SUMMARY
Eicosanoids and related oxylipins are critical, small bioactive mediators of human physiology and inflammation. While ~1100 distinct species have been predicted to exist, to date, less than 150 of these molecules have been measured in humans, limiting our understanding of their role in human biology. Using a directed non-targeted mass spectrometry approach in conjunction with chemical networking of spectral fragmentation patterns, we find over 500 discrete chemical signals highly consistent with known and putative eicosanoids and related oxylipins in human plasma including 46 putative molecules not previously described. In plasma samples from 1500 individuals, we find members of this expanded oxylipin library hold close association with markers of inflammation, as well as clinical characteristics linked with inflammation, including advancing age and obesity. These experimental and computational approaches enable discovery of chemical entities and will shed important insight into the role of bioactive molecules in human health and disease.
Graphical Abstract
eTOC
Watrous et. al. describe the integration of directed non-targeted mass spectrometry and computational chemical networking to discover hundreds of previously unrecognized inflammatory oxylipins metabolites in human plasma, providing a foundation for new insight into the role of oxylipins in human biology.
INTRODUCTION
Eicosanoids and related oxylipins, hereafter referred to simply as oxylipins, represent small polar lipid compounds produced through extensive and variable oxidation of mostly 18 to 22 carbon polyunsaturated fatty acids (PUFA’s), including omega-6 fatty acids, such as arachidonic acid, linoleic acid, and adrenic acid, and omega-3 fatty acids, such as docosahexaenoic acid (DHA), docosapenaenoic acid (DPA), and eiocosapentaenoic acid (EPA) (Astarita et al., 2015; Buczynski et al., 2009; Capra et al., 2015; Dennis and Norris, 2015; Funk, 2001; Khanapure et al., 2007; Quehenberger and Dennis, 2011). Given their chemical diversity, hundreds to thousands of theoretically possible oxylipin chemical compounds have been catalogued in databases worldwide, subdivided into chemical and functional families including prostaglandins, leukotrienes, and resolvins, among others (Fahy et al., 2009) and are highly conserved among species, originating in elementary microbes and present in fungi, plants and mammals (Forn-Cuni et al., 2017; Minderhout, 2000; Noverr et al., 2003; Stanley and Kim, 2014). By signaling through cell surface G-protein coupled receptors, tyrosine kinase receptors and intracellular nuclear receptors, oxylipins mediate a number of diverse homeostatic functions in humans including host immune activation, cellular development, ion transport, muscle contraction, thrombosis, and vasomotor tone, as well as likely many yet undiscovered processes (Gilroy et al., 1999; Lynch et al., 1999; Rocha and Fernandez-Alonso, 2001; Yokomizo et al., 1997; Yokomizo et al., 2000). In the setting of acute infection, these compounds may directly regulate the classic inflammatory triad of fever, edema and pain and provide early benefit by promoting clearing of invasive pathogens and wound healing (Calder, 2010; Dennis and Norris, 2015; Harizi et al., 2008; Levy et al., 2001; Norris and Dennis, 2014; Schirmer et al., 2016; Slatter et al., 2016; Tilley et al., 2001). They have also been implicated in the setting of chronic aseptic systemic inflammation, as is associated with a number of human diseases including obesity, diabetes, cardiovascular disease, cancer, and autoimmunity (Dalli et al., 2017; Dreisbach et al., 2014; Greene et al., 2011; Imig, 2008; Shinomiya et al., 2002; Simopoulos, 2008; Vona-Davis and Rose, 2013). Given their profound biological effects, oxylipins have been extensively targeted for therapeutic purposes with pharmaceutical agents, including non-sterol anti-inflammatory drugs (NSAIDs) and acetylsalicylic acid (aspirin), being among the most widely utilized medications in medicine today (Dennis and Norris, 2015; Khanapure et al., 2007).
Current understanding of the biology of oxylipins is limited, however, by technical challenges related to their measure, owing to their low abundance, dynamic nature, and extensive isometry in chemical structure (Puppolo et al., 2014; Quehenberger et al., 2010; Tsikas and Zoerner, 2014). To date, measurement of these molecules has required highly sensitive, liquid chromatography-mass spectrometry (LC-MS) systems using triple-quadrupole (QQQ) based targeted analysis of known chemical entities. Given the limited availability of commercial standards, much of their biology has focused on a well-established subset of 50-150 compounds, with typical studies reporting ~50 oxylipins per sample, therefore capturing only a small fraction their total biology (Brose et al., 2011; Dickinson and Murphy, 2002; Nakamura et al., 1997; Norris and Serhan, 2018; Wang et al., 2014). These targeted approaches have limited the study of oxylipins as well as notably preclude discovery of related compounds that may be mediating human biology.
Herein, to greatly expand the repertoire of oxylipins assayed in humans, we have developed a “directed non-targeted mass spectrometry” approach using high mass accuracy LC-MS for measurement of bioactive lipid species. Using chemical networking of MS/MS spectral fragments and system analysis of chemical patterning we find over 500 distinct oxylipin entities in human plasma, including 362 compounds not previously documented in humans, and 46 unknown related compounds. In plasma samples across 1500 individuals, we find a number of these oxylipins as highly associated with markers of systemic inflammation and common clinical characteristics linked to inflammatory diseases, including advancing age and increasing obesity. These analytical approaches greatly expand the spectrum of oxylipins observable in complex organisms, enable discovery of compounds in humans, and provide a foundation for new insight into the role of bioactive lipids in mediating human health and disease.
RESULTS
Chemical Characterization of Eicosanoids and Related Oxylipins
Comprehensive detection of oxylipins in a complex milieu, particularly for those chemical species for which no commercial standards or reference data exist, requires distinguishing chemical features. We therefore first sought to pinpoint common chemical characteristics unique to oxylipins. Review of the LIPID MAPS chemical database (www.lipidmaps.org, March 2017 version), which contains over 40,000 entries, revealed 1125 theoretically and experimentally curated, structurally distinct molecules classified as oxylipins (Table S-1) (Fahy et al., 2009). Virtually all compounds were found to lie within a narrow mass range of 300-400 daltons and contain high degrees of oxidation and unsaturation relative to other biomolecules within this same mass range, resulting in an abnormally high mass defect, or the non-whole number portion of a compounds mass. Given the combinatorial nature of oxylipin biosynthesis wherein only a handful of PUFAs are extensively modified through both enzymatic and non-enzymatic biosynthetic mechanisms (Dennis and Norris, 2015; Khanapure et al., 2007), these compounds were found to exhibit extensive isomerism with the 1125 theoretical compounds being derived from only 168 unique chemical formulas (Figure 1A), and 786 of the 1125 compounds stemming from only 23 unique chemical formulas. To determine if these 168 chemical formulas occupy unique masses relative to other biomolecules, the accurate mass for each formula was searched against the Human Metabolome Database (HMDB) entries of endogenous origin using a ±2 ppm mass error window (Wishart et al., 2018). Of the 168 formulas, only 9 chemical formulas (~5%) returned additional matches to non-oxylipin compounds, with the majority of these mapping to sterol, bile acid and free fatty acid metabolites, thereby suggesting that oxylipins exhibit fairly specific and unique mass characteristics.
Figure 1: Directed non-targeted LC-MS based measure of eicosanoids and related oxylipins in humans.
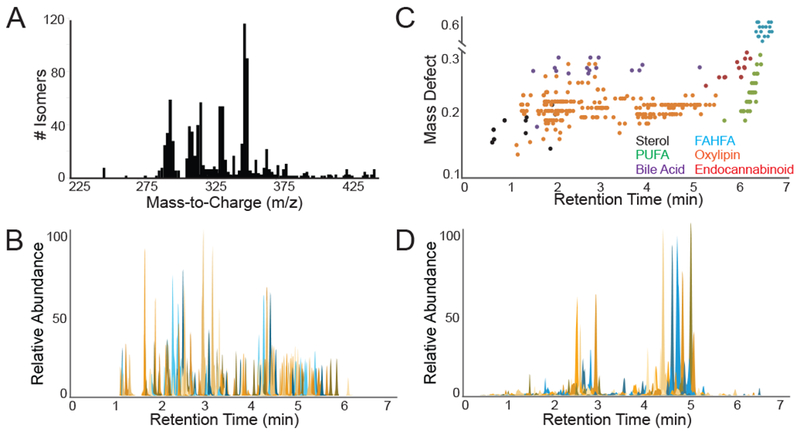
(A) Histogram illustrating the high degree of isomerism amongst compounds with number of database entries for each mass to charge (m/z). (B) Extracted ion chromatograms for all 225 oxylipin chemical standards. (C) Plot of mass defect versus retention time showing elution segregation of compound classes. (D) Extracted ion chromatograms for all 134 spectral peaks observed in pooled plasma that were matched to commercial oxylipin standards. See Figure S-1 and Table S-1.
