Structural and functional studies of the coat protein regions of potato virus Y reveal crucial roles in viral infectivity.
Abstract
Potato virus Y (PVY) is among the most economically important plant pathogens. Using cryoelectron microscopy, we determined the near-atomic structure of PVY’s flexuous virions, revealing a previously unknown lumenal interplay between extended carboxyl-terminal regions of the coat protein units and viral RNA. RNA–coat protein interactions are crucial for the helical configuration and stability of the virion, as revealed by the unique near-atomic structure of RNA-free virus-like particles. The structures offer the first evidence for plasticity of the coat protein’s amino- and carboxyl-terminal regions. Together with mutational analysis and in planta experiments, we show their crucial role in PVY infectivity and explain the ability of the coat protein to perform multiple biological tasks. Moreover, the high modularity of PVY virus-like particles suggests their potential as a new molecular scaffold for nanobiotechnological applications.
INTRODUCTION
Potato virus Y (PVY) is ranked as fifth in the top 10 most economically important plant viruses (1) and is the most important viral pathogen of potato worldwide (2). The virus causes potato tuber necrotic ringspot disease, which can result in up to 70% yield reduction, and severely affects other economically important solanaceous crops (3, 4). PVY belongs to the genus Potyvirus (family Potyviridae). It exists as a complex of strains that have evolved by accumulation of mutations and recombination, which enables the virus to efficiently adapt to new potato cultivars. Plant-to-plant PVY transmission is mainly achieved by aphids, as well as mechanically or vegetatively via seed tubers (4). The PVY genome is a 9.7-kb positive-sense single-stranded RNA (ssRNA) encoding a polyprotein, which is cleaved into 10 mature proteins by virus-specific proteases (5). In addition, another essential protein P3N-PIPO is produced from an overlapping coding sequence (6). Most of the potyviral proteins are multifunctional. Stages of viral infection including virion uncoating, translation, replication, suppression of host defenses, movement, and virion assembly all involve dynamic interactions between the viral proteins, viral genomic RNA, and the hijacked host components (5). In the later stages of infection, more than 2000 copies of a single potyviral protein, the coat protein (CP), are recruited to form a protein capsid around the viral genome, resulting in formation of flexuous filaments (i.e., virions). Beside its principal role as a sole structural protein in encapsidation of the viral RNA, the potyviral CP has also been reported to be involved in transmission by aphids, movement, and regulation of viral RNA amplification (5).
Despite extensive availability of data on PVY’s genome and pathogenicity, there has been no high-resolution structural information for this virus. Because of the extreme economic importance of PVY, and the urgent need for structural data to better understand mechanisms of viral infectivity, we have examined in detail the structure of the PVY virion and its CP. Here, we report the high-resolution cryoelectron microscopy (cryo-EM) structures of the PVY virion and a recombinant PVY-based RNA-free virus-like particle (VLP). This provides a detailed insight into the RNA-supported helical viral capsid architecture featuring an extended C-terminal region of CP, which is tightly packed in a unique fashion in the virion lumen. In addition, using extensive biochemical, biophysical, and computational characterization, as well as structure-based mutagenesis, we identified regions of CP that affect VLP filament assembly. Moreover, the biological activities of the CP’s N- and C-terminal regions for virus infectivity were explored by measuring the accumulation of viral RNA and systemic movement of selected PVY mutants in planta.
RESULTS
PVY filaments of left-handed helical symmetry contain unique lumenal structure
PVY was purified from infected tobacco plants, and its cryo-EM structure was determined at 3.4-Å resolution using single-particle–based helical reconstruction (fig. S1 and table S1). PVY virions with the diameter of 130 Å comprise a left-handed helical arrangement of CPs assembled around viral ssRNA, with 8.8 CPs per turn (Fig. 1A), confirming architectural conservation between genetically unrelated poty- and potexviruses (7–9). The polypeptide chain of an individual CP unit is well defined in the cryo-EM map from Val44 to Met267 (Fig. 1B and fig. S1, G to I).The extended N- and C-terminal regions are devoid of secondary structure elements (Fig. 1B and fig. S2). In contrast, the globular core subdomain contains seven α helices and one β-hairpin and closely resembles the corresponding regions in the potyvirus watermelon mosaic virus (WMV) [Protein Data Bank (PDB) ID 5ODV; root mean square deviation (RMSD) 1.24 Å, 76% identity] and the potexvirus pepino mosaic virus (PepMV) (PDB ID 5FN1; RMSD 2.46 Å, 19% identity). Helically arranged cores assemble into a protein layer protecting the viral ssRNA from the environment (Fig. 1C). The surface-located N-terminal region Val44-Gln77 acts as a clip between neighboring cores, further contributing to the capsid’s compactness (Fig. 1, C and D). The poorly defined cryo-EM density for residues preceding Val44 was shown to be mainly due to the flexibility of this region and only partially due to proteolysis (fig. S3, A to C). Degradation of these flexible regions may be of biological relevance, as it has been proposed that proteolysis occurring in the region between the highly conserved N-terminal Asp-Ala-Gly (DAG) motif (fig. S2) and the core could help release infectious potyviral particles from aphid stylets during viral transmission (10).
Fig. 1. Structural features of PVY.
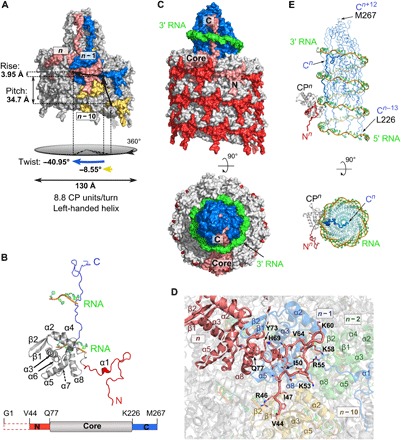
(A) Two turns of the PVY filament, surface representation. Colored units: CPn (pink), CPn−1 (blue), and CPn−10 (yellow). (B) Structure of PVY CP (N-terminal region, red; core, gray; C-terminal region, blue) showing two ssRNA interaction sites. Bottom: A linear representation of the CP; the colors are as in the CP model. The dashed box at the N terminus depicts a structurally disordered part. (C) Side and top views of the PVY filament (three turns of CPs and four turns of ssRNA), surface representation; the colors are as in (B). The position of one CP unit (pink) is shown in the filament. (D) Interface between the N-terminal region of CPn unit (pink; N-terminal residues are shown as sticks), CPn−1 (blue), CPn−10 (yellow), and CPn−2 (green). (E) Positioning of C-terminal regions (Leu226-Met267; blue) in the filament lumen, surrounded by ssRNA, side and top views. The same part of the PVY filament is shown as in (C). One CP unit is shown; the color code is as in (B). In all figures showing ssRNA, the nucleotides are presented in green with orange representing the phosphate backbone.
We were able to trace the C-terminal region of PVY CP to completeness. This revealed unique structural features to PVY virions in comparison to flexuous virions studied to date (7–9). The C-terminal region is completely buried in the lumen of the viral filament and interwoven in a distinct helical manner along the longitudinal axis, forming a compact cone-shaped structure (Fig. 1, C and E). As opposed to the previously postulated location with both potyviral termini at the surface (11–13), our PVY structure reveals that C-terminal regions are completely shielded from the environment, and the only region where C termini could be exposed is in the vicinity of the RNA 3′ terminus or in damaged virions (fig. S1A).
Virion structure is supported by extensive CP-CP and CP-RNA interaction network
In contrast to rigid rod-like virions, which contain wedge-shaped α-helical CP units devoid of extended structural elements at the termini (fig. S4) (14, 15), the virions of flexuous filamentous viruses contain extended terminal regions, which have been already suggested to contribute to their flexibility (7–9). Our PVY structure reveals that longer terminal extensions in potyviral CP result in an increased PVY CP-CP interaction network, involving 12 neighboring CP units (Fig. 2A) compared to only 8 in potexviruses (fig. S4) (8). Thus, the filament packing is more extensive in PVY, which explains the higher flexibility observed for potyviruses (16, 17). The protein-protein interactions in the PVY virion are mainly of electrostatic nature (details in Supplementary text S1 and figs. S5 and S6).
