Abstract
Objectives:
One of the standard of care regimens for advanced pancreatic cancer is gemcitabine-based chemotherapy. The efficacy of gemcitabine is limited by dose-limiting hematologic toxicities especially neutropenia. Uncovering the variability of these toxicities attributed to germline DNA variation is of great importance.
Methods:
CALGB 80303 was a randomized study in advanced pancreatic cancer patients treated with gemcitabine with or without bevacizumab. The study protocol included genotyping of genes of gemcitabine disposition (CDA, DCTD, SLC29A1, SLC28A1, SLC29A2), as well as a genome-wide analysis. The clinical phenotype was time to early high-grade neutropenia event accounting for progression or death, or other treatment-terminating adverse events as competing informative events. The inference was conducted based on the association between genotype and cause-specific hazard of a neutropenic event.
Results:
The primary analyses were conducted on the basis of 294 genetically-estimated European pancreatic cancer patients. For CDA rs2072671 (A>C), AC and CC patients had a lower risk of neutropenia than AA patients (P-value 0.01, HR 0.61, 95% CI 0.41–0.89). For SLC28A1 rs3825876 (G>A), AA patients have a higher risk of neutropenia than GA and GG patients (P-value 0.02, HR 1.51, 95% CI 1.06–2.16). CDA rs2072671 was associated with increased mRNA expression in whole blood in three studies (P-values 2.7e-14, 6.61e-62, 9.70e-65). In the genome-wide analysis, variants in TGFB2 were among the top hits (lowest P-value 1.62e-06) but had no effect in luciferase assays.
Conclusion:
The first genetic analysis of gemcitabine-induced neutropenia using a competing risk model in a prospective randomized clinical study has proposed a potentially novel mechanism of the protective effect of the CDA rs2072671 variant. Further confirmation is needed.
Keywords: Pancreatic cancer, Gemcitabine, genotyping, variant, genome-wide, neutropenia, association, clinical trial, single nucleotide polymorphism
Introduction
CALGB 80303 was a randomized double-blind phase III study in patients with metastatic or locally advanced pancreatic cancer. Patients were randomized to gemcitabine chemotherapy plus placebo or gemcitabine plus bevacizumab. Its primary clinical endpoint was overall survival and adding bevacizumab to the chemotherapy agent did not improve survival [1].
As part of the CALGB 80303 protocol, we proposed to investigate the association of gemcitabine-induced high-grade neutropenia with common variants in candidate genes of gemcitabine pharmacology. As described in the PharmGKB pathway for gemcitabine (https://www.pharmgkb.org/pathway/PA2036), the nucleoside transporters SLC29A1, SLC28A1, and SLC29A2 are involved in gemcitabine uptake into the cells, while CDA and DCTD are involved in gemcitabine inactivation through conversion of gemcitabine to dFdU or dFdUMP, respectively. Due to the importance of these genes in gemcitabine transport and metabolism, alterations in expression levels or function of the coded proteins could lead to gemcitabine accumulation and ultimately gemcitabine toxicity. Discovering variants in these genes that associate with gemcitabine-induced neutropenia will provide a better understanding of the DNA variation driving this toxicity. The study protocol was later amended to allow the interrogation of thousands of additional variants in other genes through a genome-wide genotyping approach.
In this paper, we report the results from a genetic analysis conducted in CALGB 80303 identifying germline variants associated with the occurrence of gemcitabine-induced high-grade neutropenia. Because treatment-terminating events might affect the onset of neutropenia, we performed this genetic analysis under a competing risk model with informative censoring.
Methods
Patient Population
A total of 602 patients in CALGB 80303 (Protocol ID: CR_Pro00009631) were randomized, 302 to the gemcitabine plus bevacizumab versus 300 to gemcitabine plus placebo. The randomization was stratified by extent of disease (metastatic versus locally advanced), ECOG performance status (0–½), and prior radiation (yes/no). Key baseline characteristics for these patients are summarized in Table 1.
Table 1. Clinical characteristics and overall survival of patients in CALGB 80303 investigated in this study.