To determine whether oxylipins may be further experimentally distinguished particularly from isobaric, non-oxylipin species including sterol, bile acid and free fatty acid metabolites, we utilized a ‘directed, non-targeted’ LC-MS approach with offline solid phase extraction for enrichment of oxylipin species, reverse phase chromatography, and high mass accuracy mass spectrometry operating in full scan mode (see Methods). Commercial standards for 225 oxylipins, 14 bile acids, 8 sterols, 29 fatty acids, and 13 endocannabinoids (given that their in-source fragments may be isobaric with oxylipins) were injected as neat solutions (Table S-1). LC-MS analysis revealed that all 225 oxylipin standards eluted between 1.0 and 5.75 minutes on the LC gradient (Figure 1B), exhibited consistent peak shapes (FWHM <0.1 minutes and tailing factor 0.8-1.2), and were chromatographically resolved from sterols (elution between 0.5 and 1.25 minutes), as well as fatty acids and endocannabinoids (elution between 6.0 to 7.0 minutes) (Figure 1C). Bile acids exhibited chromatographic overlap, though they exhibited noticeably wider peak widths and increased peak tailing (>1.5), particularly tauro-variants (Figure 1C). Collectively this suggests that oxylipins may be distinguished from other known metabolites using chemical characteristics, including accurate mass features, chromatographic retention, and peak shape.
Monitoring of Eicosanoids and Related Oxylipins in Human Plasma
In humans, oxylipins are produced locally in tissues, leak into the vasculature, and enter the systemic circulation (Dennis and Norris, 2015; Funk, 2001; Harizi et al., 2008). To determine the full catalog of oxylipins present in circulation, commercial pooled human plasma prepared with EDTA anticoagulant and stored at −80 °C until analysis was assayed using our directed, non-targeted LC-MS method. Human plasma revealed 134 distinct spectral peaks definitively identified as known oxylipins by precise matching of retention time, accurate mass, and tandem MS fragmentation pattern to one of 225 commercially available standards (Figure 1D), including well described members of prostaglandin, HETE, DHET, HODE and additional families. Human plasma also revealed an additional 745 spectral peaks across 56 distinct chemical formulas consistent with oxylipins based on accurate mass (<±2 ppm) matching to one of 1125 theoretical oxylipins in the LIPID MAPS database (Table S-1), retention time range (1.0 – 5.75 minutes), and peak shape (FWHM < 0.1 minutes and tailing factor 0.8-1.2) characteristics. These 745 spectral peaks include a number of entities found to be isobaric with the 134 definitively identified compounds (Figure 2A). For example, the extracted ion chromatogram (XIC) for m/z 353.2333 exhibited 18 isobaric chromatographic peaks with commercial standards accounting for 15, XIC for m/z 375.2177 exhibited 32 isobaric chromatographic peaks with commercial standards accounting for only 3, and XIC for m/z 313.2384 exhibited 20 isobaric chromatographic peaks with commercial standards accounting for only two (Figure 2A). Due to oxylipins occupying a unique chemical space, these isobaric species very likely represent additional compounds isomeric to oxylipins for which chemical standards are not currently available. For instance, search of the LIPID MAPS database for m/z 313.2384 revealed a possible 29 theoretical oxylipin species that potentially correspond to the 20 observed features at that mass.
Figure 2: Identification of oxylipins in human plasma.
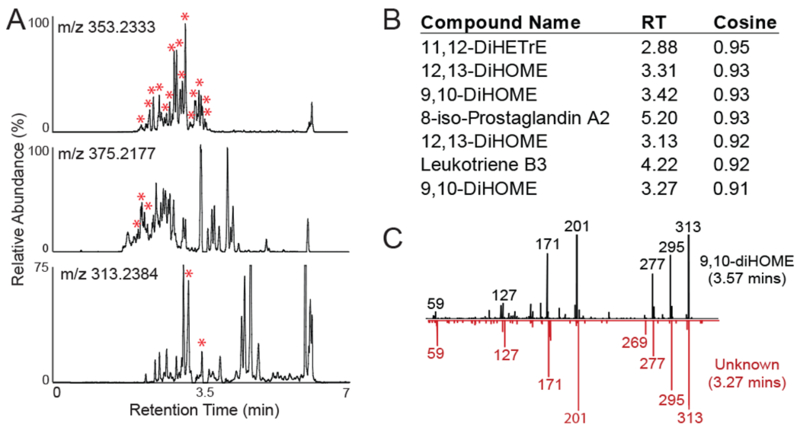
(A) Extracted ion chromatograms for m/z’s 353.2333, 375.2177 and 313.2384 with the red asterisks indicating peaks for which commercial standards are available. (B) Tandem mass spectra for all peaks present at m/z 313.2384, excluding the two peaks for which standards were available. Tandem mass spectra were clustered against known oxylipins, with top 7 correlations shown. (C) Comparison of tandem mass spectra from m/z 313.2384 chromatographic peak at 3.27 minutes with that of 9,10-DiHOME, revealing similar chemical structures.
Chemical Networking for Classification of Putative Known Oxylipins
The lack of commercial standards or reference tandem mass spectra have to date precluded identification or even chemical classification of unknown metabolites, including oxylipins. To overcome this limitation, we reasoned that chemical similarity between known and unknown molecules through comparison of tandem mass spectral fragmentation patterns may allow for putative classification of some of these unknown molecules as oxylipins. To perform this analysis, we utilized chemical networking of spectral fragmentation patterns for which tandem mass spectra for each unknown signal were compared to reference spectra from known compounds and searched for the presence of both identical mass fragments as well as similar mass shifts between fragment peaks (Wang et al., 2016; Watrous et al., 2012). This approach allows experimental tandem mass spectra to be matched to identical spectra from reference compounds for definitive identification, as well as in the absence of definitive identification, to be matched to related but non-identical compounds for putative chemical classification. To determine the utility of chemical networking for chemical classification, tandem MS fragmentation patterns were obtained for 225 oxylipin commercial standards and 600 diverse, non-oxylipin standards (Table S-1). Fragmentation patterns from these standards were chemically networked against over 50,000 metabolites for which spectral fragmentation data were publicly available, which included additional publicly-deposited tandem mass spectra for the same 225 commercial oxylipin standards previously collected by other laboratories. From this analysis, experimentally derived tandem mass spectra for 205 of the 225 oxylipin standards were found to chemically network to corresponding spectra for the same compound present in the public library suggesting a sensitivity for matching of 86%. For those 33 of the 225 standards that did not match, manual inspection of the spectra revealed poor fragmentation of the parent ion where often only the parent ion and loss of water were present in the fragmentation spectra. This precluded chemical matching due to a minimum of 6 matched fragment peaks being required for a match. Among the 600 non-oxylipin standards, none were found to chemically network with oxylipin spectra and were instead found to be chemically similar to one of 50,000 other molecules present in the public libraries, suggesting near perfect specificity for this approach. This specificity for chemical matching is consistent with the fact that oxylipins, particularly cyclized variants, exhibit unique fragmentation mechanisms compared to other classes of compounds (Brose et al., 2011; Dickinson and Murphy, 2002; MacMillan and Murphy, 1995; Murphy, 2015; Murphy et al., 2005; Wheelan et al., 1996). Importantly, this high specificity is critical in limiting false positive matches when extending the chemical networking to unknown molecules for classification as oxylipins.
