Abstract
Objective:
The grainyhead-like-2 (GRHL2) genetic variants were reported in age-related hearing impairment (ARHI) susceptibility in several case–control studies. However, their conclusions are conflicting; it is difficult to precisely assess the disease risk associated with the variants. Therefore we conduct the meta-analysis to discover the association of GRHL2 polymorphisms and the risk of ARHI.
Methods:
A related literature search was conducted in on-line databases, such as Wanfang database, China National Knowledge Infrastructure (CNKI), EMBASE, Web of Science, and PubMed (updated to August 30, 2018). We use Review Manager 5.0 and Stata SE 12.0 software to reckon the odds radio (OR), 95% confidence interval (CI) and P value in random- or fixed-effects model according to the I2 value in the heterogeneity test.
Results:
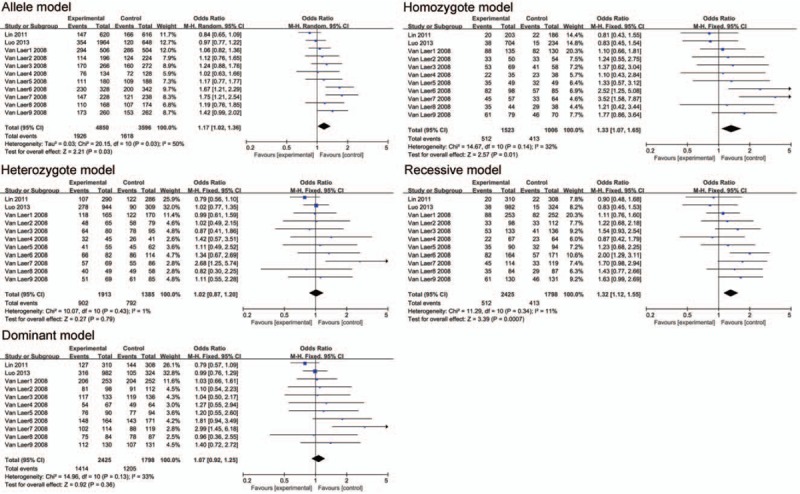
2762 cases and 2321 controls in 5 articles were provided data to the meta-analysis. The pooled ORs (95% CI) of the rs10955255 polymorphism were 1.26 (1.05–1.50, P = .01), 1.33 (1.07–1.65, P = .01), and 1.32 (1.12–1.55, P = .0007) in the allele, homozygote and recessive model separately. Besides, a significant association was detected between rs1981361 in mixed population and the ARHI risk in the allele, heterozygote, and dominant genetic model respectively. Then subgroup analyses was performed by ethnicity, for rs10955255 meaningful associations were detected for the allele model, homozygote model, dominant model and recessive model in the Caucasian population but no relations in any of the 5 genetic models in Asian population.
Conclusion:
The meta-analysis indicated that the rs10955255 polymorphism could be an important risk factor for ARHI, especially in the Caucasians. The rs1981361 polymorphism may be a risk factor for ARHI in Asians. Larger scale researches are needed to further bring the consequences up to date.
Keywords: age-related hearing loss, grainyhead-like-2 gene polymorphism, meta-analysis
1. Introduction
Age-related hearing loss (AHL), also known as presbycusis is a complex and multifactorial disorder, characterized by the progressive deterioration of auditory sensitivity associated with aging.[1] As a natural consequence of a society developing process and along with the intensifying of the population aging, the number of people with AHL in our country is accumulates and has become a severe health and social problem.[2–4] Most of the time, hearing loss initially involves high-frequency tones, results in communication impairment, particularly in reverberant and/or noisy environments. Epidemiologically, some medical comorbidity related to AHL, such as social isolation, depression,[5–8] cognitive impairment, and dementia,[9] placing heavy burden on the society. Although AHL usually could be caused, aggravated or associated by the common risk factors such as hypertension,[10,11] exposure to the noise, smoking, ototoxic drugs,[12] and so on, growing number of evidences indicate genetic factors play a significant role in the etiology of AHL through their interactions with the environmental components.[13–15] As a result, it is essential to identify the gene variants that contribute to AHL pathogenesis. The knowledge of the genetic component that influences AHL susceptibility could be crucial in creating efficient therapeutic strategies and improving disease risk's prediction, based on targeted approach.
