Abstract
Background:
TP53 gene polymorphism could increase risks of several kinds of cancer. But it remained controversial whether TP53 gene codon72 polymorphism was associated with the susceptibility to prostate cancer. Thus, we conducted a meta-analysis that evaluated the association between TP53 gene codon72 polymorphism and prostate cancer risk.
Method:
A comprehensive research was performed from PubMed, Embase, Web of Science and China National Knowledge Infrastructure (CNKI) up to December 31, 2018. A random effect model was used to evaluate the effect of the outcome. The statistical analyses were performed with Review Manager 5.3.0 and Stata 14.0. The sensitivity analysis and publication bias tests were also performed to confirm the reliability of this meta-analysis.
Results:
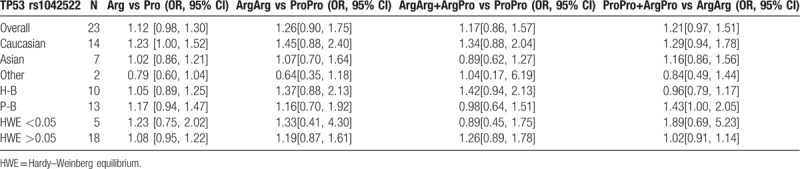
22 studies included 3146 cases and 4010 controls were involved in this meta-analysis. Overall, no association was observed between TP53 gene codon72 polymorphism and prostate cancer risk (Arg vs Pro: odds ratio [OR] = 1.12, 95% confidence interval [CI] = 0.98–1.30; ArgArg vs ProPro: OR = 1.26, 95% CI = 0.90–1.75; ProPro vs ArgArg+ ArgPro: OR = 1.17, 95% CI = 0.86–1.57; ArgPro+ ProPro vs ArgArg: OR = 1.21, 95% CI = 0.97–1.51). Subgroup analyses, based on ethnicity, source of control and Hardy–Weinberg equilibrium (HWE) status, showed consistent results.
Conclusion:
The meta-analysis we performed showed that there was no association of TP53 gene codon72 polymorphism with prostate cancer risk.
Keywords: meta-analysis, polymorphism, prostate cancer, TP53 gene
1. Introduction
Prostate cancer is the third most common cancer in the world, and it is also the second most common cancer among men.[1] It is also the second leading reason of cancer death in American males.[2] In addition to some risk factors like age, inflammation and food factor,[3,4] previous studies showed that heritable susceptibility also played an important role in the development of prostate cancer, and several gene mutations have been reported to be associated with the development and prognosis of prostate cancer.[5–7] Some studies also suggested that TP53 gene polymorphism was a possible risk factor of prostate cancer.
TP53 gene is located on chromosome 17p13 and it consists of 11 exons.[8,9] P53 protein, the product of TP53 gene, is a tumor suppressor protein that can induce cell cycle arrest and apoptosis in response to genotoxic stress.[10] It also controls some other cellular processes, including self-renewal of stem cells, autophagy, and reprogramming of differentiated cells into metastasis, immune system or stem cells.[11,12] TP53 gene mutations were associated with several kinds of cancer, such as lung cancer, breast cancer, and colon cancer.[13–15] TP53 codon72 polymorphism (rs1042522) is an important functional polymorphic form that encodes amino acids arginine (CGC) or proline (CCC).[16] Moreover, previous studies have shown that Arg72 and Pro72 variants may lead to different biochemical and biological properties of the p53 protein.[17,18] Meanwhile, studies also reported the possible association of TP53 gene polymorphism with prostate cancer risk.
To date, there are several studies that evaluate the association between TP53 codon72 polymorphism and prostate cancer. However, most of these studies did not include large patient samples, and the results are inconclusive rather than consistent. Although there were several meta-analyses that had investigated the association, results were also inconclusive.[19–21] Therefore, in this article, we conducted a comprehensive meta-analysis from all relevant scientific literatures.
2. Methods and materials
2.1. Searching strategy
Two authors independently performed a comprehensive search, using PubMed, Embase, Web of Science and China National Knowledge Infrastructure (CNKI) up to December 31, 2018. Search terms were as follows: “P53,” “TP53,” “polymorphism, mutation or variant,” “prostate cancer.” Besides, the references of reviews and several retrieved articles were also reviewed to identify other eligible studies that could be missed by the search. The search was limited to human subjects only. The search strategy flow chart is shown in Figure 1.
Figure 1.
