Abstract
In mammals, most molecular and cellular processes show circadian changes, leading to daily variations in physiology and ultimately in behaviour. Such daily variations induce a temporal coordination of processes that is essential to ensure homeostasis and health. Thus, it is of no surprise that pharmacokinetics (PK) and pharmacodynamics (PD) of many drugs are also subject to circadian variations, profoundly affecting their efficacy and tolerability. Understanding how circadian rhythms influence drug PK, PD, and toxicity might significantly improve treatment efficacy and decrease related side effects. Therefore, it is essential to take circadian variations into account and to determine circadian parameters in pharmacological studies, especially when drugs have a short half‐life or target rhythmic processes. This review provides an overview of the current knowledge on circadian rhythms and their relevance to the field of pharmacology. Methodologies to evaluate circadian rhythms in vitro, in rodent models and in humans, from experimental to computational approaches, are described and discussed. Lastly, we aim at alerting the scientific, medical, and regulatory communities to the relevance of the physiological time, as a key parameter to be considered when designing pharmacological studies. This will eventually lead to more successful preclinical and clinical trials and pave the way to a more personalized treatment to the benefit of the patients.
Abbreviations
- ABCC1
ATP‐binding cassette subfamily C member 1
- ADME
Absorption, Distribution, Metabolism and Excretion
- Bmal1
brain and muscle ARNT‐like protein‐1
- BRRE
ROR regulatory element
- CCGs
clock‐controlled genes
- Clock
circadian locomotor output cycles kaput
- Cry
cryptochrome
- DBP
D‐box‐binding PAR BZIP transcription factor
- DD
dark–dark
- FDA
Food and Drug Administration
- LD
light–dark
- luc
luciferase
- MRTF
Myocardin‐related transcription factor
- NAD+
Nicotinamide adenine dinucleotide
- NF‐κB
Nuclear Factor kappa
- NAMPT
nicotinamide phosphoribosyltransferase
- NPAS2
neuronal PAS domain protein 2
- PAR bZip
PAR‐domain basic leucine zipper
- PD
pharmacodynamics
- Per
period
- PK
pharmacokinetics
- Rev‐Erb
nuclear receptor reverse strand of ERBA
- Ror
RAR‐related orphan receptor
- SCN
suprachiasmatic nucleus
- Sirt1
sirtuin 1
- SRF
Serum Response Factor
- TEF
Thyrotroph embryonic factor
- TFs
transcription factors
1. INTRODUCTION
Biological rhythms have been observed, and quantified in detail, in a variety of species, from bacteria to humans (Fuhr, Abreu, Pett, & Relógio, 2015). Throughout evolution, most organisms have developed an endogenous clock system, which allows for the entrainment to external cues (zeitgebers) and accounts for the adaption to the geophysical time. Based on its period length, rhythms can be classified into ultradian (<24 hr), circadian (~24 hr), or infradian (>24 hr; Smolensky & Peppas, 2007). The circadian system regulates temporal physiology and behaviour, allocating specific biochemical processes to specific daytimes while temporally separating incompatible ones. In this way, the circadian clock system promotes optimal organism activity, ensuring the temporal regulation of defence mechanisms, energy conservation, and internal homeostasis (Asher & Schibler, 2011).
In mammals, almost all cellular functions and physiological systems are subjected to circadian control (see review in Pilorz, Helfrich‐Förster, & Oster, 2018). At the cellular level, specific genes encode for a set of transcription factors (TFs) that work in an interconnected manner, building up a molecular core‐clock network (CCN). These TFs, via transcriptional and translational feedback loops, regulate their own transcription, which leads to ≈24‐hr oscillations in their gene and protein expression in all cells, tissues, and organs. In addition, the CCN elements coordinate the transcription of many other genes, clock‐controlled genes (CCGs), in a circadian‐ and organ‐specific manner, regulating a variety of biological processes, at both the cell and tissue levels, such as cell division, bioenergetics, autophagy, metabolism, redox balance, hormone regulation, and immune responses (El‐Athman et al., 2017; Fuhr et al., 2018; Relógio et al., 2014; Scrima et al., 2016; Zhang, Lahens, Ballance, Hughes, & Hogenesch, 2014; reviewed in Pilorz et al., 2018). These cellular peripheral clocks, distributed throughout the body, are further coordinated by a central pacemaker, located at the suprachiasmatic nuclei (SCNs) in the hypothalamus (Albrecht, 2012).
Given its widespread location and pleiotropic roles, the study of circadian timing at the cellular and organismal levels has gained increasing relevance beyond the so‐called field of chronobiology. As the majority of cellular functions in all organs is subject to circadian regulation, it is of no surprise that the circadian clock system also affects the metabolism (pharmacokinetics—PK) and efficacy (pharmacodynamics—PD) of many drugs. For example, drug absorption, metabolism and excretion are influenced by the circadian clock (see Ballesta, Innominato, Dallmann, Rand, & Lévi, 2017; Musiek & FitzGerald, 2013). Therefore, the circadian time should be considered when developing drug dosing regimens, measuring drug levels in the blood, and evaluating drug efficacy. Indeed, randomized clinical trials have shown that circadian‐based treatments (chronotherapy) contribute to optimized treatment outcomes (Ballesta et al., 2017). Hence, understanding the molecular mechanisms through which the circadian clock system influences processes relevant for drug development is essential to improve drug discovery and treatment optimization.
Accordingly, in this article, we provide an overview of the current knowledge of the biology of circadian rhythms and the available methodologies to evaluate circadian parameters in different experimental models, covering in vitro and in vivo models, as well as in humans. We focus in mammals as the most relevant models to elucidate the influence of circadian rhythms in pharmacological studies.
2. IMPACT OF CIRCADIAN RHYTHMS ON PHARMACOLOGICAL STUDIES
Circadian regulation of molecular processes not only affects health and disease, but it may also affect the outcome of therapeutic approaches. Many conditions and diseases show acute events (e.g., myocardial infarction) and symptoms (e.g., rheumatoid arthritis) at specific times of the day. These observations provide windows of opportunity to align treatments with disease flares. In addition, biological time strongly influences drug responses. Absorption, distribution, metabolism and excretion, along with efficacy, and/or toxicity of drugs, all have pronounced diurnal fluctuations (Levi & Schibler, 2007; Ruben et al., 2018). The effects in the PK of drugs result from rhythmic changes in absorption rates, drug‐protein binding, pH, membrane viscosity, levels of liver enzymes, transport proteins and ion channels, peripheral, liver, and renal blood flow, GFR and urine flow, and pH, which influences the degree of urine acidification (Dallmann, Okyar, & Lévi, 2016; Levi & Schibler, 2007). Similarly, PD also shows circadian variations. The efficacy and tolerability of nearly 500 medications varied by up to fivefold, according to circadian scheduling, in both experimental models and patients (Ballesta et al., 2017). Based on knowledge regarding circadian changes in the processes described above, the administration route, dose, kinetics, dynamics, and elimination/toxicity of many drugs may be further optimized in order to achieve their full therapeutic potential. Still, the molecular mechanisms by which the circadian clock regulates PK and PD processes are not completely understood. Gachon et al. reported that the circadian expression of many enzymes and regulators involved in detoxification and drug metabolism, such as cytochrome P450 enzymes, carboxylesterases, and constitutive androstane receptor, is controlled by PAR‐domain basic leucine zipper (PAR bZip) transcription factors (DBP, thyrotroph embryonic factor, and hepatic leukaemia factor) whose expression is under the control of core‐clock genes (Gachon, Olela, Schaad, Descombes, & Schibler, 2006). Specific drug targets (receptors, transporters and enzymes, intracellular signalling systems, and gene transcription machinery) may also show circadian variations, with a potential effect on drug efficacy or toxicity (Musiek & FitzGerald, 2013). In this context, some treatments may benefit from a chronopharmacological approach. Chronopharmacology investigates the influence of circadian rhythms on drug PK, PD, and toxicity. Accordingly, chronotherapy applies chronopharmacological studies to clinical treatments, to optimize treatment times in order to achieve the highest efficacy and diminish treatment‐related toxicity. Chronopharmacology becomes especially important if the symptoms of a disease vary predictably over time; the drug absorption, distribution, metabolism, and excretion presents high interindividual and intraindividual variability; the drug target varies along the circadian time; and drug plasma concentrations are well correlated with therapeutic or toxic effects and are circadian‐phase dependent (Ballesta et al., 2017; Baraldo, 2008). However, more than half of the best selling drugs worldwide are known to target core‐clock genes or CCGs, but most of them have not been associated with circadian rhythms and the influence of the time of administration was not taken into account during clinical trials (Zhang et al., 2014).
In addition, different studies have highlighted large interpatient differences in the endogenous clock, resulting in significant variability of therapeutic responses (Baraldo, 2008). Circadian clocks in different individuals entrain differently to zeitgebers, especially to light, which may result in different chronotypes. Such variation may largely derive from genetic polymorphisms in clock genes. Therefore, chronotherapy should be tailored to a single patient, a single pathology, and a single drug. Assessment of circadian parameters may help design clinical trials by identifying optimal times for drug administration, depending on the patient's particular chronotype and pathology (Roenneberg & Merrow, 2016).
The effects of circadian rhythms and the importance of their characterization should thus be taken into account in biomedicine. Different methodologies have been described to quantify circadian rhythms in cell cultures, rodents, and humans, which can be applied to pharmacological studies, from drug discovery to preclinical and clinical trials.
3. THE CIRCADIAN CLOCK SYSTEM
Several major characteristics define the circadian clock system: It is endogenous and self‐sustained, freely running/generating rhythms even in the absence of zeitgebers; it is entrainable, meaning that it can be adjusted to external time cues and it is temperature compensated as, on exposure to different temperatures within the organism's physiological range, the periods of the oscillations change only slightly (Roenneberg, Kantermann, Juda, Vetter, & Allebrandt, 2013). In order to do so, the mammalian circadian clock system can be stratified in three main components: input signalling pathways, which provide time cues to the second component, the central clock, and output signalling pathways, which are the effector pathways through which the central clock generates and maintains rhythms (see Albrecht, 2012; Figure 1).
Figure 1.
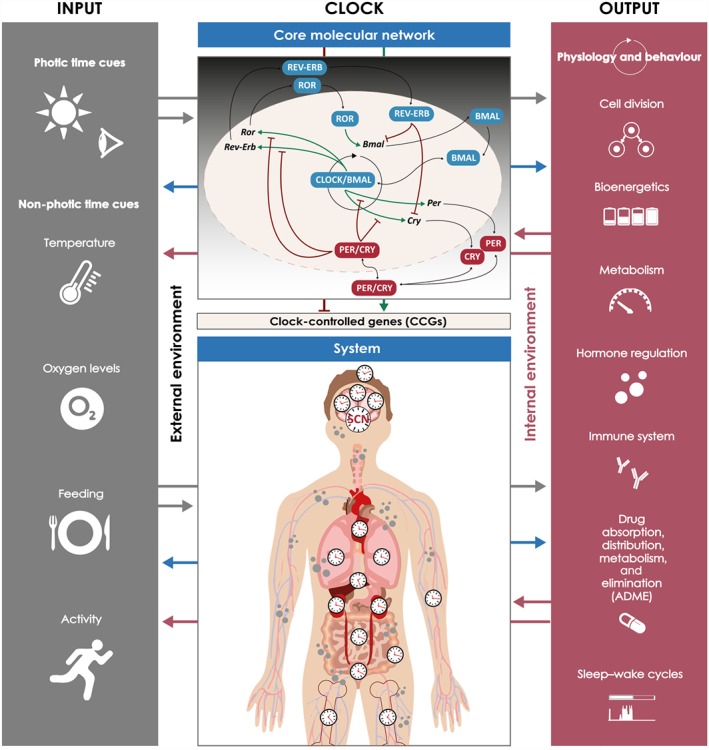
The circadian clock system. The mammalian circadian system can be stratified into three main components: inputs from the external environment to clocks (photic and non‐photic time cues), clock mechanisms (at the cellular and systemic level), and output signalling pathways that modulate physiology and behaviour. The three components are interconnected (arrows in both directions): the effects of input on the clock is time dependent, determined by the clock itself; peripheral cellular clocks and the main pacemaker, in the suprachiasmatic nucleus, are synchronized; activity influences body temperature; and sleep–wake cycles determine feeding–fasting cycles. In this way, external inputs, individual cellular clocks and subnetworks, and outputs are integrated into a coherent functional system/network
The input pathway entrains clocks according to temporal information obtained from the external environment. Light is the strongest zeitgeber, but also, oxygen levels, food, or exercise can act as time cues (Roenneberg & Merrow, 2016). The central clock coordinates the circadian system, being formed by two clusters of neurons (~100,000 neurons in each cluster in humans) located in the SCN (Ralph, Foster, Davis, & Menaker, 1990). Communication and synchronization among its neurons is essential for the SCN to produce robust rhythmic outputs (Ramkisoensing & Meijer, 2015). These are then transmitted to local clocks in the brain and to peripheral clocks (Dibner, Schibler, & Albrecht, 2010), via direct output pathways such as synaptic communication, gap junctions, and hormone secretion (Ramkisoensing & Meijer, 2015; Welsh, Takahashi, & Kay, 2010). In addition, communication via extracellular vesicles may also contribute to the temporal regulation of the system (Tao & Guo, 2018). The SCN is, thus, responsible for coordinating all clocks within the body (Albrecht, 2012). Peripheral clocks exist within all different tissues and organs throughout the organism. These can also be entrained to time cues, such as body temperature, feeding time, and physical activity rhythms, in addition to SCN‐derived messengers, being able to function independently of the SCN (Asher et al., 2010). Central and peripheral clocks may show phase differences, but the degree to which clocks in different organ systems may co‐exist out of phase remains unknown (Hughey & Butte, 2016).