Fig. 2. Protein-protein and protein-RNA interaction networks in PVY.
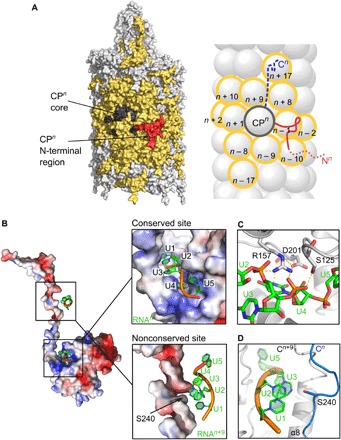
(A) Surface (left) and schematic (right) representation of the CPn unit interacting with neighboring CP units (yellow). N-terminal CP region, red; CP core, dark gray; extended CP C-terminal region inside the filament, blue dashed line. N- and C-terminal regions in the scheme are depicted only for CPn. (B) Left: Electrostatic surface of CPn (red, negative; blue, positive) with two ssRNA interaction sites. Top right: Close-up view showing NUC1 to NUC5 nucleotides bound to the conserved site in the CPn core. Bottom right: Nucleotides binding to CPn+9 core also binding to Ser240 in CPn C terminus. (C) Conserved residues Ser125, Arg157, and Asp201 in the ssRNA binding site of the core. Electrostatic interactions are shown as dashed lines. For residues shown in sticks, protein/ssRNA: carbon, gray/green; oxygen, red; and nitrogen, blue; phosphorus, orange. (D) Nucleotides NUC1 to NUC3 binding to CPn+9 core facing Ser240 of CPn.
Additional stabilization of the PVY virion comes from substantial protein-ssRNA interactions. To date, only one interaction site of CP and ssRNA has been reported for flexible filamentous viruses (7–9), located in the armpit-shaped groove between the core and the C-terminal region of CP, harboring five nucleotides (Fig. 2B). Nucleotide NUC4 faces the bottom of the groove, while bases NUC1 to NUC3 and NUC5 face the opposite direction, in a similar way to the interaction site in WMV (9) and with opposite polarity to those reported for PepMV and bamboo mosaic virus (7, 8). However, the architecture of this RNA binding site is conserved between filamentous virions and is lined with polar residues that enable tight interaction with ssRNA, including the three main conserved RNA binding residues (9), which are Ser125, Arg157, and Asp201 in PVY (Fig. 2C). However, our PVY structure reveals that each CP interacts with ssRNA twice. This second nonconserved interaction site is between Ser240 from the C-terminal region of CPn and nucleotides NUC1 to NUC3, which belong to the pentanucleotide bound into the armpit groove of CPn+9(Fig. 2, B and D).
RNA-free VLP structure reveals the importance of RNA for the helical symmetry of virions
To determine how viral ssRNA affects PVY virion architecture, we prepared the RNA-free VLPs. While the absence of viral ssRNA did not substantially affect the diameter of self-assembled VLP filaments, the lengths of VLP filaments varied from 30 nm to 3.5 μm (fig. S7A), as opposed to the approximately 730-nm-long PVY virions (fig. S1A). The cryo-EM structure of PVY-based VLP filaments at 4.1-Å resolution revealed stacked-ring architecture (fig. S7 and table S1). This nonhelical assembly of filamentous flexuous VLPs, as already indicated by negative-stain transmission electron microscopy (TEM) micrographs (fig. S7A) (18), is unique compared to potexviral Alternanthera mosaic virus VLPs, which resemble the helical architecture of an authentic virus (19). PVY VLPs are assembled from octameric CP rings of 125-Å diameter, stacked along the longitudinal axis of filaments devoid of nucleic acid. Two neighboring rings are 42.7 Å apart with a −31.8° twist angle between them (Fig. 3A). The cryo-EM density of each CP unit is well defined from Thr43 to Ile218 (Fig. 3B and fig. S7, G to I). The C-terminal region beyond Ile218 was not defined by cryo-EM density, which must be due to the flexibility of this region, since most of the CP units were shown to be at full length (fig. S3, A to C).
Fig. 3. Structural features of VLP.
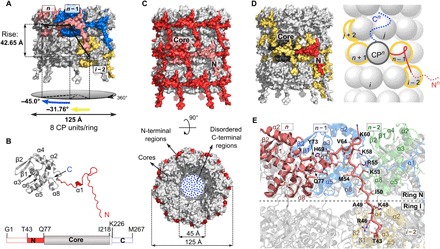
(A) Surface representation of two rings in a VLP filament with colored units CPn (pink), CPn−1 (blue), and CPi−2 (yellow). (B) Structure of VLP CP (N-terminal region, red; core, gray). Bottom: A linear representation of the CP, using the same colors as in the model above. Dashed boxes at N- and C-terminal regions depict structurally disordered parts. (C) Side and top views of the VLP filament; the colors are as in (B). Position of one CP unit (pink) is shown in the filament. (D) Protein-protein interactions in the VLP filament. Surface (left) and schematic (right) representation of central CPn unit (N-terminal region, red; core, dark gray; C-terminal region, blue) interacting with four neighboring CP units (yellow). N- and C-terminal regions in the scheme are shown only for the unit CPn. The dotted lines in N- and C-terminal regions indicate disordered parts. (E) Interface between the N-terminal region of the CPn unit (pink) and neighboring units CPn−1 (blue) and CPi−2 (yellow). CPn−2 (no interactions with CPn) is shown in green.
CP-CP interaction network is largely reduced in VLP filaments
In contrast to a large number of interconnected CP units in PVY virions, only five CPs are interconnected in VLPs (Fig. 3, C and D), aside from potential random interactions between the flexible C-terminal regions. Similar to authentic virions, the N-terminal region acts as a clip between neighboring CPs (Supplementary text S2 and Fig. 3, C to E) combining electrostatic and hydrophobic interactions (fig. S8).
To further evaluate differences in interaction networks, we performed an in silico comparison of the frequency distributions of axial vibrations between PVY and VLP filaments (Fig. 4, A and B). Results indicate softer and more flexible internal dynamics in the VLP relative to the authentic virus, shown by the significant shift of the VLP frequency distribution toward lower frequencies (Fig. 4C). Moreover, the flexibility of the virion was increased if the CP C-terminal region was removed and even more so in the absence of both the C-terminal region and ssRNA. This additionally confirmed the pivotal role of the ssRNA in the helical architecture of PVY virions and revealed the importance of the complex interplay between C-terminal regions and ssRNA for stable packing of PVY filaments.
Fig. 4. Impact of the viral RNA on filament stability and structure of CP.
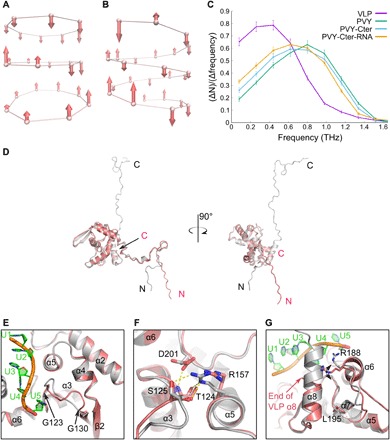
(A to C) Molecular dynamics of filaments. Axial displacements of protein unit centers of mass for the lowest vibrational modes: (A) VLP (ω = 0.0066 THz) and (B) PVY (ω = 0.0444 THz), as calculated by PCA (principal components analysis). Each protein unit is represented by a center of mass of the Val44-Ile218 region. (C) Frequency distributions of protein units’ vibrations along the fibril axis for different structural models: VLP, PVY, PVY without the C-terminal region (PVY-Cter), and PVY without C-terminal region and ssRNA (PVY-Cter-RNA). Error bars represent scattering in different time sections of the trajectory. (D) Superposition of PVY CP (gray) and VLP CP (pink) on core regions, shown at two angles. (E to G) Structural differences in CP core regions of PVY and VLP; the color code is as in (A). (E) The loop between Gly123 and Gly130. (F) Highly conserved residues Ser125, Arg157, and Asp201 involved in PVY ssRNA binding. Electrostatic interactions are shown as dashed lines. For clarity, the C-terminal region and ssRNA were removed from the picture. (G) Residues Arg188-Leu195 and α8 helix. The black arrow indicates flexibility of Arg188 in VLP in comparison to the fixed Arg188 in PVY CP.