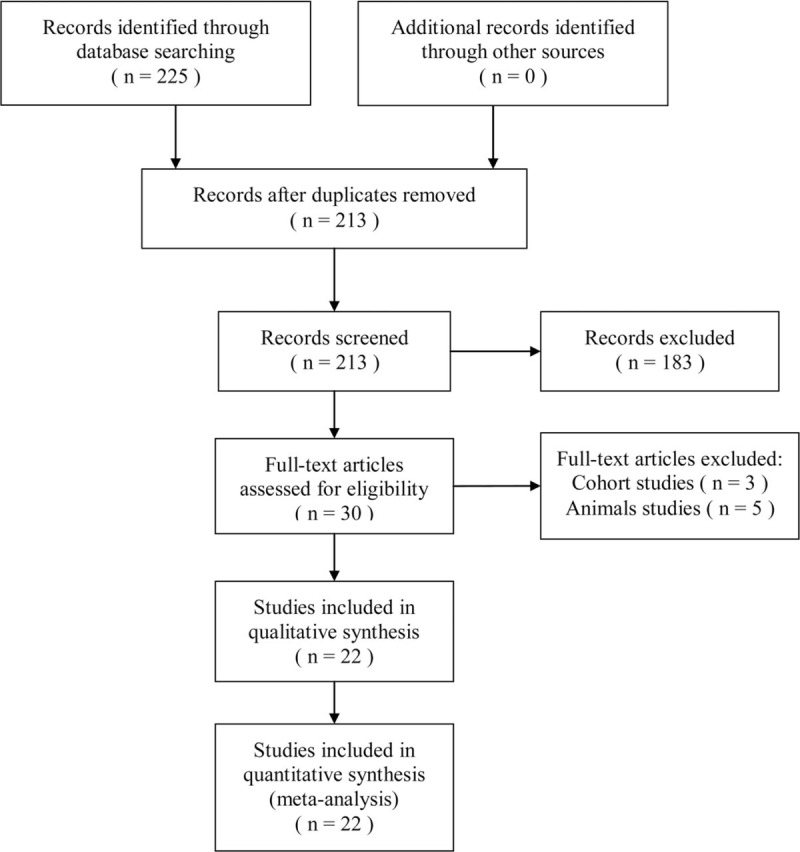
Flow chart of the study selection.
2.2. Inclusion criteria and exclusion criteria
Only the studies according to the following inclusion criteria were included:
-
(a)
studies with full-text articles;
-
(b)
case–control studies that evaluated the relationship between TP53 codon72 gene polymorphism and the susceptibility to prostate cancer;
-
(c)
the genotype distributions were available for both cases and controls;
-
(d)
no overlapping data.
Studies were excluded if meeting any of the following exclusion criteria:
-
(a)
not for the association between TP53 codon72 gene polymorphism and the risk of prostate cancer;
-
(b)
studies with partial unusable or undefined data;
-
(c)
animal studies, review articles, meta-analyses, conference abstracts, or editorial articles.
2.3. Quality assessment
We used the Newcastle–Ottawa Scale (NOS) to assess the quality of the included studies.[22] The NOS contains 8 parts for cohort or case–control studies. It is categorized into 3 parts including selection, comparability, and exposure for case–control studies. Selection has a maximum of 4 points, Comparability has a maximum of 2 points and Exposure has a maximum of 3 points. Scores ranged from 0 (worst) to 9 (best), and the quality of each study was graded as low (0–3), moderate (4–6), and high (7–9). Inconsistent opinions were solved by discussion and consensus.
2.4. Data extraction
Two authors reviewed the eligible scientific reports and extracted the relevant data independently according to the inclusion criteria. Then, extracted data were collected into a collection form and checked by a third author. Discrepancy was solved by discussion and consensus finding. In the meta-analysis, we collected the following information for each study:
-
(a)
the first author's name, year of publication, country, genotyping method, races, source of control;
-
(b)
(b)the number of people that were included in the case and control groups;
-
(c)
the results of the Hardy–Weinberg equilibrium (HWE) test
-
(d)
the scores evaluated by NOS.
2.5. Statistical analysis
The strength of the association between TP53 codon72 gene polymorphism and prostate cancer risk was measured by using odds ratio (OR) and corresponding 95% confidence interval (CI). The ORs were performed for 4 models, which are allele model, additive model, recessive model, and dominant model. Heterogeneity assumption was tested by the chi-square-based Q test. The heterogeneity was considered significant when P <.10, and I2 values of 25%, 50%, and 75% referred to low, medium and high levels of heterogeneity, respectively. A random-effect model was used in the analysis. The significance of the pooled OR was determined by the Z-test, and when P <.05, it was regarded as statistically significant. The statistical analysis was performed with Reviewer Manager 5.3.0 and Stata 14.0. The potential publication bias was evaluated applying Begg test, Egger test and funnel plots. We also performed sensitivity analysis to assess the reliability of the results. The pooled ORs were estimated by removing 1 study each time to evaluate the impact of an individual study.