AHL susceptibility is affected by numerous environmental and genetic factors.[16] What cannot be overlooked is that the susceptibility to AHL among individuals is apparently various, some people are more vulnerable to AHL than others. Several genetic association studies reported a number of genes involved in neurotransmission, cell adhesion, oxidative stress, apoptosis, immunity, cell morphology, development, and so on. could be implicated in pathophysiology of age-related hearing loss, but the reasons for all individual differences still cannot be given by these genetic variations. Thus it is required to discover more AHL-associated genes and genetic polymorphisms to further enhance genetic predictive system of AHL.
Recent researches illustrated that the t grainyhead-like 2 (GRHL2) gene, also known as TFCP2L3, expressing in diverse epithelial tissues, is expressed in cochlear duct that lines different cell populations, including hair cells, the lateral wall spiral ligament, SGNs and supporting cells.[16] Their deficiencies often cause progressive dominant hearing loss in a zebrafish model of DFNA28.[17] GRHL2 gene is part of the grainyhead family, which is crucial for several developmental processes in Drosophila, most prominently during embryonic stages. Carrying mutations of GRHL2 gene in flies is often lethal in embryonic stage. GRHL2 takes part in the maintenance and differentiation of epithelial cells throughout life. The most reasonable pathological explanation for late-onset hearing impairment is damaged epithelial cell integrity.[17]
What is notable is that some studies suggested an intimate connection between some SNPs in GRHL2 genes, including rs10955255, and rs2127034, and AHL susceptibility, while no positive consequence for rs10955255 was detected in Lin's study. Due to the limitations of sample scale, research design, ethnicity, and numerous other factors, even for each SNP site, the outcome in different studies is various, while meta-analysis exhibits an advantage in getting over the limitations and evaluating the sources of Heterogeneity. At present, there are many studies conducted to evaluate the association between the role of GRHL2 gene polymorphism and AHL susceptibility. However, the published results are inconsistent and controversial. Here, a meta-analysis including subgroup analysis was executed based on all eligible studies to assess the association of all known GRHL2 gene polymorphisms and susceptibility to AHL in aging people.
2. Material and methods
2.1. Search strategy
The systematic review and meta-analysis were developed and conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklists (Supplementary Table 1). Ethical approval was unnecessary in this study because it was a meta-analysis analyzing existing articles and did not need handle individual patient data. Literature review about the associations between the polymorphisms of GRHL2 and AHL was performed in:[1] Wanfang database;[2] CNKI;[3] EMBASE;[4] Web of Science, and[5] PubMed. Keyword combinations, such as “GRHL2,” “grainyhead like transcription factor 2,” “Age-related hearing impairment,” “Age-Related Hearing loss,” and “Presbycusis” were imported as Medical Subject Headings (MeSH terms) or Entry Terms. All studies we searched were reported before August 26, 2018. Other articles that list in the reference lists of the retrieved literatures were likewise manually checked. The detailed steps of the literature review are shown in Figure 1. Inclusion and exclusion criteria. We selected studies according to the following inclusion and exclusion criteria in the meta-analysis: inclusion criteria:[1] investigating the relationship between GRHL2 gene polymorphisms and susceptibility to AHL.[2] have sufficient genotype frequencies for calculating odds ratios (ORs);[3] different independent sample sets in 1 article were included respectively;[4] case–control or cohort studies;[5] published in English or Chinese. Exclusion criteria:[1] duplicated studies;[2] reviews or cases;[3] not about gene polymorphisms;[4] unavailable data.
Figure 1.