| All Patients | Genotyped Patients | Genotyped Patients of European Ancestry | ||
|---|---|---|---|---|
| Sample Size | 602 | 338 | 294 | |
| Sex | Male | 329 | 185 | 159 |
| Female | 273 | 153 | 135 | |
| Age | Mean (SD) | 64.0 (10.6) | 63.6 (10.4) | 64.4 (10.5) |
| Median (95%CI) |
64.1 (63.2–65.1) |
64 (62.8–65.2) |
64.3 (62.8–66.4) |
|
| Race | White | 529 (87.9%) | 301 (89.0%) | 289 (98.3%) |
| Black | 49 (8.1%) | 26 (7.7%) | 0 | |
| Asian | 10 (1.7%) | 2 (0.6%) | 0 | |
| Native Hawaiian | 1 (0.2%) | 1 (0.3%) | 0 | |
| American Indian | 3 (0.5%) | 0 | 0 | |
| Multiple | 1 (0.2%) | 1 (0.3%) | 0 | |
| Unknown | 9 (1.5%) | 7 (2.1%) | 5 (1.7%) | |
| Overall Survival Time | Median (months) | 5.85 | 5.88 | 5.95 |
| Mean (months) | 7.71 | 7.75 | 7.82 | |
|
Extent of Disease |
Metastatic | 520 (86.4%) | 299 (88.5%) | 250 (85.0%) |
| Locally Advanced | 69 (11.5%) | 39 (11.5%) | 44 (15.0%) | |
|
Prior Radiotherapy |
No | 526 (87.4%) | 302 (89.3%) | 262 (89.1%) |
| Yes | 65 (10.8%) | 36 (10.7%) | 32 (10.9%) | |
|
Performance Status |
0 or 1 | 537 (89.2%) | 305 (90.2%) | 267 (90.8%) |
| 2 | 65 (10.8%) | 33 (9.8%) | 27 (9.2%) | |
| Arm | Placebo | 300 (49.8%) | 157 (46.4%) | 140 (47.6%) |
| Bevacizumab | 302 (50.2%) | 181 (53.6%) | 154 (52.4%) |
Gemcitabine was administered intravenously over thirty minutes on days 1, 8, and 15 of a 28-day cycle at the dose of 1000 mg/m2. Bevacizumab or placebo was administered intravenously after chemotherapy on days 1 and 15 of each cycle at the dose of 10 mg/kg. Per protocol, patients with evidence of response or stable disease remained on therapy until disease progression, unacceptable toxicity, or the patient’s decision to withdraw. Patients were evaluated for response according to the Response Evaluation Criteria in Solid Tumors (RECIST) [2] criteria every two cycles.
The study utilized Common Terminology Criteria for Adverse Events (CTCAE) version 3.0 for adverse event reporting. In the CTCAE system, each adverse event report is given a treatment attribution grade ranging from 1, indicating that the event was unlikely to be related to treatment, to 5, indicating that the event was definitely related to treatment. An adverse event was qualified as treatment-related if the attribution code was at least 3. Additionally, high-grade adverse events were reported through the Adverse Event Expedited Reporting System. Adverse event forms were submitted every cycle while patients actively received therapy, then every three months for one year, or until progression or death. The follow up and response forms were submitted every cycle while patients actively received therapy, then every three months for one year, followed by every six months for three years, or until progression or death. Additional information about the design, clinical characteristics and outcome for the clinical study has been previously reported [1].
Genotyping
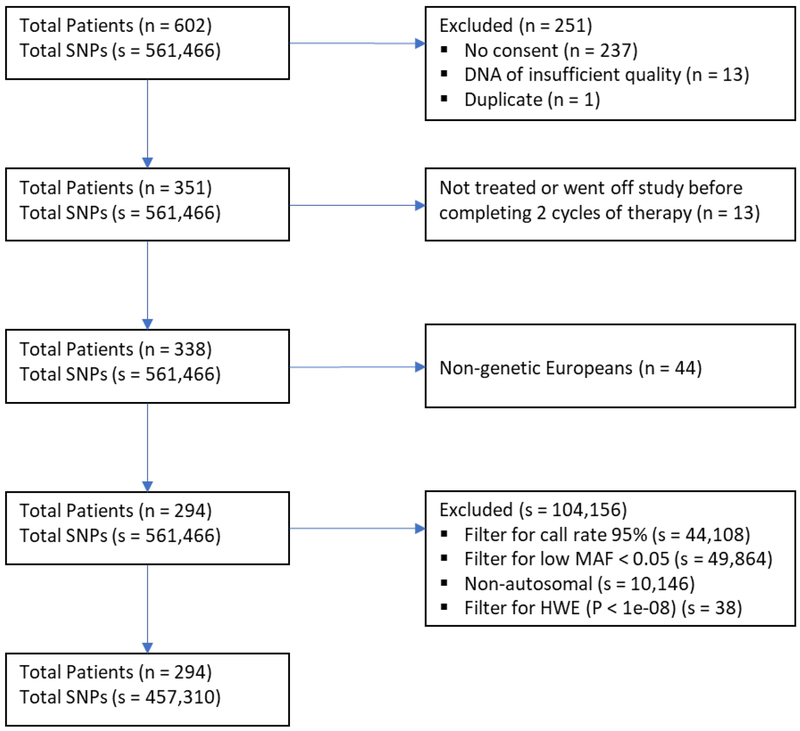
Genotyping was performed using the Illumina HapMap 550v3 beadchip on samples from 351 CALGB 80303 patients who had consented for participation in this study. This platform typed 561,466 single-nucleotide polymorphisms (SNPs). Following a comprehensive quality control and the use of ordination methodology, a genetic European subpopulation consisting of 294 patients was identified. Additional details about the quality control and filtering methodology were previously described [3]. The analyses presented in this report were based on the aforementioned subset of 294 patients, and SNPs were filtered based on a relative minor allelic frequency (MAF) cut-off of 0.05. The final number of SNPs passing quality control was 457,310. Out of these, 71 typed SNPs located in the genes CDA, DCTD, SLC28A1, SLC29A1, and SLC29A2 are reported (see Table, Supplemental Digital Content 1, http://links.lww.com/FPC/B344).
