Abstract
Rationale.
Biological studies are conducted at ever increasing rates by relying on proteomic workflows. Although MS data acquisition is highly automated and rapid, sample preparation continues to be the bottleneck of developing high-throughput workflows. Enzymatic protein processing, in particular, involves time-consuming protocols that can extend from one day to another. To address this gap, we developed and evaluated simple, in-solution tryptic enzymatic reactions that unfold within a few minutes, and demonstrate the utility of the methodology for the rapid analysis of proteins originating from cancer cell extracts.
Methods.
Tryptic enzymatic reactions were conducted for 7–60 min, and the results were compared to that of a routine approach conducted for 18 h. No other reaction conditions were changed relative to the 18 h procedure. The reaction products were analyzed by nano-HPLC/MS/MS, and the quality of the products was assessed in terms of peptide/protein identifications, sequence coverage, peptide length, missed-cleavage sites, quality of generated ions, and peptide hydrophilic/hydrophobic properties.
Results.
The results demonstrate that brief, and therefore incomplete, enzymatic processes lead to a large number of peptide fragments that improve protein sequence and proteome coverage, that the tandem mass spectra produced from these peptides are of high quality for reliable protein identifications, and that the physical properties of peptides are prone to supporting the development of alternative multi-dimensional separations and middle-down proteomics analysis strategies. The reproducibility of generating the same peptides within a few minutes of enzymatic digestion was remarkably close to that obtained from 18 h long reactions, and the combined results of short and long reactions increased proteome coverage by ~40 %.
Conclusions.
We demonstrate that partial enzymatic reactions conducted on short time-scales represent a valuable asset to proteomic studies, and propose their implementation either as simple, cost-effective, stand-alone protocols for substantially streamlining the analysis of biological samples, or as complementary protocols, for improving protein sequence and proteome coverage.
Keywords: proteolytic digestion, fast analysis, proteomics, mass spectrometry
Introduction
Enzymatic digestion of cellular proteins is conducted according to protocols that seek complete and reproducible protein cleavage at particular amino acid sites. This is typically accomplished by first denaturing the protein samples with chaotropic agents (e.g., urea, guanidinium chloride]), breaking the disulfide bridges with a reducing agent [e.g., 1,4-dithiothreitol (DTT), Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl), tributylphosphine (TBP), or 2-mercaptoethanol], alkylating the products with an agent that blocks the released cysteine residues (e.g., iodoacetamide), and digesting the proteins with enzymes that produce peptides of optimal length for MS analysis (e.g., trypsin, Lys-C). Trypsin, a commonly used enzyme in proteomics experiments, will cleave the proteins at the carboxyl terminus of Lys (K) and Arg (R). Typically, the enzymatic digestion process is allowed to unfold overnight in basic buffer solutions (pH~7–8), at a substrate:enzyme ratio of (50–100):1. After the removal of salts, additives and cell lysing detergents, the generated peptides are separated by a well-controlled HPLC gradient prior to subsequent electrospray ionization (ESI)-MS analysis. For fast screening applications that enable high-throughput analysis, the entire process must be streamlined and reduced in length. This can be accomplished most effectively by shortening the proteolytic digestion process, or, if possible, by eliminating certain sample preparation steps.
A variety of approaches have been proposed for accelerating the enzymatic digestion of proteins, most importantly, by immobilizing the enzyme on diverse substrates, by designing efficient microreactors that facilitate a close interaction between the substrate and the enzyme, and by assisting the process with various physical stresses such as heat, ultrasound, pressure, or electrical fields [1–13]. For example, Yin et al. developed an approach to synthesize an enzyme-inorganic hybrid nanoflower that was able to perform rapid proteolytic digestion of bovine and human serum albumins within 2 min with greater than 40 % sequence coverage [4]. Yuan et al. reported an immobilized trypsin-based strategy that enabled not only simultaneous rapid digestion and 18O labeling, but also online integration with nano-HPLC-ESI-MS/MS. A sequence coverage of 60 % and labeling efficiency of 98.5 % were obtained for bovine serum albumin within a reaction time of 2.5 min [5]. Ning et al. proposed a rapid digestion method by integrating a protease-modified membrane at the end of pipette tips, and demonstrated 100 % peptide coverage, within 30 s, for the monoclonal antibody Herceptin [6]. Although most of such immobilized trypsin reactors are suitable for rapid digestion, their practical applications continue to be limited primarily because of time-consuming fabrication protocols and the possibility of introducing contaminants in the reaction products. Nevertheless, other methods that utilize physical stresses, have been also developed for accelerating the digestion process. For example, by using microwave irradiation and a stable, immobilized enzyme, Kim et al. obtained 75 % sequence coverage for bovine serum albumin within 10 min digestion time [11]. Guo et al demonstrated that microwave- and ultrasound-assisted digestion enable fast enzymatic reactions at the expense of higher missed-cleavage rates [12], while Ge at al. reported a method that relied on trypsin-immobilized miniature incandescent bulbs and infrared radiation to enable the digestion of standard proteins within 5 min, with a sequence coverage of ~50 %−90 % [13]. Although the digestion process could be accelerated in all these procedures, the trypsin had to be modified to shift its optimum operating temperature to 50–60 °C, and additional equipment such as microwave ovens or sonicators were necessary for completing the enzymatic reactions. Moreover, the applicability of many of the above described methods was limited to the analysis of simple mixtures of proteins. Recently developed commercial kits address some of these problems with specialized trypsin preparations (Promega products) that enable proteolytic digestion at 70 °C within one hour.
