Abstract
Protein stability in detergent or membrane-like environments is the bottleneck for structural studies on integral membrane proteins (IMP). Irrespective of the method to study the structure of an IMP, detergent solubilization from the membrane is usually the first step in the workflow. Here, we establish a simple, high-throughput screening method to identify optimal detergent conditions for membrane protein stabilization. We apply differential scanning fluorimetry in combination with scattering upon thermal denaturation to study the unfolding of integral membrane proteins. Nine different prokaryotic and eukaryotic membrane proteins were used as test cases to benchmark our detergent screening method. Our results show that it is possible to measure the stability and solubility of IMPs by diluting them from their initial solubilization condition into different detergents. We were able to identify groups of detergents with characteristic stabilization and destabilization effects for selected targets. We further show that fos-choline and PEG family detergents may lead to membrane protein destabilization and unfolding. Finally, we determined thenmodynamic parameters that are important indicators of IMP stability. The described protocol allows the identification of conditions that are suitable for downstream handling of membrane proteins during purification.
Subject terms: Molecular biophysics, Membrane proteins
Introduction
Numerous studies have addressed the gap between the large number of sequences representing integral membrane proteins (IMPs) in sequenced genomes (up to 30 percent)1 and the 863 unique known membrane protein structures reported up-to-date (http://blanco.biomol.uci.edu/mpstruc/). This highlights the difficulties along the path to structure determination such as low expression levels, solubilization from the native lipid bilayer and stability in a new membrane mimetic2–4. Most of these known unique structures were determined by X-ray crystallography, however the “resolution revolution” in cryo-electron microscopy5,6 will lead to a rapidly growing share of IMP structures determined by single-particle cryo-EM in the future (so far 5% of the PDB entries for all deposited “not unique” membrane proteins are already determined by EM). Soon after the mesophase crystallization technique has been introduced, lipidic cubic phase (LCP) crystallization has been positioned amongst the leading methods to be chosen for membrane protein crystallography7, complementing the use of bicelles8 and traditional vapour diffusion of protein-detergent complexes3. Furthermore, different lipoprotein nanoparticle systems have been developed to allow the reconstitution of membrane proteins into a lipid mimic environment suitable for EM, NMR, SAXS and functional studies. IMPs can be stabilized by a scaffold of saposin A proteins9,10 or inserted into nanodiscs composed of MSP1 scaffold proteins11–14 with positive consequences for its structural and functional characterization. However, for most methods investigating the structure of an IMP, the need to extract and solubilize IMPs from the membrane using detergents initially remains. Despite the increasing number of commercially available detergents, membrane protein crystal structures in the PDB are represented by a subset of detergents15. This might be due to biased selection of the detergents based on previous successes in the extraction of other membrane proteins or due to historical protocols in experienced membrane protein labs. The most commonly used detergent families in membrane protein crystallography are maltosides and glucosides, followed by amine oxides and polyoxyethylene glycols3. Altogether, crystallographic data support the use of n-Dodecyl β-D-maltoside (DDM), n-Decyl-β-D-Maltopyranoside (DM), Octyl-beta-glucoside (OG) and Lauryldimethylamine-N-oxide (LDAO). Besides crystallography, CYMAL and Fos-choline detergents have been shown to be very efficient in extracting inner membrane proteins, DM for outer membrane proteins and LDAO used for solubilization of transport proteins15. In summary, as a rule of thumb, detergents with longer acyl chains seem to be more efficient in solubilization and stabilization but shorter chain detergents form smaller micelles sizes, leading to a tighter packing in the crystal lattice and better diffraction16,17. The question would be, beside the purpose of crystallographic studies, are we on the right path by repeatedly employing these detergents? Thermal denaturation assays might help us to evaluate the most commonly used detergents over the last decade in membrane protein research.
While the link between protein thermodynamic stability and crystallization success for soluble globular proteins is not indeed absolute (many very stable proteins will never crystallize and some more unstable ones form crystals overnight), mono-dispersity and stability of a membrane protein has been postulated as a critical point for crystallization18. Instability of integral membrane proteins is indeed the bottleneck for structural and functional studies. α-helical membrane proteins could be partially unfolded when solubilized with detergents, maintaining the helical structure of the transmembrane domains but losing the tertiary inter-helix interactions19. In addition, the removal of specific lipids during solubilization and SEC, a process called delipidation, can severely affect protein stability and function20–22. Therefore, the initial condition used to extract an IMP from the membrane will mostly determine its half-life and stability throughout the entire downstream process.
Several methods exist for assessment of protein stability of IMPs. GFP-tagged proteins can be monitored by fluorescent-detection SEC to address their aggregation state during expression and purification23,24. In addition, stability in detergents has been studied using analytical size exclusion chromatography18,25. Slotboom et al. have addressed the behavior of IMPs in detergent micelles and lipid/detergent micelles in SEC-LS (light-scattering) analysis compared to sedimentation equilibrium centrifugation experiments26. Other methods like differential filtration and ultracentrifugation assays4 address the stability by quantifying the remaining protein in solution. However, the lack of aggregation is not necessarily an indication for a properly folded protein. The thermal denaturation assay using the thiol-specific fluorochrome N-[4-(7-diethylamino-4-methyl-3-coumarinyl) phenyl-maleimide (CPM)27 links stability of the IMP to the accessibility of free cysteine residues that become exposed to the solvent upon denaturation. However, not all membrane proteins are suitable for this analysis due to the lack of compatible Cys residues. Thermal denaturation experiments provide information whether an IMP displays a cooperative unfolding transition following a two-state model28, providing evidence of the folded state. The inflection point in a standard denaturation curve, Tm, has been a recurrent parameter to compare protein thermodynamic stability. Other studies have evaluated the stability of membrane proteins following the change of intrinsic fluorescence during thermal denaturation using a differential scanning fluorimetry device for the screening of lipid-like peptides29 and ligands30–33. Recently, Nji et al. have developed an engineered thermal-shift screen for detecting IMP lipids preferences based on fluorescence-detection size-exclusion chromatography (FSEC-TS). By using GFP-fusion IMPs the authors compare the stabilization of bacterial and eukaryotic membrane proteins before and after purification, and after addition of selected lipids to the purified IMPs. The authors conclude that eukaryotic membrane proteins appear to be more sensitive to the presence of lipids than prokaryotic ones21.
In this work, we use the nanoDSF technology (DSF) to study the unfolding of membrane proteins, following the intrinsic fluorescence of tryptophan residues during a thermal ramp in a detergent screen with 94 different detergents. In addition to the melting temperature, this device allows to follow the onset of aggregation by monitoring static light scattering. We describe a fast, high-throughput and quantitative methodology for screening detergents based on dilution rather than buffer exchange. The samples are diluted from its original extraction condition to 94 commercially available detergents with no exchange needed to observe a stabilization or destabilization effect. This will guide the selection of the appropriate detergent for further downstream processing of the IMP.
Results
Stability of selected integral membrane proteins in DDM
We have selected nine membrane proteins belonging to different families and originating from different species (see Table 1). Four IMPs belong to the major facilitator superfamily of transporters; DtpA, DgoT, and MdfA involved in nutrient and drug transport33–38, and LacY is the well-characterised lactose permease39. Ij1 is an ABC-transporter involved in ion transport. Kv1 is an IMP of unknown function and structure from the pathogenic bacterium Pseudomonas aeruginosa and Im1 is a prokaryotic kinase with two transmembrane domains. As an eukaryotic example, we have included the human P2X4 receptor from the P2X ionotropic receptors family, a regulator in neuropathic pain40. In addition, we screened bacteriorhodopsin (BR) from Halobacterium salinarum as a control example of a well-characterized membrane protein41.
Table 1.
IMPs used for the stability assay.
| Protein | Organism | Family | Function | Number of Trp residues | PDB ID |
|---|---|---|---|---|---|
| DgoT | E. coli | MFS transporters | putative galactonate transporter | 14 | 6E9N, 6E9O |
| MdfA | E. coli | MFS transporter | multi drug resistance | 9 | 4ZP0, 4ZOW, 4ZP2, 6GV1, 6EUQ |
| DtpA | E. coli | MFS transporter | peptide transporter | 10 | 6GS1, 6GS4, 6GS7 |
| Kv1 | Pseudomonas aeruginosa | unknown | unknown | 17 | — |
| Ij1 | E. coli | ABC-Transporter | ion transport | 22 | — |
| P2X4 | Homo sapiens | P2X ionotropic receptors | regulator in mediating neuropathic pain | 6 | 4DW0, 4DW1 (zebrafish) |
| BR | Halobacterium salinarum | 7TM receptor | proton pump | 8 | 4MD1, 4MD2, 4XXJ |
| LacY | E. coli | MFS transporter | transport of beta-galactosides | 5 | 1PV6 |
| Im1 | E. coli | HisKA | Kinase | 2 | — |
We have solubilized the isolated membranes in 1–2% DDM, usually the first choice detergent of most labs working with IMPs due to its rather mild denaturation properties. All proteins were expressed and purified as described in Material and Methods using DDM as starting detergent, with size exclusion chromatography (SEC) as last purification step, concentrated using centrifugal filter devices (Supplementary Fig. 1), and then diluted tenfold to the detergent screen covering various detergent families used for membrane solubilization and purification (Fig. 1 and Table S1). Importantly, for the proteins Kv1 and P2X4 concentrators with a cutoff smaller than the DDM micelle size (59.5 KDa) have been used, leading to samples with increased detergent concentration. In the case of BR, 1% OG was used as the starting condition as an external control due to its compatibility with crystallization42,43.
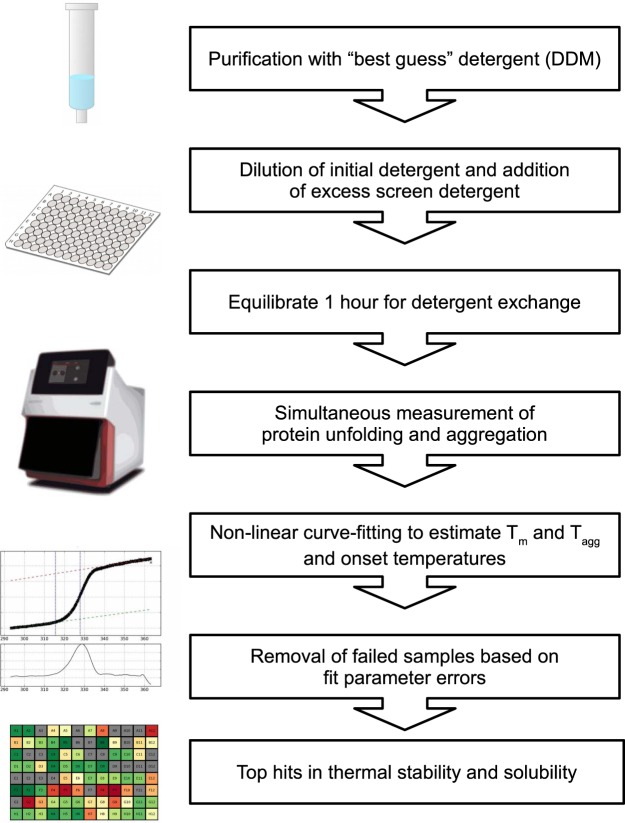
Figure 1.