The generation of circadian rhythms has been shown to occur at the single‐cell level. In cells, rhythms are driven by a complex transcriptional/translational regulatory network involving interlocked feedback loops (Figure 1). A set of 14 genes is known to form the CCN of the mammalian circadian clock system (Relógio et al., 2011). These genes are necessary for the robust generation of approximately 24‐hr oscillations, which justifies the designation of core‐clock genes (Relógio et al., 2011; Takahashi, 2017). The set includes members of the Per (period, Per1, Per2, and Per3), Cry (cryptochrome, Cry1 and Cry2), Bmal (brain and muscle ARNT‐like protein, Bmal1 and Bmal2), Clock (circadian locomotor output cycles kaput), NPAS2 (neuronal PAS domain protein 2) in neuronal tissue, Ror (RAR‐related orphan receptor, Rora, Rorb, and Rorc), and Rev‐Erb (nuclear receptor, reverse strand of ERB, Rev‐Erbα and Rev‐Erbβ) gene and protein families. This network can be seen as the interconnection of two loops, the PER/CRY and the REV‐ERB/Bmal1/ROR loops (Relógio et al., 2011). During the early time of the circadian day, CLOCK and BMAL1 associate to form a heterodimer complex that regulates the transcription of all other genes within the CCN. The CLOCK/BMAL1 complex acts as a TF binding to E‐Box sequences within the promoter regions of the target genes Ror, Rev‐Erb, Per, and Cry (Relógio et al., 2011). Following transcriptional activation of Per1, Per2, and Per3 and Cry1 and Cry2 genes and translation of the respective proteins, PER and CRY family members assemble to form PER/CRY complexes. After a series of post‐translational modifications, PER/CRY translocates to the nucleus and inhibits CLOCK/BMAL1‐mediated transcription of target genes, thus affecting the expression of all core‐clock genes. During the night, the PER/CRY complex is degraded, which releases its inhibitory action on CLOCK/BMAL1 and allows a new cycle of transcription/translation to take place. CLOCK/BMAL1 also binds to promoter regions of Rev‐Erbα and Rev‐Erbβ and Rorα, Rorβ, and Rorγ. The resulting proteins compete for ROR regulatory element‐binding sites in the promoter region of Bmal1 and regulate its transcription. While ROR acts as an activator of Bmal1 expression, REV‐ERB acts as an inhibitor, resulting in a fine tuning of Bmal1 transcription (Figure 1; Relógio et al., 2011; Takahashi, 2017).
Although the core proteins of the mammalian circadian clock have been known for nearly two decades, little is yet known about their cellular distribution. In 2017, Aryal and colleagues showed that clock proteins of the primary loop are incorporated in several complexes in the cytoplasm, while in the nucleus, they are assembled in larger nuclear complexes, suggesting an assembly pathway. Still, the number and nature of circadian clock protein complexes is still unclear (Aryal et al., 2017).
Clock proteins act as TFs for several other genes, rhythmically driving their expression (Lehmann et al., 2015). CCGs harbour specific regulatory motifs in the proximity of their promoters, such as E‐boxes and ROR regulatory elements, or cAMP response elements and D‐boxes. These can be recognized by particular core‐clock elements, leading to the circadian expression of target CCGs (Albrecht, 2012; Takahashi, 2017; Ueda et al., 2005). Nonetheless, transcriptional regulation is not the only mechanism responsible for the generation of circadian oscillations. The period, amplitude, and phase of such oscillations can be further modulated by epigenetic mechanisms, post‐transcriptional alterations, and post‐translational modifications. This allows for a tight regulation of the period, phase, amplitude, and eventually the function of circadian clocks (Takahashi, Hong, Ko, & McDearmon, 2008).
4. THE CIRCADIAN CLOCK SYSTEM IN HEALTH AND MOLECULAR MECHANISMS OF DISEASE
As previously mentioned, a variety of essential biological processes exhibit circadian rhythms. Internal timekeeping also manifests itself in behaviour, as shown by sleep–wake cycles and feeding and fasting patterns. The alignment of all internal clocks is thus crucial for the maintenance of good health.
Circadian rhythm dysfunction can be triggered by a multitude of factors, including disease states, ageing, genetic alterations, night‐shift work, jet lag, abnormal lighting conditions, feeding schedules, and social interactions (Damiola et al., 2000; Roenneberg & Merrow, 2016). In addition, many existing drugs may also influence circadian rhythms. Tamai et al. (2018) screened over 1,000 molecules from a Food and Drug Administration‐approved drug library and found that 5% had effects on the circadian clocks of human osteosarcoma epithelial cells (U2OS cell line). Disruptions in circadian rhythms have been associated with several disorders, including sleep (Vanselow et al., 2006), metabolic (Marcheva et al., 2010), neurodegenerative (Musiek, 2015), cardiovascular (Chen & Yang, 2015), autoimmune/inflammatory disorders (Hand et al., 2016), and cancer (El‐Athman, Fuhr, & Relógio, 2018). In this context, the circadian clock itself may become a therapeutic target for these disorders, along with other conditions associated with impaired circadian rhythms, namely, age‐related diseases. Indeed, behavioural and pharmacological strategies that re‐establish clock gene expression (“clock drugs”) show potential to restore circadian rhythms in disease. As long as the side effects of such drugs are not more hazardous than circadian rhythm disruption, they may mitigate disease symptoms and age‐related decline (see Sulli, Manoogian, Taub, & Panda, 2018).
The importance of circadian oscillations in neuronal, cardiovascular, endocrine, immune, metabolic, renal, respiratory, and digestive functions is thus well established (Pilorz et al., 2018). Still, our understanding of the individual contribution of the SCN and of the peripheral clocks to the regulation of each of these physiological systems and whole organism integration remains superficial. Even though much progress has been made in the identification of the molecular players driving circadian oscillations in different cells and tissues and their specific roles in health and disease, much more remains to be elucidated, especially regarding the crosstalk with other signalling pathways and regulatory mechanisms. For instance, several studies have shown an important interaction between specific clock components and metabolic regulators, like the NAD+‐dependent protein deacetylase, sirtuin 1 (SIRT1; Nakahata, Sahar, Astarita, Kaluzova, & Sassone‐Corsi, 2009; Wang et al., 2016). SIRT1 was shown to deacetylate the clock proteins BMAL1 and PER2, contributing to maintain functional clock oscillations. In turn, circadian oscillations of NAD + and thus SIRT1 activity are dependent on the levels of the enzyme nicotinamide phosphoribosyltransferase (NAMPT or visfatin), which were found to be rhythmically regulated by BMAL1:CLOCK. Moreover, SIRT1 was found to associate with BMAL1:CLOCK at the NAMPT promoter, contributing to the rhythmic circadian expression of this enzyme and, therefore, to circadian oscillations on its own activity. Additionally, SIRT1 and PER2 negatively regulate each other, SIRT1 by deacetylating histone H4 on Lys16 in the promoter of Per2, and PER2 by binding to the Sirt1 promoter at the CLOCK/BMAL1 site, suppressing its transcription (Wang et al., 2016). Thus, the reciprocal interaction between clock components and Sirt1 establishes a mutually regulated transcriptional–enzymic feedback loop that links cellular metabolism and the molecular clock (Nakahata et al., 2009). Indeed, disruption of either one of these systems leads to premature ageing (Sato et al., 2017; Wang et al., 2016). The relevance of this crosstalk is further emphasized by the role of both systems on modulating another important pathway, the NF‐κB activation pathway. This signalling cascade is involved in the cellular response to stress, inflammation, and immune responses and plays a significant role in cell senescence, ageing, and age‐related diseases (Tilstra, Clauson, Niedernhofer, & Robbins, 2011). Circadian control of NF‐κB activity involves various components of the clock system, such as CLOCK, BMAL1, and RORα (Delerive et al., 2001; Spengler et al., 2012). Furthermore, Sirt1 also interacts with NF‐κB, by deacetylating its RelA/p65 subunit, which decreases its transcriptional activity and contributes to the limitation of inflammation and its deleterious consequences (Yeung et al., 2004). Since SIRT1 activity is regulated by the clock system, so is its ability to counteract NF‐κB activity during the course of a day. On the other hand, NF‐κB also seems to have profound effects on the clock system, not only by affecting Sirt1 expression and activity but also by mediating inhibitory effects of inflammatory cytokines, such as IL‐1β, on the expression of clock genes. These affect mostly the negative feedback loop, namely, Cry, Per, and Rev‐erb genes, altering their circadian oscillations (Guo et al., 2015; Hong et al., 2018). Furthermore, acute and chronic activation of NF‐κB was recently shown to drive CLOCK:BMAL1 to sites convergent with those bound by NF‐κB, highlighting NF‐κB involvement on circadian disruption in response to inflammation (Hong et al., 2018). Surprisingly, Hong et al. (2018) also found that under non‐stimulated conditions, NF‐κB contributes to the normal circadian oscillations of clock genes, ensuring normal rhythmic behaviours in mice.
The crosstalk between these three systems, circadian clocks, metabolism, and stress response, couples the cellular ability to respond to stress and to metabolic needs with circadian rhythms, which allows the organism to meet its needs and challenges throughout the day. However, this equilibrium can be lost by impairment of any of these systems. In this regard, NF‐κB activation by low‐grade inflammation can be especially relevant as it has been reported in virtually all chronic age‐related diseases, from metabolic disturbances, like diabetes mellitus, obesity, and the metabolic syndrome, to neurodegenerative, musculoskeletal, renal, cardiovascular, pulmonary, intestinal, and even behavioural diseases (Rosa, Judas, Lopes, & Mendes, 2008; Scarpellini & Tack, 2012; Speer, Upton, Semple, & McKune, 2018; Zhu, Armstrong, Tchkonia, & Kirkland, 2014). Interestingly, ageing and age‐related diseases have also been associated with impaired circadian expression of core‐clock genes and CCGs and impaired SIRT1 activity (Kondratov, Kondratova, Gorbacheva, Vykhovanets, & Antoch, 2006; Musiek et al., 2018; Wang et al., 2018). These observations further emphasize the importance of the interplay between inflammatory, metabolic, and circadian rhythm pathways in health and disease. In the end, it is also crucial to understand the effects of a variety of physiological and non‐physiological stimuli, not only on each component of the molecular clock system but also on the overall function of the central and peripheral clocks and their mutual integration.
5. EVALUATION OF CIRCADIAN RHYTHMS: FROM CELLS TO HUMANS
5.1. Circadian studies in in vitro cell models
Clock properties (period length, phase, amplitude, and robustness of oscillations) and related disturbances can be measured in different cells and tissues.
Circadian studies have been performed in a multitude of in vitro cell models, ranging from rodent and human immortalized cell lines,such as NIH/3T3‐immortalized mouse embryo fibroblasts (Akashi & Nishida, 2000) and rat‐1 fibroblasts (Balsalobre, Damiola, & Schibler, 1998; Izumo, Sato, Straume, & Johnson, 2006), or human cancer lines, as U2OS osteosarcoma cells (Relógio et al., 2014); primary cultures, as mouse neonatal cardiomyocytes (du Pré et al., 2017) and human fibroblasts (Brown et al., 2005), blood monocytes (Keller et al., 2009), hair follicle cells (Akashi et al., 2010), or keratinocytes (Sandu et al., 2012); to organoids (Fuhr et al., 2018) and tissue explants, as mouse fetal adrenal gland (Ungar & Halberg, 1962) or SCN explants (Hughes et al., 2008).
Data from human primary cultures show that disease susceptibility is correlated with the donor chronotype, showing potential to indicate important endophenotypes (quantitative biological traits genetically correlated with disease liability) based on clock interindividual differences (Brown et al., 2005; Saini, Brown, & Dibner, 2015). This is relevant as the characterization of human circadian properties in vivo under laboratory conditions can be quite laborious and expensive. Nevertheless, when looking for processes regulated solely by the circadian clock, it is important that the donor is under controlled conditions (e.g., regular meal, exercise, and bedtimes) in order to better detect endogenous rhythms (Hughes et al., 2017). Quantitative data on clock properties can inform about dose‐ and/or time‐dependent effects of chemicals on cellular circadian rhythms (Fang, Kang, Park, Estrella, & Zarbl, 2017).
To assess clock properties, oscillations, and disturbances, rhythmic expression/levels of core‐clock genes/proteins can be evaluated. For example, the oscillatory mRNA expression/protein levels of components of the PER/CRY loop and the REV‐ERB/Bmal1/ROR loop, such as Bmal1 and Per2, should be almost anti‐phasic (El‐Athman et al., 2017; Fuhr et al., 2015). In addition, as a control, the clock may be perturbed by the use of several compounds, including resveratrol and Ex‐527, which should result in loss of functional rhythmicity (du Pré et al., 2017). Such properties can be easily observed in individual neurons from SCN explants that mimic self‐sustained oscillators, although quite resistant to perturbations with little, if any, damping across several days (Yamaguchi et al., 2003). However, the amplitude of mRNA/protein cycles in peripheral cell and tissue cultures dampens rapidly after a few cycles (Nagoshi et al., 2004). This effect is not attributed to the loss of individual cellular rhythms but instead results from the loss of synchronization between cells. In the absence of synchronization cues, cells start to run independent and freely (Nagoshi et al., 2004). This poses a limitation to the study of peripheral clocks in vitro, but in vitro cellular clocks were demonstrated to be sensitive to a large range of input signals that allow for their transient synchronization. Thus, being able to synchronize clocks in vitro is of great interest for chronotherapeutics, as it may help defining the optimal timing for drug treatments (Levi & Schibler, 2007).
