Abstract
The famed physicist-turned-biologist, Max Delbrück, once remarked that, for physicists, “the field of bacterial viruses is a fine playground for serious children who ask ambitious questions.” Early discoveries in that playground helped establish molecular genetics, and half a century later, biologists delving into the same field have ushered in the era of precision genome engineering. The focus has of course shifted—from bacterial viruses and their mechanisms of infection to the bacterial hosts and their mechanisms of immunity—but it is the very same evolutionary arms race that continues to awe and inspire researchers worldwide. In this review, we explore the remarkable diversity of CRISPR–Cas adaptive immune systems, describe the molecular components that mediate nucleic acid targeting, and outline the use of these RNA-guided machines for biotechnology applications. CRISPR–Cas research has yielded far more than just Cas9-based genome-editing tools, and the wide-reaching, innovative impacts of this fascinating biological playground are sure to be felt for years to come.
Bacterial viruses, or bacteriophages (phages for short), have been the focus of scientific exploration and inspiration for just over a century, beginning with Twort and d'Herelle's landmark phage discoveries published during World War I.1 In the ensuing years, researchers pursued the development of phages as a new way to treat pathogenic bacterial infections because of their remarkable ability to selectively kill bacterial hosts, but phage therapy quickly took a backseat to the rise of cheap and effective antibiotic treatments.2 Yet phages would eventually transform biology and medicine in a far more momentous way: by serving as incredibly versatile model systems that, together with their bacterial hosts, shed light on some of biology's most fundamental questions. Beyond providing direct evidence that DNA is the genetic material (Hershey and Chase's famous “Waring blender experiment”), phage research revealed the fine structure of genes and the processes of genetic recombination and transduction, helped prove the triplet nature of the genetic code, revealed fundamental mechanisms of gene regulation, and catalyzed the rise of the biotechnology industry through the discovery of phage- and host-encoded DNA-manipulating enzymes, among many other breakthroughs.3,4
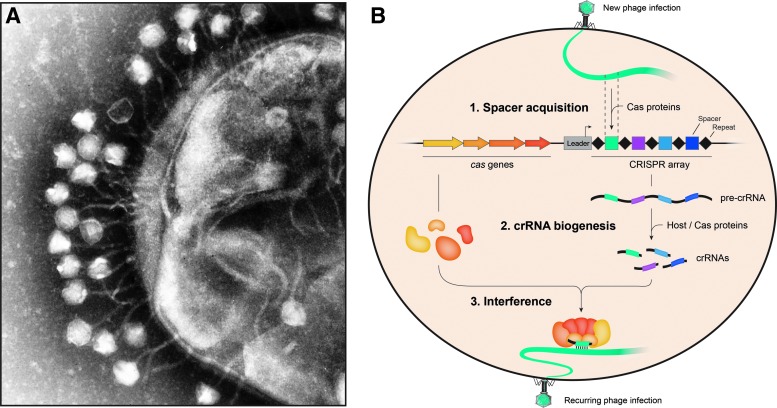
Beyond their utility for laboratory experiments, phages are the most abundant biological entity in the biosphere.5 With an estimated 1031 total phage particles, one can calculate that there are ∼1 trillion phages for every grain of sand on Earth.3 They are also expected to outnumber bacteria on the order of 10-to-1 and can be isolated from every environment in which bacteria exist. In the oceans, phages play major roles in carbon and nitrogen cycling by killing somewhere between 20% and 40% of all bacteria every day,6 highlighting the immense ecological and environmental pressures they exert. On top of that, phages are known to contribute to bacterial pathogenicity and evolution by functioning as efficient gene transfer vectors, alongside plasmids, transposons, and other mobile genetic elements. From this incessant genetic battle (Fig. 1A), an arms race ensued, in which prokaryotes and phages evolved numerous elaborate and orthogonal strategies to protect themselves from viral infection and to combat antiviral mechanisms, respectively. The impressive extent of this genetic pressure is clearly visible in prokaryotic genomes, of which a substantial fraction (up to 10%) is dedicated to their own defense.7
FIG. 1.
(A) Transmission electron micrograph of bacteriophages infecting Escherichia coli. The evolutionary arms race between viruses and their microbial hosts has resulted in the adaptation of numerous defense mechanisms, including CRISPR–Cas systems, as well as counter-attack strategies. ©Graham Beards (https://upload.wikimedia.org/wikipedia/commons/5/52/Phage.jpg). (B) Schematic overview of the three stages of CRISPR–Cas immunity. During spacer acquisition, protospacers are excised from foreign nucleic acids and integrated at the leader end of the growing CRISPR array. crRNA biogenesis involves transcription of the entire array, followed by enzymatic processing of the pre-crRNA into mature crRNAs, often by dedicated Cas proteins. In the interference stage, the crRNA and Cas protein(s) form an effector complex that targets complementary nucleic acids for degradation. Class 1 systems rely on multiple Cas proteins during interference (depicted), whereas Class 2 systems require only a single protein effector.
The resulting range of bacterial and archaeal defense mechanisms (reviewed in Refs.7,8) can be broadly classified into three functionalities: (1) impeding virus adsorption to the host cell, primarily by alteration of host receptors, (2) dormancy induction and/or programmed cell death, which protect the population, and (3) nucleic acid-targeting immune systems. This last category can be further divided into innate immunity—including the well-characterized restriction-modification systems9 and the recently discovered system involving prokaryotic Argonaute proteins10—and the adaptive (acquired) immunity provided by CRISPR*–Cas systems.
It is remarkable that, although most archaea and almost half of sequenced bacterial species contain CRISPR–Cas systems,11,12 detection of these pervasive modules eluded researchers for decades. CRISPR–Cas research is now advancing at an incredible pace, leading to thousands of CRISPR-related articles being published in 2017 alone. Within the past decade, CRISPR–Cas biology has been in the scientific limelight time and time again, and “CRISPR” has all but become a household term due to its development for revolutionary genome engineering applications.13,14 (As a testament to its growing popularity, CRISPR is already at risk of losing its privileged acronym status in the popular media, much like laser and radar.) The focus has of course largely been on Cas9-containing immune systems, but recent studies have highlighted the incredible diversity of CRISPR–Cas systems, both in terms of their distinguishing mechanistic features and their potential for novel biotechnological uses.
The scope and pace of contemporary CRISPR research are astounding, and it is impossible to adequately review the field in a single article. Nevertheless, we attempt to broadly summarize the current understanding of CRISPR–Cas biology while focusing on mechanisms of RNA-guided nucleic acid targeting and the development of CRISPR-based tools. Wherever possible, we include more focused review articles for readers interested in particular topics, and we offer a sincere apology to the many colleagues whose work we could not mention due to space constraints.
General Mechanism of CRISPR–Cas Immunity
CRISPR refers to specialized genomic regions found in bacteria and archaea that consist of alternating repeat-spacer units, often neighboring conserved sets of CRISPR-associated (cas) genes.15 Observations in 2005 that CRISPRs harbor some spacers matching extrachromosomal elements, namely viruses and plasmids, led to the first hypotheses of an immune system function.16–18 Barrangou et al. were the first to provide experimental evidence for CRISPR–Cas-mediated antiviral immunity,19 and subsequent work highlighted the essential role of noncoding CRISPR RNAs (crRNAs).20
Research in the ensuing years has greatly expanded our understanding of the molecular mechanisms of CRISPR–Cas adaptive immunity (reviewed in Refs.21–23), which can be broadly grouped into three distinct stages: adaptation, crRNA biogenesis, and interference (Fig. 1B). During adaptation,24 also known as spacer acquisition, foreign DNA segments called protospacers are processed and integrated into one end of the growing CRISPR array, accompanied by duplication of the terminal repeat. CRISPR arrays are flanked by a leader sequence, which both helps define the site of spacer integration and also serves as a promoter for the transcription of precursor crRNAs that span the length of the entire array. During crRNA biogenesis,25 these pre-crRNAs are enzymatically processed into libraries of short mature crRNAs, each of which contains a single spacer (or guide) sequence flanked by fragments of the repeat sequence. Mature crRNAs are then bound by one or more Cas proteins to form large ribonucleoprotein (RNP) effector complexes, which target complementary nucleic acids for degradation during the interference stage.
Although most well-studied CRISPR–Cas systems obey this general paradigm, as with much else in biology, exceptions are the norm. For example, some systems encode reverse transcriptase fusions that promote acquisition directly from RNA precursors,26 diverse effector complexes can cleave DNA, RNA, or both DNA and RNA targets during interference,27 and some effectors not only cleave foreign nucleic acids identified through complementary base-pairing, but also unleash potent, nonspecific “collateral” cleavage upon target binding.28–31 And beyond mediating prokaryotic antiviral immunity, CRISPR–Cas systems have been co-opted by bacterial hosts to promote pathogenesis through endogenous gene regulation,32,33 by selfish genetic elements to possibly facilitate transposition,34 and by phages themselves to evade host innate immunity.35
CRISPR–Cas Diversity and Classification
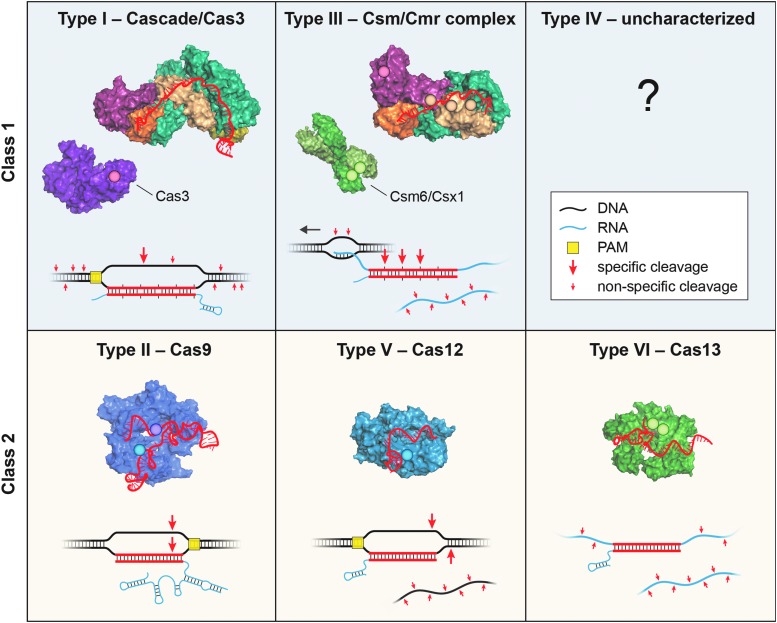
Most steps during adaptation and interference are carried out by dedicated Cas proteins expressed from cas genes that neighbor the CRISPR array and typically number on the order of 4–8. Efforts to define a general mechanism for adaptive immunity were initially hampered by the fact that, although certain cas gene cassettes exhibit highly conserved arrangements, cassettes can differ almost entirely when comparing CRISPR–Cas systems between organisms or even within single organisms. To better understand this diversity, Makarova et al. introduced a polythetic classification scheme in 2011 that combined phylogenetic, structural, and comparative genomic analyses to arrive at three major system types (I, II, and III), each of which exhibits a signature gene, cas3, cas9, and cas10, respectively.36 The contemporary classification defines two broad classes of CRISPR–Cas systems and six major types (I–VI),11,37 which are presently delineated into roughly 30 subtypes in total.38
Class 1 and Class 2 systems are differentiated based on the nature of the protein–RNA effector complex involved in nucleic acid targeting. Class 1 effector complexes39 contain crRNA and multiple protein subunits, typically encoded by three to six cas genes, whereas Class 2 effector complexes40 comprise a single Cas protein together with crRNA, sometimes accompanied by an additional trans-activating crRNA molecule known as tracrRNA. Both classes are further divided into three different types: types I, III, and IV (Class 1) and types II, V, and VI (Class 2).