3. Results
3.1. Study characteristics
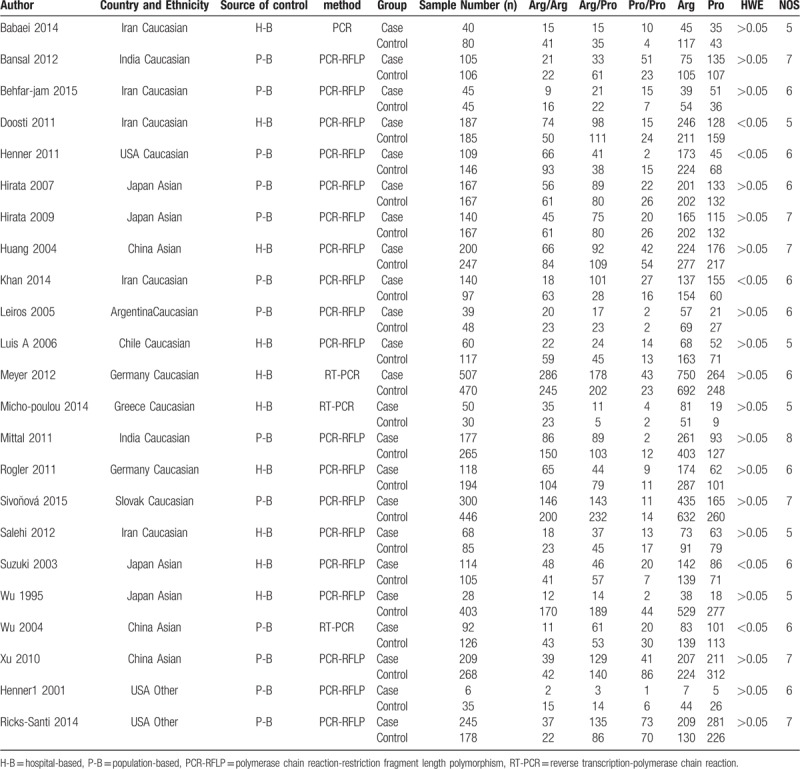
The 225 articles were retrieved after the first search in PubMed, Embase, Web of Science and CNKI. The 202 articles were excluded, according to the inclusion and exclusion criteria. Finally, after the careful selection, 22 case–control studies involving 3146 cases and 4010 controls were included in this meta-analysis.[23–44] All these studies were published between 1995 and 2015. Of these, 7 studies were based on Asian, 14 studies based on Caucasian and another 2 studies were based on other races. We also conducted the HWE test for these studies, and HWE was violated in 5 studies. As for the source of control, 10 studies were hospital-based (H-B), and others were population-based (P-B). Every study's scores were moderate or better, based on NOS. The detailed characteristics of included studies were listed in Table 1. All analyses were based on previous studies, thus no ethical approval and patient consent are required.
Table 1.
Characteristics of studies included in the meta-analysis.
3.2. Meta-analysis results
The influence of TP53 codon72 polymorphism on prostate cancer was totally evaluated by 22 case–control studies including 7156 individuals. Figures 2–5 show the results of the allele model (Arg vs Pro), additive model (ArgArg vs ProPro), recessive model (ProPro vs ArgArg+ ArgPro) and dominant model (ArgPro+ ProPro vs ArgArg). Overall, the result showed that there was no significant association between TP53 codon72 polymorphism and prostate cancer risk. (Arg vs Pro: OR = 1.12, 95% CI = 0.98–1.30; ArgArg vs ProPro: OR = 1.26, 95% CI = 0.90–1.75; ProPro vs ArgArg+ ArgPro: OR = 1.17, 95% CI = 0.86–1.57; ArgPro+ ProPro vs ArgArg: OR = 1.21, 95% CI = 0.97–1.51).
Figure 2.
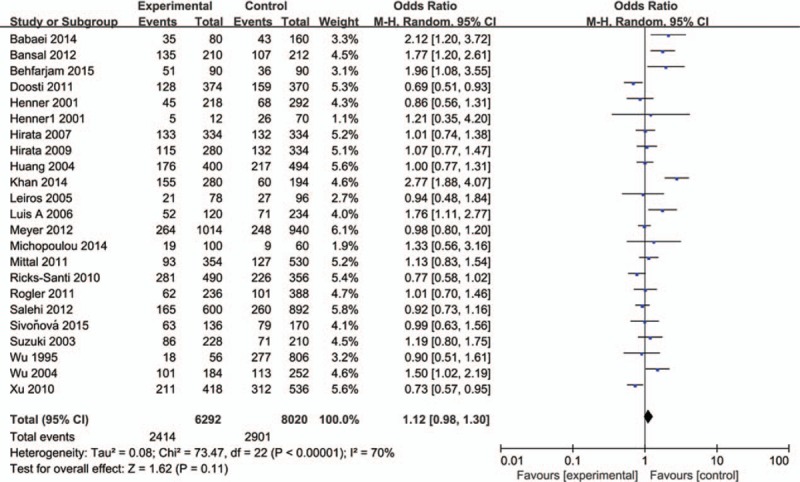
Forest plot of the studies evaluating the association of TP53 codon72 polymorphism with prostate cancer risk (Allele model: Pro vs Arg).
Figure 5.
