Abstract
Genetic variants have been associated with a myriad of molecular phenotypes that provide new insight into the range of mechanisms underlying genetic traits and diseases. Identifying any particular genetic variant’s cascade of effects, from molecule to individual, requires assaying multiple layers of molecular complexity. We introduce the Enhancing GTEx (eGTEx) project that extends the GTEx project to combine gene expression with additional intermediate molecular measurements on the same tissues to provide a resource for studying how genetic differences cascade through molecular phenotypes to impact human health.
Introduction
Identifying the molecular and cellular basis of human genetic disease provides new opportunities for disease prevention and treatment. Genome-wide association studies (GWAS) have already resulted in thousands of genetic associations, localizing regions of the genome that confer disease risk. However, within disease-associated regions, the causal variants and the mechanism of action often remains poorly understood. To address this challenge, the Genotype-Tissue Expression (GTEx) project has generated a systematic, multi-tissue reference for identifying genetic variants associated with changes gene expression (or, expression quantitative trait loci, eQTLs). This resource supports research into potential mechanisms of action for disease-associated variants1,2. Beyond gene expression, a rapidly increasing array of molecular and sequencing-based assays is identifying genetic variants associated with a diversity of intermediate molecular phenotypes. Integration of multiple layers of molecular measurements will clarify the causes and consequences of changes in gene expression as well as identify new mechanisms underlying human disease.
Among the multiple molecular phenotypes recently used in QTL-based analyses are measurements of histone modification, chromatin accessibility, allele-specific expression, alternative splicing, DNA methylation, and protein expression. QTL-based analyses of histone modifications and chromatin accessibility provide insight into variants that influence transcription factor binding and nucleosome positioning3,4. Allele-specific expression QTLs (ase-QTLs) have used allelic ratios as a quantitative phenotype to increase the power of eQTL analyses5–7, and can aid the localization of causal variants8. Alternative splicing QTLs (sQTLs) have been a major focus of multiple eQTL studies using RNA-seq and have been implicated as important contributors to human disease9. Methylation QTLs (meQTLs) have uncovered complex relationships between the genetic, DNA methylation, and expression variation10–12. Ongoing and rapid advances in high-throughput protein quantification have enabled the detection of protein QTLs (pQTLs), which expose variants influencing both transcriptional and post-transcriptional mechanisms13,14.
Beyond studies of QTL types in isolation, integrative or multi-omic analyses offer to elucidate the cascade of molecular effects of disease variants. For example, intersection of DNase I sensitivity quantitative trait loci (dsQTLs) and eQTLs established that over half of eQTLs are also associated with changes in chromatin accessibility3. Intersection of meQTLs and eQTLs identified variants with complex causal relationships depending on CpG and genic contexts15,16. Intersection of eQTLs and pQTLs exposed buffered effects, protein-specific effects, and overlap with disease-associated variants13,14,17,18. Recent integration of eight cellular phenotypes across the regulatory cascade from transcription factor binding to protein expression demonstrated essential contributions of splicing to disease-associated variation9. Such increased accessibility to multi-omics data offers new opportunities to develop and test holistic or ‘systems genomics’ approaches. These approaches offer to provide new opportunities for predictive modeling and enrich our understanding of the multitude of effects for disease-associated variants and their interplay across diverse -omics layers19. .
A major challenge to integration of multi-omics data in the study of human disease is that many multi-omics analyses have been conducted in cell lines instead of primary tissues. While GTEx has demonstrated the value of multi-tissue data to identify tissue-specific mechanisms of disease-associated variants, there remains a need to obtain multi-omics reference data to study the effects of genetic variation across multiple tissues and multiple layers of molecular complexity. In addition to complementing studies of complex genetic diseases, expanding multi-tissue molecular data from ‘normal’ individuals can enhance cancer studies20,21 (which currently comprise 28% of all requests for GTEx data utilization), by distinguishing cancer-specific alterations and elucidating the tissue-specificity of certain cancers and their mutations22,23.