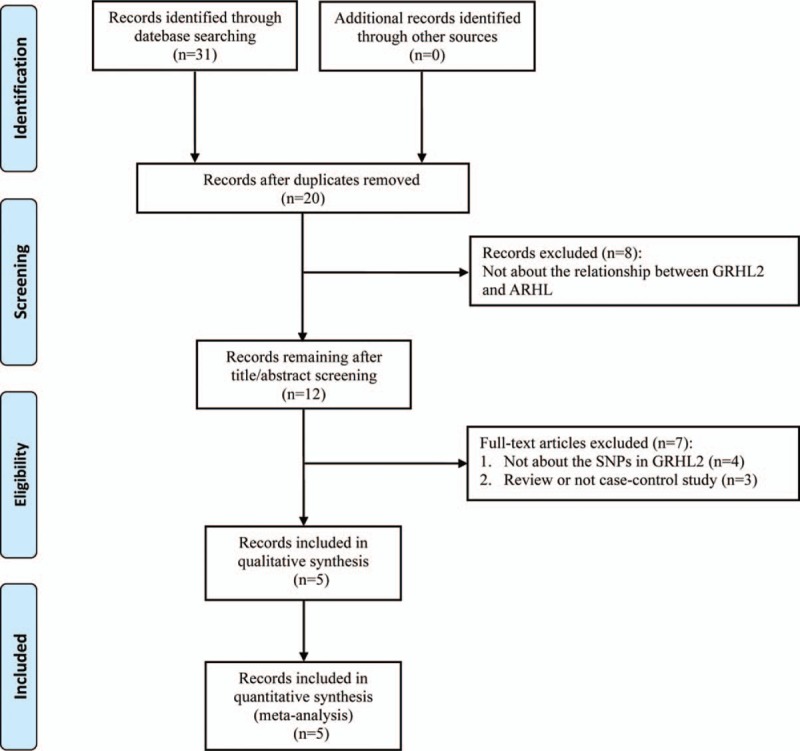
PRISMA flow chart of study selection process.
2.2. Data extraction strategy and quality assessment
Original data were extracted from the included studies by 2 investigators (BH and HS) independently. The following data were collected from the included publications: the first author, year of publication, journal, sample size of cases and controls, study population demographics (country, ethnicity, gender, and age) of AHL and controls, methods of genotyping, genotype distributions and relevant SNP polymorphisms, and Hardy–Weinberg Equilibrium (HWE) in control group. Providing that the information for meta-analysis were lacking or simply expressed graphically, we attempted to contact the authors to get more information first, and where a reply was not got, then we gauged data from the diagrams by digital ruler package or exclude. We also used the Newcastle–Ottawa scale (NOS) to assess each included study's quality. Studies that scored over 7 were considered as high-quality studies. Discrepancies were resolved by coming to an agreement after a serious discussion.
2.3. Statistical methods
We measured the SNP sites mentioned in 2 or more studies in the meta-analysis. The OR, 95% confidence interval (CI), and P value were reckoned for each study by using genotype comparison, which included these 5 gene models in total: the allele (B vs A), homozygote (BB vs AA), heterozygote (AB vs AA), dominant (BB+AB vs AA), and recessive (BB vs AB+AA) models. The purpose of this comparison is to reduce the chance of the first type of error. P value of <.05 was considered statistically significant association between AHL risks and GRHL2 gene polymorphisms. The Q-statistic test and I-square statistic were used to measure heterogeneity among the studies. A fixed-effect model by the Mantel–Haenszel method was used when I2 values <50% or P value of heterogeneity >.10, Otherwise, a random-effect model by the Mantel–Haenszel method was applied. Subgroup analyses were stratified by ethnicity (Asian or Caucasian) and research quality to investigate the potential sources of heterogeneity. Sensitivity analysis was used to forecast the pooled results’ stability. We also evaluated the HWE by the chi-square test in the control group. Studies with control groups not in HWE were still regarded for this meta-analysis, but they were omitted in the sensitivity analysis. The publication bias was measured by Egger test and Begg funnel plot. All statistical analyses were conducted using Review Manager 5.0 and Stata SE 12.0 software.
