Abstract
The chemokine receptor CXCR3 and its ligands CXCL10 and CXCL11 have been suggested to give rise to the most relevant chemokine axis able to facilitate the entrance of immune cells into inflamed tissues and be activated in different inflammatory disorders, such as celiac disease (CD).
The aim of this study was to investigate the expression level of CXCR3, CXCL10, and CXCL11 genes in celiac patients compared to healthy controls. Both cohorts have been recruited from the Iranian population.
In this case–control study, biopsy specimens were collected from 71 celiac patients (60.5% female) and 90 control subjects (57% female) during 2016. Total RNA was extracted and mRNA expression levels of CXCR3, CXCL10, and CXCL11 genes were investigated by SYBR green qPCR.
Based on qPCR and relative quantification method, the mRNA expression levels of CXCR3, CXCL10, and CXCL11 were significantly higher in duodenal biopsies of celiac patients compared to healthy controls in the study population (P = .038, P = .021, and P = .012 respectively).
The result of this study showed that CXCR3/CXCL10/CXCL11 signaling axis is overexpressed in the small intestinal mucosa of CD patients compared to controls. This finding might explain the specific enrollment of the main cell populations that infiltrate the epithelium.
Keywords: celiac disease, chemokines, intraepithelial lymphocytes
1. Introduction
Celiac disease (CD) is an autoimmune disorder and the chronic inflammation of the small intestine is associated with T-cell activation of immune system that occurs in genetically predisposed people because of the increased sensitivity to gluten.[1] Villous atrophy, crypt hyperplasia, and lymphocytic infiltration are the consequences of the inflammatory cascades in the small intestine.[2] The removal of gluten from the diet leads to a complete or nearly complete clinical and histological remission.[2,3] In the past, several studies have shown that the immune system is the main culprit of the response to gluten and of the pathological complications originated as a consequence of it.[4,5] Moreover, despite the importance of adaptive immune system during the pathogenic processes of CD, also innate immune system plays a significant role in initiating these immunological responses. In CD, among the mechanisms involved in the innate immune system, the chemokine signaling pathway is crucial in the development of the disease.[6]
Chemokines are divided into 4 groups: CXC, CC, CX3C, and XC. All of them have similar structural features, such as a small size of 8 to 10 kDa, and the presence of 4 cysteine molecules at key structural positions.[7]CXCL10 (IP-10) and CXCL11 (I-TAC or IP-9) are chemokines of the family CXC that bind their receptor CXCR3 as a member of family G-protein. Many studies have shown that CXCR3 and its ligands are the main cause of recruiting cells to the inflamed tissue and their expression have a strong association with autoimmune diseases such as CD.[8–10] In fact, chemokine receptors and their ligands trigger cellular signaling in response to gluten and ultimately cause the rearrangement of the cytoskeleton and the recruitment of cells to the site of inflammation.[10] In particular, CXCR3 and its specific ligands CXCL10 and CXCL11 are among the main determinants in the recruitment of immune cells to intestinal lamina propria.[10,11] Many lines of evidences strongly emphasized that the binding of gliadin to CXCR3 leads to an increase in Zonulin secretion.[12] This protein binds to the receptor on the surface of the intestinal epithelial cells causing the opening of intercellular junctions and facilitates the entry of gluten peptides into the intestinal lamina propria, facilitating severe mucosal damages.[12] Previous studies reported high expression levels of CXCR3, CXCL10, and CXCL11 in untreated celiac patients.[10] As the genetic background between Iranian population and the Eastern population is different and the expression of these genes has not been studied in the Iranian population, in this work we were aimed at investigating the expression level of CXCR3 and its ligands CXCL10 and CXCL11 in untreated Iranian CD patients compared to healthy controls.