Neutropenia Phenotyping Under a Competing Risk Model
For each of the 294 patients, the individual follow-up and adverse event forms were reconstructed by querying the CALGB clinical database (CALGB is now part of Alliance for Clinical Trials in Oncology). The data for survival and progression were obtained from the clinical data set [1]. Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center. For each patient, we ascertained the date of the first reported neutropenic event (MedDRA code 10029363) of grade 2 or higher.
The phenotype of interest for this study is early high-grade (grade 3 or higher) treatment-related neutropenia. We defined early high-grade neutropenia as occurring within the five weeks after start of treatment. Patients who did not experience any such event within five weeks of start of therapy were uninformatively censored at five weeks. Patients, for whom no high-grade neutropenic event were reported, the date and cause of treatment termination were ascertained. Causes of treatment termination (leading to the exit of the patients from the study) were categorized as death or progression, treatment-terminating adverse event (TTAE) other than neutropenia, or other (e.g., withdrawal of consent or unknown). Complicated cases were adjudicated by additional review by a medical oncologist. The time to event was calculated as time from the date of treatment start to the date of a high-grade neutropenic event, a TTAE, or other events of treatment termination, whichever occurred first. Patients lost to follow-up prior to the occurrence of high-grade neutropenia or any event of treatment termination were considered censored at date of last follow-up. For fourteen solicited adverse events, including neutropenia, all grades were reported, while for unsolicited events, only events graded 3 to 5 were reported. Additional technical details on the phenotype data are available in Supplemental Digital Content 2, http://links.lww.com/FPC/B345.
Statistical Considerations
The primary objective of the study was to test the association of SNPs in CDA, DCTD, SLC28A1, SLC29A1, and SLC29A2 with time to early high-grade neutropenia. The time to event distributions for this outcome are non-identifiable as they are subject to informative censoring mechanisms, including treatment termination due to progression, death, or TTAEs. Inference was conducted on the basis of the score test for cause-specific hazard within the framework of a Cox model [4] for time to early high-grade neutropenia. We also considered analyses adjusted for clinically relevant covariates within the framework of a multivariate additive Cox model. The covariates used in these analyses were prior radiotherapy, age, and baseline neutropenia levels, where age and baseline neutropenia levels were log-transformed (base 10 and base e, respectively) to address skewness of the distributions and reduce the influence of outlier values. The covariates were selected based on their putative clinical relevance to the primary outcome. These were selected due to the putative relationship between radiation-induced marrow suppression and neutropenia, and the putative association of risk of chemotherapy-induced neutropenia in older patients and those with low neutrophil counts at the start of therapy. For analyses accounting for baseline covariates, the Wald test was used. The analyses were powered for additive genetic effects. The null distribution for the marginal P-values was approximated under an asymptotic framework. Top hits were selected based on unadjusted P-values from the score test.
A secondary objective included analyzing the association between the genome-wide SNPs and time to early high-grade neutropenia. Cumulative incidence plots of the phenotype, stratified by genotypes, were used for visualizing the genetic effects. Quantile-quantile plots of the marginal asymptotic P-values for the genome-wide data were evaluated for inflation of significance levels. The reported P-values have not been adjusted for multiple testing.
All statistical analyses were carried out using the R statistical environment [5] using extension packages survival [6] for proportional hazard test, cmprsk [7] for estimating the cumulative incidence curves, GenABEL [8] for genotype data management and filtering, and to obtain Hardy-Weinberg P-values, and SNPassoc [9] for computing linkage disequilibrium (LD).
SNP Annotation
The chromosome and position information for the reported variants were obtained from an annotation file provided by Illumina (HumanHap550v3_A.csv) built against build 36 of the human genome. Additional genomic annotation, including gene and function information, was obtained from the SCAN database [10]. To assess the extent of the signals relative to genomic position and LD within a region (e.g., gene or around a SNP), the LocusZoom [11] software (version 1.3) was used. The 1,000 Genomes Pilot 1 CEU panel data (March 2012 release) based on human genome build hg19 was used as the reference data. SNPs in the candidate genes (including those up to 1kb upstream of the 5’ end or 1kb downstream of the 3’ end) were queried from the SCAN database [10] and confirmed from dbSNP database.
