRK2 gene locus. In these studies, we compared LRRK2 mRNA in DCs from patients with CD or LRRK2 mRNA and protein in lymphoblastoid cell lines (LCLs) from individuals without CD, who were heterozygous for the LRRK2 risk allele (G/A) at rs11564258, with that in control cells (G/G) from patients without the risk allele at this locus. Homozygous A/A cells were unavailable because of the low frequency of risk allele A. The heterozygous cells exhibited increased LRRK2 mRNA expression both in DCs from patients with CD and in LCLs from individuals without CD (Fig. 1, A and B, and tables S1 to S3; Fig. 1A, P = 0.01; Fig. 1B, P = 0.04). LRRK2 protein expression in LCLs was measured by Western blot and was found to be elevated in cells with the rs1156258 SNP compared to those without (Fig. 1C and fig. S1; Fig. 1C, P = 0.01). These results indicated that the risk allele was associated with increased LRRK2 expression even in individuals without intestinal inflammation. On the basis of this finding, we pursued the study of LRRK2 function in both Lrrk2 transgenic (Tg) mice expressing about eight times more LRRK2 than wild-type (WT) mice (Fig. 2H) and Lrrk2 knockout (KO) mice that lacked LRRK2 expression. The Tg mice consisted of mice bearing a bacterial artificial chromosome (BAC) containing the endogenous promoter/enhancer regions of the Lrrk2 gene and producing FLAG-tagged LRRK2.
Fig. 1. An increase in LRRK2 boosts inflammation in Lrrk2 Tg mice treated with DSS.
(A) LRRK2 mRNA expression in DCs from patients with CD. (B) LRRK2 mRNA expression in LCLs from individuals with genotype G/G or G/A at SNP rs11564258 [a nonrisk allele (G) and a risk allele (A)]. (C) LRRK2 protein relative to β-actin in CD patients with the same genotypes as in (B). (A and B) LRRK2 mRNA was measured by real-time polymerase chain reaction (PCR). (C) LRRK2 protein relative to β-actin was measured by Western blotting (fig. S1). (A) G/G genotype, n = 13; G/A genotype, n = 7, P = 0.0104; (B) G/G genotype, n = 9; G/A genotype, n = 9, P = 0.04; (C) G/G genotype, n = 9; G/A genotype, n = 9, P = 0.0098. (D) Lrrk2 Tg mice and littermate control mice were fed 2% DSS for 8 days, and body weight was measured. The data represent the average value for each group. n = 12 Lrrk2 Tg mice (Lrrk2 Tg) and n = 14 control mice (day 10, P = 0.0283; day 11, P = 0.0186). (E) Histological score of colon inflammation for Lrrk2 Tg and control mouse groups analyzed in (C). n = 12 Lrrk2 Tg mice and n = 14 control mice (P = 0.0054). Asterisk (*) indicates P < 0.05. (F) Representative photomicrographs of hematoxylin and eosin (H&E)–stained colon tissue on day 11 from Lrrk2 Tg mice and littermate controls. Data are presented as means ± SEM. Statistical significance was determined using an unpaired two-tailed Student’s t test.
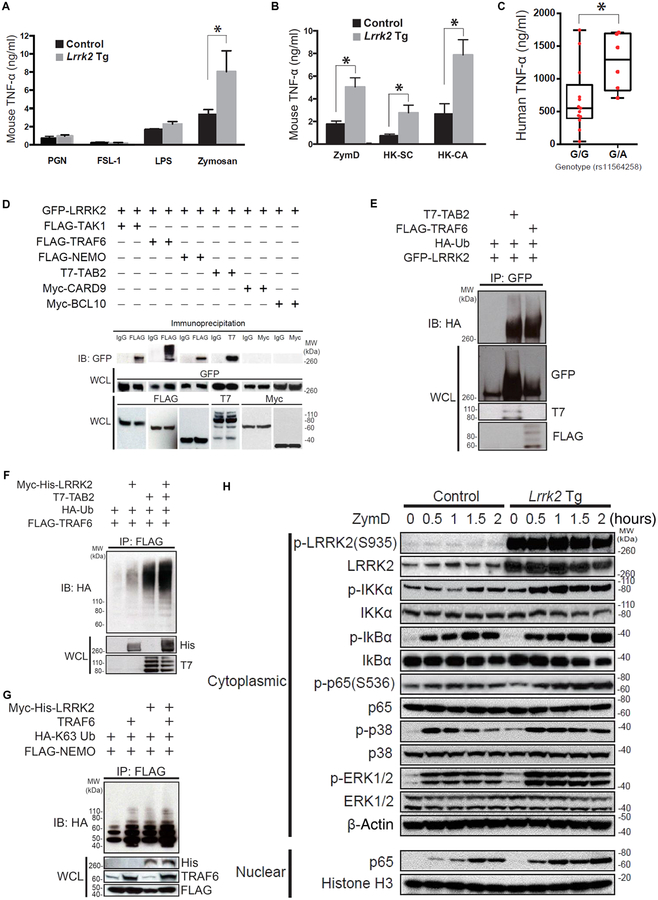
Fig. 2. LRRK2 positively regulates Dectin-1 signaling and interacts with the TAK1 complex.
(A and B) BMDCs from Lrrk2 Tg and control mice were stimulated for 24 hours with the indicated ligands in vitro. The amount of TNF-α in the culture supernatant was measured by enzyme-linked immunosorbent assay (ELISA). (A) Lrrk2 Tg mice, n = 3; control mice, n = 3, P = 0.0263; (B) Lrrk2 Tg mice, n = 3; control mice, n = 3; P = 0.0179 (ZymD), P = 0.0431 [heat-killed S. cerevisiae (HKSC)], and P = 0.0327 [heat-killed Candida albicans (HK-CA)]. (C) DCs from patients with CD (with the G/G or G/A genotype at SNP rs11564258) were stimulated for 24 hours with ZymD in vitro. The production of TNF-α in the culture super-natant was measured by ELISA (G/G genotype, n = 13; G/A genotype, n = 6, P = 0.026). (D) Whole-cell lysates (WCLs) of HEK293T cells cotransfected with the indicated plasmids were subjected to immunoprecipitation with anti- FLAG antibody followed by immunoblotting (IB) with anti– green fluorescent protein (GFP) antibody. IgG, immunoglobulin G. (E) Whole-cell lysates of HEK293T cells cotransfected with the indicated plasmids were subjected to immunoprecipitation (IP) with anti- GFP anti body, followed by immunoblotting with anti- hemagglutinin (HA) antibody. (F and G) Whole-cell lysates of HEK293T cells cotransfected with the indicated plasmids were subjected to immunoprecipitation with anti- FLAG antibody, followed by immunoblotting with anti- HA antibody. (H) Nuclear or cytoplasmic lysates of BMDCs from Lrrk2 Tg mice and littermate control mice were stimulated with ZymD in culture and then were subjected to immunoblotting with antibodies specific to the indicated ligands. Each of the studies is representative of at least three replicates. *P < 0.05 was considered a statistically significant difference. Statistical significance was determined with a Student’s t test. LPS, lipopolysaccharide; PGN, peptidoglycan; IKKα, IκB kinase α; MW, molecular weight.
Lrrk2 Tg mice exhibit more severe DSS-induced colitis than littermate control mice
In initial studies, to evaluate global LRRK2 function during inflammation of the colon, we subjected Lrrk2 Tg mice as well as Lrrk2 KO mice to dextran sodium sulfate (DSS) that induced colitis. Lrrk2 Tg mice exhibited more severe DSS-induced colitis associated with enhanced proinflammatory cytokine secretion compared to littermate control mice or control mice from parents not bearing the transgene and reared in the same environment (Fig. 1, D to F, and fig. S2A; Fig. 1D, day 10, P = 0.028; day 11, P = 0.019; Fig. 1E, P = 0.005). This was evident both in comparisons of weight loss and in histological evidence of colitis. In contrast, DSS-induced colitis in Lrrk2 KO mice was slightly less severe than in control mice (fig. S2, B to D). We verified these findings with multiple experiments inducing colitis in Tg and KO mice given that our results differed from a previous study that reported increased severity of DSS-induced colitis in Lrrk2 KO mice (9). This discrepancy was likely to be the result of differences in the control mice used in the two studies and the fact that assessment of the severity of colitis required utilization of control mice with identical genetic and microbiological features to the experimental mice (10). We found that the littermate control mice used in our studies of colitis in Lrrk2 KO mice manifested a different degree of colitis than did those mice directly obtained from the mouse supplier.