3. Results
3.1. Search results and characteristics of the eligible studies
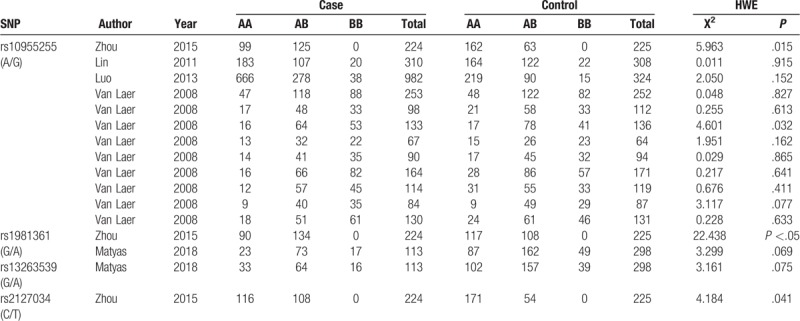
Fig. 1 shows the flow chart of the study selection in the meta-analysis. After primary literature search, 31 possibly related articles were found. Twenty articles retained for screening the title and abstract after erasing duplicate records. Eight articles were excluded as the studies were not about the relationship between GRHL2 and AHL. The remaining 12 articles were evaluated through full-text screening for qualification. Seven articles were further omitted for the reason that they are reviews (n = 3) or not about the SNPs in GRHL2 genes (n = 4). Finally, 5 articles on 13 independent studies (2762 cases and 2321 controls) satisfied the inclusion criteria and were covered in our meta-analysis. The elementary information, consisting of the NOS scores of the 13 qualified studies were summarized and showed in Table 1. Table 2 shows the genotype distribution of SNP sites in GRHL2 gene in the 13 qualified studies.
Table 1.
Summary of included studies.
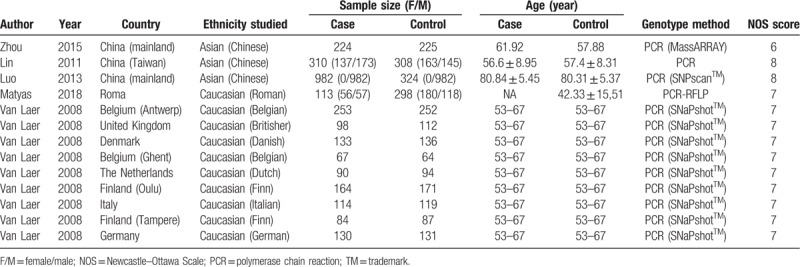
Table 2.
Genotype distribution of GRHL2 SNPs.
3.2. Meta-analysis of the SNPs in GRHL2 gene and AHL
Two SNPs (rs10955255 and rs1981361) in GRHL2 gene were selected for the meta-analysis. We chose 5 genetic models for the genotype comparison. There were 2649 cases and 2023 controls included in 9 studies on Caucasian populations and 3 studies on Asian populations for SNP rs10955255. The pooled ORs (95% CI) were 1.26 (1.05–1.50, P = .01), 1.33 (1.07–1.65, P = .01), and 1.32 (1.12–1.55, P = .0007) in the allele, homozygote, and recessive model separately. (Fig. 2 and Table 3). The evidence suggested that there may be a close relationship between the SNP site rs10955255 in the GRHL2 gene and AHL susceptibility. Regarding SNP site rs1981361, the pooled ORs (95% CI) in mixed population were 1.26 (1.02–1.56, P = .04), 1.64 (1.21–2.23, P = .001), and 1.61 (1.19–2.19, P = .02) in the allele, heterozygote, and dominant models separately (Fig. 3 and Table 3). For rs2127034 authors did not provide the original dates, so we could not include it in our study, but in the Oulu region of Finland, it is associated with AHL (P = .0009, OR = 0.58). Allele homozygote GG appears to be more common in people with hearing impairment. Homozygote AA may be a protective factor in AHL in older patients. The detailed information of the overall analysis of the association between the 2 SNP sites in GRHL2 gene and AHL susceptibility are listed in Table 3. (Figs. 4 and 5)
Figure 2.