Because Class 2 systems exhibit simpler cas gene cassettes and a more streamlined effector complex architecture than Class 1 systems, they have been more rapidly adopted for biotechnology applications involving heterologous expression or delivery. Nevertheless, Class 2 systems are far less abundant in nature, comprising roughly 10% of CRISPR–Cas loci in sequenced bacterial genomes and being almost completely absent in archaea.11,41 In contrast, Class 1 systems are widespread, with type I alone making up ∼50% of the loci in both bacteria and archaea.
Based on both the relative prevalence of Class 1 and Class 2 systems as well as phylogenetic analyses of the cas1 gene, which is present in all autonomous CRISPR–Cas systems, the current scenario for the evolution of prokaryotic adaptive immune systems posits that the ancestral CRISPR–Cas system was of the Class 1 type.37 After insertion of a Cas1-encoding transposon (casposon)42 next to a primitive innate immune system, further duplication of ancient RNA recognition motif domains led to ancestral type I and III systems containing multisubunit targeting complexes. Type II, V, and VI systems then evolved independently, largely through the co-option of genes and enzymatic domains from transposons.37 The observation that Class 2 systems appear to derive almost completely from different mobile genetic elements beautifully highlights the extent to which enzymes have been co-opted as a means of both offense and defense within the evolutionary “guns for hire” paradigm.43
It is worth noting that the precise annotation of CRISPR–Cas subtypes continues to evolve as new cas genes are discovered and validated. New discoveries will undoubtedly follow from the availability of novel genomic or metagenomic sequences,41 as well as from new bioinformatics strategies to search for CRISPR-associated or CRISPR-linked genes.37,44,45
Mechanisms of RNA-Guided Nucleic Acid Targeting
In the following sections, we review our current understanding of nucleic acid-targeting mechanisms across different CRISPR–Cas immune systems. Given most readers' familiarity with Cas9, we begin by discussing type II systems, followed by type I systems wherein many hallmark features of RNA-guided targeting were first described. After reviewing the remaining four types, we highlight the various ways in which Cas enzymes have been harnessed for tool development.
Type II – Cas9: Facile double-strand breaks
Type II systems feature the well-known Cas9 endonuclease,46 a protein that formerly went by the monikers Cas5 and Csn1.19,36,47,48 Guided by a dual-RNA substrate comprising crRNA and tracrRNA,49 Cas9 introduces double-strand breaks (DSBs) in DNA targets that contain a flanking protospacer-adjacent motif (PAM)50 and harbor complementarity to the ∼20-nucleotide crRNA guide sequence49,51 (Fig. 2). The PAM requirement ensures that Cas9 avoids cleaving complementary targets within the CRISPR array itself, because these sequences lack flanking PAMs. Interestingly, Cas9-mediated PAM recognition is also critical for the acquisition of new spacer sequences.52,53 After R-loop formation, cleavage of the target strand (base-paired to the crRNA) and nontarget strand (displaced within the R-loop complex) is mediated by HNH and RuvC nuclease domains of Cas9, respectively.49,51
FIG. 2.
A schematic of RNA-guided nucleic acid-targeting mechanism and effector complex structure for each CRISPR–Cas type. In the schematic, crRNAs are shown 5′-to-3′ (left-to-right), and large and small red arrows indicate specific and nonspecific cleavage sites, respectively. For type III, the direction of transcription is indicated with a black arrow. In the structures, nuclease active sites are indicated with opaque circles, target nucleic acids are omitted, and crRNAs are highlighted in red for clarity. PDB accession numbers by type: 4QYZ and 4QQW (I), 4OO8 (II), 3X1L/4W8Y and 5FSH (III), 5B43 (V), and 5XWP (VI).
The apo form of Streptococcus pyogenes Cas9 (SpyCas9) is conformationally flexible, and guide RNA (gRNA) binding drives large-scale conformational rearrangements that lock Cas9 into a bilobed architecture.54–56 Once complexed with gRNA—either a natural crRNA-tracrRNA dual-RNA guide or an engineered single-RNA guide49—the Cas9–gRNA RNP complex interrogates double-stranded DNA for potential target sites matching the guide sequence. Numerous experimental studies point to a model for target search, in which PAM recognition acts as an obligate first step during protospacer recognition and leads directly to local DNA unwinding.57–60 The search is largely three-dimensional,57,61,62 and sampling times at off-target sites are a function of the degree of gRNA-DNA complementarity.63–65 Targeting by Cas9–gRNA is most sensitive to mismatches in the PAM-proximal region of the DNA target site, a phenomenon that results at least, in part, from preordering of the gRNA “seed” sequence.55,66
Numerous in vitro and in vivo experiments have revealed that DNA binding is significantly more promiscuous than cleavage, and recent biophysical studies have highlighted the conformational rearrangements that are required for driving DNA-bound SpyCas9 into a catalytically active state.67–70 Target-strand cleavage by the HNH domain precedes nontarget-strand cleavage by the RuvC domain,71,72 and mutations that modulate either DNA interactions or Cas9 conformational dynamics can significantly enhance the fidelity of RNA-guided DNA cleavage.69,73–76
Two other observations regarding DNA cleavage deserve emphasis. First, although Cas9 is often described as an enzyme that generates blunt DSBs, biochemical experiments with SpyCas9 clearly demonstrate that the RuvC nuclease domain catalyzes additional “trimming” of the nontarget strand in the 3′→5′ direction, upstream of the cleavage site.49,77,78 The production of asymmetric, recessed ends may explain, in part, the observation that DNA repair outcomes of cellular Cas9-mediated cleavage often involved templated insertions.79,80 Second, Cas9 lacks multiple-turnover catalytic activity in biochemical cleavage assays, due to its high affinity for DNA cleavage products57,71,72,81 and may persist at cleaved sites in cells as well; whether and how this impacts both adaptive immunity in bacteria and genome-editing outcomes in eukaryotic cells has not been systematically explored, although kinetic analyses indicate that in vivo repair of DSBs occurs more rapidly than the long Cas9–DSB half-life measured in vitro.82
In addition to DNA, Cas9 can also be naturally or artificially programmed to bind and cleave RNA targets. Whereas SpyCas9 has been specifically manipulated to enable RNA targeting in vitro and in vivo,83–85 other Cas9 orthologs possess the intrinsic capability to target single-stranded RNA molecules,86–88 but the biological import of this activity is unclear. In at least one case, though, Cas9 from Francisella novicida has been shown to directly downregulate an endogenous transcript using tracrRNA and an alternative CRISPR-associated RNA, as a mechanism for promoting pathogenesis during host infection.32
Although type II CRISPR–Cas systems provide robust protection against viral infection, phages can escape interference using multiple evasion strategies. Spontaneous mutations in the PAM or protospacer preclude efficient targeting,89 and DNA modifications such as glucosylation can impair DNA recognition and/or lead to enhanced mutation frequencies that enable escape.90,91 Multiple phage-encoded inhibitors of Cas9 (“anti-CRISPRs,” reviewed in Ref.92) have also been discovered, which arrest nucleic acid targeting at various stages either by preventing DNA binding or by blocking the conformational rearrangements required for DNA cleavage.
Type I – Cascade/Cas3: DNA cut and run
In contrast to single-effector type II systems, type I CRISPR–Cas systems—the most abundant in sequenced genomes—encode multisubunit effector complexes93 (Fig. 2). Many mechanistic discoveries on interference, such as the existence of a seed sequence,94,95 the nature of R-loop formation,96 and inhibition by anti-CRISPRs,97 were first described for type I. An incredible variety of type I systems is known, spanning at least six different subtypes (A–F), all of which utilize a DNA binding complex known as Cascade and harbor the signature protein, Cas3.38
The effector complex in type I systems requires a mature crRNA that is formed upon pre-crRNA processing. A designated ribonuclease (usually Cas6) cleaves a specific stem-loop structure formed by portions of the repeat sequences,98,99 resulting in a spacer sequence flanked by two repeat fragments. The 5′ end of the crRNA lies at the “base” of Cascade, where it binds Cas5 and neighbors the large subunit (Cas8) that is responsible for PAM recognition.100,101 The crRNA spacer sequence is spanned by multiple copies of Cas7 that define the helical “backbone” of the overall architecture, whereas small subunit proteins (Cas11, or a Cas8-fusion thereof) constitute the “belly” and help stabilize the crRNA–DNA heteroduplex upon target binding. In some systems, Cas6 remains bound to the 3′ end of the crRNA after processing and caps the “top” of the RNP complex.20 Beyond the general overview presented here, multiple groups have published high-resolution X-ray crystal and cryoelectron microscopy structures of Cascade in different functional states,101–107 providing a wealth of information about complex assembly, gRNA presentation, target DNA binding, Cas3 nuclease recruitment, and subtype-specific variations.