CP adopts distinct structural features in PVY virions and VLP filaments
While the overall fold of CP units in PVY and VLP is highly conserved, with RMSD of 0.84 Å for their cores, several notable differences can be observed. Structural plasticity of the N-terminal region allows different conformations of this region between the PVY virion and VLP (Figs. 1D, 3E, and 4D). Furthermore, in the absence of RNA in VLPs, the loop of the armpit-like ssRNA binding site moves closer to the center of the core, compensating for the loss of protein-RNA interactions (Fig. 4, E and F). The loop Arg188-Leu195 (Fig. 4G), which is important for core-core and ssRNA-CP interactions in virions, is adjacent to the empty space between the rings in VLPs. Moreover, because of the absence of ssRNA in VLPs, the final α8 helix in VLPs has only half of the length found in PVY virions, instead continuing into a structurally disordered C terminus (Fig. 4G). The intrinsic plasticity of CP thus enables structural adaptation to the absence or presence of a binding partner, such as viral RNA. This plasticity is most pronounced within extended terminal regions, while the conserved fold of the central core allows only small but distinct structural changes. Because of a large number of CP units present in filaments, the multiplication effect of these structural modifications leads to considerable differences in the overall architectural arrangement between PVY and VLP filaments.
The N-terminal CP region is crucial, while the C-terminal is dispensable for VLP assembly
To assess the importance of the CP extended N and C termini in VLP filament assembly and stability, we performed systematic mutational analysis by stepwise deletion of N- or C-terminal residues, as well as simultaneous deletions at both termini (Supplementary texts S3 and S4, Fig. 5, and fig. S3, C to E). As expected from the VLP interaction network (Fig. 3E), deletion of 49 N-terminal residues prevents filament assembly, resulting in the formation of separated octameric rings, while deletion of 69 N-terminal residues results in monomeric CP units (Fig. 5B and fig. S3D). On the other hand, deletion of up to 60 C-terminal residues did not affect filament assembly but rather resulted in the formation of hollow flexuous nanotubes (Fig. 5C). Thus, CP deletion mutants revealed the importance of the N-terminal region for inter- and intra-ring connections in VLP filaments and the dispensability of the C-terminal region for VLP filament assembly.
Fig. 5. Analysis of VLPs formed from deletion CP mutants.
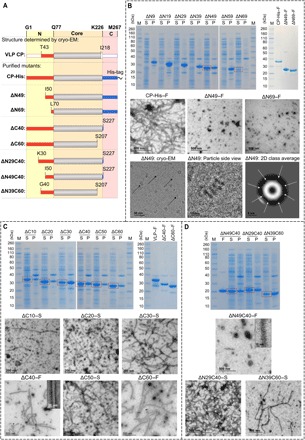
(A) Linear schematic representation of purified deletion mutants, based on the structure of VLP CP (N-terminal region, red; core, gray; C-terminal region, blue). SDS-PAGE (SDS–polyacrylamide gel electrophoresis) gels were obtained from soluble fractions (S) and pellets (P) after bacterial lysis and final purified soluble samples (F) of (B) N-terminal deletion mutants, (C) C-terminal deletion mutants, and (D) double deletion mutants. Negative-stain TEM micrographs show the presence or absence of filaments in various samples. The bottom part of (B) shows cryo-EM micrograph of ΔN49 (left), close-up side view of four stacked rings of ΔN49 (middle), and two-dimensional (2D) class average of the top view of ΔN49 (right).
Both extended CP terminal regions are crucial for PVY infectivity
To analyze the biological effects of deletions of extended terminal regions, we prepared three PVY clones, with CP lacking 50 N-terminal resides (ΔN50-CP) and 40 or 60 C-terminal residues (ΔC40-CP and ΔC60-CP). Both C-terminal deletion mutants showed extremely low accumulation of viral RNA in inoculated tissue of Nicotiana clevelandii plants (Fig. 6A and data S1), indicating decreased viral RNA replication and/or stability due to restricted interaction with the CP. We detected particles resembling VLPs in leaves bombarded with either ΔC40-CP or ΔC60-CP (Fig. 6B), showing that C-terminal CP truncations still allow assembly of CP into filaments, even in the absence of RNA accumulation. Viral RNA accumulation with ΔN50-CP was notably lower than for wild type but significantly higher than in the case of the C-terminal mutants (Fig. 6A and data S1). However, as predicted from PVY structure (Fig. 1D), no filamentous structures were observed (Fig. 6B). The absence of N-terminal residues allowed the replication signal to be observed but most likely hindered if not completely abolished viral intercellular movement. While the wild-type infectious clone successfully systemically infected plants, as evidenced by stunting and rugosity symptoms, neither of the mutants was able to systemically infect plants (Fig. 6C and data S1). Thus, interaction of the CP C-terminal region with PVY RNA is essential for establishment of efficient viral replication, while the N terminus is involved in assembly and movement of the virus, both being indispensable for successful systemic infection.
Fig. 6. Functional analysis of PVY CP N- and C-terminal deletions in planta.
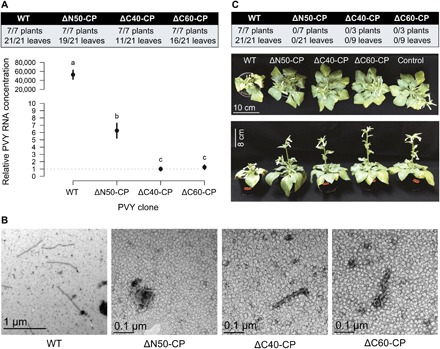
The effect of deletion mutants ΔN50-CP, ΔC40-CP, and ΔC60-CP was evaluated by bombardment of N. clevelandii with respective PVY clones. (A) Local multiplication in bombarded leaves: Relative concentration of PVY RNA was measured in individual leaves 7 days postbombardment (dpb). Results are given as the frequency of leaves with detected RNA and as a comparison of viral RNA quantities measured in the bombarded leaves. Differences were statistically evaluated using one-way analysis of variance (ANOVA) with Bonferroni post hoc analysis. Means not sharing the same letter are significantly different (P < 0.05). Error bars represent SE. (B) Local multiplication in bombarded leaves: VLPs were detected by negative-stain TEM of samples from bombarded leaves. (C) Long-distance movement: Relative concentration of viral RNA was measured in upper nonbombarded leaves (table; 14 dpb). Only wild-type (WT) PVY caused typical disease symptoms in N. clevelandii plants (bottom; 22 dpb). (Photo credit: Ion Gutierrez-Aguirre, National Institute of Biology, Slovenia.)
DISCUSSION
Results of this study offer a detailed insight into the high-order and efficient packing of the PVY filamentous virion, with its capsid built of CP units assembled around the viral ssRNA in a helical arrangement. Numerous CP-CP and CP-ssRNA interactions provide flexibility and stability in PVY filaments, resulting in long-term infectivity of this economically important virus. Our study now sets out the first evidence for the structural plasticity of PVY CP, which may explain its capability of involvement in the various stages of infection. Namely, beyond its role as the protector of the viral genome, the potyviral CP is also involved in viral transmission by aphids, genome replication, and translation as well as cell-to-cell and long-distance movement of the virus (13, 20–23), demonstrating the multitasking nature of this protein during all stages of the viral life cycle.