In prior work we described a capillary microreactor that enabled fast proteolytic digestion reactions (1–10 min) by immobilizing not the enzyme, but the proteins, through adsorption on reversed phase C18 particles, and flowing the enzyme solution over the adsorbed proteins. The performance of the microreactor was characterized with standard protein digests [14,15]. At the expense of a somewhat incomplete enzymatic reaction, this simple procedure generated a larger number of tryptic peptides and better sequence coverage than conventional overnight digestion protocols. Given the substantial reduction in analysis time enabled by shortening the enzymatic processing step, and the associated benefits of increased protein sequence coverage, the aim of the present study was to evalaute the applicability of digestion reactions performed with trypsin within a few minutes to the characterization of whole, complex proteomes. Specifically, the study was aimed at conducting an in-depth evaluation of the quality and properties of peptides generated through standard, in-solution enzymatic reactions quenched within a few minutes from start, and of the obtainable reproducibility in identifying the same subset of peptides in replicate analyses. There were no other attempts made to enhance the reaction rates. Comparisons of proteolysis reactions performed within minutes, to a reference overnight protocol, were performed. The results of this study demonstrate that partial enzymatic reactions performed on short time-scales can provide quality results for comprehensive proteomic characterizations of biological samples, and can be used as stand-alone or complementary approaches with straightforward implementation in high-throughput analysis work-flows.
Experimental
Materials.
SKBR3 breast cancer cells, phosphate buffered saline (PBS) and trypsin/EDTA were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). Fetal bovine serum (FBS) was from Gemini-Bio Products (West Sacramento, CA, USA), McCoy’s 5A cell culture medium from Life Technologies (Carlsbad, CA, USA), human epidermal growth factor (hEGF) from PeproTech (Rocky Hill, NJ), Normocin from InvivoGen (San Diego, CA, USA), and sequencing grade modified trypsin and Lys-C from Promega Corporation (Madison, WI, USA). Urea, dithiothreitol (DTT), acetic acid, trifluoroacetic acid (TFA), ammonium bicarbonate, sodium chloride, Trizma base and hydrochloride buffer, protease inhibitor solution and phosphatase inhibitor cocktails 2 and 3 were obtained from Sigma-Aldrich (St. Louis, MO, USA). Zorbax SB-C18/5 μm particles, SPEC-PTC18 and SPEC-PTSCX solid-phase extraction pipette tips were purchased from Agilent Technologies (Santa Clara, CA, USA), and fused silica capillaries from Polymicro Technologies. HPLC-grade acetonitrile and methanol were from Fisher Scientific (Fair Lawn, NJ, USA), and DI water was prepared with a MilliQ Ultrapure water system (Millipore, Bedford, MA, USA).
Cell culture and processing.
The SKBR3 breast cancer cells were cultured in McCoy’s 5A cell culture medium, with 10 % FBS, by incubation at 37 °C with 5 % CO2. The cell cultures were either arrested by serum deprivation for 48 h (sample S1), arrested and released with EGF (10 ng/mL/15 min, 20 ng/mL/10 min, 150 ng/mL/36 h) in the culture medium (samples S2, S3, S4), or just simply allowed to proliferate in the culture medium (sample S5). Normocin (0.1 mg/mL) was added to the cell culture to protect against bacteria, fungi and mycoplasma. At full confluence, the cells were harvested by trypsinization, washed with cold PBS, and stored at −80 °C. For further processing, the cells were suspended in a lysis buffer prepared from 50 mM Tris (pH~8), 75 mM NaCl, 8 M urea, 1–2 mM DTT, and protease (lysis buffer:protease inhibitor solution 100:1 v/v) and phosphatase inhibitor cocktails 2 and 3 (lysis buffer:phosphatase inhibitor solution 50:1 v/v). Lysis was performed through intermittent sonication for 10 min (5 × 1 min sonication bursts followed by 1 min pause) in an ice-cooled sonic bath. The lysis buffer-to-packed cell volume ratio was 5:1. The cells were centrifuged at 20,000xg and the protein concentration in the supernatant was measured with the Bradford assay (SmartSpec Plus spectrophotometer, Bio-Rad, Hercules, CA, USA).
Enzymatic digestion.
The SKBR3 protein extracts were denatured and reduced at 56 °C for 1 h in the lysis buffer that already contained denaturing and reducing agents, diluted 10-fold with 50 mM NH4HCO3, and digested with trypsin (50:1 substrate:enzyme ratio) in solution at 37 °C, for various times (7 min, 15 min, 30 min, 60 min, 18 h). Alkylation was not performed to avoid the presence of incomplete reactions and side-products [16]. The protein digestion solution was quenched with glacial CH3COOH (10 µL per 1 mL protein digest), and sample cleanup was performed with SPEC C18/SCX cartridges. The Lys-C proteolysis of SKBR3 cells was performed by following the same procedure. All peptide samples were brought to dryness in a vacuum centrifuge, dissolved in a solution of H2O/CH3CN/TFA 98:2:0.01 v/v to a final concentration 2 μg/μL, and further analyzed by nano-HPLC-MS.
LC-MS/MS analysis and data processing.
LC-MS/MS analysis and data processing protocols were described in detail in previous work [17]. In short, MS analysis was performed with linear ion trap LTQ or LTQ-XL mass spectrometers (Thermo Electron Corp., San Jose, CA, USA), operated in positive-ion electrospray mode at ~2 kV. Nano-LC was performed with Agilent 1100 or 1260 micro-LC separation systems and in-house prepared separation columns (100 μm i.d. × 360 µm o.d., 10–12 cm long, packed with 5 µm/C18 Zorbax particles, 300 Å pore size), with the eluent flow rate set at 180–200 nL/min. The flow was generated by the micro-LC pumps at 10 µL/min, and split to the desired level with an in-house built split/splitless injector. Sample injections (8 µL) were performed in splitless mode. The eluent was prepared from H2O/CH3CN/TFA, and the concentration gradient was from 96:4:0.01 v/v (solvent A) to 10:90:0.01 v/v (solvent B). For global proteomic profiling of cell extracts, a 4 h long HPLC gradient was used, and the samples were analyzed via a data-dependent analysis (DDA) method by performing zoom/MS2 scans on the top 5 most intense peaks from each MS scan (produced by averaging 5 scans), with the data being acquired over a mass range of 500–2000 m/z. The collision induced dissociation (CID) parameters were 3 m/z ion isolation width, normalized collision energy 21 % (LTQ-XL) or 30–35 % (LTQ), 0.25 activation Q, 30 ms activation time, and threshold for triggering MS2 scans 100 counts. Conditions for data dependent analysis included: ±5 m/z zoom scan width, ±1.5 m/z exclusion mass width, dynamic exclusion at repeat count 1, repeat duration of 30 s, exclusion list size 200, and exclusion duration 60 s.
