Abstract
Drug repurposing techniques allow existing drugs to be tested against diseases outside their initial spectrum, resulting in reduced cost and eliminating the long time-frames of new drug development. In silico drug repurposing further speeds up the process either by proposing drugs suitable to invert the transcriptomic profile of a disease or by indicating drugs based on their common targets or structural similarity with other drugs with similar mode of action.
Such methods usually return a number of potential repurposed drugs that need to be tested against the disease in in vitro, pre-clinical and clinical studies. Thus, it is crucial to have a more sophisticated candidate drug ranking in order to start testing from the most promising chemical substances. As a means to enhance the above decision process, we present CoDReS (Composite Drug Reranking Scoring), a drug (re-)ranking web-based tool, which combines an initial drug ranking (i.e. repurposing score or hypothesis/potentiality score) with a functional score of each drug considered in conjunction with the disease under study as well as with a structural score derived from potential drugability violations. Furthermore, a structural similarity clustering is applied on the considered drugs and a handful of structural exemplars are suggested for further in vitro and in vivo validation. The user is able to filter the results further, through structural similarity examination of the candidate drugs with drugs that have failed against the queried disease where related clinical trials have been carried out.
CoDReS is publicly available online at http://bioinformatics.cing.ac.cy/codres.
Keywords: Drug discovery, Drug ranking, Data mining, Cheminformatics
1. Introduction
Transcriptomic-based computational drug repurposing (DR) tools, such as Connectivity Map [1] and L1000CDS2 [2], compare a disease-related gene expression profile with a number of stored existing expression profiles corresponding to cellular responses against a number of perturbations. Existing tools return lists of candidate repurposed drugs, which can be ordered by their inhibition score. The inhibition score describes the potentiality of a chemical substance to alter the perturbed gene signature state of a disease back to its “normal-healthy” values. Although the inhibition score might give insight onto the potency of a drug against a disease, it alone cannot guarantee success in a clinical trial. On the other hand, cheminformatics tools, such as ChemMine Tools [3] and programming packages such as Rcpi [4] and ChemmineR [5] can suggest drugs with similar structure and possibly similar mode of action to drugs with a-priori knowledge regarding their effectiveness either against a specific disease-related mechanism or against diseases with phenotypic similarity to the targeted disease. However, the derived similarity score is often not enough to deem a drug an appropriate candidate against a disease.
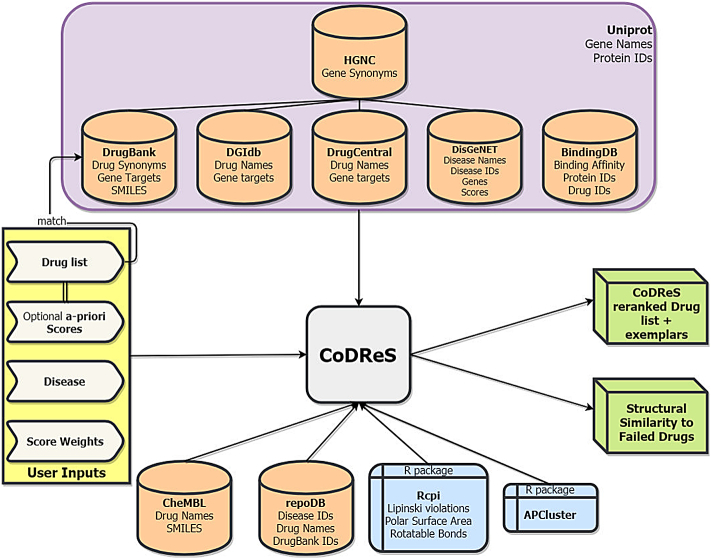
Other types of drug information are ought to be examined, like the candidate drug's functional relation to the disease and its binding affinity to any related-to-the-disease gene target as well as its drug-likeness evaluation based on structural rules that might categorize the drug inappropriate for clinical trials. In order to attain both the scoring implementation for these different drug aspects and provide a more meaningful ranking of the candidate repurposed drugs, we have developed the CoDReS (Composite Drug Reranking Score) web-based tool based on- and extending the initial methodology introduced in [6] in the following ways; CoDReS integrates information from updated biological databases, incorporates binding affinity scores between ligands and proteins, evaluates drug-likeness and presents structural similarities between input drugs and possible failed drugs that have already been tested against the queried disease in clinical trials. A summary figure of the CoDReS pipeline is depicted in Fig. 1.