Analysis of Gene Expression and Bioinformatics
To visualize the P-value for association in each of the candidate genes, the LocusZoom [11] software (version 1.3) was employed using a 50 Kb window left and right from the gene limits. SNPs in candidate genes that had a P-value <0.05 for association, and variants in high LD with them (r2 ≥0.8 in Europeans from the 1,000 Genomes Project) were further evaluated for functional effects using bioinformatics (RegulomeDB) [12]. The same variants were also tested for association with mRNA gene expression in the human liver [13] and blood [14–16] as expression quantitative trait loci (eQTL). Candidate gene expression localization and single-tissue eQTLs were analyzed using the GTEx portal [14] (https://www.gtexportal.org/home/). The NESDA NTR Conditional eQTL Catalog [16] (https://eqtl.onderzoek.io/index.php?page=info) was used to observe eQTLs in whole blood. SIFT [17] and PolyPhen-2 [18] were interrogated for evaluating the functionality of the CDA rs2072671 (79A>C) missense variant.
Luciferase Assays of SNPs in TGFB2
Three SNPs rs2799083, rs2799090, and rs11118109 in TGFB2 were selected for functional evaluation in luciferease assays based on their associations with neutropenia. The 500 bp regions spanning the SNPs were PCR amplified and cloned into pGL4.26 in the forward and reverse direction (for luciferase assay primer sequences, see Table, Supplemental Digital Content 3, http://links.lww.com/FPC/B346). Bases in bold align with the pGL4.26 luciferase vector sequence for In-Fusion cloning at the SacI and XhoI restriction enzyme sites. Underlined bases indicate the SNP change in mutagenesis. Variant alleles for each SNP were generated using site-directed mutagenesis. The empty pGL4.26 vector served as the negative control, and regions of 20q13.2 and 19p13.2, previously reported to have enhancer activity, were used as the positive controls [19, 20].
PANC-1 and K-562 cells were co-transfected with 1050 ng pGL4.26 enhancer construct and 1 ng pNL1.1TK plasmid using Lipofectamine 2000. After 48 hours, luminescence was measured. Three independent clones were assayed for each enhancer construct. Relative luciferase activity was calculated using the ratio of luminescence from the enhancer construct to the negative control construct. A relative luciferase reading of greater than 2 was considered enhancer activity.
Luciferase activity (normalized by blank control and log base e transformed) of the variant constructs for rs11118109, rs2799090, and rs2799083, were compared to the activity of the reference constructs in both the forward and reverse orientations. Statistical significance was assessed using the framework of a linear mixed effect model with reference genotypes as fixed effects, and clone and day of the experiment as two independent random effects. Results from 3 clones for each construct with varying replicates are reported as the log transformation of normalized luciferase activity values. The lmer function from the lme4 [21] R extension package was used to implement this model.
SNPs in TGFB2 and Association with Plasma Levels of the TGFβ2 Protein
Plasma concentrations of the TGFβ2 protein were measured at baseline in 216 patients of CALGB 80303, as previously reported [22]. Associations between rs2799083 (T>C), rs2799090 (A>G), and rs11118109 (T>G) in TGFB2 with TGFβ2 protein levels were tested using the Jonckheere-Terpstra (JT) test [23–25] for ordered alternatives.
Results
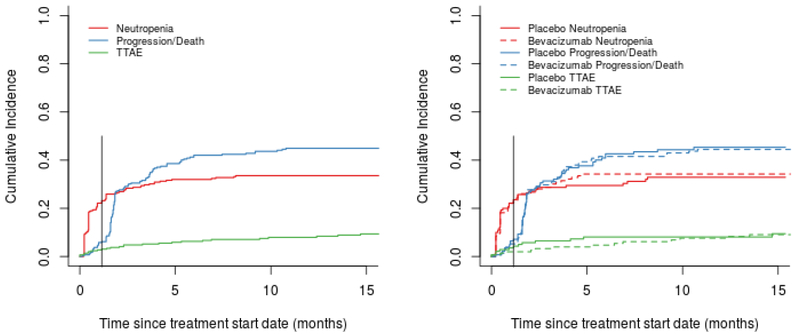
The CONSORT chart shown in Figure 1 summarizes the patient and SNP selection process. With the exception of one case of grade 3 neutropenia, all high-grade neutropenic events in this study were qualified by the treating physicians as treatment-related. Figure 2 shows the estimated cumulative incidence curves for neutropenia (at any time). The two competing risks are treatment termination due to progression/death, and TTAE, with death or progression occurring most frequently. The observed toxicities that resulted in a TTAE were hypertension, proteinuria, and thrombosis.
Figure 1. CONSORT chart.
Figure 2. Cumulative incidence of high-grade neutropenia and competing events.
The graph on the right shows the stratification by arm (gemcitabine plus placebo and gemcitabine plus bevacizumab). The vertical line represents the cut-off for the definition of early neutropenia (5 weeks).