2. Patients and methods
In this case–control study, all untreated CD patients or treated (under gluten-free diet [GFD] for 6 months) and resulted positive for anti-tTG (IgA) were invited to the clinic. After obtaining informed consent, they underwent endoscopy and biopsy specimens were collected. Seventy-one CD patients (43 females and 28 males) with a mean age of 32.4 ± 7.54 years referred to the Celiac Department at the Research Institute for Gastroenterology and Liver, Shahid Beheshti University of Medical Sciences during 2016. The disease was diagnosed by serological test (anti-tTG IgA and/or anti-EMA IgA) and confirmed by endoscopical and histopathological analyses. Demographic and clinical data were collected using a dedicated questionnaire. The number of treated patients was very low; therefore, we decided to exclude them from further analysis. In addition, 90 healthy subjects (51 females and 39 males) with a mean age of 34.41 ± 9.47 years who had no history of CD in their family and were negative for anti-tTG (IgA) were matched with cases and enrolled as controls. Six biopsy specimens were taken from duodenal tissue (D2) of each individual using sampling forceps; 2 were immediately placed in RNA Later solution (Yekta Tajhiz Azma Co., Iran) and kept at −80°C until molecular study and 4 samples were transferred to the department of pathology for CD diagnosis according to the Marsh classification.[13] RNA was extracted from the specimens by a commercially available kit (Yekta Tajhiz Azma Co., Iran) according to manufacturer's instructions. The quantity and quality of extracted RNA were evaluated using Nanodrop and 1.5% agarose gel electrophoresis, respectively. Then, complementary DNA (cDNA) was synthesized from the extracted RNA by a RT kit (Takara kit) according to the manufacturer's instructions. Reaction products were stored at −20°C until the analysis.
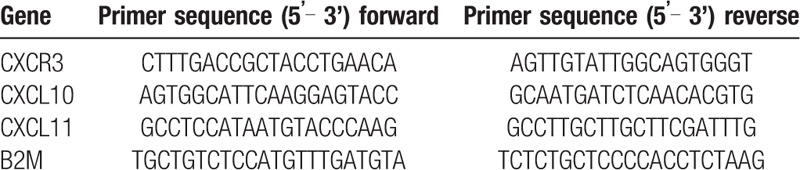
Primers for CXCR3, CXCL10, CXCL11 genes and B2MG (β2-microglobulin as the reference gene) were designed using the GeneRunner software and checked in the Primer Blast section of the NCBI website. The sequences of these primers are listed in Table 1. Applied Biosystems 7500 Real Time PCR (ABI) and SYBR GREEN method were employed to conduct the realtime qPCR reaction. In this study, the Relative Quantification (RQ) method was employed to evaluate and compare the expression levels of CXCR3, CXCL10, and CXCL11 genes in the duodenal samples of CD patients compared to healthy individuals. Samples were analyzed by LightCycler system (model 3.5) using the following protocol: an initial denaturation at 95°C for 30 seconds, 40 cycles of amplification including denaturation at 95°C for 5 seconds then primer annealing at 60°C for 34 seconds and extension steps at 95°C for 15 seconds.
Table 1.
Primer pairs used for real-time PCR.
For HLA genotyping, genomic DNA was extracted using salting out method and DQ2/DQ8 haplotypes were genotyped by real-time PCR using SYBR Green as described previously.[14]
Demographic data and clinical information were collected for patients and healthy people. The study was approved by ethic committee of Research Institute for Gastroenterology and Liver, Shahid Beheshti University of Medical Sciences, Tehran, Iran, and all participants signed the informed consent form.
In this study, the data analysis and statistics were performed by using SPSS software (version 21, Co Ltd, Tokyo, Japan) and data with P < .05 were considered statistically significant. In addition, the curve of real-time PCR was plotted using GraphPad, Prism (v.3). Correlation analysis of HLA and severity of mucosal damage with the expression of the studied genes was carried out by using Spearman correlation analysis.