Flow chart of the thermostability assay. Solubilized and purified proteins in DDM (see Supplementary Table 1 for DDM concentrations for each protein) are diluted tenfold to a 94 detergent screen followed by DSF and scattering measurements. Apparent melting and mid-aggregation temperatures (Tm and Tagg respectively) are estimated after curve fitting. Successful measurements for the thermodynamic parameters are plotted using a color code (red “denatured” and green “native”) for visual identification of stabilization conditions.
Detergent screening
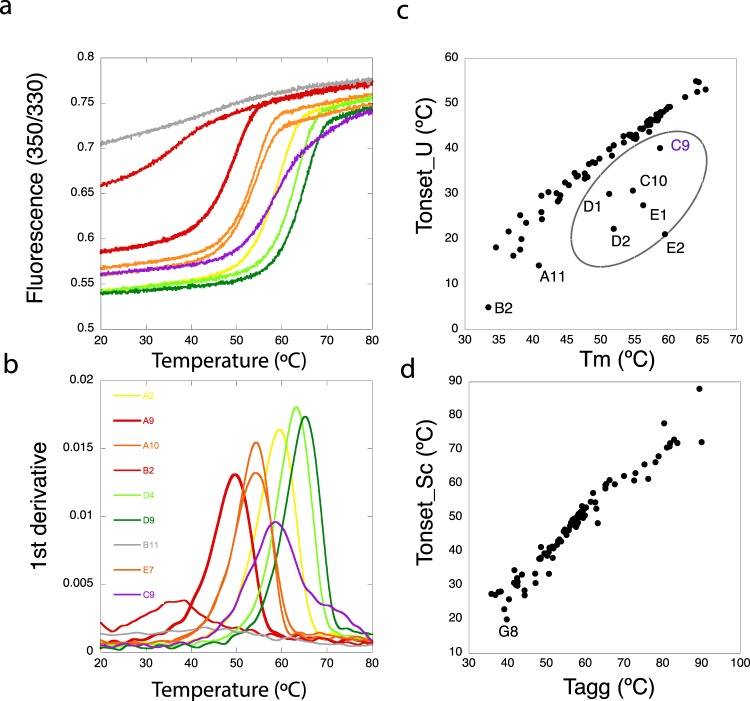
In order to test the thermodynamic stability of IMPs after being isolated from their natural environment and solubilized in detergents, we have performed a high-throughput analysis by DSF and light scattering. In IMPs, tryptophan residues (Trp) are either buried in the hydrophobic core of the protein, or exposed to the micelle belt after solubilization with detergents (Fig. 2a and Supplementary Fig. 2a). Thus, upon denaturation, IMPs could display shifts of the center of mass of the Trp fluorescence spectrum in both directions (blue and red)44. Fluorescence transition curves measured at 330 nm and 350 nm (and the ratio 350/330) result in “S curves” with positive slopes, or “Z curves” with negative slopes (Supplementary Fig. 3), from which the thermodynamic parameters like the melting temperature (Tm) and the onset of unfolding (Tonset_U) can be extracted (Fig. 2b,c; and Supplementary Fig. 2b). For the high-throughput analysis we used the fluorescence ratio (350/330) to obtain Tm (see Material and Methods) with the exception of LacY and BR where the ratio was less informative than the fluorescence measured at 330 nm (Supplementary Fig. 3). Figure 3 shows the first derivatives from the DSF curves obtained for the thermal denaturation at the starting condition, prior to dilution into the detergent screen. The selected nine IMPs display different thermal stabilities (represented by different Tm) at the starting point of the assay with a range between 38.9 °C and 59.3 °C (Table 2). Most proteins show a clear transition between the native and the unfolded state (Supplementary Fig. 3 and Fig. 3, black curves). In the case of LacY, the Ratio curve contained no unfolding transtions as judged by the absence of a peak in the smoothened first derivative curve. At the same time F330 and F350 of LacY showed a clear transition with a similar shape, so we used F330 due to higher signal strength (Supplementary Fig. 3). Finally, Im1 seems either to be unfolded after purification or its two Trp residues are not suited to report the unfolding of the protein.
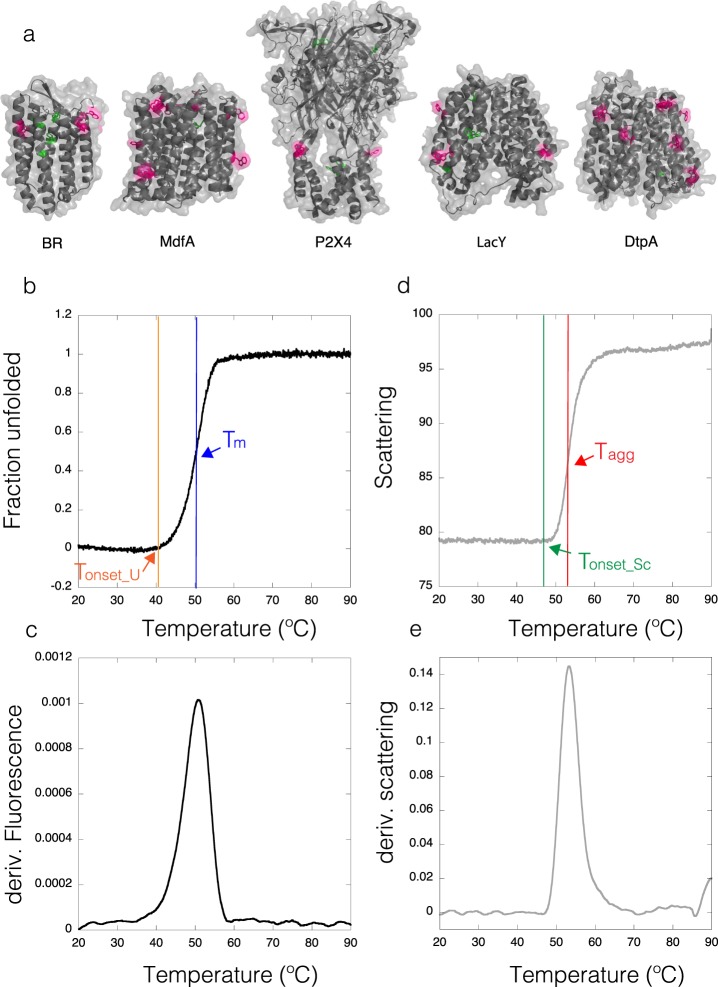
Figure 2.
Thermal denaturation of IMPs. (a) Cartoon representations for Halobacterium salinarum Bacteriorhodopsin (BR) PDB:4XXJ, peptide transporter DtpA (DtpA) PDB: 6GS1, multidrug transporter MdfA (MdfA) PDB:4ZP0, human P2X4 ion channel (P2X4) based on PDB:4DW1 and lactose permease (LacY) PDB:1PV6. Surface exposed Trp residues are highlighted in pink while buried ones are highlighted in green. (b) Example of a thermal denaturation curve (baseline corrected) and the visualization of Tm at the inflection point and the change in the slope reporting Tonset_U. (c) First derivative of the DSF curve with the maximum reporting Tm. (d) Scattering curve and the visualization of Tagg and Tonset_Sc. (e) First derivative from the scattering curve with the maximum reporting Tagg (comparable to 50% of the molecules in an aggregated state).
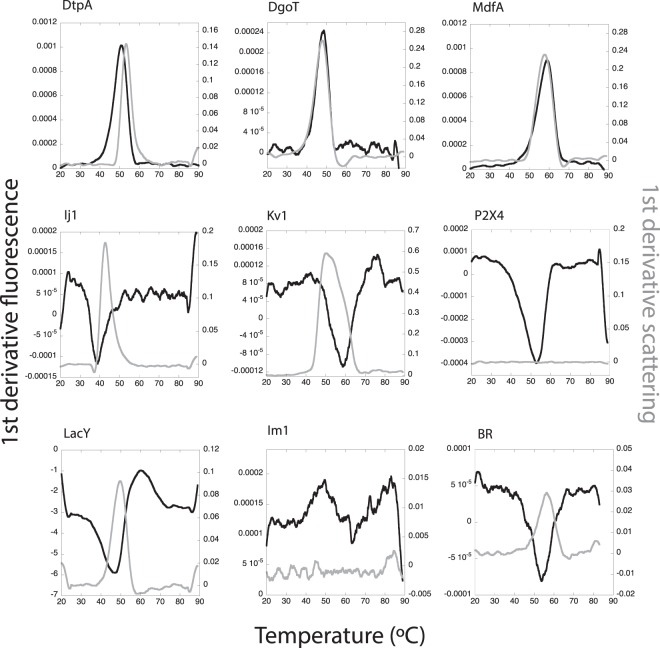
Figure 3.
Stability of all IMPs at the starting condition. First derivatives of the DSF (black) and scattering (grey) curves are shown as a function of temperature for condition A2. All proteins display clear DSF unfolding transitions with exception of Im1. Scattering data for P2X4 and Im1 did not display transitions.
Table 2.
Melting and aggregation temperatures for the standard condition (A2) before dilution to the detergent screen.
| Protein | Tm | Tagg | Tonset_U | Tonset_Sc | n DSF | n Sc |
|---|---|---|---|---|---|---|
| DtpA | 50.1 ± 0.2 | 52.9 ± 0.9 | 40.9 ± 0.2 | 46.0 ± 1.4 | 2 | 2 |
| DgoT | 48.1 ± 0.01 | 47.6 ± 0.1 | 40.0 ± 0.1 | 38.7 ± 0.1 | 2 | 2 |
| LacY | 46.2 ± 0.1 | 49.6 ± 0.1 | 37.0 ± 0.3 | 41.0 ± 0.1 | 1 | 2 |
| Kv1 | 59.3 ± 0.2 | 53.5 ± 0.2 | 47.6 ± 0.5 | 37.7 ± 0.3 | 2 | 2 |
| Ij1 | 38.9 ± 0.2 | 43.9 ± 0.2 | 27.2 ± 0.5 | 35.9 ± 0.2 | 2 | 2 |
| MdfA | 58.5 ± 0.1 | 57.9 ± 0.1 | 46.6 ± 0.3 | 48.0 ± 0.1 | 2 | 2 |
| P2X4 | 51.5 ± 1.4 | NA | 36.8 ± 1.0 | NA | 4 | 0 |
| BR | 54.9 ± 0.1 | 57.2 ± 0.1 | 40.3 ± 0.1 | 46.0 ± 0.1 | 4 | 4 |
Where Tm is the melting temperature and Tagg the mid-aggregation point obtained by curve fitting from the DSF and scattering curves respectively. Tonset_U relates to the change in the slope, equivalent to the temperature where 1% of protein becomes unfolded (U) or aggregated (Sc). n indicates the number of replicates. Average and standard deviation were computed from the replicates.
In parallel to the DSF measurements, static light scattering has been recorded in our Prometheus device equipped with a backscattering module45 which enables monitoring the onset of aggregation (Tonset_Sc) of the protein and the calculation of an aggregation mid-point (Tagg) (Figs 2d,e and 3, grey curves). Aggregation transitions were not detected for two proteins from the dataset (P2X4 and Im1). The absence of a scattering transition for Im1 would agree with either the hypothesis of an unfolded protein, or due to the protein concentration used because of limited material available. For the P2X4 receptor, experiments were performed at 0.15 and 0.25 mg/ml and no scattering transition could be detected for unknown reasons.
In some cases, as for DgoT, both the unfolding and scattering transitions, Tm, and Tagg, are similar (Fig. 3) reporting coupled unfolding and aggregation. In other cases, however, e.g. for Ij1, the Tm is lower than Tagg indicating that the protein starts to unfold, leading to macroscopic aggregation.
