Abstract
Semen fertilizing potential is dependent upon the morphological, functional and molecular attributes of sperm. Sperm microRNAs (miRNAs) were recently shown to hold promise regarding their association with different fertility phenotypes. However, their role in fertility regulation remains to be determined. We postulated that sperm miRNAs might regulate early embryonic development. From this perspective, sperm quality and 380 sperm miRNAs were investigated in frozen–thawed semen from high (HF; 54.3 ± 1.0% pregnancy rate) and low (LF; 41.5 ± 2.3%) fertility bulls. Out of nine miRNAs that showed different levels in sperm cells, miR-216b was present at lower levels in HF sperm cells and zygotes. Among miR-216b target genes (K-RAS, BECN1 and JUN), K-RAS, related to cell proliferation, revealed a higher level in HF two-cell embryos. First cleavage rate, blastocyst cell number and division number were also higher in HF. In addition, by using a model based on polyspermy embryos, we demonstrated an increase in miR-216b levels in zygotes associated with sperm cell entry. Our results shed light on a possible mechanism of paternal contribution involving sperm-borne miR-216b that modulates levels of miR-216b in zygotes and K-RAS in two-cell embryos. This modulation might regulate early development by interfering with the first cleavage and blastocyst quality.
Subject terms: Embryology, Development, Gene expression, miRNAs
Introduction
All sperm attributes, including morphological, functional and molecular characteristics, are important for male fertility1,2. As a result of the complexity of these attributes, recording pregnancy rates is the most satisfactory procedure that allows the assessment of semen fertilization potential; however, this is expensive and laborious3. Thus, an investigation into the efficiency of each sperm attribute as a predictor of male fertility is essential to further our understanding of their relative contributions. In this context, sperm cells should have the ability to: (1) reach the fertilization site; (2) fertilize the ovum; and (3) contribute to early embryonic development2. Sperm morphological and functional aspects, referred to herein as sperm quality attributes (SQA), are directly related to the sperm’s ability to reach the oviduct and fertilize the ovum. Sperm motility, abnormalities, membrane integrity, mitochondria function and DNA status are examples of SQA that often show high correlation with male fertility. As such, SQA are frequently used to assess the fertility potential of frozen–thawed semen batches1,4–6. However, since male fertility potential is dependent upon several sperm characteristics7, restricting evaluation to these features might be insufficient to predict fertility for some semen samples8. Molecular sperm signatures are frequently linked to their contribution to early embryonic development, which involves delivery of the following to the embryo: sperm DNA, mRNAs, proteins and non-coding RNAs such as microRNAs (miRNAs) that appear to be related to healthy sperm and male fertility potential2,9,10.
MicroRNAs are small non-coding RNA molecules (~22 nt) that show a high degree of conservation among species and are involved in modulation of protein translation11. The canonical biogenesis pathway consists of RNA polymerase II-mediated transcription of primary miRNA transcripts (pri-miRNA; ~200 nt) from DNA, which are cleaved by Drosha and DGCR8 to produce precursor miRNAs (pre-miRNA; ~70 nt). Pre-miRNA is transported from the nucleus to the cytoplasm by Exportin 5, where it undergoes cleavage by Dicer and RNA-binding protein TRBP to form a miRNA duplex of ~22 nt. Only one strand binds to a complex of Argonaute proteins to form the RISC complex (RNA-induced silencing complex) whose activity is dependent on binding of the miRNA to the 3′ untranslated region (3′ UTR) of its target gene: perfect binding induces degradation of the target mRNA; while imperfect binding prevents translation of the target mRNA11. MiRNAs are currently known for their roles in spermatogenesis12, sperm maturation13 and embryo development14. Human sperm cell miRNAs were first described by Ostermeier et al. (2005)15,16 and exhibit different profiles according to fertility status17,18. Specific “sperm-borne” miRNAs such as miR-34c and miR-449b are important for the first cleavage in mouse and bovine embryos, respectively10,19,20.
Based on (1) the dependence on morphological, functional and molecular sperm attributes for male fertility success, and, (2) the potential of sperm-borne miRNAs to play a role in fertility by regulating embryo development, we hypothesized that initial steps of embryo development are under the control of specific miRNAs delivered by sperm cells. Thus, the aims of this study were to: (1) determine the SQA and miRNA profiles of semen samples from high and low fertility bulls; (2) investigate the relative levels of sperm miRNAs and target genes in embryos from high and low fertility sperm samples; (3) investigate how alterations in miRNAs could regulate embryo development; and (4) demonstrate that sperm cells are capable of delivering miRNAs to the zygote.
Results
Experiment 1: Lower level of miR-216b in sperm cells and zygotes is associated with a high level of K-RAS in two-cell embryos and an increase in first cleavage rate and blastocyst cell number
HF and LF semen samples display similarity regarding sperm quality attributes
Sperm quality characterization was first performed on three commercial semen batches from each bull to give a total of 18 batches. The results showed that the majority of the evaluated characteristics were similar for the two different groups of samples— refer to Supplementary Table S1. Selection of the best semen batch from each bull was performed according to Supplementary Table S2 to give a total of three batches from HF bulls (High Fertility; n = 3) and three batches from LF bulls (Low Fertility; n = 3). In that regard, the following formula was used:
where PROG was progressive motility, MAJ was major defects, MIN was minor defects and PIAIH was sperm with plasma and acrosome membrane integrity and high mitochondrial membrane potential. The individual characteristics of each batch are described in Supplementary Tables S3 and S4. The morphological and functional analyses of the selected batches showed that sperm cells from HF and LF bulls were similar with regard to sperm motility, abnormalities, kinetic, plasma and acrosome membrane integrity, mitochondrial function, production of ROS, lipid peroxidation and DNA fragmentation according to data presented in Tables 1 and 2.
Table 1.