Dectin-1–mediated cytokine responses in DCs from Lrrk2 Tg mice
Given that several of the susceptibility genes in IBD (for example, NOD2 and CARD9) are likely to be causing disease through an abnormal innate immune response (2, 11), we reasoned that increased LRRK2 expression might enhance a pathogen- associated molecular pattern (PAMP)–driven inflammatory response. To examine this possibility, we stimulated bone marrow–derived DCs (BMDCs) obtained from Lrrk2 Tg mice with various Toll-like receptor (TLR) ligands including zymosan, a yeast wall extract that also signals via Dectin-1 (a β-glucan receptor). We found that whereas TLR2-and TLR4- specific ligands did not elicit an increased response in Lrrk2 Tg mice compared to control mice, zymosan did induce significant increases in TNF-α and IL-23 synthesis (Fig. 2A, fig. S3A, and table S3; Fig. 2A, P = 0.026; fig. S3A, P = 0.035). To further define the increased responsiveness of Lrrk2 Tg mouse BMDCs, we stimulated BMDCs with various Dectin-1 agonists including a form of zymosan [zymosan depleted (ZymD)] that lacked TLR- stimulating properties. We found that ZymD, as well as heat-killed Saccharomyces cerevisiae and heat-killed C. albicans also induced increased TNF-α and IL-23 production in Lrrk2 Tg mouse BMDCs compared to WT BMDCs (Fig. 2B, fig. S3B, and table S3). This increased responsiveness of Lrrk2 Tg BMDCs correlated with the responsiveness of DCs from CD patients with the G/A genotype. These DCs also exhibited significantly increased production of TNF-α when stimulated with ZymD compared to DCs from CD patients with the G/G genotype (Fig. 2C and table S3, P = 0.026). In contrast to these findings with Lrrk2 Tg mouse BMDCs, TNF-α production in BMDCs from Lrrk2 KO mice was similar to that of BMDCs from Lrrk2 WT mice (fig. S4 and table S3).
LRRK2 expression after NF-κB activation in DCs from Lrrk2 Tg mice
The augmented Dectin-1–specific induction of proinflammatory cytokines by Lrrk2 Tg mouse BMDCs reported above as well as previous data suggested a role for LRRK2 in nuclear factor κB (NF-κB) activation (12, 13). To investigate this possibility, we first determined the downstream binding partners of LRRK2 and their interactions using human embryonic kidney (HEK) 293T cells transfected with a LRRK2-GFP–expressing vector along with one or more tagged vectors encoding possible downstream-interacting molecules. We found that LRRK2 bound to various components of the NF-κB activation complex including TRAF6, TAK1, TAB2, and NEMO (Fig. 2D). However, LRRK2 did not bind to the components of the Malt1-BCL10-CARD9 complex (Fig. 2D). Given that TRAF6 is an E3 ubiquitin ligase that mediates Lysine 63 (K63)–linked polyubiquitination and activation of interacting molecules, we next determined the ubiquitination status of LRRK2 after its binding to TRAF6. We found that LRRK2 interactions with TRAF6 or with TRAF6 and TAB2 resulted in robust LRRK2 polyubiquitination (Fig. 2E). Unexpectedly, we also found that LRRK2 in the presence of TAB2 induced TRAF6 polyubiquitination, suggesting that LRRK2 interactions with downstream components may have initiated a positive polyubiquitination feedback loop (Fig. 2F). Next, we tested whether LRRK2 interacted with NEMO, the proximal regulator of NF-κB activation. We found that LRRK2 either alone or acting synergistically with TRAF6 augmented K63-linked polyubiquitination of NEMO (Fig. 2G). Finally, we found that ZymD stimulation of BMDCs from Lrrk2 Tg mice enhanced the phosphorylation of NF-κB–related components (IKKα, IκBα, and p65) and, to a lesser extent, that of mitogen-activated protein kinase (MAPK)–related molecules as well (p38 and ERK1/2) (Fig. 2H). These data cannot be attributed to possible effects of increased LRRK2 on total protein of the various NF-κB components because stimulated cells from control and Lrrk2 Tg mice contained equivalent amounts of NF-κB component total protein (Fig. 2H).
LRRK2 regulates NFAT transcriptional activity in mouse BMDCs
Previous studies have reported that LRRK2 affects proinflammatory responses by down-regulating NFAT (nuclear factor of activated T cells) translocation and transcriptional activity (9, 14, 15). Re-examining this possibility, we found that an NFAT luciferase reporter construct transfected into HEK293T cells exhibited an increase in NFAT activity after cotransfection of a LRRK2-expressing plasmid (fig. S5A and table S3). In addition, we found that either zymosan or ZymD stimulation of BMDCs from Lrrk2 Tg mice led to significantly greater production of IL-2 (an NFAT-dependent cytokine) than similarly stimulated cells from littermate control mice (Zymosan, P = 0.039; ZymD, P = 0.023) (fig. S5B and table S3). Finally, we found that culturing ZymD-stimulated cells from control and Lrrk2 Tg mice in the presence of several inhibitors of LRRK2 led to decreased IL-2 production (figs. S7B and S8 and table S3). Together, these studies suggest that, contrary to a previous report, increased LRRK2 enhanced both NFAT translocation and transcriptional activity.
LRRK2 negatively regulates autophagy in mouse BMDCs
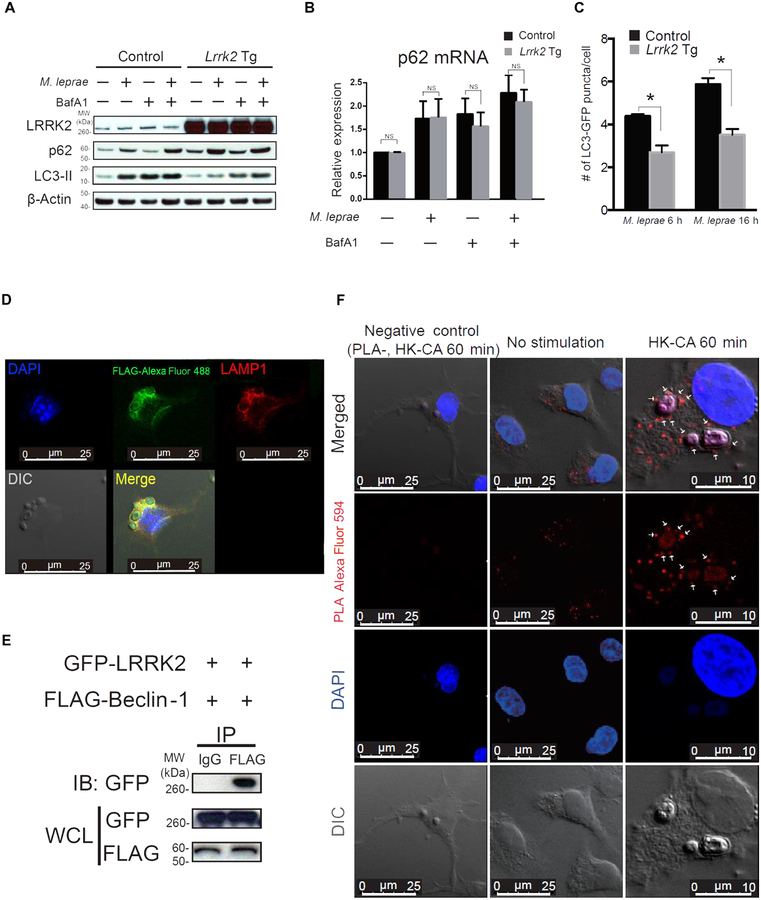
The aberrant NF-κB activation associated with increased LRRK2 could be related to an accompanying defect in autophagy because the latter has been shown to affect NF-κB signaling (16–18). In studies addressing this question, we assessed autophagy in BMDCs from both Lrrk2 Tg mice and Lrrk2 KO mice after exposure of these cells to irradiated Mycobacterium leprae. This bacterial stimulus was chosen for several reasons including the fact that LRRK2 is a susceptibility gene for leprosy (7) and mycobacteria express molecules that activate human macrophages and mouse DCs through Dectin-1 (19, 20). M. leprae, like Mycobacterium tuberculosis, is targeted for autophagy and auto-phagic elimination after uptake into macrophages through the action of guanosine triphosphatase (GTPase), the ubiquitin ligase parkin, and other components (21, 22). Western blotting showed that conversion of LC3-I to LC3-II, an indicator of the induction of autophagy (23), was decreased in BMDCs from Lrrk2 Tg mice (Fig. 3A) and increased in BMDCs from Lrrk2 KO mice (fig. S3A) compared to control mice. Furthermore, the decreased LC3-II in BMDCs from Lrrk2 Tg mice was not due to increased autophagic degradation given that LC3-II expression was reduced in BMDCs from Lrrk2 Tg mice compared to cells from control mice even after autophagic degradation was impaired by the addition of Bafilomycin A1 (BafA1) to the culture. In concert with this finding, p62 (an autophagic protein that was up- regulated in Lrrk2 Tg cells at baseline) underwent an equivalent up- regulation at both the mRNA (Fig. 3B and table S3) and protein level (Fig. 3A) after cells were exposed to M. leprae. The bacteria were phagocytosed less by BMDCs from Lrrk2 Tg mice than by BMDCs from control mice. This was shown by blockade of autophagic degradation in BMDCs using BafA1, which resulted in less of a decrease in p62 in BMDCs from Lrrk2 Tg mice compared to littermate control cells (Fig. 3A).