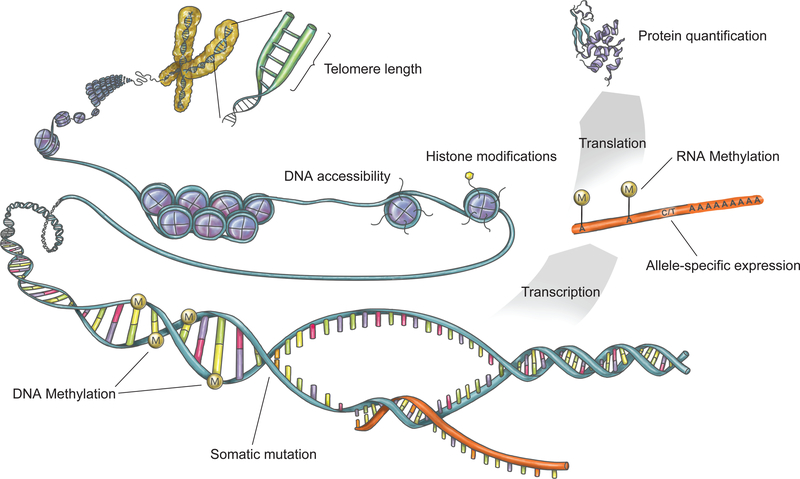
Here, we introduce the NIH Common Fund’s Enhancing GTEx (eGTEx) project, which seeks to complement the gene expression phenotypes determined in the GTEx project with intermediate phenotypes across the same tissues and individuals (Fig. 1). These additional data types will provide a more complete reference of how genetic differences cascade through molecular and cellular phenotypes to impact organismal phenotypes. To achieve this goal, eGTEx is applying diverse molecular assays to the GTEx sample collection, including DNase I hypersensitivity, ChIP-sequencing, DNA and RNA methylation, allele-specific expression, protein expression, somatic mutation, and telomere length assays. Together, the eGTEx reference aims to enable high-resolution identification of the mechanistic impacts of genetic variants and their role in human diseases, and will serve as an enabling resource that will facilitate novel integrative and holistic computational methods development and biological insights.
Figure 1. Quantifying layers of molecular and cellular phenotypes.
The eGTEx Project plans to study telomere length, DNA accessibility, histone modifications, DNA and RNA methylation, somatic mutation, allele-specific expression, and protein quantification across individuals and tissues.
The eGTEX project: Study design and assays
The goal of the GTEx project is to establish a national multi-tissue cohort for molecular phenotypes. The current release (dbGaP accession phs000424.v7.p2) of GTEx provides 11688 transcriptomes from 714 individuals and 53 tissues (median of 17 tissues per individual, 173 samples per tissue). The next release, v8, is expected to include 17,500 transcriptomes from ~850 individuals, and final data production for the project is targeted for late in 2017. In addition to molecular data, GTEx includes pathology reports, histology images and reports, and donor characteristics, including ethnicity, age and sex. Within GTEx, tissues are obtained from deceased donors with next-of-kin consent to permit the collection and banking of anonymized samples for scientific research24. Two existing strengths of the GTEx project are the large number of tissues collected from each donor, facilitating characterization of gene expression across a wide variety of tissues, and the relatively large size of the donor population, allowing one to evaluate the contribution of individual genetic variation. The first steps of assaying genetic variation and its impact on gene expression is the focus of several accompanying consortium papers25,26. However, fully understanding how a genetic variant regulates gene expression, such as through changes in DNA methylation or the binding affinity of a transcription factor, and subsequently connecting the downstream effects of differential gene expression through to protein abundance requires additional molecular assays.
The goal of the eGTEx initiative is to enhance our understanding of gene regulation by performing additional molecular analyses on the same tissues that underwent gene expression analysis. Because of the large size of the GTEx tissue collection (over 25,000 samples), the variable quality across collected samples, and the relatively small aliquot remaining for each sample, the eGTEx initiative will analyze a subset of the entire collection. The study design for eGTEx activities was allocated across 2 “dimensions” of analysis: Phase I, involving a relatively small number of donors (~15) analyzed for a large number of different tissues (>20); and Phase II, involving a relatively small number of tissues (4–6) analyzed in a larger number of donors (150–200). eGTEx has planned to use the same tissues from the same individuals for as many assays as possible. However, because available aliquots are limited, some assays require frozen tissue as input, and the throughput differs by assay, the extent of overlap and the number of phenotypes that will be generated from each individual sample will vary. The molecular phenotypes being studied are shown in Table 1 and described in the following sections.
Table 1. eGTEx study design.