Fig. 1.
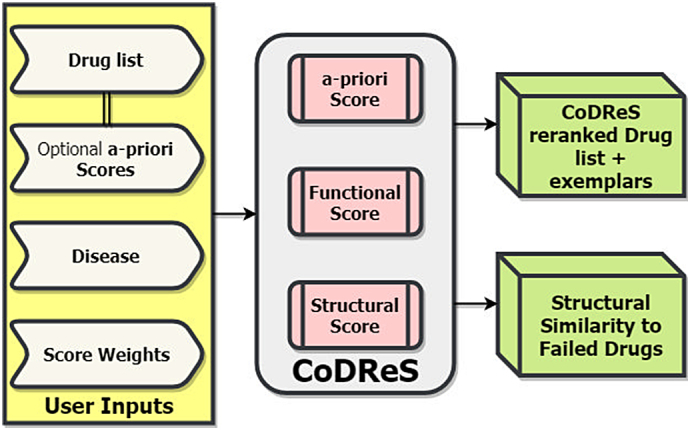
CoDReS summary figure.
2. Tool Description
2.1. Scoring Scheme
A composite score (from here on referred to as CoDReS) is calculated, for each drug, as the normalized weighted sum of the initial a-priori score (aS) with a functional (FS) and a structural score (StS) as introduced below:
The weights waS, wFS and wStS are user-defined parameters that determine the desired influence of each part (a-priori, functional and structural scores respectively) to the final score and have equal default values. The a-priori scores can be uploaded by the user and are automatically normalized in the unit interval [0, 1] by dividing with the absolute maximum a-priori score.
The functional score requires the calculation of two different parameters:
-
(i)
the Confidence Score, which reflects the gene-disease association and
-
(ii)
the Ki, which is an inhibitory constant, measured in nM, and represents the reciprocal of the binding affinity between the inhibitor (drug) and the enzyme (target) [7]. The smaller the Ki, the greater the binding affinity.
The FS for each drug is calculated as the sum of the products of Confidence Score with the inverse value of Ki, for each gene target of the drug that has been related to the queried disease. Each drug's FS is finally normalized in [0, 1] by dividing with the maximum FS.
The structural score calculates a substance's drug-likeness based on the Lipinski “rules of 5” [8] and Veber's rule [9]. According to the Lipinski rules, in order for a drug to be orally active in humans, it should conform to the following rules: (i) have ≤5 hydrogen bond donors, (ii) have ≤10 hydrogen bond acceptors, (iii) weigh <500 Da and (iv) have an octanol-water partition coefficient (log P) ≤5. The Veber's rule further requires that the chemical substance (v) contains ≤ 10 rotatable bonds and (vi) its polar surface area does not exceed 140 Ǻ2 (angstrom2). The final StS for each drug is a value within the range [0, 1] calculated in the following way:
where “6” is the maximum number of structural rules that a drug might violate.
2.2. Development
The static components of the user interface (UI) of the CoDReS web-based application are developed in php, html, css (bootstrap) and javascript (ajax), while the dynamic components of the UI are refreshed via php and back-end R scripts. Several data-repositories have been downloaded, parsed and integrated into a MySQL database, which in turn serves the CoDReS web-based application. Information regarding the database releases, versions and links can be found on Table 1.
Table 1.
Information regarding resources integrated to CoDReS.