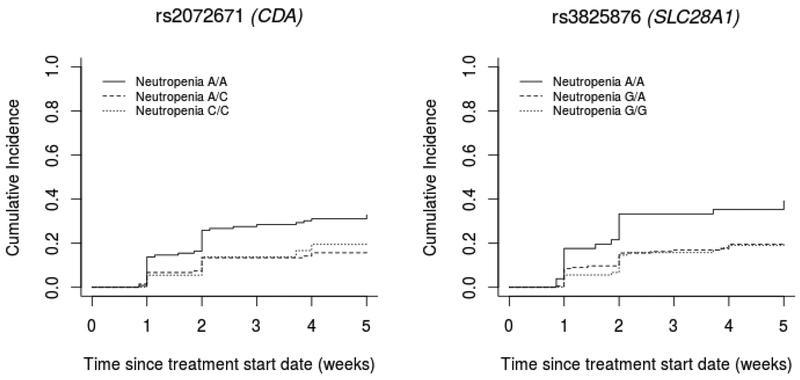
A total of 71 candidate SNPs from CDA, DCTD, SLC28A1, SLC29A1 and SLC29A2 (MAF>0.05) were tested for association with time to early high-grade neutropenia. We performed additional downstream analyses on the SNPs in CDA and SLC28A1 (Table 2). Because of the high LD between the two pairs of SNPs (CDA rs2072671 and rs471760: r2 0.89; SLC28A1 rs3825876 and rs12148896: r2 0.67), Figure 3 shows the genotype effects of CDA rs2072671 and SLC28A1 rs3825876. For CDA rs2072671 (A>C), patients with AC and CC genotypes have a lower risk of neutropenia than patients with the AA genotype (P-value 0.01, HR 0.61, 95% CI 0.41–0.89). For SLC28A1 rs3825876 (G>A), patients with AA genotypes have a higher risk of neutropenia than patients with the GA and GG genotypes (P-value 0.02, HR 1.51, 95% CI 1.06–2.16). Neither of these two variants (or any other interrogated variants in high LD with them) were eQTLs in the human liver (results not shown). When repositories of eQTL data from whole blood were interrogated for the CDA and SLC28A1 variants, the only association observed was for the CDA variants (see Table 3). The C allele of rs2072671 and the A allele of rs471760 were associated with an increase in CDA mRNA expression in whole blood (Table 3). There was minimal evidence for binding of SLC28A1 rs3825876 according to RegulomeDB (score 4) [12] and ENCODE [26], and there was no strong evidence that SNPs in LD with an r2>0.5 were located in regulatory regions. Since CDA rs2072671 was more likely to be located in a regulatory region (RegulomeDB score 3a) [12, 26], and associated with CDA expression in blood, this variant appeared to have more functional relevance.
Table 2. Association of candidate gene SNPs with early high-grade neutropenia.
Both unadjusted and adjusted (for prior radiotherapy, log base 10 transformed age, and natural log transformed baseline neutropenia levels) results are shown. For the unadjusted results, score test P-values are reported. For the adjusted results, Wald test P-values are reported. MAF = minor allele frequency; HWE = Hardy-Weinberg equilibrium P-value; HR = hazard ratio; LCL = lower 95% confidence limit; UCL = upper 95% confidence limit.
| SNP | Gene | MAF | HWE | Alleles | P-value | HR | LCL | UCL | Adj. P-value | Adj. HR | Adj. LCL | Adj. UCL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2072671 | CDA | 0.360 | 0.898 | A>C | 0.010 | 0.61 | 0.41 | 0.89 | 0.110 | 0.72 | 0.49 | 1.08 |
| rs471760 | CDA | 0.347 | 0.607 | G>A | 0.022 | 0.64 | 0.43 | 0.94 | 0.096 | 0.72 | 0.48 | 1.16 |
| rs3825876 | SLC28A1 | 0.435 | 0.287 | G>A | 0.023 | 1.51 | 1.06 | 2.16 | 0.026 | 1.51 | 1.05 | 2.18 |
| rs12148896 | SLC28A1 | 0.466 | 0.907 | G>A | 0.034 | 0.68 | 0.48 | 0.97 | 0.023 | 0.65 | 0.45 | 0.94 |
Figure 3. Effect of genotypes of CDA and SLC28A1 variants associated with early high-grade neutropenia.
Table 3. Association of CDA variants with CDA mRNA expression in whole blood.
A comparison of P-values and effect size for rs2072671 C and rs471760 A associations with CDA mRNA expression in three separate whole blood eQTL studies. *The G allele was assessed for rs471760 in Fehrmann et al.15.
As an exploratory analysis to select novel variants for further investigation, we conducted a genome-wide analysis on the basis of 457,310 SNPs to test for association with early high-grade neutropenia. The resulting quantile-quantile (Q-Q) and Manhattan plots are shown in Supplemental Digital Content 4 and 5, http://links.lww.com/FPC/B347 and http://links.lww.com/FPC/B348 respectively.