Molecular assays, targeted tissues and sample number for eGTEx.
| Molecular phenotypes | Primary assay(s) | Targeted tissues (Phase II) | Targeted sample number |
|---|---|---|---|
| DNA accessibility | DNase I hypersensitivity | Brain regions, Heart, Lung Muscle, Esophagus, Breast, Prostate, Skin | ~1,135 |
| Histone modifications | Chromatin immunoprecipitation sequencing (ChIP-seq) | Brain regions, Heart, Lung, Muscle | ~600 |
| DNA methylation | Whole genome bisulfite sequencing (WGBS) and capture bisulfite sequencing | Brain regions, Heart, Lung, Muscle, Thyroid | ~2,000 |
| Allele-specific expression | Microfluidic multiplex PCR followed by deep sequencing (mmPCR-seq) | All tissues | ~2,000 |
| Post-transcriptional RNA modifications | m6a methylation capture sequencing | Brain regions, Heart, Lung, Muscle | ~300 |
| Proteomic variation | Mass-spectrometry, targeted arrays for transcription factors and cell signaling proteins | Brain, Heart, Lung, Muscle, Thyroid, Colon, Liver, Prostate, Pancreas, Ovary, Testis, Breast | ~1,000 (MS) ~2,500 (array) |
| Somatic variation | Deep Exome Sequencing, RNA-seq, SNP arrays, probe-based telomere length assay | ~20–25 tissues | ~800 |
| Telomere length | Luminex-based assay for telomere repeat abundance | ~20 tissues | ~5,000 |
DNA accessibility
Systematic understanding of the impact of genetic variation on gene expression requires both comprehensive delineation of regulatory DNA, and an understanding of the degree to which individual regulatory regions vary at the population level. DNA is tightly packaged into chromatin inside our cells, with 147-nucleotide segments of DNA wrapped around each histone octomer (themselves separated by ~50 nucleotide linkers). Displacement of nucleosomes through the binding of transcriptional regulators results in accessible regions of ‘open chromatin’, which can be mapped using endonucleases such as DNaseI27,28. Past work has shown that disease and trait associations are highly concentrated in accessible elements29 and that allelic variation in DNA accessibility can be precisely map the effects of sequence variation on transcription factor activity3,30,31. In eGTEx, we will examine DNA accessibility using both the DNaseI hypersensitivity assay (DHS) and the higher-resolution DNaseI footprinting assay to map transcription factor occupancy within regulatory DNA at nucleotide-level resolution. This assay is highly unbiased and captures variation in diverse regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions.
Histone modifications
Each histone protein in the chromatin fiber has a long amino-acid tail that can be post-transcriptionally modified, thus serving both structural and informational roles32. An ever-growing multitude of histone modifications has been described, whose combinations mark diverse chromatin functions, including active enhancers, active promoters, poised promoters, repressed regions, heterochromatin, or transcribed functions33. Among diverse chromatin states that have been broadly surveyed, enhancer regions marked by histone H3 lysine 27 acetylation (H3K27ac) were shown to be the most variable across tissues and cell types33,34, the most variable across individuals35, and the most highly enriched for disease-associated genetic variants34. Thus, to characterize the protein-DNA interactions through which gene regulation is mediated, eGTEx will perform chromatin immunoprecipitation sequencing (ChIP-seq) to profile the enhancer- and promoter-associated histone modification H3K27ac.
Because DNA accessibility and ChIP-seq assays require frozen material, they cannot be performed on as wide a spectrum of GTEx tissues as most other assays, and instead will focus on the brain and the tissues that were collected frozen on a subset of GTEx donors. Availability of DNaseI and H3K27ac data will aid in fine-mapping causal variants and improving localization and interpretation of tissue-specific and shared regulatory variants.
DNA methylation
DNA methylation of cytosine residues throughout the human genome is an important element of gene regulation and a key component of eGTEx. DNA methylation influences the binding of regulatory elements (such as transcription factors) to DNA36 and is involved in imprinting37,38, X-chromosome inactivation39, and gene silencing40. eGTEx will utilize two complementary methods (whole genome bisulfite sequencing (WGBS) and capture bisulfite sequencing) to characterize the DNA methylation landscape of GTEx tissues, with a particular focus on distinct brain regions implicated in mental health. In addition, as disease-driven changes in cell type composition (specifically neuron loss in many mental health disorders) have become increasingly appreciated, eGTEx will aim to account for this source of variation. To address this issue, eGTEx will perform Fluorescence Activated Nuclei Sorting (FANS) to specifically isolate neuronal nuclei from GTEx brain tissues for WGBS analysis. These analyses will help identify variable methylated regions (VMRs), methylation quantitative trait loci (meQTLs) and regions of allele-specific methylation (ASM).