DNA targeting depends on PAM recognition to avoid self-targeting of the host CRISPR array, although PAM specificity is far more relaxed in type I systems than type II.108 As with Cas9, PAM binding leads to local dsDNA melting and directional unwinding of the DNA upon formation of the growing crRNA–DNA heteroduplex,58,107,109,110 with complementarity being most critical within the seed sequence. Interestingly, because type I crRNAs contain repeat fragments flanking both ends of the spacer, Cascade is sterically prevented from forming a continuous RNA–DNA double helix upon target binding. This results in a characteristic protein-induced kink at every sixth base-pair.102 The impact of this organization on target specificity throughout the target and PAM sequence has been revealed in a number of high-throughput experiments.108,111–113
Unlike most other CRISPR–Cas types, in which the effector complex possesses intrinsic nuclease activity, Cascade generally functions as an RNA-guided DNA binding complex only; PAM and protospacer recognition lead to downstream, Cas8-mediated recruitment of Cas3.104,114 Cas3 is essential for type I immunity and harbors both helicase activity and a nucleolytic HD domain.20,115,116 Recruitment to the target leads to initial nicking of the nontarget strand, followed by extensive DNA degradation outside of the target region through ATP-dependent DNA unwinding and concomitant ssDNA nuclease activity117–119 (Fig. 2). Recent single-molecule experiments have demonstrated that, in contrast to earlier models, Cascade remains tightly associated with Cas3 during DNA unwinding and translocation, leading to a looped intermediate.109,120,121 In addition, type I interference creates a positive feedback loop that facilitates the acquisition of new spacers against the same invader, a process known as priming, by recruiting the adaptation machinery and providing degradation products that serve as spacer precursors.24,120,122
Type I systems may be the most diverse CRISPR–Cas grouping, and one generalized description does not adequately encompass their multiplicity. Apart from having a characteristic large subunit protein, each subtype possesses additional particularities, including the presence of a split Cas3 organization (I-A); the functional substitution of Cas6 with Cas5 or the presence of their fusion (I-C, I-U); the functional replacement of Cas8 with Cas5 (I-F variant); and fusions between Cas2 and Cas3 (I-F), and between Cas8 and Cas3 (some I-E systems). The level of variation among these complexes highlights the plasticity of CRISPR–Cas immune systems and demonstrates the many possibilities that nature has explored over evolutionary time.
Type III – Csm/Cmr: Targeting by triple threat
Nucleic acid targeting within type III systems (reviewed in Ref.27) is arguably the most complicated among CRISPR–Cas systems, involving three distinct nuclease modalities (Fig. 2). The interference mechanism appeared inconsistent for years, with in vivo experiments on a type III-A system reporting DNA targeting123 whereas in vitro experiments on a type III-B system demonstrated RNA targeting.124 Only recently has this conundrum been resolved125,126 and further extended by remarkable reports on an additional nuclease activity activated by the production of second messengers.29,30
Type III systems are divided into four subtypes (A–D), and the multisubunit effectors are known as Csm (III-A, III-D) or Cmr (III-B, III-C) complexes. The type III effector complex architecture shows striking similarities to the type I effectors. Both systems require a mature crRNA that is processed by Cas6127; however, type III systems require further processing of the 3′ end by non-Cas nucleases128 and as such do not maintain Cas6 as a subunit of the mature complex. The “base” of the complex is again composed of a Cas5 family member that accommodates the 5′ end of the crRNA and a characteristic large subunit, which in this case is Cas10. The helical “backbone” filament is likewise composed of multiple Cas7 family proteins that span the spacer sequence but also cap the 3′ end of the crRNA. The “belly” of the complex is composed of multiple small subunit proteins that belong to the Cas11 family.129–131
Type III complexes function foremost as RNA-guided RNA-targeting effectors by identifying single-stranded target RNAs through base-pairing with the crRNA. However, the ensuing RNA duplex is systematically disrupted by a conserved β-hairpin present in Cas7, resulting in the outward flipping of a nucleotide at 6-nt intervals.131,132 These specific nucleotides are thereby positioned for cleavage, which is facilitated by a conserved aspartate residue present in Cas7.131,132 Rigid positioning of the crRNA handle within the effector complex orchestrates cleavage at fixed distances from the 5′ end, resulting in a characteristic, ruler-like degradation pattern.132–135
Elegant experiments revealed that target RNA binding (but not necessarily cleavage) activates orthogonal deoxyribonuclease activity of the HD domain in Cas10, which nonspecifically cleaves ssDNA.123,136–138 Targeting does not depend on the presence of a PAM, as previously described for types I and II, but rather on the absence of complementarity between the crRNA 5′ handle and the sequence flanking the RNA target.139 Although Cas10 can cleave ssDNA within mismatched bubble regions in vitro,136 the primary substrate in vivo is likely to be the nontemplate strand of DNA within transcription bubbles, where the effector complex is spatially restricted during targeting in cis of the nascent transcript. This DNase activity is also temporally controlled, because rapid cleavage of target RNA returns Cas10 to an inactive state.138
Two recent studies29,30 uncovered perhaps the most remarkable twist on type III interference. Beyond triggering DNase activity of Cas10, target RNA binding by the Csm complex also induces Cas10-mediated synthesis of cyclic oligoadenylates, an activity catalyzed by the Palm domain. These oligoadenylates function as signaling molecules that bind the CARF domains of homodimeric Csm6 or Csx1, Cas proteins often encoded within type III CRISPR–Cas loci but that do not associate with the effector complex.140,141 Binding to the CARF domains allosterically activates the nonspecific RNA degradation activity of the HEPN ribonuclease domains also present in Csm6/Csx1. In addition to revealing an ingenious adaptation that allows type III systems to target foreign nucleic acids on three fronts, these studies also discovered the first examples of oligoadenylate molecules functioning as second messengers in prokaryotes, and reveal a fascinating parallel with eukaryotic innate immune systems that similarly synthesize oligoadenylate second messengers in response to viral RNA detection.142
Apart from employing an impressive collection of weapons for stand-alone defense, type III immune systems have been shown to target escape mutants from other CRISPR systems.143 In addition, they differentiate between lysogenic and lytic infections,144,145 thereby allowing potentially beneficial traits of lysogenic infection to be enjoyed by the host. Collectively, these observations underscore the marvelous polishing effect of natural selection on the composition and function of CRISPR–Cas systems.
Type V – Cas12: Staggering cuts
Type V systems were first assigned as a putative grouping by Makarova et al. in 2015,11 based on the signature gene cpf1 that had been detected in several prokaryotic genomes adjacent to adaptation genes and a CRISPR array.146,147 Experiments published soon thereafter by Zetsche et al. firmly substantiated this new assignment within Class 2 systems, demonstrating that, like Cas9, Cpf1—now known as Cas12a—functions as a single-effector, RNA-guided endonuclease that catalyzes double-stranded DNA cleavage148 (Fig. 2).
Yet, there are critical mechanistic features that distinguish Cas9 and Cas12a.148 Rather than utilizing a dual-RNA guide, Cas12a, like Cascade and Csm/Cmr complexes, naturally functions with just crRNA and no tracrRNA. DNA targets are recognized in a reversed orientation as compared with Cas9, with a T-rich PAM located upstream (5′) of the target region and the seed sequence located at the 5′ end of the crRNA guide region. Instead of cleaving target DNA toward the PAM-proximal end, at similar positions on both strands, Cas12a introduces staggered cuts toward the PAM-distal end, leaving 5′ overhangs. In addition to possessing the associated deoxyribonuclease cleavage activity for target DNA cleavage, Cas12a also contains a ribonuclease domain that is responsible for enzymatic pre-crRNA processing.149
Cas12a possesses a RuvC-like nuclease domain that is homologous to the respective Cas9 nuclease domain; however, it lacks the HNH nuclease domain. Early structural work identified a novel domain sandwiched between discontinuous RuvC motifs, which was assigned putative nuclease function, and initial biochemical experiments suggested that the novel nuclease domain and RuvC domains were responsible for cleaving the target and nontarget strands, respectively.150 More recently, additional structures have provided tantalizing evidence that both strands within the DNA-bound R-loop are sequentially threaded through and cleaved by the very same RuvC active site,151,152 a conclusion that is well supported by careful mutational studies.153 Perhaps most intriguing, target DNA binding also activates potent, single-stranded DNase activity,31 a behavior that is reminiscent of the collateral damage effect first observed with Cas13a28 (see Type VI). Whether target-activated ssDNA shredding is biologically meaningful during an adaptive immune response, such as to degrade ssDNA phages or to target exposed ssDNA regions during replication or transcription, is not known.
Type V systems recently expanded to include subtypes A–E, which encode Cas12a–e, previously referred to in the literature as Cpf1 (A), C2c1 (B), C2c3 (C), CasY (D), and CasX (E).37,38,41 All Cas12 homologs share a RuvC nuclease domain (but otherwise show low sequence similarity), and, with the exception of Cas12c, have been shown to mediate biochemical DNA cleavage and/or DNA interference activity in Escherichia coli. Lastly, the Type V group also includes uncharacterized loci (subtype V-U), which encode Class 2 candidate proteins that are substantially smaller than other Cas12 family members.154 These putative RNA-guided effectors possess RuvC-like nuclease domains and are predicted to be active, based on the observation that they flank CRISPR arrays that often contain spacers matching phage genomes. It will be interesting to learn whether they provide the same degree of adaptive immunity as their larger counterparts, and how the molecular mechanism of DNA targeting is similar or different.