5.1.1. Methodologies to synchronize clocks in vitro
There are many published methodologies to synchronize clocks in vitro, ranging from cell treatment with systemic blood‐borne signals (Balsalobre et al., 1998; Gerber et al., 2013) or with different chemical compounds (dexamethasone, forskolin, growth factors, among others; Akashi & Nishida, 2000; Balsalobre et al., 2000; Yagita & Okamura, 2000), medium change (Hirota et al., 2002; Relógio et al., 2014), light pulses (Mazuski & Herzog, 2015), temperature shock (Brown, Zumbrunn, Fleury‐Olela, Preitner, & Schibler, 2002; Saini, Morf, Stratmann, Gos, & Schibler, 2012), and oxygen cycles (Adamovich, Ladeuix, Golik, Koeners, & Asher, 2017) to mechanical stimulation (Rogers, Fawcett, Pekovic‐Vaughan, & Hunt, 2017).
Table 1 presents different methodologies shown to induce clock synchronization in vitro. Among these, systemic blood‐borne signals, typically serum, are probably among the strongest zeitgebers for peripheral oscillators not only in vitro (Esnault et al., 2014; Gosselin, Rando, Fleury‐Olela, & Schibler, 2016) but also in vivo (Gerber et al., 2013). While for some of the synchronizing agents the respective pathways are well known, for others, the underlying mechanisms are not fully understood. Different synchronization methods may involve different molecular mechanisms, most likely due to different communication pathways. For example, while systemic blood‐borne signals were shown to modulate Per2 expression through the Rho‐actin–serum response factor–myocardin‐related transcription factor pathway, forskolin and dexamethasone‐bound glucocorticoid receptors also induce Per genes transcription but by promoting the activation of the PKA pathway and cAMP response element‐binding protein (CREB) phosphorylation (Gerber et al., 2013; Izumo et al., 2006; Yagita & Okamura, 2000). Other signalling pathways have also been implicated in in vitro cell synchronization, including PKC and MAPK either via transmembrane GPCRs or nuclear hormone receptors (Balsalobre et al., 1998; Balsalobre, Brown, et al., 2000; Yagita & Okamura, 2000). Furthermore, it is also incompletely understood whether clock synchronization in vitro jumpstarts dormant oscillations or synchronizes active but out of phase oscillations. Synchronization agents probably act by desynchronizing clocks first and then resynchronizing them, in a heterogeneous but consistent manner (Roberts et al., 2015).
Table 1.
Methodologies to synchronize circadian clocks in vitro, as described in the literature
| Synchronization methoda | Specificationsb | Final concentration | Treatment duration | In vitro model | Validation | References |
|---|---|---|---|---|---|---|
| Systemic blood‐borne signals | Serum shock | 50% | 2 hr | Rat‐1, rat fibroblasts cells H35, rat hepatoma cells | RNase protection assays—clock genes mRNA levels at defined time points | Balsalobre et al. (1998) |
| Addition of sera or plasma dialysed against PBS | 1%, 2% or 5% | N/A |
Rat‐1, rat fibroblasts cells U2OS, human bone osteosarcoma epithelial cells |
RT‐PCR—clock genes mRNA levels at defined time points Real‐time bioluminescence assay—clock genes‐driven expression along time |
Gerber et al. (2013) | |
| Cell culture medium change | Medium change to serum‐free medium | N/A | N/A | Rat‐1, rat fibroblasts cells | RT‐PCR—clock genes mRNA levels at defined time points | Hirota et al. (2002) |
| Medium change to regular cell culture medium | N/A | N/A | Rat‐1, rat fibroblasts cells | Real‐time bioluminescence assay—clock genes‐driven expression along time | Welsh, Yoo, Liu, Takahashi, and Kay (2004) | |
| Chemical compounds | Melatonin hormone | 1 nM | 1 hr | Rat SCN hypothalamic slices | Electrophysiological recording—neuronal activity | Mcarthur, Hunt, and Gillette (1997) |
| Dex glucocorticoid hormone analogue | 100 nM | 15 min–2 hr | Rat‐1, rat fibroblasts cells | RNase protection assays—clock genes mRNA levels at defined time points | Balsalobre, Brown, et al. (2000) and Balsalobre, Marcacci, and Schibler (2000) | |
| Forskolin diterpenoid | 10 μM | 15 min–2 hr | Rat‐1, rat fibroblasts cells |
‐ Northern blot analysis—clock genes mRNA levels at defined time points (Yagita & Okamura, 2000) ‐ Immunoblot—clock genes protein levels at defined time points (Yagita & Okamura, 2000) ‐ RNase protection assays—clock genes mRNA levels at defined time points (Balsalobre, Marcacci, & Schibler, 2000) |
Balsalobre, Marcacci, and Schibler (2000), Morgan and Williams (1996), and Yagita and Okamura (2000) | |
| PMA, also known as TPA diester of phorbol | 10 nM–1 μM | 15 min–2 hr |
‐ Rat‐1, rat fibroblasts cells (Balsalobre, Marcacci, & Schibler, 2000) ‐ NIH/3T3, mouse embryo fibroblast cells (Akashi & Nishida, 2000) |
‐ RNase protection assays—clock genes mRNA levels at defined time points (Balsalobre, Marcacci, & Schibler, 2000) ‐ RNase protection assays—clock genes mRNA levels (Akashi & Nishida, 2000) |
Akashi and Nishida (2000) and Balsalobre, Marcacci, and Schibler (2000) | |
| Calcimycin calcium ionophore | 1 μg·ml−1 | 15 min | Rat‐1, rat fibroblasts cells | RNase protection assays—clock genes mRNA levels at defined time points | Balsalobre, Marcacci, and Schibler (2000) | |
| Sp‐5,6‐DCl‐cBiMPS cAMP analogue | 100 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative PCR—clock genes mRNA levels at defined time points Bioluminescence assay—clock genes‐driven expression at defined time points | Balsalobre, Marcacci, and Schibler (2000) | |
| GABA inhibitory amino acid neurotransmitter | 100 μM | 1 hr | Mouse SCN‐dissociated neurons | Multielectrode plate recordings—neuronal activity | Liu and Reppert (2000) | |
| FGF | 25 ng·ml−1 | 70 min | NIH/3T3, mouse embryo fibroblast cells | RNase protection assays—clock genes mRNA levels | Akashi and Nishida (2000) | |
| EGF | 30 nM | 70 min | NIH/3T3, mouse embryo fibroblast cells | RNase protection assays—clock genes mRNA levels | ||
| PDGF | 30 ng·ml−1 | 70 min | NIH/3T3, mouse embryo fibroblast cells | RNase protection assays—clock genes mRNA levels | ||
| ET‐1 vasoconstrictor peptide | 30 nM | 2 hr | Rat‐1, rat fibroblasts cells |
‐ Northern blot analysis—clock genes mRNA levels at defined time points ‐ Immunocytochemistry—clock genes protein levels at defined time points |
Yagita, Tamanini, Van Der Horst, and Okamura (2001) | |
| Glucose sugar | 5.6 mM | During entire experiment | Rat‐1, rat fibroblasts cells | RT‐PCR—clock genes mRNA levels at defined time points | Hirota et al. (2002) | |
| Haem protein‐bound prosthetic group | 30 μM | 2 hr | NIH/3T3, mouse embryo fibroblast cells | Northern blot analysis—clock genes mRNA levels at defined time points | Kaasik and Lee (2004) | |
| Fenofibrate PPAR‐α activator | 50 μM | 2 hr | Rat‐1, rat fibroblasts cells | RT‐PCR—clock genes mRNA levels at defined time points | Canaple et al. (2006) | |
| 8‐Bromo‐cAMP analogue | 100 μM | 2 hr | Rat‐1, rat fibroblasts cells | Real‐time bioluminescence assay—clock genes‐driven expression along time | Izumo et al. (2006) | |
| B27 medium supplementation‐optimized serum‐free supplement | 2% | After medium change | Rat‐1, rat fibroblasts cells | Real‐time bioluminescence assay—clock genes‐driven expression along time | ||
| Ionomycin ionophore | 1 μM | 2 hr | Rat‐1, rat fibroblasts cells | Real‐time bioluminescence assay—clock genes‐driven expression along time | ||
| PGJ 2 bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | Nakahata, Akashi, Trcka, Yasuda, and Takumi (2006) | |
| Δ12‐PGJ 2 bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| 15d‐PGJ 2 bioactive lipid—natural ligand of PPAR‐γ | 1 or 10 μM | 1 hr |
‐ Rat‐1 fibroblast line ‐ NIH/3T3, mouse embryo fibroblast cells |
Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| Enantio‐PAF C16 bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| 1‐Acyl‐PAF bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| N‐Palmitoyl dopamine bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| N‐Arachidonoyl dopamine bioactive lipid | 1 or 10 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| 6‐Formylindolo[3,2‐b]carbazole peptide | 1 μM | 1 hr | Rat‐1, rat fibroblasts cells | Quantitative real‐time RT‐PCR—clock genes mRNA levels at defined time points | ||
| TNF‐α proinflammatory cytokine | 50 ng·ml−1 | 2 hr | Human synovial fibroblasts primary cultures | Quantitative real‐time PCR—clock genes mRNA levels at defined time points | Becker et al. (2014) | |
| Temperature | Square waves temperature cycles 12 hr at 33°C and 12 hr at 37°C | N/A | 6 days | Rat‐1, rat fibroblasts cells | RNase protection, real‐time PCR, and northern analyses—clock genes mRNA levels at defined time points | Brown et al. (2002) |
| Oxygen | Oxygen cycles 12 hr 5% O2 and 12 hr 8% O2 (mimicking physiological O2 rhythms) | N/A | 3 days and subsequently released to constant 8% O2 (free running) |
‐ NIH/3T3, mouse embryo fibroblast cells ‐ Hepa‐1c1c7, mouse liver hepatoma cells |
‐ RT‐PCR—clock genes mRNA levels at defined time points ‐ Immunoblot—clock genes protein levels at defined time points |
Adamovich et al. (2017) |
| Mechanical stimulation | Flexible silicone chambers coated in fibronectin for 1 hr before cells were seeded into the chambers. Once confluent, the chambers were loaded into a unique uniaxial stretch rig and stretched for three consecutive days in a rhythmic manner (frequency 1 Hz, 6.66% stretch, 12‐hr on/12‐hr off) | N/A | After 3 days, cells were allowed to rest for 16 hr |
‐ Primary human postnatal human DPSCs ‐ Primary human BMSCs |
Quantitative RT‐qPCR—clock genes mRNA levels at defined time points | Rogers et al. (2017) |
The methods described here were suggested by each author as in vitro synchronizing agents, based on the induction of circadian clock mRNA/protein oscillations.
Cells were grown to confluence, treated for the indicated times with the indicated substances, and kept in serum‐free or serum‐containing medium thereafter to record circadian gene expression.
Abbreviations: 15d‐PGJ2, 15‐deoxy‐Δ12,14‐prostaglandin J2; BMSCs, bone marrow stromal cells; Dex, dexamethasone; DPSCs, dental pulp stem cells; ET‐1, endothelin 1; N/A, not applicable; PMA, phorbol 12‐myristate 13‐acetate; TPA, 12‐O‐tetradecanoylphorbol‐13‐acetate.
In 2006, Izumo et al. compared rhythm‐inducing properties of 10 synchronizing treatments, including drugs, growth factors, and serum, in rat‐1 fibroblasts. The authors compared the presence and significance level of rhythmicity and the amplitude of the resulting oscillations, through a computational model. Among the 10 drug/hormone treatments, the applied model identified 50% horse serum, dexamethasone, forskolin, and EGF as being the most effective in generating high‐amplitude rhythms in the selected in vitro model (Izumo et al., 2006). However, the efficacy of synchronizing agents may vary according to the cellular model used. As an example, bone marrow‐derived mesenchymal stem cells appear to synchronize more readily in response to chemical than to mechanical stimulation, whereas dental pulp‐derived mesenchymal‐like stem cells were shown to be more responsive to entrainment by mechanical means (Rogers et al., 2017). Thus, the most appropriate method must be defined based on the cellular model (maturity, presence/absence of specific receptors/proteins, and cell response) and the aim of the study (it should not affect targets of the study). Independently of the synchronization method, to quantify the clock properties of in vitro cultures, measurements should only start one cycle (~24 hr) after cessation of the synchronizing stimulus, in order to minimize the efffects of immediate early gene expression. Indeed, synchronization methods induce a transient burst followed by a decay in the expression of clock genes within the first 24 hr of treatment that might erroneously be interpreted as part of the circadian cycle (Hughes et al., 2017).