Allele-specific expression
Within individuals, allele-specific expression (ASE) can validate cis-eQTLs or identify and characterize rare and private cis-regulatory effects41. However, the number of reads mapping to the coding heterozygous sites within a gene of interest limits the power to detect ASE. When using RNA-seq data, this is directly related to the gene’s expression level, which varies from tissue to tissue. eGTEx will apply microfluidic multiplex PCR followed by deep sequencing (mmPCR-seq) to bypass this problem and provide high depth ASE measurements42. This high-throughput, targeted approach decouples the power to detect ASE from the gene’s expression level. Therefore, mmPCR-seq can provide ASE data from a wider range of tissues than RNA-seq. The assay works effectively with low quality or low quantity RNA, which makes it perfectly suited to process the limited quantities of RNA available from GTEx samples. Previously, mmPCR-seq has been applied to study the impact of imprinting and loss-of-function variants across multiple tissues43–45. As part of eGTEx, mmPCR-seq data will be generated for a few hundred genes across all available tissue samples from ~80 individuals. Generated data will validate and complement eQTLs identified by the GTEx project through joint analysis of allele-specific and total expression data5–7. In addition, these data will aid assessment of tissue-specificity and detection of changes in the magnitude of effects across tissues46.
Post-transcriptional RNA modifications
m6A methylation recently emerged as an important post-transcriptional modification of RNA, affecting more than 7,000 protein-coding and non-coding genes, and influencing protein translation, transcriptional gene regulation, stability, alternative splicing, microRNA targeting, circadian rhythms, and overall gene function47,48. m6A methylation is present in a wide variety of tissues, varies in abundance across tissues and across development. However, much remains unknown, including the variation of m6A methylation across individuals in different tissues, the role that genetic variants play in guiding methylation changes, and the role of m6A methylation QTLs in human disease association. Thus, a systematic survey of the inter-individual and inter-tissue variation in m6A methylation within the GTEx cohort can have important implications for the study of human disease, by revealing tissue-specific m6A methylation patterns, inter-individual differences in m6A methylation, and individual genetic variants that act as m6A-QTLs, all in the context of a deeply profiled cohort that benefits from genomic, transcriptomic, epigenomic, and proteomic measurements. To address this opportunity, eGTEx will carry out m6A-seq experiments along two dimensions, the first exploring tissue diversity for a small set of individuals, and the second exploring inter-individual variation for a small set of tissues. Along the tissue dimension, we will profile 20 tissues in 8 individuals, across a total of ~100 samples as not all individuals have a sufficient sample quantity or quality. Along the individual dimension, we will profile 4 tissues across 100 individuals, for a total of ~300 samples, once more based on sample quality and availability in sufficient quantity. We will use a combination of two m6A profiling technologies: ‘location analysis’, by MeRIP-seq, which uses an additional RNA fractionation step prior to m6A profiling, thus enabling us to pinpoint the locations in the transcript where m6A methylation occurs, and ‘level analysis’, by m6A-LAIC-seq, which does not include a fractionation step and provides a more quantitative readout of overall m6A methylation levels for each transcript. We will seek to profile these data matrices sufficiently densely to enable imputation across technology platforms, across tissues, and across individuals in the context of the larger GTEx and eGTEx data matrices.
Protein abundance
Proteins provide a critical link that allows us to fill the gap between RNA expression and phenotype. On one hand, protein levels can be considered as biomarkers for phenotypes; at the same time, they are heritable molecular phenotypes whose genetic basis can be linked to genotype or RNA expression. Preliminary studies have indicated that there is an imperfect correspondence between eQTL and pQTL: in particular, not all pQTLs coincide with eQTLs13,14,17,18. Measurement of protein expression in the GTEx cohort will allow further validation of genome-transcriptome relationships while simultaneously characterizing novel genome-proteome relationships and their relationship to transcriptome biology. eGTEx will characterize protein expression using two complementary strategies: isobaric tandem mass tag (TMT)-based quantitative mass spectrometry (MS) 13 and a custom panel set of antibodies targeting ~400 transcription factors and cell signaling proteins, using the high-throughput, robust, microwestern technology49. Preliminary studies show that using a customized sample preparation protocol that maximizes the protein yield of PAXgene preserved tissue samples and running each tissue sample in duplicate, we can additionally identify ~7,500 proteins per sample using MS. The microwestern assay has been optimized to use small input quantities and will allow quantification across the full spectrum of abundance levels (albeit on a subset of all proteins). Importantly, the microwestern assay allows for quantification of low abundance proteins that are not typically captured using MS, thus rendering the two approaches highly complementary. Generated data will support detection of pQTLs from approximately 200 individuals per tissue, and will enhance studies of tissue-specificity, network relationships, and multi-omics integrative analyses.