Type VI – Cas13: Collateral cleavage
Before 2016, Csm/Cmr complexes (type III) were the only crRNA-guided effectors known to naturally target RNA during an immune response. Then, Abudayyeh et al. demonstrated that C2c2—now known as Cas13a—functions as a crRNA-guided RNA-targeting nuclease,28 expanding single-effector Class 2 systems to encompass a new type VI grouping. There are presently four distinct subtypes within type VI (A–D) that encode Cas13a–d.155,156
Cas13 family members possess two HEPN domains with predicted single-stranded RNase activity, and experiments in which Type VI systems were heterologously expressed in E. coli demonstrated that these systems provide resistance against RNA phages. Instead of cleaving within the targeted sequence itself, Cas13 becomes activated upon target RNA binding, unleashing multiple-turnover nonspecific RNA cleavage activity in trans (Fig. 2). Targeting is stimulated by the presence of a specific protospacer flanking sequence,28,157 although not in all scenarios,158,159 and collateral, nonspecific RNA cleavage is both sensitive to secondary structure and has specific nucleotide preferences, depending on the Cas13 homolog.160
A set of recent structural studies have provided elegant insights into the mechanisms of RNA targeting and RNA cleavage for Cas13a.161–163 Similar to Cas9 and Cas12, Cas13a adopts a bilobed architecture in which the nuclease domains form part of a lobe that is distinct from the crRNA recognition lobe. Target RNA binding drives large-scale conformational changes that bring the two HEPN domains into proximity, forming a composite catalytic center that is competent for RNA cleavage. This active site is exposed on the exterior of the protein, explaining how RNAs in solution, other than the target-bound RNA, can be bound and cleaved in trans after Cas13 activation. Interestingly, Cas13, like Cas12, employs an additional ribonuclease domain for pre-crRNA processing,164 and inactivation of either catalytic activity does not impair the other.
Although Cas13 members can confer specific protection against RNA phages when expressed heterologously in E. coli,28,157 the collateral, nonspecific cleavage of other cellular RNAs impedes bacterial growth and may have evolved as a strategy to induce cell dormancy and/or programmed cell death. Understanding the physiological consequences of interference in Type VI systems will require additional studies, ideally including experiments in which Cas13 function is tested in native organisms.
Type IV: To be determined
Type IV systems are a recent addition to Class 1 and encode known components of multisubunit protein–crRNA complexes but often lack adaptation genes, a candidate DNA nuclease, and even CRISPR arrays.11 They have not been experimentally studied, and their function is unknown.
Expanding the CRISPR–Cas Toolbox
The mechanisms employed in CRISPR–Cas immunity have proven to be nothing less than a goldmine for biotechnological tool development, with researchers utilizing machinery from all three different stages—adaptation, crRNA biogenesis, and interference—for a wide range of applications. As early as 1993, microbiologists appreciated the inherent value of using CRISPR arrays, one of the fastest evolving regions of bacterial and archaeal genomes,165 for high-resolution genotyping and strain differentiation.166 More recently, processing ribonucleases involved in the crRNA biogenesis stage have been employed for applications ranging from tagged RNA isolation167,168 to multiplexed gRNA processing,169,170 owing to their exquisite sequence specificity for repeat-derived RNA substrates. Yet, the interference stage of CRISPR–Cas immune systems has clearly been the richest source of biotechnology tool development, with RNA-guided Cas proteins proving invaluable for next-generation methods to control cellular genotype and phenotype. Many of these advances would not have been possible without the solid foundations laid by heroic efforts to develop earlier genome-editing platforms.171,172
In the sections that follow, we summarize some of the major areas of CRISPR tool development, providing reviews where possible for expanded content. We refer interested readers elsewhere for work on gene drives,173 model organism-specific applications,174,175 multiplexing,176 genome-wide screening,177 in vivo delivery methods,178 and development of human therapeutics.179
CRISPR-based genome editing
S. pyogenes Cas9 has seen the widest use as a genome engineering platform since it was first harnessed for DNA editing in eukaryotic cells.180–184 Together with a natural dual-RNA guide or engineered single-RNA guide,49 Cas9 can be programmed to introduce DSBs at DNA target sites, leading to DNA repair outcomes that fall into two major classes: nonhomologous end joining (NHEJ) and homology-directed repair (HDR)185 (Fig. 3A). Repair outcomes from NHEJ, although not random,79 typically result in small insertions or deletions at the target site that often create loss-of-function phenotypes when introduced within exons. Precise genetic alterations through HDR can be accessed through the combined use of Cas9–gRNA and a donor template. Yet, NHEJ is generally the dominant repair pathway in mammalian cells, and significant ongoing efforts are aimed at increasing HDR efficiency.186
FIG. 3.
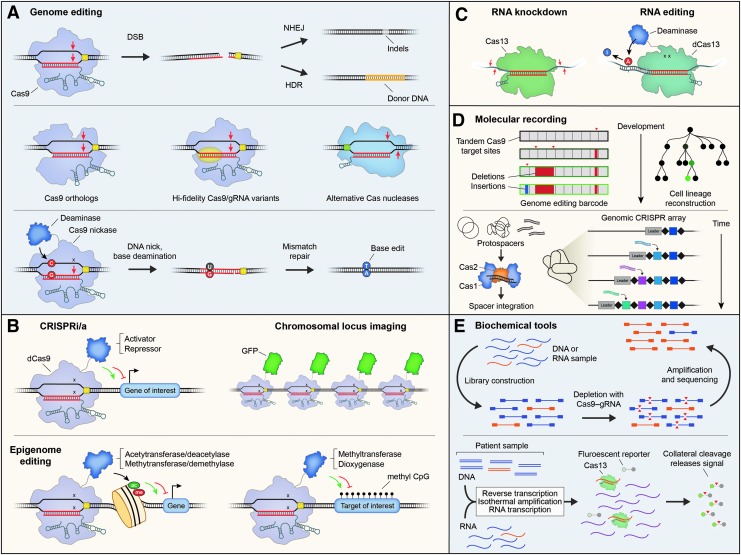
Major tool categories in the CRISPR–Cas toolbox. (A) DSBs are introduced by Cas9–gRNA complexes, leading to permanent genomic edits through repair by either NHEJ or HDR. Cas9 orthologs, Cas9/gRNA variants, and alternative Cas nucleases can increase the fidelity of editing and expand targetable space in the genome. Base editing is achieved using nickase Cas9 variants fused to either cytidine or adenosine deaminases (bottom). (B) CRISPR-mediated transcriptional repression and activation (CRISPRi/CRISPRa) rely on deactivated Cas9 nucleases (dCas9) fused to various activator or repressor domains. Fluorescent protein fusions allow for imaging of specific chromosomal loci, and recruitment of histone- and DNA-modifying enzymes enables epigenome editing (bottom). (C) Cas13 has been harnessed for both targeted RNA knockdown and RNA editing, wherein the latter tool utilizes deactivated nucleases fused to adenosine deaminase domains. (D) Lineage tracing and molecular recording have been achieved using both Cas9-based strategies (top) and CRISPR arrays themselves (bottom). (E) Cas enzymes have proven useful outside of the cell, such as for Cas9-mediated removal of undesirable molecules within high-throughput DNA sequencing libraries (top), and for Cas13-mediated fluorescence detection of specific nucleic acid molecules in complex mixtures (bottom). DSBs, double-strand breaks; gRNA, guide RNA; HDR, homology-directed repair; NHEJ, nonhomologous end joining.
A major challenge of genome-editing applications is mitigating off-target effects. Cas9 readily cleaves genomic sequences that differ from the gRNA at one or more positions,187–189 and the development of systematic approaches to interrogate off-target effects in an unbiased way, at the genome-wide scale, has been the focus of numerous laboratories.190 Available methods use high-throughput sequencing and may broadly be grouped into two categories, cell-based methods and in vitro methods. In parallel, considerable efforts are underway to generate higher fidelity Cas9–gRNA variants, either by modifying the gRNA191–193 or by creating improved Cas9 mutants through rational engineering or directed evolution.69,74–76,194 Multiple strategies have improved accuracy through general destabilization of Cas9–gRNA DNA binding affinity, and recent theoretical models provide compelling kinetic explanations for the origins of this enhancement.65,195
SpyCas9 continues to be improved as a genome-editing reagent, but so too are other crRNA-guided nucleases being harnessed as alternatives (Fig. 3A). Some Cas9 orthologs are smaller and more amenable to viral packaging,196 others may have higher intrinsic targeting fidelity,197 and still others derive from thermophilic bacteria and exhibit elevated stability.198,199 The recent discovery that human serum contains antibodies against two of the most commonly used Cas9 variants200 is sure to stimulate additional Cas9 ortholog screening. Beyond Cas9, Cas12—the other family of single-effector Cas deoxyribonucleases—has been rapidly adopted for genome-editing experiments.148,201 In addition to recognizing a distinct PAM and exhibiting high specificity,202,203 initial studies suggested that the staggered nature of DNA cleavage products generated by Cas12 might naturally lend themselves to enhanced repair by the homologous recombination machinery; thus far, supporting data are lacking. Finally, efforts to engineer or evolve modified Cas9 and Cas12 variants that recognize novel PAM sequences have succeeded in substantially expanding the available “targetable” space of the genome.194,204
In recent years, an alternative approach to introduce single base-pair changes has been developed by Komor et al., termed base editing205 (Fig. 3A). Rather than relying on host machinery for the repair of DSBs, base editors—a fusion of a Cas9 nickases to a nucleoside deaminase—achieve direct chemical conversion of one nucleobase to its deaminated counterpart, ultimately resulting in a permanent mutation. C·G-to-T·A base editors have been constructed using various cytidine deaminases,206,207 including APOBEC and AID, and more recently, adenine base editors that mediate A·T-to-G·C mutations were created through extensive directed evolution and protein engineering.208 In addition to enabling high-efficiency edits without the semistochastic outcomes characteristic of NHEJ, base editing also avoids adverse consequences of introducing mutagenic DSBs.