We next analyzed the behavior of the different IMPs after tenfold dilution to other detergents to see whether we are able to monitor the stabilizing and destabilizing properties of particular detergents by dilution instead of purifying the protein in individual detergents from the beginning. We have incubated the IMPs in the screening conditions for one hour. Our control experiments for addressing the equilibration of the detergent mix indicate that this time is sufficient to obtain consistent and reproducible results (Supplementary Fig. 4). To address how dilution into a new detergent condition compares to methods for detergent exchange, we have performed experiments after chromatographic detergent exchange and compared the inflection points from these DSF curves with those obtained after the 10-fold dilution (Supplementary Fig. 5). Our experiments show that dilution is a good indicator for whether the IMP would be stabilized or destabilized in the presence of the “new detergent”.
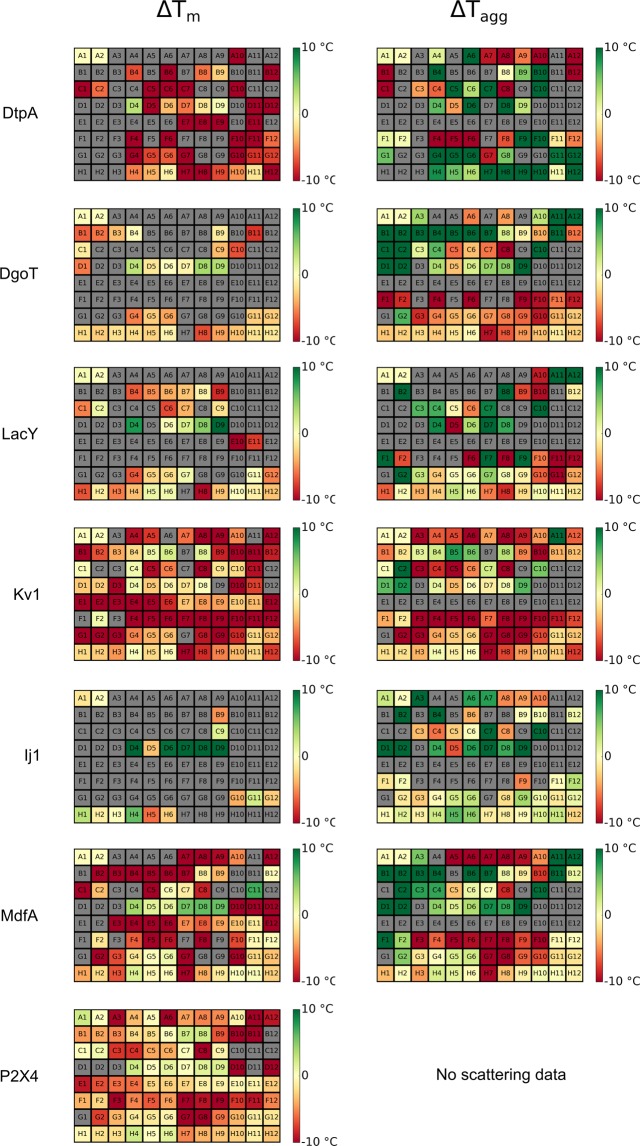
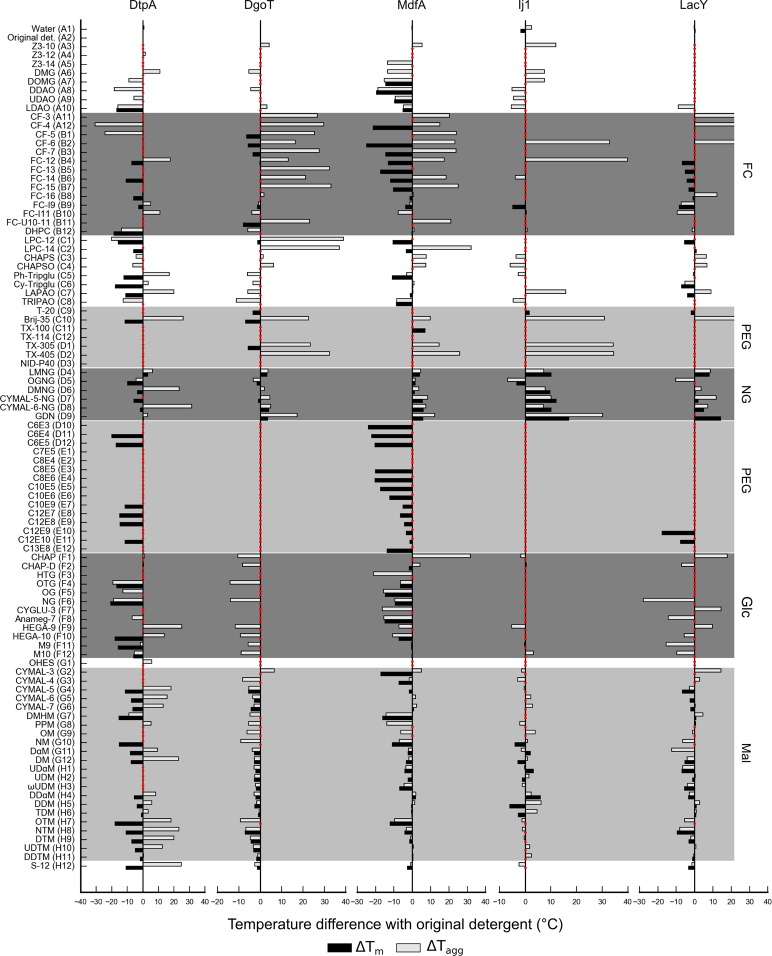
Figure 4 represents heat maps for both DSF and scattering data. The data are presented as changes in Tm and Tagg compared to the stability of the starting point (position A2). Our high-throughput stability analysis (initially) compares the observed Tm values in different detergents.
Figure 4.
Heat-maps for delta Tm and Tagg. Heat-maps for delta Tm and Tagg. Red/yellow/green wells are colored according to the difference between the sample and original detergent (screen position A2) with difference 10 °C and above being green and −10 °C and below being red. Yellow corresponds to 0 °C (no difference compared to the the original detergent). Grey wells indicate samples that could not be analyzed.
As a control experiment, we tested if the detergent screen per-se could introduce background fluorescence or scattering upon heating that could be misinterpreted as an unfolding or aggregation transition. Even though detergent fluorescence contributed up to 25% of the overall signal in some cases, none of the detergents showed a sigmoidal curve in the F350/F330 ratio in the absence of protein. However, strong scattering signal similar to aggregation transitions were detected for PEG derivatives, ethylene glycol alkyl ethers, Tween-20, NID-P40, Triton X100 and Triton X115 (Supplementary Table 2). Consequently, these conditions were removed from the subsequent analysis when it has not been possible to distinguish whether the protein or the detergent contributed to the scattering signal.
Targets stabilized by long chain maltoside detergents
All transporter proteins analyzed in this study (DtpA, DgoT, MdfA, LacY and Ij1) are stabilized by detergents from the maltose-NG family (Figs 4 and 5). Specifically, the detergent Lauryl Maltose Neopentyl Glycol (LMNG) (Table 3), represented in position D4, generated positive shifts in both Tm and Tagg for all five transporters. In addition, detergent GDN-101 (position D9) has been the second best stabilizing detergent. DtpA has been the most destabilized protein after its dilution in the detergent screen, indicating that the IMP was already very stable in DDM. Consistently, OGNG (Glucose-NG; position D5) destabilized all analyzed transporters. For MdfA and Ij1, n-Dodecyl-α-D-Maltopyranoside (α-DDM) seems to be a better stabilizer than the mostly used and cheaper version DDM (n-Dodecyl-β-D-maltoside). Indeed, alpha isomers of the maltosides display positive hits (DαM, UDαM and DDαM) for Ij1 (Fig. 5). Finally, detergents belonging to the PEG family have shown a major destabilizing effect resulting in lower Tm or curves without transitions indicating that the proteins are likely denatured at room temperature in this detergent class (Fig. 5).
Figure 5.
Bar graphs representing changes in Tm (black) and Tagg (grey) for transporters. Tm is calculated from the fluorescence ratio F350/F330 data with exception of LacY, where fluorescence at 330 nm was used. Red dots correspond to conditions that could not be fitted. Several prominent detergent families are highlighted: fos-cholines (FC), polyethyleneglycol (PEG), neopentyl-glycol (NG), glucose (Glc) and maltose (Mal) based detergents.
Table 3.
Detergent screen.