3. Results
3.1. Demographic and clinical data
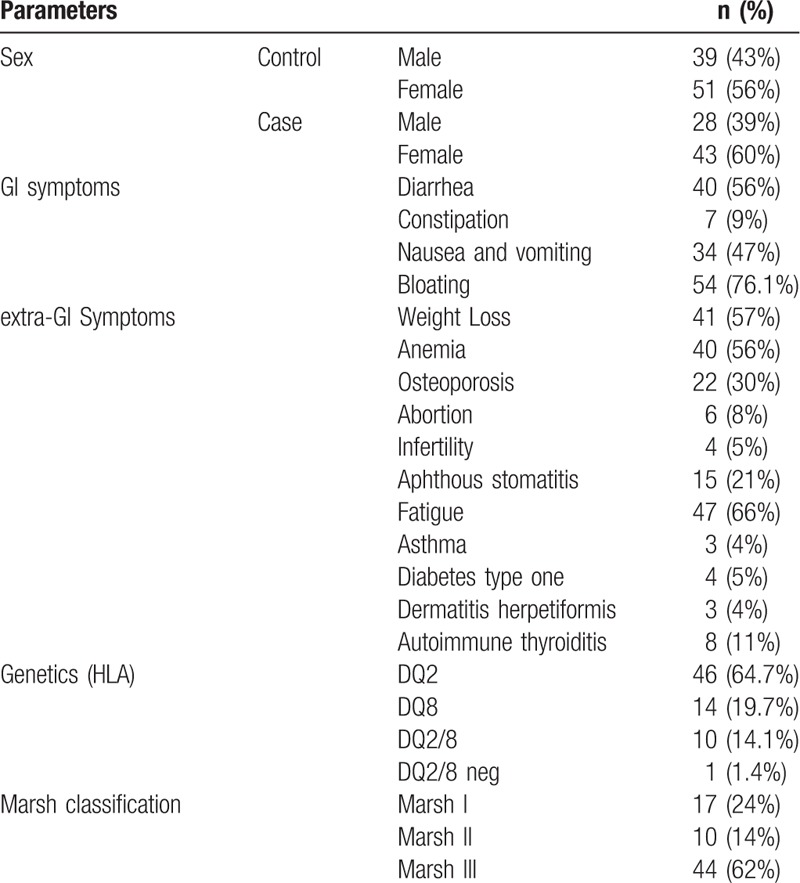
Seventy-one patients with CD including 28 males (39%) and 43 females (60%) and 90 control individuals consisting in of 39 males (43%) and 51 females (56%) were enrolled in this study. As reported in Table 2, the most common gastrointestinal (GI) symptoms among the 71 studied CD patients were bloating (76.1%) and diarrhea (56%), and the most prevalent non-GI symptoms were fatigue (66%), weight loss (57%), and anemia (56%). For HLA genotypes, most of the patients were HLA-DQ2-positive (64.7%) and the pathological status was mainly Marsh III (62%).
Table 2.
Demographic characteristics of study population.
3.2. Expression of CXCR3, CXCL10 and CXCL11 in duodenal biopsies
The RQ method was used to determine the changes in the expression level of these genes between the case and the control group. In this method, the rate of gene expression changes is measured as fold change (FC). Figure 1 shows the mRNA expression level of CXCR3, CXCL10, and CXCL11 in the duodenal mucosa of patients with CD compared with controls.
Figure 1.