Bioinformatics.
Raw data files were analyzed with the Proteome Discoverer 1.4 software package, using the Sequest HT search engine (Thermo Electron Corp.) for performing searches against a Homo sapiens protein database from UniProt (January 2015 download) comprising 20,198 reviewed/non-redundant sequences (500–5000 mass range, minimum/maximum peptide length of 6/144 amino acids, S/N threshold 1.5, precursor ion tolerance 2 Da, fragment ion tolerance 1 Da, b/y/a ion fragments only, fully tryptic fragments with up to maximum 4 missed cleavages allowed, no PTMs). Three LC-MS/MS technical replicates were performed for each sample and combined in a multiconsensus report that was used for comparison. This ensured the identification of a larger number of peptides, and increased sequence coverage and detection reliability for proteins. For the two extreme data-points that were critical for assessing the performance of the fast digestion process, (i.e., for the 7 min to 18 h comparison), the reproducibility of the enzymatic reactions was assessed based on triplicate measurements performed with various SKBR3 cell cultures. The peptide false discovery rate (FDR) settings were 3 % (medium confidence) and 1 % (high confidence) for relaxed and stringent database searches, respectively, and the search results were combined for analysis. FDRs were calculated with the Target Decoy PSM Validator node based on Xcorr vs. charge state values. The GRAVY (grand average of hydropathy) index for tryptic peptides was calculated with tools provided by Bioinformatics.org [18]. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [19] partner repository with the dataset identifiers PXD010306.
Results and Discussion
To assess the effectiveness of partial enzymatic reactions performed in solution within a limited time-frame, and their value for MS proteomics experiments, SKBR3 breast cancer cell extracts were subjected to enzymatic digestion with trypsin for 7 min, 15 min, 30 min, 60 min and 18 h. The results of the 18 h long enzymatic process were used as a reference. A detailed comparison of the critical 7 min to the 18 h reaction is provided, and intermediate time-points (15 min, 30 min and 60 min) were evaluated to highlight the trends. The reaction performance and quality of products were assessed by measuring the total number of identifiable peptides and proteins, the number of missed cleavage sites, the reproducibility of detection, the detection of low abundance proteins, the peptide Xcorr scores, and the GRAVY index. Independent cell batches cultured under various conditions were used to assess the reproducibility of the enzymatic process in terms of missed cleavage sites, and independent digest pools of the same cell culture were used to assess the reproducibility of detecting the same proteolytic peptide fragments in a sample.
Peptide identification and protein sequence coverage.
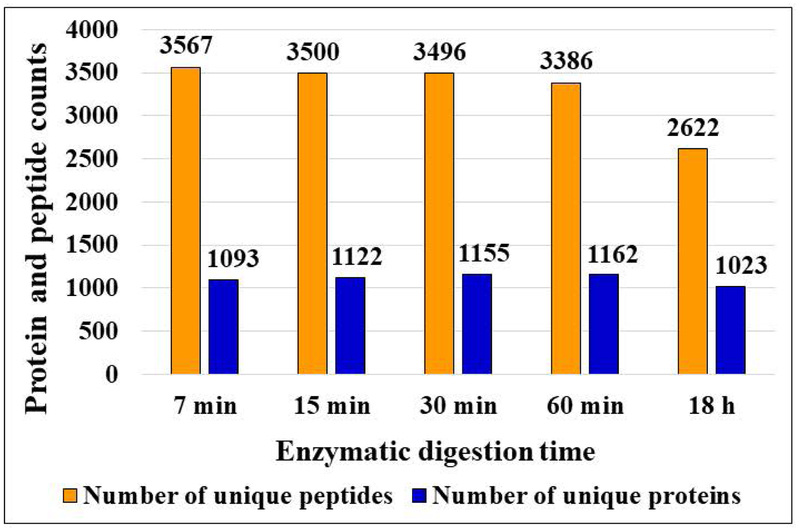
An effective proteomics protocol aimed at profiling complex cellular extracts leads to the identification of thousands of proteins, matched, ideally, by multiple peptides per protein to ensure unambiguous identifications. The proteomic profiling of the SKBR3 cell extracts subjected to short proteolysis reaction times (7–60 min) led to very similar results in terms of protein IDs (e.g., groups), with numbers ranging from 1,093 to 1,162, but with matching unique peptides increasing progressively from 3,386 to 3,567, as the reaction time decreased from 60 min to 7 min (Figure 1). Both numbers exceeded the results for the 18 h reference (1,023 protein groups matched by 2,622 peptides). Overall, for these samples, the range of unique peptides and sequence coverage per protein dropped from 3–30 to 0–15, and from 5–77 % to 0–45 %, respectively, when extending the digestion time from 7 min to 18 h (Supplemental Figures 1A and B). The sequence coverage was less at 18 h due to the loss of short, typically 2–6 amino acid long sequences, some with multiple K and R residues, that flanked longer sequence stretches with no missed cleavage sites. Many of these short peptides fell in a range of m/z<500 that was not included in the data acquisition process due to the presence of background ions that interfered with, and reduced the overall effectiveness of the DDA process. Therefore, the short peptides could not be detected after enzymatic cleavage, but their presence was observable in longer peptides with multiple missed cleavages where they contributed to an increased protein sequence coverage (Supplemental Figure 1C). Being also more hydrophilic, such short peptides are expected to be lost during the sample cleanup and enrichment steps. As a result, none of the proteolytic digestion products in these datasets contained peptides with <8 amino acid residues. Supplemental Table 1 provides the database search results for each individual LC-MS run of the time-point experiment, including the Xcorr and the high/medium confidence scores for peptides, and the sequence coverage, number of unique peptides, and peptide spectrum matches (PSMs) for proteins.
Figure 1.