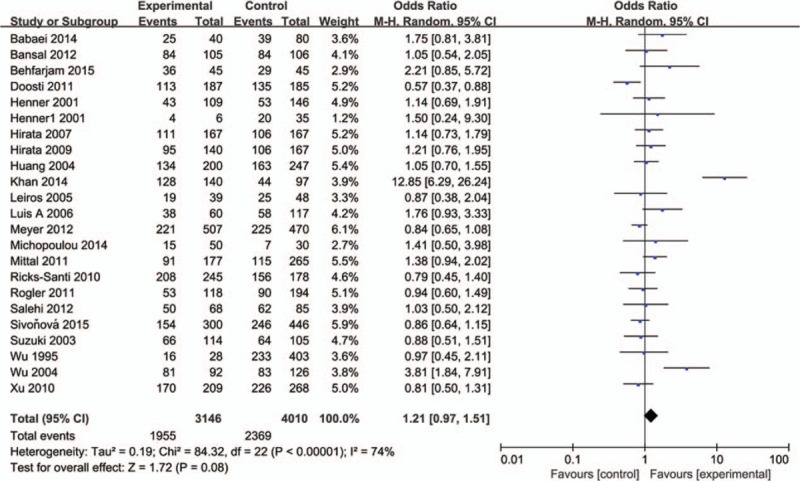
Forest plot of the studies evaluating the association of TP53 codon72 polymorphism with prostate cancer risk (Dominant model: ProPro+ArgPro vs ArgArg).
Figure 3.
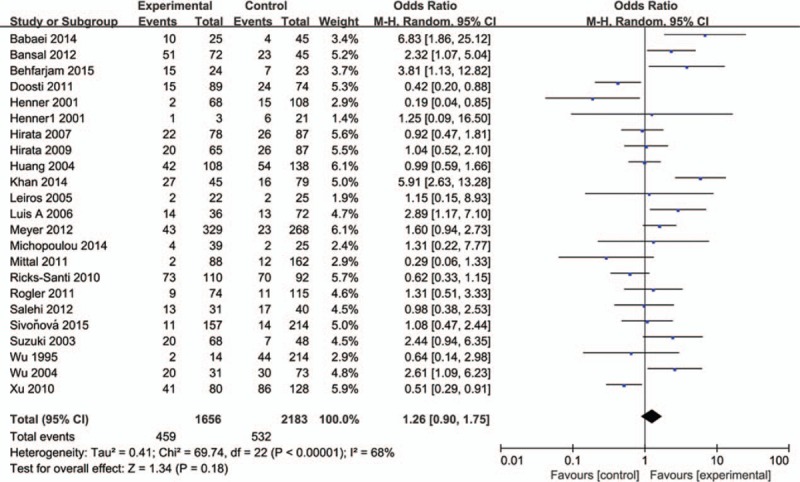
Forest plot of the studies evaluating the association of TP53 codon72 polymorphism with prostate cancer risk (Additive model: ProPro vs ArgArg).
Figure 4.
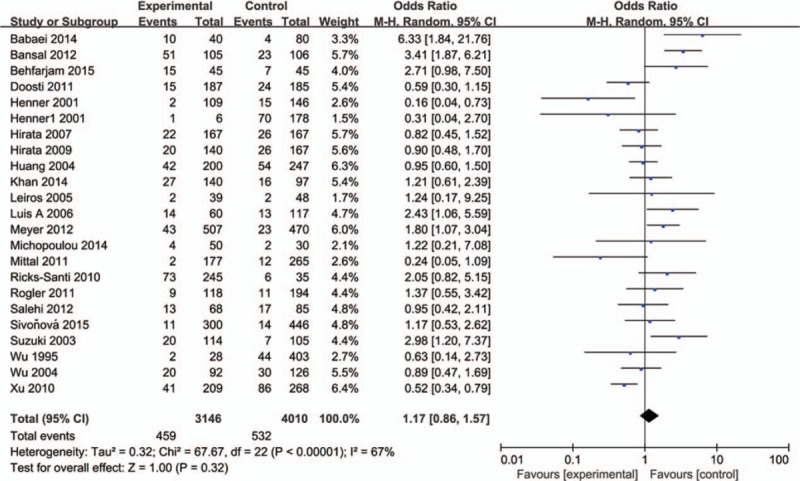
Forest plot of the studies evaluating the association of TP53 codon72 polymorphism with prostate cancer risk (Recessive model: ProPro vs ArgArg+ArgPro).
In the subgroup analysis by HWE status, no significant association between TP53 codon72 polymorphism and prostate cancer risk was observed in 4 models (Table 2). A weak association of TP53 gene codon72 polymorphism and prostate cancer risk was observed in the allele model in Caucasians (OR = 1.23, 95% CI = 1.00–1.52). No association was found among Asian in 4 models. (Table 2) A possible weak association was also observed in the dominant model in the population-based group (OR = 1.43, 95% CI = 1.00–2.05). In the hospital-based group, we found that TP53 gene codon72 polymorphism was not associated with prostate cancer susceptibility (Arg vs Pro: OR = 1.07, 95% CI = 0.90–1.29; ArgArg vs ProPro: OR = 1.37, 95% CI = 0.88–2.13; ProPro vs ArgArg+ ArgPro: OR = 1.42, 95% CI = 0.94–2.13; ArgPro+ ProPro vs ArgArg: OR = 0.96, 95% CI = 0.79–1.17). After all, we found no association of TP53 gene codon72 polymorphism with prostate cancer risk based on subgroup analysis. The results were shown in Table 2.