Fig. 3. LRRK2 interacts with Beclin-1 and suppresses autophagy.
(A and B) BMDCs from Lrrk2 Tg mice (Lrrk2 Tg) or littermate control mice were cultured with or without BafA1 (300 nM) for 30 min and then were stimulated with M. leprae for 2 hours. The cell lysates were subjected to immunoblotting to examine autophagic flux. Total RNA was extracted from BMDCs, and p62 mRNA expression was determined using real-time PCR. (B) Lrrk2 Tg mice, n = 3; control mice, n = 3; P values from left to right: P = 0.6819, P = 0.9586, P = 0.3713, and P = 0.3073). NS, not significant. (C) BMDCs from Lrrk2 Tg mice and littermate control mice that had been crossed with GFP-LC3 Tg mice were stimulated with M. leprae and then were evaluated for the number of GFP-LC3 puncta by confocal laser scanning microscopy. The data shown consist of pooled data from three independent experiments. (C) Lrrk2 Tg mice, n = 3; control mice, n = 3; M. leprae: 6 hours, P = 0.0010; M. leprae: 16 hours, P < 0.0001. *P < 0.05 was considered a statistically significant difference. Statistical significance was determined using a Student’s t test. (D) BMDCs from Lrrk2 Tg mice were stimulated with HK-CA for 60 min in culture and then were stained with anti-FLAG antibody (green) and anti-LAMP1 antibody (red). DIC, differential interference contrast. (E) Whole-cell lysates of HEK293T cells cotransfected with GFP-LRRK2 and FLAG–Beclin-1 plasmids were subjected to immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-GFP antibody. (F) BMDCs from Lrrk2 Tg mice cultured with or without HK-CA were subjected to a proximity ligation assay (PLA) to detect LRRK2/Beclin-1 complexes (red). PLA-, no PLA probe, negative control; DAPI, 4′,6-diamidino-2-phenylindole.
To verify these findings, we next measured autophagy in M. leprae– stimulated BMDCs from Lrrk2 Tg or Lrrk2 KO mice that were crossed with GFP-LC3 Tg mice. This allowed us to quantitate the development of autophagy-associated LC3 puncta in primary BMDCs in the absence of secondary autophagy triggered by transfected plasmid DNA commonly used in autophagy assays. Using confocal laser scan microscopy, we found that Lrrk2 Tg BMDCs exhibited a decreased number of LC3 puncta, whereas cells from Lrrk2 KO mice exhibited an increased number of LC3 puncta (Fig. 3C and fig. S6B).
Mechanism of LRRK2 inhibition of autophagy in mouse BMDCs
To study the mechanism of LRRK2 inhibition of autophagy, we first determined whether LRRK2 was associated with an autophagosomal membrane, the usual location of autophagy-associated proteins after stimulation of BMDCs with a phagosome/autophagosome- inducing stimulus. Using confocal microscopy, we found that after stimulation with heat-killed C. albicans, LRRK2 colocalized with the endosomal/lysosomal membrane marker LAMP1 (Fig. 3D). Next, we determined whether LRRK2 interacted with Beclin-1, a key initiator of the autophagy cascade that previously was shown to be inhibited by TAB2 (24, 25), a protein that interacts with LRRK2. Immunoblotting studies conducted in HEK293T cells revealed that LRRK2 bound to Beclin-1 (Fig. 3E). In addition, using a proximity ligation assay (26), we showed that in unstimulated BMDCs from Lrrk2 Tg mice, Lrrk2 interacted with endogenous Beclin-1, and LRRK2/Beclin-1 complexes were found in the cytosol. When Lrrk2 Tg mouse BMDCs were stimulated with heat-killed C. albicans, the LRRK2/Beclin-1 complexes were also found on membranes surrounding the bacteria (Fig. 3F).
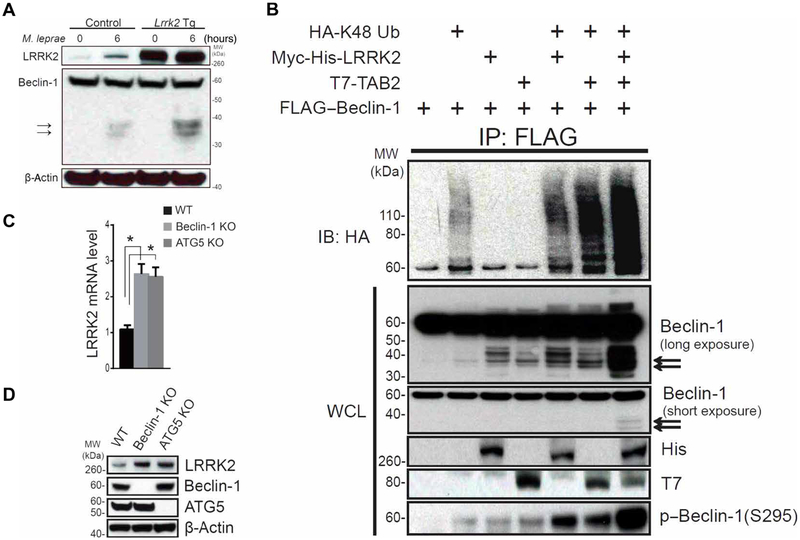
In further exploration of the mechanism of LRRK2 inhibition of autophagy, we measured LRRK2-induced Beclin-1 inactivation. Beclin-1 is known to be degraded and inactivated by caspases 3 and 8 in a process that results in Beclin-1 cleavage (27). Thus, we stimulated BMDCs from Lrrk2 Tg mice and littermate control mice with M. leprae and then subjected whole-cell lysates to Western blotting using anti–Beclin-1 antibodies. We found that cell lysates from Lrrk2 Tg mice exhibited more cleavage bands than did cell lysates from littermate control mice (Fig. 4A). To determine whether LRRK2 alone or LRRK2 in association with TAB2 promoted Beclin-1 degradation, we analyzed immunoblots for Beclin-1 degradation. We found that Lrrk2 transfection alone or in combination with Tab2 transfection gave rise to Beclin-1 degradation bands and K48 polyubiquitination bands (Fig. 4B). It is known that a second or concomitant mechanism of Beclin-1 inactivation consists of Akt-mediated phosphorylation of this molecule at its Ser234 and Ser295 sites (28). To investigate this mechanism of inactivation, we transfected Beclin-1 along with Lrrk2 or Tab2 into HEK293T cells and then determined Beclin-1 phosphorylation at Ser295. We found that both LRRK2 and TAB2 induced phosphorylation of Beclin-1 at Ser295 and that these proteins acted synergistically to increase the phosphorylation of Beclin-1 (Fig. 4B).