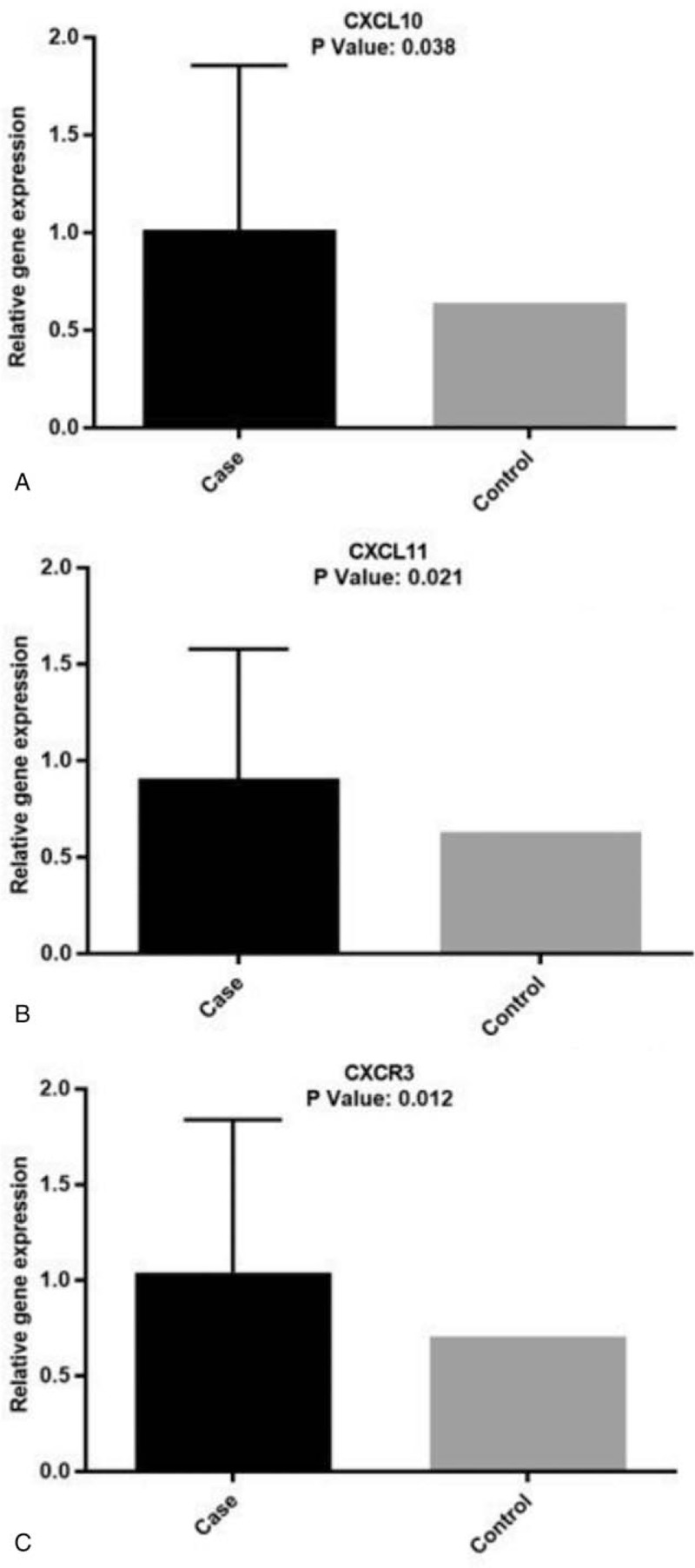
CXCL10, CXCL11, and CXCR3 mRNA levels in the duodenum. (A) Expression levels of CXCL10 (P = .038); (B) expression levels of CXCL11 (P = .021); (C) expression levels of CXCR3 (P = .012).
We found that CXCL10 (P = .038) (Fig. 1A), CXCL11 (P = 0.021) (Fig. 1B), and CXCR3 (P = .012) (Fig. 1C) mRNA levels in the duodenal mucosa of untreated patients with CD was significantly increased compared to controls. This behavior seems not correlated to sex or age of the patients as shown in Figure 2. No statistically significant differences were shown when we compared the expression levels of CXCL10 with CXCL11 (P > .05).
Figure 2.