Table 2.
Meta-analysis of the association of TP53 codon72 polymorphism with prostate cancer risk.
3.3. Sensitivity analysis and publication bias
Sensitivity analysis was performed by omitting 1 study each time in 4 models; the results showed that the overall pooled ORs were not influenced by any individual study, indicating the results of this meta-analysis are stable. (Fig. 6)
Figure 6.
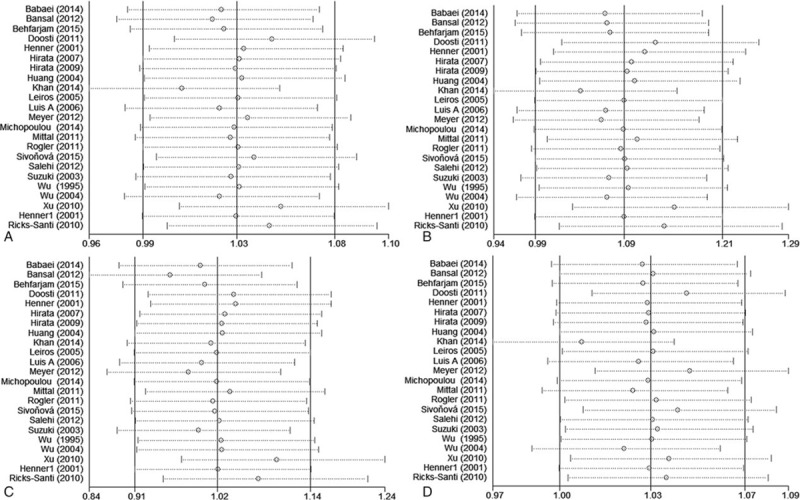
Sensitivity analysis diagram for each study used to evaluate the relative risk estimated for the TP53 codon72 polymorphism and prostate cancer risk in all of the included studies (A. allele model: Pro vs Arg; B. additive model: ProPro vs ArgArg; C. recessive model: ProPro vs ArgArg+ArgPro; D. dominant model: ProPro+ArgPro vs ArgArg).
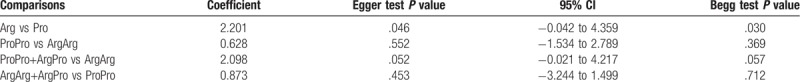
Begg test, Egger test, and funnel plots were conducted to assess the publication bias on TP53 codon72 polymorphism.(Fig. 7) No publication bias was observed based on funnel plots or according to Begg and Egger test for prostate cancer risk in additive model, recessive model, and dominant model. In addition, some publication bias was observed in the results of allele model according to Begg and Egger tests (Begg test: P = .030; Egger test P = .046). The results were shown in Table 3.
Figure 7.
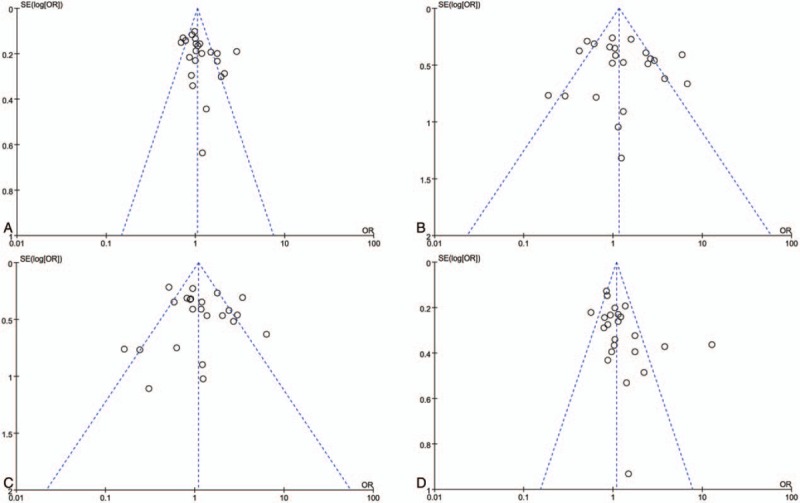
Funnel plots for the TP53 codon72 polymorphism and prostate cancer risk. (A. allele model: Pro vs Arg; B. additive model: ProPro vs ArgArg; C. recessive model: ProPro vs ArgArg+ArgPro; D. dominant model: ProPro+ArgPro vs ArgArg).
Table 3.
Publication bias tests for the TP53 codon72 polymorphism.