Fig. 4. LRRK2 augments Beclin-1 degradation blocking autophagy and increasing LRRK2 expression.
(A) BMDCs from Lrrk2 Tg mice or littermate control mice were stimulated for 6 hours with M. leprae. The cell lysates were then subjected to immunoblotting with the anti–Beclin-1 antibody H-300. (B) Whole-cell lysates of HEK293T cells cotransfected with the indicated plasmids were sonicated and subjected to immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA antibody. Immunoblotting using anti–Beclin-1 antibody (clone: 20/Beclin) demonstrated degradation bands (black arrows). (C and D) LRRK2 mRNA and protein expression was determined using real-time PCR (C) and immunoblotting (D) in HCT116 WT cells, Beclin-1 KO cells, and ATG5 KO cells. Data are representative of at least three identical experiments. (C) HCT116 WT versus Beclin-1 KO, P = 0.0009; HCT116 WT versus ATG5 KO, P = 0.0015. *P < 0.05 was considered a statistically significant difference. Statistical significance was determined by analysis of variance (ANOVA) for multiple comparisons.
In previous studies of neural tissue from Atg7 conditional KO mice and from ATG5-deficient mouse embryonic fibroblasts, it was shown that defective autophagy was associated with increased LRRK2 (29). Thus, we compared LRRK2 mRNA and protein expression in HCT116 cells deficient in Beclin-1 or ATG5 with LRRK2 mRNA and protein expression in WT HCT116 cells. We found that ATG5-deficient cells exhibited increased LRRK2 expression compared to WT cells (Fig. 4, C and D, and table S3).
LRRK2 inhibitors abrogate the effects of LRRK2 on inflammation and autophagy
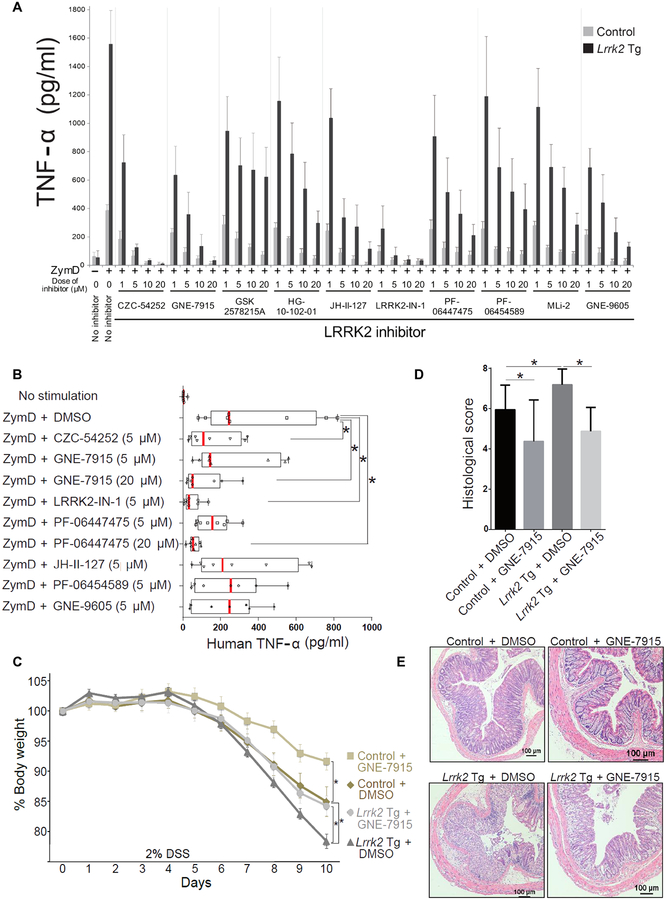
LRRK2 inhibitors provide protection from the development of LRRK2- induced neurodegeneration in both in vitro and in vivo models of PD (30). Furthermore, in rat microglia, LRRK2 inhibitors block TLR4- induced TNF-α and inducible nitric oxide synthase (iNOS) production (31). We therefore screened 12 different LRRK2 inhibitors for their capacity to suppress ZymD-induced induced TNF-α and IL-2 production by BMDCs from littermate control mice and found that 10 of the inhibitors exhibited dose-dependent suppression of TNF-α and IL-2 production, whereas two of the inhibitors (GNE-0877 and FX2149) showed little or no effect on production of these cytokines (Fig. 5 and fig. S7). The 10 effective LRRK2 inhibitors also suppressed the increased ZymD-induced TNF-α and IL-2 production by BMDCs from Lrrk2 Tg mice (Fig. 5A, fig. S8, and table S3). Furthermore, two of the LRRK2 inhibitors (LRRK2- IN-1 and CZC54252) that suppressed ZymD-induced production of cytokines by BMDCs from Lrrk2 Tg mice also suppressed production of a number of proinflammatory cytokines by these BMDCs (fig. S9 and table S3). Finally, we determined whether the suppressive effects of LRRK2 inhibitors on mouse BMDC cytokine production were relevant to DCs from patients with CD. We found that human DCs derived from the peripheral blood mononuclear cells (PBMCs) of randomly selected CD patients also exhibited suppression of ZymD-induced TNF-α production when cultured in the presence of four of seven LRRK2 inhibitors (Fig. 5B and table S3). Finally, we investigated the effect of a LRRK2 inhibitor on autophagy and found both by Western blotting and quantitation of LC3 puncta that the inhibitor LRRK2-IN-1 reversed inhibition of autophagy in M. leprae– stimulated BMDCs from Lrrk2 Tg mice (fig. S10, A and B) (32). Then, we determined whether a LRRK2 inhibitor could affect the intensity of DSS-induced colitis in Lrrk2 Tg mice and littermate control mice. We found that administration of LRRK2-IN-1 to control mice ameliorated DSS-induced colitis (fig. S11, A to C). In addition, administration of a second LRRK2 inhibitor GNE-7915 inhibited DSS-induced colitis in both Lrrk2 Tg mice and littermate control mice (Fig. 5, C to E).
Fig. 5. LRRK2 inhibitors ameliorate gut inflammation in Lrrk2 Tg mice with DSS-induced colitis.
(A) BMDCs from Lrrk2 Tg mice or littermate control mice were cultured with dimethyl sulfoxide (DMSO) vehicle or the indicated LRRK2 inhibitor for 30 min in vitro and then stimulated with ZymD for 24 hours. Mouse TNF-α concentrations in culture supernatants were determined using ELISA. (B) DCs derived from PBMCs from eight CD patients were cultured with DMSO vehicle or the indicated LRRK2 inhibitor for 30 min in vitro and then stimulated with ZymD for 24 hours. Human TNF-α in culture supernatants was measured by ELISA. Inhibition of TNF-α was observed with four of seven LRRK2 inhibitors (P < 0.03, unpaired one-tailed t test). (C to E) Lrrk2 Tg mice or littermate control mice treated with DMSO vehicle or the LRRK2 inhibitor GNE-7915 (20 mg/kg) were administered 2% DSS for 8 days. (C)Changes in body weight as a percentage of untreated animals, (D) histological score, and (E) representative H&E staining of mouse colon epithelial tissue are shown. Control + DMSO, n = 19; control + GNE-7915, n = 8; Lrrk2 Tg + DMSO, n = 16; Lrrk2 Tg + GNE-7915, n = 16. All experiments were repeated at least three times. (C) Control + DMSO, n = 19; control + GNE-7915, n = 8; Lrrk2 Tg mice + DMSO, n = 16; Lrrk2 Tg mice +GNE-7915, n = 16; P values determined on day 10: control + DMSO versus control + GNE-7915, P = 0.0356; control + DMSO versus Lrrk2 Tg mice + DMSO, P = 0.0070; Lrrk2 Tg mice + DMSO versus Lrrk2 Tg mice + GNE-7915, P = 0.0208. (D) Control + DMSO versus control + GNE-7915, P = 0.0127; control + DMSO versus Lrrk2 Tg mice + DMSO, P = 0.0150; Lrrk2 Tg mice + DMSO versus Lrrk2 Tg mice + GNE-7915, P = 0.0001. Statistical significance was determined using ANOVA for multiple comparisons. Asterisk (*) indicates P < 0.05.