Mean and SEM of sperm quality attributes (sperm motility, concentration, abnormalities and kinetic) evaluated in the selected commercial semen batches.
| Sperm quality analyses of the selected batches | Fertility groups | |
|---|---|---|
| HFa (n = 3) | LFb (n = 3) | |
| Sperm motility, concentration and abnormalities | ||
| Straw volume (mL) | 0.25 | 0.25 |
| Subjective motility (%) | 68.3 ± 0.8 | 60.8 ± 3.6 |
| Vigor (1–5) | 3.5 ± 0.1 | 3.3 ± 0.2 |
| Sperm concentration (×106 sperm/mL) | 90.0 ± 11.3 | 88.3 ± 11.6 |
| Major defects (%) | 16.5 ± 3.9 | 24.3 ± 1.5 |
| Minor defects (%) | 6.5 ± 1.8 | 4.3 ± 0.8 |
| Total defects (%) | 23.0 ± 5.7 | 28.6 ± 2.3 |
| Total of sperm/straw (×106) | 22.5 ± 2.8 | 22.1 ± 2.9 |
| Total of motility sperm/straw (×106) | 15.3 ± 1.9 | 13.5 ± 2.4 |
| Sperm kinetic | ||
| Total motility (%) | 82.2 ± 1.4 | 73.9 ± 5.3 |
| Progressive motility (%) | 57.2 ± 5.7 | 58.5 ± 4.4 |
| Rapid cells (%) | 75.6 ± 0.2 | 69.3 ± 4.6 |
| Curvilinear velocity (VCL; µm/s) | 144.7 ± 8.0 | 172.0 ± 10.7 |
| Progressive velocity (VSL; µm/s) | 75.7 ± 6.7 | 87.4 ± 3.4 |
| Path velocity (VAP; µm/s) | 95.0 ± 4.3 | 104.7 ± 5.6 |
| Linearity (LIN; %) | 52.5 ± 4.9 | 51.0 ± 2.1 |
| Straightness (STR; %) | 79.3 ± 4.2 | 83.5 ± 1.3 |
| Wobble (WOB; %) | 65.9 ± 2.7 | 61.1 ± 2.1 |
| Lateral head displacement (ALH; µm) | 2.6 ± 0.1 | 3.3 ± 0.3 |
| Beat cross frequency (BCF; Hz) | 21.8 ± 0.7 | 24.2 ± 1.1 |
Differences were considered significant when P < 0.05. aHigh fertility; bLow fertility. SEM: standard error of the mean.
Table 2.
Mean and SEM of sperm quality attributes (sperm membranes integrity and function, production of ROS, lipid peroxidation and DNA fragmentation) evaluated in the selected commercial semen batches.
| Sperm quality analyses of the selected batches | Fertility groups | |
|---|---|---|
| HFa (n = 3) | LFb (n = 3) | |
| Sperm membranes integrity, mitochondrial membrane function and ROS production by fluorescence microscopy | ||
| PIAIHc (%) | 48.9 ± 4.7 | 45.2 ± 6.1 |
| Plasma membrane integrity (%) | 51.5 ± 5.1 | 48.5 ± 6.4 |
| Acrosome membrane integrity (%) | 60.7 ± 3.6 | 59.4 ± 4.1 |
| High mitochondrial membrane potential (%) | 62.6 ± 7.9 | 54.7 ± 7.2 |
| ROSd (%) | 5.2 ± 0.9 | 10.7 ± 4.9 |
| Sperm membranes integrity, mitochondrial membrane function, ROS production, lipid peroxidation and DNA fragmentation by flow cytometry | ||
| Plasma membrane integrity (%) | 49.5 ± 0.3 | 45.6 ± 8.2 |
| Acrosome membrane integrity (%) | 83.9 ± 2.3 | 78.3 ± 3.7 |
| Plasma and acrosome membrane integrity (%) | 49.3 ± 0.3 | 45.4 ± 8.3 |
| Plasma membrane integrity and high mitochondrial membrane potential (%) | 24.4 ± 2.1 | 26.3 ± 2.9 |
| Plasma membrane integrity and mitochondrial positive ROS (%) | 5.3 ± 1.6 | 4.1 ± 1.0 |
| Mitochondrial positive ROS (ua) | 621.0 ± 76.2 | 678.3 ± 23.5 |
| Lipid peroxidation (ua) | 2,502.0 ± 395.6 | 2,764.9 ± 921.8 |
| DNA fragmentation (%) | 5.8 ± 0.1 | 5.7 ± 0.1 |
Differences were considered significant when P < 0.05. aHigh fertility; bLow fertility; cSperm with plasma and acrosome membrane integrity and high mitochondrial potential; dProduction of ROS (reactive oxygen species). SEM: standard error of the mean.
Nine miRNAs are present at a different relative level between sperm cells from HF and LF
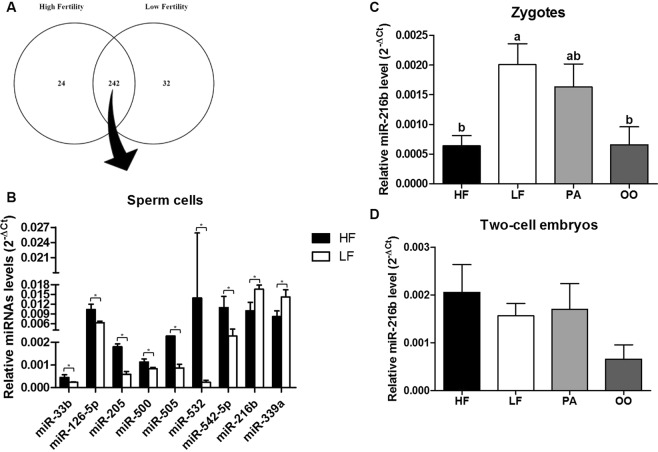
A panel of 380 miRNAs was evaluated in sperm cells from selected batches. Among these, 24 miRNAs were exclusively detected in sperm cells from HF bulls, 32 miRNAs were exclusively detected in sperm cells from LF bulls and 242 miRNAs were detected in both groups, as shown in Supplementary Table S5 and Fig. 1A. Nine out of 242 miRNAs showed different relative levels between HF and LF sperm samples with a P value of 0.10 (Fig. 1B). Different relative levels of bta-miR-205, -505 and -532 showed a statistical significance of P < 0.05; bta-miR-33b, -126-5p, -216b, -339a, -500 and -542-5p presented a P value between 0.05 and 0.10. Bta-miR-205, -505, -532, -33b, -126-5p, -500 and -542-5p were found in higher abundance in sperm cells from HF bulls, whereas bta-miR-216b and -339a were found in higher abundance in sperm cells from LF bulls (Fig. 1B).