| Database Name | Link | File | Current Version | Last Update |
|---|---|---|---|---|
| BindingDB | https://www.bindingdb.org/bind/chemsearch/marvin/SDFdownload.jsp?all_download = yes | BindingDB_All_2019m1.tsv.zip | – | 2019/02 |
| CheMBL | ftp://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/latest/ | chembl_24_1_mysql.tar.gz | 24.1 | 2018/06 |
| DGIdb | http://www.dgidb.org/downloads | Interactions TSV | 3.0.2 | 2018/01 |
| DisGeNET | http://www.disgenet.org/downloads | ALL gene-disease associations | 6.0 | 2019/02 |
| DrugBank | https://www.drugbank.ca/releases/latest#open-data | DrugBank Vocabulary | 5.1.2 | 2018/12 |
| https://www.drugbank.ca/releases/latest#protein-identifiers | Drug Target Identifiers - All | 5.1.2 | 2018/12 | |
| https://www.drugbank.ca/releases/latest#structures | Structural External Links - All | 5.1.2 | 2018/12 | |
| DrugCentral | http://drugcentral.org/download | Drug-target interaction | 10.4 | 2018/08 |
| HGNC | https://www.genenames.org/cgi-bin/download | Approved Symbol, Synonyms | – | 2019/02 |
| repoDB | http://apps.chiragjpgroup.org/repoDB/ | full repoDB dataset | 1.2 | 2017/07 |
| Uniprot | ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/by_organism | HUMAN_9606_idmapping.dat | – | 2019/02 |
CoDReS works with drug synonyms that have been downloaded from DrugBank [10]. DrugBank is a drug-centric online database that provides detailed information on drugs and their gene targets. The rest of the databases that were used in CoDReS that include drug names have been parsed and have had their drug names translated to DrugBank's usual names. DrugBank identifiers were also assigned to each input drug where applicable or an “unassigned” value was given otherwise. At the gene level, CoDReS works on gene synonyms that derive from the HGNC [11] database and every other database that contains gene identifiers, is parsed and translated according to the HGNC gene synonyms.
The backend of CoDReS is developed in R. The FS's first parameter, namely Confidence Score, is taken from DisGeNET [12] which is an online database linking genes to diseases by integrating information of various biological databases and giving a score to each interaction. The Ki value of a drug-protein pair is queried from BindingDB [13] which is another online database that contains data from experimentally validated binding affinities between proteins and ligands. To achieve the proper linking between the databases, we convert genes to proteins through Uniprot [14] and by querying BindingDB the proteins are linked to drug identifiers either from DrugBank or CheMBL [15]. If no Ki value is found for a drug-protein pair, the application uses the median Ki value (184) of BindingDB, instead of the average value (120523.41) which results from the database's outliers. Uniprot is an online knowledgebase that hosts annotated sequences of over 120 million proteins as well as provides protein visualization methods. CheMBL is another drug related database similar to DrugBank. The gene targets of the input drugs are found in the parsed DrugBank, DrugCentral [16] and DGIdb [17] databases.
The StS of each drug is calculated via the Rcpi package of R and requires a drug's molecular structure as input. Each molecular structure is extracted by its respective simplified molecular-input line-entry system (SMILES) type, which is a specification in form of a line notation for describing the structure of chemical species using short ASCII strings. CoDReS tries to either map DrugBank identifiers or CheMBL drug names to SMILES. For every violated rule (aforementioned Lipinski and Veber rules), a drug receives a “plus 1” violation score, with the lowest possible score being six violations. In case there was no SMILES for a specific drug, the candidate drug is assigned a “zero” StS score acting in a conservative manner by adopting the worst-case scenario presenting the max number of violations.
Another important aspect of the CoDReS tool, is that it highlights the highest ranked drugs of structural clusters, as exemplars, by applying an affinity propagation clustering via the R package APCluster [18] on the similarity matrix of the fingerprints of the input drugs. Specifically, the calcDrugFPSim function of the Rcpi package is used in order to calculate the similarity matrix with a compact E-State fragments fingerprint type and a tanimoto metric as arguments. The structural exemplars are presented as a good choice of disease inhibitors for further investigation, since different structural properties might target different biological mechanisms of a disease phenotype.
Finally, if there are clinical trials carried out for a disease that have led to failed drugs against a disease, the structural similarity between these compounds and the input list of drugs is measured. For this purpose, the online dataset of repoDB [19] has been parsed, keeping the suspended, terminated and withdrawn drugs for each disease identifier. The execution pipeline together with all the integrated databases and packages are depicted in Fig. 2.
Fig. 2.
CoDReS integration scheme.