Somatic mutation
It is generally assumed that the trillions of cells in a human body share identical DNA sequences, but in reality we are a mosaic of genomes. In addition to many known epigenetic differences, we are made up of subpopulations of cells with genetic differences, such as point mutations, structural changes, and differences in telomere length. The extent of this mosaicism is largely unknown, but recent studies suggest that the extent of somatic variability in humans is considerable and contributes to disease phenotypes50. However, there have been few systematic and comprehensive studies of human somatic variability, particularly outside of readily obtainable tissues like blood. This gap in knowledge is a significant impediment to many ongoing and future studies of human phenotypic variation and disease susceptibility, such as the interpretation of somatic variability in cancer genome sequencing projects. The eGTEx project will systematically study somatic variability by performing several new assays in addition to leveraging the existing RNA-sequencing data. Somatic point mutations will be evaluated by deep exome sequencing using NimbleGen’s SeqCap EZ Human Exome Library v2.0 and sequencing to an average depth of 150x on an Illumina HiSeq 2000. Somatic structural variability will be analyzed using the deep exome sequencing data and SNP arrays for copy number variation (CNV) and copy-neutral loss of heterozygosity (LOH) events.
Telomere length
Telomere length (TL) plays a central role in maintaining cellular proliferative potential and genome stability, and telomere shortening over the life course may be a critical mechanism underlying many age-related health conditions, including cardiovascular disease51. In contrast, longer telomeres may increase risk for some types of cancer52. Most epidemiological studies of the association between TL and disease risk are difficult to interpret, in part because it is not clear how well TL in peripheral blood cells reflects TL disease-relevant tissues. We are addressing this gap in eGTEx by measuring average TL (i.e., telomere repeat abundance) across many tissue types using a high-throughput, Luminex-based method53–55. We will characterize the relationships among TL measures taken from different tissues and determine if tissue-specific TL is associated with the frequency of somatic events in the same tissue (i.e., copy number variation (CNV) and loss of heterozygosity (LOH) events, detected using exome sequencing and SNP array data). In addition, we will determine whether SNPs known to affect leukocyte TL (based on prior GWAS) also influence TL in other tissues. Knowledge of tissue-specific effects of SNPs on TL will enable Mendelian randomization studies to estimate the effects of TL on disease in a tissue-specific fashion. The results from this eGTEx project will provide a foundation for interpreting epidemiological findings and guide the design of future studies of the effect of TL on human health.
Integrative multi-omics analyses
Integrative analyses that combine across both GTEx and eGTEx data will complement the collection of molecular phenotype data (Box 1). These efforts will aim to (1) determine the extent to which different molecular phenotypes vary across tissues, and which factors mediate the levels of each phenotype. For example, it may be found that the variation of mRNA abundance of a particular gene largely occurs between individuals, while that of the protein abundance occurs between tissues, suggesting tissue-specific regulatory steps that occur at post-transcriptional levels. Integrative analyses will also aim to (2) establish the similarity of covariation and coexpression networks across tissues, and (3) identify which loss-of-function mutations are expressed at the RNA and protein level and to what degree this expression varies across tissues. Recent evidence suggests that a subset of loss-of-function effects are compensated at the protein level, highlighting the utility of multi-omics data in personal genome interpretation56.
Box 1: Examples of integrative analyses across tissues using eGTEx data.
| ● Determine the relative variability of molecular phenotypes. ● Compare covariation networks across molecular phenotypes. ● Determine if genes/proteins with loss of function mutations are expressed. E.g. examine regulatory features, mRNA levels, proteins levels. ● Map QTLs for each molecular phenotype to determine where most functional genetic variation resides. ● Construct integrative regulatory networks using ‘systems genomics’ approaches. ● Connect regions of allele-specific chromatin accessibility, allele-specific methylation and allele-specific gene expression. ● Integrated analysis of patterns of X-inactivation. ● Quantify tissue-specific levels of somatic mutations and their relationship to heterogeneity in gene expression levels ● Associate levels of methylation and expression at telomere maintenance genes (e.g., TERC, TERT, DKC1) with telomere length measurements. ● Multi-omics enrichments of trait-associated variation. ● Support holistic predictive modeling across molecular phenotypes. |
Studies using eGTEx data will integrate and enhance diverse external projects and resources. As an example, characterization of tissue-specific methylation and expression patterns will benefit from the expanding catalog of chromatin modifications (H3K4me1, H3K27ac, H3K9me3, etc.), transcription factors, DNaseI hypersensitivity sites, and other regulatory DNA binding factors from ENCODE and NIH Epigenomics Roadmap resources. The Roadmap Consortium in particular has an extensive dataset including many of the same brain regions being profiled by eGTEx and will allow identification of brain-specific regulatory regions. To better enable these types of integrated studies eGTEx and ENCODE have developed the ENTEX collaborative project to focus on deeply profiling a small subset of directly overlapping tissues using shared technologies.