| Screen | Chemistry | Detergent | Abbreviation | CMC (mM) | Conc (mM) | Micelle size (kDa) | |
|---|---|---|---|---|---|---|---|
| A | 1 | — | Water | ||||
| A | 2 | — | Blank (for solubilizing detergent) | ||||
| A | 3 | Propanesulfonate | Anzergent 3-10 | Z3-10 | 39 | 78 | 13 |
| A | 4 | Propanesulfonate | Anzergent 3-12 | Z3-12 | 2.8 | 8.4 | 23.5 |
| A | 5 | Propanesulfonate | Anzergent 3-14 | Z3-14 | 0.2 | 10 | 39 |
| A | 6 | Dimethylglycine | n-Decyl-N,N-Dimethylglycine | DMG | 19 | 38 | |
| A | 7 | Dimethylglycine | n-Dodecyl-N,N-Dimethylglycine | DOMG | 1.5 | 4.5 | |
| A | 8 | Dimethylamine-oxide | n-Decyl-N,N-Dimethylamine-N-Oxide | DDAO | 10.5 | 21 | 1.4098 |
| A | 9 | Dimethylamine-oxide | n-Undecyl-n,n,-Dimethylamine-Oxide | UDAO | 3.2 | 9.6 | |
| A | 10 | Dimethylamine-oxide | n-Dodecyl-N,N-Dimethylamine-N-Oxide | LDAO | 1 | 3 | 17.4 |
| A | 11 | Phosphocholine | CyclofosTM-3 | CF-3 | 43 | 86 | |
| A | 12 | Phosphocholine | Cyclofos-4 | CF-4 | 14 | 28 | |
| B | 1 | Phosphocholine | Cyclofos-5 | CF-5 | 4.5 | 13.5 | |
| B | 2 | Phosphocholine | Cyclofos-6 | CF-6 | 2.68 | 8.04 | |
| B | 3 | Phosphocholine | Cyclofos-7 | CF-7 | 0.62 | 6.2 | |
| B | 4 | Phosphocholine | Fos-Choline-12 | FC-12 | 1.5 | 4.5 | 19 |
| B | 5 | Phosphocholine | Fos-Choline-13 | FC-13 | 0.75 | 7.5 | 32 |
| B | 6 | Phosphocholine | Fos-Choline-14 | FC-14 | 0.12 | 6 | 44 |
| B | 7 | Phosphocholine | Fos-Choline-15 | FC-15 | 0.07 | 7 | 52 |
| B | 8 | Phosphocholine | Fos-Choline-16 | FC-16 | 0.013 | 1.3 | 73 |
| B | 9 | Phosphocholine | Fos-Choline-ISO-9 | FC-I9 | 32 | 64 | |
| B | 10 | Phosphocholine | Fos-Choline-ISO-11 | FC-I11 | 26.6 | 53.2 | |
| B | 11 | Phosphocholine | Fos-Choline-UNSAT-11-10 | FC-U10-11 | 6.2 | 15.5 | |
| B | 12 | Phosphocholine | 1,2-Diheptanoyl-sn-Glycero-3-Phosphocholine | DHPC | 1.4 | 4.2 | |
| C | 1 | Phosphocholine_lyso | LysoPC-12 | LPC-12 | 0.7 | 7 | |
| C | 2 | Phosphocholine_lyso | LysoPC-14 | LPC-14 | 0.036 | 3.6 | |
| C | 3 | Propanesulfonate | CHAPS | CHAPS | 8 | 20 | 7 |
| C | 4 | Propanesulfonate | CHAPSO | CHAPSO | 8 | 20 | 7 |
| C | 5 | Glycotripod | Ph-Tripglu | Ph-Tripglu | 3.6 | 10.8 | |
| C | 6 | Glycotripod | Cy-Tripglu | Cy-Tripglu | 1.8 | 5.4 | |
| C | 7 | Dimethylamine-oxide | LAPAO | LAPAO | 1.6 | 4.8 | 37.8 |
| C | 8 | Dimethylamine-oxide | Tripao | TRIPAO | 4.5 | 13.5 | |
| C | 9 | Polyoxyethylene | Anapoe-20 (Tween 20) | T-20 | 0.059 | 5.9 | |
| C | 10 | Polyoxyethylene | Anapoe-35 (Brij-35) | Brij-35 | 0.091 | 9.1 | 47.92 |
| C | 11 | Polyoxyethylene | Anapoe-X-100 | TX-100 | 0.23 | 11.5 | 80 |
| C | 12 | Polyoxyethylene | Anapoe-X-114 | TX-114 | 0.2 | 10 | |
| D | 1 | Polyoxyethylene | Anapoe-X-305 | TX-305 | 0.65 | 6.5 | |
| D | 2 | Polyoxyethylene | Anapoe-X-405 | TX-405 | 0.81 | 8.1 | |
| D | 3 | Polyoxyethylene | [Octylphenoxy]Polyethoxyethanol | NID-P40 | 0.3 | 15 | 60–90 |
| D | 4 | Maltose_NG | Lauryl Maltose Neopentyl Glycol | LMNG | 0.01 | 1 | 93 |
| D | 5 | Glucose_NG | Octyl Glucose Neopentyl Glycol | OGNG | 1.02 | 3.06 | |
| D | 6 | Maltose_NG | Decyl Maltose Neopentyl Glycol | DMNG | 0.036 | 3.6 | |
| D | 7 | Maltose_NG | CYMAL-5 Neopentyl Glycol | CYMAL-5-NG | 0.058 | 5.8 | |
| D | 8 | Maltose_NG | CYMAL-6 Neopentyl Glycol | CYMAL-6-NG | 0.02 | 2 | |
| D | 9 | Maltose_NG | GDN-101 | GDN | 0.018 | 1.8 | 80 |
| D | 10 | Polyoxyethylene | Triethylene Glycol Monohexyl Ether | C6E3 | 23 | 46 | |
| D | 11 | Polyoxyethylene | Tetraethylene Glycol Monohexyl Ether | C6E4 | 30 | 60 | |
| D | 12 | Polyoxyethylene | Pentaethylene Glycol Monohexyl Ether | C6E5 | 37 | ||
| E | 1 | Polyoxyethylene | Pentaethylene Glycol Monoheptyl Ether | C7E5 | 21 | 42 | |
| E | 2 | Polyoxyethylene | Tetraethylene Glycol Monooctyl Ether | C8E4 | 8 | 20 | 25 |
| E | 3 | Polyoxyethylene | Pentaethylene Glycol Monooctyl Ether | C8E5 | 7.1 | 17.75 | |
| E | 4 | Polyoxyethylene | Hexaethylene Glycol Monooctyl Ether | C8E6 | 10 | 25 | 13 |
| E | 5 | Polyoxyethylene | Pentaethylene Glycol Monodecyl Ether | C10E5 | 0.81 | 8.1 | 28 |
| E | 6 | Polyoxyethylene | Hexaethylene Glycol Monodecyl Ether | C10E6 | 0.9 | 9 | |
| E | 7 | Polyoxyethylene | Polyoxyethylene(9)decyl Ether | C10E9 | 1.3 | 3.9 | |
| E | 8 | Polyoxyethylene | Heptaethylene Glycol Monododecyl Ether | C12E7 | 0.069 | 6.9 | |
| E | 9 | Polyoxyethylene | Octaethylene Glycol Monododecyl Ether | C12E8 | 0.09 | 9 | 67 |
| E | 10 | Polyoxyethylene | Polyoxyethylene(9)dodecyl Ether | C12E9 | 0.05 | 5 | 83 |
| E | 11 | Polyoxyethylene | Polyoxyethylene(10)dodecyl Ether | C12E10 | 0.1 | 10 | |
| E | 12 | Polyoxyethylene | Polyoxyethylene(8)tridecyl Ether | C13E8 | 0.1 | 10 | |
| F | 1 | Gluconamidopropyl | Big CHAP | CHAP | 2.9 | 8.7 | 9 |
| F | 2 | Gluconamidopropyl | Big CHAP, Deoxy | CHAP-D | 1.4 | 4.2 | 13.5 |
| F | 3 | Thioglucopyranoside | n-Heptyl-b-D-Thioglucopyranoside | HTG | 29 | 58 | 8 |
| F | 4 | Thioglucopyranoside | n-Octyl-b-D-Thioglucopyranoside | OTG | 9 | 12.5 | |
| F | 5 | Glucopyranoside | n-Octyl-b-D-Glucopyranoside | OG | 18 | 36 | 19 |
| F | 6 | Glucopyranoside | n-Nonyl-b-D-Glucopyranoside | NG | 6.5 | 16.25 | 41 |
| F | 7 | Glucopyranoside | CYGLU-3 | CYGLU-3 | 28 | 56 | |
| F | 8 | Glucopyranoside | HECAMEG | Anameg-7 | 19.5 | 39 | 30.9 |
| F | 9 | Glucamide | Hega-9 | HEGA-9 | 39 | 78 | 1.8275 |
| F | 10 | Glucamide | Hega-10 | HEGA-10 | 7 | 17.5 | |
| F | 11 | Methylglucamide | Mega-9 | M9 | 25 | 50 | |
| F | 12 | Methylglucamide | Mega-10 | M10 | 6 | 6 | |
| G | 1 | Hydroxyethylsulfoxide | 2-Hydroxyethyloctylsulfoxide | OHES | 24.3 | 48.4 | |
| G | 2 | Maltoside | CYMAL-3 | CYMAL-3 | 30 | 60 | 3 |
| G | 3 | Maltoside | CYMAL-4 | CYMAL-4 | 7.6 | 19 | 12 |
| G | 4 | Maltoside | CYMAL-5 | CYMAL-5 | 2.4 | 7.2 | 23 |
| G | 5 | Maltoside | CYMAL-6 | CYMAL-6 | 0.56 | 5.6 | 53 |
| G | 6 | Maltoside | CYMAL-7 | CYMAL-7 | 0.19 | 9.5 | 80 |
| G | 7 | Maltoside | 2,6-Dimethyl-4-Heptyl-b-D-Maltoside | DMHM | 27.5 | 55 | |
| G | 8 | Maltoside | 2-Propyl-1-Pentyl-b-D-Maltopyranoside | PPM | 42.5 | 85 | |
| G | 9 | Maltoside | n-Octyl-b-D-Maltopyranoside | OM | 19.5 | 39 | 22 |
| G | 10 | Maltoside | n-Nonyl-b-D-Maltopyranoside | NM | 6 | 15 | 26 |
| G | 11 | Maltoside | n-Decyl-a-D-Maltopyranoside | DαM | 1.6 | 4.8 | |
| G | 12 | Maltoside | n-Decyl-b-D-Maltopyranoside | DM | 1.8 | 5.4 | 33 |
| H | 1 | Maltoside | n-Undecyl-a-D-Maltopyranoside | UdαM | 0.58 | 5.8 | |
| H | 2 | Maltoside | n-Undecyl-b-D-Maltopyranoside | UDM | 0.59 | 5.9 | 36 |
| H | 3 | Maltoside | ω-Undecylenyl-b-D-Maltopyranoside | ωUDM | 1.2 | 3.6 | |
| H | 4 | Maltoside | n-Dodecyl-a-D-Maltopyranoside | DdαM | 0.15 | 7.5 | 50 |
| H | 5 | Maltoside | n-Dodecyl-b-D-Maltopyranoside | DDM | 0.17 | 8.5 | 59.5 |
| H | 6 | Maltoside | n-Tridecyl-b-D-Maltopyranoside | TDM | 0.03 | 1.5 | 100 |
| H | 7 | Thiomaltoside | n-Octyl-b-D-Thiomaltopyranoside | OTM | 8.5 | 21.25 | |
| H | 8 | Thiomaltoside | n-Nonyl-b-D-Thiomaltopyranoside | NTM | 3.2 | 9.6 | |
| H | 9 | Thiomaltoside | n-Decyl-b-D-Thiomaltopyranoside | DTM | 0.9 | 9 | 37 |
| H | 10 | Thiomaltoside | n-Undecyl-b-D-Thiomaltopyranoside | UDTM | 0.21 | 10.5 | 55 |
| H | 11 | Thiomaltoside | n-Dodecyl-b-D-Thiomaltopyranoside | DDTM | 0.05 | 5 | 66 |
| H | 12 | Sucrose | Sucrose-12 | S-12 | 0.3 | 15 |
CMC: critical micelle concentration (mM). Conc: final concentration in the assay (mM).
Figure 6a,b show selected DSF curves (raw data) and first derivatives for MdfA in selected detergents showing different stabilization/destabilization examples. Besides general effects reported by Tm (Figs 4 and 5), this study also enabled us to detect a linear relationship between inflection points (Tm) and onset of unfolding temperatures (calculated from slopes). We have observed cases of DSF curves that exhibit a relative high Tm combined with low Tonset_U (Fig. 6). Samples displaying such behavior can be detected by a scatter plot of Tonset_U as a function of Tm. In most cases these values are linearly correlated, while samples with low Tonset_U can be identified as outliers in this analysis (Fig. 6c). In such cases Tm would not be the best reporter for overall protein stability and Tonset_U should be taken into account. Interestingly, when analyzing the scattering signal, the linear relationship between Tagg and Tonset_Sc contained no outliers (Fig. 6d).
Figure 6.
Comparison of Tm and Tonset_U as stability reporters. MdfA DSF transitions, fluorescence ratio (a) and first derivatives (b), in selected detergents: GDN-101 (green), LMNG (light green), 0.03% DDM (yellow, initial condition), LDAO and APO109 (orange), UDAO and cyclofos-6 (red), fos-choline U10-11 (grey) and Tween 20 (violet). (c) Scatter plot of Tm vs Tonset_U. The circle highlights samples with atypically low Tonset_U. (d) Scatter plot for Tagg vs Tonset_Sc.