DISCUSSION
A number of IBD-related risk polymorphisms have been identified in the chromosome 12q12 region that includes the LRRK2/MUC19 locus that confers varying degrees of increased risk for CD and, to a lesser extent, UC. The risk polymorphism rs11564258—studied here to define its effect on LRRK2 expression—is located downstream of LRRK2 and is a noncoding SNP that is in linkage disequilibrium with a coding SNP at rs33995883. Both SNPs confer an equal and substantially increased risk for the development of CD (OR, 1.7) (33). The LRRK2 risk polymorphism studied here was shown to be associated with increased LRRK2 expression in LCLs obtained from individuals without GI inflammation and in DCs from individuals with CD who were heterozygous for the risk allele. This suggests that the increased LRRK2 expression was not secondary to the presence of CD-associated inflammation. Notably, the risk polymorphism studied here is associated with a substantially greater risk for CD than reported previously by Trabzuni et al. (34) and Liu et al. (9) (OR, 1.7 versus 1.1).
Previous studies of LRRK2 in IBD have shown that LRRK2 is increased in the inflamed gut of CD patients possibly because interferon-γ (IFN-γ) is an inducer of LRRK2 (12) or because LRRK2 exhibits increased phosphorylation and membrane localization in response to stimulation by TLR ligands up-regulated during inflammation (35). These findings are consistent with those in the present study in which we found that mice with increased LRRK2 expression displayed increased Dectin-1–mediated cytokine responses and that LRRK2 exhibited interactions with NF-κB signaling components associated with NF-κB and MAPK activation. We found that although Lrrk2 Tg mice did not exhibit spontaneous gut inflammation, they exhibited increased inflammation and colitis when exposed to DSS. Lrrk2 Tg mice are thus similar to mice with loss-of-function Nod2 mutations that only show susceptibility to gut inflammation after exposure to inflammatory stress (36). Finally, treating Lrrk2 Tg mice with LRRK2 inhibitors decreased the various proinflammatory responses including DSS-induced colitis due to increased LRRK2.
An interesting aspect of the current study is that although DCs from Lrrk2 Tg mice responded to stimulation by a Dectin-1 ligand (ZymD) with increased production of proinflammatory cytokines and increased cell signaling leading to activation of NF-κB, they did not mount increased responses to the several TLR ligands tested. This may be due to an as yet poorly understood specific relation of LRRK2 to the Dectin-1 signaling pathway that does not apply to TLR signaling pathways. This observation is consistent with previous studies showing that cells from patients with CD mount increased proinflammatory cytokine responses compared to cells from control individuals when stimulated by several Dectin-1 stimuli (37).
Our observations indicating that LRRK2 increased inflammatory responses such as DSS-induced colitis in Lrrk2 Tg mice are in conflict with other studies reporting that LRRK2 reduced inflammatory responses (9). These previous studies suggested that LRRK2 is an inhibitor of NFAT nuclear translocation and that loss of LRRK2 resulted in increased NFAT translocation and an increased inflammatory cytokine response mediated by NFAT. Thus, these studies suggest that decreased LRRK2 rather than increased LRRK2 expression underlies the increased proinflammatory cytokine responses in CD (9). However, we could not verify these findings in our study. For example, we found that DCs from patients with and without CD, who were heterozygous for the LRRK2 polymorphism, exhibited increased rather than decreased LRRK2 expression. In addition, we found that whereas DCs from Lrrk2 Tg mice exhibited increased cytokine responses, those from Lrrk2 KO mice exhibited diminished cytokine responses. These cytokine data in Lrrk2 KO mice differed from findings reported by Liu et al. (9) in which bone marrow–derived macrophages (BMDMs) from Lrrk2 KO mice showed increased cytokine production. However, our findings are supported by previous reports showing that BMDMs from Lrrk2 KO mice exhibited no changes in cytokine production or NF-κB expression when subjected to TLR ligand stimulation (38, 39) and that microglial cells subjected to Lrrk2 knockdown exhibited decreased TLR ligand–induced cytokine production compared to control cells (13, 31). Finally, in contrast to Liu et al. (9), we found that Lrrk2 KO mice did not exhibit increased DSS-induced colitis, whereas Lrrk2 Tg mice expressing increased LRRK2 did. We showed that inhibition of LRRK2 led to decreased ZymD-induced cytokine responses in human DCs and that LRRK2 inhibitors ameliorated DSS-induced colitis both in Lrrk2 WT and Lrrk2 Tg mice.
We conducted an analysis of LRRK2 function in relation to autophagy in human DCs from patients with and without CD. Previous insight into this relation came from studies of the effects of LRRK2 mutations on neuronal cell function in PD, which suggested that such mutations boost autophagy. However, the interpretation of these data with respect to normal (nonmutated) LRRK2 was equivocal (8). For example, in a study of the role of LRRK2 in the regulation of rapamycin-induced autophagy in Raw264.7 cells, a mouse macrophage cell line, it was shown that endogenous LRRK2 supported autophagy upon its recruitment to intracellular membranes, whereas either silencing Lrrk2 or blocking the LRRK2 kinase domain inhibited rapamycin-induced autophagy (35). In contrast, another study showed that transfected human neuronal cells overexpressing LRRK2 exhibited inhibition of autophagy and that LRRK2 silencing increased autophagy and prevented cell death induced by BafA1 under starvation culture conditions (40). A potential problem with the interpretation of autophagy studies involving LRRK2 overexpression is that cells transfected with plasmids expressing LRRK2 are subjected to LRRK2-mediated apoptosis and thus secondary autophagy induced by survival signals. We therefore studied autophagy in mouse BMDCs exposed only to an autophagy-inducing microbe M. leprae. With this approach, we showed that cells from Lrrk2 Tg mice stably overexpressing LRRK2 or cells from mice lacking LRRK2 exhibited decreased and increased autophagy, respectively. These studies provide support for the notion that LRRK2 is primarily an inhibitor of autophagy. In addition, our data support those of a recent study showing that Lrrk2 KO mice had greater LC3-II in renal tissue at young ages (that is, increased autophagy) but lower amounts at older ages (41). This finding suggested that although LRRK2 primarily suppresses autophagy, continuous dysregulation of autophagy over long time periods may override this effect.
The notion that LRRK2 inhibits autophagy is also supported by our data exploring the underlying mechanism, namely the ability of LRRK2 to bind to and then to induce K48 polyubiquitination and degradation of Beclin-1, an initiator of autophagy (42). In addition, we demonstrated that the interaction of LRRK2 with Beclin-1 led to phosphorylation of Beclin-1, a known mechanism of Beclin-1 inactivation (28).