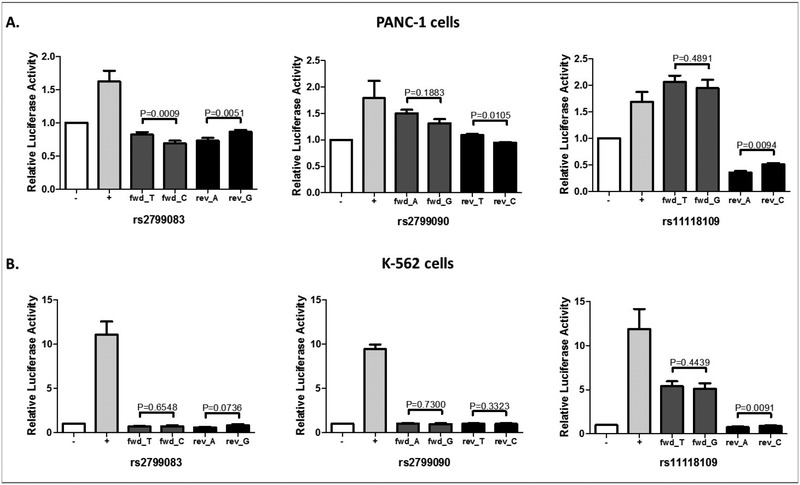
The two most highly ranked variants, according to the unadjusted P-values, were rs2799083 (T>C, intron 2 in TGFB2) and rs11118109 (T>G, 15 Kb 3’ of TGFB2) (see table, Supplemental Digital Content 6, http://links.lww.com/FPC/B349 for the top 50 SNPs based on P-value). We performed bioinformatic analyses and functional assays on these two TGFB2 variants plus an additional one (rs2799090 A>G, intron 1), (see 3 variants in bold, Supplemental Digital Content 6, http://links.lww.com/FPC/B349) in two human cell lines. RegulomeDB has shown that rs11118109 is located in a region where DNA is accessible to transcription factor binding (score 4), while rs2799083 and rs2799090 are less likely to be located in a regulatory region (score 5). In agreement with this finding, enhancer activity (relative luciferase activity >2.0) has been detected for the forward orientation of the construct containing rs11118109, but not for the other two variants. However, there was no significant difference in luciferase activity between the two alleles of rs11118109 (Figure 4).
Figure 4. Enhancer luciferase assays of TGFB2 variants.
A) PANC-1. B) K-562. Results are reported as the mean of 3 individual clones with n=3 experiments, with the exception of rs2799090 which had n=4 experiments in K-562 cells. The enhancer activity of each SNP was measured in both the forward and reverse orientation. Error bars represent standard error of the mean (SEM).
Because TGFβ2 protein levels were measured before treatment in a subset of patients in CALGB 80303 [22], we tested the hypothesis that TGFB2 variants might affect the secretion of soluble TGFβ2. There was no evidence for association between these three SNPs and TGFβ2 protein levels (P-value 0.54, 0.64, and 0.47, respectively) (see Box plots, Supplemental Digital Content 7, http://links.lww.com/FPC/B350).
Discussion
This is the first and largest comprehensive interrogation of the genetic risk of high-grade neutropenia in genetically-determined European pancreatic cancer patients treated with gemcitabine.
The analysis of common variants in CDA and DCTD (gemcitabine metabolism), and SLC29A1, SLC28A1, SLC29A2 (gemcitabine transport) and their association with high-grade neutropenia points towards CDA rs2072671 as a marker of clinical validity. CDA rs2072671 (A>C) is the most extensively studied variant for gemcitabine pharmacogenetics, and this study proposes a new hypothesis of its mechanism and clinical relevance to high-grade, gemcitabine-induced neutropenia, as described below.
CDA codes for cytidine deaminase, the enzyme that inactivates gemcitabine through deamination to its inactive metabolite 2’,2’-difluorodeoxyuridine, as previously described [27]. In our study, patients with the rs2072671 AA genotype have a higher risk for early high-grade neutropenia than patients with the AC or CC genotypes (Figure 3). In the literature, the associations of the rs2072671 variant allele with gemcitabine-induced neutropenia reported so far are inconclusive, with decreased risk [28, 29], increased risk [30], or no identifiable risk [31–33]. To our knowledge, our study is the first to provide evidence that the 79A>C variant in CDA may confer protection from early high-grade neutropenia in pancreatic cancer patients treated with gemcitabine.