We first applied this chemical networking approach to m/z 313.2384 which exhibited two identifiable oxylipin peaks and 18 unknown isobaric features (Figure 2A). When tandem mass spectra from these chromatographic features at were subjected to the chemical networking strategy, 16 of the 18 features were specifically matched to one of the 225 oxylipin standards (Figure 2B). Matched tandem mass spectral pairs were manually inspected and found to be consistent with sharing a common base structure. Figure 2C shows one such comparison between the tandem mass spectra for 9,10-diHOME and an unknown isobaric peak at retention time 3.13 minutes for which nearly identical fragmentation patterns are observed, indicating a very similar chemical structure. As such, matching of unknown compounds based on accurate mass and analogous fragmentation patterns to a known oxylipin may allow such molecules to be classified as putative known oxylipins. This approach suggests that chemical networking of unknown molecules to known chemical entities may be utilized to identify and study additional, high confidence, putative compounds in human circulation.
We next extended this chemical networking to all 745 spectral features identified as potential oxylipins, based on accurate mass match to LIPID MAPS database entries, but for which no chemical standard or reference spectra was available. The results from the search were visualized as a mass spectral network (Figure 3A), where top scoring, high confidence, chemical matches between experimental and reference fragmentation data were clustered together and displayed in order to represent chemical relationships among compound families. Of the 745 unknown experimental tandem mass spectra, 362 spectra were chemically networked with at least one oxylipin standard and did not form secondary connections with any of the ~50,000 non-oxylipin compounds present in the library repository, thereby classifying these 362 metabolites as putative known oxylipins (Table S-2). The remaining 383 tandem mass spectra that did not chemically network with a known standard were mostly clustered with either non-oxylipin compounds (such as glucuronides, sterols, bile acids, and fatty acid ethyl esters) or clustered within themselves. The lack of similarity in tandem MS fragmentation patterns with any reference spectra thereby suggest that these 383 remaining chromatographic peaks cannot be considered as oxylipin in nature without further evidence.
Figure 3: Chemical networking for discovery of oxylipins.
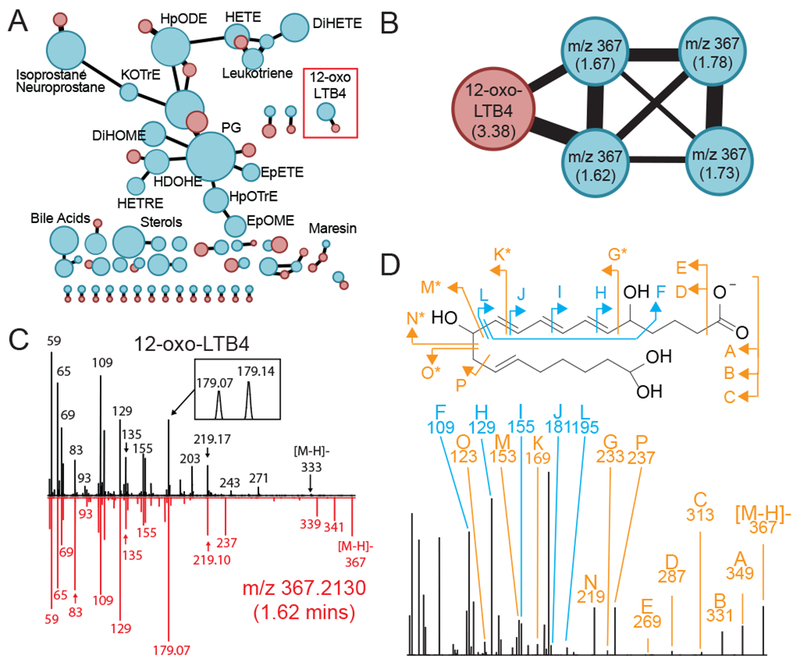
(A) Tandem mass spectra for the 745 chromatographic features matching to chemical formulas of known oxylipins was clustered against tandem mass spectra for the 225 commercial standards with the resulting correlations displayed as a nested molecular network. Here, red nodes indicate tandem mass spectra for commercial standards with blue nodes indicating tandem mass spectra from the 745 unknown compounds, with the size of the nodes reflecting number of unique spectra contained within each node. (B) Expanded view of the 12-oxo-LTB4 node (red box from 3A) showing the observed m/z and retention time in minutes within parenthesis. (C) Overlay of tandem mass spectra from 12-oxo-LTB4 and m/z 367 at 1.62 minutes showing high degree of similarity. (D) Manual annotation of spectral fragments of m/z 367 at 1.62 minutes and how they relate to the structure of 12-oxo-LTB4 with blue arrows indicating identical fragments and orange arrows indicating a shift of +CH4O between carbons 11 and 13. See also Data S-1.
Tandem mass spectral pairs from the chemical networking of 362 putative knowns with the 225 reference standards were manually inspected and found to be consistent with both compounds sharing a common key substructure. For instance, in the molecular tandem mass spectral network, 12-oxo LTB4 was clustered with spectra from four other unknown signals (Figure 3B). When the spectrum for 12-oxo LTB4 and one of these unknowns was compared, it was observed that their patterns were highly similar thus indicating a common chemical structure (Figure 3C–D). Importantly, algorithms for automated in silico fragmentation, such as CFM-ID (Allen et al., 2014) and MetFrag (Ruttkies et al., 2016), were not able to accurately predict fragments for these oxylipins, especially cyclized variants such as prostaglandins (Figure S-2), given the unpredictable nature of their fragmentation (Brose et al., 2011; Cui et al., 2008; Dangi et al., 2009; MacMillan and Murphy, 1995; Murphy, 2015; Murphy et al., 2005; Song et al., 2013; Wheelan et al., 1996; Yi et al., 2007). However, these irregular fragmentation mechanisms favor a chemical networking approach as they allow for far fewer false positive matches to non-oxylipin compounds. All 362 spectral matched were manually inspected to confirm similarity with 100 of these matches plotted in Data S-1. Collectively, these analyses suggest that directed non-targeted LC-MS/MS in conjunction with chemical networking may be utilized to identify an additional 362 putative known oxylipins in human circulation, thereby greatly extending the analytical breath of current approaches.
Systems Chemistry for Identification of Unknown Oxylipins
In addition to the ~500 known and putative known oxylipins found in human plasma, we sought to determine if unknown oxylipins, yet undescribed in any chemical database including LIPID MAPS, were also present in human circulation. Given their unique chemical properties, we reasoned additional systems chemistry approaches may prove useful in identification of unknown oxylipins. Across the 1125 theoretical compounds in the LIPID MAPS database, the extent of these modifications is nearly combinatorial where compounds exist at almost every Hn+2 and/or On+1 formula increment. These chemical patterns within compound families may be visualized by plotting the nominal mass versus the mass defect, here defined as the non-whole number value of the monoisotopic mass, for each compound within the family.(Sleno, 2012) For instance, among the prostaglandin family of eicosanoids, this type of plot reveals four distinct chemical series (Figure 4A), where each series is incremented by addition of two hydrogens and the distance between each series represents the addition of a single oxygen. When extending this analysis beyond prostaglandins, distinct gaps in these formulaic patterns were readily observed. Among docosanoids starting at formula C22H34O2, database entries were present for every addition of oxygen up to C22H34O6 with the lone exception of C22H34O3 (Figure 4B). When this formula’s calculated nominal mass of m/z 345.2435 was searched within the non-targeted LC-MS data collected from human plasma, a spectral peak at 4.72 minutes was observed and produced a tandem MS pattern that was nearly identical with that of the known docosanoid 10-HDoHE (Figure 4C). Manual interpretation of the fragments showed that while 10-HDoHE has four double bonds between carbons 11 and 22, this potentially novel oxylipin only has three. This result indicated that the gaps in chemical series within oxylipin database entries may be utilized to identify putative novel oxylipins.
Figure 4: Systems chemistry for identification of unknown oxylipins.