Understanding of the molecular mechanism of the complex in planta cycle of potyviruses and, in particular, the virion assembly is still very scarce. The origin of assembly in potyviruses was shown to be located near the 5′ end of genomic RNA (24) and was recently suggested for potyvirus plum pox virus (PPV) to be in its 5′ noncoding region (25). For PPV, it was also shown that the multifunctional potyviral protein helper component proteinase (HcPro) contributes to the assembly of stable viral particles (26) and that other viral and host factors are also required for the formation of stable virions (25). Besides virion stabilization, HcPro is also involved in polyprotein processing, suppression of host antiviral RNA silencing, and aphid transmission (5). Another multitasking viral protein, called the viral protein genome-linked (VPg), was found directly linked to potyviral virions via a covalent bond to the 5′ end of viral RNA, thereby playing a major role in viral RNA replication and translation (5). HcPro was colocalized with VPg in protruding tips on one side of a smaller fraction (<10%) of PVY virions, presumably at the 5′ end. However, these tips could only be observed using atomic force microscopy and were not discernible by negative-stain electron microscopy, probably due to specificity of sample preparation (27). Therefore, the structural knowledge of PVY virions contributed by our study is an important basis for further studies of these complex interactions between potyviruses and other viral, plant, or aphid components at high resolution. Moreover, the structural features we discovered open new possibilities for functional studies of different CP regions, which will facilitate design of more efficient plant protection strategies. In addition, our study offers the near-atomic insight into potyviral VLPs. In contrast to tight packing of PVY filaments, which leaves only a very narrow 1-nm solvent channel along the filament lumen, the inner channel of the engineered VLP filaments can be wider, offering many possibilities for reshaping (fig. S9). Hence, the structure of potyviral flexuous VLPs may be exploited as a new scaffold for the design and bioengineering of future nanoparticles or nanotubes.
MATERIALS AND METHODS
Purification of PVY
PVY (NTN strain; GenBank accession no. KM396648) from the virus collection at the National Institute of Biology (Slovenia) was purified to homogeneity following the procedure from Rupar et al. (28). Briefly, symptomatic leaves from infected tobacco plants (Nicotiana tabacum cv Xanthi) were harvested and ground in extraction buffer, centrifuged at low speed (4,500g) to remove plant debris, treated with the detergent Triton X-100, and further centrifuged at high speed (47,900g) to pellet the virus-rich fraction. Virus was then further purified by consecutive centrifugations in sucrose and CsCl gradients. The purified virus was dialyzed, pelleted, and resuspended in 5 mM disodium tetraborate buffer (pH 8.0). The concentration of the purified virus suspension calculated spectrophotometrically (28) was 3.7 mg/ml with A260/A280 ratio of 1.27, typical for pure PVY particles. The homogeneity of the suspension and the integrity of virus particles were further confirmed by negative-stain TEM.
Cloning, expression, and purification of VLPs
The gene fragment encoding PVY CP was cloned from a complementary DNA (cDNA) of PVY-NTN strain (GenBank accession no. KM396648) and inserted into the vector pT7-7 (Addgene). The vector was used to transform Escherichia coli BL21(DE3) cells, which were grown to an optical density at 600 nm of 0.8 to 1.0 in 2× YT medium [tryptone (16 g/liter) (BD Biosciences), yeast extract (10 g/liter) (BD Biosciences), and NaCl (5 g/liter)] supplemented with 5 mM MgCl2 and 2 mM CaCl2. Protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside for 20 hours at 20°C. Cells were lysed by sonication in phosphate-buffered saline (PBS) buffer [1.8 mM KH2PO4, 10.1 mM Na2HPO4 (pH 7.4), 140 mM NaCl, and 2.7 mM KCl], and cell debris was removed by centrifugation. The lysate was incubated for 20 min in a mixture of 4% PEG 8000 (Sigma-Aldrich) and 0.5 M NaCl to precipitate VLPs. The VLP suspension was centrifuged for 20 min at 14,000g, and the pellet was resuspended in PBS buffer by gentle overnight shaking. Remaining solid material was removed by 20 min of centrifugation at 35,000g. The supernatant was loaded on 20 to 60% sucrose density gradient and ultracentrifuged at 117,000g for 6 hours in a Beckman 50 Ti rotor. All fractions of the gradient were collected and analyzed with SDS–polyacrylamide gel electrophoresis (SDS-PAGE) to identify fractions containing VLPs. Selected fractions were pooled, dialyzed for 24 hours against PBS buffer, and concentrated using Amicon Ultra centrifugal filters with a 100,000 molecular weight cutoff (Millipore) to the final concentration of 1 to 7 mg/ml. All steps of the purification procedure including VLP storage were done at 4°C.
Cloning, expression, and purification of CP mutants
The following CP mutants were cloned and expressed in pET28a vector (Addgene): N-terminal deletion mutants (ΔN9, ΔN19, ΔN29, ΔN39, ΔN49, ΔN59, and ΔN69), C-terminal deletion mutants (ΔC10, ΔC20, ΔC30, ΔC40, ΔC50, and ΔC60), and double deletion mutants (ΔN49C40, ΔN39C60, and ΔN29C40). Sequences were verified by nucleotide sequencing (Eurofins Genomics). Since we have shown that a hexahistidine (His)–tag at the C terminus of the wild-type CP does not affect filament formation (Fig. 5B), mutants ΔN49, ΔN69, and ΔN49C40 were prepared with C-terminal His-tags, followed by a tobacco etch virus (TEV) protease cleavage site to simplify purification in the absence of filaments. All CP mutant proteins were expressed in BL21(DE3) cells under the same conditions as described for VLPs. After expression, cells were lysed using an EmulsiFlex-C5 high-pressure homogenizer (Avestin) in PBS buffer (pH 7.4) containing 5% glycerol because sonication in some cases resulted in insoluble proteins. The presence of filaments in lysates was monitored by negative-stain TEM. CP mutant proteins ΔC40 and ΔC60 were further purified following the VLP purification procedure described above. His-tagged mutant proteins (ΔN49, ΔN69, and ΔN49C40) were purified on Ni–NTA (nitrilotriacetic acid) columns (Qiagen), and bound proteins were eluted with PBS buffer (pH 7.4) containing 5% glycerol and 300 mM imidazole. Eluted samples were dialyzed against PBS buffer (pH 7.4) containing 5% glycerol. For further purification, ΔN49 and ΔN69 were dialyzed in the presence of TEV protease to cleave the His-tag. ΔN49 and ΔN69 mutants were subjected to a second Ni-NTA chromatography with the cleaved protein eluting in the unbound fraction. ΔN49 and ΔN69 from unbound fractions or ΔN49C40 with intact C-terminal His-tag after dialysis was further purified by size exclusion chromatography using Superdex 200 10/300 GL (24 ml) or Superdex 200 16/60 PG (120 ml) columns (GE Healthcare). The running buffer for ΔN49 was 15 mM tris-HCl, 130 mM NaCl, and 5% glycerol (pH 7.4), whereas the running buffer for ΔN69 and ΔN49C40 was PBS and 5% glycerol (pH 7.4). After chromatography, fractions containing pure proteins were pooled and concentrated in Amicon Ultra centrifugal filters (10,000 or 30,000 molecular weight cutoff) (Millipore). The final solutions of ΔN69, ΔC40, and ΔC60 with their respective concentrations of 0.9, 0.7 to 0.8, and 0.8 to 1.0 mg/ml were stored at 4°C. ΔN49 and ΔN49C40 were concentrated to 10 mg/ml and stored at −80°C.
Negative-stain TEM
Suspensions of viruses and VLPs were applied on Formvar- and carbon-coated copper grids for 5 min and stained with 1% (w/v) uranyl acetate (SPI Supplies, USA). Grids were examined with Philips CM100 TEM (FEI) operating at 80 kV and equipped with a ORIUS SC200 camera (Gatan Inc.).