CDA rs2072671 (79A>C in exon 1) creates a missense mutation leading to a lysine to glutamine (K27Q) change. Biochemical results on the effect of this variant on enzyme affinity and Vmax for the deamination of gemcitabine are conflicting [34–36]. According to our bioinformatic analysis, the K27Q change is benign. When we tested the association of rs2072671 with gene expression data in our human liver data sets, no significant associations were observed, for either rs2072671 or other variants in high LD with it. Instead, a consistent association was observed between the C allele of rs2072671 (and variants in LD, including rs471760) and increased mRNA expression of CDA in whole blood from three datasets (Table 3). Among all human tissues, whole blood is where CDA has the highest expression [14], (see GTEx diagram, Supplemental Digital Content 8, http://links.lww.com/FPC/B351 for CDA mRNA expression across tissues). We hypothesize that the rs2072671 variant acts through a local effect in either the bone marrow or in circulating neutrophils (or both), by protecting neutrophils from the anti-proliferative effects of gemcitabine. Active, phosphate metabolites of gemcitabine accumulate rapidly in mononuclear cells during the infusion, with a peak of tri-phosphate gemcitabine reached within 30 minutes [37]. The action of the rs2072671 variant is probably indirect, as a proxy of other alleles in moderate-high LD with it (and their haplotypes) which increased CDA expression, as detected from the whole blood studies. A systemic effect of whole blood as a clearing system for gemcitabine is ruled out by the marginal quantitative contribution of plasma cytidine deaminase activity to gemcitabine clearance [37].
The exploratory analysis of common variants in the genome has selected TGFB2 as a novel candidate gene. TGFB2 codes for the a secreted ligand of the TGF-beta (transforming growth factor-beta) superfamily of proteins. Its biology could be related to neutrophil function via its regulatory role on hematopoietic stem cells and subsequent progenitor cells [38]. However the luciferase assays of rs2799083 (and other gene variants also associated with neutropenia in our study) did not show evidence of functionality for this variant (Figure 4), as well as no association with circulating levels of the TGFβ2 protein in a subset of the patients (Supplemental Digital Content 7, http://links.lww.com/FPC/B350). These experimental approaches failed to link a functional mechanism of TGFB2 variants to the clinical effect. Nevertheless, other mechanisms should be explored, including observing the roles of the TGFB2 variants in an earlier progenitor cell type within the hematopoietic pathway.
As a limitation to this study, we acknowledge that the phenotyping was conducted solely on the basis of data provided on clinical research forms. Information provided on these forms may be incomplete, for example, they do not report dose omissions or reductions, the impact of which we have tried to minimize by investigating early events within cycle 1. The associations in CDA, SLC28A1, and TGFB2 do not meet threshold of statistical significance based upon corrected P-values. We also recognize that our selection of covariates is not entirely impartial and consequently may not fully account for the baseline hazard. Despite these limitations, we intend to disseminate these discoveries into the public scientific domain, so that other studies could be built on the basis of our data.
We are reporting the first comprehensive genomic analysis of gemcitabine-induced neutropenia in pancreatic cancer in patients of European origin. This study has generated a novel mechanistic hypothesis on the protective effect of the CDA rs2072671 on early high-grade neutropenia that should be tested in other studies of gemcitabine and other pyrimidine analogs inactivated by cytidine deaminase. All genotype and phenotype data will be made available in dbGaP, as per NIH guidelines of NIH-sponsored studies and upon permission from the Alliance for Clinical Trials in Oncology. Because a validation set was not available, reproduction of the signals presented in this study will be key to provide evidence of clinical validity of novel markers of gemcitabine neutropenia.
Supplementary Material
Footnotes
ClinicalTrials.gov identifier: NCT00088894
References
- 1.Kindler HL, Niedzwiecki D, Hollis D, Sutherland S, Schrag D, Hurwitz H, et al. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol. 2010;28(22):3617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 3.Innocenti F, Owzar K, Cox NL, Evans P, Kubo M, Zembutsu H, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18(2):577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Therneau TM, Grambsch PM (2000). Modeling Survival Data: Extending the Cox Model. New York: Springer. [Google Scholar]
- 5.R Core Team. R: A Language and Environment for Statistical Computing. 2018. Vienna, Austria: [cited 2018]. Available from: www.R-project.org. [Google Scholar]
- 6.Therneau T A Package for Survival Analysis in S. 2015. v2.38. Available from: https://CRAN.R-project.org/package=survival.
- 7.Gray B cmprsk: Subdistribution Analysis of Competing Risks. 2014. v2.2–7. Available from: https://CRAN.R-project.org/package=cmprsk.
- 8.GenABEL project developers. GenABEL: genome-wide SNP association analysis. 2013. v1.8–0. Available from: https://CRAN.R-project.org/package=GenABEL.