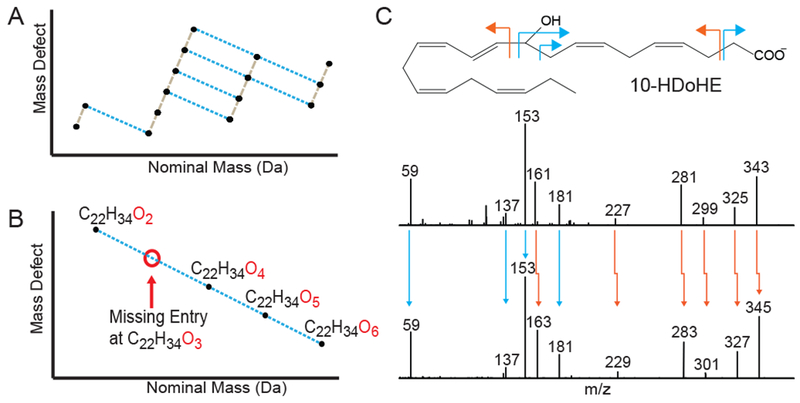
(A) Mass defect versus nominal mass plot of 15 chemical formulas, which compose the prostaglandin series of eicosanoids, illustrating common chemical patterns within compound families. (B) Example of a series of chemical formulas that exhibit a gap, or missing formula entry, in the overall pattern. Nontargeted MS data was searched for these formula masses to determine if chromatographic features were present. (C) Tandem mass spectra from one of these missing entries (C22H34O3) was found to be highly similar to the tandem mass spectra for 10-HDoHE with both sharing common fragments (blue arrows) as well as analogous fragments that were shifted by the mass of two hydrogens (orange arrows). Manual annotation indicates that the potentially novel oxylipin is identical to 10-HDoHE with the exception that it contains 3 instead of 4 double bonds between carbons 11 and 22. See also Figure S-3 and Data S-2.
To determine if additional unknown oxylipins were present in human plasma, all 1125 theoretical compounds listed in the LIPID MAPS database were plotted as nominal mass versus their mass defect and computationally analyzed for missing values along Hn+2 and On+1 series (Figure S-3). This analysis was extended to include one entry preceding the lowest formula value in the series as well as one entry past the final value as long as the number of oxygens was constrained between 3 and 8 and the number of hydrogens was between 24 and 40, which represents the maximum and minimum values observed for known oxylipins of 18 to 22 carbons. This analysis revealed 214 missing unique formulas spread across almost all compound subfamilies with each one representing a potential unknown oxylipin. When the non-targeted LC-MS data from human plasma was searched for compounds matching the accurate masses for these 214 missing chemical formulas, 184 distinct chromatographic peaks present at 45 of the 214 chemical formulas were found with each of these peaks having accurate mass (< ±2 ppm) matching to a theoretical formula, a retention time range (1.0 – 5.75 minutes), and a peak shape (FWHM < 0.1 minutes and tailing factor 0.8 – 1.2) consistent with known oxylipins. To determine if these peaks were structurally related to known compounds, targeted tandem mass spectra was collected for each of the 184 chromatographic peaks and chemical networking was performed. The resulting analysis revealed that 46 of the 184 putative compounds clustered with at least one known oxylipin standard and none of the 50,000 non-oxylipin metabolites, indicating structural similarity is present between these putative compounds with known members of oxylipin families (Table S-2). Each of the 46 spectral pairs resulting from the chemical networking search were manually inspected and found to be consistent with those of the reference standards with five examples shown in Data S-2. One such example is an unknown compound m/z 347.2600, which corresponds to the chemical formula C22H36O3, and was spectrally matched as an analog to the octadecanoid 13-HoTrE (Data S-2). Comparison of the tandem MS fragmentation patterns between the unknown and known compounds revealed that all carboxyl-containing fragments from carbons C7 to C1 were identical between the two compounds while carboxyl-containing fragments from carbons C12/C13 to C1 were shifted by 54.0472 daltons. This shift indicates that between carbons C12 and C7 there is an additional C4H6 present in the unknown compound. This systems chemistry approach thereby enables discovery of potentially novel oxylipins, identifying an additional 46 putative compounds in human circulation.
Oxylipins may potentially undergo non-enzymatic oxidation during sample isolation and storage, as well as during metabolite extraction and analysis (Dorow et al., 2016; Goodfriend et al., 2007; Kamlage et al., 2014; Slatter et al., 2016). To ensure that observed known and putative oxylipins were not the result of non-enzymatic oxidation, fresh human plasma was collected in EDTA tubes with great care to avoid cell lysis, processed for plasma isolation within minutes, immediately flash frozen, and analyzed for oxylipins within twenty-four hours of collection. Additionally, samples were prepared both with and without the addition of the antioxidant butylated hydroxytoluene (BHT) throughout the sample preparation process (see Methods). Resulting data showed 313 known and putative oxylipins were observed in the fresh plasma sample with minimal change in signal presence and stability due to the addition of BHT during the sample preparation process. Only four oxylipins were observed solely in the non-BHT sample and eight were observed solely in the BHT treated sample, with the median percent change between BHT and non-BHT treated samples of 4.6% (Figure S-4). Additionally, preparation and analysis of 96 replicate fresh plasma samples revealed a median correlation of variance (CV) of 7.9% over the sample run (Figure S-4). Collectively, this data suggests that non-enzymatic oxidation during sample collection, preparation, or analysis is unlikely contributing significantly to plasma oxylipin formation.
Association of Eicosanoids and Related Oxylipins with Human Inflammation
Oxylipin lipid mediators play critical roles in regulating systemic pro- and anti-inflammatory responses in the setting of both acute infection and chronic inflammation (Dennis and Norris, 2015; Khanapure et al., 2007; Levy et al., 2001). To date, however, biological functions have only been described for a fraction of the ~150 commonly studied oxylipins found in human plasma. To determine whether the hundreds of proposed putative known and putative novel oxylipins observed herein may have a role in inflammation, plasma samples previously collected from 1500 fasting, community-dwelling individuals were assayed using our directed, non-targeted LC-MS approaches (Table 1). Definitive known, putative known, and putative novel oxylipins were mapped to plasma samples by matching of retention time, accurate mass, and tandem MS fragmentation patterns, with inclusion of metabolites previously observed in pooled plasma and present at above signal-to-noise thresholds in >95% of individuals. Multiple hypothesis correction was performed using standard false discovery rate (FDR) (Benjamini and Hochberg, 1995), and oxylipins were statistically associated with common clinical features linked to systemic inflammation, including advancing age, obesity (measured by body mass index [BMI]), and circulating high sensitivity C-reactive protein (CRP) levels, a well-established marker of systemic inflammation (Figure 5) (Pasceri et al., 2000; Pepys, 1981). For both clinical features and CRP measures, dozens of both known and putative oxylipins were found to be highly related, at ‘metabolome-wide’ significance thresholds (Figure 5). Results were highly similar when dichotomized variables were used for age, BMI and CRP (Figure S-5 and Table S-3). For each clinical phenotype, both positive and negative associations were observed (Table S-3), suggesting both pro- and anti-inflammatory activities, consistent with the described function of oxylipins. For instance, production of arachidonic acid metabolites (e.g. HETE’s and HpETE’s) showed positive correlation with BMI and CRP levels, suggesting a pro-inflammatory role for these molecules, while DiHDPA and DiHETrE metabolites showed negative correlations, suggesting an anti-inflammatory role; similar patterns have been observed in cellular systems upon acute activation of inflammatory pathways (Askari et al., 2014). Moreover, particular putative compounds, such as m/z 327.2176 were found to be commonly associated with multiple correlates of inflammation (FDR-corrected p-value 1e−31 for BMI, 3e−24 for CRP), whereas other putative compounds, such as m/z 349.2020 (FDR-corrected p-value 1e−35), m/z 327.2162 (FDR-corrected p-value 4e−18), and m/z 347.2590 (FDR-corrected p-value 5e−11) were specifically associated with age, BMI or CRP levels, respectively, suggesting both universal and specific mediators of inflammation. Interestingly, for all three clinical phenotypes examined, a number of putative known and putative novel compounds were found to be highly associated, highlighting the discovery potential of these directed, non-targeted approaches.
Table 1: Clinical demographics for human studies.
Values are displayed as means ± SD, frequencies, or median and 25th, 75th percentiles.
| Characteristic | Total Sample (N=1500) |
|---|---|
| Age, years | 66.1 ± 8.8 |
| Women, % | 52.7 |
| Body mass index, kg/m2 | 28.5 ± 5.6 |
| C-reactive protein | 1.57 (0.76, 3.29) |
Figure 5: Oxylipin correlates of human inflammation.