Cryo-EM sample preparation and data acquisition
PVY (3.7 mg/ml) and VLP (6.7 mg/ml) samples were diluted 1:3 in 5 mM sodium tetraborate buffer (pH 8.0) and PBS buffer (pH 7.4), respectively, and 4 μl was applied to Quantifoil R2/1 holey carbon grids followed by vitrification in Vitrobot Mark IV (FEI, Eindhoven). Both datasets were collected on Titan Krios TEM (FEI, Eindhoven) operated at 300 kV and equipped with a Falcon2 (FEI, Eindhoven) direct electron detector with the defocus range between −2.2 and −1.0 μm. Micrographs were collected with a pixel size of 1.061 Å per pixel, and 16 frames were collected within 1-s exposure giving a total dose of 48 e−/Å2. Frames were aligned using MotionCor2 (29) to minimize the effect of beam-induced motion, and the parameters of the contrast transfer function (CTF) of the micrographs were estimated using Gctf program (30).
Deletion mutant ΔN49 (16 mg/ml) was diluted in PBS buffer (pH 7.4) and applied to Quantifoil R2/1 holey carbon grids, followed by vitrification in Vitrobot Mark IV (FEI, Eindhoven). The dataset was collected using Tecnai F20 (FEI, Eindhoven) operated at 200 kV and equipped with a 4k charge-coupled device detector FEI Eagle. Micrographs were collected with a pixel size of 1.79 Å and total dose of 30 e−/Å2. Frames were aligned using MotionCor2 (29), and CTF parameters were estimated using Gctf program (30).
Cryo-EM image processing
PVY image processing
A total number of 726 micrographs of the PVY sample were collected, and 235 micrographs were subsequently selected for determination of PVY CP structure. Straight segments of the filaments were manually picked using e2helixboxer.py in the EMAN2.1 package (31). The subsequent steps of the data analysis workflow were performed in the RELION2.1 package (32). The data analysis comprised the following steps: A subset of 215,123 segments of PVY was extracted from the micrographs (boxes of 360 pixels × 360 pixels). Two rounds of reference-free two-dimensional (2D) classification with 25 iterations were performed, and only class averages showing high-resolution structural features were selected after each round. Initial estimate of the helical parameters was obtained from indexing the power spectra (fig. S1D). The helical parameters were found to be very similar to those of PepMV (7). The helical_toolbox program (RELION2.1) was then used to generate the initial geometrical model of the one-start left-handed helix with rise of 4 Å and twist of 40°. The 3D classification using 162,213 segments revealed minimal structural variability at 4 Å resolution. Therefore, all these segments were used for the final 3D refinement of the model, with a local search of helical parameters, which yielded values of −40.95° for helical twist and 3.95 Å for helical rise. The reconstructed 3D map of PVY was sharpened using the relion_postprocess program (RELION2.1), and helical symmetry was imposed using the relion_helix_toolbox program (RELION2.1). The overall resolution of 3.4 Å was determined on the basis of gold-standard fourier shell correlation (FSC) at 0.143 criterion (fig. S1F) (33). Local resolution was calculated using MonoRes (34), and cryo-EM densities were visualized in UCSF Chimera (35).
VLP image processing
A total number of 5344 micrographs of the VLP sample were collected, and 3759 micrographs were subsequently used for data analysis. Straight segments of VLP filaments were manually picked from the micrographs using e2helixboxer.py in the EMAN2.1 package (31). Subsequently, helical reconstruction based on single-particle analysis was achieved using the RELION2.0 and RELION2.1 packages (32). A subset of 244,988 segments of VLPs was extracted from the micrographs (boxes of 360 pixels × 360 pixels). Two rounds of reference-free 2D classification with 25 iterations were performed, and only class averages showing high-resolution structural features were selected after each round.
Additional analysis of the micrographs revealed the population of circular particles corresponding to the orthogonal projections (top views) of the single VLP ring. Reference-free 2D classification revealed the presence of eightfold symmetry within the ring. Therefore, C8 symmetry was imposed during subsequent steps of structure refinement. A featureless cylinder was used as the initial 3D model for 3D refinement. No particles were removed on the basis of 3D classification; therefore, 148,876 segments of VLPs were used in 3D autorefinement with an applied mask with a diameter of 184 Å. A local search of helical parameters during 3D autorefinement yielded values of 13.24° for helical twist and 42.65 Å for helical rise. The reconstructed 3D map of VLP was sharpened using the relion_postprocess program (RELION2.1), and helical symmetry was imposed using the relion_helix_toolbox program (RELION2.1). Overall, resolution of 4.1 Å was determined on the basis of gold-standard FSC at 0.143 criterion (fig. S7F) (33). The local resolutions were calculated using MonoRes (34), and cryo-EM densities were visualized in UCSF Chimera (35).
ΔN49 deletion mutant image processing
A total of 23 micrographs of ΔN49 deletion mutant were collected to obtain 2D class averages of the oligomer. In total, 2132 particles were extracted using e2boxer.py in EMAN2.1 package (31). After reference-free 2D classification with 25 iterations and three classes in RELION2.1 (36), the best 2D class image with 1907 particles indicated octameric ring assembly of the ΔN49 CPs.
Model building
PVY model building
A 3D model of PVY CP for virion model building was obtained by homology modeling in Phyre2 (37). To facilitate model reconstruction in the cryo-EM density, a smaller segment representing approximately one CP unit was extracted from the whole cryo-EM map using the Bsoft package (38). The predicted 3D model was used to create a mask for extraction of specific segments, one or more CP units in size. Well-defined density in the core region of the CP allowed a separate fit of four main helices from the predicted 3D model using UCSF Chimera (35). The rest of the CP was built de novo in Coot (39) after converting cryo-EM maps into .mtz files with mrc2mtz.sh script from CCP-EM software suite (40). The density of the first 43 amino acid residues on the N terminus was not detected; therefore, the model was reconstructed from the amino acid residue Val44 onward. On the other hand, the density of the map at C terminus was well defined, which allowed us to build the C terminus to the final Met267. After the CP model was fitted into a cryo-EM density map, an obvious density corresponding to viral ssRNA was observed. The density of each nucleotide position was well defined; however, because of helical averaging, all residues were assigned the uracil base. De novo building of RNA in Coot was performed by inserting a segment of 10 ideal uracils into the empty density of the armpit-like groove of CPn. Namely, each PVY CP harbors five nucleotides; therefore, we started model building with five nucleotides of CPn. Additional nucleotides were placed on both sides to ensure correct fitting and refinement of the central region. The model of the virus (CPn with 10 uracils, surrounded by neighboring copies of CP) was subjected to several iterative cycles of manual refinement using Coot and real-space refinement with secondary structure and geometry restraints in Phenix package (41). MolProbity (42) was used for validation of individual models after each cycle. Refinement was always carried out on one CP, surrounded by at least 12 neighboring CP copies and an ssRNA molecule to mimic all intermolecular interactions and prevent clashes between subunits. When good fitting and geometry of the RNA segment containing 10 uracils was obtained, central 5 uracils from the RNA model were extracted and copied to all 35 copies of CP in the final model of the virus. Segments of five uracils were merged together to form one single ssRNA molecule.
VLP model building
The reconstructed 3D model of the PVY CP unit was fitted into cryo-EM density map of VLP. The density of the same length of the N-terminal region as in the virus was defined, but it had a different way of binding the neighboring subunit. Therefore, this part (Thr43-Tyr73) was rebuilt de novo, while there was no density for residues preceding Thr43. Because of the missing density for the C-terminal region, it was not possible to build this part of the polypeptide chain; therefore, the model of VLP CP ends with the residue Ile218. Several iterative cycles of manual refinement in Coot and real-space refinement in Phenix were carried out on CPn surrounded by at least eight copies of neighboring CPs, and the model was validated in MolProbity each cycle.
Final 3D models were visualized and analyzed in PyMOL (43). The surface electrostatic potential of the CP of virus and VLP was calculated using PDB2PQR code (44) and APBS approach (45) using default settings. Cutoff of −5 kTe−1 was used for negative potential and +5 kTe−1 for positive potential.