Figure 1.
MiRNA analysis in sperm cells and in in vitro-produced embryos. (A) Venn diagram of 298 detected miRNAs in sperm cells from high fertility (HF) or low fertility (LF) bulls from a profile of 380 miRNAs. (B) Relative levels of the differentially abundant miRNAs (P < 0.10) between sperm cells from high fertility and low fertility bulls. (C) MiR-216b relative level in zygotes from high fertility (HF), low fertility (LF), parthenogenetic (PA) and mature oocyte (OO) groups. (D) miR-216b relative level in two-cell embryos from high fertility (HF), low fertility (LF), parthenogenetic (PA) and mature oocyte (OO) groups. Asterisks (*) indicate difference between the groups with a significance level of P < 0.10. a,bDifferent letters indicate statistical difference (P < 0.05) between groups. All quantitative data are presented as means and SEM.
Higher first cleavage rate is detected in HF embryos
After evaluation of sperm quality and miRNA profile, sperm from selected HF and LF semen batches (characteristics are described in Tables 1 and 2) were used to produce IVF embryos; parthenogenetic embryos were also produced and used as controls. In each replicate of embryo production, pools of 20 to 25 oocytes were previously divided into drops intended specifically for evaluation and collection of zygotes, two-cell embryos and blastocysts at a specific time interval. Fertilized zygotes were classified and collected according the rate of second polar body extrusion evaluated 16 to 18 hours post insemination (hpi); this rate was found to be similar between HF and LF zygotes as shown in Table 3. However, the first cleavage rate, evaluated 28 to 30 hpi in HF and LF, and 20 to 22 hours post activation in parthenogenetic embryos, was higher in embryos from HF bulls and parthenogenesis than in embryos from LF bulls (Table 3). The cleavage and blastocyst rates were evaluated from the same group of oocytes/embryos. The cleavage rate assessed on day-4 of development were similar among the three groups. Blastocyst rate evaluated on day-7 showed a higher percentage in the parthenogenetic embryo group compared with HF and LF, as shown in Table 3.
Table 3.
Pre-implantation development rates of bovine embryos produced in vitro.
| Fertility Groups | 2nd PBa | 1st cleavageb | Cleavage and blastocystsc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oocytes | Cleavage | Blastocysts | |||||||||
| Oocytes | N | Mean ± SEM | Oocytes | N | Mean ± SEM | N | Mean ± SEM | N | Mean ± SEM | ||
| HFd | 329 | 160 | 48.6 ± 2.1 | 273 | 75 | 27.5 ± 3.8a | 335 | 251 | 74.9 ± 3.6 | 111 | 33.1 ± 2.9b |
| LFe | 322 | 139 | 43.2 ± 4.2 | 278 | 52 | 18.7 ± 3.7b | 375 | 290 | 77.3 ± 2.9 | 115 | 30.7 ± 4.1b |
| PAf | — | — | — | 227 | 70 | 30.8 ± 3.7a | 300 | 228 | 76.0 ± 2.9 | 126 | 42.0 ± 2.9a |
aSecond polar body extrusion (PB; evaluated 16 to 18 hpi), bfirst cleavage (evaluated 28 to 30 hpi or 20 to 22 hpa), ccleavage on day-4 and blastocysts on day-7 were evaluated in embryos produced in vitro using high and low fertility semen samples and embryos produced in vitro by parthenogenesis. a,bDifferent letters on the same column indicate statistical difference (P < 0.05) between groups. dHigh fertility; eLow fertility; fParthenogenetic. Hpi: hours post insemination. Hpa: hours post activation. Mean and SEM are presented as percentages. SEM: standard error of the mean.