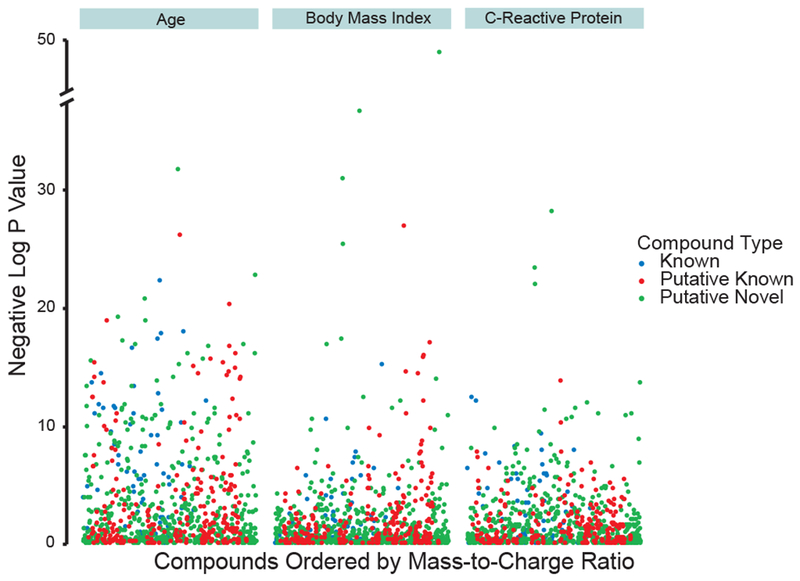
Associations of known (blue), putative known (red), and putative novel (green) circulating plasma oxylipins with age, body mass index (BMI), and C-reactive protein (CRP) across 1500 individuals. Data points are visualized using a modified Manhattan plot, with FDR-corrected p-values detailing association between metabolite abundance and clinical features. See Figure S-5.
DISCUSSION
In this report, we describe approaches for the assay and identification of oxylipins in human plasma using directed, non-targeted LC-MS coupled to computational chemical networking. While traditional targeted methods for studying these compounds can produce very sensitive and quantitative measurements, directed, non-targeted analysis greatly expands upon the spectrum of species measurable in complex organisms, enables discovery of hundreds of unknown oxylipins in humans, and provide a foundation for new insight into the role of bioactive lipids in mediating human health and disease. This approach makes use of directed extraction whereby samples were prepared using methods designed specifically for isolation of oxylipins (versus a general Bligh-Dyer lipid extraction), chromatographic separation designed to separate the extensive array of isomers, non-targeted mass spectrometric measured allowing capture of all possible species, and MS/MS chemical networking allowing for identification of previously unreported compounds. Application of these approaches to human plasma revealed hundreds of putative compounds, advancing upon the current state of the art analytical techniques and greatly expanding the current chemical repertoire, revealing 134 definitively known, 362 putative known, and 46 putative novel oxylipins observable in human plasma. These molecules were validated by a number of methodologies, including extensive chemical networking of mass spectral fragmentation patterns and manual annotation of a subset of putative oxylipins. These methods were also found to have high specificity for the target compound class given the unique and irregular mechanisms by which they fragment. Many of the putative compounds described herein were found to be highly associated with both pro- and anti-inflammatory clinical characteristics and measures in a cohort of 1500 individuals. Tandem mass spectral data for these compounds as well as LC-MS parent mass data for the 1500 plasma samples have been provided in a database as a resource to the scientific community for further study and discovery (ftp://massive.ucsd.edu/MSV000082852). Additionally, as many other chemical compound classes exhibit similar unique chemical patterning as observed in oxylipins, such as PUFAs, endocannabinoids, FAHFAs and sterols, this approach may be adapted to perform similar discovery based work in these other chemical families, thereby potentially expanding their number of measurable entities.
While greatly expanding upon the number of compounds reported in human plasma, the current experimental and computational approaches will continue to shed even more insight into oxylipin biology as they are applied in complementary systems. While the current study focuses on human plasma, undoubtedly additional oxylipins will be found localized to particular cells or tissues, or induced under particular biological conditions. Moreover, it is important to note that since our approach relies on comparing tandem MS fragmentation patterns to those of known oxylipins, our approach excludes putative compounds with completely unique fragmentation patterns and it is therefore likely that a number of additional oxylipins await discovery. Similarly, while human studies across 1500 individuals focused upon those metabolites that were initially observed in pooled human plasma, a number of additional potential oxylipins were also observed in specific individuals and are currently being investigated. Future human studies and further samples will continue to validate these additional signals.
Importantly, oxylipins are susceptible to non-enzymatic processing, including oxidation, and are therefore sensitive to specific collection, storage, processing and analysis methodologies (Dorow et al., 2016; Goodfriend et al., 2007; Slatter et al., 2016). For all samples included in these studies, only EDTA anticoagulant was used during blood collection and plasma storage. Moreover, care was taken using standardized collection protocols to ensure rapid blood processing and plasma isolation following collection with appropriate long term cold storage at −80 °C until analysis. Notably, comparison of previously collected and frozen plasma samples (Figure 5) with fresh plasma preparations revealed that 76% of observed putative oxylipins in frozen samples were also observed in fresh samples, suggesting that assayed oxylipin metabolites were not due non-enzymatic oxidation during years of storage (data not shown). Furthermore, addition of antioxidants minimally influenced the presence or levels of oxylipins (Figure S-4). Similarly, repeat sampling and analysis over a 96-well microtiter plate revealed minimal decrement in metabolite signals or production of oxidized byproducts over time (Figure S-4). While these data collectively suggest that artefactual, non-enzymatic modification of oxylipins during sample collection, storage, processing and analysis does not likely contribute significantly to the species observed herein, such processes cannot be ruled out completely and must be considered with follow-up biological experiments.
As commercial standards are only available for a fraction of unique theoretical oxylipins, targeted studies detailing the role of these molecules in inflammation have largely been confined to this subset of metabolites, despite potentially hundreds more species present in human circulation. As many of these compounds are likely modulated under particular conditions such as acute systemic inflammation or during development and wound repair or may occur in particular tissues and cells in a localized manner, their study is critical for advancing our understanding of these complex signaling processes. Furthermore, as a number of these putative metabolites were found to be highly associated with clinical characteristics and markers of inflammation, future studies will aim to further characterize these oxylipins including identifying the biochemical pathways responsible for their production and degradation, localizing individual species to specific tissues of origin, establishing causal pro- and anti-inflammatory molecules, and elucidating their downstream signaling mechanisms. Importantly, the studies described herein highlight the discovery potential of directed, non-targeted LC-MS and corresponding computational chemical networking approaches, underscore the breath, complexity and diversity of oxylipins present in humans, and provide a foundation for new insight into the role of bioactive lipids in mediating human health and disease.
Significance
There is great interest in mapping of the small molecule signals that modulate cellular processes. Key among these signaling molecules are eicosanoids and related oxylipin metabolites that derive from polyunsaturated fatty acids and mediate a number of diverse homeostatic functions in humans including systemic inflammation, cellular development, ion transport, muscle contraction, vascular tone, and thrombosis, among many others. Despite more than 1000 oxylipins predicted to exist, only a small fraction have been monitored in complex organisms to date, owing to technical limitations with targeted measurement strategies. Herein, using a directed non-targeted mass spectrometry approach in conjunction with computational chemical networking of spectral data, we greatly expand the breadth of oxylipins observable in humans, discovering hundreds of previously undescribed oxylipin species. Moreover, we find that a number of these oxylipins are strongly associated with markers of systemic inflammation in humans or clinical characteristics linked to inflammation, including advancing age and increasing obesity. These analytical approaches greatly expand the spectrum of oxylipin metabolites monitored in humans and provide a foundation for new insight into the role of bioactive lipids in mediating health and disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mohit Jain (mjain@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fasting, EDTA prepared plasma specimens collected in 2005-2008 from N=1500 participants (mean age 66.2±9.0 years, 55% women) of the community-based Framingham Heart Study Offspring Cohort (Table 1) (Kannel and McGee, 1979), were used for analyses of oxylipins in human inflammation. All participants provided written informed consent. All study protocols were approved by the Brigham and Women’s Hospital, Boston University School of Medicine, National Institute for Health and Welfare (Finland) and UC San Diego Institutional Review Boards. At the time of specimen collection, the participants underwent a medical history, physical, and laboratory assessment for high sensitivity C-reactive protein (CRP). CRP was measured using an immunoturbidometric method on a Roche Cobas 501.