4. Discussion
Overall, in our meta-analysis, we found no association of TP53 gene polymorphism with prostate cancer risk in 4 models (Arg vs Pro: OR = 1.12, 95% CI = 0.98–1.30; ArgArg vs ProPro: OR = 1.26, 95% CI = 0.90–1.75; ProPro vs ArgArg+ ArgPro: OR = 1.17, 95% CI = 0.86–1.57; ArgPro+ ProPro vs ArgArg: OR = 1.21, 95% CI = 0.97–1.51). In subgroup analyses by ethnicity, source of control and HWE status, no significant association was observed between prostate cancer risk and TP53 gene polymorphism. (Table 2)
Studies showed that TP53 gene mutations could have an impact on 50% of human cancers,[45] and several studies have been taken to study the underlying mechanism of the association between TP53 gene and prostate cancer, as well. For example, Ashkari et al reported that p53 may translocate to the cytoplasm by androgen-mediated induction of G3BP2, a newly described direct target gene of androgen receptor, which played a central role in prostate cancer progression.[46] Potential gene–gene interaction could also play a vital role in the association of TP53 gene polymorphism and prostate cancer risk. Wang et al demonstrated that ELL Associated Factor 2 (EAF2) gene and TP53 gene may functionally interact in prostate tumor suppression and the simultaneous inactivation of EAF2 and TP53 may drive prostate carcinogenesis, based on their findings on mice.[47] However, the association between TP53 gene codon72 polymorphism and prostate cancer risk remained unclear. To draw a better comprehensive understanding, we conducted this meta-analysis to evaluate the association of TP53 gene codon72 polymorphism with prostate cancer risk. And the result of our meta-analysis implied no association between TP53 gene codon72 polymorphism and the risk of prostate cancer.
In previous studies, some of the researchers thought that TP53 codon72 polymorphism was significantly associated with prostate cancer risk. Mittal et al[35] observed that individuals with heterozygous genotype of TP53 codon72 polymorphism demonstrated prostate cancer risk (OR = 1.5, 95% CI = 1.00–2.199). Similarly, Xu et al[44] also found that the frequencies of TP53 codon72 between the case group and control group were significantly different (P <.01), after adjusting some potential covariates.
However, some other studies’ conclusions were inconsistent. For example, Huang et al,[30] one of the studies included, found no significant association between p53 polymorphism and risk of prostate cancer. Michopolou et al[34] observed no statistically significant association between the HPV presence and TP53 codon 72 polymorphism, and in that case, they did not think that TP53 polymorphism status at codon 72 was associated with prostate cancer. It is possible to lead to the inconsistence, because of the scale of samples or other environmental factors that were not considered.
Thus, these conclusions needed further validation based on a larger population. Meanwhile, other factors, like ethnicity, should be taken in consideration. Therefore, we conducted this meta-analysis. And the results showed no association between TP53 polymorphism and prostate cancer risk, which was consistent with a previous study.[19] However, in this meta-analysis, we included more studies than before. In addition, we also used NOS to evaluate the methodological quality of the studies included, and it helped us to pick out and evaluate eligible articles.
I2 statistics and Q test were performed to evaluate the significance of heterogeneity in this meta-analysis. Significant heterogeneity among the including studies was found in all 4 models. After subgrouped by ethnicity, source of control, and HWE status, the heterogeneity remained obvious. Therefore, we considered the heterogeneity may result from the variety of countries that studies were published and other confounding factors. Moreover, some limitations of this meta-analysis should be taken in consideration. First, we did not estimate some latent hereditary factors, like the potential gene-gene and gene-environment interactions, because of the lack of information available in the original studies included. Second, subject age, sample quality, and some other clinical data, were not considered here, due to the lack of information. Third, publication bias existed in the allele model, which indicated that more studies should be taken and included.
5. Conclusion
In conclusion, this meta-analysis suggested no association between TP53 codon72 polymorphism and prostate cancer risk. Nevertheless, more large and representative case-control studies are needed for the validation of our conclusion.
Author contributions
Conceptualization: Zheng-Ju Ren.
Data curation: Xue-Ling Zhang.
Formal analysis: Pei-Zhen Han.
Funding acquisition: Qiang Wei.
Investigation: Pei-Zhen Han.
Methodology: Xue-Ling Zhang.
Resources: Zheng-Ju Ren.
Software: Zheng-Ju Ren.
Supervision: De-Hong Cao, Qiang Wei.
Visualization: Xue-Ling Zhang.
Writing – original draft: Pei-Zhen Han.
Writing – review & editing: Pei-Zhen Han.
Footnotes
Abbreviations: CIs = confidence intervals, CNKI = China National Knowledge Infrastructure, EAF2 = ELL Associated Factor 2, HWE = Hardy–Weinberg equilibrium, NOS = Newcastle–Ottawa Scale, ORs = odds ratios.
This research was funded by the National Natural Science Foundation of China (Grant no. 81770756).