Last but not least, Class 1 systems—thus far not leveraged outside of prokaryotes, but by far the more abundant class in sequenced prokaryotic genomes—also show potential for certain genome-editing applications. In E. coli, types I and III systems have been harnessed for targeted plasmid elimination and programmable removal of specific strains from mixed populations.209–211 In addition, by programming Cascade (together with Cas3) or Csm/Cmr complexes with self-targeting spacers in native hosts, researchers have achieved genome editing of bacterial, archaeal, and even phage genomes.212–215 Whether similar strategies can be ported into eukaryotic cells remains to be determined, although one effort currently underway aims to treat antibiotic-resistant bacterial infections in human hosts by harnessing the programmable cell-killing activity of Cascade/Cas3.216
Deactivated nucleases for genome manipulation
As revolutionary as the Cas9 nuclease has been for genome editing, the utility of nuclease-deactivated Cas9 (dCas9) as a programmable DNA-binding protein has made nearly as large an impact on the research community. By virtue of its ability to associate with nearly any payload imaginable—either through protein–protein fusions or via RNA aptamers fused to gRNA scaffolds—dCas9–gRNA can achieve a wide variety of cellular outputs by ferrying diverse machinery to specific genomic loci (Fig. 3B; reviewed in Ref.217).
At the transcriptional level, dCas9 was first leveraged for repression, also known as CRISPR interference (CRISPRi), by simply occluding transcriptional machinery in bacteria218 or by recruiting repressor domains like KRAB to promoter regions in mammalian cells.219 CRISPR-mediated transcriptional activation (CRISPRa) was developed soon thereafter, employing either dCas9–activator fusions, the recruitment of tagged activators to gRNAs through RNA aptamers, or a combination of these approaches.220–222
The same basic recruitment strategies have allowed researchers to transform dCas9–gRNA into a robust system for editing the epigenome.223 dCas9 fusions to histone demethylases like LSD1 or histone acetyltransferases like p300 have enabled up- and downregulation of gene expression through altered epigenetic marks at the histone level.224,225 Parallel approaches have succeeded in achieving exquisite control over DNA epigenetic marks as well, whereby dCas9 fusions to either Tet1 or Dnmt3a have led to sustainable DNA demethylation or methylation and corresponding activation or silencing of gene expression, respectively.226,227 The recent demonstration that methylation-edited cells can lead to persistent changes in heterochromatin status that reverses a disease phenotype suggests that epigenome editing will have therapeutic promise.228
dCas9 also offers a powerful new approach to image the three-dimensional organization of chromosomes in living cells through RNA-guided recruitment of fluorescent proteins to specific genomic loci.229 As with CRISPRi/CRISPRa, various molecular strategies for recruitment have been explored, and the use of orthogonal reagents has allowed for multiplexed imaging of multiple loci simultaneously.230 Although signal improvement remains a challenge for imaging-based approaches, particularly for nonrepetitive loci, a recent study developed a tandem gRNA assembly strategy to tile dCas9–GFP along a dozen or more sites within genomic regions of interest.231
Beyond chromosome imaging and gene expression control, the possibilities for recruiting other factors using dCas9–gRNA seem limitless. Large gRNA fusions have enabled locus-specific targeting of long noncoding RNAs;232 dCas9–Spo11 fusion proteins have been harnessed to stimulate meiotic recombination at novel sites;233 orthogonal dCas9 heterodimers have been constructed to enforce DNA looping in E. coli;234 and dCas9–recombinase fusion proteins have been developed toward the eventual goal of programmable, recombinase-based genome editing.235
RNA-targeting applications
Two of the six immune system types (III and VI) are presently known to recognize RNA as their primary target, and effectors from two others (I and II) retain the capability to target RNA in vivo.32,33 Naturally, then, there has been eager interest in exploring applications that harness crRNA-programmed Cas proteins for RNA manipulation.
The realization that SpyCas9 could be engineered to bind and cleave RNAs in vitro83 led to some of the earliest demonstrations of programmable RNA targeting in mammalian cells, first for live cell RNA imaging84 and later for the specific elimination of toxic RNA species directly in primary patient cells.85 And although the multisubunit nature of Csm/Cmr complexes has thus far precluded biotechnological applications in eukaryotic cells, a recent preprint describes the use of recombinant Csm complexes to achieve targeted RNA knockdown in zebrafish embryos.236
Cas13, which naturally targets RNA within type VI systems, is proving to be a far more powerful enzyme for RNA-specific applications (Fig. 3C). In addition to harnessing Cas13 for nucleic acid detection (see CRISPR-based biochemical tools), the Zhang laboratory has achieved targeted knockdown of endogenous transcripts in mammalian and plant cells using Cas13, with comparable specificity and potency as RNA interference; fluorescent tracking of RNA transcripts using nuclease-inactive Cas13 (dCas13); and direct adenosine-to-inosine editing of RNA targets using ADAR2 fusions to an improved dCas13 variant from subtype VI-B.158,159 It will be fascinating to see future applications of Cas13 for genome-wide RNA knockdown screens, direct manipulation of RNA splicing and protein synthesis, isolation and characterization of cellular RNA–protein complexes, and interrogation of lncRNA function.
Lineage tracing and molecular recording
Cas enzymes have also enabled new methods of molecular recording and cell lineage tracing (Fig. 3D). Rather than perturbing RNA or DNA sequences as a means of understanding the resulting impact on a cell or organism, these approaches instead seek to reveal the history of a cell through some form of genetic barcoding. Subsequent analysis reveals the comprehensive lineage relationships between cells within a population, and/or the past stimuli that a given cell and its ancestors have experienced.
Genome editing of synthetic target arrays for lineage tracing, developed by McKenna et al., leverages Cas9 to stochastically introduce indels within arrays of tandem target sequences.237 Specific barcodes are inherited during each cell division, allowing for reconstruction of cell lineages along a developmental pathway by either high-throughput sequencing or, in an alternative approach, by in situ single-molecule FISH.238 An alternative method for cellular recording leverages self-targeting gRNAs whose encoding loci can themselves be edited in a manner that reveals lineage history or specific biological events of interest, such as lipopolysaccharide-induced inflammation.239,240 More recently, an analog recording strategy known as CAMERA, which harnesses either Cas9 or Cas9 base editors for DNA removal or DNA editing, respectively, has allowed multiple stimuli to be recorded, including exposure to viruses, light, and nutrients.241
Remarkably, molecular recording has also been achieved with actual CRISPR arrays, aided not by interference enzymes but by Cas1 and Cas2, the adaptation machinery responsible for spacer integration. After all, CRISPR arrays are nothing but a molecular memory of past infections that grow directionally, thus lending themselves naturally to function like a “biological tape recorder.” Shipman et al. were the first to apply a Cas1–Cas2 expression system242 for molecular recording and CRISPR-archived data storage, which they used to encode a digital movie in a living bacterial population.243,244 Whereas these studies required user-defined protospacers to be electroporated into cells, Sheth et al. have since engineered a system whereby the availability and selection of intracellular protospacers for CRISPR integration directly reflect past biological stimuli, such as the availability of metabolites in the growth medium.245 Future improvements in CRISPR-based molecular recording methods will not only advance our understanding of development and cellular differentiation but may also shed light on the emergence and spread of cancer and other diseases.
CRISPR-based biochemical tools
The last category in the CRISPR toolbox we discuss revolves around the biochemical use of Cas enzymes (Fig. 3E). In an analogous manner to restriction enzymes, Cas9 has been combined with Gibson assembly and other DNA manipulations to facilitate molecular cloning of recombinant plasmids with nucleotide precision.246–248 Potentially more broadly useful are molecular biology applications that harness Cas9 for the targeted depletion of abundant, unwanted sequences from high-throughput sequencing library preparations. For example, Wu et al. removed mitochondrial DNA sequences from ATAC-seq libraries in their study of chromatin accessibility,249 and Gu et al. adopted a similar approach while further demonstrating that Cas9 could deplete wild-type alleles of KRAS from patient cancer samples, facilitating the detection of rare mutant KRAS alleles.250 Cas9 may also be useful to directly enrich for desired sequences, either through dCas9-based affinity purification or through the excision and subsequent isolation of genomic regions of interest.251
Cas9 has even proven useful for physical mapping of genomic DNA. In one study, a Cas9 mutant was used to nick specific genomic sequences, followed by DNA polymerase-mediated local incorporation of fluorescent nucleotides; the resulting sites could then be directly visualized within nanochannel arrays using fluorescence microscopy to identify structural variants.252 Unlabeled, DNA-bound Cas9–gRNA particles have also been directly imaged by high-speed atomic force microscopy, enabling a new form of “nanomapping” that can fill gaps not addressable by traditional sequencing.253
An exciting development within just the past year has revolved around the use of Cas12 and Cas13 enzymes for the in vitro detection of target nucleic acid sequences with attomolar (10−18 M) sensitivity and single-mismatch specificity. The two related detection platforms, SHERLOCK254,255 and DETECTR,31 take advantage of the target-activated, nonspecific nucleic acid degradation activity that was first observed for Cas13 with ssRNA and later for Cas12 with ssDNA. After the demonstration that quenched fluorescent reporter substrates could be employed for optical detection of specific RNA transcripts,164 Gootenberg et al. adopted a similar approach but, importantly, incorporated isothermal amplification, reverse transcription, and in vitro transcription steps to both increase the sensitivity and generalize the approach to allow for both RNA and DNA detection.254 The latest developments in SHERLOCK technology include the use of orthogonal enzymes for multiplexing and lateral flow read-out, such that Zika and Dengue viral ssRNAs, as well as specific cancer mutations, could be detected without the requirement of any additional instrumentation beyond the SHERLOCK cocktail itself.255
Collectively, these methods highlight the remarkable functional diversity and utility of CRISPR–Cas proteins, as well as their robustness and stability in biochemical assays. In light of the growing availability of recombinant Cas enzymes and gRNAs from commercial vendors, in vitro tool development is sure to continue growing alongside cell-based applications in the coming years.