Effect of enzymatic reaction time on the identification of peptides and proteins generated from SKBR3 cell extracts. Conditions: SKBR3 cells were lysed through sonication, digested with trypsin for 7 min, 15 min, 30 min, 60 min or 18 h, subjected to C18/SCX cleanup, and analyzed by nano-HPLC-MS/MS; sample concentration 2 μg/μL, HPLC injection volume 8 µL; nano-LC gradient 4 h long, from 4 % to 96 % CH3CN (TFA 0.01 %). The number of unique peptides and protein groups per time-point represent each the combined results of 3 LC-MS/MS analyses.
Reproducibility and complementarity.
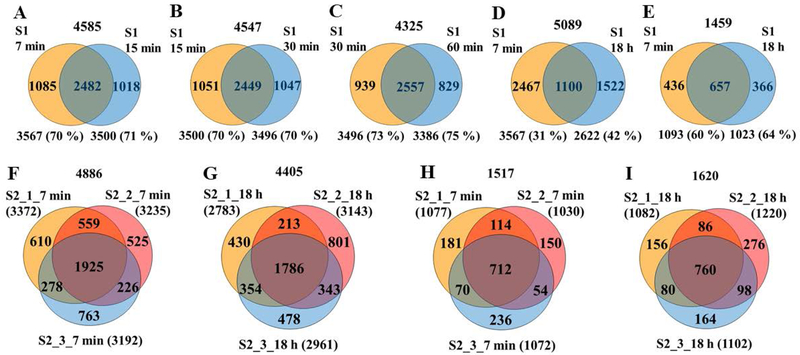
The ability to detect more peptides per protein when using short enzymatic digestion times has a number of benefits, most importantly, increased confidence in protein identification and quantitation. To explore the level of reproducibility that can be achieved within a few minutes of proteolytic digestion, the overlap between unique peptides generated at different time-points, and from different digest replicates at the same time-point, was assessed. Between successive time-points (sample S1, 7 min to 60 min), the reproducibility of detecting the same peptides was consistently preserved at 70–75 % (Figure 2A–2C), but it dropped to ~31–42 % when comparing the 7 min to the 18 h experiment (Figure 2D). If only peptides containing zero missed cleavages were compared, the reproducibility was only marginally better, i.e., 70–80 %. Proteolytic digestion experiments conducted in triplicate with a different cell batch (S2) confirmed that the % overlaps within the same sample were very similar for the 7 min and the 18 h time-points, indicating that the short enzymatic process did not result in loss of reproducibility and performance (two way overlaps: 64–77 % peptide and 70–80 % protein levels; three way overlaps: ~40 % peptide and ~47 % protein levels, respectively, Figures 2F–2I). Supplemental Tables 2 and 3 provide the database search results for the replicate analysis of the 7 min and 18 h reactions. The peptide-level FDRs were also very similar between the two time-points, with 83–84 % and 83–87 % of the identified peptides in the 7 min and 18 h experiments, respectively, qualifying in the high-confidence group with FDR<1 %. Interestingly, however, neither the peptides nor the proteins detected in the 18 h experiment were a subset of the 7 min one, but the two sets were rather complementary (Figures 2D and 2E), with larger numbers in combined results than those that would have arisen from experiments conducted using identical conditions. Altogether, on a global level, the results indicate that the detectability of proteins was not hampered by the short enzymatic reaction, but rather diversified. Therefore, an analysis strategy that would combine the products of short and long proteolytic digestion times could be used to increase the peptide and protein identification rates, to lead to a more complete characterization of complex cellular extracts.
Figure 2.
Venn diagrams of unique peptide and protein overlaps from the 7 min–18 h time-point experiments. Number of peptide or protein IDs in each enzymatic replicate and two-way % overlaps are indicated in each figure. Numbers on the top indicate the combined IDs in the enzymatic replicates. Conditions: (A-D) Peptide overlaps between the 7 min, 15 min, 30 min, 60 min and 18 h enzymatic reaction products (sample S1); (E) Protein overlaps between the 7 min and 18 h enzymatic reaction products (sample S1); Peptide (F and G) and protein (H and I) overlaps between three tryptic digest pool replicates (7 min and 18 h) (sample S2).
Missed cleavage sites.
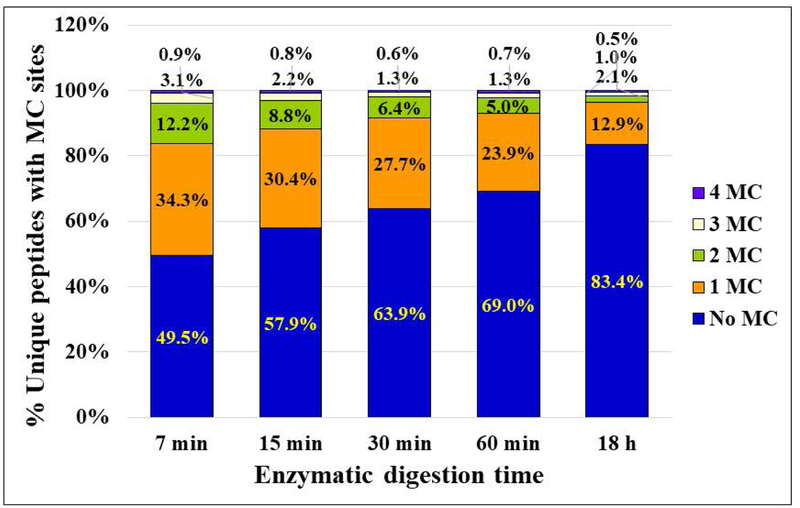
A count of the missed cleavages at internal K and R amino acid residues provided very clear trends over the time period taken under study (Figure 3A). The peptides with no missed cleavage sites increased from ~ 49 % to ~83 %, mainly at the expense of peptides with 1 or 2 missed cleavages, as the proteolysis reaction was increased from 7 min to 18 h. Peptides with one missed K experienced the largest change, followed by peptides with one missed R and two missed K and R residues (Figure 4A). Peptides with two missed R residues were generally low in number, and peptides with 3 or 4 missed cleavages, K or R, represented each only <1.6 % (mostly <0.5 %) of the total number of identified sequences at both time-points. Experiments conducted with five independent cell batches confirmed these results (Figure 3B and Supplemental Table 2), the fraction of peptides with no missed cleavages being in the range of 42–53 % for the 7 min digestion reactions (Mean=49 %, RSD=9.6 %), and 77–85 % for the 18 h digestions (Mean=83 %, RSD=4 %).