One consequence of autophagy inhibition mediated by LRRK2 signaling is that it leads to increased LRRK2 and thus the potential enhancement of inflammation. The mechanism of increased LRRK2 secondary to inhibition of autophagy is unclear; however, it may be related to the fact that autophagy inhibition decreases LRRK2 degradation that would otherwise result from its propensity to bind to autophagic membranes (43). A second consequence of LRRK2-mediated inhibition of autophagy relating to its proinflammatory effect arises from the effect of decreased autophagy on p62, an autophagy acceptor protein. Although Lrrk2 Tg mice and control mice both exhibited equivalent p62 mRNA expression upon stimulation with M. leprae, p62 protein was higher in Lrrk2 Tg mice presumably because of decreased consumption of p62 by autophagy. This increase in p62 could also contribute to LRRK2’s effects on inflammation given that p62 can activate NF-κB (16). Finally, it should be noted that there is evidence that NF-κB can inhibit autophagy via up-regulation of NEDD4, a signaling component that can cause Beclin-1 cleavage as does LRRK2 (44, 45). Thus, NF-κB activation and autophagy have reciprocal effects on one another, and it is presently unknown whether the two effects of LRRK2 on NF-κB activation and autophagy are independent or linked.
Our demonstration that Dectin-1 signaling led to increased NF-κB activity via activation of LRRK2 coupled with the fact that such activation suppressed autophagy suggests that Dectin-1 signaling can down-regulate autophagy via LRRK2 or the LRRK2 binding partner TAB2. Notably, LC3 is not a specific marker of autophagy in that it can be recruited to single-membrane phagosomes during LC3-associated phagocytosis (46, 47). Because LC3-associated phagocytosis can be induced by zymosan (46), LRRK2 may negatively regulate LC3-associated phagocytosis as well as autophagy upon stimulation with ZymD. This seems to be the case given that we found that ZymD stimulation of BMDCs from Lrrk2 Tg mice also inhibited LC3-I conversion to LC3-II in the same manner as M. leprae.
The fact that LRRK2 activation links proinflammatory function with autophagy (and possibly LC3-associated phagocytosis) places this signaling molecule in a unique position vis-à-vis mucosal responses to the gut microbiome. On the one hand, LRRK2 activation initiates innate immune responses resulting in the production of cytokines such as IL-17 that drive IBD inflammation. On the other hand, LRRK2 inhibits cellular functions that play a role in the clearance of gut bacteria (fig. S13). Thus, increased LRRK2 expression resulting from a risk polymorphism may have a dual effect on the enhancement of innate immunological responses that underlie inflammation in IBD. This finding may shed light on the pathogenesis of PD given recent data showing that the rs11564258 WT G allele that is highly protective in CD was also significantly protective in PD (OR, 0.69; P = 5.49 × 10−5) (48). Finally, the finding that blockade of LRRK2 function with a low–molecular weight inhibitor can ameliorate the proinflammatory responses mediated by LRRK2 suggests that such inhibitors could be an effective treatment option for IBD patients, even for those IBD patients lacking the LRRK2 risk polymorphism that nevertheless up-regulate LRRK2 during the normal course of inflammation. However, in view of reports showing that LRRK2 inhibition may affect pulmonary lysosome function, the various LRRK2 inhibitors must first be subjected to extensive long-term toxicity studies in nonhuman primates (49).
Our findings suggest the possibility that agents that block LRRK2 function could ameliorate colitis in patients with IBD; however, the data set presented is incomplete and needs to be augmented by additional studies. First, it is important to show that patients bearing one of the major LRRK2 risk polymorphisms, particularly as homozygotes, do have elevated LRRK2 expression in the absence of active gut inflammation that can secondarily increase such expression via IFN-γ secretion. Furthermore, it will be important to show that such increased LRRK2 expression gives rise to increased proinflammatory responses in these patients compared to patients not bearing the polymorphism. Second, our data show that increased responses are restricted to those elicited by Dectin-1 ligands and therefore may be irrelevant to proinflammatory responses induced by other stimuli. Thus, more studies are necessary to define the range of stimuli influenced by LRRK2 expression. Third, although we show that LRRK2 blocking agents ameliorate colitis in the DSS-induced colitis mouse model, this model is not equivalent to CD in human patients. This mandates that LRRK2 blockers be tested in a variety of other mouse models of gut inflammation. Finally, given that LRRK2 occupies an important signaling position vis-à-vis both inflammation and autophagy, it is conceivable that LRRK2 inhibitors would have side effects that preclude their use in patients.
MATERIALS AND METHODS
Study design
This study was designed to determine the effect of risk polymorphisms in the LRRK2/MUC19 gene region associated with CD on LRRK2 expression and the functional consequences of such effects. To this end, LRRK2 expression was assessed in DCs from patients with CD. Lrrk2 Tg mice overexpressing LRRK2 and Lrrk2 KO mice were evaluated for proinflammatory responses and autophagy. LRRK2 mRNA expression in human DCs from 20 patients with CD and in LCLs was measured by real-time PCR. Subsequently, the severity of DSS-induced colitis was determined in Lrrk2 Tg mice by analyzing body weight loss, histological score, and mRNA cytokine expression in Lrrk2 Tg mice and littermate control animals. In addition, Dectin-1–stimulated BMDCs from Lrrk2 Tg mice and littermate controls were assessed for cytokine production, NF-κB activation, NFAT responses, and autophagy induction in culture. Finally, the capacity of LRRK2 inhibitors to block cytokine and autophagic responses in Lrrk2 Tg mice and littermate control mice was determined.
Mice
Lrrk2 KO (JAX012453) (50), BAC FLAG-Lrrk2 Tg (JAX012466) (51), and C57BL/6J mice were purchased from The Jackson Laboratory. The nominally C57BL/6 Lrrk2 KO mice thus obtained had a mixed genetic background containing FVB and 129/SvJ strain; they were therefore crossed with C57BL/6J mice in our animal facility for two or more generations to obtain Lrrk2 heterozygous mice (N10 backcross generation). These Lrrk2 heterozygous mice were then intercrossed with Lrrk2 heterozygous mice to obtain Lrrk2 WT and Lrrk2 KO mice, which were then maintained in the same animal facility for at least three generations to achieve both genetic and intestinal microbial parity (52). In some experiments, Lrrk2 KO mice derived from Lrrk2 heterozygous breeders were compared with WT control mice arising in the same litter (littermate controls). However, because the frequency of Lrrk2 KO mice derived from heterozygous breeders was low, it was usually difficult to obtain a sufficient number of Lrrk2 KO and WT littermate control mice to conduct studies with littermate control mice in a timely manner; for example, even if a large number of heterozygous breeder pairs were set up to obtain 30 pups per month, this population only contained one or two sex-matched pairs of Lrrk2 KO mice and WT littermates. Thus, in most experiments, Lrrk2 KO mice derived from homozygous breeders were compared with Lrrk2 WT control mice derived from Lrrk2 heterozygous breeders, that is, control mice closely related to the KO mice that were raised in the same environment. These WT controls proved to have a similar phenotype to WT littermates derived from Lrrk2 heterozygous breeders in that BMDCs from both WT mice exhibited similar TNF-α production. We did not use C57BL/6J mice (JAX) as control mice because we found that they had different susceptibility to DSS-induced colitis compared with our Lrrk2 WT mice derived and raised as described above (fig. S12). For studies of Lrrk2 Tg mice, the mice were repeatedly crossed with C57BL/6J mice to obtain a pure C57BL/6J background. Tg mice obtained after 14 backcross generations were subjected to DSS-induced colitis in comparison to littermate control mice. GFP-LC3 Tg mice were obtained from N. Mizushima (Tokyo University) and were crossed with Lrrk2 WT, Lrrk2 KO, or Lrrk2 Tg mice. The animal studies were approved by National Institutes of Health (NIH) Animal Care and Use Committee and performed under NIH animal care guidelines.