Lower level of miR-216b and higher level of its target gene K-RAS are detected in HF zygotes and two-cell embryos, respectively
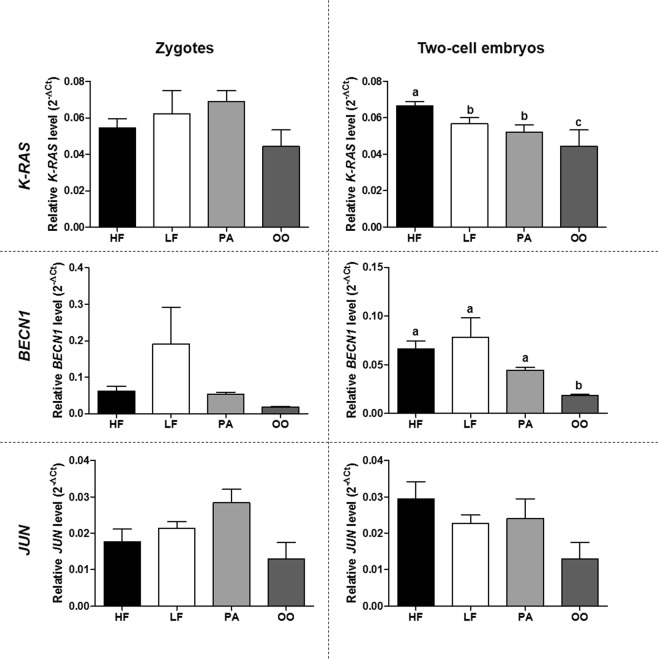
The relative levels of nine miRNAs (Fig. 1B) that were different between HF and LF sperm samples were investigated in zygotes and two-cell embryos produced from HF and LF bulls. Parthenogenetic embryos and mature oocytes were used as controls for this analysis. Among the miRNAs evaluated, it was observed that the relative level of miR-216b was lower in HF zygotes and oocytes (Fig. 1C). In two-cell embryos, none of the miRNAs, including miR-216b, showed a difference in relative levels between HF and LF embryos (Fig. 1D). The relative levels of the nine miRNAs investigated in embryos are listed in Supplementary Tables S6 and S7. After establishing that miR-216b presented the same differential pattern in both sperm cells and zygotes from HF and LF groups, the relative levels of miR-216b target genes were investigated in embryos. In order to determine miR-216b target genes, we first performed a homology analysis of bta-miR-216b with hsa-miR-216b-5p, which showed that the miRNAs shared the same nucleotide sequence (aaaucucugcaggcaaauguga), revealing 100% homology. Target genes of hsa-miR-216b were then investigated and selected according to two criteria: (1) strong validated mRNA-miR216b interaction (according to the miRTarBase platform); and (2) genes that play a relevant role in embryo development. Thus, K-RAS, BECN1 and JUN target genes were selected21–24. Furthermore, the 3′ UTR was conserved between K-RAS, BECN1 and JUN sequences from humans and cattle. The target gene levels were investigated in mature oocytes as well as in zygotes and two-cell embryos from the HF, LF and parthenogenetic groups as shown in Fig. 2. Relative levels of K-RAS, BECN1 and JUN showed no difference between zygotes and oocytes. However, the K-RAS gene was present at a higher relative level in two-cell embryos from the HF group as shown in Fig. 2. The BECN1 gene was present at a lower relative level in mature oocytes, but showed the same relative level in embryos from the HF and LF groups.
Figure 2.
Analyses of miR-216b target genes in mature oocytes and in in vitro-produced embryos. K-RAS, BECN1 and JUN relative level in zygotes and two-cell embryos from high fertility (HF), low fertility (LF), parthenogenetic (PA) and mature oocyte (OO) groups. a,b,cDifferent letters indicate statistical difference (P < 0.05) between groups. All quantitative data are presented as means and SEM.
HF embryos display a higher cell number in blastocysts and a higher number of cell divisions up to the day-7 blastocyst stage
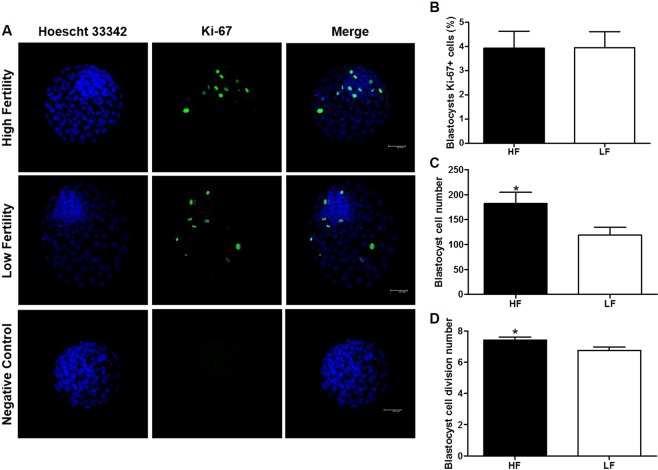
Since K-RAS gene expression is associated with cell proliferation, embryo quality was investigated with respect to blastocyst proliferation, diameter, stage and blastocyst cell number as well the number of cell divisions up to the day-7 blastocyst stage. Immunostaining was performed with Ki-67 to investigate the proliferation rate of blastocyst embryos. This revealed that HF and LF blastocysts proliferated at a similar rate (Fig. 3A,B). In a similar manner, the diameter of blastocysts from HF (235.4 ± 18.3 µm) and LF (218.4 ± 16.7 µm) groups were similar. Moreover, the percentage of early blastocysts (HF: 3.3 ± 2.0%; LF: 6.8 ± 3.5%), blastocysts (HF: 28.3 ± 5.1%; LF: 25.2 ± 5.7%), expanded blastocysts (HF: 51.7 ± 5.9%; LF: 43.7 ± 3.9%) and hatched blastocysts (HF: 16.7 ± 5.7%; LF: 24.3 ± 5.1%) showed no difference between the groups. However, the cell number and number of cell divisions were higher in HF blastocysts than in LF blastocysts (Fig. 3C,D). In HF blastocysts the number of cells and the number of cell divisions were 182.4 ± 22.4 and 7.4 ± 0.2, respectively; in LF blastocysts they were 119.3 ± 15.5 and 6.8 ± 0.2, respectively. Furthermore, developmental kinetic parameters were investigated in embryos produced by IVF performed for 8 hours, instead of the 18 hours performed previously, in order to investigate the possibility of a fertilization delay with LF sperm cells. Although cleavage rates for four-cell embryos (HF: 25.4 ± 4.2%; LF: 26.8 ± 3.2%) were similar in this study, HF (35.1 ± 6.8%) showed a tendency (P = 0.07) to produce a higher percentage of two-cell embryos than LF (28.5 ± 6.7%) as shown in Supplementary Table S8.
Figure 3.
Blastocyst qualitative analysis. (A) Confocal representative photomicrographs of Ki-67 immunostaining in blastocysts from high fertility and low fertility groups and negative control. Scale bar: 50 μm. (B) Percentage of cells in high fertility (HF) and low fertility (LF) blastocysts that were marked with Ki-67 antibody. (C) Cell numbers in high fertility (HF) and low fertility (LF) blastocysts stained with Hoechst 33342. (D) Cell division number in high fertility (HF) and low fertility (LF) blastocysts. Asterisks (*) indicate difference between the groups with a significance level of P < 0.05. All quantitative data are presented as means and SEM.