Integrative eGTEx analyses will continue to focus on methodologies that enhance GWAS interpretation. Such methods will take advantage of recent efforts to impute molecular phenotypes57, test colocalization of trait and molecular association signals58,59 and jointly model multi-omics data60–62. The specific focus on brain tissues in GTEx and eGTEx is particularly useful for contextualizing GWAS of neurological and neuropsychiatric traits. Combinatorial analyses of GWAS associations and diverse molecular factors can reveal important disease-associated SNPs that may have fallen below standard association thresholds or help identify the likely driver SNP from a group of associated and tightly-linked variants. Increasingly, as whole-genome-sequenced cohorts become available, integration with eGTEx is expected to enhance new methods for predicting and identifying the tissue context of diverse classes of genetic variation.
Data release and community impact
GTEx and the eGTEx projects are community resources committed to rapid and complete data release. Thus far, wet lab analysis of germ line genetic variation and RNA-seq-based mRNA expression measurements of the main GTEx project have been generated at the Broad Institute, serving as the Laboratory, Data Analysis, and Coordinating Center (LDACC) for the project. The entire Consortium has contributed to all aspects of the analysis pipeline, but the fact that the vast majority of the data has been generated at a single lab has facilitated its integration and coordinated release. eGTEx experiments, on the other hand, will be generating a heterogenous collection of data in more than seven laboratories, making data integration more challenging. Some of the data generated (for example, protein expression), will have little to no privacy concerns and can be made available without access restrictions. Other molecular phenotypes, such as DNA methylation, will be similar to RNA-seq expression, generating both raw sequence data with individual sequence variation information that will be deposited into the controlled access database dbGaP, while much of the processed data and results can be made available without access controls through the GTEx portal (see URLs). The LDACC, who are continuing to release the primary GTEx data, will also serve as the coordination center for the release of the controlled access data for the eGTEx assays. This will ensure that data ID’s and metadata are harmonized across the entire project, and will enable eGTEx data to be included with the major GTEx releases. Additionally, to the extent possible, open-access eGTEx data will be integrated and made available through the GTEx portal alongside the gene expression and eQTL results.
Given the heterogenous nature of the data, the differing capacity of the participating labs, and differences in how samples will need to be batched across the various assays for quality control purposes, we expect components of eGTEx data to be released intermittently over the next several years with this publication serving as a guide to the overall eGTEx effort. The Phase I data, representing analysis of a wide range of tissues from a relatively small number of donors, will be generated first by most groups and thus should be released before Phase II data, representing a smaller number of tissues analyzed over a larger number of individuals. We plan initial data deposition in late 2017/early 2018 with no publication embargo after data release.
Impact of eGTEx on research community and future directions
The tissues from GTEx donors are collected without focus on a disease state and are representative of a US-based human population24. With detailed molecular data being collected across diverse tissues by eGTEx, the resource provides a snapshot of ‘normal’ (or non-diseased state) genetic and genomic variation among individuals and across tissues. As such, the resource serves as a reference for disease-focused research, where investigators can compare eGTEx data with genomic data characterized from disease cohorts. Much in the way that eQTL studies are being used to generate hypotheses regarding causal genes and mechanisms underlying GWAS trait associations, these novel eGTEx genomic data are expected to be widely used to elucidate additional mechanisms contributing to diverse human disease.
The diversity of genetic and genomic data types being surveyed by eGTEx will facilitate statistical methods development in the area of data integration. Although analysis methods for specific pairs of data types (e.g., genetic and transcriptomic, eQTLs) have become relatively standardized by the community, methods development for holistic analysis of diverse data types remains an active research area. The eGTEx population-based genomics data characterizing multiple modalities of genome function from genetic to epigenetic to transcriptomic and proteomic variation will provide a rich primary tissue-based resource for development of ‘best practices’ for integrative data analysis.
As new computational and experimental approaches continue to elucidate the function of the genome, eGTEx aims to provide a rich data source that enables the integration of multi-omics data in the interpretation of health and disease. These efforts will help pave the route towards an increased understanding of genome function, the elucidation of novel molecular therapeutics and the integration of high throughput molecular diagnostics in individualized patient care.