Evaluation of fos-choline and and PEG family detergents for IMP solubilization
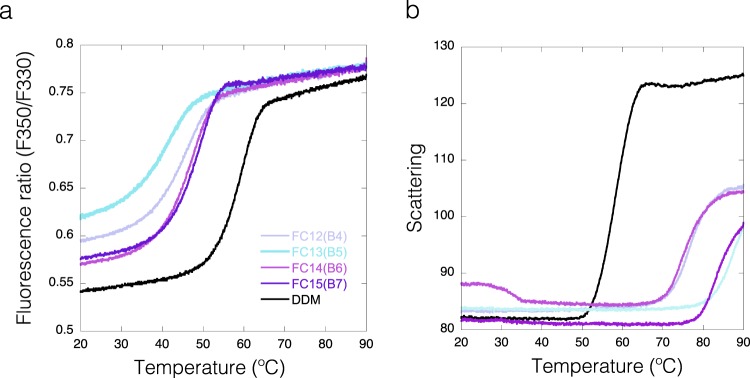
Currently it is a debate in the field whether fos-choline detergents, known for being very efficient in solubilization, have denaturing effects on membrane proteins46,47. Indeed, based on our scattering data, detergents from the fos-choline family have a general positive effect in preventing aggregation (see Tagg heatmaps in Fig. 4 and grey bars in Fig. 5). But despite its improved solubility the fluorescence data on IMPs in fos-choline detergents give a different picture on the folding status of the protein. The derived Tm of unfolding is much lower than Tagg or in many cases an unfolding transition cannot be monitored (Fig. 5 black bars, Fig. 7). This indicates that IMPs in fos-choline detergents are strongly destabilized and only represent partially folded or unfolded states. This observation is demonstrated in Fig. 7, showing that MdfA starts to aggregate in fos-choline detergents at temperatures above 70 °C while the fluorescence data show the protein is already unfolded. In summary, fos-choline detergents can efficiently solubilize unfolded IMPs. A clear example would be observing conditions B5 to B8 for DgoT on the DSF and Scattering heatmaps (Fig. 4). After dilution to Fos-Choline 13, 14, 15 and 16 detergents; DgoT did not present DSF transitions (grey wells) while the scattering profiles for these conditions showed increased solubility with late aggregation profiles (green wells).
Figure 7.
Fos-Choline enhances solubility of an unfolded MdfA. (a) DSF curves for MdfA in DDM (black) and in different Fos-choline detergents (Fos-Choline 12, 13, 14, 15 and 16; in the violet-cyan range). (b) Scattering curves, same colour code as for A.
In addition, our assay shows that dilution to PEG family detergents retrieved destabilized or unfolded proteins displaying no DSF or Scattering transitions (Fig. 5). Remarkably, some members of this family (conditions C10, D1 and D2) enhanced the solubility of the IMPs while highly destabilizing them similarly to what was previously described for the fos-choline family (Figs 4 and 5).
Correlation between IMP stability and micelle size
It has been previously addressed that smaller detergent micelle sizes are better suited for crystallographic studies of IMPs23,48, but at the same time the change to detergents with shorter acyl chains destabilizes IMPs and often leads to precipitation. Therefore, it is crucial to balance the length of the acyl chain of the used detergent with the stability of membrane proteins. We have calculated Spearman correlation coefficients to test if there is a monotonic relationship between the melting or aggregation temperatures and the size of the micelle (Table 4 and Supplementary Fig. 6). This coefficient does not make any assumptions about the underlying dependence between the variables, but shows how consistently one variable increases as the other one increases or decreases49. The number of data points used to calculate these correlation coefficients depends on the available information on micelle size in kDa for that particular detergent (Table 3) and reliable unfolding data for a given IMP in that detergent. For 5 out of 9 proteins the coefficient was >0.7, so the reported Tm is predominantly increasing with the micelle size. For 3 proteins the coefficient was around 0.4, which implies a weaker though still valid correlation. For 1 protein (due to incomplete dataset) no correlation was found. Overall, our results indicate that there is a positive correlation between the size of the detergent micelle and the thermodynamic stability of the IMP.
Table 4.
Correlation between micellar size and Tm or Tagg.
| Sample | Readout | n | Spearman’s ρ |
|---|---|---|---|
| DtpA | Ratio | 21 | 0.70 |
| Scattering | 27 | 0.24 | |
| DgoT | Ratio | 15 | 0.43 |
| Scattering | 33 | 0.25 | |
| LacY | F330 | 8 | 0.00 |
| Scattering | 27 | -0.04 | |
| Kv1 | Ratio | 38 | 0.46 |
| Scattering | 37 | 0.66 | |
| Ij1 | Ratio | 8 | 0.45 |
| Scattering | 29 | 0.65 | |
| Im1 | Ratio | 9 | 0.78 |
| MdfA | Ratio | 33 | 0.74 |
| Scattering | 36 | 0.20 | |
| P2X4 | Ratio | 42 | 0.80 |
| BR | F330 | 28 | 0.84 |
| Scattering | 28 | 0.73 |
Where n in the number of data points used to calculate the correlation coefficient. Coefficients approaching zero show no correlation between variables while those approaching 1 indicate a positive correlation (Y values increase as the X values increase).
Implications for sample optimization
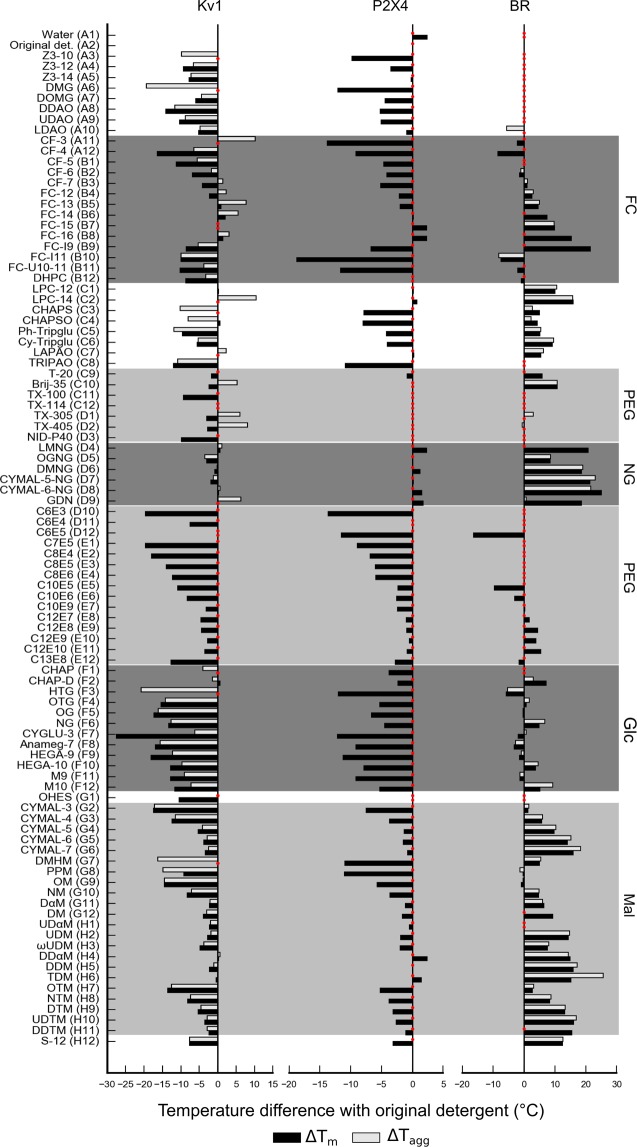
Here we compare cases of IMPs that show improved, intermediate or poor general stabilization upon dilution in the detergent screen. How the protein will behave in the assay mainly depends on the stability in the starting condition. Kv1 is already stable in DDM and none of the screened detergents stabilizes the protein further (Fig. 8). Furthermore, the plot Tm vs Tonset_U does not show any outliers (Supplementary Fig. 7a). P2X4 shows positive hits for Maltose-NG detergents, DDαM and TDM, suggesting that an exchange of detergent for further downstream processing could be beneficial. BR (only IMP originally kept in OG) presents the largest positive changes in Tm after dilution to the detergent screen (Fig. 8). It is known that BR is less stable in OG compared to DDM, however OG is a good choice for crystallization due to its small micelle size. Once the starting condition of BR is changed to DDM, the result of the detergent screening approach changes, displaying a general destabilization effect (Supplementary Fig. 7b).
Figure 8.
Bar graphs representing changes in Tm (black) and Tagg (grey) for Kv1, P2X4 and BR.Tm is calculated from the fluorescence ratio F350/F330 data with exception of BR, where fluorescence at 330 nm was used. Red dots correspond to conditions that could not be fitted properly. Several prominent detergent families are highlighted: fos-cholines (FC), polyethyleneglycol (PEG), neopentyl-glycol (NG), glucose (Glc) and maltose (Mal) based detergents. Note that the concentration used of P2X4 was not high enough to record scattering data.
An interesting observation is the case of Im1 that does not have a transition of unfolding detectable by DSF (Fig. 3), most likely because the protein is misfolded. However, our assay shows some conditions displaying DSF transitions after detergent dilution (see Supplementary Fig. 8). These results suggest that a detergent substitution in an early stage of the purification could be beneficial. However, the sample so far has not been suitable for performing structural studies and different strategies for protein expression and solubilization need to be considered.
Discussion
In this study we have measured the thermal stability of nine IMPs against a panel of 94 detergents by monitoring the intrinsic fluorescence of each IMP and its scattering properties. We conclude that the Tm calculated from the DSF measurements is a more stringent selection criterion than Tagg from the recorded scattering curves. We observe that in a number of cases, especially for PEG and fos-choline detergents, stabilization hits for Tagg are not accompanied by a higher Tm. As a general observation, to obtain reliable scattering data higher protein concentrations (>0.15 mg/ml) are required, while for most of the IMPs tested, the fluorescence ratio obtained at 0.1 mg/ml produced reliable and reproducible unfolding data. In addition, whenever the ratio (F350/F330) cannot be used for stability determination, the fluorescence at 330 nm or 350 nm could be used to extract the thermodynamic parameters. Trp residues are often present in transmembrane domains of IMPs, and are excellent fluorophores in reporting changes in the environment upon changes in the folding state. Therefore, no additional dyes as used in other applications are necessary. In addition, analyzing Tm vs. Tonset_U could reveal conditions with similar melting temperatures but an earlier onset of unfolding indicates reduced stability.
During sample preparation, both Tm and Tagg should be taken into account and proteins should be kept below the lowest temperature at which anything deleterious happens.
Our workflow starts with a membrane protein that has been purified in DDM that is subsequently 10-fold diluted into the test detergent buffer. The fact that our workflow does not require additional chromatographic or filtration detergent exchange allows faster and less expensive screening. However, the dilution step brings residual DDM (bound to the IMP) that remains in the assay. Bound residual DDM could have different stabilizing or destabilizing effects on the IMP, which are translated into small perturbances in the observed Tm. However, the scope of the assay is not to obtain the absolute melting temperatures in a given detergent but a reference system to compare detergents. Our results suggest that it is not necessary to go through a complete detergent exchange but a 10-fold dilution into a new detergent is sufficient to monitor the influence of other detergents on the protein stability. Our screening protocol allowed us to measure the stability and solubility of IMPs in a high-throughput manner and to determine the detergent or class of detergents that could be used (or should be avoided) when working with each system. Residual detergent concentration from the starting condition can affect the output of the assay. Therefore, we recommend researches to perform experiments considering the residual detergent form the dilution present in the assay to determine what would be the best working condition for each IMP. The ideal detergent would solubilize the IMP from the membrane in its native conformation and form a stable complex throughout purification. However, it has been discussed whether membrane protein stability is an intrinsic feature of the membrane protein and not detergent specific23. This would mean a stable IMP would be robust in more detergents compared to an unstable IMP. In our study, we have observed that when a membrane protein is quite stable in DDM it would be most likely destabilized by diluting it in many others. In contrast, a stable protein like BR could be further stabilized by other detergents when the starting condition has not been as favorable.