Experiment 2: Higher level of miR-216b is associated with polyspermic zygotes
Sperm concentration of 8 × 106 sperm/mL per IVF drop is efficient at increasing the polyspermic zygote rate
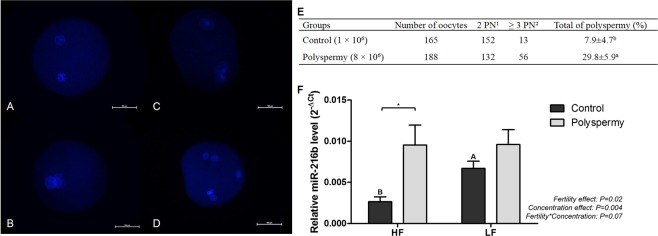
In order to investigate whether sperm cells are able to deliver miR-216b to zygotes, we proposed a new model based on polyspermic embryos. Our intent with this model was to show that by increasing the number of sperm cells per oocyte, the level of sperm-borne miR-216b in zygotes might be increased. With this aim, we first conducted a study to determine the sperm concentration that could increase polyspermic zygote formation. Previous studies conducted in our lab have shown that a sperm concentration of 8 × 106 sperm/mL per IVF drop results in enhanced induction of polyspermy in a physiological and consistent manner compared with 4 × 106 sperm/mL and 16 × 106 sperm/mL (see Supplementary Fig. S1). Thus, to verify polyspermy induction rates, a sperm concentration of 1 × 106 sperm/mL per IVF drop was considered the control and a concentration of 8 × 106 sperm/mL was considered polyspermy induced. Control and polyspermy induced sperm concentrations were evaluated with regard to the rate of polyspermic zygote formation (Fig. 4A–D). The percentage of polyspermic embryos was increased 3.8 times by the polyspermy induced sperm concentration as shown in Fig. 4E. The polyspermy rate was 7.9 ± 4.7% for the control group and 29.8 ± 5.9% for the polyspermy induced group. The percentage of polyspermic zygotes presenting with exactly three pronuclei in the polyspermy group (17.5 ± 4.6%) was 2.9 times more than in the control group (6.1 ± 4.7%). In addition, the rate of polyspermic zygotes presenting four or more pronuclei was markedly lower in the control group (1.8 ± 0.4%), while the polyspermy induced group was 6.8 times higher (12.2 ± 3.1%).
Figure 4.
Polyspermic embryo validation and analysis of miR-216b relative level. (A–D) Representative fluorescence microscopy photomicrographs of pronuclei from zygotes of control and polyspermic groups. Scale bar: 100 μm. In (A,B), zygotes with two pronuclei. In (C,D), zygotes with more than three pronuclei. (E) Evaluation of pronucleus (PN) and polyspermy rate in bovine zygotes produced in vitro with control IVF (sperm concentration of 1 × 106 sperm/mL) or polyspermy IVF (sperm concentration of 8 × 106 sperm/mL). 12 PN: two pronuclei; 2 ≥ 3 PN: more than three pronuclei. (F) MiR-216b relative level in zygotes produced in vitro with control IVF (sperm concentration of 1 × 106 sperm/mL) or polyspermy IVF (sperm concentration of 8 × 106 sperm/mL) using semen samples from high (HF) and low (LF) fertility bulls. Capital letters indicate difference (P < 0.05) between groups (HF vs. LF) with the same treatment (Control). Asterisk (*) indicates difference (P < 0.05) between the treatments (control vs. polyspermy) in the same group. All quantitative data are presented as means and SEM.
MiR-216b is present at a higher level in polyspermic embryos
Since our data suggest that the levels of miR-216b might be modulated by sperm cells in zygotes, we expected that polyspermic embryos would have a higher level of miR-216b. Thus, after verifying polyspermy induction, the relative level of miR-216b was investigated in polyspermic and control embryos produced with selected HF and LF semen samples. The results showed that polyspermy increased the miR-216b relative level (Sperm concentration effect: P = 0.004) as shown in Fig. 4F. Furthermore, comparison between controls revealed that the relative level of miR-216b was higher in the LF group than in the HF group as shown previously.
Discussion
Fertility success depends on many factors including female, male and environmental influences. Among the male factors, the presence of healthy sperm cells is crucial for fertility2. Sperm quality attributes (SQA; morphological and functional attributes) and sperm molecular aspects are directly related to healthy sperm determination. Sperm quality attributes are usually highly correlated with male fertilization potential and are frequently a useful predictor of the latter4,25,26. However, some semen samples exhibit satisfactory SQA coupled with unexpected low fertility27. Thus, sperm molecular characteristics have been studied in order to better understand these instances. A particular sperm molecular characteristic that has been described concerns sperm miRNAs. Indeed, miRNAs are important for spermatogenesis12,28, sperm maturation13,29,30 and also embryo development10,14,20. Furthermore, we now know, in addition to delivering DNA to embryos, sperm cells are also able to deliver RNAs and miRNAs2,10,13,20,31,32. Therefore, in the present study, we investigated the SQA from semen samples obtained from HF and LF bulls as well the effect of sperm miRNAs contribution to pre-implantation embryonic development. Of interest is that differences between SQA in semen samples from HF and LF might not explain the difference in fertility rates. In addition, we demonstrated that the miR-216b level in zygotes is related with the level in sperm, and that this might be related to the transcriptional changes in the relative K-RAS gene levels found in two-cell embryos. Moreover, the reduced level of the K-RAS gene in LF two-cell embryos could be associated with the reduction in LF first cleavage rate. Of note is that while blastocyst rates were unaffected, we detected a lower number of cells in blastocysts derived from the LF group suggesting a reduction in the number of cell cycles up to the day-7 blastocyst stage. In addition, we observed that polyspermy zygotes displayed a higher level of miR-216b. Thus, our results provide, for the first time, a probable regulation of early embryonic development by sperm-borne miR-216b and reveal the modulation of its levels by producing polyspermic embryos.