Concluding Thoughts
The surge of innovative technologies harnessing CRISPR–Cas—which can no longer even be confined within the “genome engineering” umbrella term—is proceeding unabated, and it is clear that biologists, regardless of model system or research problem, will increasingly turn to CRISPR-based tools for help. Yet, while the toolbox expands and grows in complexity, so too are the adaptive immune systems that inspired it in the first place becoming ever richer and more interesting than previously appreciated. Indeed, far from the study of CRISPR–Cas biology decelerating or approaching a point of saturation, new discoveries continue to lurk just around the corner. From the discovery of novel effectors155 and virally encoded CRISPR–Cas inhibitors,256 to the identification of entirely new families of putative, CRISPR-linked accessory genes,44,45 there will undoubtedly be ample subject matter for future experimental exploration and exploitation.
In the grander scheme of the microbial defense arsenal, though, CRISPR–Cas adaptive immune systems and other well-studied innate systems may be just the tip of the iceberg. Inspiring work from Sorek and colleagues have uncovered entirely new families of defense systems,257–259 many of which have unknown or poorly understood mechanisms of action. In addition, the future mining of metagenomic data sets is sure to turn up more novelties buried within the genomes of uncultivated species.41 Given that the present sampling of microbes and viruses is many orders of magnitude below the true diversity in our biosphere,260,261 plenty remains to be discovered.
Although our understanding of phage biology and host defenses has exploded in recent years, it is remarkable that one of the fundamental assays used time and time again—infect a bacterial culture with virus and count plaques or resistant colonies—is hardly different than it was nearly a century ago. Luria and Delbrück used this approach with great success in their famous Fluctuation Test experiment in the 1940s, which provided support for the “hypothesis of mutation to immunity” and confirmed Darwin's theory of natural selection.262 Now imagine if they had been using a bug other than the immunocompromised E. coli B strain (which lacks cas genes): would a fully functioning CRISPR–Cas system have provided evidence for the “hypothesis of acquired immunity” instead, and thus supported a Lamarckian model of evolution?263,264 How might this fictitious outcome have affected the trajectory of molecular genetics research?
Delbrück was never one to shy away from new experimental problems, and from his own writings, one can envision how he might have tackled the mystery of CRISPRs: “I could not do it in a few months. Perhaps it will take a few decades, and perhaps it will take the help of a few dozen other people. But listen to what I have found, perhaps you will be interested to join me.”265
Acknowledgments
We would like to thank Joseph Bondy-Denomy, Mitchell O'Connell, Rodolphe Barrangou, Kevin Davies, and the anonymous reviewers for providing helpful feedback and thoughtful comments.
Author Disclosure Statements
S.H.S. is an inventor on patent applications related to CRISPR–Cas systems and uses thereof, a scientific advisor to Dahlia Biosciences, and an equity holder in Dahlia Biosciences and Caribou Biosciences. S.H.S. is an associate editor of The CRISPR Journal.
Clustered Regularly Interspaced Short Palindromic Repeats.
References
- 1.Duckworth DH. Who discovered bacteriophage? Bacteriol Rev. 1976;40:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roach DR, Debarbieux L. Phage therapy: Awakening a sleeping giant. Emerg Top Life Sci. 2017;1:93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keen EC. A century of phage research: Bacteriophages and the shaping of modern biology. Bioessays. 2015;37:6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmond GPC, Fineran PC. A century of the phage: Past, present and future. Nat Rev Microbiol. 2015;13:777–786 [DOI] [PubMed] [Google Scholar]
- 5.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, et al. . Uncovering Earth's virome. Nature. 2016;536:425–430 [DOI] [PubMed] [Google Scholar]
- 6.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361 [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS, Wolf YI. Evolutionary genomics of defense systems in Archaea and Bacteria. Annu Rev Microbiol. 2017;71:233–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327 [DOI] [PubMed] [Google Scholar]
- 9.Pingoud A, Wilson GG, Wende W. Type II restriction endonucleases—A historical perspective and more. Nucleic Acids Res. 2014;42:7489–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur JK, Olovnikov I, Aravin AA. Prokaryotic Argonautes defend genomes against invasive DNA. Trends Biochem Sci. 2014;39:257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Wolf YI, Alkhnbashi OS, et al. . An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burstein D, Sun CL, Brown CT, et al. . Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 2016;7:1061–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:125809–6. [DOI] [PubMed] [Google Scholar]
- 15.Jansen R, Embden JDAV, Gaastra W, et al. . Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 16.Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. . Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182 [DOI] [PubMed] [Google Scholar]
- 17.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663 [DOI] [PubMed] [Google Scholar]
- 18.Bolotin A, Quinquis B, Sorokin A, et al. . Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561 [DOI] [PubMed] [Google Scholar]
- 19.Barrangou R, Fremaux C, Deveau H, et al. . CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 20.Brouns SJJ, Jore MM, Lundgren M, et al. . Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature's toolbox for genome engineering. Cell. 2016;164:29–44 [DOI] [PubMed] [Google Scholar]
- 22.Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:1709–2. [DOI] [PubMed] [Google Scholar]
- 23.Hille F, Richter H, Wong SP, et al. . The biology of CRISPR-Cas: Backward and forward. Cell. 2018;172:1239–1259 [DOI] [PubMed] [Google Scholar]
- 24.Jackson SA, McKenzie RE, Fagerlund RD, et al. . CRISPR-Cas: Adapting to change. Science. 2017;356:eaal505–6. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66 [DOI] [PubMed] [Google Scholar]
- 26.Silas S, Mohr G, Sidote DJ, et al. . Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science. 2016;351:aad423–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamulaitis G, Venclovas Č, Siksnys V. Type III CRISPR-Cas immunity: Major differences brushed aside. Trends Microbiol. 2017;25:49–61 [DOI] [PubMed] [Google Scholar]
- 28.Abudayyeh OO, Gootenberg JS, Konermann S, et al. . C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf557–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazlauskiene M, Kostiuk G, Venclovas Č, et al. . A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609 [DOI] [PubMed] [Google Scholar]
- 30.Niewoehner O, Garcia-Doval C, Rostøl JT, et al. . Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548 [DOI] [PubMed] [Google Scholar]
- 31.Chen JS, Ma E, Harrington LB, et al. . CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;546:eaar624–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampson TR, Saroj SD, Llewellyn AC, et al. . A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R, Fang L, Tan S, et al. . Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 2016;26:1273–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters JE, Makarova KS, Shmakov S, et al. . Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A. 2017;114:E7358–E7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seed KD, Lazinski DW, Calderwood SB, et al. . A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova KS, Haft DH, Barrangou R, et al. . Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmakov S, Abudayyeh OO, Makarova KS, et al. . Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova KS, Zhang F, Koonin EV. SnapShot: Class 1 CRISPR-Cas systems. Cell. 2017;168:946–946.e1 [DOI] [PubMed] [Google Scholar]
- 40.Makarova KS, Zhang F, Koonin EV. SnapShot: Class 2 CRISPR-Cas systems. Cell. 2017;168:328–328.e1 [DOI] [PubMed] [Google Scholar]
- 41.Burstein D, Harrington LB, Strutt SC, et al. . New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krupovic M, Béguin P, Koonin EV. Casposons: Mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr Opin Microbiol. 2017;38:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-Cas systems: All the way there and back. Genome Biol Evol. 2017;9:2812–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shmakov SA, Makarova KS, Wolf YI, et al. . Towards comprehensive characterization of CRISPR-linked genes. BioRxiv. 2018;1–32 [Google Scholar]
- 45.Shah SA, Alkhnbashi OS, Behler J, et al. . Conserved accessory proteins encoded with archaeal and bacterial Type III CRISPR-Cas gene cassettes that may specifically modulate, complement or extend interference activity. BioRxiv. 2018;1–36 [Google Scholar]
- 46.Chen JS, Doudna JA. The chemistry of Cas9 and its CRISPR colleagues. Nat Rev Chem. 2017;1:1–15 [Google Scholar]
- 47.Garneau JE, Dupuis M-È, Villion M, et al. . The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71 [DOI] [PubMed] [Google Scholar]
- 48.Deltcheva E, Chylinski K, Sharma CM, et al. . CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jinek M, Chylinski K, Fonfara I, et al. . A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. . Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740 [DOI] [PubMed] [Google Scholar]
- 51.Gasiunas G, Barrangou R, Horvath P, et al. . Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heler R, Samai P, Modell JW, et al. . Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jinek M, Jiang F, Taylor DW, et al. . Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:124799–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang F, Zhou K, Ma L, et al. . A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481 [DOI] [PubMed] [Google Scholar]
- 56.Shibata M, Nishimasu H, Kodera N, et al. . Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat Commun. 2017;8:143–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sternberg SH, Redding S, Jinek M, et al. . DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. . Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders C, Niewoehner O, Duerst A, et al. . Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mekler V, Minakhin L, Severinov K. Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proc Natl Acad Sci U S A. 2017;114:5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knight SC, Xie L, Deng W, et al. . Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826 [DOI] [PubMed] [Google Scholar]
- 62.Globyte V, Lee SH, Bae T, et al. . CRISPR Cas9 searches for a protospacer adjacent motif by one-dimensional diffusion. BioRxiv. 2018;1–23 [Google Scholar]
- 63.Singh D, Sternberg SH, Fei J, et al. . Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun. 2016;7:1277–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyle EA, Andreasson JOL, Chircus LM, et al. . High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci U S A. 2017;114:5461–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein M, Eslami-Mossallam B, Arroyo DG, et al. . Hybridization kinetics explains CRISPR-Cas off-targeting rules. Cell Rep. 2018;22:1413–1423 [DOI] [PubMed] [Google Scholar]
- 66.Gorski SA, Vogel J, Doudna JA. RNA-based recognition and targeting: Sowing the seeds of specificity. Nat Rev Mol Cell Biol. 2017;18:215–228 [DOI] [PubMed] [Google Scholar]
- 67.Sternberg SH, LaFrance B, Kaplan M, et al. . Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dagdas YS, Chen JS, Sternberg SH, et al. . A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci Adv. 2017;3:eaao002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JS, Dagdas YS, Kleinstiver BP, et al. . Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palermo G, Miao Y, Walker RC, et al. . CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci U S A. 2017;114:7260–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gong S, Yu HH, Johnson KA, et al. . DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 2018;22:359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raper AT, Stephenson AA, Suo Z. Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J Am Chem Soc. 2018;140:2971–2984 [DOI] [PubMed] [Google Scholar]
- 73.Kleinstiver BP, Pattanayak V, Prew MS, et al. . High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slaymaker IM, Gao L, Zetsche B, et al. . Rationally engineered Cas9 nucleases with improved specificity. Science. 2015;351:84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu JH, Miller SM, Geurts MH, et al. . Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Casini A, Olivieri M, Petris G, et al. . A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36:265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuo Z, Liu J. Cas9-catalyzed DNA cleavage generates staggered ends: Evidence from molecular dynamics simulations. Sci Rep. 2016;5:3758–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stephenson AA, Raper AT, Suo Z. Bidirectional degradation of DNA cleavage products catalyzed by CRISPR/Cas9. J Am Chem Soc. 2018;140:3743–3750 [DOI] [PubMed] [Google Scholar]