DSS-induced colitis
Six- to 8-week-old mice were used for DSS-induced colitis. DSS (molecular weight, 36,000 to 50,000) was purchased from MP Biomedicals. DSS was added in the drinking water bottle, given for the indicated days, and then replaced with regular animal facility water for 2 days. We repeated DSS colitis three times in Figs. 1 and 5 and fig. S11 and five times in fig. S2. Total number of the mice used in DSS colitis is as follows: Fig. 1 (Lrrk2 Tg, n = 12; control, n = 14), fig. S11 (control, n = 15; inhibitor-treated mice, n = 15), and fig. S2 (Lrrk2 WT, n = 23; Lrrk2 KO, n = 22). After mice were sacrificed, colon tissues were fixed in 10% formalin and sent to Histoserv Inc. for H&E staining. For epithelium, histology was scored as follows: 0, normal morphology; 1, loss of goblet cells; 2, loss of goblet cells in large areas; 3, loss of crypts; and 4, loss of crypts in large areas. For infiltration, scoring was as follows: 0, no infiltrate; 1, infiltrate around crypt basis; 2, infiltrate reaching to lamina muscularis mucosae; 3, extensive infiltration reaching the lamina muscularis mucosae and thickening of the mucosa with abundant edema; and 4, infiltration of the lamina submucosa. The total histological score is given as epithelium plus infiltration (53).
Western blotting
Whole-cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer (Santa Cruz Biotechnology) with cOmplete Mini Protease Inhibitor Cocktail and PhosSTOP Phosphatase Inhibitor Cocktail (Roche). The lysate was vortexed every 5 min and cooled on ice for 30 min. After centrifugation (15,000 rpm, 15 min, 4°C), the supernatant was collected, and the protein concentration was measured with a BCA Protein Assay Kit (Pierce). The samples were incubated with LDS sample buffer and reducing reagent (Life Technologies) for 10 min at 70°C and subjected to SDS–polyacrylamide gel electrophoresis with NuPAGE Bis-Tris gels and MOPS or MES buffer (Life Technologies). For high–molecular weight protein, tris-acetate gels and buffer were used. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membrane using Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). The membranes were blocked with SuperBlock (TBS) Blocking Buffer (Thermo Fisher Scientific) or 5% nonfat dry milk and immunoblotted with the indicated primary antibodies and then horseradish peroxidase–conjugated secondary antibodies. The signals were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific), followed by exposure to Kodak XAR film in a dark room. Nuclear and cytoplasmic extractions were prepared with NE-PER Nuclear and Cytoplasmic Extraction Kit according to the manufacturer’s protocol (Pierce). When required, the membrane was stripped with Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific).
Immunoprecipitation
HEK293T cells were transfected with indicated plasmids. Whole-cell lysates of these cells were prepared with cell lysis buffer (Cell Signaling Technology) 18 to 24 hours after transfection. The protein was incubated with the complex of antibody and Dynabeads Protein G for 60 min. The Dynabeads Protein G was washed four times with the buffer that is included in Dynabeads Protein G Immunoprecipitation Kit (Life Technologies). For FLAG-tagged protein, the lysate was incubated with EZview Red anti-FLAG M2 Affinity Gel and washed four times with tris-buffered saline with Tween 20 buffer.
Statistical analysis
A two-tailed Student’s t test was used to evaluate the differences between two groups. One-way ANOVA, followed by Dunnett’s or Holm-Sidak’s multiple comparisons test, was used for multiple group comparisons. Data were analyzed with Prism 6 (GraphPad Software). P < 0.05 was considered statistically significant. All experiments were repeated at least three times and presented as means ± SEM.
Supplementary Material
Acknowledgments:
We thank H. Cai of National Institute on Aging (NIH) for providing LRRK2 KO mice. Funding: This work was supported by Grant-in-Aid for Researchers, Hyogo College of Medicine 2013 (to T.T.), the New York Crohn’s Foundation and the Leona M. and Harry B. Helmsley Charitable Trust (to I.P.), and NIH grant DK062431 (to S.R.B).
Footnotes
Competing interests: T.T., A.K., I.F., and W.S. are co-inventors on patent application no. US 20170079987 A1 “Treatment or prevention of an intestinal disease or disorder.” The other authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
http://www.sciencetranslationalmedicine.org/cgi/content/full/10/444/eaan8162/DC1
Fig. S1. LRRK2 protein expression in representative LCLs determined by Western blots.
Fig. S2. Additional information about DSS-induced colitis in Lrrk2 Tg and Lrrk2 KO mice.
Fig. S3. BMDCs from Lrrk2 Tg mice exhibit higher production of IL-23 than BMDCs from control mice in response to Dectin-1 agonists.
Fig. S4. BMDCs from Lrrk2 KO mice do not produce increased TNF-α compared to BMDCs from
Lrrk2 WT mice.
Fig. S5. LRRK2 positively regulates NFAT activity.
Fig. S6. Increased autophagy in BMDCs from Lrrk2 KO mice.
Fig. S7. Inhibitory effects of 12 LRRK2 inhibitors.
Fig. S8. IL-2 production by Lrrk2 Tg mouse and control BMDCs was suppressed by LRRK2 inhibitors.
Fig. S9. LRRK2 inhibitors suppressed a broad range of cytokines produced by mouse BMDCs. Fig. S10. BMDCs from Lrrk2 Tg mice exhibited increased autophagy when cultured with a LRRK2 inhibitor (LRRK2-IN-1).
Fig. S11. The LRRK2 Inhibitor LRRK2-IN-1 ameliorated DSS-induced colitis in C57BL/6J mice.
Fig. S12. Severity of DSS-induced colitis in control mice on a C57BL/6J background varied with housing conditions.
Fig. S13. Summary diagram.
Table S1. Characteristics of CD patients.
Table S2. Eighteen LCLs used in this study.
Table S3. Individual level data.
Reference (54)
REFERENCES AND NOTES
- 1.Kiesler P, Fuss IJ, Strober W, Experimental models of inflammatory bowel diseases. Cell. Mol. Gastroenterol. Hepatol 1, 154–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strober W, Asano N, Fuss IJ, Kitani A, Watanabe T, Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn’s disease. Immunol. Rev 260, 249–260 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Sarin R, Wu X, Abraham C, Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc. Natl. Acad. Sci. U.S.A 108, 9560–9565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer MJ, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T, Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Krüger R, Federoff M, Klein C, Goate AM, Perlmutter JS, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T, Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet 41, 1308–1312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar J-P, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards CM, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart AL, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield JC, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; The International IBD Genetics Consortium (IIBDGC); Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH, Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F-R, Huang W, Chen S-M, Sun L-D, Liu H, Li Y, Cui Y, Yan X-X, Yang H-T, Yang R-D, Chu T-S, Zhang C, Zhang L, Han J-W, Yu G-Q, Quan C, Yu Y-X, Zhang Z, Shi B-Q, Zhang L-H, Cheng H, Wang C-Y, Lin Y, Zheng H-F, Fu X-A, Zuo X-B, Wang Q, Long H, Sun Y-P, Cheng Y-L, Tian H-Q, Zhou F-S, Liu H-X, Lu W-S, He S-M, Du W-L, Shen M, Jin Q-Y, Wang Y, Low H-Q, Erwin T, Yang N-H, Li J-Y, Zhao X, Jiao Y-L, Mao L-G, Yin G, Jiang Z-X, Wang X-D, Yu J-P, Hu Z-H, Gong C-H, Liu Y-Q, Liu R-Y, Wang D-M, Wei D, Liu J-X, Cao W-K, Cao H-Z, Li Y-P, Yan W-G, Wei S-Y, Wang K-J, Hibberd ML, Yang S, Zhang X-J, Liu J-J, Genomewide association study of leprosy. N. Engl. J. Med 361, 2609–2618 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Wallings R, Manzoni C, Bandopadhyay R, Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 282, 2806–2826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ, The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol 12, 1063–1070 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberl G, Addressing the experimental variability associated with the microbiota. Mucosal Immunol. 8, 487–490 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Sokol H, Conway KL, Zhang M, Choi M, Morin B, Cao Z, Villablanca EJ, Li C, Wijmenga C, Yun S-H, Shi HN, Xavier RJ, Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology 145, 591–601.e3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ, Podolsky DK, LRRK2 is involved in the IFN-γ response and host response to pathogens. J. Immunol 185, 5577–5585 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim B, Yang M-S, Choi D, Kim J-H, Kim H-S, Seol W, Choi S, Jou I, Kim E-Y, Joe E.