N-terminal Edman sequencing
PVY and VLP samples were analyzed under reducing conditions with SDS-PAGE using NuPAGE 4 to 12% bis-tris protein gel (Invitrogen). Wet electroblotting was carried out to transfer proteins from the gel to the polyvinylidene difluoride membrane. The transfer sandwich (filter paper–gel–membrane–filter paper) was placed into the transfer box (XCell SureLock, Invitrogen) at constant voltage of 30 V at 5°C for 60 min in NuPAGE transfer buffer with 10% methanol. After the transfer, the membrane was washed for 5 min in 50% methanol; then stained for 1 min in 0.1% Coomassie Blue R-250, 50% methanol, and 5% acetic acid; and then destained in 50% methanol until bands were visible. The membrane was left to air dry, and the protein bands were cut out and submitted for N-terminal Edman sequencing on Procise protein sequencing system 492A (PE Applied Biosystems at Jožef Stefan Institute, Ljubljana, Slovenia). Edman chemistry using pulsed-liquid blot was performed for preparation of phenylthiohydantoin amino acid derivatives. Analysis of the derivatives was performed on high-performance liquid chromatography (HPLC) system 140C (PE Applied Biosystems) using RP C18 column Spheri-5 (5 μm, 220 mm by 2.1 mm) (Brownlee).
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Measurements were carried out on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). First, the buffer and salt components from the samples of PVY virus and VLP constructs were exchanged with pure water, and samples were concentrated to 1.5 to 3.0 mg/ml with centrifugation through 10-kDa cutoff membranes. Samples were then mixed with saturated solution of sinapinic acid in a mixture of acetonitrile and 0.1% trifluoroacetic acid (1:1, v/v) in a 1:1 (v/v). One microliter of this solution was spotted on the target plate (dried-droplet method). The linear positive ion mode was used to acquire the mass spectra of the samples. The calibration was done externally using protein calibration standards I and II (Bruker Daltonics). Sample preparation procedure for the standards was the same as that for other samples. The standards were spotted on the nearest neighboring positions.
Size-exclusion chromatography coupled to a multidetection system
Separation of samples by size-exclusion chromatography was carried out in PBS buffer solution (pH 7.4) using an Agilent 1260 HPLC chromatograph (Agilent Technologies, USA) and Superdex 200 10/300 GL (24 ml) analytical column (GE Healthcare). For detection, successively online connected detectors were used: diode array detector Infinity 1260 detector operating at 280 nm (Agilent Technologies, USA), multiangle light scattering (MALS with 18 angles) photometer with GaAs linearly polarized laser (λ0 = 661 nm; DAWN HELEOS II), and Optilab rEX interferometric refractometer operating at the same wavelength as the photometer (both instruments from Wyatt Technology Corp., USA). The nominal eluent (PBS buffer) flow rate was 0.5 ml/min.
Circular dichroism spectroscopy
Far ultraviolet circular dichroism spectra of PVY-derived VLPs and mutant CPs (all samples at 0.2 mg/ml) in 10 mM phosphate buffer (pH 7.4) were recorded on a Chirascan spectrometer (Applied Photophysics) in a 0.1-cm path length quartz cuvette at 20°C. Spectra were measured in a 200- to 250-nm wavelength range with a step size of 0.5 nm and integration time of 0.5 s. Each spectrum was the average of 10 scans. All spectra were processed, baseline-corrected, averaged, smoothed, and converted with the Chirascan software. Spectral units were expressed as the mean molar ellipticity per residue.
Atomistic modeling, molecular dynamics simulations, and principal components analysis
On the basis of structural information, four atomistic models were prepared: one of VLP (24 protein units, residues Val44-Ile218) and three variants of the PVY virion composed as well of 24 protein units: (i) full protein unit, residues Val44-Met267 including ssRNA; (ii) C-terminal truncated protein unit, residues Val44-Ile218 including ssRNA; and (iii) C-terminal truncated protein unit, Val44-Ile218, without ssRNA. These systems were made periodic in one dimension along the longitudinal axis by implementing skew-symmetry operations of the unit cell: translation by 94.9 Å (24 × ~3.95 Å) in PVY and by 127.9 Å (3 × 42.65 Å) in VLP, followed by rotation by 86.6° in PVY and by 39.7° in VLP, respectively. In case of the ssRNA molecule, a proper chemical bond was introduced between the terminal phosphate groups of the unit cell and its image, translated along the filament axis to assure the 1D periodicity of the system. Molecular assemblies were hydrated by immersing them in cylinders of explicit water with a padding distance of 10 Å from the protein surface. The proper amount of sodium counterions was added to the solvent to neutralize each system (192 Na2+ ions for PVY, 144 Na2+ ions for truncated PVY, 24 Na2+ ions for truncated PVY without RNA, and 48 Na2+ ions for VLP). Solvent molecules were not allowed to exit the cylinder of 81-Å radius with the axis aligning the fibril axis of the system during the simulation to provide the padding of the bulk solvent around the fibril. We applied a cylindrical tube constraint of the force constant 50 kcal mol−1 Å−2.
Molecular dynamics (MD) simulations were run using the CHARMM program (46) with a time step of 2 fs and CHARMM36 force field (47). The transferable intermolecular potential with 3 points (TIP3P) water model was used. All systems, each composed of approximately 200,000 atoms, were first minimized using the steepest descent method for 500 steps followed by 1000 steps of the adaptive Newton-Raphson method (46) and then heated and equilibrated at 300 K for 5 ns. The system temperature was maintained constant by using velocity rescale for temperature coupling. Nonbonded interactions were cut at 14 Å. Trajectories of 30 ns were generated for each system in production runs with the saving frequency of 1 ps.
It has been shown that normal modes or principal modes provide a suitable basis for representing domain motions in biological macromolecules (48), while comparison of the frequency distributions of these modes between different systems can be used efficiently to describe elastic properties of the systems (49). For example, the propensity of a filament for stretching and bending deformation should be associated with an increased population of vibrational modes displaying motion along the filament axis at lower frequencies, as the lower frequency vibrations correspond to the weaker force constants. Here, we performed principal components analysis (PCA) (50) of molecular dynamics production trajectories by using in-house computer programs (49). We focused on dynamical fluctuations of protein building unit centers of mass represented as beads, which are quantified in terms of effective force constants within the framework of PCA. The results of the PCA are frequencies and displacements of beads for each individual dynamical/vibrational mode that the system undergoes. The lowest vibrational modes with projected displacements along the filament axis are shown in Fig. 4 (A and B).
In planta functional analysis of PVY CP N- and C-terminal deletion mutants
The PVY cDNA clone PVY-N605(123) (51) was used for construction of PVY clones with mutant CP. This PVY clone was chosen as, among the few available clones, it is similar to PVY-NTN and it is easy to manipulate (52).
Deletion of 50 amino acid residues at the N terminus (ΔN50-CP) and deletions of 40 or 60 residues at the C terminus (ΔC40-CP and ΔC60-CP, respectively) of the CP protein were introduced into the plasmid using in vitro site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit, Agilent Technologies) according to the manufacturer’s instructions with the following modifications: The plasmid was amplified in 25-μl polymerase chain reaction (PCR) reaction using a touch-down cycling program with annealing temperature from 65° to 55°C for 10 cycles followed by 8 cycles with constant annealing temperature of 55°C and an elongation step of 28 min. Dpn I (Agilent Technologies) digestion was performed by adding 40 U of enzyme to each amplification reaction and incubating it at 37°C for 2 hours. Afterward, 2 μl of the digested DNA was used to transform XL10-Gold Ultracompetent Cells (Agilent Technologies). The plasmids were isolated with a Monarch plasmid miniprep kit (New England BioLabs), and mutations were confirmed by automated Sanger sequencing (Eurofins Scientific).
The wild-type PVY clone and the three CP mutant clones (ΔN50-CP, ΔC40-CP, and ΔC60-CP) were used to infect N. clevelandii at the 6 to 10 leaf stage. For each construct, seven plants (five leaves per plant) were inoculated by biolistic bombardment using Helios Gen Gun system (Bio-Rad), as previously described (53). Bombardment with a yellow fluorescent protein–expressing plasmid that lacked any PVY-related sequence served as negative bombardment control to assess for any background signal due to PVY carry over during the bombardment procedure.