Different fertility phenotypes have already been associated with specific sperm miRNA profiles in cattle18,33 and humans17,34. In the present study, we used commercial frozen semen samples from bulls that showed different pregnancy rates in field trials and similar sperm morphological and functional characteristics. Although the blastocyst rate for in vitro embryo production was similar between the HF and LF groups according to our previous results, a parallel prospective study was performed using the same batches used in the present study and revealed a difference of ~13% on the pregnancy rate between HF and LF semen samples35. The goal of this prospective study was to improve the use of low fertility semen samples in fixed-time artificial insemination (FTAI) programs. In this regard, 522 suckled cows from three different herds were subjected to FTAI. It was hypothesized that the delay in time of artificial insemination in FTAI would improve pregnancy rates from LF semen samples, but this was not confirmed. Hence, the results showed that pregnancy rates were not impacted by FTAI protocol modifications. In addition, our results revealed that blastocyst rates were similar between the fertility groups. Thus, we hypothesize that differences in fertility rates were probably due to functional characteristics related to early embryo development that could involve, among other factors, molecular components/miRNAs delivered by HF and LF sperm cells.
In the present study, we detected nine miRNAs that showed a difference in their relative levels of expression between LF and HF with a statistical significance of 10%. Of interest is that a large number of other miRNAs showed differences in relative abundance between high and low fertility bulls e.g. miR-19b-3p, -27a-5p, -34c-3p, -148b-3p, -320a -502-5p and -124918. Investigation of the profiles of nine sperm miRNAs revealed that one miRNA (miR-216b) was present at a higher relative level in sperm cells and also in zygotes from LF compared with HF. However, HF and LF two-cell embryos showed similar relative levels of this miRNA. Although hsa-miR-216b-5p is described in the SpermBase platform (http://spermbase.org/), to the best of our knowledge, our study is the first to report the importance of miR-216b to male fertility and early embryo development. However, in different types of cancer tumors, the role of miR-216b in inhibiting cell growth and proliferation is well described36,37.
MiR-216b has a limited number of target genes that have been conclusively identified. K-RAS, BECN1 and JUN were selected from among these for the present study due to their importance in embryo development21–24. Only K-RAS showed high relative levels in HF two-cell embryos compared with LF embryos. K-RAS is a member of the ras proto-oncogene family and plays a role in cell proliferation and differentiation38. It is the only member of the ras family with an important role in embryo development22,23. Depletion of K-RAS in mice results in embryo developmental deficiency and increased embryo mortality22,23. However, there is a lack of information regarding the importance of K-RAS to embryo development in cattle. Of interest is that the 3′ UTR of K-RAS is completely conserved among cattle, humans and mice. Moreover, it is well described that miR-216b targets K-RAS in many cell types. It has been documented in pancreatic39,40, nasopharyngeal41 and renal tumors42 that decreasing the abundance of the K-RAS gene consequently leads to low cellular proliferation and differentiation. Thus, we propose that lower levels of K-RAS in LF two-cell embryos is a temporal effect resulting from the higher level of miR-216b in LF zygotes provided by LF sperm cells. In fact, we showed here that sperm cells from LF bulls exhibited a higher level of miR-216b than sperm cells from HF bulls. Furthermore, we showed that the miR-216b level was higher when more than one sperm cell fertilized the oocyte to produce polyspermic embryos.
Since K-RAS is related to cell proliferation, we investigated the rates of first cleavage, cleavage on day-4, blastocyst stage on day-7, blastocyst proliferation, blastocyst cell number and number of cell divisions during initial development. We also evaluated the two-cell and four-cell rates using a developmental kinetic model. We hypothesized that HF embryos would show more proliferation. Indeed, with respect to these parameters, first cleavage rate, blastocyst cell number and blastocyst cell division number were higher in HF embryos compared with LF. Of interest is that HF embryos underwent an additional cell cycle compared with LF embryos (see Fig. 3D). The higher cell number and the higher number of cell divisions in HF blastocysts could indicate some beneficial effect to the establishment of pregnancy. In humans, a higher number of cells in transferred embryos is highly associated with pregnancy success43. In fact, blastocysts are able to produce and secrete factors, called embryotropins44, involved in modulating endometrium transcripts mediated by interferon-tau, prostaglandin metabolism and aquaporin channels45. In light of this, we propose that embryos with a high first cleavage rate and high cell number could release an increased quantity of embryotropins44 that are then able to modulate and signal the presence of the embryo to the endometrium45, thereby influencing in vivo fertility success. However, this hypothesis is yet to be confirmed and further studies are necessary to confirm this association.
In addition to the importance of miR-216b to embryo development, we showed, using a polyspermy induction model, that we can modulate the miR-216b level in zygotes by increasing the sperm cell concentration per IVF drop. Indeed, we revealed that the increasing sperm cell entry into oocytes at fertilization is able to increase the levels of miR-216b in zygotes. We produced polyspermic zygotes by increasing the number of sperm cells per IVF drop while maintaining the oocyte physiology as intact as possible to verify that the alteration was caused by increasing the number of sperm cells entering each oocyte. The zygotes were evaluated in terms of the relative level of miR-216b. In polyspermic embryos, miR-216b showed increased relative levels, demonstrating that sperm cells might modulate miR-216b levels in the embryo. However, the relative levels of miR-216b in HF and LF polyspermy embryos were unexpectedly similar. This might be attributed to the fact that we were unable to sort the polyspermic embryos prior to PCR, and we were also unable to control the number of pronuclei generated in these embryos. In addition, the miR-216b level might be modulated by sperm-related factors that stimulate miR-216b transcription in LF embryos or that inhibit it in HF embryos; this hypothesis could justify the similar levels of miR-216b in HF and LF polyspermy embryos since the transcription process might reach a maximum rate. However, further studies are necessary to verify this probable new mechanism of sperm-borne modulation.