- 79.van Overbeek M, Capurso D, Carter MM, et al. . DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol Cell. 2016;63:633–646 [DOI] [PubMed] [Google Scholar]
- 80.Lemos BR, Kaplan AC, Bae JE, et al. . CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci U S A. 2018;115:E2040–E2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richardson CD, Ray GJ, DeWitt MA, et al. . Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344 [DOI] [PubMed] [Google Scholar]
- 82.Rose JC, Stephany JJ, Valente WJ, et al. . Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat Methods. 2017;14:891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Connell MR, Oakes BL, Sternberg SH, et al. . Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nelles DA, Fang MY, O'Connell MR, et al. . Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Batra R, Nelles DA, Pirie E, et al. . Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell. 2017;170:899–912.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strutt SC, Torrez RM, Kaya E, et al. . RNA-dependent RNA targeting by CRISPR-Cas9. Elife. 2018;7:e3272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rousseau BA, Hou Z, Gramelspacher MJ, et al. . Programmable RNA cleavage and recognition by a natural CRISPR-Cas9 system from Neisseria meningitidis. Mol Cell. 2018;69:906–914.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dugar G, Leenay RT, Eisenbart SK, et al. . CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell. 2018;69:893–905.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deveau H, Barrangou R, Garneau JE, et al. . Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vlot M, Houkes J, Lochs SJA, et al. . Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res. 2018;46:873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tao P, Wu X, Rao V. Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci Adv. 2018;4:eaar413–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bondy-Denomy J. Protein inhibitors of CRISPR-Cas9. ACS Chem Biol. 2018;13:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plagens A, Richter H, Charpentier E, et al. . DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 2015;39:442–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiedenheft B, van Duijn E, Bultema JB, et al. . RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011;108:10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Semenova E, Jore MM, Datsenko KA, et al. . Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jore MM, Lundgren M, van Duijn E, et al. . Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536 [DOI] [PubMed] [Google Scholar]
- 97.Bondy-Denomy J, Pawluk A, Maxwell KL, et al. . Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haurwitz RE, Jinek M, Wiedenheft B, et al. . Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687 [DOI] [PubMed] [Google Scholar]
- 100.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hayes RP, Xiao Y, Ding F, et al. . Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mulepati S, Héroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jackson RN, Golden SM, van Erp PBG, et al. . Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hochstrasser ML, Taylor DW, Bhat P, et al. . CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chowdhury S, Carter J, Rollins MF, et al. . Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169:47–57.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pausch P, Müller-Esparza H, Gleditzsch D, et al. . Structural variation of type I-F CRISPR RNA guided DNA surveillance. Mol Cell. 2017;67:622–632.e4 [DOI] [PubMed] [Google Scholar]
- 107.Xiao Y, Luo M, Hayes RP, et al. . Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell. 2017;170:48–60.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fineran PC, Gerritzen MJH, Suárez-Diez M, et al. . Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014;111:E1629–E1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Redding S, Sternberg SH, Marshall M, et al. . Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell. 2015;163:854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rutkauskas M, Sinkunas T, Songailiene I, et al. . Directional R-loop formation by the CRISPR-Cas surveillance complex Cascade provides efficient off-target site rejection. Cell Rep. 2015;10:1534–1543 [DOI] [PubMed] [Google Scholar]
- 111.Leenay RT, Maksimchuk KR, Slotkowski RA, et al. . Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell. 2016;62:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu BXH, Wainberg M, Kundaje A, et al. . High-throughput characterization of Cascade type I-E CRISPR guide efficacy reveals unexpected PAM diversity and target sequence preferences. Genetics. 2017;206:1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jung C, Hawkins JA, Jones SK, et al. . Massively parallel biophysical analysis of CRISPR-Cas complexes on next generation sequencing chips. Cell. 2017;170:35–47.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xue C, Whitis NR, Sashital DG. Conformational control of Cascade interference and priming activities in CRISPR immunity. Mol Cell. 2016;64:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jackson RN, Lavin M, Carter J, et al. . Fitting CRISPR-associated Cas3 into the Helicase family tree. Curr Opin Struct Biol. 2014;24C:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huo Y, Nam KH, Ding F, et al. . Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014;21:771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sinkunas T, Gasiunas G, Fremaux C, et al. . Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sinkunas T, Gasiunas G, Waghmare SP, et al. . In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brown MW, Dillard KE, Xiao Y, et al. . Assembly and translocation of a CRISPR-Cas primed acquisition complex. BioRxiv. 2017;1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Loeff L, Brouns SJJ, Joo C. Repetitive DNA reeling by the Cascade-Cas3 complex in nucleotide unwinding steps. BioRxiv. 2017;1–10 [DOI] [PubMed] [Google Scholar]
- 122.Künne T, Kieper SN, Bannenberg JW, et al. . Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell. 2016;63:852–864 [DOI] [PubMed] [Google Scholar]
- 123.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science. 2008;322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hale CR, Zhao P, Olson S, et al. . RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Samai P, Pyenson N, Jiang W, et al. . Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peng W, Feng M, Feng X, et al. . An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hale CR, Cocozaki A, Li H, et al. . Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Walker FC, Chou-Zheng L, Dunkle JA, et al. . Molecular determinants for CRISPR RNA maturation in the Cas10-Csm complex and roles for non-Cas nucleases. Nucleic Acids Res. 2017;45:2112–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Staals RHJ, Taylor DW, Barendregt A, et al. . RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taylor DW, Zhu Y, Staals RHJ, et al. . Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osawa T, Inanaga H, Sato C, et al. . Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–430 [DOI] [PubMed] [Google Scholar]
- 132.Benda C, Ebert J, Scheltema RA, et al. . Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54 [DOI] [PubMed] [Google Scholar]
- 133.Tamulaitis G, Tamulaitis G, Kazlauskiene M, et al. . Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–517 [DOI] [PubMed] [Google Scholar]
- 134.Ramia NF, Spilman M, Tang L, et al. . Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014;9:1610–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu X, Ye K. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015;43:1257–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Elmore JR, Sheppard NF, Ramia N, et al. . Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 2016;30:447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Estrella MA, Kuo F-T, Bailey S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016;30:460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kazlauskiene M, Tamulaitis G, Kostiuk G, et al. . Spatiotemporal control of type III-A CRISPR-Cas immunity: Coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62:295–306 [DOI] [PubMed] [Google Scholar]
- 139.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Makarova KS, Anantharaman V, Grishin NV, et al. . CARF and WYL domains: Ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014;5:10–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang W, Samai P, Marraffini LA. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell. 2016;164:710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hornung V, Hartmann R, Ablasser A, et al. . OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Silas S, Lucas-Elio P, Jackson SA, et al. . Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. Elife. 2017;6:e2760–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goldberg GW, Jiang W, Bikard D, et al. . Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goldberg GW, McMillan EA, Varble A, et al. . Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat Commun. 2018;9:6–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schunder E, Rydzewski K, Grunow R, et al. . First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60 [DOI] [PubMed] [Google Scholar]
- 147.Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014;11:156–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. . Cpf1 is a single RNA-guided Endonuclease of a Class 2 CRISPR-Cas system. Cell. 2015;163:759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fonfara I, Richter H, Bratovič M, et al. . The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521 [DOI] [PubMed] [Google Scholar]
- 150.Yamano T, Nishimasu H, Zetsche B, et al. . Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang H, Gao P, Rajashankar KR, et al. . PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167:1814–1828.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stella S, Alcon P, Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546:559–563 [DOI] [PubMed] [Google Scholar]
- 153.Swarts DC, van der Oost J, Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell. 2017;66:221–233.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shmakov S, Smargon A, Scott D, et al. . Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Konermann S, Lotfy P, Brideau NJ, et al. . Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018. [Epub ahead of print]; DOI: 10.1016/j.cell.2018.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yan WX, Chong S, Zhang H, et al. . Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018. [Epub ahead of print]. DOI: 10.1016/j.molcel.2018.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smargon AA, Cox DBT, Pyzocha NK, et al. . Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65:618–630.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. . RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cox DBT, Gootenberg JS, Abudayyeh OO, et al. . RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.East-Seletsky A, O'Connell MR, Burstein D, et al. . RNA Targeting by functionally orthogonal type VI-A CRISPR-Cas enzymes. Mol Cell. 2017;66:373–383.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu L, Li X, Wang J, et al. . Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168:121–134.e12 [DOI] [PubMed] [Google Scholar]
- 162.Liu L, Li X, Ma J, et al. . The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726.e10 [DOI] [PubMed] [Google Scholar]
- 163.Knott GJ, East-Seletsky A, Cofsky JC, et al. . Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat Struct Mol Biol. 2017;24:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.East-Seletsky A, O'Connell MR, Knight SC, et al. . Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207 [DOI] [PubMed] [Google Scholar]
- 166.Groenen PM, Bunschoten AE, van Soolingen D, et al. . Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065 [DOI] [PubMed] [Google Scholar]
- 167.Lee HY, Haurwitz RE, Apffel A, et al. . RNA-protein analysis using a conditional CRISPR nuclease. Proc Natl Acad Sci U S A. 2013;110:5416–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Salvail-Lacoste A, Di Tomasso G, Piette BL, et al. . Affinity purification of T7 RNA transcripts with homogeneous ends using ARiBo and CRISPR tags. RNA. 2013;19:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nissim L, Perli SD, Fridkin A, et al. . Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tsai SQ, Wyvekens N, Khayter C, et al. . Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439 [DOI] [PubMed] [Google Scholar]