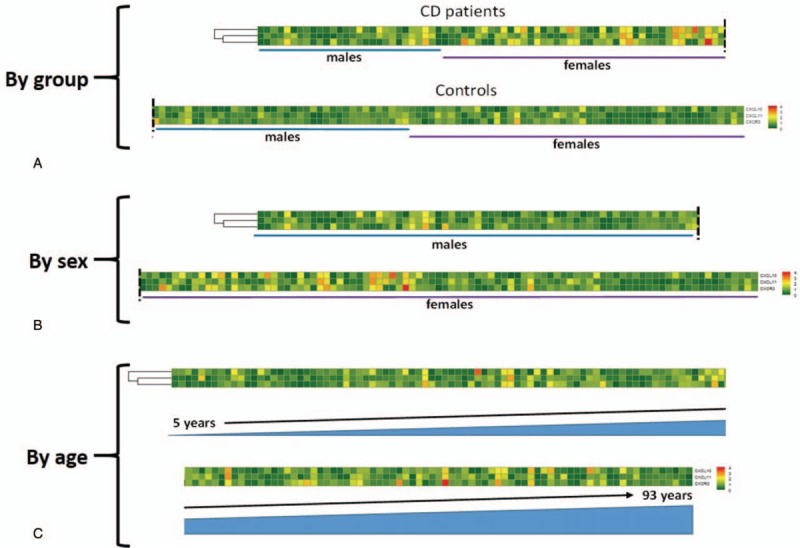
Heatmaps of CXCL10, CXCL11, and CXCR3 mRNA levels in the duodenum. Patients are ordered by groups (A), sex (B) and age (C).
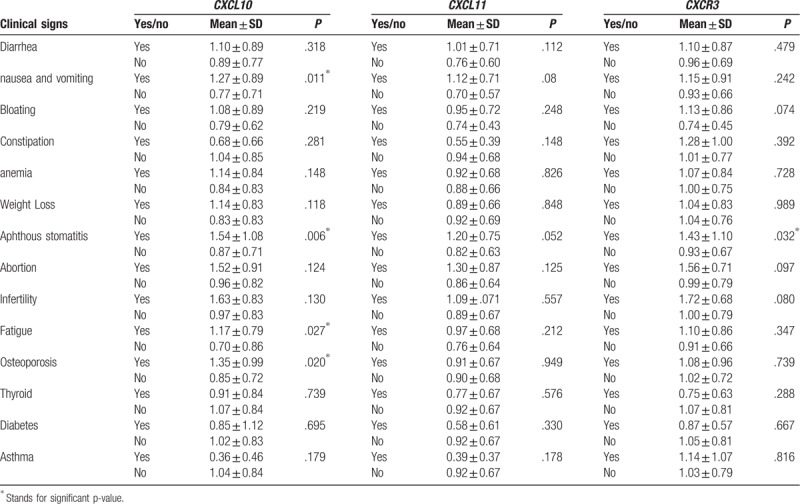
3.3. Many clinical symptoms correlate with the expression of the studied genes
As reported in Table 3, there was a significant relationship between CXCL10 gene expression and nausea and vomiting (P = .011), aphthous stomatitis (P = .006), fatigue (P = .027), and osteoporosis (P = .020), as well as CXCR3 and aphthous stomatitis (P = .032). No clinical symptoms resulted significantly correlated with CXCL11.
Table 3.
Relationship between clinical signs and expression of the studied genes.
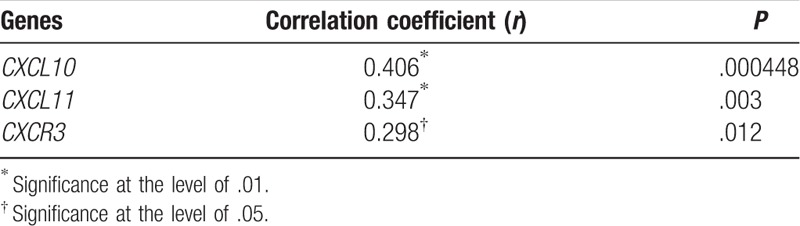
3.4. Severity of mucosal damage correlate with the expression of the studied genes
Spearman correlation analysis showed a positive correlation between the severity of the mucosal damage and mRNA expression levels of CXCR3, CXCL10, and CXCL11 in celiac patients. According to the results (Table 4), we observed a direct relationship between tissue degradation in CD and increased expression of these chemokines, especially for CXCL10 gene.
Table 4.
Relationship between the severity of mucosal damage and the expression of the genes studied.