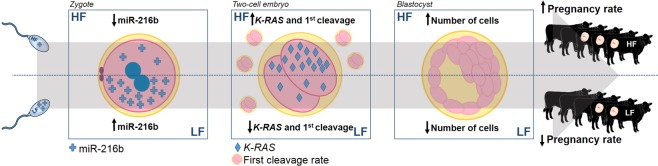
In conclusion, our results shed light on a probable mechanism by which sperm cells are able to contribute miR-216b to zygotes, thereby modulating early embryo development. In summary, our data showed that miR-216b was present at a lower level in sperm cells from HF bulls. This was associated with a lower relative level of miR-216b in HF zygotes and with a higher relative level of its target gene, K-RAS, in HF two-cell embryos, implicating an increase in first cleavage rate and blastocyst cell number (Fig. 5). Finally, our data provide a better understanding of how paternal contributions could modulate early embryo development. The present study may open new avenues for the implications of sperm miRNAs in fertility regulation and establishment of healthy pregnancy in cattle.
Figure 5.
Hypothetical schematic model. Schematic figure demonstrating that sperm cells from high fertility (HF) bulls might deliver a lower level of miR-216b to zygotes than sperm from low fertility (LF) bulls. Thus, zygotes from HF may display a lower miR-216b level than zygotes from LF and its target gene, K-RAS, may exhibit a higher level in two-cell embryos from HF than LF. Furthermore, this might result in a higher first cleavage rate in HF embryos. These changes are probably reflected in the higher HF blastocyst cell number, which might be beneficial to in vivo development and thereby could increase pregnancy rates in cattle.
Methods
Ethics statement
All procedures performed during this study were conducted in accordance with ethical guidelines of research and animal care and were approved by the Ethics Committee on Animal Use of the School of Veterinary Medicine and Animal Science of the University of São Paulo (CEUA/FMVZ) under protocol number 1912050516. We adopted the International Guiding Principles for Biomedical Research Involving Animals (Society for the Study of Reproduction) as well. All schematic images were made by the authors or obtained from the Mind the Graph platform (https://mindthegraph.com/).
Reagents and chemicals
Unless otherwise stated, all reagents and chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA). Fluorescent probes used were: Hoechst 33342 (Hoechst; reference number H1399), 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1; T3168), CellROXTM Deep Red (CellROX Red; C10422), SYTO™ 59 Red (Syto59; S11341), MitoSOXTM Red (MitoSOX, M36008), YO-PROTM-1 iodide (YoPro, Y3603), C11-BODIPYTM 581/591 (Bodipy, D3861) and Acridine Orange (AO; A1301) purchased from Thermo Fisher Scientific (Waltham, Massachusetts, USA); and propidium iodide (PI; P4170), fluorescein isothiocyanate-conjugated Pisum sativum agglutinin (FITC-PSA; L0770) and fluorescein isothiocyanate-conjugated Arachis hypogaea (FITC-PNA, L7381) purchased from Sigma-Aldrich.
Experiment 1: Lower level of miR-216b in sperm cells and zygotes is associated with a high level of K-RAS in two-cell embryos and an increase in first cleavage rate and blastocyst cell number
Experimental design
Six commercial batches of frozen–thawed semen from high fertility (HF; n = 3; 54.3 ± 1% pregnancy rate) and low fertility (LF; n = 3; 41.5 ± 2.3%) Aberdeen Angus (Bos taurus) bulls were used. The bulls’ field fertility rates were different (P = 0.007) and determined by the Concept Plus® program developed by Alta Genetics® (Uberaba, MG, Brazil). High and low fertility bulls were evaluated respectively in 396 herds and 52,259 services and in 22 herds and 3,405 services. First, all batch samples were evaluated for sperm quality. Then, one batch from each bull was selected, analysed for sperm quality and for a profile of 380 miRNAs, and then used for in vitro embryo production.
Sperm quality assessment and semen batch selection
All sperm quality analyses were conducted in duplicate by the same technician. Two semen straws (0.25 mL) from the same batch were thawed (37 °C/30 seconds) and the total volume (~500 µL) was placed in a pre-warmed tube and homogenized before proceeding with sperm quality assessment. Semen evaluation was performed as soon the semen was thawed and homogenized.
Assessment of sperm motility, concentration and abnormalities
Contrast phase microscopy (100× magnification) was used to evaluate (Nikon, 80i model, Tokyo, Japan) subjective sperm motility (%) and vigor (1 to 5, whereby 1 corresponds to immotile sperm cells and 5 corresponds to sperm cells with progressive and fast movement). Sperm concentration was evaluated by diluting semen in formaldehyde 4% (1:100) using a Neubauer chamber under magnification at 400×. The percentage of abnormalities was evaluated by fixing the semen sample in formaldehyde 4% and classifying 200 sperm cells as having major, minor and total defects (major plus minor)46 (Supplementary Table S9) using differential interference contrast microscopy (Nikon, 80i model, Tokyo, Japan) under 1,000× magnification.
Assessment of sperm kinetic
Sperm kinetic parameters were evaluated by Computer-Assisted Sperm Analyser (CASA) using Sperm Class Analyser (SCA, Microptics, Barcelona, Spain) software. Set up was adjusted for bovine sperm (Supplementary Table S10). The sperm concentration was adjusted to 10 × 106 sperm/mL in Tyrode’s albumin lactate pyruvate medium (TALP sperm: 4.2 mg/mL sodium chloride, 1.87 mg/mL potassium chloride, 2.1 mg/mL sodium bicarbonate, 50 µg/mL sodium phosphate, 290 µg/mL calcium chloride monohydrate, 80 µg/mL magnesium chloride hexahydrate and 6.5 mg/mL Hepes, supplemented with 10 mg/mL bovine serum albumin - pH adjusted to 7.4 with NaOH 1N). Then, 10 µL of diluted semen was placed on a Makler® chamber (Selfi-Medical Instruments, Haifa, Israel) and evaluated for total motility (%), progressive motility (%), rapid cells (%), curvilinear velocity (VCL, μm/s), path velocity (VAP, μm/s), progressive velocity (VSL, μm/s), linearity (LIN, %), straightness (STR, %), wobble (WOB, %), lateral head displacement (ALH, μm) and beat cross frequency (BCF, Hz).