- 172.Urnov FD. Genome editing B.C. (before CRISPR): Lasting lessons from the “old testament.” CRISPR J. 2018;1:34–46 [DOI] [PubMed] [Google Scholar]
- 173.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159 [DOI] [PubMed] [Google Scholar]
- 174.Tan W, Proudfoot C, Lillico SG, et al. . Gene targeting, genome editing: From Dolly to editors. Transgenic Res. 2016;25:273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yin K, Gao C, Qiu J-L. Progress and prospects in plant genome editing. Nat Plants. 2017;3:1710–7. [DOI] [PubMed] [Google Scholar]
- 176.Thompson DB, Aboulhouda S, Hysolli E, et al. . The future of multiplexed eukaryotic genome engineering. ACS Chem Biol. 2018;13:313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Doench JG. Am I ready for CRISPR? A user's guide to genetic screens. Nat Rev Genet. 2017;5:e1976–0. [DOI] [PubMed] [Google Scholar]
- 178.Wilson RC, Gilbert LA. The promise and challenge of in vivo delivery for genome therapeutics. ACS Chem Biol. 2018;13:376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fellmann C, Gowen BG, Lin P-C, et al. . Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16:89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cho SW, Kim S, Kim JM, et al. . Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232 [DOI] [PubMed] [Google Scholar]
- 183.Hwang WY, Fu Y, Reyon D, et al. . Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jinek M, East A, Cheng A, et al. . RNA-programmed genome editing in human cells. Elife. 2013;2:e0047–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst). 2016;44:6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pawelczak KS, Gavande NS, VanderVere-Carozza PS, et al. . Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol. 2018;13:389–396 [DOI] [PubMed] [Google Scholar]
- 187.Hsu PD, Scott DA, Weinstein JA, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pattanayak V, Lin S, Guilinger JP, et al. . High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fu Y, Foden JA, Khayter C, et al. . High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tsai SQ. Discovering the genome-wide activity of CRISPR-Cas nucleases. ACS Chem Biol. 2018;13:305–308 [DOI] [PubMed] [Google Scholar]
- 191.Fu Y, Sander JD, Reyon D, et al. . Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ryan DE, Taussig D, Steinfeld I, et al. . Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46:792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yin H, Song C-Q, Suresh S, et al. . Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol. 2018;14:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kleinstiver BP, Prew MS, Tsai SQ, et al. . Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bisaria N, Jarmoskaite I, Herschlag D. Lessons from enzyme kinetics reveal specificity principles for RNA-guided nucleases in RNA interference and CRISPR-based genome Editing. Cell Syst. 2017;4:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ran FA, Cong L, Yan WX, et al. . In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Amrani N, Gao XD, Liu P, et al. . NmeCas9 is an intrinsically high-fidelity genome editing platform. BioRxiv. 2017;1–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Harrington LB, Paez-Espino D, Staahl BT, et al. . A thermostable Cas9 with increased lifetime in human plasma. Nat Commun. 2017;8:142–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mougiakos I, Mohanraju P, Bosma EF, et al. . Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat Commun. 2017;8:164–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Charlesworth CT, Deshpande PS, Dever DP, et al. . Identification of pre-existing adaptive immunity to Cas9 proteins in humans. BioRxiv. 2018;1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kim Y, Cheong S-A, Lee JG, et al. . Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016;34:808–810 [DOI] [PubMed] [Google Scholar]
- 202.Kim D, Kim J, Hur JK, et al. . Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–868 [DOI] [PubMed] [Google Scholar]
- 203.Kleinstiver BP, Tsai SQ, Prew MS, et al. . Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gao L, Cox DBT, Yan WX, et al. . Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Komor AC, Badran AH, Liu DR. Editing the genome without double-stranded DNA breaks. ACS Chem Biol. 2018;13:383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Komor AC, Kim YB, Packer MS, et al. . Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nishida K, Arazoe T, Yachie N, et al. . Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:1–14 [DOI] [PubMed] [Google Scholar]
- 208.Gaudelli NM, Komor AC, Rees HA, et al. . Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gomaa AA, Klumpe HE, Luo ML, et al. . Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5:e00928–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Caliando BJ, Voigt CA. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun. 2015;6:698–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ichikawa HT, Cooper JC, Lo L, et al. . Programmable type III-A CRISPR-Cas DNA targeting modules. PLoS One. 2017;12:e017622–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vercoe RB, Chang JT, Dy RL, et al. . Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e100345–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li Y, Pan S, Zhang Y, et al. . Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pyne ME, Bruder MR, Moo-Young M, et al. . Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep. 2016;6:2566–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bari SMN, Walker FC, Cater K, et al. . Strategies for editing virulent Staphylococcal phages using CRISPR-Cas10. ACS Synth Biol. 2017;6:2316–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Reardon S. Modified viruses deliver death to antibiotic-resistant bacteria. Nature. 2017;546:586–587 [DOI] [PubMed] [Google Scholar]
- 217.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264 [DOI] [PubMed] [Google Scholar]
- 218.Qi LS, Larson MH, Gilbert LA, et al. . Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gilbert LA, Larson MH, Morsut L, et al. . CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chavez A, Scheiman J, Vora S, et al. . Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tanenbaum ME, Gilbert LA, Qi LS, et al. . A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Konermann S, Brigham MD, Trevino AE, et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pulecio J, Verma N, Mejía-Ramírez E, et al. . CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell. 2017;21:431–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hilton IB, D'Ippolito AM, Vockley CM, et al. . Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kearns NA, Pham H, Tabak B, et al. . Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Choudhury SR, Cui Y, Lubecka K, et al. . CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7:46545–46556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vojta A, Dobrinić P, Tadić V, et al. . Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu XS, Wu H, Krzisch M, et al. . Rescue of Fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chen B, Guan J, Huang B. Imaging specific genomic DNA in living cells. Annu Rev Biophys. 2016;45:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ma H, Tu L-C, Naseri A, et al. . Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol. 2016;34:528–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gu B, Swigut T, Spencley A, et al. . Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 2018;359:1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Shechner DM, Hacisuleyman E, Younger ST, et al. . Multiplexable, locus-specific targeting of long RNAs with CRISPR-display. Nat Methods. 2015;12:664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sarno R, Vicq Y, Uematsu N, et al. . Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 2017;45:e16–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hao N, Shearwin KE, Dodd IB. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat Commun. 2017;8:162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chaikind B, Bessen JL, Thompson DB, et al. . A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic Acids Res. 2016;44:9758–9770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fricke T, Tamulaitis G, Smalakyte D, et al. . Targeted RNA knockdown by crRNA guided Csm in zebrafish. BioRxiv. 2017;1–18 [Google Scholar]
- 237.McKenna A, Findlay GM, Gagnon JA, et al. . Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Frieda KL, Linton JM, Hormoz S, et al. . Synthetic recording and in situ readout of lineage information in single cells. Nature. 2017;541:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Perli SD, Cui CH, Lu TK. Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science. 2016:353;pii: DOI: 10.1126/science.aag0511 [DOI] [PubMed] [Google Scholar]
- 240.Kalhor R, Mali P, Church GM. Rapidly evolving homing CRISPR barcodes. Nat Methods. 2017;14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tang W, Liu DR. Rewritable multi-event analog recording in bacterial and mammalian cells. Science. 2018;eaap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Shipman SL, Nivala J, Macklis JD, et al. . Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353:aaf117–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shipman SL, Nivala J, Macklis JD, et al. . CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017;547:345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sheth RU, Yim SS, Wu FL, et al. . Multiplex recording of cellular events over time on CRISPR biological tape. Science. 2017;eaao0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang J-W, Wang A, Li K, et al. . CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques. 2015;58:161–170 [DOI] [PubMed] [Google Scholar]
- 247.Jiang W, Zhao X, Gabrieli T, et al. . Cas9-assisted targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun. 2015;6:810–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wang H, Li Z, Jia R, et al. . ExoCET: Exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 2018;46:e2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wu J, Huang B, Chen H, et al. . The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657 [DOI] [PubMed] [Google Scholar]
- 250.Gu W, Crawford ED, O'Donovan BD, et al. . Depletion of abundant sequences by hybridization (DASH): Using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:4–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gabrieli T, Sharim H, Michaeli Y, et al. . Cas9-assisted targeting of CHromosome segments (CATCH) for targeted nanopore sequencing and optical genome mapping. BioRxiv. 2017;1–11 [Google Scholar]
- 252.McCaffrey J, Sibert J, Zhang B, et al. . CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double strand DNA for whole genome mapping and structural variation analysis. Nucleic Acids Res. 2016;44:e1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mikheikin A, Olsen A, Leslie K, et al. . DNA nanomapping using CRISPR-Cas9 as a programmable nanoparticle. Nat Commun. 2017;8:166–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gootenberg JS, Abudayyeh OO, Lee JW, et al. . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. . Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018. [Epub ahead of print]; DOI: 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.He F, Bhoobalan-Chitty Y, Van LB, et al. . Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol. 2018;71:23–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Goldfarb T, Sberro H, Weinstock E, et al. . BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ofir G, Melamed S, Sberro H, et al. . DISARM is a widespread bacterial defence system with broad anti-phage activities. Nature Microbiol. 2017;8:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Doron S, Melamed S, Ofir G, et al. . Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359:eaar412–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proc Natl Acad Sci U S A. 2016;113:5970–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ofir G, Sorek R. Contemporary phage biology: From classic models to new insights. Cell. 2018;172:1260–1270 [DOI] [PubMed] [Google Scholar]
- 262.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Holmes C, Ghafari M, Anzar A, et al. . Luria-Delbrück, revisited: The classic experiment does not rule out Lamarckian evolution. ArXiv. 2017;1–13 [DOI] [PubMed] [Google Scholar]
- 264.Koonin EV, Wolf YI. Just how Lamarckian is CRISPR-Cas immunity: The continuum of evolvability mechanisms. Biol Direct. 2016;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Delbrück M. Experiments with bacterial viruses (Bacteriophages). Harvey Lect. 1946;41:161–187 [PubMed] [Google Scholar]