In our assay we detected detergents with general stabilizing effects. DDM, empirically the first chosen detergent in most labs working on IMPs, seemed to be a very good starting condition for extraction, solubilization and purification of folded membrane proteins. Also, LMNG seems to be a good stabilizer for all the cases tested in our study, and the maltose-NG family stabilized all transporters tested. For the detergent screen used, the diversity of the tested detergent chemistries showed that members of the PEG family do not improve the stabilization of our selected targets. We have now reported cases of detergents that contribute to the scattering signal and that fos-choline detergents keep unfolded IMPs in solution.
Our assay also quantified the correlation between thermal stability of an IMP in a detergent with its micelle size. Regarding crystallization, shorter chain detergents are preferred as they allow for better crystal packing and better diffracting crystals50. The goal is to find the shortest possible detergent that does not cause the protein to unfold16,17. Although shorter detergents would be more suitable for crystallography, a more stabilized protein in longer detergents could be beneficial for performing activity tests or reconstitution into scaffold systems such as nanodiscs or SapNPs. Lipids play a major role in preserving the native environment of membrane proteins. It has been shown for example that basal ATPase activity in ABC transporters purified in DDM micelles is lost but regained when reconstituted in liposomes or lipid bilayers22. It is of general knowledge that eukaryotic membrane proteins are on average less stable than their bacterial homologues after extraction with detergents. This is probably mainly due to delipidation. In our case, P2X4 is quite stable in DDM (Tm around 51 °C) however it could only be purified in the presence of 0.005% cholesteryl hemisuccinate (CHS). Therefore, we are working with a mix of detergent and CHS (see Material and Methods).
Finally, we propose that the described assay should be applied as an iterative process for membrane protein stabilization during purification. Once the IMP is solubilized in an appropriate detergent, addition of lipids could play a critical role in further stabilization. We propose to start with a first trial purification in DDM for a new IMP and then apply the described protocol with the most commonly used detergents (no need to screen all 94 detergents). The goal is to obtain a properly folded protein with sufficient stability to succeed in structural and functional studies.
Methods
Protein constructs
DgoT and DtpA
The DgoT and DtpA genes were cloned into the pNIC-CHTF vector (Addgene plasmid-ID Plasmid #26105)51 using LIC cloning52,53.
DgoT protein sequence including tags
MDIPVNAAKPGRRRYLTLVMIFITVVICYVDRANLAVASAHIQEEFGITKAEMGYVFSAFAWLYTLCQIPGGWFLDRVGSRVTYFIAIFGWSVATLFQGFATGLMSLIGLRAITGIFEAPAFPTNNRMVTSWFPEHERASAVGFYTSGQFVGLAFLTPLLIWIQEMLSWHWVFIVTGGIGIIWSLIWFKVYQPPRLTKGISKAELDYIRDGGGLVDGDAPVKKEARQPLTAKDWKLVFHRKLIGVYLGQFAVASTLWFFLTWFPNYLTQEKGITALKAGFMTTVPFLAAFVGVLLSGWVADLLVRKGFSLGFARKTPIICGLLISTCIMGANYTNDPMMIMCLMALAFFGNGFASITWSLVSSLAPMRLIGLTGGVFNFAGGLGGITVPLVVGYLAQGYGFAPALVYISAVALIGALSYILLVGDVKRVGAENLYFQ\SHHHHHHDYKDDDDK.
DtpA protein sequence including tags
MSTANQKPTESVSLNAFKQPKAFYLIFSIELWERFGYYGLQGIMAVYLVKQLGMSEADSITLFSSFSALVYGLVAIGGWLGDKVLGTKRVIMLGAIVLAIGYALVAWSGHDAGIVYMGMAAIAVGNGLFKANPSSLLSTCYEKNDPRLDGAFTMYYMSVNIGSFFSMIATPWLAAKYGWSVAFALSVVGLLITIVNFAFCQRWVKQYGSKPDFEPINYRNLLLTIIGVVALIAIATWLLHNQEVARMALGVVAFGIVVIFGKEAFAMKGAARRKMIVAFILMLEAIIFFVLYSQMPTSLNFFAIRNVEHSILGLAVEPEQYQALNPFWIIIGSPILAAIYNKMGDTLPMPTKFAIGMVMCSGAFLILPLGAKFASDAGIVSVSWLVASYGLQSIGELMISGLGLAMVAQLVPQRLMGFIMGSWFLTTAGANLIGGYVAGMMAVPDNVTDPLMSLEVYGRVFLQIGVATAVIAVLMLLTAPKLHRMTQDDAADKAAKAAVAAENLYFQ\SHHHHHHDYKDDDDK.
MdfA
The mdfA gene (NCBI GenBank accession No. AAC73929.1 for E. coli K-12 substrain MG1655) was cloned upstream of the TEV cleavage-site sequence (TEVcs) of the pWaldo-GFPe vector54 via the XhoI and KpnI restriction sites, allowing expression of the MdfA-(TEVcs)-GFP-His8 fusion protein. The cloned vector was a kind gift from Mikio Tanabe from the KEK/High Energy Accelerator Research Organization in Japan.
MdfA protein sequence
MQNKLASGARLGRQALLFPLCLVLYEFSTYIGNDMRQPGMLENVEQYQAGIEWVPTSMNAYLAGGMFIQWLLGPLSDRIGRRPVMLAGVVWFIVTCLAILLAQNIEQFTLLRFLHGISLCFIGAVGYDAIQESFEEAVCIKITALMANVALIAPLLGPLVGASWIHVLPWEGMFVLFAALAAISFFGLQRAMPETATRIGEKLSLKELGRDYKLVLKNGRFVAGALALGFLSLPLLAWIAQSPIIIITGEQLSSYEYGLLQVPIFGALIAGNLLLARLTSRRTVRSLIIMGGWPIMIGLLVAAAATVISSHAYLWMTAGLSIYAFGIGLANAGLVRLTLFASVMSKGTVSAAMGMLQMLIFTVGIEISKHAWLNGGNGLFNLFNLVNGILLLSLMVIFLKDKQMGNSHEG.
Kv1
The Kv1 gene (NCBI GenBank accession No. PA3789) was cloned into the NdeI - BamHI sites of the pnEK–vH expression vector55 as a fusion protein with an N-terminal His6-tag.
Ij1
The Ij1 gene (NCBI GenBank accession No. CAQ31984.1) was cloned into the NcoI - XhoI sites of the pET28a-TEV-His vector (Novagen) as a fusion protein with a C-terminal His6-tag.
P2X4
The P2X4 orf was cloned into the EcoRI - BamHI sites of the pOET2 vector (Oxford Expression Technologies).
P2X4 protein sequence
MAGCCSALAAFLFEYDTPRIVLIRSRKVGLMNRAVQLLILAYVIGWVFVWEKGYQETDSVVSSVTTKVKGVAVTRTSKLGFRIWDVADYVIPAQEENSLFVMTNVILTMNQTQGLCPEIPDATTVCKSDASCTAGSAGTHSNGVSTGRCVAFNGSVKTCEVAAWCPVEDDTHVPQPAFLKAAERFTLLVKNNIWYPKFNFSKRNILPNITTTYLKSCIYDAKTDPFCPIFRLGKIVENAGHSFQDMAVEGGIMGIQVNWDCNLDRAASLCLPRYSFRRLDTRDVEHNVSPGYNFRFAKYYRDLAGNTQRTLIKAYGIRFDIIVFGKAGKFDIIPTMINIGSGLALLGMATVLCDIIVLYCMKKRLYYREKKYKYVEDYEQGLASELDQGSSGTETSQVAPA.
BR protein sequence
mlellptavegvsqaqitgrpewiwlalgtalmglgtlyflvkgmgvsdpdakkfyaittlvpaiaftmylsmllgygltmvpfggeqnpiywaryadwlfttplllldlallvdadqgtilalvgadgimigtglvgaltkvysyrfvwwaistaamlyilyvlffgftskaesmrpevastfkvlrnvtvvlwsaypvvwligsegagivplnietllfmvldvsakv gfglillrsraifgeaeapepsagdgaaatsd.
LacY
The LacY gene was cloned into the pWaldo-GFP plasmid (a pET28 derived system) downstream the TEV site as a GFP-his tagged fusion.
LacY protein sequence
MYYLKNTNFWMFGLFFFFYFFIMGAYFPFFPIWLHDINHISKSDTGIIFAAISLFSLLFQ PLFGLLSDKLGLRKYLLWIITGMLVMFAPFFIFIFGPLLQYNILVGSIVGGIYLGFCFNA GAPAVEAFIEKVSRRSNFEFGRARMFGCVGWALCASIVGIMFTINNQFVFWLGSGCALILAVLLFFAKTDAPSSATVANAVGANHSAFSLKLALELFRQPKLWFLSLYVIGVSCTYDVFDQQFANFFTSFFATGEQGTRVFGYVTTMGELLNASIMFFAPLIINRIGGKNALLLAGTIMSVRIIGSSFATSALEVVILKTLHMFEVPFLLVGCFKYITSQFEVRFSATIYLVCFCFFKQLAMIFMSVLAGNMYESIGFQGAYLVLGLVALGFTLISVFTLSGPGPLSLLRRQVNEVA.
Im1
The Im1 gene (NCBI GenBank accession No. EF1051) was cloned into a pET28-derived vector with a C-terminal His8-tag.
Protein expression
The DgoT, DtpA and MdfA proteins were expressed in E. coli C41(DE3) cells. For protein expression, cells were grown in terrific broth (TB) media supplemented with 30 μg/ml kanamycin. Cultures were grown at 37 °C and protein expression was induced with 0.2 mM IPTG at an OD600nm of 0.6–0.8. After induction, culture growth continued at 18 °C for 16–18 hours. Cells were harvested by centrifugation (9379 rcf, 15 minutes, 4 °C using a JLA 8.1 rotor in an Avanti JXN-26 centrifuge, Beckman Coulter), and the pellet was stored at −20 °C until further use. Typically, around 20 g of biomass (wet weight) per litre of culture were obtained.
The Kv1 protein was expressed in E. coli C43(DE3) cells. Cultures were grown in TB media at 37 °C until an OD600nm of 2, cooled down to 20 °C and induced with 0.1 mM IPTG. After induction, culture growth continued at 20 °C for over-night expression. Cells were harvested by centrifuging (4000 rcf, 25 min, 6 °C) and the pellet was stored at −20 °C until further usage.
The Ij1 protein was expressed in E. coli LEMO21(DE3) cells56. Cultures were grown in ZY media with 0.3 mM rhamnose at 37 °C until an OD600nm of 1, cooled down to 20 °C and induced with 0.1 mM IPTG. After induction, culture growth continued at 20 °C for over-night expression. Cells were harvested by centrifuging (5000 rcf, 25 min, 6 °C) and the pellet was stored at −20 °C until further usage.
The P2X4 protein was expressed in Trichoplusia ni (T.ni) insect cells. Cells with expressed Rho-tagged P2X4 were obtained from Cube Biotech GmbH.
The LacY and Im1 proteins were expressed in E. coli C43(DE3) cells in LB media; 20 mL of inoculation culture were added to 1 L flask LB media with kanamycin and incubated at 37 °C, shaking at 260 rpm. When the OD600nm reached 0.5 the temperature was reduced to 25 °C and induced with 0.4 mM IPTG. The culture was left growing for further 16 hours.