2.3. User Execution
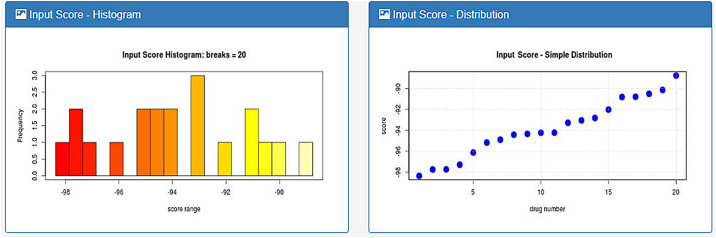
The user is required to upload a file containing the drug names and, optionally their respective a-priori scores, as might be acquired from a drug-repurposing tool. As soon as the input file is uploaded, a histogram and a distribution diagram of the input scores are generated (Fig. 3). The weights denoting the importance of the aS, FS and StS are user-selected and have equal default values. The user must then choose a disease from a select box with auto-complete functionality that hosts all DisGeNET diseases.
Fig. 3.
Input score diagrams are drawn after the user uploads a drug list with their respective scores as returned by any drug repurposing tool.
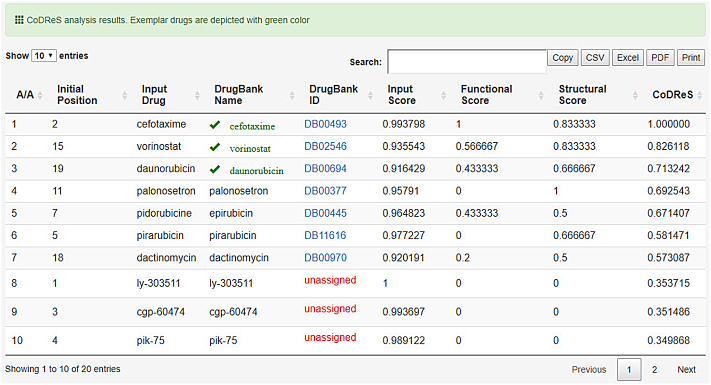
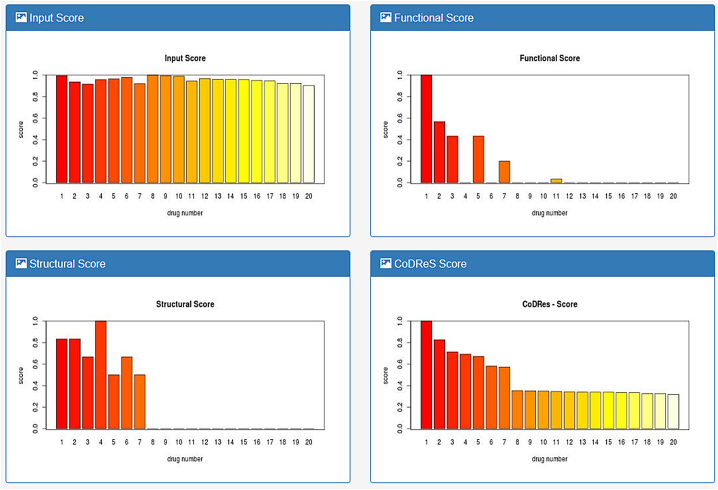
The output of the CoDReS function is then rendered in tabular form and can be sorted, printed and downloaded either as plain text, csv, spreadsheet or pdf file. The main CoDReS output table consists of the CoDReS rank, the initial position of the input drugs, their input names, their DrugBank usual names and identifiers (or input name again and “unassigned” identifier respectively, if not found in DrugBank's synonyms list), their normalized score per category and their normalized CoDReS, by which they are sorted in descending order (Fig. 4). A drug-score diagram for each scoring parameter is also printed at the bottom of the page after the execution (Fig. 5).
Fig. 4.
Main CoDReS output matrix.
Fig. 5.
Drug score diagrams for each scoring parameter.