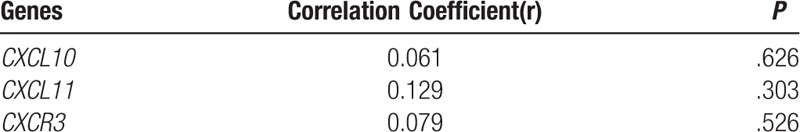
3.5. HLA genotypes did not correlate with the expression of the studied genes
According to Table 5, the Spearman correlation analysis showed no correlation between the HLA genotypes of patients in the studied population and the expression of CXCR3, CXCL10, and CXCL11 genes.
Table 5.
Relationship between the Type of HLA and the expression of the genes studied.
4. Discussion
Many papers have recently focused on the importance of finding biomarkers and determinants for a better understanding of the underlying causes of CD. According to the literature, the environment and genetics are known to be the main causes of CD, similarly to many other autoimmune diseases.[4,5] Recent studies on the functioning and involvement of the immune system in the pathogenesis of CD suggested that the innate immune system has a significant role in the response against the intestinal mucosa.[15] One of the most exclusive features of the immune system, compared to other tissue systems, is the completely regular migration of cells from blood to tissues and vice versa. This is allowed by proteins, classified as chemokines, that have common structural features, such as small size (8–10 kDa) and 4 cysteine molecules at key structural positions.[16] Many chemokines and their receptors have key roles in inflammatory responses in autoimmune diseases.[17]
In this study, to assess the involvement of CXCL10 and CXCL11 chemokines and their receptor CXCR3 in CD, the gene expression of CXCR3, CXCL10, and CXCL11 was assessed in duodenal tissues of CD patients and controls. Pathways where CXCR3 and its ligands, particularly CXCL10 and CXCL11, are involved in have been recommended to be crucial for the attraction of immune cells into tissues. Therefore, the evaluation of the expression levels of these chemokines could be an effective mean to identify potential biomarkers and to predict the inflammation of the intestinal tissue (obtained by biopsies). The results of this study showed that the expressions of all of these genes in CD patients are significantly increased compared to controls.
Immune cell differences depend on the expression patterns of chemokine receptors on leucocytes and the production of different chemokines in tissues that allow leucocytes to enter selectively into tissues. As a result, the expression of chemokines and their receptors is distinct in different cells. In the last few years, the expression of CXCR3/CXCL10 on 26 untreated and 6 treated celiac patients from Argentina were evaluated.[10] The results of that study showed an increased expression of CXCR3 and CXCL10 genes, which is in agreement with our study. Authors concluded that the production of CXCL10 in untreated celiac patients’ epithelium and lamina propria has been increased by enterocyte and plasma cells. They also found that the expression of CXCR3 in intraepithelial lymphocytes, lamina propria lymphocytes, and plasma cells was increased.
In a study on a specific mouse model, the authors concluded that gliadin was able to bind CXCR3 in CD patients and, as a consequence, Zonulin protein was released.[11] They also showed that the CXCR3 expression level in the epithelium and in the intestinal lamina propria of CD patients is higher than that expressed by the group without CD. In agreement with this study, we found that the expression level of this gene in CD patients was significantly higher than in controls.
Rashtak et al in 2010 demonstrated that the gluten protein in cereals could stimulate blood monocytes to produce more chemokines, such as CXCL10.[18] The authors examined the role of this chemokine in CD patients and suggested that CXCL10 has a crucial role in recruiting T-cell lymphocytes in the epithelium. The authors suggested that this chemokine could be used as a novel therapeutic target for CD patients and even nonceliac gluten-sensitive patients.[18] In line with this study, we observed a higher expression of CXCL10 in our CD patients compared to controls.