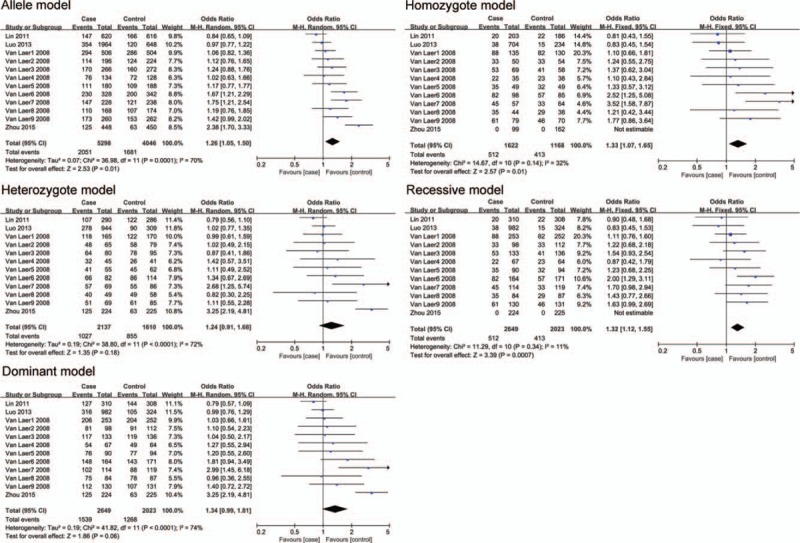
The association between polymorphism rs10955255 and age-related hearing impairment (AHL) in different genetic models. The results indicated that there is a close association in the allele, homozygous and recessive model between the SNP rs10955255 in the GRHL2 gene and AHL susceptibility.
Table 3.
Overall analysis of the association between GRHL2 SNPs and AHL susceptibility.
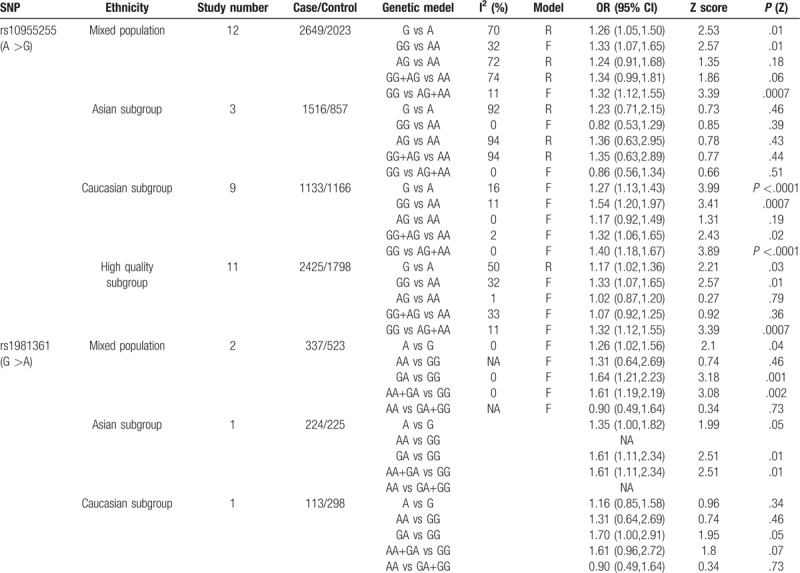
Figure 3.
The association between polymorphism rs1981361 and AHL in different genetic models. The results indicated that there may be an association in the allele, heterozygote and dominant genetic models between the SNP rs1981361 in the GRHL2 gene and AHL susceptibility.
Figure 4.
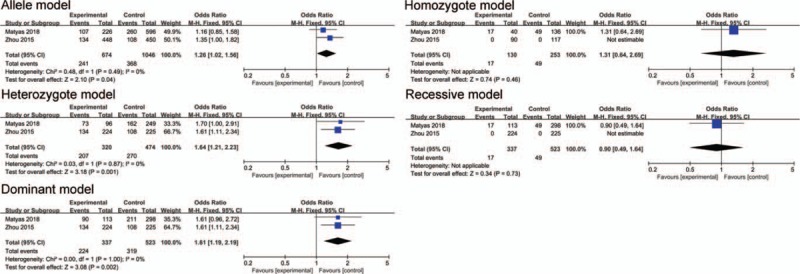
The association between polymorphism rs10955255 and AHL in 5 genetic models in Asian subgroup analysis. The results indicated that there is no statistical significance in each gene model of Asian population.