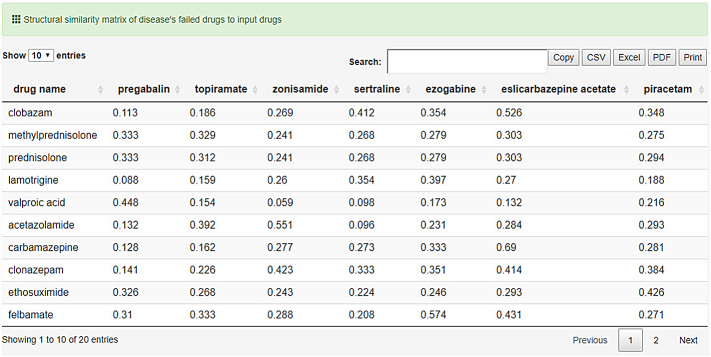
In case there are stated failed clinical trials in repoDB for the selected disease, a similarity matrix of all input drugs against the failed drugs is returned to the user, where the column names represent the failed drugs and the row names the input drugs (Fig. 6).
Fig. 6.
Structural similarity of input drugs (rows) to clinically failed drugs of input disease (columns) as found in repoDB.
3. Results and Validation
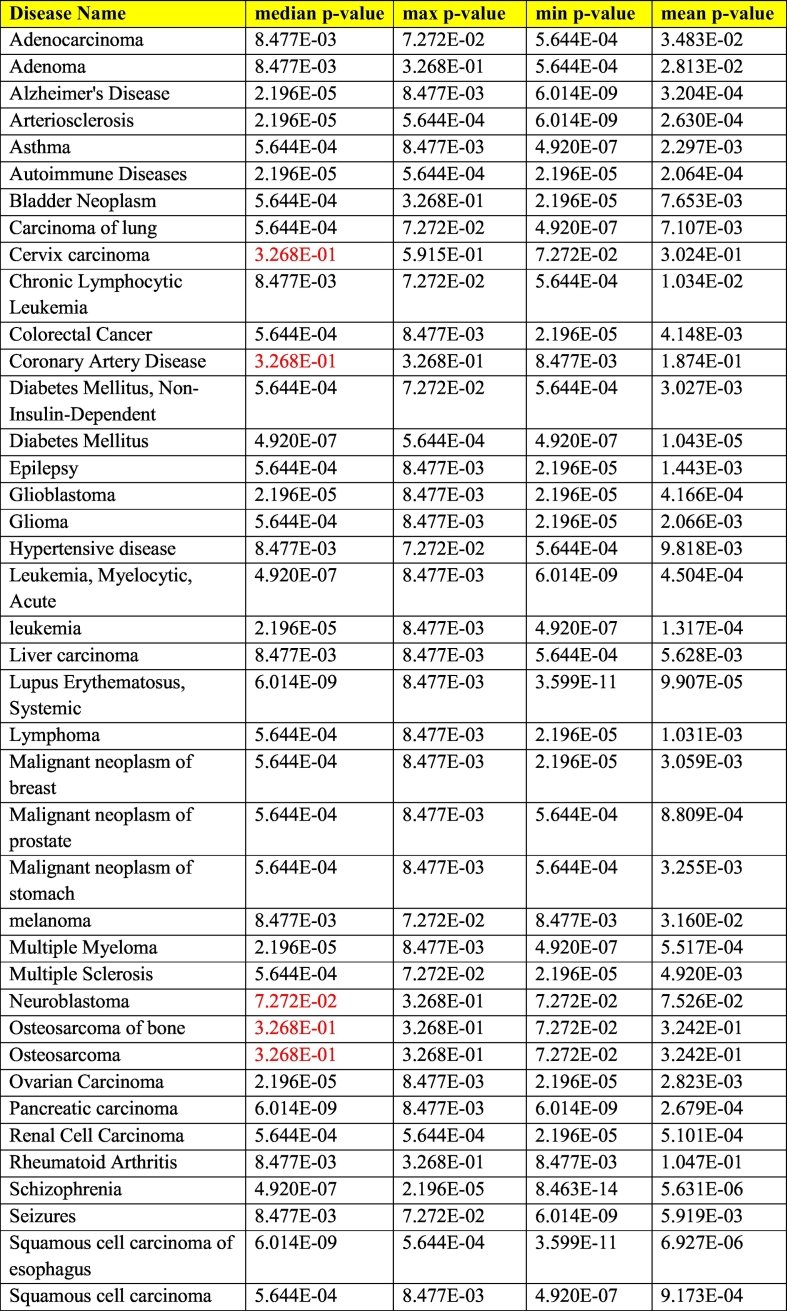
To check the validity of the CoDReS results, we considered examples disregarding a-priori scores. We chose the top forty diseases from DisGeNET with the most correlated genes that have at least twenty drug candidates in Malacards [20]. These diseases are listed in Table 2. For each disease, we created a mixture list of two hundred drugs: 95% randomly selected from DrugBank and 5% of the top drugs reported from Malacards repository as developed/used for the selected disease. After executing CoDReS for each experiment, we counted the number of the actual disease-related drugs that were found in the top 5% of the ranked drugs, based on their CoDReS along with a p-value calculated through a hypergeometric distribution test. We repeated this procedure a hundred times for each disease and then calculated the median, maximum, minimum and average p-value metrics for each disease. CoDReS ranked effectively (median p-value <.05) the input drugs in 35/40 diseases. CoDReS failed to rank drugs correctly in five out of 40 diseases but this failure can be partially explained since the top ten drugs corresponding to most of these diseases contain abstract substances or generic categories such as “Anti-Inflammatory Agents”, “Cytochrome P-450 Enzyme Inhibitors”, “Immunologic Factors” or drugs with close to zero gene targets participating in the disease. The statistical results are presented in Table 3.