-h., Impaired inflammatory responses in murine Lrrk2-knockdown brain microglia. PLOS ONE 7, e34693(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aramburu J, García-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG, Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 1, 627–637 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A, Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. U.S.A 108, 11381–11386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J, The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell 13, 343–354 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Kim HR, Quinley C, Kim J, Gonzalez-Navajas J, Xavier RJ, Raz E, Autophagy suppresses interleukin-1β (IL-1β) signaling by activation of p62 degradation via lysosomal and proteasomal pathways. J. Biol. Chem 287, 4033–4040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul S, Kashyap AK, Jia W, He Y-W, Schaefer BC, Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity 36, 947–958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothfuchs AG, Bafica A, Feng CG, Egen JG, Williams DL, Brown GD, Sher A, Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J. Immunol 179, 3463–3471 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Lee H-M, Yuk J-M, Kim K-H, Jang J, Kang G, Park JB, Son J-W, Jo E-K, Mycobacterium abscessus activates the NLRP3 inflammasome via Dectin-1–Syk and p62/SQSTM1. Immunol. Cell Biol 90, 601–610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS, The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Chen J, Zhang L, Cha Z, Han S, Shi W, Ding R, Ma L, Xiao H, Shi C, Jing Z, Song N, Mycobacterium leprae upregulates IRGM expression in monocytes and monocyte-derived macrophages. Inflammation 37, 1028–1034 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Choi AMK, Ryter SW, Levine B, Autophagy in human health and disease. N. Engl. J. Med 368, 651–662 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Mariño G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, Rain J-C, Ninomiya-Tsuji J, Fuentes JM, Lavandero S, Galluzzi L, Maiuri MC, Kroemer G, Inhibition of autophagy by TAB2 and TAB3. EMBO J. 30, 4908–4920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaesu G, Kobayashi T, Yoshimura A, TGFβ-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. J. Biochem 151, 157–166 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, Landegren U, Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Cho D-H, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC, Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 274, 95–100 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B, Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z, Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J. Neurosci. 32, 7585–7593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee J-D, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS, Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat. Chem. Biol 7, 203–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moehle MS, Webber PJ, Tse T, Sukar N, Standaert DG, DeSilva TM, Cowell RM, West AB, LRRK2 inhibition attenuates microglial inflammatory responses. J. Neurosci 32, 1602–1611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manzoni C, Mamais A, Roosen DA, Dihanich S, Soutar MPM, Plun-Favreau H, Bandopadhyay R, Hardy J, Tooze SA, Cookson MR, Lewis PA, mTOR independent regulation of macroautophagy by Leucine Rich Repeat Kinase 2 via Beclin-1. Sci. Rep 6, 35106(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, Hsu N-Y, Chuang L-S, Carmi S, Villaverde N, Li X, Rivas M, Levine AP, Bao X, Labrias PR, Haritunians T, Ruane D, Gettler K, Chen E, Li D, Schiff ER, Pontikos N, Barzilai N, Brant SR, Bressman S, Cheifetz AS, Clark LN, Daly MJ, Desnick RJ, Duerr RH, Katz S, Lencz T, Myers RH, Ostrer H, Ozelius LJ, Payami H, Peter Y, Rioux JD, Segal AW, Scott WK, Silverberg MS, Vance JM, Ubarretxena-Belandia I, Foroud T, Atzmon G, Pe’er I, Ioannou YA, McGovern DPB, Yue Z, Schadt EE, Cho JH, Peter I, Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med 10, eaai7795(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trabzuni D, Ryten M, Emmett W, Ramasamy A, Lackner KJ, Zeller T, Walker R, Smith C, Lewis PA, Mamais A, de Silva R, Vandrovcova J; International Parkinson Disease Genomics Consortium (IPDGC); Hernandez D, Nalls MA, Sharma M, Garnier S, Lesage S, Simon-Sanchez J, Gasser T, Heutink P, Brice A, Singleton A, Cai H, Schadt E, Wood NW, Bandopadhyay R, Weale ME, Hardy J, Plagnol V, Fine-mapping, gene expression and splicing analysis of the disease associated LRRK2 locus. PLOS ONE 8, e70724(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schapansky J, Nardozzi JD, Felizia F, LaVoie MJ, Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum. Mol. Genet 23, 4201–4214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amendola A, Butera A, Sanchez M, Strober W, Boirivant M, Nod2 deficiency is associated with an increased mucosal immunoregulatory response to commensal microorganisms. Mucosal Immunol. 7, 391–404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baram L, Cohen-Kedar S, Spektor L, Elad H, Guzner-Gur H, Dotan I, Differential stimulation of peripheral blood mononuclear cells in Crohn’s disease by fungal glycans. J. Gastroenterol. Hepatol 29, 1976–1984 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Dzamko N, Inesta-Vaquera F, Zhang J, Xie C, Cai H, Arthur S, Tan L, Choi H, Gray N, Cohen P, Pedrioli P, Clark K, Alessi DR, The IκB kinase family phosphorylates the Parkinson’s disease kinase LRRK2 at Ser935 and Ser910 during Toll-like receptor signaling. PLOS ONE 7, e39132(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hakimi M, Selvanantham T, Swinton E, Padmore RF, Tong Y, Kabbach G, Venderova K, Girardin SE, Bulman DE, Scherzer CR, LaVoie MJ, Gris D, Park DS, Angel JB, Shen J, Philpott DJ, Schlossmacher MG, Parkinson’s disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J. Neural Transm 118, 795–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alegre-Abarrategui J, Christian H, Lufino MMP, Mutihac R, Venda LL, Ansorge O, Wade-Martins R, LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet 18, 4022–4034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J, Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol. Neurodegener 7, 2(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine B, Liu R, Dong X, Zhong Q, Beclin orthologs: Integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 25, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orenstein SJ, Kuo S-H, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, Raya A, Sulzer D, Cuervo AM, Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci 16, 394–406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kachaeva EV, Shenkman BS, Various jobs of proteolytic enzymes in skeletal muscle during unloading: Facts and speculations. J. Biomed. Biotechnol 2012, 493618(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H, Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J 441, 399–406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanjuan MA, Milasta S, Green DR, Toll-like receptor signaling in the lysosomal pathways. Immunol. Rev 227, 203–220 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Boyle KB, Randow F, Rubicon swaps autophagy for LAP. Nat. Cell Biol 17, 843–845 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Nalls MA, Saad M, Noyce AJ, Keller MF, Schrag A, Bestwick JP, Traynor BJ, Gibbs JR, Hernandez DG, Cookson MR, Morris HR, Williams N, Gasser T, Heutink P, Wood N, Hardy J, Martinez M, Singleton AB; International Parkinson’s Disease Genomics Consortium (IPDGC); The Wellcome Trust Case Control Consortium 2 (WTCCC2); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC), Genetic comorbidities in Parkinson’s disease. Hum. Mol. Genet 23, 831–841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuji RN, Flagella M, Baca M, Baptista MAS, Brodbeck J, Chan BK, Fiske BK, Honigberg L, Jubb AM, Katavolos P, Lee DW, Lewin-Koh S-C, Lin T, Liu X, Liu S, Lyssikatos JP, O’Mahony J, Reichelt M, Roose-Girma M, Sheng Z, Sherer T, Smith A, Solon M, Sweeney ZK, Tarrant J, Urkowitz A, Warming S, Yaylaoglu M, Zhang S, Zhu H, Estrada AA, Watts RJ, Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med 7, 273ra215(2015). [DOI] [PubMed] [Google Scholar]
- 50.Parisiadou L, Xie C, Cho HJ, Lin X, Gu X-L, Long C-X, Lobbestael E, Baekelandt V, Taymans J-M, Sun L, Cai H, Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J. Neurosci 29, 13971–13980 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Tan Y-C, Poulose S, Olanow CW, Huang X-Y, Yue Z, Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J. Neurochem 103, 238–247 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR, Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W, Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J. Clin. Invest 118, 545–559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fogel AI, Dlouhy BJ, Wang C, Ryu S-W, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ, Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol 33, 3675–3688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.