Figure 5.
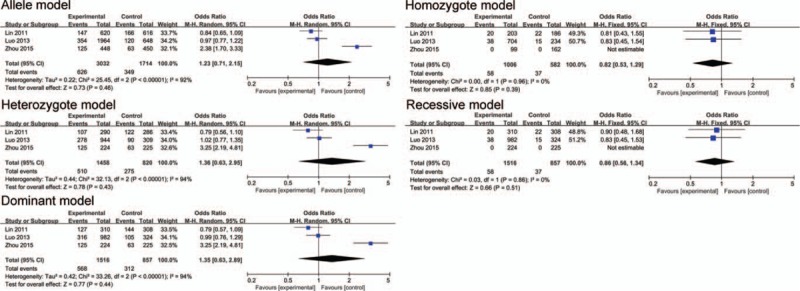
The association between polymorphism rs10955255 and AHL in 5 genetic models in Caucasian subgroup analysis. The results indicated that there is a close association in the allele model, homozygote model, dominant model and recessive model between the SNP rs10955255 in the GRHL2 gene and AHL susceptibility.
3.3. Publication bias and sensitivity analysis
As shown in Fig. 7 there is no publication bias was found in all 5 genetic models of SNP site rs10955255 via Egger test. Due to lack of sufficient data, we did not check the publication bias for rs1981361. The stability of the pooled ORs of the SNP site rs10955255 was calculated by removing each of the covered studies by turns. The results show no matter which of the covered studies was excluded, no significant change of the pooled ORs was discovered (data not shown). The consequences suggest that the relevant ORs were comparatively sturdy. For rs1981361, we did not execute the sensitivity analysis due to insufficient number of included studies.
Figure 7.
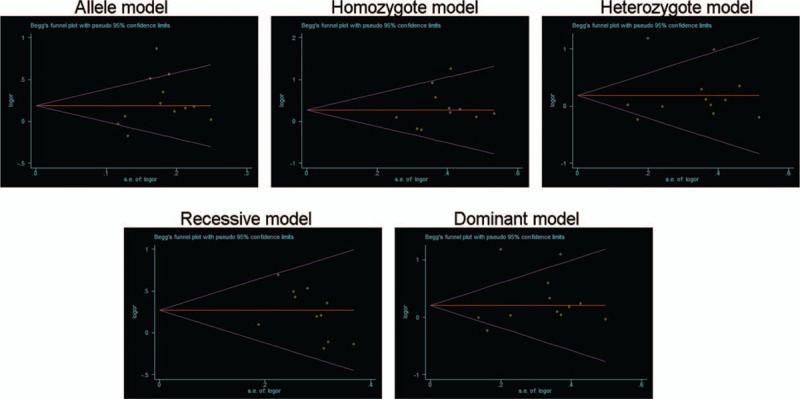
Publication bias in different genetic models of SNP rs10955255. The results indicated that no publication bias was found in all 5 genetic models of SNP rs10955255 via Bigg’ test and Egger's test.