Table 2.
information on the diseases used for the validation; the two first columns present the disease's name and umls id respectively as found in disgenet, the third column the total genes that participate in the disease and the fourth column the disease's name as returned from malacards.
| Disease name | UMLS ID | Gene count | Malacards name |
|---|---|---|---|
| Malignant neoplasm of breast | C0006142 | 5053 | Breast Cancer |
| Liver carcinoma | C2239176 | 3592 | Hepatocellular Carcinoma |
| Colorectal Cancer | C1527249 | 3298 | Colorectal Cancer |
| Malignant neoplasm of prostate | C0376358 | 3238 | Prostate Cancer |
| Carcinoma of lung | C0684249 | 2475 | Lung Cancer |
| melanoma | C0025202 | 2453 | Melanoma |
| Malignant neoplasm of stomach | C0024623 | 2397 | Gastric Cancer |
| Glioma | C0017638 | 2210 | Glioma |
| Ovarian Carcinoma | C0029925 | 2202 | Ovarian Cancer |
| Alzheimer's Disease | C0002395 | 1981 | Alzheimer Disease |
| leukemia | C0023418 | 1940 | Leukemia |
| Glioblastoma | C0017636 | 1936 | Glioblastoma |
| Schizophrenia | C0036341 | 1922 | Schizophrenia |
| Squamous cell carcinoma | C0007137 | 1875 | Squamous Cell Carcinoma |
| Pancreatic carcinoma | C0235974 | 1868 | Pancreatic Cancer |
| Rheumatoid Arthritis | C0003873 | 1832 | Rheumatoid Arthritis |
| Adenocarcinoma | C0001418 | 1711 | Adenocarcinoma |
| Leukemia, Myelocytic, Acute | C0023467 | 1702 | Leukemia, Acute Myeloid |
| Neuroblastoma | C0027819 | 1698 | Neuroblastoma |
| Diabetes Mellitus, Non-Insulin-Dependent | C0011860 | 1671 | Diabetes Mellitus, Noninsulin-Dependent |
| Diabetes Mellitus | C0011849 | 1506 | Diabetes Mellitus |
| Renal Cell Carcinoma | C0007134 | 1347 | Renal Cell Carcinoma, Papillary, 1 |
| Asthma | C0004096 | 1312 | Asthma |
| Multiple Myeloma | C0026764 | 1311 | Myeloma, Multiple |
| Hypertensive disease | C0020538 | 1309 | Hypertension, Essential |
| Lymphoma | C0024299 | 1306 | Lymphoma |
| Bladder Neoplasm | C0005695 | 1216 | Bladder Cancer |
| Epilepsy | C0014544 | 1176 | Epilepsy |
| Seizures | C0036572 | 1173 | Seizure Disorder |
| Chronic Lymphocytic Leukemia | C0023434 | 1119 | Leukemia, Chronic Lymphocytic |
| Lupus Erythematosus, Systemic | C0024141 | 1112 | Systemic Lupus Erythematosus |
| Multiple Sclerosis | C0026769 | 1105 | Multiple Sclerosis |
| Cervix carcinoma | C0302592 | 1104 | Cervix carcinoma |
| Osteosarcoma | C0029463 | 1102 | Osteogenic Sarcoma |
| Arteriosclerosis | C0003850 | 1086 | Arteriosclerosis |
| Autoimmune Diseases | C0004364 | 1059 | Autoimmune Disease |
| Osteosarcoma of bone | C0585442 | 1041 | Bone Osteosarcoma |
| Squamous cell carcinoma of esophagus | C0279626 | 1022 | Esophagus Squamous Cell Carcinoma |
| Adenoma | C0001430 | 999 | Adenoma |
| Coronary Artery Disease | C1956346 | 980 | Coronary Artery Anomaly |
Table 3.