5.1.2. Methodologies to evaluate in vitro circadian oscillations
Most studies focusing on the evaluation of circadian oscillations have relied on serially sampling cells/tissues to examine circadian variation in mRNA/protein levels. Sample collection is recommended to be performed every 2 hr (Hughes et al., 2017). If the constraints of the experimental system do not allow this frequency, additional efforts should be made in follow‐up experiments to validate any findings (Hughes et al., 2017). This recommendation is based on down‐sampling simulations of real data and on simulations using synthetic data (Hughes et al., 2017; Hughes, Hogenesch, & Kornacker, 2010). Fluorescence and bioluminescence measurements are amongst the most commonly used techniques to assess mRNA/protein levels over several days. Circadian promoters that drive the expression of fluorescent‐ or enzyme‐based reporter genes allow rapid and high‐resolution quantification of circadian rhythms in cultured cells. With this approach, it is possible to know where, when, and how much a gene or protein is expressed. Expression cassettes, in which a circadian promoter is fused with the reporter gene, can be introduced into cells via transient transfection (Ueda et al., 2005) or stable transduction (Zhang et al., 2009). Stable transduction is known to be highly efficient and versatile as it permits efficient delivery and stable integration into the host genome of both dividing and non‐dividing cells (Tiscornia, Singer, & Verma, 2006). Alternatively, a reporter cassette harbouring a reporter gene can be introduced by knock‐in, via homologous recombination, into the resident gene. Such an approach constitutes the most reliable method because all the regulatory elements are conserved in this assay (Yoo et al., 2004). Once a reporter cell line is established, the dynamics of clock oscillations can be examined through optical imaging or bioluminescence recording. A potential limitation in real‐time monitoring of either enzymic or fluorescent circadian reporters is that circadian promoters can have low activity, leading to extremely low signals. Recording fluorescence or bioluminescence for circadian studies requires a stable environment with no environmental light contamination or temperature fluctuations (Beaulé, Granados‐Fuentes, Marpegan, & Herzog, 2011). Again, the choice of the methodology is dependent on the characteristics of the experiment and the desired spatial and temporal resolutions.
Fragments of the promoters of the clock genes Per1 and Per2 driving the expression of variants of fluorescent reporters, such as the GFP, have been commonly used (LeSauter et al., 2003). Fluorescent reporters allow to identify the cells in which a specific gene is being expressed, as well as the cellular location of proteins. This method can be combined with optical imaging and electrophysiological studies, allowing the simultaneous monitoring of multiple genes (e.g., Per1 and Per2), by using distinct fluorophores that can be discriminated with optical filters (Beaulé et al., 2011). However, the fluorescence background and the increased half‐life of fluorescent reporters decrease the sensitivity of the method. This can be overcome by integrating degradation sequences in the reporter proteins that recruit the cellular proteolysis machinery, thus causing reporters to be quickly degraded and allowing a more faithful tracking of gene expression changes (Li et al., 1998). Still, photobleaching and phototoxicity constitute significant problems of real‐time imaging, as reporters need to be repeatedly exposed to excitation light for the pattern of expression over time to be observed (White & Stelzer, 1999).
Bioluminescent reporter genes, although dim, are highly quantitative and suitable to quantify the expression and peak phase of a particular gene or protein. In the presence of an adequate substrate and co‐factors, bioluminescent reporters catalyse a chemical reaction that results in the emission of visible light that can be easily quantified directly from cells (Ramanathan, Khan, Kathale, Xu, & Liu, 2012). Bioluminescent reporters have usually lower backgrounds and shorter half‐lives than fluorescent ones, thus being generally more sensitive and less toxic (Welsh & Noguchi, 2012), which makes them more suitable for circadian studies. Although a variety of reporter genes has been described, the high sensitivity, excellent dynamic range, and low toxicity have made luciferase (luc) the most commonly used bioluminescent reporter for the analysis of clock gene expression (Welsh, Imaizumi, & Kay, 2005). Glow‐type luciferases generate stable long‐lasting bioluminescence, being more suitable for long‐term recording of circadian rhythms. Depending on the species origin and chemical conditions, luciferases can also exhibit different bioluminescence colours, which allows tracking the expression patterns of more than one clock gene at the same time (Noguchi, Ikeda, Ohmiya, & Nakajima, 2012). Luc fused to Bmal1 and Per1/2 promoters is commonly used as a readout of circadian oscillations in a variety of cell lines (Relógio et al., 2014; Zhang et al., 2009). In addition, circadian clock oscillations can also be traced at the protein level by collecting cells and tissue explants from luc knock‐in mice in which a clock gene–luc fusion protein is expressed through activation of a clock gene promoter (Mohawk, Green, & Takahashi, 2012). Technological advances have allowed longitudinal measurements in real‐time living cells with a higher temporal resolution, allowing assessment of the persistence and dynamics of molecular rhythms. Automated luminometer devices with photomultiplier tubes, as LumiCycle, are commonly used for real‐time recording (Fuhr et al., 2018). These systems employ photomultiplier tubes as light detectors, providing extremely high sensitivity and low‐noise data, particularly suitable for data acquisition of extremely dim luc‐based bioluminescence (Ramanathan et al., 2012). In addition, besides quantity and temporal information, the addition of highly sensitive, low‐noise CCD cameras has also allowed the acquisition of spatial information (Welsh & Noguchi, 2012). Still, the spatial resolution of bioluminescent images is lower than that of fluorescent images.
The study of circadian changes in the electrical activity of neurons, in vitro, provides another strategy to chase circadian rhythms, here based on firing rates (Belle, Diekman, Forger, & Piggins, 2009; Kuhlman & McMahon, 2006). Electrophysiology can reveal circadian oscillations with single‐cell resolution in tissue explants or cell dispersals shortly after removal from the animal or after weeks to months in culture (Beaulé et al., 2011). Intracellular patch or voltage clamp can record single neurons for up to 1 hr, while extracellular approaches allow to record single to multi‐unit action potentials for up to 30 days. Multielectrode arrays allow recording large numbers of individual neurons (Welsh, Logothetis, Meister, & Reppert, 1995).
Independently of the technique chosen, it is important to collect data from at least two complete circadian cycles to detect rhythmicity (i.e., 48 hr under constant conditions). Simulation studies show that collecting fewer than 2 cycles in a time series increases the likelihood of detecting outliers, which affects the number of false‐negative results (Hughes et al., 2017).
The progress made in cell culture synchronization methods and approaches to evaluate circadian clock properties and disturbances allows the strict control of experimental conditions and the reduction of a number of variables. This has enabled experimental research on the effects of circadian clocks on intracellular pathways involved in drug response, and, in turn, on the effects of drugs on circadian clocks. For example, El‐Athman et al. have recently shown that clock dysregulation in human colorectal carcinoma cell lines derived from primary and metastatic sites of the same patient lead to reprogramming of the circadian behaviour of ABCC1 expression. ABCC1 encodes the membrane transporter multidrug resistance‐associated protein 1 (MRP1), and different splice variants of the gene have been found to confer a drug‐resistant phenotype to ovarian cancer cells (El‐Athman et al., 2018). ABCC1 showed phase‐shifted circadian oscillations in both cell lines at the transcriptome level, and circadian oscillations in its splicing pattern were lost in the metastatic cell line (El‐Athman et al., 2018). Their findings on the circadian regulation of ABCC1 strengthen the relevance of the gene as a promising drug target in cancer and highlight the potential of circadian clock modulation for therapy optimization (El‐Athman et al., 2018).
5.2. Evaluation of circadian rhythms in vivo
Although flies constitute the best studied system in chronobiology, in this review, we focus on mammals, and particularly on rodents, due to their central importance in pharmacological studies. In this regard, mouse models are the best studied systems in terms of circadian rhythm monitoring (Bains et al., 2018; Eckel‐Mahan & Sassone‐Corsi, 2015). Accordingly, this section provides an overview of available mouse models to investigate the circadian system and methodologies to evaluate circadian rhythms in vivo.
5.2.1. Mouse models to study circadian rhythms (dys)regulation
A variety of mouse models displaying circadian alterations has been generated over the past years. Mice depict robust circadian rhythms, which are preserved under constant darkness conditions (dark–dark [DD]; Oliverio & Malorni, 1979). However, the knockout or loss of function of core‐clock genes such as Bmal1, Clock, Per (Per1 −/− Per2 −/−), or Cry (Cry1 −/− Cry2 −/−) induces arrhythmicity under DD conditions (see Husse, Eichele, & Oster, 2015). Interestingly, these mice can partially recover circadian behaviour and physiology rhythms when maintained under 24‐hr light–dark (LD) cycles probably due to the effect of light as a strong zeitgeber. Although these models may present some changes in the amplitude and period of clock genes, published data point to a readjustment of the clock to substitute the missing clock element (see Husse et al., 2015). These mouse models are relevant to study the specific effect of each clock gene on overall physiology. Indeed, several studies have shown correlations with predisposition for cancer (Gery & Koeffler, 2007), mood disorders (Roybal et al., 2007), and metabolic disorders (Turek et al., 2005) in these mouse models.
Additional in vivo models include mice with lesions on the central clock used to study the mechanisms of circadian rhythm generation, namely, the interactions between central and peripheral clocks (see Husse et al., 2015). Complete bilateral lesion of the SCN abolishes rhythmicity of locomotor activity, drinking behaviour, body temperature, and hormone release (Refinetti, Kaufman, & Menaker, 1994; Stephan & Zucker, 1972). These observations are accompanied by an abolishment of clock gene rhythmicity in tissues (Sakamoto et al., 1998). In lesioned SCN models, Tahara et al. (2012) found that peripheral clocks maintain some rhythmicity for a short period of time without a functioning SCN or external cues. In free running, restricted food access and LD conditions, their peripheral clocks (in the liver, kidney, and submandibular gland) maintain a period of oscillations of about 24 hr. However, when kept in an ad libitum regimen under DD conditions, the amplitude of the oscillations dampens with time, supporting the idea that the SCN is in fact necessary for the regulation of peripheral rhythms, in the absence of external zeitgebers (food and light; Saini et al., 2013; Tahara et al., 2012). In this context, mice with lesions in the central clock constitute a relevant model to dissect central and peripheral clocks and to study the specific effect of drugs on peripheral clocks without external zeitgebers. Animal models allow the separation of several mechanisms involved in the circadian regulation at the organismal level, which allows for a clearer perspective on the effects of alterations of circadian rhythms in health and treatment outcomes.
5.2.2. Evaluation of circadian rhythms in mouse models
In mouse models, several parameters can be used to assess circadian rhythms, including temperature, cortisol, and melatonin. However, their measurement can be affected by several external factors, which may lead to confounding results.
Currently, the most straightforward in vivo approach to study circadian‐derived behaviour is to measure physical activity. As the majority of rodents are nocturnal, their activity is increased during the night. Thus, studying behaviour or activity of an animal under different conditions, namely, under different light–dark cycles, can provide important insights regarding the circadian phenotypes (Eckel‐Mahan & Sassone‐Corsi, 2015). Considering photic entrainment as the main synchronizer of the central and peripheral clocks, subjecting mice to DD conditions reveals their endogenous circadian period. Accordingly, most in vivo studies with mice, aiming to investigate circadian rhythms, are performed under constant darkness (Eckel‐Mahan & Sassone‐Corsi, 2015).
Mouse activity is typically monitored by voluntary wheel running, a technique that dates back to the 19th century (Stewart, 1898). The measurements of activity on running wheels provide direct access to mice activity and allow to infer certain circadian parameters, such as the free running period (usually monitored in DD conditions) and the phase of entrainment (comparing the phase of activity relative to the LD cycles; Bains et al., 2018; Eckel‐Mahan & Sassone‐Corsi, 2015). These parameters can be correlated with circadian rhythm mis‐alignments (Bains et al., 2016) and have been used to explain drug effects on the circadian system (Kitanaka et al., 2012). Despite the beauty of this system reflecting its simplicity, mouse behaviour is dynamic and can be influenced by a wide range of variables, from genetics to motivation or environmental factors (Bains et al., 2018). In addition or as an alternative to wheel running monitoring, home‐caging video systems have also been used to characterize circadian parameters (e.g., ANYmaze or COMPASS; Bains et al., 2018). These systems allow the observation of additional parameters that may also be correlated with circadian rhythms, such as anxiety‐like behaviour, feeding patterns, and sleep. Moreover, video systems facilitate the investigation of the treatment effect on behaviour along the course of several weeks or months, which allows discrimination between observations in behaviour that may occur either sporadically, as a side effect of the treatment, or frequently, as a direct and more profound effect of the treatment on circadian rhythms and behaviour. However, such systems are rather complex and require an unbiased analysis from the researcher.
Besides behaviour analysis, circadian rhythms can also be monitored by evaluating circadian expression of clock genes or proteins. Bioluminescence approaches are the gold standard for measuring in vivo circadian expression of genes or proteins. The production of a transgenic mice carrying a luc reporter under the control of a clock gene promoter was first applied to ex vivo cultures of mice tissues (Yamaguchi et al., 2000). Soon after, it started to be applied in living mice for circadian rhythm monitoring (Yamaguchi et al., 2001). In vivo bioluminescence recording was used for the first time in SCN monitorization, and mPer1–luc mice were implanted with an optical fibre above the SCN, connected to a photon‐counting device equipped with a photo multiplier tube. Luciferin, the substrate of luc, was then delivered through continuous injection to the third ventricle (Yamaguchi et al., 2001). With this approach, the authors were able to record bioluminescence parallel to Per1 activation, which allowed for the quantification of circadian fluctuations directly from the SCN (Yamaguchi et al., 2001). This technique was then used by many other authors that further improved luciferin delivery, using osmotic pumps (Yamaguchi et al., 2016). More recently, researchers have also developed double transgenic mice (Per1–luc and Bmal1–Eluc), taking advantage of two different types of luc that produce light of different wavelengths (Noguchi et al., 2012). Through bioluminescence approaches, in vivo monitoring of circadian rhythms in specific organs is also possible. In 2012, Tahara and colleagues crossed Per2–luc mice with an albino strain (ICR) and introduced an osmotic mini pump in the interscapular region, to deliver luciferin. Mice were then anaesthetized and placed in a black box, and bioluminescence was captured for each organ (the kidney, liver, and submandibular gland; Tahara et al., 2012). Also in 2013, Saini and colleagues were able to perform in vivo real‐time recording in the liver of awake mice (Saini et al., 2013). Since then, these and similar models have been used to study the effects of diseases, such as diabetes (Hou, Su, Guo, & Gong, 2018), and drugs, like glucocorticoids (Kamagata et al., 2017), on circadian rhythm fluctuations in peripheral organs. The main disadvantages of this type of approach are the need for specific and expensive equipment and the fact that it requires conditions of total darkness for bioluminescence readings. Recently, a novel approach to avoid the need of constant darkness was proposed. By using a Cry1 promoter, Mei et al. (2018) drove the expression of the fluorescent protein Venus. In this study, the authors delivered the fluorescent reporter as a viral vector directly to the SCN, and following injection, an optical fibre was inserted in the same location, covered in black nail polish (to protect from external light) and connected to a homemade recording system to measure fluorescence spikes. Readouts were similar to bioluminescence assays, showing rhythmic expression of Cry, shown by a rhythmic fluorescence pattern (Mei et al., 2018). This technique presents advantages over previous bioluminescence approaches by not requiring an osmotic pump to deliver the substrate, for overcoming the need for surgery and its associated variability, and for allowing experiments to be carried out in normal LD conditions, under normal physiological conditions of light and dark. This model is thus very promising for translational studies of drugs and other treatments.