4. Discussion
The general aim of this research was to discover the positive correlation between AHL susceptibility and 2 SNP sites (rs10955255, rs1981361) of GRHL2 gene. In our results, a significant relationship was observed between the SNP sites (rs10955255, rs1981361) and AHL, which indicated that the A allele is a potential risk factor to AHL. We first analyzed the mixed population to assess the relationship between rs10955255 and AHL. The pooled ORs showed there were significant correlations in the allele, homozygous, and recessive model, respectively. But with a high heterogeneity, to explore where it came from, we conducted a subgroup analysis according to 9 case–control studies in Caucasians and 3 case–control studies in Asians. The outcomes revealed the heterogeneity between-study was decreased to accredited level after analysis by ethnicity, suggesting that the source of heterogeneity may mainly come from ethnicity. Subgroup analysis revealed that a significant association was observed between rs10955255 and AHL in the allele model, homozygote model, dominant model, and recessive model in the Caucasian population, but no statistical significance in each gene model of Asian population. In groups of Caucasians, a significant association was observed between rs10955255 and AHL in the allele model, homozygote model, dominant model, and recessive model, while no statistical significance in each gene model of Asians. In addition, Caucasians with the G allele were especially vulnerable to AHL (allele model, G versus A: OR (95% CI) = 1.27 (1.13–1.43)). Also, the NOS scale was employed to validate the quality of these studies (Fig. 6 and Table 3) and eventually confirmed it maybe one of the sources of heterogeneity. For rs1981361 polymorphism, an increased risks of AHL was found under allele model (A vs G), heterozygote model (GA vs GG) and dominant model (AA+GA vs GG), but not homozygote model (AA vs GG) and recessive model (AA vs GA+GG), suggesting that the risk of AHL is increased in general population with A allele. However, there was only 1 study in Asian and 1 study in Caucasian population separately, so we were unable to do subgroup analysis.
Figure 6.
The association between polymorphism rs10955255 and AHL in 5 genetic models in high quality subgroup analysis. The results indicated that there is a close association in the allele model, homozygote model and recessive model between the SNP rs10955255 in the GRHL2 gene and AHL susceptibility.
The present study has several restrictions that should be acknowledged. The sample size included in the meta-analysis is relatively small. In our meta-analysis, there were only 2 studies included for SNP site rs1981361, its funnel plots were highly asymmetric, which suggested there is a significant publication bias (data not shown). Limited studies also increase the heterogeneity. For rs2127034 and 13263539, there is only 1 related research respectively, which restricted our next analysis. In addition, Zhou's study showed that the distribution of rs10955255, rs2127034, rs1981361 did not comply with the HWE, the same to Van Laer, L’ study of rs10955255 conducted in Denmark. We need a larger sample of reports to further update our conclusion, though 2321 controls and 2762 cases are included in this meta-analysis. The majority of the included studies were about Caucasians, and only a few about Asians, which could make our findings less relevant for the Asian population. We only detected the studies that published in Chinese and English. Maybe there are studies in other languages not covered in this meta-analysis. Therefore, the results could increase random mistake, publication bias, and so on.
As far as we know, fairly complex natural environmental factors and multiple genetic factors, as well as other unknown factors, together led to the occurrence and development of AHL. At present, AHL has become a severe health and social problem. Accumulating testimony showed the allele G in rs10955255 and the allele A rs1981361 (simply in Caucasians) may be the genetic predisposition factor of AHL. However, given the above limitations, larger studies and further meta-analyses based on populations and other factors should be performed to clarify the association of GRHL2 gene polymorphisms and the risks of AHL.
Author contributions
Formal analysis: Baoai Han, Xiuping Yang, Yongqin Li, Yaqin Tu, Yaodong Dong, Kai Zhang.
Software: Haiying Sun, Baoai Han, Zuhong He, Jie Yuan, Hua Cai.
Supervision: Haiying Sun, Tao Zhou.
Writing – original draft: Baoai Han, Xiuping Yang, Yongqin Li, Davood K Hosseini, Xiujuan Zhang.
Writing – review & editing: Haiying Sun, Tao Zhou.
Haiying Sun orcid: 0000-0001-8112-4803.
Footnotes
Abbreviations: ARHI = age-related hearing impairment, CI = confidence interval, CNKI = China National Knowledge Infrastructure, GRHL2 = grainyhead-like-2, HWE = Hardy–Weinberg Equilibrium, NOS = Newcastle–Ottawa scale, OR = odds radio.
BH, XY, and YL contribute the equal work to this article.
This study was supported by the National Natural Science Foundation of China (No, 81600801, 81771005, 81502527). National Science Foundation of Hubei Province, China (No, 2015CFB527).
The authors have no funding, financial relationships, or conflicts of interest to disclose.