Figure 3.
Trends in missed cleavage sites. (A) Stacked column chart illustrating the % change in peptides with various numbers of missed cleavage sites, as the enzymatic digestion progressed from 7 min to 18 h; (B) Reproducibility results for the 7 min and 18 h time-points. Conditions: the same as in Figure 1.
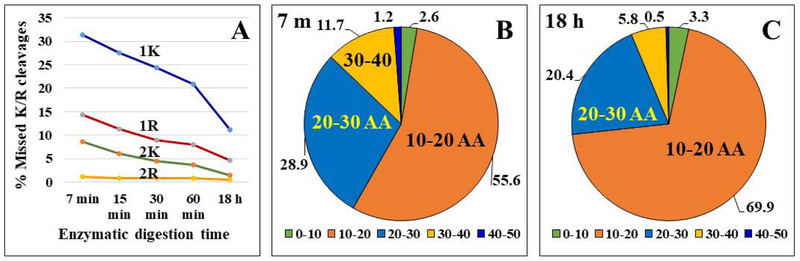
Figure 4.
Trends in missed cleavage sites and peptide length as a function of enzymatic reaction time. (A) Line chart illustrating the % change in missed K and R cleavage sites with the progression of the proteolytic digestion reaction. (B) and (C) Pie charts illustrating the % change in peptide length, in terms of number of amino acids, for enzymatic reactions conducted for 7 min and 18 h. Conditions: the same as in Figure 1.
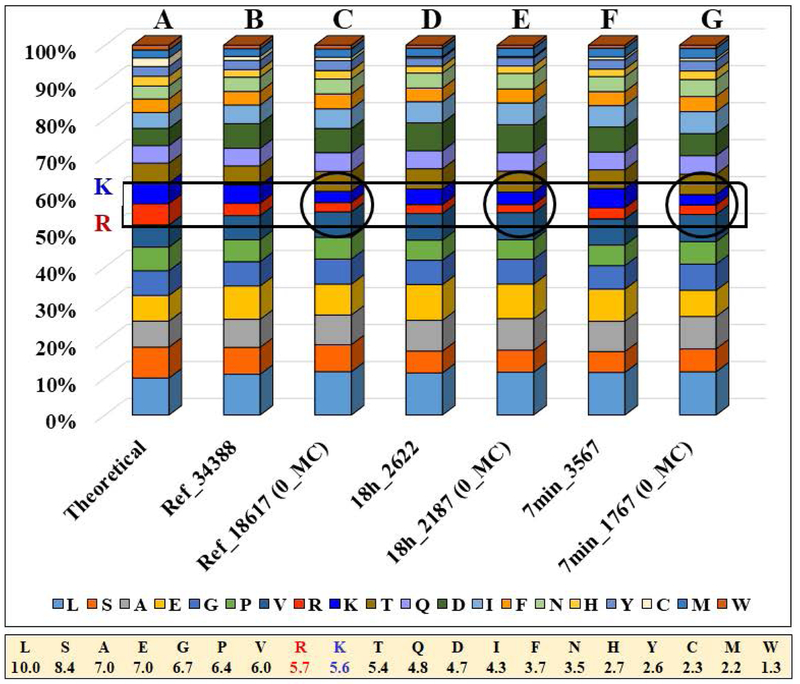
It is worth to note that the highest contribution to missed cleavages was provided by one missed K, and this was much larger than that contributed by one missed R. Given that the frequency of K and R in the Homo sapiens proteome is roughly the same, 5.6 % and 5.7 % [20], respectively, the observed results led to the conclusion that either there was a bias in the detection of peptides with multiple missed R residues, or that the enzymatic digestion was more effective at R than at K, or both. The frequencies of amino acid detection in the 7 min and 18 h experiment were therefore compared to the theoretical frequency of amino acids in the Homo sapiens proteome and to a reference set of 34,288 peptides generated from a variety of proteomic experiments in our laboratory from MCF7, MCF10 and SKBR3 cell lines. Entire datasets, or subsets containing only peptides with zero missed cleavages, were compared (Figure 5). Overall, for the entire set of 20 amino acids, the experimental results closely matched the theoretical predictions. The largest discrepancies were found for K and R. For the full datasets, the frequency of K detection (4.2–5.2 %) came close to the theoretical value (5.6 %), while the frequency of R (3.0–3.4 %) was lower than the theoretical one (5.7 %). When only peptides with zero missed cleavage sites were counted, the frequencies dropped even more, to 2.8–3.4 % for K, and to 2.2–2.5 % for R (encircled areas in the bar graph from Figure 5).
Figure 5.
Stacked column chart illustrating amino acid frequency distributions. (A) Theoretical distribution of amino acids in the Homo sapiens proteome. (B) Reference set of 34,388 peptides generated from a combination of proteomic experiments conducted on various human cell lines. (C) Reference set from above, including only 18,617 peptides with zero missed cleavage sites. (D) Full set of 2,622 peptides from the 18 h experiment. (E) Sub-set of 2,187 peptides with zero missed cleavage sites from the 18 h experiment. (F) Full set of 3,567 peptides from the 7 min experiment. (G) Sub-set of 1,767 peptides with zero missed cleavage sites from the 7 min experiment. The lower bar indicates the theoretical frequency of amino acids in the Homo sapiens proteome.