At 7 days postbombardment (dpb), samples of three inoculated leaves per plant were collected. Following the same procedure, three upper nonbombarded leaves were sampled at 14 dpb. TEM grids were prepared with sections of lower leaves for each treatment, at 7 dpb. The experiment was repeated twice for all three CP mutant clones. The homogenization, RNA isolation, deoxyribonuclease treatment and quality control of RNA were performed, as described (54). A one-step reverse transcription PCR (RT-PCR) kit (Qiagen) was used according to the manufacturer’s protocol to confirm that introduced mutations were maintained in the viral progeny. Relative PVY RNA concentration was analyzed by single-step quantitative RT-PCR (RT-qPCR; AgPath-ID One-Step RT-PCR, Thermo Fisher Scientific), and primers are shown in table S2. The cytochrome oxidase transcript was used as the endogenous control. All samples with Cq values above the background (Cq = 32) were considered negative. To check for the contribution of plasmid-derived signal to the concentration of PVY RNA, all samples were analyzed using the same assay and kit but omitting the RT step by preheating up the master mix (5 min, 95°C). The quantGenius software (55) was used for relative quantification of PVY RNA using the standard curve. The copy numbers calculated from qPCR without RT (presence of plasmid DNA in the sample) were subtracted from copy numbers calculated from RT-qPCR. One-way analysis of variance (ANOVA) with Bonferroni post hoc analysis was performed to statistically evaluate differences in virus accumulation between the PVY clones (data S1).
Supplementary Material
Acknowledgments
We thank J. Lenarčič, M. Srnko, L. Mourrain, M. Kisovec, A. Blejec, B. Dušak, and R. Vollmeier for support and F. Cillo for PVY clone. Funding: This work was supported by the Slovenian Research Agency (P1-0391, J7-7248, P4-0165, P2-0145, and P1-0010, PhD grants for A.K.), CIISB research infrastructure project LM2015043 by MEYS CR, and the National Grid Infrastructure MetaCentrum (CESNET LM2015042). Author contributions: M. Pod., A.K., K.G., M.R., I.G.-A., and G.A. conceived the project and analyzed data. A.K., L.K., M. Pol., J.N., I.G.-A., M.T.Ž., A.C., K.S., D.P., E.Ž., and F.M. performed the experiments. M. Pod. and A.K. wrote the manuscript with contribution from all the authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Coordinates are deposited in the PDB under accession codes 6HXX and 6HXZ for PVY and VLP, respectively. Cryo-EM reconstructions of the PVY and VLP are deposited in the EM Data Bank under accession codes EMD-0297 and EMD-0298. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaaw3808/DC1
Supplementary Text S1. Detailed description of protein-protein interactions in PVY
Supplementary Text S2. Detailed description of protein-protein interactions in VLP
Supplementary Text S3. Importance of the CP N terminus for longitudinal assembly of VLPs
Supplementary Text S4. Redundancy of CP C termini for VLP filament or octameric ring
Fig. S1. Cryo-EM data collection, image processing, and model building for the PVY virion.
Fig. S2. Consensus sequence alignment of potyviral CPs.
Fig. S3. Biophysical characterization of PVY virions and various VLP constructs.
Fig. S4. Structural comparison of PVY to other filamentous viruses, flexible and rod shaped.
Fig. S5. Details of the PVY interaction network.
Fig. S6. Electrostatic interactions are crucial for PVY virion assembly.
Fig. S7. Cryo-EM data collection, image processing, and model building for VLP.
Fig. S8. Surface electrostatic potential of VLPs.
Fig. S9. Water channels in different types of filamentous particles.
Table S1. Cryo-EM data collection and refinement statistics of PVY and VLP.
Table S2. Primers and probes used for in planta functional analysis of PVY mutants and their properties.
Data S1. Relative concentration of PVY RNA as measured in the leaves of N. clevelandii including tables S3 to S5:
Table S3. Relative concentration of PVY RNA as measured in bombarded leaves of N. clevelandii 7 dpb (experiment 1).
Table S4. Relative concentration of PVY RNA as measured in upper nonbombarded leaves of N. clevelandii 14 dpb (experiment 1).
Table S5. Relative concentration of PVY RNA as measured in bombarded leaves of N. clevelandii (experiment 2).
REFERENCES AND NOTES
- 1.Scholthof K.-B. G., Adkins S., Czosnek H., Palukaitis P., Jacquot E., Hohn T., Hohn B., Saunders K., Candresse T., Ahlquist P., Hemenway C., Foster G. D., Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. P. T. Valkonen, Viruses: Economical losses and biotechnological potential, in Potato Biology and Biotechnology: Advances and Perspectives, D. Vreugdenhil, J. Bradshaw, C. Gebhardt, F. Govers, D. K. L. Mackerron, M. A. Taylor, H. A. Ross, Eds. (Elsevier, ed. 1, 2007), pp. 619–641. [Google Scholar]
- 3.C. Kerlan, B. Moury, Potato virus Y, in Encyclopedia of Virology, B. W. J. Mahy, M. H. V. Van Regenmortel, Eds. (Academic Press, ed. 3, 2008), pp. 287–296. [Google Scholar]
- 4.Karasev A. V., Gray S. M., Continuous and emerging challenges of potato virus Y in potato. Annu. Rev. Phytopathol. 51, 571–586 (2013). [DOI] [PubMed] [Google Scholar]
- 5.F. Revers, J. A. García, Molecular biology of potyviruses, in Advances in Virus Research (Academic Press, ed. 1, 2015), vol. 92, pp. 101–199. [DOI] [PubMed] [Google Scholar]
- 6.Chung B. Y.-W., Miller W. A., Atkins J. F., Firth A. E., An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U.S.A. 105, 5897–5902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agirrezabala X., Méndez-López E., Lasso G., Sánchez-Pina M. A., Aranda M., Valle M., The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. eLife 4, e11795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMaio F., Chen C.-C., Yu X., Frenz B., Hsu Y.-H., Lin N.-S., Egelman E. H., The molecular basis for flexibility in the flexible filamentous plant viruses. Nat. Struct. Mol. Biol. 22, 642–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamora M., Méndez-López E., Agirrezabala X., Cuesta R., Lavín J. L., Sánchez-Pina M. A., Aranda M. A., Valle M., Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 3, eaao2182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison B. D., Robinson D. J., Molecular variation in vector-borne plant viruses: Epidemiological [and Discussion] significance. Philos. Trans. R. Soc. Lond. B 321, 447–462 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Shukla D. D., Strike P. M., Tracy S. L., Gough K. H., Ward C. W., The N and C termini of the coat proteins of potyviruses are surface-located and the N terminus contains the major virus-specific epitopes. J. Gen. Virol. 69, 1497–1508 (1988). [Google Scholar]
- 12.Anindya R., Savithri H. S., Surface-exposed amino- and carboxy-terminal residues are crucial for the initiation of assembly in Pepper vein banding virus: A flexuous rod-shaped virus. Virology 316, 325–336 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Seo J.-K., Vo Phan M. S., Kang S.-H., Choi H.-S., Kim K.-H., The charged residues in the surface-exposed C-terminus of the Soybean mosaic virus coat protein are critical for cell-to-cell movement. Virology 446, 95–101 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Ge P., Zhou Z. H., Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc. Natl. Acad. Sci. U.S.A. 108, 9637–9642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clare D. K., Pechnikova E. V., Skurat E. V., Makarov V. V., Sokolova O. S., Solovyev A. G., Orlova E. V., Novel inter-subunit contacts in barley stripe mosaic virus revealed by cryo-electron microscopy. Structure 23, 1815–1826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall A., McDonald M., Bian W., Bowles T., Baumgarten S. C., Shi J., Stewart P. L., Bullitt E., Gore D., Irving T. C., Havens W. M., Ghabrial S. A., Wall J. S., Stubbs G., Structure of flexible filamentous plant viruses. J. Virol. 82, 9546–9554 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stubbs G., Kendall A., McDonald M., Bian W., Bowles T., Baumgarten S., McCullough I., Shi J., Stewart P., Bullitt E., Gore D., Ghabrial S., Flexible filamentous virus structures from fiber diffraction. Powder Diffract. 23, 113–117 (2008). [Google Scholar]