The authors have no conflicts of interest to disclose.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Gomella LG. Prostate cancer statistics: anything you want them to be. Canad J Urol 2017;24:8603–4. [PubMed] [Google Scholar]
- [3].Gathirua-Mwangi WG, Zhang J. Dietary factors and risk for advanced prostate cancer. Eur J Cancer Prevent Off J Eur Cancer Prevent Organ (ECP) 2014;23:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Platz EA, Kulac I, Barber JR, et al. A prospective study of chronic inflammation in benign prostate tissue and risk of prostate cancer: linked PCPT and SELECT cohorts. Cancer Epidemiol Biomarkers Prevent Publ Am Assoc Cancer Res Cosponsored Am Soc Prevent Oncol 2017;26:1549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Giri VN, Beebe-Dimmer JL. Familial prostate cancer. Seminars Oncol 2016;43:560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lecarpentier J, Silvestri V, Kuchenbaecker KB, et al. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J Clin Oncol Off J Am Soc Clin Oncol 2017;35:2240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Walsh PC. The search for the missing heritability of prostate cancer. Eur Urol 2017;72:657–9. [DOI] [PubMed] [Google Scholar]
- [8].Hashemi M, Amininia S, Ebrahimi M, et al. Association between polymorphisms in TP53 and MDM2 genes and susceptibility to prostate cancer. Oncol Lett 2017;13:2483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu Y, Chen C, Xu Z, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 2016;531:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tonnessen-Murray CA, Lozano G, Jackson JG. The regulation of cellular functions by the p53 protein: cellular senescence. Cold Spring Harbor Perspect Med 2017;7:7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kamada R, Toguchi Y, Nomura T, et al. Tetramer formation of tumor suppressor protein p53: Structure, function, and applications. Biopolymers 2016;106:598–612. [DOI] [PubMed] [Google Scholar]
- [12].Muller PA, Vousden KH. p53 mutations in cancer. Nature Cell Biol 2013;15:2–8. [DOI] [PubMed] [Google Scholar]
- [13].Labbe C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer (Amsterdam, Netherlands) 2017;111:23–9. [DOI] [PubMed] [Google Scholar]
- [14].Schon K, Tischkowitz M. Clinical implications of germline mutations in breast cancer: TP53. Breast Cancer Res Treat 2018;167:417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yurgelun MB, Masciari S, Joshi VA, et al. Germline TP53 mutations in patients with early-onset colorectal cancer in the colon cancer family registry. JAMA Oncol 2015;1:214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rodriguez C, Sobrino T, Agulla J, et al. Neovascularization and functional recovery after intracerebral hemorrhage is conditioned by the Tp53 Arg72Pro single-nucleotide polymorphism. Cell Death Differ 2017;24:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Singamsetty GK, Malempati S, Bhogadhi S, et al. TP53 alterations and colorectal cancer predisposition in south Indian population: a case-control study. Tumour Biol J Int Soc Oncodevelop Biol Med 2014;35:2303–11. [DOI] [PubMed] [Google Scholar]
- [18].Szymanowska A, Jassem E, Dziadziuszko R, et al. Increased risk of non-small cell lung cancer and frequency of somatic TP53 gene mutations in Pro72 carriers of TP53 Arg72Pro polymorphism. Lung Cancer (Amsterdam, Netherlands) 2006;52:9–14. [DOI] [PubMed] [Google Scholar]
- [19].Fan S, Hao ZY, Zhang M, et al. Association between the rs1042522 polymorphism in TP53 and prostate cancer risk: an updated meta-analysis. Chronic Dis Transl Med 2017;3:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li MS, Liu JL, Wu Y, et al. Meta-analysis demonstrates no association between p53 codon 72 polymorphism and prostate cancer risk. Genet Mol Res GMR 2011;10:2924–33. [DOI] [PubMed] [Google Scholar]
- [21].Zhang J, Zhuo WL, Zheng Y, et al. Polymorphisms of TP53 codon 72 with prostate carcinoma risk: a meta-analysis. Med Oncol (Northwood, Lond, Engl) 2010;27:540–6. [DOI] [PubMed] [Google Scholar]
- [22].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [23].Abbas D, Payam GD. The p53 codon 72 polymorphism and association to prostate cancer in Iranian patients. African J Biotechnol 2011;10:12821–5. [Google Scholar]
- [24].Babaei F, Ahmadi SA, Abiri R, et al. The TP53 codon 72 polymorphism and risk of sporadic prostate cancer among iranian patients. Iranian J Public Health 2014;43:453–9. [PMC free article] [PubMed] [Google Scholar]
- [25].Bansal A, Soni A, Rao P, et al. Implication of DNA repair genes in prostate tumourigenesis in Indian males. Indian J Med Res 2012;136:622–32. [PMC free article] [PubMed] [Google Scholar]