Valerii et al[19] in 2015 reported that peripheral blood mononuclear cells in nonceliac gluten-sensitive patients greatly increase the pre-inflammatory chemokine CXCL10 in the presence of wheat gluten and cause recruitment of cells to the inflammatory tissue. This study emphasized the important role of this chemokine in inflammation and its relationship with gluten-related diseases. In fact, owing to the fact that the expression of this chemokine is higher in the nonceliac gluten-sensitive patients, it can be hypothesized that the increased expression of CXCL10 may be directly related to an increased gluten consumption.[19]
Other studies have shown that CXCL10 is essential in the modulation and increase of inflammation in autoimmune diseases.[17,20] In our study, we showed a correlation between the increased expression of the CXCL10 chemokine and the severity of the disease (according to histological findings that showed that most of the patients were Marsh III).
However, other indirect data support the role of chemokines production in CD disease. In fact, IL-15 is one of the most prominent cytokines formed as a consequence of the immune response against gluten,[21] whereas another study reported that IL-15 induces the production of CXCL10 in the small intestine of CD patients.[22]
5. Conclusions
In CD, among the mechanisms involved in the innate immune system, the chemokine signaling pathway is involved in the development of this disease.[8] The alteration of CXCL10, CXCL11, and CXCR3 gene expression in small intestinal biopsies of CD patients suggests that these chemokines are important factors in the development of the disease and able to induce an innate immune response. In this study, we found a significant upregulation of CXCL10, CXCL11, and CXCR3 mRNAs in the duodenal mucosa of CD patients compared to controls (Iranian population). Here, we suggest that CXCL10 and CXCL11 chemokines and their receptor (CXCR3) have potential role in the pathogenesis of CD and could be an important player in the recruitment of immune cells into the intestinal mucosa of CD patients. Therefore, they might be considered as candidate biomarkers of gluten-induced intestinal inflammation in CD patients.
Acknowledgments
The authors thank Prof. Knut Lundin from Faculty of Medicine, University of Oslo, Oslo, Norway for precise and thoughtful comments and constructive criticism which has led to a better manuscript.
Author contributions
Conceptualization: Mohammad Rostami-Nejad.
Sample collection: Amir Sadeghi.
Formal analysis: Flora Forouzesh, Kamran Rostami.
Funding acquisition: Andrea Masotti.
Investigation: Mahrokh Haghbin, Elham Aghamohammadi.
Methodology: Mahrokh Haghbin.
Project administration: Mohammad Reza Zali.
Software: Flora Forouzesh.
Supervision: Mohammad rostami-Nejad, Kamran Rostami, Hamid Asadzadeh-Aghdaei, Andrea Masotti, Mohammad Reza Zali.
Validation: Mohammad rostami-Nejad, Flora Forouzesh, Andrea Masotti.
Writing – review & editing: Mohammad rostami-Nejad, Flora Forouzesh, Kamran Rostami, Hamid Asadzadeh-Aghdaei, Andrea Masotti, Mohammad Reza Zali.
Footnotes
Abbreviations: anti-tTG Ab = anti-tissue transglutaminase antibodies, CD = celiac disease, cDNA = complementary DNA, CXCL = C-X-C Motif Chemokine Ligand, CXCR3 = chemokine (C-X-C motif) receptor 3, RQ = relative quantification.
This study was financially supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran.
The authors report no conflicts of interest.