- 18.McDonald J. G., Bancroft J. B., Assembly studies on potato virus Y and its coat protein. J. Gen. Virol. 35, 251–263 (1977). [Google Scholar]
- 19.Donchenko E. K., Pechnikova E. V., Mishyna M. Y., Manukhova T. I., Sokolova O. S., Nikitin N. A., Atabekov J. G., Karpova O. V., Structure and properties of virions and virus-like particles derived from the coat protein of Alternanthera mosaic virus. PLOS ONE 12, e0183824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolja V. V., Haldeman R., Robertson N. L., Dougherty W. G., Carrington J. C., Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13, 1482–1491 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolja V. V., Haldeman-Cahill R., Montgomery A. E., Vandenbosch K. A., Carrington J. C., Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 206, 1007–1016 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Merits A., Puurand U., Merits A., Rabenstein F., Järvekülg L., Valkonen J. P., Potyvirus helper component-proteinase and coat protein (CP) have coordinated functions in virus-host interactions and the same CP motif affects virus transmission and accumulation. J. Gen. Virol. 80, 1133–1139 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Besong-Ndika J., Ivanov K. I., Hafrèn A., Michon T., Mäkinen K., Cotranslational coat protein-mediated inhibition of potyviral RNA translation. J. Virol. 89, 4237–4248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X., Shaw J. G., Evidence that assembly of a potyvirus begins near the 5′ terminus of the viral RNA. J. Gen. Virol. 79, 1525–1529 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Gallo A., Valli A., Calvo M., García J. A., A functional link between RNA replication and virion assembly in the potyvirus plum pox virus. J. Virol. 92, e02179-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valli A., Gallo A., Calvo M., Pérez J. d. J., García J. A., A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J. Virol. 88, 9808–9818 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torrance L., Andreev I. A., Gabrenaite-Verhovskaya R., Cowan G., Mäkinen K., Taliansky M. E., An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357, 1–8 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Rupar M., Ravnikar M., Tušek-Žnidarič M., Kramberger P., Glais L., Gutiérrez-Aguirre I., Fast purification of the filamentous Potato virus Y using monolithic chromatographic supports. J. Chromatogr. A 1272, 33–40 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Zheng S., Palovcak E., Armache J.-P., Cheng Y., Agard D., Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. bioRxiv 061960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell J. M., Chen M., Baldwin P. R., Ludtke S. J., High resolution single particle refinement in EMAN2.1. Methods 100, 25–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilas J. L., Gómez-Blanco J., Conesa P., Melero R., Miguel de la Rosa-Trevín J., Otón J., Cuenca J., Marabini R., Carazo J. M., Vargas J., Sorzano C. O. S., MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heymann J. B., Belnap D. M., Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burnley T., Palmer C. M., Winn M., Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrödinger, ThePyMOL Molecular Graphics System, Version 1.8 (2015).
- 44.Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A., PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks B. R., Brooks C. L. III, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Best R. B., Zhu X., Shim J., Lopes P. E. M., Mittal J., Feig M., MacKerell A. D. Jr., Optimization of the additive CHARMM All-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. Fuchigami, H. Fujisaki, Y. Matsunaga, A. Kidera, Protein functional motions: Basic concepts and computational methodologies, in Advancing Theory for Kinetics and Dynamics of Complex, Many-Dimensional Systems: Clusters and Proteins, T. Komatsuzaki, R. S. Berry, D. M. Leitner, Eds. (John Wiley & Sons, 2011), pp. 35–82. [Google Scholar]
- 49.Balog E., Perahia D., Smith J. C., Merzel F., Vibrational softening of a protein on ligand binding. J. Phys. Chem. B 115, 6811–6817 (2011). [DOI] [PubMed] [Google Scholar]
- 50.A. Kitao, Extension of the normal mode concept: Principal component analysis, jumping-among-minima model, and their applications to experimental data analysis, in Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems, Q. Cui, I. Bahar, Eds. (CRC Press, 2005), pp. 233–251. [Google Scholar]
- 51.Bukovinszki Á., Götz R., Johansen E., Maiss E., Balázs E., The role of the coat protein region in symptom formation on Physalis floridana varies between PVY strains. Virus Res. 127, 122–125 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Rupar M., Faurez F., Tribodet M., Gutiérrez-Aguirre I., Delaunay A., Glais L., Kriznik M., Dobnik D., Gruden K., Jacquot E., Ravnikar M., Fluorescently tagged Potato virus Y: A versatile tool for functional analysis of plant-virus interactions. Mol. Plant-Microbe Interact. 28, 739–750 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Dobnik D., Baebler Š., Kogovšek P., Pompe-Novak M., Štebih D., Panter G., Janež N., Morisset D., Žel J., Gruden K., β-1,3-glucanase class III promotes spread of PVYNTN and improves in planta protein production. Plant Biotechnol. Rep. 7, 547–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baebler S., Krečič-Stres H., Rotter A., Kogovšek P., Cankar K., Kok E. J., Gruden K., Kovač M., Žel J., Pompe-Novak M., Ravnikar M., PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol. Plant Pathol. 10, 263–275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baebler Š., Svalina M., Petek M., Stare K., Rotter A., Pompe-Novak M., Gruden K., quantGenius: Implementation of a decision support system for qPCR-based gene quantification. BMC Bioinformatics 18, 276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall T. A., BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- 58.Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T., The MIQE guidelines: Minimum information for publication of quantitative real-Time PCR experiments. Clin. Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Kogovšek P., Gow L., Pompe-Novak M., Gruden K., Foster G. D., Boonham N., Ravnikar M., Single-step RT real-time PCR for sensitive detection and discrimination of Potato virus Y isolates. J. Virol. Methods 149, 1–11 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Weller S. A., Elphinstone J. G., Smith N. C., Boonham N., Stead D. E., Detection of ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66, 2853–2858 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaaw3808/DC1
Supplementary Text S1. Detailed description of protein-protein interactions in PVY
Supplementary Text S2. Detailed description of protein-protein interactions in VLP
Supplementary Text S3. Importance of the CP N terminus for longitudinal assembly of VLPs
Supplementary Text S4. Redundancy of CP C termini for VLP filament or octameric ring
Fig. S1. Cryo-EM data collection, image processing, and model building for the PVY virion.
Fig. S2. Consensus sequence alignment of potyviral CPs.
Fig. S3. Biophysical characterization of PVY virions and various VLP constructs.
Fig. S4. Structural comparison of PVY to other filamentous viruses, flexible and rod shaped.
Fig. S5. Details of the PVY interaction network.
Fig. S6. Electrostatic interactions are crucial for PVY virion assembly.
Fig. S7. Cryo-EM data collection, image processing, and model building for VLP.
Fig. S8. Surface electrostatic potential of VLPs.
Fig. S9. Water channels in different types of filamentous particles.
Table S1. Cryo-EM data collection and refinement statistics of PVY and VLP.
Table S2. Primers and probes used for in planta functional analysis of PVY mutants and their properties.
Data S1. Relative concentration of PVY RNA as measured in the leaves of N. clevelandii including tables S3 to S5:
Table S3. Relative concentration of PVY RNA as measured in bombarded leaves of N. clevelandii 7 dpb (experiment 1).
Table S4. Relative concentration of PVY RNA as measured in upper nonbombarded leaves of N. clevelandii 14 dpb (experiment 1).
Table S5. Relative concentration of PVY RNA as measured in bombarded leaves of N. clevelandii (experiment 2).