References
- [1].Huang Q, Tang J. Age-related hearing loss or presbycusis. Eur Arch Oto-rhino-laryngol Off J Eur Federation Oto-Rhino-Laryngol Soc EUFOS Affil German Soc Oto-Rhino-Laryngol Head Neck Surg 2010;267:1179–91. [DOI] [PubMed] [Google Scholar]
- [2].Uchida Y, Sugiura S, Nishita Y, et al. Age-related hearing loss and cognitive decline - The potential mechanisms linking the two. Auris, nasus, larynx 2019;46:1–9. [DOI] [PubMed] [Google Scholar]
- [3].Shukla A, Nieman CL, Price C, et al. Impact of hearing loss on patient-provider communication among hospitalized patients: a systematic review. Am J Med Qual Off J Am Coll Med Qual 2019;34:284–92. [DOI] [PubMed] [Google Scholar]
- [4].Punch R, Horstmanshof L. Hearing loss and its impact on residents in long term care facilities: a systematic review of literature. Geriatr Nurs 2019;40:138–47. [DOI] [PubMed] [Google Scholar]
- [5].Stern D, Hilly O. The relationship between hearing loss and cognitive decline in the elderly and the efficiency of hearing rehabilitation in preventing cognitive decline. Harefuah 2018;157:374–7. [PubMed] [Google Scholar]
- [6].Brewster KK, Ciarleglio A, Brown PJ, et al. Age-related hearing loss and its association with depression in later life. Am J Geriatr Psychiatry Off J Am Assoc Geriatr Psychiatry 2018;26:788–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang F, Yao L, Peng YC, et al. Application of dysphonia severity index in laryngeal reflux related voice diseases. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi J Clin Otorhinolaryngol Head Neck Surg 2017;31:1745–8. [DOI] [PubMed] [Google Scholar]
- [8].Xing YZ, Pei JJ, Sun YT, et al. Clinical research of anxiety and depression state among patients suffered from sudden deafness with vertigo. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi J Clin Otorhinolaryngol Head Neck Surg 2017;31:1919–22. [DOI] [PubMed] [Google Scholar]
- [9].Mamo SK, Nirmalasari O, Nieman CL, et al. Hearing care intervention for persons with dementia: a pilot study. Am J Geriatr Psychiatry Off J Am Assoc Geriatr Psychiatry 2017;25:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li-Korotky HS. Age-related hearing loss: quality of care for quality of life. Gerontologist 2012;52:265–71. [DOI] [PubMed] [Google Scholar]
- [11].Fernandez-D’ Pool J, Butron J, Colina-Chourio J. Effect of noise on blood pressure in workers of a Venezuelan oil company. Invest Clin 2010;51:301–14. [PubMed] [Google Scholar]
- [12].Prendergast G, Millman RE, Guest H, et al. Effects of noise exposure on young adults with normal audiograms II: behavioral measures. Hearing Res 2017;356:74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yu P, Jiao J, Chen G, et al. Effect of GRM7 polymorphisms on the development of noise-induced hearing loss in Chinese Han workers: a nested case-control study. BMC Med Genet 2018;19:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vuckovic D, Mezzavilla M, Cocca M, et al. Whole-genome sequencing reveals new insights into age-related hearing loss: cumulative effects, pleiotropy and the role of selection. Eur J Hum Genet EJHG 2018;26:1167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Soliman SE, D'Silva CN, Dimaras H, et al. Clinical and genetic associations for carboplatin-related ototoxicity in children treated for retinoblastoma: a retrospective noncomparative single-institute experience. Pediatr Blood Cancer 2018;65:e26931. [DOI] [PubMed] [Google Scholar]
- [16].Sliwiniska-Kowalska M, Pawelczyk M, Kowalski TJ. Genetic factors in susceptibility to age- and noise-related hearing loss. Polski Merkuriusz Lekarski: Organ Polskiego Towarzystwa Lekarskiego 2006;21:384–8. [PubMed] [Google Scholar]
- [17].Han Y, Mu Y, Li X, et al. Grhl2 deficiency impairs otic development and hearing ability in a zebrafish model of the progressive dominant hearing loss DFNA28. Hum Mol Genet 2011;20:3213–26. [DOI] [PubMed] [Google Scholar]