Protein purification
DgoT, DtpA and MdfA
Purification was carried out similarly as described for other transporters33–35. The cell pellet was resuspended in lysis buffer (20 mM NaP at pH 7.5, 300 mM NaCl, 5% (v/v) glycerol, 15 mM imidazole, 5 ml of lysis buffer per gram of wet weight pellet), supplemented with lysozyme (1 mg/ml final concentration), DNase (5 units/ml) and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP). The cells were lysed by three cycles using an EmulsiFlex-C3 (Avestin) at 10.000–15.000 psi. Recovered material was centrifuged to remove non-lysed cells (9379 rcf, 15 minutes, 4 °C using a JLA 8.1 rotor in an Avanti JXN-26 centrifuge, Beckman Coulter and the supernatant was subjected to ultracentrifugation to collect the total membrane fraction (95834 rcf, 1 hour, 4 °C using a 45 Ti rotor in an Optima XE-90 centrifuge, Beckman Coulter). Membranes were resuspended in lysis buffer supplemented with protease inhibitors (one tablet per 100 ml lysis buffer, Roche), and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), and solubilized by adding 1% n-dodecyl-β-D-maltoside (DDM) detergent. Solubilized protein was first purified by immobilized-metal affinity chromatography (IMAC) on a gravity column. The beads were pre-equilibrated in Wash 1 buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 5% (v/v) glycerol, 15 mM imidazole 0.5 mM TCEP, 0.03% DDM) and incubated with the solubilized membrane proteins for one hour at 4 °C on a rotating wheel. Loaded beads were extensively washed with wash buffer with increasing imidazole concentrations (20 mM HEPES at pH 7.5, 300 mM NaCl, 5% glycerol, 15–40 mM imidazole, 0.5 mM TCEP, 0.03% DDM). The protein was eluted from the column with a buffer containing high imidazole concentration (20 mM HEPES at pH 7.5, 150 mM NaCl, 5% glycerol, 300 mM imidazole, 0.5 mM TCEP, 0.03% DDM) and combined with 1 ml of TEV protease at 1 mg/ml to cleave the His-tag during dialysis overnight at 4 °C. Typically, 1 mg of TEV protease was sufficient to cleave the tag from the purified protein from 3 liters of culture. The dialysis buffer contained 20 mM HEPES at pH 7.5, 150 mM NaCl, 5% glycerol, 0.5 mM TCEP, 0.03% DDM. Cleaved protein was recovered by negative IMAC. A second purification step was done by size-exclusion chromatography (SEC). The cleaved protein was concentrated to 5 ml using a 100 kDa concentrator (Corning® Spin-X® UF concentrators) and run on an ÄKTA Pure system (GE Healthcare Life Sciences), using a HiLoad 16/600 Superdex 200 (S200) column (GE Healthcare Life Sciences) in SEC buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 5% glycerol, 0.5 mM TCEP, 0.03% DDM). Fractions containing the protein were pooled and concentrated to 5 mg/ml, flash frozen and stored at −80 °C until further use.
Kv1
The cell pellet was resuspended in lysis buffer (30 mM TrisHCL pH 7.5; 300 mM NaCl; 10% glycerol, 2 mM MgCl2, 2 µg/ml DNase I; 200 µg/ml lysozyme). The cells were lysed by passing three times through an EmulsiFlex-C3 (Avestin); the lysate was centrifuged (25000 × g; 30 min; 6 °C) supernatant was taken and spun again at 150000 × g; 1 h; 6 °C. The membrane pellets were collected and resuspended in high salt buffer (30 mM TrisHCl pH 7.5 800 mM NaCl; 10% glycerol) and ultracentrifuged (150000 rcf; 1:35 h; 6 °C). Membrane pellets were resuspended in 30 mM TrisHCl pH 7.5; 300 mM NaCl, 10% glycerol, flash frozen in liquid nitrogen and stored at −20 °C. Protein was solubilized by adding 1% DDM and followed by centrifugation at 50000 rcf for 30 min at 6 °C. Solubilized protein was first purified by a nickel-IMAC beads in a gravity column; washed with 10 mM and 30 mM imidazole in 30 mM TrisHCl pH 7.5; 300 mM NaCl, 10% glycerol; 0.03% DDM, and eluted with 250 mM imidazole. Elution fractions containing the protein were concentrated and loaded onto a gel filtration 10/300 S200 column in 20 mM HEPES pH 7.4, 150 mM NaCl and 0.03% DDM. Fractions containing the protein (5 ml) were concentrated in an Amicon® Ultra 4 ml centrifugal filter devices (50 kDa MWCO) to 0.5 ml, final concentration of 9 mg/ml and stored at 4 °C.
Ij1
The cell pellet was resuspended in lysis buffer (30 mM TrisHCL pH 7.5; 200 mM NaCl; 5% glycerol, 2 mM MgCl2, 2 µg/mL DNase I; 200 µg/ml lysozyme). The cells were lysed by passing three times through an EmulsiFlex-C3 (Avestin); the lysate was centrifuged (25000 × g; 30 min; 6 °C) supernatant was taken and spun again at 150000 × g; 1 h; 6 °C. The membrane pellets resuspended in buffer (30 mM TrisHCl pH 7.5; 200 mM NaCl, 5% glycerol), flash frozen in liquid nitrogen and stored at −20 °C. Protein was solubilized by adding 1% DDM and followed by centrifugation at 50000 rcf for 30 min at 6 °C. Solubilized protein was first purified by nickel-IMAC in a gravity column; washed with 40 mM imidazole in 30 mM TrisHCl pH 7.5; 200 mM NaCl; 5% glycerol; 0.03% DDM and eluted with 300 mM imidazole. Eluted fractions were combined with 1 mg of TEV protease/mg of protein to perform the His-tag cleavage during dialysis overnight at 4 °C. Protein solution was added to gravity nickel-NTA column to perform a reverse IMAC; the flow-through was collected, concentrated and loaded onto a gel filtration 10/300 S200 column in 20 mM HEPES pH 7.4, 150 mM NaCl and 0.03% DDM. Fractions containing the protein were concentrated in an Amicon® Ultra 4 ml centrifugal filter devices (100 kDa MWCO) to a final concentration of 2.5 mg/ml and stored at 4 °C.
P2X4
Protein was purified using PureCube Rho1D4 MagBeads following manufacturer’s protocol. Membrane solubilization was achieved by using 2% DDM. Elution fractions were concentrated and loaded onto a gel filtration HiLoad 16/600 Superdex 200 pg column in SEC buffer (150 mM NaCl, 50 mM Tris, 0.05% (w/v) DDM, 0,005% (w/v) CHS Anatrace (CAS# 102601-49-0), 5% v/v glycerol, pH 7). Fractions containing protein (6 ml) were pooled and concentrated in a Millipore device (30 kD MWCO) to 250 μl, final concentration of 0.95 mg/ml and flash frozen using liquid nitrogen. Note that the theoretical MW for P2X4 is around 44.5 kDa and the other bands correspond to monomer/dimer/trimer on the SDS PAGE (see Supplementary Fig. 1).
LacY
The resuspended membranes were solubilized in 1x PBS, 150 mM NaCl, 1% DDM and incubated at 4 °C for 2 hours, followed by centrifugation at 100,000 rcf at 4 °C for 45 min. The supernatant was mixed with Ni-NTA (IMAC) resin pre-equilibrated in x1 PBS, 150 mM NaCl, 0.1% DDM and 10 mM imidazole. The protein was eluted with an elution buffer containing a final concentration of 250 mM imidazole. After the first IMAC, the eluted protein was dialyzed overnight at 4 °C against a large volume of buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.03% DDM). An equimolar amount of TEV protease was added for a complete overnight digest at 4 °C. The dialyzed fraction was submitted to a reverse-IMAC to remove the His-tagged TEV protease, cleaved GFP-His8-tag and co-eluting contaminating proteins. A final SEC was run in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.03% DDM. The protein was concentrated using a 100-kDa centrifugal concentrator (Vivaspin) in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.03% DDM and stored at −80 °C (after snap-freeze in liquid nitrogen) at 10 mg/ml. Note that the theoretical MW for LacY is around 47 kDa but this construct runs on the SDS gels between 38 and 28 kDa. Two bands are always visible with the upper band corresponding to the dimer (see Supplementary Fig. 1).
Im1
The resuspended membranes were solubilized in 1x PBS, 250 mM NaCl, 1% DDM and incubated at 4 °C for 2 hours, followed by centrifugation at 100,000 rcf at 4 °C for 45 min. The supernatant was mixed with Ni-NTA (IMAC) resin in x1 PBS, 250 mM NaCl, 0.2% DDM and 10 mM imidazole. The protein was eluted with an elution buffer containing a final concentration of 250 mM imidazole. A final SEC was run in 20 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.02% DDM. The protein was concentrated using a 100-kDa centrifugal concentrator (Vivaspin) in 20 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.02% DDM and stored at −80 °C (after snap-freeze in liquid nitrogen) at 14 mg/ml.
Detergent exchange
Purified DtpA (in 0.03% DDM) was loaded onto SD200 Increase 10/300 GL gel filtration column in the following SEC buffer: 20 mM HEPES 7.5, 50 mM NaCl, 5% glycerol, 0.01% LMNG, 0.5 mM TCEP.
Screen sample preparation
Each condition from the detergent screen “Analytic Selector Kit” (Anatrace, product number AL-SEL) (12.5 μl) was pipetted into a PCR plate and mixed with 10 μl of 2x protein buffer (without any detergent and glycerol). Protein stock (2.5 μl) was added to obtain a final protein concentration in the range of 0.1–0.5 mg/ml and thoroughly mixed by pipetting. The plate was briefly spun down in a swing-bucket centrifuge and incubated for 1 hour at room temperature prior to the thermal denaturation assay. Fluorescence and scattering of detergents without proteins were measured in 50 mM Tris, pH 7.5 and 200 mM NaCl (see Supplementary Table 2).
Thermal denaturation assay
Each sample was used to fill two standard grade NanoDSF capillaries (Nanotemper) and loaded into a Prometheus NT.48 device (Nanotemper) controlled by PR.ThermControl (version 2.1.2). Excitation power was pre-adjusted to get fluorescence readings above 2000 RFU for F330 and F350, and samples were heated from 20 °C to 90 °C with a slope of 1 °C/min. An XLSX file with “processed data” was exported from PR. ThermControl and used for further analysis.
Data analysis
The fluorescent readouts used in the screening (F330, F350 and F350/F330 Ratio) are redundant, and in most cases analyzing the Ratio data was sufficient. In case of LacY F330 was chosen based on the higher signal strength. In case of BR an unfolding transition was observed in all readouts, however, the Ratio data appeared noisier and F330 was used instead.
A two-state unfolding model is only applicable for reversible unfolding events not compatible with large molecular weight IMPs. Therefore the fitted parameters described below as Tm, Tagg and Tonset are only apparent rather than absolute thermodynamic parameters. Raw curves were fit to the equation described by Santoro and Bolen56 with Gibbs free energy expressed as a function of temperature57, and protein heat capacity change assumed to be zero:
where ΔG is the Gibbs free energy of unfolding, ΔHm is the apparent enthalpy of unfolding at Tm and Tm is the melting temperature. Tonset_U was calculated from ΔHm as follows:
where R is the universal gas constant.