The approaches described not only allow the evaluation of the effects of specific drugs on circadian rhythms, but they also enable the exploration of the circadian rhythmicity of genes that may be targeted by specific drugs, by adapting the system to their target promoters. Mouse models have indeed been crucial for the current knowledge of circadian biology and still have great potential for preclinical trials and to enhance the design of clinical trials. However, mice are nocturnal animals, and as a consequence, their rhythms differ substantially from those of humans. Moreover, knowing that each individual chronotype can affect treatment response (Innominato et al., 2018), there is a need to find approaches that can be used to accurately measure circadian rhythms in humans.
5.3. Monitoring of circadian rhythms in humans
Several studies have correlated circadian mis‐alignments, for example, in shift workers, with diseases, such as cancer (Haus & Smolensky, 2013) and metabolic disorders (Lin, Hsiao, & Chen, 2009). These observations, together with the fact that chronotype has a significant effect on treatment outcome, lead to an urgent need to find better procedures to evaluate circadian rhythms in humans. Chronotypes could be used to distinguish responders and non‐responders to drug treatments and therapies. Once the individual intrinsic circadian variations are understood and defined, this type of approach may be useful in establishing doses and defining the optimum timing for drug administration. Current methods to evaluate these parameters in humans open new avenues for personalized medicine. In this context, circadian rhythm monitoring in humans is highly relevant to improvements in therapeutic approaches, under a personalized healthcare perspective.
Circadian rhythms in humans can be assessed in biological samples, such as saliva, hair, and blood, collected at several time points along the day, in order to evaluate circadian gene and/or protein expression. For example, studies in hair follicle cells showed that rotating shift workers suffer from a serious time lag between circadian gene expression rhythms and lifestyle (Akashi et al., 2010; Watanabe et al., 2012).
In addition, there are other markers that can be easily measured and that can reflect circadian phases as well, such as the core body temperature, melatonin, and cortisol secretion (reviewed in Evans & Davidson, 2013). Several recent studies have selected body temperature as a determinant parameter for circadian rhythm evaluation (Garrido et al., 2017; Martinez‐Nicolas et al., 2018). Body temperature can be easily measured through wristbands, usually in the non‐dominant arm. Cortisol can be measured at home by collecting saliva at the awakening time. However, collection of samples within 15 min of awakening is necessary for the sample to be translatable and correlated with circadian rhythm activity onset. Nonetheless, this can only be achieved if complemented with electronic activity monitoring (Smyth, Thorn, Hucklebridge, Clow, & Evans, 2016). Similar in‐home salivary measurements can be adapted to measure melatonin at awakening, a couple of hours before bedtime and at bedtime (Keijzer et al., 2011).
Sleep–wake cycles have also been assessed to establish patient chronotypes. These can easily be assessed through patient‐reported outcome measures, but such questionnaires are rather subjective. Wristbands are currently the most common device used for measuring activity in humans and have been validated as an objective biomarker of circadian function (Ancoli‐Israel et al., 2003; Innominato et al., 2018). Other wearable biosensors have also been correlated with the circadian rhythms of patients (Innominato et al., 2018). For example, a recent study studied the circadian rhythms in individuals of different ages and measured several parameters such as distal skin temperature, activity, body position, light exposure, and environmental temperature, using various wearable biosensors (Martinez‐Nicolas et al., 2018). Recordings allowed the calculation of a coefficient (temperature activity body position) that is highly predictive of the age of the individual (Martinez‐Nicolas et al., 2018). These parameters correlate with circadian activity and can be measured in a non‐invasive manner.
Internal time can, however, be influenced by many factors, such as age, environmental light levels, or season, which may cause difficulties in classifying chronotypes. Thus, efforts have been made in order to more objectively assess internal time. Omics approaches and sophisticated computational and bioinformatics methodologies are becoming relevant in circumventing such limitations.
6. COMPUTATIONAL APPROACHES TO INVESTIGATE CIRCADIAN RHYTHMS
To investigate and characterize in detail the circadian system, it is essential to carry out dynamic measurements of gene and protein expression, at an omics level, across at least 24 hr. Most circadian studies are based on transcriptomics data, and only a reduced set uses proteomics data sets (Wang et al., 2017; Figure 2, left panels). Sequencing and array technologies allow quantification of the expression of transcripts in regular intervals over the course of 1 day or longer. The analysis of circadian rhythmicity can be carried out using algorithms like the detection of differential rhythmicity method available in R‐packages, which detects altered rhythmicity by quantifying phase shifts and amplitude changes in circadian data sets (Thaben & Westermark, 2016). These tools can be used to characterize temporal changes in biological samples (using cell lines and animal models), which via further functional analysis can then be linked to specific cellular decisions (Ruben et al., 2018; Zhang et al., 2014).
Figure 2.
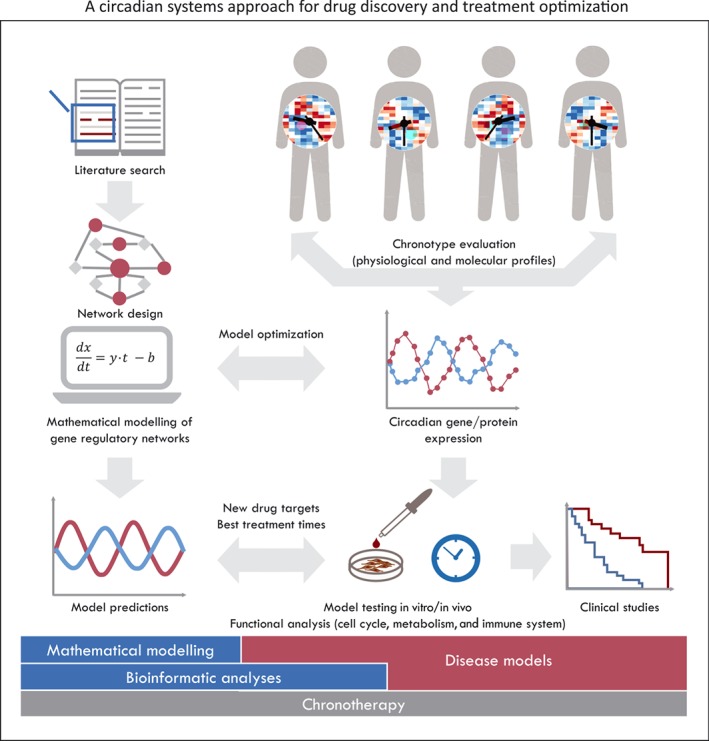
A circadian systems approach for drug discovery and treatment optimization. Circadian regulatory networks can be assembled via iterative steps including automatic literature search (text mining) to determine interactions between predefined elements of the network and de novo interaction discovery, based on the analysis of transcription‐binding sites of clock‐controlled genes. The subsequent co‐expression analysis of elements in the network and manual curation are used to establish the type of existing interactions (Lehmann et al., 2015). The use of patient‐derived expression data (RNA or protein), who have been stratified for clinical trials based on their chronotype, is needed to refine the network and to model different biological scenarios. Sophisticated computational and bioinformatics methodologies are required to analyse and integrate the large sets of data. Ultimately, particular elements of the regulatory networks can be identified for potential use as drug targets or disease markers. Computational models can be further used to establish chronomodulated treatment schedules, which allow for an improvement of treatment outcome and a reduction of toxic effects
El‐Athman et al. reported a circadian fine tuning of cell cycle decisions by using high‐throughput bioinformatics analysis of regulatory networks. Accurate predictions of the effects of the clock on cell cycle decisions were obtained using machine learning approaches and validated experimentally (El‐Athman et al., 2017). Also, metabolic alterations resulting from a dysregulated clock were detected with a computational/experimental high‐throughput approach using a human colon cancer model. The authors identified well‐known metabolic genes to be under circadian control and further expanded their findings to the analysis of metabolic alterations due to the action of different drugs in a time‐dependent manner (Fuhr et al., 2018). Another large‐scale study, which characterized the variation in expression of clock genes across 32 cancer types, demonstrated that alterations of core‐clock genes are associated with key oncogenic pathways and tumour stage and correlate with patient survival (Ye et al., 2018). Though this study was based on expression data from one single time point, the authors combined several types of omics data to identify interactions between clock genes and clinically actionable genes and demonstrated a correlation between altered clock expression, drug sensitivity, and patient survival.
One major drawback of circadian investigation is the need of time course data that can be easily obtained from cellular models but which is more difficult to acquire at the organismal level. There are only a few existing data sets for mammals across tissues (Mure et al., 2018; Zhang et al., 2014), and it becomes even more challenging when dealing with patient samples, which besides being costly and labour intensive, are cumbersome to the patient. To circumvent these problems, in 2016, Hughey et al. developed a machine learning‐based computational tool to determine the subject's circadian time from blood collected at a single time point—the ZeitZeiger algorithm (Hughey, Hastie, & Butte, 2016). While the authors used the most comprehensive multi‐organ circadian data set available at the time (Zhang et al., 2014) as the training set, the method can be applied to any data set and has recently been used in the analysis of the core clock in cancer models (El‐Athman et al., 2018). Similar models have further been developed such as BodyTime or TimeSignature (Braun et al., 2018; Wittenbrink et al., 2018). These methods can be used to determine the chronotype of patients and can aid in both stratifying patients and optimizing treatment times based on circadian clock phenotypes (Ballesta et al., 2017; El‐Athman & Relógio, 2018; Zhang et al., 2014). Pioneering studies by the group of Francis Lévi point to both a decrease of toxic effects and an increase in survival when delivering chemotherapy to cancer patients, in a time‐modulated manner. These studies highlight the promising role of the circadian clock in therapy and pave the way for the integration of chronotherapy into clinical practice (Ballesta et al., 2017).
To better determine treatment windows and to establish personalized times for treatment regimens, mathematical models of the circadian clock can also be used. Particular parts of the regulatory networks discussed above can be simulated in greater detail using ordinary differential equations mathematical models (Karlebach & Shamir, 2008). Similar to the large regulatory networks, also for ordinary differential equation models time series, measurements of transcripts and protein expression can be used to fit the model to particular biological scenarios. A well‐known and widely used mathematical model of the circadian clock was developed by the Relógio group and was successfully used to investigate the influence of the Ras oncogene on the circadian clock (Relógio et al., 2014), and the coupling of the circadian clock to the cell cycle, pointing to a significant cross‐talk between both systems (El‐Athman et al., 2017). The authors further showed that the timing at which perturbations (e.g., caused by therapeutic agents) are applied to the system leads to differential effects, which ultimately regulate cell cycle decisions. Analogous computational models can be used to determine the time‐dependent effect of drug administration and also to estimate optimal times for administering treatments, according to the specific clock of the patient (Dulong, Ballesta, Okyar, & Levi, 2015). As already described for cancer treatment, the characterization of the chronotype of the patient is likely to increase the outcome of the treatment in many other pathologies linked to circadian function, including heart diseases, sleep and metabolic disorders, inflammation, and asthma (Bass & Lazar, 2016).
Likewise, when designing clinical trials, the chronotype is a relevant additional factor to take into account in the stratification of patients. This might shed light into the sometimes variable treatment outcomes for patients categorized within the same test group but potentially having different chronotypes. Furthermore, the re‐establishment of circadian rhythms using low MW compounds can also be used to enhance therapeutic effects (El‐Athman & Relógio, 2018; Gloston, Yoo, & Chen, 2017).
However there is still a long way to go until the considerations of the patient's chronotype for the establishment of personalized treatments become daily practice in the clinic (Figure 2, right panels). Until then, more in vitro, in vivo, and clinical studies and more powerful computational tools that enable the integration of omics data in a time‐dependent manner, including data mining algorithms, machine learning‐based approaches, and dynamic mathematical modelling of gene regulatory networks, are necessary (Hughey, 2017; Lehmann et al., 2015). Such combinations of computational and experimental approaches will allow for a full integration of circadian data and will ultimately enable the generation of predictive models for time‐dependent treatment with major benefits for patients.
7. CONCLUSIONS
Over the past years, circadian regulation has been linked to human health and disease, as shown by its cross‐talk with fundamental processes such as metabolism and stress responses. If, on one hand, specific drugs may interfere with circadian regulation, on the other hand, the circadian system has also profound effects on drug PK and PD. This body of evidence should motivate the exploitation of circadian (dys)regulation on pharmacological studies and in the clinic, for improved treatment protocols. Physiological time becomes especially relevant when designing and dosing drugs that show a rapid excretion, a narrow therapeutic range, or that target rhythmic processes. For that, as described above, there is a multitude of methodologies which can be implemented in pharmacological studies in vitro, in vivo, and in silico. Although standardized and objective methodologies to assess physiological time in humans and the chronotype of each patient are still needed, efforts have been made that pave the way for such personalization of medical treatments. More powerful computational tools will be fundamental to more accurately define and analyse the molecular clockwork.