These findings confirmed that the frequency of R detection is lower than optimal for all datasets, but this is not a bias introduced by the fast digestion process. Moreover, prolonged reaction times that led to a more complete enzymatic reaction, also led to the preferential loss of both K and R residue containing peptides - an expected outcome, as the short peptides that are cleaved by the enzyme and are overlooked by detection always carry at least one K or R residue. The analysis of the shortest detectable peptides from these datasets (i.e., peptides containing 8–10 amino acids) confirmed this assumption, revealing that the frequencies of both K and R in these peptides approached or even exceed the theoretical values (i.e., 5.8 % K and 5.0 % R for 18 h, and 7.3 % K and 6.3 % R for 7 min reactions). Such short peptide products that are missed in proteomics experiments lead therefore not just to reduced protein sequence coverage, but are also a source of K/R loss in the full proteome datasets. The presence of inhibitory effects of tryptic attack, induced by flanking or neighboring D, E, or P residues, or of RK sequences, calls for additional scrutiny [21], even though these did not appear to affect the main trends in these datasets. Peptide sequences rich in basic amino acid residues are part of protein motifs and domains that are involved in the regulation of a variety of cellular processes. Arginine-rich domains, for example, have critical roles in RNA processing, maturation and ribonucleotide assembly [22], while arginine methylation has been found to control the subcellular localization of the oncoprotein splicing factor SF2/ASF [23]. Fine-tuning the enzymatic reaction and further optimizing the sample processing and data acquisition process could therefore enable a more accurate interpretation of the biological implications of proteomic data.
Peptide size and quality of MS identifications.
While some K/R residues from short peptide sequences escaped detection, the presence of missed cleavage sites in the products of the fast proteolysis reactions led to a larger proportion of long peptides containing 20–50 amino acid residues (Figures 4B/C). The production of such peptides could support applications that use middle-down proteomic sequencing and peptide fragmentation techniques such as ETD (electron transfer dissociation). Middle–down proteomics seeks the detection of peptides in the mass range of 3,000–15,000 Da. Such peptides result in better sequence coverage and improved ability to detect protein variants, as well as a more accurate assignment to gene products [24,25]. ETD, on the other hand, is a fragmentation technique that has been developed for the analysis of large, multiply-charged peptides, to capture posttranslational modifications (PTMs) and enable the identification of the modified amino acid sites. It benefits, therefore, from the presence of basic amino acid residues in the sequence of the peptide. A number of enzymes (Lys-C, Lys-N, Arg-C, Asp-N, Glu-C, ompT) and chemically induced digestion protocols have been explored for generating peptides in this mass range [24,25]. Generally, it was found that the use of such enzymes led to only a modest increase in peptide identifications and in average peptide length (e.g, 1.9 kDa for Glu-C and Asp-N peptides, relative to 1.5 kDa for tryptic peptides) [25,26]. However, the benefit of alternative proteases manifested itself in using their combined peptide results that led to improved sequence coverage, and ability to detect low abundance proteins [26]. In comparison, the analysis of the 7 min reaction products showed that the proportion of peptides with >20 amino acid residues was markedly higher (43–47 %) than in the 18 h products (26–29 %) (Figures 4B and 5C). We also note that the combination of 7 min and 18 h tryptic peptides, when compared to all other combinations that were explored, led to the largest increase in both peptide and protein IDs (~94 % and ~40 % increase in new peptide and protein IDs relative to the 18h products) (Figures 2D and E). These values are much higher than reported by the use of combined enzymes that resulted in an average increase of only 16 % of new protein IDs per use of a new enzyme with different cleavage specificity [26]. A fast proteolysis strategy could complement, therefore, either bottom-up or middle-down approaches enabled by the use of a variety of enzymes.
To demonstrate the broader applicability of the procedure, a Lys-C digestion reaction was conducted for 7 min. The results were similar to the tryptic experiments: peptides with zero missed cleavage sites 53 %, peptides with >20 amino acid residues 46 %, K sites 7.4 %, and R sites 2.8 %, with K detectability higher, and R detectability similar to the tryptic digests. The median and maximum peptide length for all 7 min tryptic and Lys-C digestions was 19–20 and 47–51, respectively, higher than typically observed for overnight digestions (median 16–17, maximum 46–48 amino acids), with the major difference lying in the higher proportion of peptides with >20 amino acid residues (see above). Most importantly, when combining two sets of peptides produced in the 7 min Lys-C and tryptic enzymatic reactions, the increase in unique peptides reached a maximum of 5695 IDs and sequence coverage (Supplemental Figure 2).
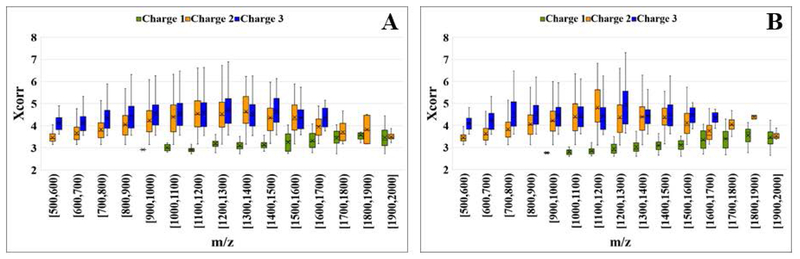
To assess the quality of higher mass peptides generated through fast enzymatic reactions, the peptide pools from the 7 min and 18 h experiments were compared based on the distribution of XCorr scores vs. m/z for charge states 1+, 2+ and 3+ (Figure 6). The Xcorr scores were very similar for the two experiments, distributed in the ranges of 2.7–3.8 for singly charged peptides, 3.3–5.6 for double charged peptides, and 3.8–5.5 for triply charged peptides, with slightly higher trends for the 7 min peptides. A similar assessment was performed for peptides originating from Lys-C digests, as well as for peptides containing only K or R residues from either the tryptic and Lys-C (7 min) digests. The ranges of the Xcorr scores were the same as above, for Lys-C or K- or R-only containing peptides, confirming that there was no bias in their detection through tandem MS. While accurate characterization of higher charge states would require high mass accuracy MS instruments, overall, the observed scores for the 7 min experiments corroborated the quality of the generated peptides, supporting the applicability of fast enzymatic processing for proteomic explorations of complex samples and integration in workflows and platforms that seek rapid sample analysis [27].