We decided to compare samples in terms of Tm and Tonset rather than ΔHm and Tm, because in the former case both curve characteristics have the same dimensionality (temperature degrees), while ΔHm is in J/mol.
For scattering curves the Gibbs free energy function was substituted to a descriptive equation, which represents the transition with two values: Tagg – aggregation temperature (50% molecules aggregated) and Tonset – onset temperature (1% molecules aggregated):
Covariance matrix of the fit parameters was used to calculate standard deviation of Tm and Tagg of individual samples and if the value exceeded 0.5 K the sample was discarded. Fit parameters for the duplicates were averaged to obtain standard deviation.
Derivative curves were obtained from experimental curves by applying a Savtizky-Golay filter with 4-order polynomial and 10 °C window size.
For correlation analysis between micelle size and Tm or Tagg we chose Spearman’s rank correlation coefficient (ρ). This coefficient quantifies how well the relationship between two can be described by a monotonic function, but does not make any assumptions about the function itself. Computation was performed using stats.spearmanr function from scipy57.
Replicates
The standard deviation for the calculated Tm and Tagg for six replicate experiments is usually below 0.3 °C due to the low intrinsic error of the nanoDSF measurements and the accuracy of replicates of dilution. As demonstrated by Malo et al.58, measuring in a duplicate reduces the imprecision by 29%. In our experience working with an n = 2 produces reliable and consistent results and would be a good compromise when dealing with scarce material.
Software
Data processing and visualization was done in MoltenProt59, which is written in Python60 using the following modules: scipy57, numpy61, pandas62, matplotlib63.
Chemicals
Selector detergent screen (Anatrace). See Table 1 and Supplementary Information.
Bacteriorhodopsin (25 mg/ml in 250 mM Na/K phosphate buffer, pH 6.5, 1% OG) was obtained from Cube Biotech.
Supplementary information
Acknowledgements
We acknowledge technical support by the SPC facility at EMBL Hamburg. We thank Ioanna Maria Nemtanu for assistance with the DSF measurements. K.V., I.J. and H.T. acknowledge support by the excellence cluster ‘The Hamburg Centre for Ultrafast Imaging - Structure, Dynamics and Control of Matter at the Atomic Scale’ of the Deutsche Forschungsgemeinschaft (DFG EXC 1074). This work was also funded by the Horizon2020 program of the European Union (iNEXT grant, project No. 653706), the Swedish Research Council (grant number 621-2013-5905), a grant from the German-Israeli Foundation (GIF Grant No: G-1288-207.9/2015) and a grant from the UK’s Department for Business, Energy and Industrial Strategy the UK National Physical Laboratory.
Author Contributions
Conceptualization, H.T., C.L. and M.G.A.; Methodology, V.K., K.B., K.V., I.J., U.K.T.S., I.M., C.G. and M.G.A.; Investigation, V.K., H.T., C.L. and M.G.A.; Writing—Original Draft, M.G.A.; Writing—Review, V.K., H.T., C.L. and M.G.A.; Editing, all authors; Resources and Funding Acquisition, T.M., C.L., H.T., I.M., J.L. and M.G.A.; Supervision, M.G.A.
Data Availability
The datasets and analysis generated in the current study are available upon request (m.garcia@embl-hamburg.de).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46686-8.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newby ZE, et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat Protoc. 2009;4:619–637. doi: 10.1038/nprot.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker JL, Newstead S. Membrane Protein Crystallisation: Current Trends and Future Perspectives. Adv Exp Med Biol. 2016;922:61–72. doi: 10.1007/978-3-319-35072-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vergis JM, Purdy MD, Wiener MC. A high-throughput differential filtration assay to screen and select detergents for membrane proteins. Anal Biochem. 2010;407:1–11. doi: 10.1016/j.ab.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinothkumar KR, Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Q Rev Biophys. 2016;49:e13. doi: 10.1017/S0033583516000068. [DOI] [PubMed] [Google Scholar]
- 6.Kuhlbrandt WB. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 7.Cherezov V, Fersi H, Caffrey M. Crystallization screens: compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophys J. 2001;81:225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 9.Frauenfeld J, et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flayhan A, et al. Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research. Structure. 2018;26:345–355 e345. doi: 10.1016/j.str.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles TJ, et al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 14.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arachea BT, et al. Detergent selection for enhanced extraction of membrane proteins. Protein Expr Purif. 2012;86:12–20. doi: 10.1016/j.pep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newstead S, Hobbs J, Jordan D, Carpenter EP, Iwata S. Insights into outer membrane protein crystallization. Mol Membr Biol. 2008;25:631–638. doi: 10.1080/09687680802526574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mancusso R, Karpowich NK, Czyzewski BK, Wang DN. Simple screening method for improving membrane protein thermostability. Methods. 2011;55:324–329. doi: 10.1016/j.ymeth.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curnow P, et al. Stable folding core in the folding transition state of an alpha-helical integral membrane protein. Proc Natl Acad Sci USA. 2011;108:14133–14138. doi: 10.1073/pnas.1012594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champeil P, et al. A robust method to screen detergents for membrane protein stabilization, revisited. Anal Biochem. 2016;511:31–35. doi: 10.1016/j.ab.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Nji E, Chatzikyriakidou Y, Landreh M, Drew D. An engineered thermal-shift screen reveals specific lipid preferences of eukaryotic and prokaryotic membrane proteins. Nat Commun. 2018;9:4253. doi: 10.1038/s41467-018-06702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann J, Rose-Sperling D, Hellmich UA. Diverse relations between ABC transporters and lipids: An overview. Biochim Biophys Acta Biomembr. 2017;1859:605–618. doi: 10.1016/j.bbamem.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Sonoda Y, et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19:17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Harris NJ, Booth PJ. Folding and stability of membrane transport proteins in vitro. Biochim Biophys Acta. 2012;1818:1055–1066. doi: 10.1016/j.bbamem.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Veith K, et al. Lipid-like Peptides can Stabilize Integral Membrane Proteins for Biophysical and Structural Studies. Chembiochem. 2017;18:1735–1742. doi: 10.1002/cbic.201700235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majd, H. et al. Screening of candidate substrates and coupling ions of transporters by thermostability shift assays. Elife7, 10.7554/eLife.38821 (2018). [DOI] [PMC free article] [PubMed]
- 31.Martinez Molledo M, Quistgaard EM, Low C. Tripeptide binding in a proton-dependent oligopeptide transporter. FEBS Lett. 2018;592:3239–3247. doi: 10.1002/1873-3468.13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez Molledo M, Quistgaard EM, Flayhan A, Pieprzyk J, Low C. Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter. Structure. 2018;26:467–476 e464. doi: 10.1016/j.str.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quistgaard EM, Martinez Molledo M, Low C. Structure determination of a major facilitator peptide transporter: Inward facing PepTSt from Streptococcus thermophilus crystallized in space group P3121. PLoS One. 2017;12:e0173126. doi: 10.1371/journal.pone.0173126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low C, Jegerschold C, Kovermann M, Moberg P, Nordlund P. Optimisation of over-expression in E. coli and biophysical characterisation of human membrane protein synaptogyrin 1. PLoS One. 2012;7:e38244. doi: 10.1371/journal.pone.0038244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low C, et al. High-throughput analytical gel filtration screening of integral membrane proteins for structural studies. Biochim Biophys Acta. 2013;1830:3497–3508. doi: 10.1016/j.bbagen.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Ural-Blimke Yonca, Flayhan Ali, Strauss Jan, Rantos Vasileios, Bartels Kim, Nielsen Rolf, Pardon Els, Steyaert Jan, Kosinski Jan, Quistgaard Esben M., Löw Christian. Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1. Journal of the American Chemical Society. 2019;141(6):2404–2412. doi: 10.1021/jacs.8b11343. [DOI] [PubMed] [Google Scholar]
- 37.Heng J, et al. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 2015;25:1060–1073. doi: 10.1038/cr.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagarathinam K, et al. Outward open conformation of a Major Facilitator Superfamily multidrug/H(+) antiporter provides insights into switching mechanism. Nat Commun. 2018;9:4005. doi: 10.1038/s41467-018-06306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 40.Suurvali J, Boudinot P, Kanellopoulos J, Ruutel Boudinot S. P2X4: A fast and sensitive purinergic receptor. Biomed J. 2017;40:245–256. doi: 10.1016/j.bj.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bratanov D, et al. An Approach to Heterologous Expression of Membrane Proteins. The Case of Bacteriorhodopsin. PLoS One. 2015;10:e0128390. doi: 10.1371/journal.pone.0128390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 43.Michel H. Characterization and crystal packing of three-dimensional bacteriorhodopsin crystals. EMBO J. 1982;1:1267–1271. doi: 10.1002/j.1460-2075.1982.tb00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baaske, P., Duhr, S., Breitsprecher, D. & Derix, J. System and method for optically measuring the stability and aggregation of particles. WO2017055583A1 (2017).
- 46.King MS, Crichton PG, Ruprecht JJ, Kunji ERS. Publisher Correction: Concerns with yeast mitochondrial ADP/ATP carrier’s integrity in DPC. Nat Struct Mol Biol. 2018;25:988. doi: 10.1038/s41594-018-0138-1. [DOI] [PubMed] [Google Scholar]
- 47.Yang Q, Bruschweiler S, Zhao L, Chou JJ. Reply to ‘Concerns with yeast mitochondrial ADP/ATP carrier’s integrity in DPC’ and ‘Dynamics and interactions of AAC3 in DPC are not functionally relevant’. Nat Struct Mol Biol. 2018;25:749–750. doi: 10.1038/s41594-018-0126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stetsenko, A. & Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 7, 10.3390/cryst7070197 (2017).
- 49.Spearman C. The proof and measurement of association between two things. By C. Spearman, 1904. Am J Psychol. 1987;100:441–471. doi: 10.2307/1422689. [DOI] [PubMed] [Google Scholar]
- 50.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savitsky P, et al. High-throughput production of human proteins for crystallization: the SGC experience. J Struct Biol. 2010;172:3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woestenenk EA, Hammarstrom M, van den Berg S, Hard T, Berglund H. His tag effect on solubility of human proteins produced in Escherichia coli: a comparison between four expression vectors. J Struct Funct Genomics. 2004;5:217–229. doi: 10.1023/B:jsfg.0000031965.37625.0e. [DOI] [PubMed] [Google Scholar]
- 54.Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/S0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 55.Busso D, et al. Expression of protein complexes using multiple Escherichia coli protein co-expression systems: a benchmarking study. J Struct Biol. 2011;175:159–170. doi: 10.1016/j.jsb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Schlegel S, et al. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3) J Mol Biol. 2012;423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 57.Jones, E., Oliphant, T., Peterson, P. & others. SciPy: Open source scientific tools for Python (2001).
- 58.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 59.Kotov V, Vesper O, Garcia Alai M, Loew C, Marlovits TC. Moltenprot: A High-Throughput Analysis Platform to Assess Thermodynamic Stability of Membrane Proteins and Complexes. Biophysical Journal. 2019;116:191a. doi: 10.1016/j.bpj.2018.11.1060. [DOI] [Google Scholar]
- 60.Python v. 2.7 (Python Software Foundation, 2019).
- 61.Oliphant, T. E. Guide to NumPy. 2nd edn, (CreateSpace Independent Publishing Platform, 2015).
- 62.McKinney, W. (eds Stéfan van der Walt & Jarrod Millman) 51–56.
- 63.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and analysis generated in the current study are available upon request (m.garcia@embl-hamburg.de).