- 9.González JR, Armengol L, Guinó E, Solé X, and Moreno V. SNPassoc: an R package to perform whole genome association studies. 2007. v1.9–2. Available from: https://CRAN.R-project.org/package=SNPassoc. [DOI] [PubMed]
- 10.Zhang W, Zhang X. SCAN database: facilitating integrative analyses of cytosine modification and expression QTL. Database (Oxford) 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etheridge AS, Jima D, Molony C, Wright F, Zhou YH, Innocenti F. Hepatic eQTL meta-analysis leads to identification of novel cis-eQTL in pharmacogenes. (Abstract, #1419). Presented at the 67th Annual Meeting of The American Society of Human Genetics, 2017, Orlando, FL. [Google Scholar]
- 14.Consortium GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7(8):e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen R, Hottenga JJ, Nivard MG, Abdellaoui A, Laport B, de Geus EJ, et al. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum Mol Genet. 2017;26(8):1444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40 (Web Server issue):W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Bonneville R, Li T, Jin VX. Transcription factor-associated combinatorial epigenetic pattern reveals higher transcriptional activity of TCF7L2-regulated intragenic enhancers. BMC Genomics. 2017;18(1):375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Q, Zhang Z, Chang KH, Qu H, Wang H, Qi H, et al. Comprehensive characterization of erythroid-specific enhancers in the genomic regions of human Kruppel-like factors. BMC Genomics. 2013;14:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using Ime4. J Stat Softw. 2015;67(1):1–48. [Google Scholar]
- 22.Nixon AB, Pang H, Starr MD, Friedman PN, Bertagnolli MM, Kindler HL, et al. Prognostic and predictive blood-based biomarkers in patients with advanced pancreatic cancer: results from CALGB80303 (Alliance). Clin Cancer Res. 2013;19(24):6957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terpstra TJ. The asymptotic normality and consistency of Kendall’s test against trend, when ties are present in one ranking. Indagationes Mathematicae. 1952;14:327–33. [Google Scholar]
- 24.Jonckheere AR. A Distribution-Free Kappa-Sample Test against Ordered Alternatives. Biometrika. 1954;41(1–2):133–45. [Google Scholar]
- 25.Hollander M and Wolfe D (1999). Nonparametric Statistical Methods. 2nd ed. New York: John Wiley & Sons, Inc. [Google Scholar]
- 26.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciccolini J, Mercier C, Dahan L, Andre N. Integrating pharmacogenetics into gemcitabine dosing-time for a change? Nat Rev Clin Oncol. 2011;8(7):439–44. [DOI] [PubMed] [Google Scholar]
- 28.Farrell JJ, Bae K, Wong J, Guha C, Dicker AP, Elsaleh H. Cytidine deaminase single-nucleotide polymorphism is predictive of toxicity from gemcitabine in patients with pancreatic cancer: RTOG 9704. Pharmacogenomics J. 2012;12(5):395–403. [DOI] [PubMed] [Google Scholar]
- 29.Tibaldi C, Giovannetti E, Vasile E, Mey V, Laan AC, Nannizzi S, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14(6):1797–803. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Zhou Y, Zhang J, Chen Y, Zhuang R, Liu T, et al. High incidence of severe neutropenia after gemcitabine-based chemotherapy in Chinese cancer patients with CDA 79A>C mutation. Clin Chim Acta. 2012;413(15–16):1284–7. [DOI] [PubMed] [Google Scholar]
- 31.Ciccolini J, Dahan L, Andre N, Evrard A, Duluc M, Blesius A, et al. Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J Clin Oncol. 2010;28(1):160–5. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Wang X, Wang X. The impact of CDA A79C gene polymorphisms on the response and hematologic toxicity in gemcitabine-treated patients: a meta-analysis. Int J Biol Markers. 2014;29(3):e224–32. [DOI] [PubMed] [Google Scholar]
- 33.Joerger M, Burgers JA, Baas P, Doodeman VD, Smits PH, Jansen RS, et al. Gene polymorphisms, pharmacokinetics, and hematological toxicity in advanced non-small-cell lung cancer patients receiving cisplatin/gemcitabine. Cancer Chemother Pharmacol. 2012;69(1):25–33. [DOI] [PubMed] [Google Scholar]
- 34.Carpi FM, Vincenzetti S, Ubaldi J, Pucciarelli S, Polzonetti V, Micozzi D, et al. CDA gene polymorphisms and enzyme activity: genotype-phenotype relationship in an Italian-Caucasian population. Pharmacogenomics. 2013;14(7):769–81. [DOI] [PubMed] [Google Scholar]
- 35.Baker JA, Wickremsinhe ER, Li CH, Oluyedun OA, Dantzig AH, Hall SD, et al. Pharmacogenomics of gemcitabine metabolism: functional analysis of genetic variants in cytidine deaminase and deoxycytidine kinase. Drug Metab Dispos. 2013;41(3):541–5. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert JA, Salavaggione OE, Ji Y, Pelleymounter LL, Eckloff BW, Wieben ED, et al. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006;12(6):1794–803. [DOI] [PubMed] [Google Scholar]
- 37.Abbruzzese JL, Grunewald R, Weeks EA, Gravel D, Adams T, Nowak B, et al. A phase I clinical, plasma, and cellular pharmacology study of gemcitabine. J Clin Oncol. 1991;9(3):491–8. [DOI] [PubMed] [Google Scholar]
- 38.Fortunel NO, Hatzfeld A, Hatzfeld JA. Transforming growth factor-beta: pleiotropic role in the regulation of hematopoiesis. Blood. 2000;96(6):2022–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.