Figure 6.
Box plots representing Xcorr score distributions for (1+), (2+) and (3+) charged peptides, as a function of m/z for (A) 7 min and (B) 18 h enzymatic digest products.
Gravy index.
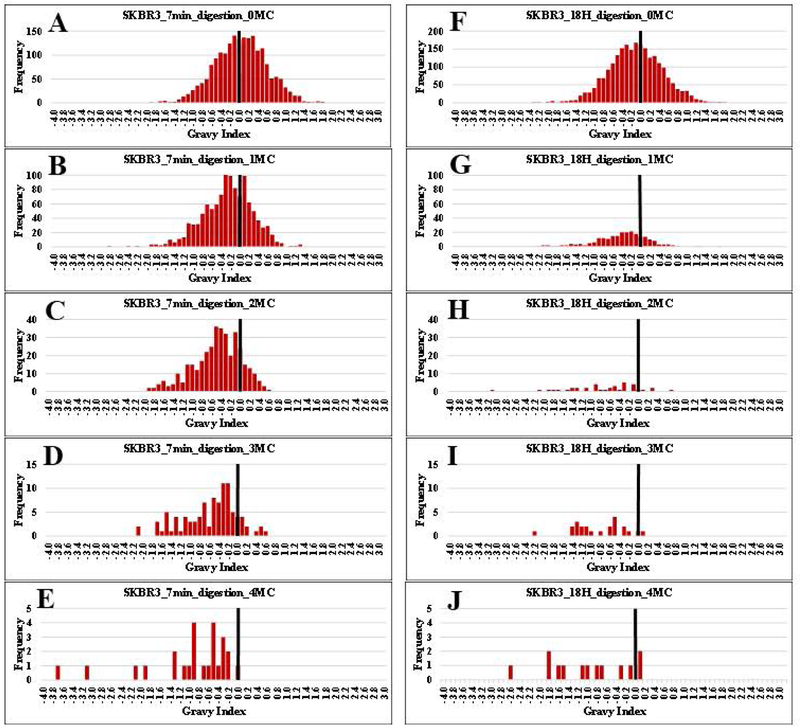
Longer peptide sequences are often associated with an increase in hydrophobic properties that impact negatively their recovery from clean-up cartridges or separation on C18/LC columns. To answer this concern, the distributions of GRAVY scores for peptides generated from the 7 min and 18 h long enzymatic reactions, were compared (Figure 7). The GRAVY index values represent the hydrophilic and hydrophobic properties of an amino acid, and cover a range from (−4) to (+4), the more negative values being characteristic of the more hydrophilic components [28]. The GRAVY histograms were generated by calculating the GRAVY score for each peptide and partitioning the values in bins of 0.1 width. Peptides with a different number of missed cleavage sites were placed in different histograms. As observed from the figure, for all peptide sub-sets at 7 min (7A-E) and 18 h (7F-J), the presence of a larger number of missed cleavages resulted in a shift toward more negative GRAVY scores, revealing that the longer peptides were, in fact, more hydrophilic, rather than hydrophobic. The shift in properties was introduced by the presence of additional K and R residues, which are the most hydrophilic amino acids with a hydropathy score of −4.5 (R) and −3.9 (K). This result brings an unintended advantage, as a broader distribution of the hydrophobic/hydrophilic properties of peptides would allow for a superior refinement of reversed phase LC separations, and a better utilization of the landscape of available techniques (ion exchange, HILIC, or high-pH reversed phase LC) for devising multi-dimensional peptide pre-fractionation strategies.
Figure 7.
Gravy index histograms for peptides with 0 to 4 missed cleavages, generated from the 7 min (A-E) and 18 h (F-J) proteolytic digestion experiments. A reference bar is drawn for an index value of zero, to help visualize the shift in GRAVY scores with an increased number of missed K/R cleavage sites.
Conclusions
To streamline proteomic experiments, in this work, we propose the use of enzymatic reactions conducted on short time-scales for processing the protein complement of cells. The performance of this approach was assessed by evaluating the impact of the enzymatic digestion time on the quality of generated peptides. We demonstrate that a rapid digestion process can produce results that are superior to established protocols in terms of achieving protein and proteome coverage, with no sacrifice in ability to perform efficient tandem mass spectrometric analysis. While the reaction products of short and long enzymatic reactions differed considerably, the reproducibility of obtaining the same results from fast replicate enzymatic processes was not affected by the short time-scale. By properly combining the reaction products generated through short (partial) and long (complete) enzymatic reactions, possibly completed with two distinct enzymes, a substantial increase in the identification of peptides and proteins can be achieved. Moreover, by fine-tuning the reaction conditions, changes in the length and physical properties of peptides can be induced, to support more effective separations and complementary analysis approaches in middle-down proteomics experiments. Altogether, while complete enzymatic reactions will continue to stay at the foundations of proteomics research, these results demonstrate the value of fast and partial enzymatic digestions, and lay the necessary premise for catalyzing the development of novel high-throughput proteomics workflows.
Supplementary Material
Supplemental Table 1. Protein- and peptide-level database search results for different proteolytic digestion time-points (S1: 7 min, 15 min, 30 min, 60 min, 18 h).
Supplemental Table 2. Peptide-level reproducibility results for the 7 min and 18 h time-points (S2), and missed cleavage data (S1, S2, S3, S4, S5).
Supplemental Table 3. Protein-level reproducibility results for the 7 min and 18 h time-points (S2).
Acknowledgments
This work was supported by awards from the NSF (DBI-1255991) and NIGMS (1R01GM121920–01A1) to IML. We thank Shreya Ahuja and Arba Karcini for providing support with sample processing.