METHOD DETAILS
Chemicals and consumables
Non-deuterated and deuterated chemical standards (See Table S-1) were purchased from Cayman Chemical and IROA Technologies. LCMS grade solvents used for sample preparation and metabolomic analysis including methanol, acetonitrile, water, and isopropanol were purchased from Honeywell International Inc while LCMS grade ethanol and acetic acid were purchased from Sigma-Aldrich. All pipetting instruments and consumables were purchased from Eppendorf. For sample preparation, 250μL V-bottom, 500μL V-bottom, and 1.2mL deep well 96-well plates were purchased from Thermo Scientific, Axygen and Greiner, respectively. LCMS amber autosampler vials and tri-layer vial caps were purchased from Agilent Technologies and 300μL glass inserts were purchased from Wheaton. Strata-X polymeric 96-well (10mg/well) solid phase extraction plates as well as Kinetex C18 1.8μm (100 × 2.1 mm) UPLC columns were purchased from Phenomenex Inc. UPLC BEH RP-18 guard columns were purchased from Waters Inc. EZ-Pierce 20μm aluminum foil was purchased from Thermo Scientific. Pooled human plasma was obtained from BioreclamationIVT.
LC-MS and spectral data handling
Directed, non-targeted LC-MS was performed using a Thermo Vanquish UPLC coupled to a Thermo QExactive Orbitrap mass spectrometer. For data processing, in-house R-scripts were used to perform initial bulk feature alignment, MS1-MS2 data parsing, pseudo DIA-to-DDA MS2 deconvolution, and CSV-to-MGF file generation. RAW to mzXML file conversion was performed using MSconvert version 3.0.9393 (Chambers et al., 2012). Feature extraction, secondary alignment, and compound identification were performed using both mzMine 2.21 (Pluskal et al., 2010). Statistical analysis was performed using R (3.3.3).
Plasma oxylipin extraction
For preparation of fresh and commercial plasma, EDTA prepared plasma samples were thawed at 4 °C over 8 hours in light free conditions. Once thawed, plasma was mixed thoroughly by orbital shaking at 500 rpm at 4 °C for 15 minutes. Cold (−20°C) ethanol containing 20 deuterated oxylipin internal standards was used to precipitate proteins and non-polar lipids as well as extract bioactive lipids from the plasma through the addition of 1:4 plasma:ethanol-standard mixture to each well, shaking at 500rpm at 4 °C for 15 minutes, and centrifugation at 4200 rpm at 4 °C for 10 minutes. To allow for more efficient SPE loading, 65μL of supernatant was transferred to an Axygen 500μL V-bottom 96-well plate where each well contained 350μL of water. A second extraction of the protein pellet was performed by gently adding 65μL of −20 °C ethanol (containing no standards), gently hand vortex mixing for 30 seconds, and transferring another 65μL aliquot of supernatant to the same Axygen V-bottom 96-well plate, at which point samples are ready for SPE plate loading. The Phenomenex Strata-X polymeric 10mg/mL 96-well SPE plate was washed by sequentially adding and vacuum assisted eluting 600 μL of methanol and 600μL of ethanol and equilibrated by addition of 1.2mL of water per well. All solvents were added while taking care not to let wells run dry between pull through. The entire volume of the water-diluted samples was added to the SPE wells and allowed to gravity elute. Once the sample volume reached the top of the SPE bed, 600μL of 90:10 water:methanol was added to each well and slowly pulled through (5 mmHg vacuum) until no liquid was visualized in the wells. Wells were further dried by increasing vacuum to 20 mmHg for 60 seconds. Bound metabolites were then eluted into an Axygen 500μL V-bottom 96-well plate by addition of 450μL of ethanol to each SPE plate well, with slow pull through (5 mmHg vacuum) after 120 second equilibration. Eluent was immediately dried in vacuo using a Thermo Savant vacuum concentrator operated at 35 °C. Once dry, samples were resuspended in a solution containing 40:60:0.1 methanol:water:acetic acid as well as 10μM CUDA internal standard by addition of 50μL of solution to each well, immediately sealing the plate, vortex mixing at 500rpm at 4 °C for 10 minutes. Samples were immediately transferred to a Greiner deep-well 96-well plate with each well containing a Wheaton 300μL glass insert (to prevent the rapid degradation of oxylipin signals typically observed when they are prepared in polypropylene plates). The sample plate was then immediately sealed and centrifuged at 500 rpm at 4 °C for 5 minutes to remove any air bubbles that might have formed within the glass inserts. Samples were stored at less than 2 hours at 4 °C until LC-MS/MS analysis. For samples prepared with BHT, 0.05% BHT was added to the ethanol used for protein precipitation, the ethanol used to elute bound oxylipins off the SPE plate, and the 40:60:0.1 methanol:water:acetic acid resuspension mixture.
LC-MS/MS data acquisition
Once prepared, 20μL of sample was injected onto a Phenomenex Kinetex C18 (1.7μm particle size, 100 × 2.1 mm) column and separated using mobile phases A (70% water, 30% acetonitrile and 0.1% acetic acid) and B (50% acetonitrile, 50% isopropanol, 0.02% acetic acid) running the following gradient: 1% B from −1.00 to 0.25 minutes, 1% to 55% B from 0.25 to 5.00 minutes, 55% to 99% B from 5.00 to 5.50 minutes, and 99% B from 5.50 to 7.00 minutes. Flow rate was set at 0.375 mL/min, column temperature and mobile phase pre-heater was set at 50 °C, needle wash was set for 5 seconds post-draw using 50:25:25:0.1 water:acetonitrile:isopropanol:acetic acid. Mass detection was performed using a Thermo QExactive orbitrap mass spectrometer equipped with a heated electrospray ionization (HESI) source and collision-induced dissociation (CID) fragmentation. HESI probe geometry was manually optimized about the x, y and z axis by infusing CUDA and PGA2 standards into the mobile phase stream at 1%, 50% and 99% B. Final geometry was probe height of 0.75 B-C (between B and C markers about 3/4 the way to C), x-offset of 1 mm left of center, and a Y-offset of 1.70 according to the micrometer. Source settings used for all samples were: negative ion mode profile data, sheath gas flow of 40 units, aux gas flow of 15 units, sweep gas flow of 2 units, spray voltage of −3.5kV, capillary temperature of 265 °C, aux gas temp of 350 °C, S-lens RF at 45. Data was collected using an MS1 scan event followed by 4 DDA scan events using an isolation window of 1.0 m/z and a normalized collision energy of 35 arbitrary units. For MS1 scan events, scan range of m/z 225-650, mass resolution of 17.5k, AGC of 1e6 and inject time of 50ms was used. For tandem MS acquisition, mass resolution of 17.5k, AGC 3e5 and inject time of 40ms was used. Once target oxylipins were discovered, additional tandem MS spectra were collected in a targeted fashion using Inclusion Lists to obtain high quality spectra for each compound.
Chemical networking of tandem MS spectra
Tandem mass spectra were collected in a targeted fashion for all observed chromatographic peaks in pooled plasma matching all known, putative known and putative novel oxylipins based on accurate mass (m/z for [M-H]-within ±5 ppm mass error). Tandem mass spectra for each chromatographic peak were averaged, extracted and saved as individual MGF files. These MGF files were uploaded to GNPS (http://gnps.ucsd.edu) for chemical networking analysis (Wang et al., 2016). Analysis was performed using both Networking and Dereplication workflows. For Networking the following thresholds: Precursor Ion Mass Tolerance of +/− 0.05 Da, Fragment Ion Mass Tolerance of +/−0.05 Da, Minimum Cosine Score of 0.60, Minimum Matched peaks of 6, TopK of 10, Maximum Connected Component Size of 500, Minimum Cluster Size of 1, Library Score Threshold of 0.85, Minimum Matched Library Peaks of 6, Analog Search turned On, Maximum Analog Mass Difference of 85 Da, Filter Precursor Window turned On, all other filters turned off. For Dereplication the following thresholds were used: Precursor Ion Mass Tolerance of +/− 0.05 Da, Fragment Ion Mass Tolerance of +/−0.05 Da, Minimum Cosine Score of 0.60, Minimum Matched peaks of 6, Analog Search turned On, Maximum Analog Mass Difference of 85Da, Filter Precursor Window turned On, all other filters turned off. All primary and third-party databases, including our in-house database from the 225 oxylipin commercial standards were used for matching. For all matches to library reference spectra, only the top scoring hit was considered. All resulting matches were manually checked for consistency in fragmentation patterns between the library reference spectra and the experimental spectra.