Acknowledgments
The Genotype-Tissue Expression (GTEx) project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (see URLs). Additional funds were provided by the National Cancer Institute (NCI), National Human Genome Research Institute (NHGRI), National Heart, Lung, and Blood Institute (NHLBI), National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH), and National Institute of Neurological Disorders and Stroke (NINDS). Donors were enrolled at Biospecimen Source Sites funded by Leidos Biomedical, Inc. (Leidos) subcontracts to the National Disease Research Interchange (10XS170) and Roswell Park Cancer Institute (10XS171). The LDACC was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through a Leidos subcontract to the Van Andel Research Institute (10ST1035). Additional data repository and project management were provided by Leidos (HHSN261200800001E). The Brain Bank was supported by a supplement to University of Miami grant DA006227. EKT is supported by a Hewlett-Packard Stanford Graduate Fellowship and a doctoral scholarship from the Natural Science and Engineering Council of Canada. NIH grant U01MH104393 supported APF, KDH, LFR, and PH. NIH grant U01HG007598 supported BES. NIH grant U01HG007599 supported JAS. NIH grant U01HG007593 supported JBL and SBM. NIH grant U01HG007591 supported JA. NIH grant U01HG007610 supported MK. NIH grant U01HG007601 supported BLP. NIH grant U01HL131042 supported MS and HT.
†
Lists of participants and their affiliations appear at the end of the paper
Competing Financial Interests Statement
MS is a cofounder of Personalis and Q bio and on the scientific advisory board of Personalis, Epinomics and Genapsys.
References
- 1.Nicolae DL et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genetics 6, e1000888, doi: 10.1371/journal.pgen.1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660, doi: 10.1126/science.1262110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degner JF et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394, doi: 10.1038/nature10808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McVicker G et al. Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–749, doi: 10.1126/science.1242429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W A statistical framework for eQTL mapping using RNA-seq data. Biometrics 68, 1–11, doi: 10.1111/j.1541-0420.2011.01654.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Geijn B, McVicker G, Gilad Y & Pritchard JK WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nature Methods 12, 1061–1063, doi: 10.1038/nmeth.3582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumasaka N, Knights AJ & Gaffney DJ Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nature Genetics 48, 206–213, doi: 10.1038/ng.3467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lappalainen T et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511, doi: 10.1038/nature12531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YI et al. RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604, doi: 10.1126/science.aad9417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs JR et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genetics 6, e1000952, doi: 10.1371/journal.pgen.1000952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell JT et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology 12, R10, doi: 10.1186/gb-2011-12-1-r10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez-Arcelus M et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2, e00523, doi: 10.7554/eLife.00523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82, doi: 10.1038/nature12223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hause RJ et al. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. American Journal of Human Genetics 95, 194–208, doi: 10.1016/j.ajhg.2014.07.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banovich NE et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genetics 10, e1004663, doi: 10.1371/journal.pgen.1004663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez-Arcelus M et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genetics 11, e1004958, doi: 10.1371/journal.pgen.1004958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle A et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science 347, 664–667, doi: 10.1126/science.1260793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cenik C et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Research 25, 1610–1621, doi: 10.1101/gr.193342.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie MD, Holzinger ER, Li R, Pendergrass SA & Kim D Methods of integrating data to uncover genotype-phenotype interactions. Nature Reviews Genetics 16, 85–97, doi: 10.1038/nrg3868 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Vucic EA et al. Translating cancer ‘omics’ to improved outcomes. Genome Research 22, 188–195, doi: 10.1101/gr.124354.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61, doi: 10.1016/j.cell.2014.12.033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Banet J et al. OASIS: web-based platform for exploring cancer multi-omics data. Nature Methods 13, 9–10, doi: 10.1038/nmeth.3692 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Kosti I, Jain N, Aran D, Butte AJ & Sirota M Cross-tissue Analysis of Gene and Protein Expression in Normal and Cancer Tissues. Scientific Reports 6, 24799, doi: 10.1038/srep24799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carithers LJ et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreservation and biobanking 13, 311–319, doi: 10.1089/bio.2015.0032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. Genetic effects on gene expression across 44 human tissues. Nature in press (2017). [DOI] [PubMed]
- 26.Li X, Kim Y, Tsang EK, Davis J & al., e. The impact of rare variation on gene expression across tissues. Nature in press (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weintraub H & Groudine M Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976). [DOI] [PubMed] [Google Scholar]
- 28.Wu C, Wong YC & Elgin SC The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell 16, 807–814 (1979). [DOI] [PubMed] [Google Scholar]