References
- 1.Hustoft HK, Malerod H, Ray S, Reubsaet L, Lundanes E, Greibrokk T. A Critical Review of Trypsin Digestion for LC-MS Based Proteomics In: Leung H, ed. Integrative Proteomics. InTech: Rijeka, Croatia, 2012:73–82. [Google Scholar]
- 2.Switzar L, Giera M, Niessen WMA. Protein digestion: An overview of the available techniques and recent developments. J. Proteome Res 2013;12:1067–1077. [DOI] [PubMed] [Google Scholar]
- 3.Safdar M, Sproß J, Jänis J. Microscale immobilized enzyme reactors in proteomics: Latest developments. J. Chromatogr. A 2014;1324:1–10. [DOI] [PubMed] [Google Scholar]
- 4.Yin Y, Xiao Y, Lin G, Xiao Q, Lin Z, Cai Z. An enzyme–inorganic hybrid nanoflower based immobilized enzyme reactor with enhanced enzymatic activity. J. Mater. Chem. B 2015;3:2295–2300. [DOI] [PubMed] [Google Scholar]
- 5.Yuan H, Zhang S, Zhao B, et al. Enzymatic Reactor with Trypsin Immobilized on Graphene Oxide Modified Polymer Microspheres to Achieve Automated Proteome Quantification. Anal. Chem 2017;89:6324–6329. [DOI] [PubMed] [Google Scholar]
- 6.Ning W, Bruening ML. Rapid Protein Digestion and Purification with Membranes Attached to Pipet Tips. Anal. Chem 2015;87:11984–11989. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Zhang L, Yang K, Liang Z, Zhang L, Zhang Y. Preparing a metal-ion chelated immobilized enzyme reactor based on the polyacrylamide monolith grafted with polyethylenimine for a facile regeneration and high throughput tryptic digestion in proteomics. Anal. Bioanal. Chem 2012;402(2):703–710. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H, Zhang L, Zhang Y. Preparation of high efficiency and low carry-over immobilized enzymatic reactor with methacrylic acid-silica hybrid monolith as matrix for on-line protein digestion. J. Chromatogr. A 2014;1371:48–57. [DOI] [PubMed] [Google Scholar]
- 9.Liu W-L, Lo S- H, Singco B, Yang C- C, Huang H- Y, Lin C- H. Novel trypsin–FITC@MOF bioreactor efficiently catalyzes protein digestion. J. Mater. Chem. B 2013;1(7):928. [DOI] [PubMed] [Google Scholar]
- 10.Starke S, Went M, Prager A, Schulze A. A novel electron beam-based method for the immobilization of trypsin on poly(ethersulfone) and poly(vinylidene fluoride) membranes. React. Funct. Polym 2013;73:698–702. [Google Scholar]
- 11.Kim H, Kim HS, Lee D, Shin D, Shin D, Kim J, Kim J. Microwave-Assisted Protein Digestion in a Plate Well for Facile Sampling and Rapid Digestion. Anal. Chem 2017;89(20):10655–10660. [DOI] [PubMed] [Google Scholar]
- 12.Guo Z, Cheng J, Sun H, Sun W. A qualitative and quantitative evaluation of the peptide characteristics of microwave- and ultrasound-assisted digestion in discovery and targeted proteomic analyses. Rapid Commun. Mass Spectrom 2017;31:1353–1362. [DOI] [PubMed] [Google Scholar]
- 13.Ge H, Bao H, Zhang L, Chen G. Immobilization of trypsin on miniature incandescent bulbs for infrared-assisted proteolysis. Anal. Chim. Acta 2014;845:77–84. [DOI] [PubMed] [Google Scholar]
- 14.Deng J, Lazar IM. Proteolytic Digestion and TiO2 Phosphopeptide Enrichment Microreactor for Fast MS Identification of Proteins. J. Am. Soc. Mass Spectrom 2016;27(4):686–698. [DOI] [PubMed] [Google Scholar]
- 15.Lazar IM, Deng J, Smith N. Fast Enzymatic Processing of Proteins for MS Detection with a Flow-through Microreactor. J. Vis. Exp 2016;110:e53564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo M, Weng G, Yin D. Identification of the over alkylation sites of a protein by IAM in MALDI-TOF/TOF tandem mass spectrometry. RSC Adv 2015;5:103662–103668. [Google Scholar]
- 17.Lazar IM, Hoeschele I, de Morais J, Tenga M. Cell Cycle Model System for Advancing Cancer Biomarker Research. Sci. Rep 2017;7:17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sequence Manipulation Suite for Protein GRAVY.http://www.bioinformatics.org/sms2/protein_gravy.html. Version 2.
- 19.Vizcaíno JA, Csordas A, del-Toro N, et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016; 44(D1):D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruess M, Apweiler R. Bioinformatics Resources for In Silico Proteome Analysis. J. Biomed. Biotechnol 2003;4:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gershon PD. Cleaved and missed sites for trypsin, Lys-C, and Lys-N can be predicted with high confidence on the basis of sequence context. J. Proteome Res 2014;13:702–709. [DOI] [PubMed] [Google Scholar]
- 22.Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol 2007;4:69–75. [DOI] [PubMed] [Google Scholar]
- 23.Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Arginine Methylation Controls the Subcellular Localization and Functions of the Oncoprotein Splicing Factor SF2/ASF. Mol. Cell. Biol 2010;30(11):2762–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Tran JC, Zamdborg L, et al. A protease for’middle-down’proteomics. Nat. Methods 2012;9(8):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cristobal A, Marino F, Post H, Van Den Toorn HWP, Mohammed S, Heck AJR. Toward an Optimized Workflow for Middle-Down Proteomics. Anal. Chem 2017;89(6):3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res 2010;9(3):1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazar IM, Rockwood AL, Lee ED, Sin JCH, Lee ML. High-speed TOFMS detection for capillary electrophoresis. Anal. Chem 1999;71(13):2578–2581. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 1982;157(1):105–132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Protein- and peptide-level database search results for different proteolytic digestion time-points (S1: 7 min, 15 min, 30 min, 60 min, 18 h).
Supplemental Table 2. Peptide-level reproducibility results for the 7 min and 18 h time-points (S2), and missed cleavage data (S1, S2, S3, S4, S5).
Supplemental Table 3. Protein-level reproducibility results for the 7 min and 18 h time-points (S2).