Analysis of oxylipins in human inflammation
The intra- and inter-assay CVs for CRP were 2.5% and 4.5%, respectively. All human plasma was collected and prepared within thirty minutes of collection by centrifugation for 15 minutes, followed by immediate aliquoting and storage at −80 °C. Plasma samples were visually inspected for evidence of cell lysis prior to preparation. Frozen plasma was prepared and LC-MS analysis performed as described above. All data was converted to 32-bit mzXML data format using MSconvert version 3.0.9393 (part of the Proteowizard software suite). Initial bulk retention time correction was performed as described previously (Watrous et al., 2017) whereby all data files for a given sample set were loaded into Mzmine 2.21 and the sample specific retention times for all deuterated internal standards as well as approximately 50 endogenous landmark peaks (defined as chromatographic features with %CV < 50% across all samples, minimum peak height of 500,000 counts and a no isobaric peaks within +/− 30 seconds of target) were exported and used to model non-linear retention time drift within each sample using cubic smoothing splines with 8 to 16 degrees of freedom within the model. Using these sample specific models, the retention times for all MS1 and MS2 scans with each mzXML file were adjusted so that the maximum drift observed was reduced from (on average) 0.15 minutes to 0.025 minutes across the 1500 patient sample injections. Parameters used for Mzmine peak extraction are as follows: Mass Detection threshold of 250k counts, Chromatogram Builder 0.025 minute min time span, min height of 500k counts and m/z tolerance of ±5 ppm, Chromatogram Deconvolution (Local Minimum Search) chromatographic threshold of 80%, search minimum RT range of 0.03 minutes, minimum relative height of 0.4%, min height of 500k counts, top-edge ratio of 1.2, peak duration of 0.025 to 0.5 minutes, Join Aligner m/z tolerance of ±5ppm, RT tolerance of 0.4 minutes, weight for retention and mass both at 90, Peak List Rows Filter retaining only peaks present in at least 50% of data files. Custom Library Search for all known, putative known and putative novel oxylipins was performed using an RT tolerance of 0.1 minutes and a mass tolerance of ±5 ppm. Resulting features were manually denoised by visual inspection using the Mzmine peak list viewer where features exhibiting abnormal/poor peak shapes, inconsistent peak shapes and/or drastic shifts in retention time were deleted.
Exported peak lists for each file group were normalized by centering the batch mean for each m/z-feature to the global mean for all batches within the run set. Once normalized, features present in the MS1 data were filtered based on signal reliability by the used of the ‘dtwclust’ R-module as described previously where essentially all features were hierarchical clustered and automatically parsed based on patterns exhibited within their intensity profile across all samples when plotted with respect to injection order with features exhibiting stable patterns being retained while those exhibiting patterns indicative of chemical instability, misalignment or highly random distributions were removed.
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical analyses of oxylipin data, we constructed linear regression models relating each of the known, putative known, and putative novel metabolites with factors previously associated with systemic inflammation in humans, including advanced age, elevated body mass index (BMI), and increased C-reactive protein levels (Log CRP). We also performed comparative analyses in which dichotomous variables were used for age (<65 versus >65 years), BMI (<30 versus >30 kg/m2). and CRP (<3 versus >3 mg/dL). For all statistical analyses, we corrected for multiple testing using false discovery rate (FDR) (Benjamini and Hochberg, 1995). Prior to entry into regression analyses, oxylipin variables were logarithmically transformed and standardized. Statistical analyses were performed using R v3.2.2 (R Foundation for Statistical Computing, Vienna, Austria).
DATA AND SOFTWARE AVAILABILITY
Mass spectrometry data represented in this study can be found online at the UCSD MassIVE repository (ftp://massive.ucsd.edu/MSV000082852)
ADDITIONAL RESOURCES
N/A
Supplementary Material
HIGHLIGHT.
Directed nontargeted LC-MS/MS analysis of oxylipins in human plasma
Mass spectral networking reveals hundreds of unreported oxylipins in plasma
Systems chemistry analysis identifies potentially unknown circulating oxylipins
Newly reported oxylipins show correlation with markers of systemic inflammation
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (K01DK116917 to J.D.W.; R01GM020501, R01DK105961, U19AI135972, P30DK063491, and U54GM069338 to E.A.D. and O.Q.; R01HL134168 to S.C.; R01DK112940, R01HL134811 and K24HL136852 to S.M.; and S10OD020025, R03AG053287, and R01ES027595 to M.J.), support from the NHLBI to the Framingham Heart Study (contracts N01HC25195; HHSN268201500001I), as well as generous awards from Foundations, including the American Heart Association (CVGPS Pathway Award to S.C., M.J.), the Doris Duke Charitable Foundation (#2015092 to M.J.), Tobacco-Related Disease Research Program (#24RT-0032 to M.J., 24FT-0010 to J.D.W.), UC San Diego Frontiers of Innovation Scholars Program (to K.A.L.), Emil Aaltonen Foundation (to T.N.), the Finnish Medical Foundation (to T.N.), and the Paavo Nurmi Foundation (to T.N.). The funders play no role in the design of the study; the collection, analysis, and interpretation of the data; and the decision to approve publication of the finished manuscript. All authors had full access to all the data (including statistical reports and tables) in the study and can take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allen F, Pon A, Wilson M, Greiner R, and Wishart D (2014). CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res 42, W94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari AA, Thomson S, Edin ML, Lih FB, Zeldin DC, and Bishop-Bailey D (2014). Basal and inducible anti-inflammatory epoxygenase activity in endothelial cells. Biochem Biophys Res Commun 446, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita G, Kendall AC, Dennis EA, and Nicolaou A (2015). Targeted lipidomic strategies for oxygenated metabolites of polyunsaturated fatty acids. Biochim Biophys Acta 1851, 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society 57, 289–300. [Google Scholar]
- Brose SA, Thuen BT, and Golovko MY (2011). LC/MS/MS method for analysis of E(2) series prostaglandins and isoprostanes. J Lipid Res 52, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski MW, Dumlao DS, and Dennis EA (2009). Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. J Lipid Res 50, 1015–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder PC (2010). Omega-3 fatty acids and inflammatory processes. Nutrients 2, 355–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra V, Rovati GE, Mangano P, Buccellati C, Murphy RC, and Sala A (2015). Transcellular biosynthesis of eicosanoid lipid mediators. Biochim Biophys Acta 1851, 377–382. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Isbell MA, Chawengsub Y, Falck JR, Campbell WB, and Nithipatikom K (2008). Structural characterization of monohydroxyeicosatetraenoic acids and dihydroxy- and trihydroxyeicosatrienoic acids by ESI-FTICR. J Am Soc Mass Spectrom 19, 569–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalli J, Colas RA, Quintana C, Barragan-Bradford D, Hurwitz S, Levy BD, Choi AM, Serhan CN, and Baron RM (2017). Human Sepsis Eicosanoid and Proresolving Lipid Mediator Temporal Profiles: Correlations With Survival and Clinical Outcomes. Crit Care Med 45, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi B, Obeng M, Nauroth JM, Teymourlouei M, Needham M, Raman K, and Arterburn LM (2009). Biogenic synthesis, purification, and chemical characterization of anti-inflammatory resolvins derived from docosapentaenoic acid (DPAn-6). J Biol Chem 284, 14744–14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, and Norris PC (2015). Eicosanoid storm in infection and inflammation. Nat Rev Immunol 15, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JS, and Murphy RC (2002). Mass spectrometric analysis of leukotriene A4 and other chemically reactive metabolites of arachidonic acid. J Am Soc Mass Spectrom 13, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Dorow J, Becker S, Kortz L, Thiery J, Hauschildt S, and Ceglarek U (2016). Preanalytical Investigation of Polyunsaturated Fatty Acids and Eicosanoids in Human Plasma by Liquid Chromatography-Tandem Mass Spectrometry. Biopreserv Biobank 14, 107–113. [DOI] [PubMed] [Google Scholar]