- 29.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195, doi: 10.1126/science.1222794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurano MT et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nature Genetics 47, 1393–1401, doi: 10.1038/ng.3432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neph S et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90, doi: 10.1038/nature11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bannister AJ & Kouzarides T Regulation of chromatin by histone modifications. Cell Res 21, 381–395, doi: 10.1038/cr.2011.22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst J et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49, doi: 10.1038/nature09906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasowski M et al. Extensive variation in chromatin states across humans. Science 342, 750–752, doi: 10.1126/science.1242510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurano MT et al. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell reports 12, 1184–1195, doi: 10.1016/j.celrep.2015.07.024 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Pervjakova N et al. Imprinted genes and imprinting control regions show predominant intermediate methylation in adult somatic tissues. Epigenomics 8, 789–799, doi: 10.2217/epi.16.8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li E, Beard C & Jaenisch R Role for DNA methylation in genomic imprinting. Nature 366, 362–365, doi: 10.1038/366362a0 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Payer B & Lee JT X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42, 733–772, doi: 10.1146/annurev.genet.42.110807.091711 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Curradi M, Izzo A, Badaracco G & Landsberger N Molecular mechanisms of gene silencing mediated by DNA methylation. Molecular and Cellular Biology 22, 3157–3173 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E & Lappalainen T Tools and best practices for data processing in allelic expression analysis. Genome Biology 16, 195, doi: 10.1186/s13059-015-0762-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R et al. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nature Methods 11, 51–54, doi: 10.1038/nmeth.2736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kukurba KR et al. Allelic expression of deleterious protein-coding variants across human tissues. PLoS Genetics 10, e1004304, doi: 10.1371/journal.pgen.1004304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivas MA et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348, 666–669, doi: 10.1126/science.1261877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baran Y et al. The landscape of genomic imprinting across diverse adult human tissues. Genome Research 25, 927–936, doi: 10.1101/gr.192278.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirinen M et al. Assessing allele-specific expression across multiple tissues from RNA-seq read data. Bioinformatics 31, 2497–2504, doi: 10.1093/bioinformatics/btv074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206, doi: 10.1038/nature11112 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Meyer KD et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646, doi: 10.1016/j.cell.2012.05.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA & Jones RB Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nature Methods 7, 148–155, doi: 10.1038/nmeth.1418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Huallachain M, Karczewski KJ, Weissman SM, Urban AE & Snyder MP Extensive genetic variation in somatic human tissues. Proceedings of the National Academy of Sciences 109, 18018–18023, doi: 10.1073/pnas.1213736109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haycock PC et al. Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ 349, g4227, doi: 10.1136/bmj.g4227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stone RC et al. Telomere Length and the Cancer-Atherosclerosis Trade-Off. PLoS Genetics 12, e1006144, doi: 10.1371/journal.pgen.1006144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kibriya MG, Jasmine F, Roy S, Ahsan H & Pierce B Measurement of telomere length: a new assay using QuantiGene chemistry on a Luminex platform. Cancer epidemiology, biomarkers & prevention 23, 2667–2672, doi: 10.1158/1055-9965.EPI-14-0610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce BL et al. Telomere length measurement by a novel Luminex-based assay: a blinded comparison to Southern blot. Int J Mol Epidemiol Genet 7, 18–23 (2016). [PMC free article] [PubMed] [Google Scholar]
- 55.Kibriya MG, Jasmine F, Roy S, Ahsan H & Pierce BL Novel Luminex Assay for Telomere Repeat Mass Does Not Show Well Position Effects Like qPCR. PloS One 11, e0155548, doi: 10.1371/journal.pone.0155548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jagannathan S & Bradley RK Translational plasticity facilitates the accumulation of nonsense genetic variants in the human population. Genome Research 26, 1639–1650, doi: 10.1101/gr.205070.116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gamazon ER et al. A gene-based association method for mapping traits using reference transcriptome data. Nature Genetics 47, 1091–1098, doi: 10.1038/ng.3367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nica AC et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genetics 6, e1000895, doi: 10.1371/journal.pgen.1000895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hormozdiari F et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. American Journal of Human Genetics 99, 1245–1260, doi: 10.1016/j.ajhg.2016.10.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Civelek M & Lusis AJ Systems genetics approaches to understand complex traits. Nature Reviews Genetics 15, 34–48, doi: 10.1038/nrg3575 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parikshak NN, Gandal MJ & Geschwind DH Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nature Reviews Genetics 16, 441–458, doi: 10.1038/nrg3934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu J et al. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biology 10, e1001301, doi: 10.1371/journal.pbio.1001301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]