In conclusion, this multidisciplinary area of chronopharmacology should promote a new era of drug development, which tailors medication to a single patient, a particular pathology, and a specific drug for safer and more effective treatments. To achieve this, it is fundamental to alert not only the scientific, medical, and regulatory communities but also undergraduate and graduate students to the relevance of the circadian clock system. This will contribute to better designed and more successful preclinical and clinical trials, with a more general goal of successful personalized medicine.
7.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander, Christopoulos et al., 2017; Alexander, Cidlowski et al., 2017; Alexander, Fabbro et al., 2017; Alexander, Kelly et al., 2017).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
Work in C.C.'s laboratory is supported by the European Regional Development Fund (ERDF), through INOVC 2020 and Centro 2020 Regional Operational Programme, under the project CENTRO‐01‐0246‐FEDER‐000017, and through the Operational Programme for Competitiveness and Internationalisation—COMPETE 2020 and Portuguese national funds via FCT—Fundação para a Ciência e a Tecnologia, under the projects POCI‐01‐0145‐FEDER‐029002, POCI‐01‐0145‐FEDER‐007440 and UID/NEU/04539/2019; and by the European Social Fund through POCH ‐ Human Capital Operational Programme and Portuguese national funds via FCT under PD/BD/135497/2018; work in A.R.'s laboratory is supported by the German Federal Ministry of Education and Research (BMBF)—eBio‐CIRSPLICE—FKZ031A316 and by the Dr Rolf M. Schwiete Stiftung.
Gaspar L, Álvaro AR, Carmo‐Silva S, Mendes AF, Relógio A, Cavadas C. The importance of determining circadian parameters in pharmacological studies. Br J Pharmacol. 2019;176:2827–2847. 10.1111/bph.14712
REFERENCES
- Adamovich, Y. , Ladeuix, B. , Golik, M. , Koeners, M. P. , & Asher, G. (2017). Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metabolism, 25, 93–101. 10.1016/j.cmet.2016.09.014 [DOI] [PubMed] [Google Scholar]
- Akashi, M. , & Nishida, E. (2000). Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes & Development, 14, 645–649. [PMC free article] [PubMed] [Google Scholar]
- Akashi, M. , Soma, H. , Yamamoto, T. , Tsugitomi, A. , Yamashita, S. , Yamamoto, T. , … Node, K. (2010). Noninvasive method for assessing the human circadian clock using hair follicle cells. Proceedings of the National Academy of Sciences, 107, 15643–15648. 10.1073/pnas.1003878107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht, U. (2012). Timing to perfection: The biology of central and peripheral circadian clocks. Neuron, 74, 246–260. 10.1016/j.neuron.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Marrion, N. V. , Peters, J. A. , … CGTP Collaborators . (2017). The Concise Guide to PHARMACOLOGY 2017/18: G protein‐coupled receptors. British Journal of Pharmacology, 174, S17–S129. 10.1111/bph.13878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Cidlowski, J. A. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators . (2017). The Concise Guide to PHARMACOLOGY 2017/18: Nuclear hormone receptors. British Journal of Pharmacology, 174, S208–S224. 10.1111/bph.13880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators . (2017). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. British Journal of Pharmacology, 174, S272–S359. 10.1111/bph.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , Harding, S. D. , … CGTP Collaborators . (2017). The Concise Guide to PHARMACOLOGY 2017/18: Transporters. British Journal of Pharmacology, 174, S360–S446. 10.1111/bph.13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancoli‐Israel, S. , Cole, R. , Alessi, C. , Chambers, M. , Moorcroft, W. , & Pollak, C. P. (2003). The role of actigraphy in the study of sleep and circadian rhythms. Sleep, 26, 342–392. [DOI] [PubMed] [Google Scholar]
- Aryal, R. P. , Kwak, P. B. , Tamayo, A. G. , Gebert, M. , Chiu, P. L. , Walz, T. , & Weitz, C. J. (2017). Macromolecular assemblies of the mammalian circadian clock. Molecular Cell, 67, 770–782. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher, G. , Reinke, H. , Altmeyer, M. , Gutierrez‐Arcelus, M. , Hottiger, M. O. , & Schibler, U. (2010). Poly(ADP‐ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell, 142, 943–953. [DOI] [PubMed] [Google Scholar]
- Asher, G. , & Schibler, U. (2011). Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metabolism, 13, 125–137. 10.1016/j.cmet.2011.01.006 [DOI] [PubMed] [Google Scholar]
- Bains, R. S. , Cater, H. L. , Sillito, R. R. , Chartsias, A. , Sneddon, D. , Concas, D. , … Armstrong, J. D. (2016). Analysis of individual mouse activity in group housed animals of different inbred strains using a novel automated home cage analysis system. Frontiers in Behavioral Neuroscience, 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains, R. S. , Wells, S. , Sillito, R. R. , Armstrong, J. D. , Cater, H. L. , Banks, G. , & Nolan, P. M. (2018). Assessing mouse behaviour throughout the light/dark cycle using automated in‐cage analysis tools. Journal of Neuroscience Methods, 300, 37–47. 10.1016/j.jneumeth.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta, A. , Innominato, P. F. , Dallmann, R. , Rand, D. A. , & Lévi, F. A. (2017). Systems chronotherapeutics. Pharmacological Reviews, 69, 161–199. 10.1124/pr.116.013441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre, A. , Brown, S. A. , Marcacci, L. , Tronche, F. , Kellendonk, C. , Reichardt, H. M. , … Schibler, U. (2000). Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science, 289, 2344–2347. [DOI] [PubMed] [Google Scholar]
- Balsalobre, A. , Damiola, F. , & Schibler, U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell, 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Balsalobre, A. , Marcacci, L. , & Schibler, U. (2000). Multiple signaling pathways elicit circadian gene expression in cultured rat‐1 fibroblasts. Current Biology, 10, 1291–1294. 10.1016/S0960-9822(00)00758-2 [DOI] [PubMed] [Google Scholar]
- Baraldo, M. (2008). The influence of circadian rhythms on the kinetics of drugs in humans. Expert Opinion on Drug Metabolism & Toxicology, 4, 175–192. [DOI] [PubMed] [Google Scholar]
- Bass, J. , & Lazar, M. A. (2016). Circadian time signatures of fitness and disease. Science, 354, 994–999. 10.1126/science.aah4965 [DOI] [PubMed] [Google Scholar]
- Beaulé, C. , Granados‐Fuentes, D. , Marpegan, L. , & Herzog, E. D. (2011). In vitro circadian rhythms: Imaging and electrophysiology. Essays in Biochemistry, 49, 103–117. 10.1042/bse0490103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T. , Tohidast‐Akrad, M. , Humpeler, S. , Gerlag, D. M. , Kiener, H. P. , Zenz, P. , … Ekmekcioglu, C. (2014). Clock gene expression in different synovial cells of patients with rheumatoid arthritis and osteoarthritis. Acta Histochemica, 116, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Belle, M. D. C. , Diekman, C. O. , Forger, D. B. , & Piggins, H. D. (2009). Daily electrical silencing in the mammalian circadian clock. Science, 326, 281–284. 10.1126/science.1169657 [DOI] [PubMed] [Google Scholar]
- Braun, R. , Kath, W. L. , Iwanaszko, M. , Kula‐Eversole, E. , Abbott, S. M. , Reid, K. J. , … Allada, R. (2018). Universal method for robust detection of circadian state from gene expression. Proceedings of the National Academy of Sciences, 115, E9247–E9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. A. , Fleury‐Olela, F. , Nagoshi, E. , Hauser, C. , Juge, C. , Meier, C. A. , … Schibler, U. (2005). The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biology, 3, e338 10.1371/journal.pbio.0030338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. A. , Zumbrunn, G. , Fleury‐Olela, F. , Preitner, N. , & Schibler, U. (2002). Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Current Biology, 12, 1574–1583. [DOI] [PubMed] [Google Scholar]
- Canaple, L. , Rambaud, J. , Dkhissi‐Benyahya, O. , Rayet, B. , Tan, N. S. , Michalik, L. , … Laudet, V. (2006). Reciprocal regulation of brain and muscle Arnt‐like protein 1 and peroxisome proliferator‐activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Molecular Endocrinology, 20, 1715–1727. [DOI] [PubMed] [Google Scholar]
- Chen, L. , & Yang, G. (2015). Recent advances in circadian rhythms in cardiovascular system. Frontiers in Pharmacology, 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann, R. , Okyar, A. , & Lévi, F. (2016). Dosing‐time makes the poison: Circadian regulation and pharmacotherapy. Trends in Molecular Medicine, 22, 430–445. [DOI] [PubMed] [Google Scholar]
- Damiola, F. , Le Minli, N. , Preitner, N. , Kornmann, B. , Fleury‐Olela, F. , & Schibler, U. (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development, 14, 2950–2961. 10.1101/gad.183500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive, P. , Monté, D. , Dubois, G. , Trottein, F. , Fruchart‐Najib, J. , Mariani, J. , … Staels, B. (2001). The orphan nuclear receptor RORα is a negative regulator of the inflammatory response. EMBO Reports, 2, 42–48. 10.1093/embo-reports/kve007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner, C. , Schibler, U. , & Albrecht, U. (2010). The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annual Review of Physiology, 72, 517–549. 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- du Pré, B. C. , Dierickx, P. , Crnko, S. , Doevendans, P. A. , Vos, M. A. , Geijsen, N. , … van Laake, L. W. (2017). Neonatal rat cardiomyocytes as an in vitro model for circadian rhythms in the heart. Journal of Molecular and Cellular Cardiology, 112, 58–63. 10.1016/j.yjmcc.2017.08.009 [DOI] [PubMed] [Google Scholar]
- Dulong, S. , Ballesta, A. , Okyar, A. , & Levi, F. (2015). Identification of circadian determinants of cancer chronotherapy through in vitro chronopharmacology and mathematical modeling. Molecular Cancer Therapeutics, 14, 2154–2164. 10.1158/1535-7163.MCT-15-0129 [DOI] [PubMed] [Google Scholar]
- Eckel‐Mahan, K. , & Sassone‐Corsi, P. (2015). Phenotyping circadian rhythms in mice. Curr. Protoc. Mouse Biol., 5, 271–281. 10.1002/9780470942390.mo140229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Athman, R. , Fuhr, L. , & Relógio, A. (2018). A systems‐level analysis reveals circadian regulation of splicing in colorectal cancer. eBioMedicine, 33, 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Athman, R. , Genov, N. N. , Mazuch, J. , Zhang, K. , Yu, Y. , Fuhr, L. , … Relógio, A. (2017). The Ink4a/Arf locus operates as a regulator of the circadian clock modulating RAS activity. PLoS Biology, 15, e2002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Athman, R. , & Relógio, A. (2018). Escaping circadian regulation: An emerging hallmark of cancer? Cell Systems, 6, 266–267. 10.1016/j.cels.2018.03.006 [DOI] [PubMed] [Google Scholar]
- Esnault, C. , Stewart, A. , Gualdrini, F. , East, P. , Horswell, S. , Matthews, N. , & Treisman, R. (2014). Rho‐actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes & Development, 28, 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. A. , & Davidson, A. J. (2013). Health consequences of circadian disruption in humans and animal models In Gillette M. U. (Ed.), Progress in molecular biology and translational science (pp. 283–323). Wageningen, the Netherlands: Academic Press. [DOI] [PubMed] [Google Scholar]
- Fang, M. , Kang, H.‐G. , Park, Y. , Estrella, B. , & Zarbl, H. (2017). In vitro bioluminescence assay to characterize circadian rhythm in mammary epithelial cells. Journal of Visualized Experiments, 127, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhr, L. , Abreu, M. , Pett, P. , & Relógio, A. (2015). Circadian systems biology: When time matters. Computational and Structural Biotechnology Journal, 13, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhr, L. , El‐Athman, R. , Scrima, R. , Cela, O. , Carbone, A. , Knoop, H. , … Relógio, A. (2018). The circadian clock regulates metabolic phenotype rewiring via HKDC1 and modulates tumor progression and drug response in colorectal cancer. eBioMedicine, 33, 105–121. 10.1016/j.ebiom.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon, F. , Olela, F. F. , Schaad, O. , Descombes, P. , & Schibler, U. (2006). The circadian PAR‐domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metabolism, 4, 25–36. 10.1016/j.cmet.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Garrido, M. , Saccardo, D. , De Rui, M. , Vettore, E. , Verardo, A. , Carraro, P. , … Montagnese, S. (2017). Abnormalities in the 24‐hour rhythm of skin temperature in cirrhosis: Sleep–wake and general clinical implications. Liver International, 37, 1833–1842. [DOI] [PubMed] [Google Scholar]
- Gerber, A. , Esnault, C. , Aubert, G. , Treisman, R. , Pralong, F. , & Schibler, U. (2013). Blood‐borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell, 152, 492–503. 10.1016/j.cell.2012.12.027 [DOI] [PubMed] [Google Scholar]
- Gery, S. , & Koeffler, H. P. (2007). The role of circadian regulation in cancer. Cold Spring Harbor Symposia on Quantitative Biology, 72, 459–464. [DOI] [PubMed] [Google Scholar]