References
- [1].Ehsani-Ardakani M, Nejad M, Villanacci V, et al. Gastrointestinal and non-gastrointestinal presentation in patients with celiac disease. Arch Iran Med 2013;16:78. [PubMed] [Google Scholar]
- [2].Karinen H, Kärkkäinen P, Pihlajamäki J, et al. Gene dose effect of the DQB1∗ 0201 allele contributes to severity of coeliac disease. Scand J Gastroenterol 2006;41:191–9. [DOI] [PubMed] [Google Scholar]
- [3].Louka A, Sollid L. HLA in coeliac disease: unravelling the complex genetics of a complex disorder. Tissue Antigens 2003;61:105–17. [DOI] [PubMed] [Google Scholar]
- [4].Ciclitira P. AGA technical review on celiac sprue. Gastroenterology 2001;120:1526–40. [DOI] [PubMed] [Google Scholar]
- [5].Green P, Cellier C. Celiac disease. N Engl J Med 2007;357:1731–43. [DOI] [PubMed] [Google Scholar]
- [6].Ghasiyari H, Nejad M, Amani D, et al. Crucial role of innate immune system in the pathogenesis of celiac disease. Arvand J Heal Med Sci 2016;1:3. [Google Scholar]
- [7].Rotondi M, Chiovato L, Romagnani S, et al. Role of chemokines in endocrine autoimmune diseases. Endocr Rev 2007;28:492–520. [DOI] [PubMed] [Google Scholar]
- [8].Schroepf S, Kappler R, Brand S, et al. Strong overexpression of CXCR3 axis components in childhood inflammatory bowel disease. Inflamm Bowel Dis 2010;16:1882–90. [DOI] [PubMed] [Google Scholar]
- [9].Lee E, Lee Z, Song Y. CXCL10 and autoimmune diseases. Autoimmun Rev 2009;8:379–83. [DOI] [PubMed] [Google Scholar]
- [10].Bondar C, Araya R, Guzman L, et al. Role of CXCR3/CXCL10 axis in immune cell recruitment into the small intestine in celiac disease. PLoS One 2014;9:e89068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lammers K, Khandelwal S, Chaudhry F, et al. Identification of a novel immunomodulatory gliadin peptide that causes interleukin-8 release in a chemokine receptor CXCR3-dependent manner only in patients with coeliac disease. Immunology 2011;132:432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fasano A. Zonulin and its regulation of intestinal barrier function: the biological door to inflammation, autoimmunity, and cancer. Physiol Rev 2011;91:151–75. [DOI] [PubMed] [Google Scholar]
- [13].Marsh M, Johnson M, Rostami K. Mucosal histopathology in celiac disease: a rebuttal of Oberhuber's sub-division of Marsh III. Gastroenterol Hepatol ed Bench 2015;8:99. [PMC free article] [PubMed] [Google Scholar]
- [14].Mashayekhi K, Nejad MR, Amani D, et al. A rapid and sensitive assay to identify HLA-DQ2/8 risk alleles for celiac disease using real-time PCR method. Gastroenterol Hepatol BedBench 2018;11:250–8. [PMC free article] [PubMed] [Google Scholar]
- [15].Kooy-Winkelaar Y, van Lummel M, Moustakas A, et al. Gluten-specific T cells cross-react between HLA-DQ8 and the HLA-DQ2α/DQ8β transdimer. J Immunol 2011;187:5123–9. [DOI] [PubMed] [Google Scholar]
- [16].Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol 1997;15:675–705. [DOI] [PubMed] [Google Scholar]
- [17].Christen U, McGavern D, Luster A, et al. Among CXCR3 chemokines, IFN-γ-inducible protein of 10 kDa (CXC chemokine ligand (CXCL) 10) but not monokine induced by IFN-γ (CXCL9) imprints a pattern for the subsequent development of autoimmune disease. J. Immunol 2003;171:6838–45. [DOI] [PubMed] [Google Scholar]
- [18].Rashtak S, Marietta E, Murray J. Gliadin stimulation of monocytes leads to increased expression of multiple T cell recruiting chemokines: a novel innate immune response. Clin Immunol 2010;135:S47. [Google Scholar]
- [19].Valerii M, Ricci C, Spisni E, et al. Responses of peripheral blood mononucleated cells from non-celiac gluten sensitive patients to various cereal sources. Food Chem 2015;176:167–74. [DOI] [PubMed] [Google Scholar]
- [20].Lee E, Lee Z, Song Y. The interaction between CXCL10 and cytokines in chronic inflammatory arthritis. Autoimmun Rev 2013;12:554–7. [DOI] [PubMed] [Google Scholar]
- [21].Di Niro R, Mesin L, Zheng N, et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med 2012;18:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Meresse B, Chen Z, Maria T, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 2004;21:357–66. [DOI] [PubMed] [Google Scholar]