- [26].Behfarjam F, Rostamzadeh J, Zarei MA, et al. Association of two polymorphic codons in P53 and ABCC1 promoter with prostate cancer. Iranian J Biotechnol 2015;13:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Henner WD, Evans AJ, Hough KM, et al. Association of codon 72 polymorphism of p53 with lower prostate cancer risk. Prostate 2001;49:263–6. [DOI] [PubMed] [Google Scholar]
- [28].Hirata H, Hinoda Y, Kikuno N, et al. CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin Cancer Res 2007;13:5056–62. [DOI] [PubMed] [Google Scholar]
- [29].Hirata H, Hinoda Y, Kikuno N, et al. Bcl2 -938C/A polymorphism carries increased risk of biochemical recurrence after radical prostatectomy. J Urol 2009;181:1907–12. [DOI] [PubMed] [Google Scholar]
- [30].Huang SP, Wu WJ, Chang WS, et al. p53 codon 72 and p21 codon 31 polymorphisms in prostate cancer. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prevent Oncol 2004;13:2217–24. [PubMed] [Google Scholar]
- [31].Khan MH, Rashid H, Mansoor Q, et al. Association of rs1042522 polymorphism with increased risk of prostate adenocarcinoma in the Pakistani population and its HuGE review. Asian Pac J Cancer Prev 2014;15:3973–80. [DOI] [PubMed] [Google Scholar]
- [32].Leiros GJ, Galliano SR, Sember ME, et al. Detection of human papillomavirus DNA and p53 codon 72 polymorphism in prostate carcinomas of patients from Argentina. BMC Urol 2005;5:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meyer A, Coinac I, Bogdanova N, et al. Apoptosis gene polymorphisms and risk of prostate cancer: a hospital-based study of German patients treated with brachytherapy. Urol Oncol 2013;31:74–81. [DOI] [PubMed] [Google Scholar]
- [34].Michopoulou V, Derdas SP, Symvoulakis E, et al. Detection of human papillomavirus (HPV) DNA prevalence and p53 codon 72 (Arg72Pro) polymorphism in prostate cancer in a Greek group of patients. Tumour Biol J Int Soc Oncodevelop Biol Med 2014;35:12765–73. [DOI] [PubMed] [Google Scholar]
- [35].Mittal RD, George GP, Mishra J, et al. Role of functional polymorphisms of P53 and P73 genes with the risk of prostate cancer in a case-control study from Northern India. Arch Med Res 2011;42:122–7. [DOI] [PubMed] [Google Scholar]
- [36].Quinones LA, Irarrazabal CE, Rojas CR, et al. Joint effect among p53, CYP1A1, GSTM1 polymorphism combinations and smoking on prostate cancer risk: an exploratory genotype-environment interaction study. Asian J Androl 2006;8:349–55. [DOI] [PubMed] [Google Scholar]
- [37].Ricks-Santi L, Mason T, Apprey V, et al. p53 Pro72Arg polymorphism and prostate cancer in men of African descent. Prostate 2010;70:1739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rogler A, Rogenhofer M, Borchardt A, et al. P53 codon 72 (Arg72Pro) polymorphism and prostate cancer risk: association between disease onset and proline genotype. Pathobiology 2011;78:193–200. [DOI] [PubMed] [Google Scholar]
- [39].Salehi Z, Hadavi M. Analysis of the codon 72 polymorphism of TP53 and human papillomavirus infection in Iranian patients with prostate cancer. J Med Virol 2012;84:1423–7. [DOI] [PubMed] [Google Scholar]
- [40].Sivonova MK, Vilckova M, Kliment J, et al. Association of p53 and p21 polymorphisms with prostate cancer. Biomed Rep 2015;3:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Suzuki K, Matsui H, Ohtake N, et al. A p53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci 2003;10:430–5. [DOI] [PubMed] [Google Scholar]
- [42].Wu HC, Chang CH, Chen HY, et al. p53 gene codon 72 polymorphism but not tumor necrosis factor-alpha gene is associated with prostate cancer. Urol Int 2004;73:41–6. [DOI] [PubMed] [Google Scholar]
- [43].Wu WJ, Kakehi Y, Habuchi T, et al. Allelic frequency of p53 gene codon 72 polymorphism in urologic cancers. Jpn J Cancer Res Gann 1995;86:730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu B, Xu Z, Cheng G, et al. Association between polymorphisms of TP53 and MDM2 and prostate cancer risk in southern Chinese. Cancer Genet Cytogenet 2010;202:76–81. [DOI] [PubMed] [Google Scholar]
- [45].Aubrey BJ, Kelly GL, Janic A, et al. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression. Cell Death Differ 2018;25:104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ashikari D, Takayama K, Tanaka T, et al. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer. Oncogene 2017;36:6272–81. [DOI] [PubMed] [Google Scholar]
- [47].Wang Y, Pascal LE, Zhong M, et al. Combined loss of EAF2 and p53 induces prostate carcinogenesis in male mice. Endocrinology 2017;158:4189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]