- Gloston, G. F. , Yoo, S. H. , & Chen, Z. (2017). Clock‐enhancing small molecules and potential applications in chronic diseases and aging. Frontiers in Neurology, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin, P. , Rando, G. , Fleury‐Olela, F. , & Schibler, U. (2016). Unbiased identification of signal‐activated transcription factors by barcoded synthetic tandem repeat promoter screening (BC‐STAR‐PROM). Genes & Development, 30, 1895–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B. , Yang, N. , Borysiewicz, E. , Dudek, M. , Williams, J. L. , Li, J. , … Meng, Q. J. (2015). Catabolic cytokines disrupt the circadian clock and the expression of clock‐controlled genes in cartilage via an NFkB‐dependent pathway. Osteoarthritis and Cartilage, 23, 1981–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand, L. E. , Hopwood, T. W. , Dickson, S. H. , Walker, A. L. , Loudon, A. S. I. , Ray, D. W. , … Gibbs, J. E. (2016). The circadian clock regulates inflammatory arthritis. The FASEB Journal, 30, 3759–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haus, E. L. , & Smolensky, M. H. (2013). Shift work and cancer risk: Potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Medicine Reviews, 17, 273–284. 10.1016/j.smrv.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Hirota, T. , Okano, T. , Kokame, K. , Shirotani‐Ikejima, H. , Miyata, T. , & Fukada, Y. (2002). Glucose down‐regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured rat‐1 fibroblasts. The Journal of Biological Chemistry, 277, 44244–44251. 10.1074/jbc.M206233200 [DOI] [PubMed] [Google Scholar]
- Hong, H. K. , Maury, E. , Ramsey, K. M. , Perelis, M. , Marcheva, B. , Omura, C. , … Bass, J. (2018). Requirement for NF‐κB in maintenance of molecular and behavioral circadian rhythms in mice. Genes & Development, 32, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, T. , Su, W. , Guo, Z. , & Gong, M. (2018). A novel diabetic mouse model for real‐time monitoring of clock gene oscillation and blood pressure circadian rhythm. Journal of Biological Rhythms, 34, 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. T. L. , Guilding, C. , Lennox, L. , Samuels, R. E. , McMahon, D. G. , & Piggins, H. D. (2008). Live imaging of altered period1 expression in the suprachiasmatic nuclei of Vipr2 −/− mice. Journal of Neurochemistry, 106, 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, M. E. , Abruzzi, K. C. , Allada, R. , Anafi, R. , Arpat, A. B. , Asher, G. , … Hogenesch, J. B. (2017). Guidelines for genome‐scale analysis of biological rhythms. Journal of Biological Rhythms, 32, 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, M. E. , Hogenesch, J. B. , & Kornacker, K. (2010). JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. Journal of Biological Rhythms, 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey, J. J. (2017). Machine learning identifies a compact gene set for monitoring the circadian clock in human blood. Genome Medicine, 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey, J. J. , & Butte, A. J. (2016). Differential phasing between circadian clocks in the brain and peripheral organs in humans. Journal of Biological Rhythms, 31, 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey, J. J. , Hastie, T. , & Butte, A. J. (2016). ZeitZeiger: Supervised learning for high‐dimensional data from an oscillatory system. Nucleic Acids Research, 44, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse, J. , Eichele, G. , & Oster, H. (2015). Synchronization of the mammalian circadian timing system: Light can control peripheral clocks independently of the SCN clock: Alternate routes of entrainment optimize the alignment of the body's circadian clock network with external time. BioEssays, 37, 1119–1128. 10.1002/bies.201500026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innominato, P. F. , Komarzynski, S. , Palesh, O. G. , Dallmann, R. , Bjarnason, G. A. , Giacchetti, S. , … Lévi, F. A. (2018). Circadian rest–activity rhythm as an objective biomarker of patient‐reported outcomes in patients with advanced cancer. Cancer Medicine, 7, 4396–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo, M. , Sato, T. R. , Straume, M. , & Johnson, C. H. (2006). Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Computational Biology, 2, e136 10.1371/journal.pcbi.0020136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik, K. , & Lee, C. C. (2004). Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature, 430, 467–471. 10.1038/nature02724 [DOI] [PubMed] [Google Scholar]
- Kamagata, M. , Ikeda, Y. , Sasaki, H. , Hattori, Y. , Yasuda, S. , Iwami, S. , … Shibata, S. (2017). Potent synchronization of peripheral circadian clocks by glucocorticoid injections in PER2::LUC‐Clock/Clock mice. Chronobiology International, 34, 1067–1082. [DOI] [PubMed] [Google Scholar]
- Karlebach, G. , & Shamir, R. (2008). Modelling and analysis of gene regulatory networks. Nature Reviews. Molecular Cell Biology, 9, 770–780. [DOI] [PubMed] [Google Scholar]
- Keijzer, H. , Smits, M. G. , Peeters, T. , Looman, C. W. N. , Endenburg, S. C. , & Gunnewiek, J. M. T. K. (2011). Evaluation of salivary melatonin measurements for dim light melatonin onset calculations in patients with possible sleep–wake rhythm disorders. Clinica Chimica Acta, 412, 1616–1620. 10.1016/j.cca.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Keller, M. , Mazuch, J. , Abraham, U. , Eom, G. D. , Herzog, E. D. , Volk, H.‐D. , … Maier, B. (2009). A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences, 106, 21407–21412. 10.1073/pnas.0906361106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanaka, N. , Kitanaka, J. , Hall, F. S. , Uhl, G. R. , Watabe, K. , Kubo, H. , … Takemura, M. (2012). A single administration of methamphetamine to mice early in the light period decreases running wheel activity observed during the dark period. Brain Research, 1429, 155–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov, R. V. , Kondratova, A. A. , Gorbacheva, V. Y. , Vykhovanets, O. V. , & Antoch, M. P. (2006). Early aging and age‐related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes & Development, 20, 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman, S. J. , & McMahon, D. G. (2006). Encoding the ins and outs of circadian pacemaking. Journal of Biological Rhythms, 21, 470–481. 10.1177/0748730406294316 [DOI] [PubMed] [Google Scholar]
- Lehmann, R. , Childs, L. , Thomas, P. , Abreu, M. , Fuhr, L. , Herzel, H. , … Relógio, A. (2015). Assembly of a comprehensive regulatory network for the mammalian circadian clock: A bioinformatics approach. PLoS ONE, 10, e0126283 10.1371/journal.pone.0126283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter, J. , Yan, L. , Vishnubhotla, B. , Quintero, J. E. , Kuhlman, S. J. , McMahon, D. G. , & Silver, R. (2003). A short half‐life GFP mouse model for analysis of suprachiasmatic nucleus organization. Brain Research, 964, 279–287. 10.1016/S0006-8993(02)04084-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi, F. , & Schibler, U. (2007). Circadian rhythms: Mechanisms and therapeutic implications. Annual Review of Pharmacology and Toxicology, 47, 593–628. [DOI] [PubMed] [Google Scholar]
- Li, X. , Zhao, X. , Fang, Y. , Jiang, X. , Duong, T. , Fan, C. , … Kain, S. R. (1998). Generation of destabilized green fluorescent protein as a transcription reporter. The Journal of Biological Chemistry, 273, 34970–34975. 10.1074/jbc.273.52.34970 [DOI] [PubMed] [Google Scholar]
- Lin, Y. C. , Hsiao, T. J. , & Chen, P. C. (2009). Persistent rotating shift–work exposure accelerates development of metabolic syndrome among middle‐aged female employees: A five‐year follow‐up. Chronobiology International, 26, 740–755. 10.1080/07420520902929029 [DOI] [PubMed] [Google Scholar]
- Liu, C. , & Reppert, S. M. (2000). GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron, 25, 123–128. 10.1016/S0896-6273(00)80876-4 [DOI] [PubMed] [Google Scholar]
- Marcheva, B. , Ramsey, K. M. , Buhr, E. D. , Kobayashi, Y. , Su, H. , Ko, C. H. , … Bass, J. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature, 466, 627–631. 10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Nicolas, A. , Madrid, J. A. , García, F. J. , Campos, M. , Moreno‐Casbas, M. T. , Almaida‐Pagán, P. F. , … Rol, M. A. (2018). Circadian monitoring as an aging predictor. Scientific Reports, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazuski, C. , & Herzog, E. D. (2015). Circadian rhythms: To sync or not to sync. Current Biology, 25, R337–R339. 10.1016/j.cub.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcarthur, A. J. , Hunt, A. E. , & Gillette, M. U. (1997). Melatonin action and signal transduction in the rat suprachiasmatic circadian clock: Activation of protein kinase C at dusk and dawn. Endocrinology, 138, 627–634. 10.1210/endo.138.2.4925 [DOI] [PubMed] [Google Scholar]
- Mei, L. , Fan, Y. , Lv, X. , Welsh, D. K. , Zhan, C. , & Zhang, E. E. (2018). Long‐term in vivo recording of circadian rhythms in brains of freely moving mice. Proceedings of the National Academy of Sciences, 115, 4276–4281. 10.1073/pnas.1717735115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk, J. A. , Green, C. B. , & Takahashi, J. S. (2012). Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience, 35, 445–462. 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, P. J. , & Williams, L. M. (1996). The pars tuberalis of the pituitary: A gateway for neuroendocrine output. Reviews of Reproduction, 1, 153–161. 10.1530/ror.0.0010153 [DOI] [PubMed] [Google Scholar]
- Mure, L. S. , Le, H. D. , Benegiamo, G. , Chang, M. W. , Rios, L. , Jillani, N. , … Panda, S. (2018). Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science, 359, eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek, E. S. (2015). Circadian clock disruption in neurodegenerative diseases: Cause and effect? Frontiers in Pharmacology, 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek, E. S. , Bhimasani, M. , Zangrilli, M. A. , Morris, J. C. , Holtzman, D. M. , & Ju, Y. E. S. (2018). Circadian rest–activity pattern changes in aging and preclinical Alzheimer disease. JAMA Neurology, 75, 582–590. 10.1001/jamaneurol.2017.4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek, E. S. , & FitzGerald, G. A. (2013). Molecular clocks in pharmacology. Handbook of Experimental Pharmacology, 217, 243–260. 10.1007/978-3-642-25950-0_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi, E. , Saini, C. , Bauer, C. , Laroche, T. , Naef, F. , & Schibler, U. (2004). Circadian gene expression in individual fibroblasts: Cell‐autonomous and self‐sustained oscillators pass time to daughter cells. Cell, 119, 693–705. 10.1016/j.cell.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Nakahata, Y. , Akashi, M. , Trcka, D. , Yasuda, A. , & Takumi, T. (2006). The in vitro real‐time oscillation monitoring system identifies potential entrainment factors for circadian clocks. BMC Molecular Biology, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, Y. , Sahar, S. , Astarita, G. , Kaluzova, M. , & Sassone‐Corsi, P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK–SIRT1. Science, 324, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T. , Ikeda, M. , Ohmiya, Y. , & Nakajima, Y. (2012). A dual‐color luciferase assay system reveals circadian resetting of cultured fibroblasts by co‐cultured adrenal glands. PLoS ONE, 7, e37093 10.1371/journal.pone.0037093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio, A. , & Malorni, W. (1979). Wheel running and sleep in two strains of mice: Plasticity and rigidity in the expression of circadian rhythmicity. Brain Research, 163, 121–133. [DOI] [PubMed] [Google Scholar]
- Pilorz, V. , Helfrich‐Förster, C. , & Oster, H. (2018). The role of the circadian clock system in physiology. Pflugers Arch. Eur. J. Physiol., 470, 227–239. 10.1007/s00424-017-2103-y [DOI] [PubMed] [Google Scholar]
- Ralph, M. R. , Foster, R. G. , Davis, F. C. , & Menaker, M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science, 247, 975–978. 10.1126/science.2305266 [DOI] [PubMed] [Google Scholar]
- Ramanathan, C. , Khan, S. K. , Kathale, N. D. , Xu, H. , & Liu, A. C. (2012). Monitoring cell‐autonomous circadian clock rhythms of gene expression using luciferase bioluminescence reporters. Journal of Visualized Experiments, 27, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkisoensing, A. , & Meijer, J. H. (2015). Synchronization of biological clock neurons by light and peripheral feedback systems promotes circadian rhythms and health. Frontiers in Neurology, 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti, R. , Kaufman, C. M. , & Menaker, M. (1994). Complete suprachiasmatic lesions eliminate circadian rhythmicity of body temperature and locomotor activity in golden hamsters. Journal of Comparative Physiology. A, 175, 223–232. 10.1007/BF00215118 [DOI] [PubMed] [Google Scholar]
- Relógio, A. , Thomas, P. , Medina‐Pérez, P. , Reischl, S. , Bervoets, S. , Gloc, E. , … Sers, C. (2014). Ras‐mediated deregulation of the circadian clock in cancer. PLoS Genetics, 10, e1004338 10.1371/journal.pgen.1004338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relógio, A. , Westermark, P. O. , Wallach, T. , Schellenberg, K. , Kramer, A. , & Herzel, H. (2011). Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Computational Biology, 7, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, L. , Leise, T. L. , Noguchi, T. , Galschiodt, A. M. , Houl, J. H. , Welsh, D. K. , & Holmes, T. C. (2015). Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Current Biology, 25, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg, T. , Kantermann, T. , Juda, M. , Vetter, C. , & Allebrandt, K. V. (2013). Light and the human circadian clock In Kramer A., & Merrow M. (Eds.), Circadian clocks. Handbook of experimental pharmacology (pp. 311–331). Berlin, Heidelberg: Springer. [DOI] [PubMed] [Google Scholar]