The median, maximum, minimum and average p-value results of 100 codres executions for each disease as calculated by hypergeometric distribution tests. The median p-values that are above 0.05 are painted red.
4. Discussion
In this article we present CoDReS, a drug (re-)ranking tool that can act as a tool for post filtering drug lists generated either by conventional drug repurposing tools or by any other drug discovery pipeline. CoDReS should be used as a means of suggesting the best candidates for in vitro or clinical studies by combining a priori knowledge with functional and structural information. The in silico validation schema of CoDReS, as presented in the previous paragraph, brought the disease-related drugs to the top of the random drug pool in almost every case. Despite the promising results, this schema is only a computational validation of the tool's capabilities. In the end, the scientists using the tool should always incorporate their knowledge, expertise and the bibliography in order to decide the best drug candidates for further experiments.
Declarations of Copmeting Interest
None declared.
Acknowledgments
George M. Spyrou holds the Bioinformatics ERA Chair Position funded by the European Commission Research Executive Agency (REA) Grant BIORISE (Num. 669026), under the Spreading Excellence, Widening Participation, Science with and for Society Framework. George Minadakis holds a postdoctoral research fellow position funded by the European Commission Research Executive Agency Grant BIORISE (No. 669026), under the Spreading Excellence, Widening Participation, Science with and for Society Framework. Evangelos S. Karatzas is a PHD student in the National and Kapodistrian University of Athens. His doctoral thesis is being funded by the IKY (State Scholarships Foundation) scholarship, funded by the Action “Strengthening Human Resources, Education and Lifelong Learning”, 2014-2020, co-funded by the European Social Fund (ESF) and the Greek State.
References
- 1.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452. doi: 10.1016/j.cell.2017.10.049. [e1417] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duan Q., Reid S.P., Clark N.R., Wang Z., Fernandez N.F. L1000CDS 2: LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2 doi: 10.1038/npjsba.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backman T.W., Cao Y., Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–W491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao D.-S., Xiao N., Xu Q.-S., Chen A.F. Rcpi: R/Bioconductor package to generate various descriptors of proteins, compounds and their interactions. Bioinformatics. 2014;31:279–281. doi: 10.1093/bioinformatics/btu624. [DOI] [PubMed] [Google Scholar]
- 5.Cao Y., Charisi A., Cheng L.-C., Jiang T., Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karatzas E., Bourdakou M., Kolios G., Spyrou G. Drug repurposing in idiopathic pulmonary fibrosis filtered by a bioinformatics-derived composite score. Sci Rep. 2017;7 doi: 10.1038/s41598-017-12849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yung-Chi C., Prusoff W.H. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 8.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 9.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 10.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2017;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S. Genenames. org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2016:gkw1033. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016:gkw943. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2006;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J. The ChEMBL database in 2017. Nucleic Acids Res. 2016;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ursu O., Holmes J., Knockel J., Bologa C.G., Yang J.J. DrugCentral: online drug compendium. Nucleic Acids Res. 2016:gkw993. doi: 10.1093/nar/gkw993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotto K.C., Wagner A.H., Feng Y.-Y., Kiwala S., Coffman A.C. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 2017;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenhofer U., Kothmeier A., Hochreiter S. APCluster: an R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–2464. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- 19.Brown A.S., Patel C.J. A standard database for drug repositioning. Scientific Data. 2017;4 doi: 10.1038/sdata.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappaport N., Twik M., Plaschkes I., Nudel R., Iny Stein T. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2016;45:D877–D887. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]