- Dreisbach AW, Smith SV, Kyle PB, Ramaiah M, Amenuke M, Garrett MR, Lirette ST, Griswold ME, and Roman RJ (2014). Urinary CYP eicosanoid excretion correlates with glomerular filtration in African-Americans with chronic kidney disease. Prostaglandins Other Lipid Mediat 113–115, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, and Dennis EA (2009). Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res 50 Suppl, S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forn-Cuni G, Varela M, Pereiro P, Novoa B, and Figueras A (2017). Conserved gene regulation during acute inflammation between zebrafish and mammals. Sci Rep 7, 41905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CD (2001). Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875. [DOI] [PubMed] [Google Scholar]
- Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, and Willoughby DA (1999). Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med 5, 698–701. [DOI] [PubMed] [Google Scholar]
- Goodfriend TL, Pedersen TL, Grekin RJ, Hammock BD, Ball DL, and Vollmer A (2007). Heparin, lipoproteins, and oxygenated fatty acids in blood: a cautionary note. Prostaglandins Leukot Essent Fatty Acids 77, 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene ER, Huang S, Serhan CN, and Panigrahy D (2011). Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat 96, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harizi H, Corcuff JB, and Gualde N (2008). Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med 14, 461–469. [DOI] [PubMed] [Google Scholar]
- Imig JD (2008). Eicosanoids and renal damage in cardiometabolic syndrome. Expert Opin Drug Metab Toxicol 4, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamlage B, Maldonado SG, Bethan B, Peter E, Schmitz O, Liebenberg V, and Schatz P (2014). Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clin Chem 60, 399–412. [DOI] [PubMed] [Google Scholar]
- Kannel WB, and McGee DL (1979). Diabetes and cardiovascular disease. The Framingham study. JAMA 241, 2035–2038. [DOI] [PubMed] [Google Scholar]
- Khanapure SP, Garvey DS, Janero DR, and Letts LG (2007). Eicosanoids in inflammation: biosynthesis, pharmacology, and therapeutic frontiers. Curr Top Med Chem 7, 311–340. [DOI] [PubMed] [Google Scholar]
- Levy BD, Clish CB, Schmidt B, Gronert K, and Serhan CN (2001). Lipid mediator class switching during acute inflammation: signals in resolution. Nat Immunol 2, 612–619. [DOI] [PubMed] [Google Scholar]
- Lynch KR, O’Neill GP, Liu Q, Im DS, Sawyer N, Metters KM, Coulombe N, Abramovitz M, Figueroa DJ, Zeng Z, et al. (1999). Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature 399, 789–793. [DOI] [PubMed] [Google Scholar]
- MacMillan DK, and Murphy RC (1995). Analysis of lipid hydroperoxides and long-chain conjugated keto acids by negative ion electrospray mass spectrometry. J Am Soc Mass Spectrom 6, 1190–1201. [DOI] [PubMed] [Google Scholar]
- Minderhout V (2000). Eicosanoids and Related Compounds in Plants and Animals (ed.s Rowley AF; Kuhn H; Schewe T). Journal of Chemical Education 77, 452. [Google Scholar]
- Murphy RC (2015). Tandem mass spectrometry of lipids : molecular analysis of complex lipids.
- Murphy RC, Barkley RM, Zemski Berry K, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, and Zarini S (2005). Electrospray ionization and tandem mass spectrometry of eicosanoids. Anal Biochem 346, 1–42. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Bratton DL, and Murphy RC (1997). Analysis of epoxyeicosatrienoic and monohydroxyeicosatetraenoic acids esterified to phospholipids in human red blood cells by electrospray tandem mass spectrometry. J Mass Spectrom 32, 888–896. [DOI] [PubMed] [Google Scholar]
- Norris PC, and Dennis EA (2014). A lipidomic perspective on inflammatory macrophage eicosanoid signaling. Adv Biol Regul 54, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PC, and Serhan CN (2018). Metabololipidomic profiling of functional immunoresolvent clusters and eicosanoids in mammalian tissues. Biochem Biophys Res Commun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Erb-Downward JR, and Huffnagle GB (2003). Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin Microbiol Rev 16, 517–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasceri V, Willerson JT, and Yeh ET (2000). Direct proinflammatory effect of C-reactive protein on human endothelial cells. Circulation 102, 2165–2168. [DOI] [PubMed] [Google Scholar]
- Pepys MB (1981). C-reactive protein fifty years on. Lancet 1, 653–657. [DOI] [PubMed] [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, and Oresic M (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppolo M, Varma D, and Jansen SA (2014). A review of analytical methods for eicosanoids in brain tissue. J Chromatogr B Analyt Technol Biomed Life Sci 964, 50–64. [DOI] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, et al. (2010). Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res 51, 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, and Dennis EA (2011). The human plasma lipidome. N Engl J Med 365, 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha JL, and Fernandez-Alonso J (2001). Acute tubulointerstitial nephritis associated with the selective COX-2 enzyme inhibitor, rofecoxib. Lancet 357, 1946–1947. [DOI] [PubMed] [Google Scholar]
- Ruttkies C, Schymanski EL, Wolf S, Hollender J, and Neumann S (2016). MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminform 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Horst RT, Jansen T, Jacobs L, Bonder MJ, et al. (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1897. [DOI] [PubMed] [Google Scholar]
- Shinomiya K, Fukunaga M, Kiyomoto H, Mizushige K, Tsuji T, Noma T, Ohmori K, Kohno M, and Senda S (2002). A role of oxidative stress-generated eicosanoid in the progression of arteriosclerosis in type 2 diabetes mellitus model rats. Hypertens Res 25, 91–98. [DOI] [PubMed] [Google Scholar]
- Simopoulos AP (2008). The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp Biol Med (Maywood) 233, 674–688. [DOI] [PubMed] [Google Scholar]
- Slatter DA, Aldrovandi M, O’Connor A, Allen SM, Brasher CJ, Murphy RC, Mecklemann S, Ravi S, Darley-Usmar V, and O’Donnell VB (2016). Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation. Cell Metab 23, 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleno L (2012). The use of mass defect in modern mass spectrometry. Journal of Mass Spectrometry 47, 226–236. [DOI] [PubMed] [Google Scholar]
- Song J, Liu X, Wu J, Meehan MJ, Blevitt JM, Dorrestein PC, and Milla ME (2013). A highly efficient, high-throughput lipidomics platform for the quantitative detection of eicosanoids in human whole blood. Analytical Biochemistry 433, 181–188. [DOI] [PubMed] [Google Scholar]
- Stanley D, and Kim Y (2014). Eicosanoid Signaling in Insects: from Discovery to Plant Protection. Critical Reviews in Plant Sciences 33, 20–63. [Google Scholar]
- Tilley SL, Coffman TM, and Koller BH (2001). Mixed messages: modulation of inflammation and immune responses by prostaglandins and thromboxanes. J Clin Invest 108, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsikas D, and Zoerner AA (2014). Analysis of eicosanoids by LC-MS/MS and GC-MS/MS: a historical retrospect and a discussion. J Chromatogr B Analyt Technol Biomed Life Sci 964, 79–88. [DOI] [PubMed] [Google Scholar]
- Vona-Davis L, and Rose DP (2013). The obesity-inflammation-eicosanoid axis in breast cancer. J Mammary Gland Biol Neoplasia 18, 291–307. [DOI] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Armando AM, Quehenberger O, Yan C, and Dennis EA (2014). Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J Chromatogr A 1359, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A 109, E1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Henglin M, Claggett B, Lehmann KA, Larson MG, Cheng S, and Jain M (2017). Visualization, Quantification, and Alignment of Spectral Drift in Population Scale Untargeted Metabolomics Data. Anal Chem 89, 1399–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan P, Zirrolli JA, and Murphy RC (1996). Electrospray ionization and low energy tandem mass spectrometry of polyhydroxy unsaturated fatty acids. J Am Soc Mass Spectrom 7, 140–149. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, et al. (2018). HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46, D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi XY, Gauthier KM, Cui L, Nithipatikom K, Falck JR, and Campbell WB (2007). Metabolism of adrenic acid to vasodilatory 1alpha,1beta-dihomo-epoxyeicosatrienoic acids by bovine coronary arteries. Am J Physiol Heart Circ Physiol 292, H2265–2274. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Izumi T, Chang K, Takuwa Y, and Shimizu T (1997). A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature 387, 620–624. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Kato K, Terawaki K, Izumi T, and Shimizu T (2000). A second leukotriene B(4) receptor, BLT2. A new therapeutic target in inflammation and immunological disorders. J Exp Med 192, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data represented in this study can be found online at the UCSD MassIVE repository (ftp://massive.ucsd.edu/MSV000082852)