- Roenneberg, T. , & Merrow, M. (2016). The circadian clock and human health. Current Biology, 26, R432–R443. [DOI] [PubMed] [Google Scholar]
- Rogers, E. H. , Fawcett, S. A. , Pekovic‐Vaughan, V. , & Hunt, J. A. (2017). Comparing circadian dynamics in primary derived stem cells from different sources of human adult tissue. Stem Cells International, 2017, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa, S. C. , Judas, F. , Lopes, M. C. , & Mendes, A. F. (2008). Nitric oxide synthase isoforms and NF‐κB activity in normal and osteoarthritic human chondrocytes: Regulation by inducible nitric oxide. Nitric Oxide—Biol. Chem., 19, 276–283. [DOI] [PubMed] [Google Scholar]
- Roybal, K. , Theobold, D. , Graham, A. , DiNieri, J. A. , Russo, S. J. , Krishnan, V. , … McClung, C. A. (2007). Mania‐like behavior induced by disruption of CLOCK. Proceedings of the National Academy of Sciences, 104, 6406–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben, M. D. , Wu, G. , Smith, D. F. , Schmidt, R. E. , Francey, L. J. , Lee, Y. Y. , … Hogenesch, J. B. (2018). A database of tissue‐specific rhythmically expressed human genes has potential applications in circadian medicine. Science Translational Medicine, 10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, C. , Brown, S. A. , & Dibner, C. (2015). Human peripheral clocks: Applications for studying circadian phenotypes in physiology and pathophysiology. Frontiers in Neurology, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, C. , Liani, A. , Curie, T. , Gos, P. , Kreppel, F. , Emmenegger, Y. , … Schibler, U. (2013). Real‐time recording of circadian liver gene expression in freely moving mice reveals the phase‐setting behavior of hepatocyte clocks. Genes & Development, 27, 1526–1536. 10.1101/gad.221374.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, C. , Morf, J. , Stratmann, M. , Gos, P. , & Schibler, U. (2012). Simulated body temperature rhythms reveal the phase‐shifting behavior and plasticity of mammalian circadian oscillators. Genes & Development, 26, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K. , Nagase, T. , Fukui, H. , Horikawa, K. , Okada, T. , Tanaka, H. , … Ishida, N. (1998). Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. The Journal of Biological Chemistry, 273(42), 27039–27042. 10.1074/jbc.273.42.27039 [DOI] [PubMed] [Google Scholar]
- Sandu, C. , Dumas, M. , Malan, A. , Sambakhe, D. , Marteau, C. , Nizard, C. , … Felder‐Schmittbuhl, M. P. (2012). Human skin keratinocytes, melanocytes, and fibroblasts contain distinct circadian clock machineries. Cellular and Molecular Life Sciences, 69, 3329–3339. 10.1007/s00018-012-1026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S. , Solanas, G. , Peixoto, F. O. , Bee, L. , Symeonidi, A. , Schmidt, M. S. , … Sassone‐Corsi, P. (2017). Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell, 170, 664–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpellini, E. , & Tack, J. (2012). Obesity and metabolic syndrome: An inflammatory condition. Digestive Diseases, 30, 148–153. [DOI] [PubMed] [Google Scholar]
- Scrima, R. , Cela, O. , Merla, G. , Augello, B. , Rubino, R. , Quarato, G. , … Capitanio, N. (2016). Clock‐genes and mitochondrial respiratory activity: Evidence of a reciprocal interplay. Biochim. Biophys. Acta—Bioenerg., 1857, 1344–1351. [DOI] [PubMed] [Google Scholar]
- Smolensky, M. H. , & Peppas, N. A. (2007). Chronobiology, drug delivery, and chronotherapeutics. Advanced Drug Delivery Reviews, 59, 828–851. 10.1016/j.addr.2007.07.001 [DOI] [PubMed] [Google Scholar]
- Smyth, N. , Thorn, L. , Hucklebridge, F. , Clow, A. , & Evans, P. (2016). Assessment of the cortisol awakening response: Real‐time analysis and curvilinear effects of sample timing inaccuracy. Psychoneuroendocrinology, 74, 380–386. 10.1016/j.psyneuen.2016.09.026 [DOI] [PubMed] [Google Scholar]
- Speer, K. , Upton, D. , Semple, S. , & McKune, A. (2018). Systemic low‐grade inflammation in post‐traumatic stress disorder: A systematic review. Journal of Inflammation Research, 11, 111–121. 10.2147/JIR.S155903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler, M. L. , Kuropatwinski, K. K. , Comas, M. , Gasparian, A. V. , Fedtsova, N. , Gleiberman, A. S. , … Antoch, M. P. (2012). Core circadian protein CLOCK is a positive regulator of NF‐κB‐mediated transcription. Proceedings of the National Academy of Sciences, 109, E2457–E2465. 10.1073/pnas.1206274109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, F. K. , & Zucker, I. (1972). Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proceedings of the National Academy of Sciences, 69, 1583–1586. 10.1073/pnas.69.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, C. C. (1898). Variations in daily activity produced by alcohol and by changes in barometric pressure and diet with a description of recording methods. The American Journal of Physiology, 1, 40–56. 10.1152/ajplegacy.1898.1.1.40 [DOI] [Google Scholar]
- Sulli, G. , Manoogian, E. N. C. , Taub, P. R. , & Panda, S. (2018). Training the circadian clock, clocking the drugs, and drugging the clock to prevent, manage, and treat chronic diseases. Trends in Pharmacological Sciences, 39, 812–827. 10.1016/j.tips.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara, Y. , Kuroda, H. , Saito, K. , Nakajima, Y. , Kubo, Y. , Ohnishi, N. , … Shibata, S. (2012). In vivo monitoring of peripheral circadian clocks in the mouse. Current Biology, 22, 1029–1034. 10.1016/j.cub.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nature Reviews. Genetics, 18, 164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, J. S. , Hong, H. K. , Ko, C. H. , & McDearmon, E. L. (2008). The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nature Reviews. Genetics, 9, 764–775. 10.1038/nrg2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai, T. K. , Nakane, Y. , Ota, W. , Kobayashi, A. , Ishiguro, M. , Kadofusa, N. , … Yoshimura, T. (2018). Identification of circadian clock modulators from existing drugs. EMBO Molecular Medicine, 10, e8724 10.15252/emmm.201708724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, S.‐C. , & Guo, S.‐C. (2018). Extracellular vesicles: Potential participants in circadian rhythm synchronization. International Journal of Biological Sciences, 14, 1610–1620. 10.7150/ijbs.26518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben, P. F. , & Westermark, P. O. (2016). Differential rhythmicity: Detecting altered rhythmicity in biological data. Bioinformatics, 32, 2800–2808. 10.1093/bioinformatics/btw309 [DOI] [PubMed] [Google Scholar]
- Tilstra, J. S. , Clauson, C. L. , Niedernhofer, L. J. , & Robbins, P. D. (2011). NF‐κB in aging and disease. Aging and Disease, 2, 449–465. [PMC free article] [PubMed] [Google Scholar]
- Tiscornia, G. , Singer, O. , & Verma, I. M. (2006). Production and purification of lentiviral vectors. Nature Protocols, 1, 241–245. 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
- Turek, F. W. , Joshu, C. , Kohsaka, A. , Lin, E. , Ivanova, G. , McDearmon, E. , … Bass, J. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science, 308, 1043–1045. 10.1126/science.1108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H. R. , Hayashi, S. , Chen, W. , Sano, M. , Machida, M. , Shigeyoshi, Y. , … Hashimoto, S. (2005). System‐level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genetics, 37, 187–192. 10.1038/ng1504 [DOI] [PubMed] [Google Scholar]
- Ungar, F. , & Halberg, F. (1962). Circadian rhythm in the in vitro response of mouse adrenal to adrenocorticotropic hormone. Science, 137, 1058–1060. 10.1126/science.137.3535.1058 [DOI] [PubMed] [Google Scholar]
- Vanselow, K. , Vanselow, J. T. , Westermark, P. O. , Reischl, S. , Maier, B. , Korte, T. , … Kramer, A. (2006). Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes & Development, 20, 2660–2672. 10.1101/gad.397006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Mauvoisin, D. , Martin, E. , Atger, F. , Galindo, A. N. , Dayon, L. , … Gachon, F. (2017). Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metabolism, 25, 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. H. , Zhao, T. , Cui, K. , Hu, G. , Chen, Q. , Chen, W. , … Deng, C. X. (2016). Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging. Scientific Reports, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Lv, D. , Liu, W. , Li, S. , Chen, J. , Shen, Y. , … Liu, C. F. (2018). Disruption of the circadian clock alters antioxidative defense via the SIRT1–BMAL1 pathway in 6‐OHDA‐induced models of Parkinson's disease. Oxidative Medicine and Cellular Longevity, 2018, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M. , Hida, A. , Kitamura, S. , Enomoto, M. , Ohsawa, Y. , Katayose, Y. , … Mishima, K. (2012). Biochemical and biophysical research communications rhythmic expression of circadian clock genes in human leukocytes and beard hair follicle cells. Biochemical and Biophysical Research Communications, 425, 902–907. 10.1016/j.bbrc.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Welsh, D. K. , Imaizumi, T. , & Kay, S. A. (2005). Real‐time reporting of circadian‐regulated gene expression by luciferase imaging in plants and mammalian cells. Methods in Enzymology, 393, 269–288. [DOI] [PubMed] [Google Scholar]
- Welsh, D. K. , Logothetis, D. E. , Meister, M. , & Reppert, S. M. (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron, 14, 697–706. [DOI] [PubMed] [Google Scholar]
- Welsh, D. K. , & Noguchi, T. (2012). Cellular bioluminescence imaging. Cold Spring Harbor Protocols, 2012, 852–866. [DOI] [PubMed] [Google Scholar]
- Welsh, D. K. , Takahashi, J. S. , & Kay, S. A. (2010). Suprachiasmatic nucleus: Cell autonomy and network properties. Annual Review of Physiology, 72, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, D. K. , Yoo, S. H. , Liu, A. C. , Takahashi, J. S. , & Kay, S. A. (2004). Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Current Biology, 14, 2289–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. , & Stelzer, E. (1999). Photobleaching GFP reveals protein dynamics inside live cells. Trends in Cell Biology, 9, 61–65. 10.1016/S0962-8924(98)01433-0 [DOI] [PubMed] [Google Scholar]
- Wittenbrink, N. , Ananthasubramaniam, B. , Münch, M. , Koller, B. , Maier, B. , Weschke, C. , … Kramer, A. (2018). High‐accuracy determination of internal circadian time from a single blood sample. The Journal of Clinical Investigation, 128, 3826–3839. 10.1172/JCI120874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita, K. , & Okamura, H. (2000). Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat‐1 fibroblasts. FEBS Letters, 465, 79–82. 10.1016/S0014-5793(99)01724-X [DOI] [PubMed] [Google Scholar]
- Yagita, K. , Tamanini, F. , van der Horst, G. T. , & Okamura, H. (2001). Molecular mechanisms of the biological clock in cultured fibroblasts. Science, 292, 278–281. 10.1126/science.1059542 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S. , Isejima, H. , Matsuo, T. , Okura, R. , Yagita, K. , Kobayashi, M. , & Okamura, H. (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science, 302, 1408–1412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S. , Kobayashi, M. , Mitsui, S. , Ishida, Y. , van der Horst, G. T. J. , Suzuki, M. , … Okamura, H. (2001). View of a mouse clock gene ticking. Nature, 409, 684 10.1038/35055628 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S. , Mitsui, S. , Miyake, S. , Yan, L. , Onishi, H. , Yagita, K. , … Okamura, H. (2000). The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Current Biology, 10, 873–876. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Okada, K. , Mizuno, T. , Ota, T. , Yamada, H. , Doi, M. , … Okamura, H. (2016). Real‐time recording of circadian Per1 and Per2 expression in the suprachiasmatic nucleus of freely moving rats. Journal of Biological Rhythms, 31, 108–111. 10.1177/0748730415621412 [DOI] [PubMed] [Google Scholar]
- Ye, Y. , Xiang, Y. , Ozguc, F. M. , Kim, Y. , Liu, C. J. , Park, P. K. , … Han, L. (2018). The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Systems, 6, 314–328. 10.1016/j.cels.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, F. , Hoberg, J. E. , Ramsey, C. S. , Keller, M. D. , Jones, D. R. , Frye, R. A. , & Mayo, M. W. (2004). Modulation of NF‐κB‐dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal, 23, 2369–2380. 10.1038/sj.emboj.7600244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.‐H. , Yamazaki, S. , Lowrey, P. L. , Shimomura, K. , Ko, C. H. , Buhr, E. D. , … Takahashi, J. S. (2004). PERIOD2::LUCIFERASE real‐time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proceedings of the National Academy of Sciences of the United States of America, 101, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, E. E. , Liu, A. C. , Hirota, T. , Miraglia, L. J. , Welch, G. , Pongsawakul, P. Y. , … Kay, S. A. (2009). A genome‐wide RNAi screen for modifiers of the circadian clock in human cells. Cell, 139, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Lahens, N. F. , Ballance, H. I. , Hughes, M. E. , & Hogenesch, J. B. (2014). A circadian gene expression atlas in mammals: Implications for biology and medicine. Proceedings of the National Academy of Sciences, 111, 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Armstrong, J. L. , Tchkonia, T. , & Kirkland, J. L. (2014). Cellular senescence and the senescent secretory phenotype in age‐related chronic diseases. Current Opinion in Clinical Nutrition and Metabolic Care, 17, 324–328. 10.1097/MCO.0000000000000065 [DOI] [PubMed] [Google Scholar]