Assessment of sperm membranes and reactive oxygen species production
Epifluorescence microscopy (Nikon, model 80i, Tokyo, Japan) at 1,000× magnification using a triple filter (D/F/R, C58420) with UV-2E/C (excitation 340–380 nm/emission 435–485 nm), B-2E/C (excitation 465–495 nm/emission 515–555 nm) and G-2E/C (excitation 540–525 nm/emission 605–655 nm) was used for analyses of plasma and acrosome membrane integrity and mitochondrial membrane potential, and reactive oxygen species (ROS) production. For assessment of sperm plasma and acrosome membrane integrity and mitochondrial membrane potential47, 2 µL of Hoechst 0.5 mg/mL, 2 µL of PI 0.5 mg/mL, 6 µL of JC-1 153 µM and 20 µL of FITC-PSA 100 µg/mL were added to 150 µL of semen diluted to 10 × 106 sperm/mL in TALP sperm. The incubation was performed at 37 °C/8 minutes. Two hundred sperm cells were then classified (percentage) according to PIAIH (sperm cells with plasma and acrosome membrane integrity, and high potential of mitochondrial membrane), integrity of plasma membrane, integrity of acrosome membrane and high potential of mitochondrial membrane. Production of ROS48 was evaluated by the addition of 1.2 µL of CellROX Red 2.5 mM and 2 µL of Hoechst 0.5 mg/mL to 150 µL diluted semen (10 × 106 sperm/mL). The incubation was performed at 37 °C/30 minutes. Samples were then centrifuged at 600 g for 5 minutes and resuspended in TALP sperm. Two hundred sperm cells were then classified (percentage) as either positive or negative for ROS production.
Semen batch selection
The selection of one semen batch per bull was performed according to the highest value obtained using the formula:
where PROG was progressive motility, MAJ was major defects, MIN was minor defects and PIAIH was sperm with plasma and acrosome membrane integrity and high mitochondrial membrane potential.
Sperm quality assessment of selected-semen batches
The selected semen batches were evaluated by all the previously described analyses and also by flow cytometry using the BD Accuri C6TM (Becton-Dickinson, San Jose, CA, USA) equipped with an argon laser (488 nm) and red laser (635 nm) both used to excite the samples. The following four filters were used in the analyses: 533/30 nm, 585 nm/40 nm, 675/25 nm and >670 nm. Particles and cellular debris were excluded from the acquisition using size and scatter properties and also by staining sperm samples with 2 µL of the nuclear dye Syto59 750 mM. Manual compensation analysis was performed adding controls: sperm samples were submitted to flash-freezing for plasma, acrosome and mitochondrial membrane controls47; for superoxide sperm generation controls, sperm samples were incubated with Antimycin A (20 µM) and DPI (diphenyleneiodonium chloride; 10 µM) at 37 °C for 1 hour49; for lipid peroxidation controls, sperm samples were incubated with ROS generator system (iron sulfate 4 mM, sodium ascorbate 20 mM and hydrogen peroxide 4 mM) at 37 °C for 30 minutes48; finally, for DNA fragmentation controls, sperm samples were exposed to hydrogen peroxide 10 mM for 1 hour50.
Acquisition rate was approximately 600 to 1,000 events/second, totalling 5,000 to 10,000 cells per analysis. Sperm concentration was adjusted to 5 × 106 sperm/mL with TALP sperm for the incubation and adjusted to 2.5 × 106 sperm/mL for flow cytometry analysis. For sperm plasma and acrosome membrane integrity, 2 µL of Syto59 750 mM, 1 µL of PI 0.5 mg/mL and 1 µL of FITC-PNA 37.5 µg/mL were added to 150 µL of diluted semen and incubated at 37 °C/10 minutes. For mitochondrial membrane potential evaluation, 2 µL of Syto59 750 mM, 1 µL of PI 0.5 mg/mL and 1 µL of JC-1 153 µM were added to 150 µL of diluted semen and incubated at 37 °C/10 minutes. For production of superoxide anion by mitochondria, 1.5 µL of MitoSOX 2 µM was added to 150 µL of diluted semen and incubated at 37 °C/30 minutes. After a 10-minute incubation, 1 µL of YoPro 7.5 µM was added; and at 20 minutes, 1.5 µL of Syto59 750 mM was added. For lipid peroxidation analysis, 1 µL of BODIPY 2 mM was added to 150 µL of diluted semen and incubated at 37 °C/30 minutes. At 20 minutes, 1 µL of PI 0.5 mg/mL and 2 µL of Syto59 750 mM were added. For DNA fragmentation, AO was used according to the protocol described by Simões et al.51.
Sperm miRNA levels
Two semen straws (0.25 mL) from the same batch were thawed (37 °C/30 seconds) and each 200 µL sample was placed into a Percoll gradient (45% and 90%) and centrifuged at 3,600 g for 7 minutes. The pellet was resuspended in PBS (10 mg/mL sodium chloride, 0.2 mg/mL potassium chloride, 1.44 mg/mL sodium phosphate dibasic, and 0.24 mg/mL potassium phosphate) and centrifuged twice at 520 g for 5 minutes. RNA extraction and purification were performed with miRNeasy Mini Kit® (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA concentration and purity were determined by NanoDrop™ 1000 (Thermo Scientific, Massachusetts, USA) spectrophotometer. Complementary DNA (cDNA) was generated using miScript RT Kit® (Qiagen, Hilden, Germany) by adding a volume of approximately 100 ng of total RNA to HiFlex buffer, 10 × nucleic acid mix, reverse transcriptase enzyme and RNAse/DNAse-free water followed by incubation at 37 °C/1 hour and 95 °C/5 minutes. Next, the abundance of a customized panel of 380 bovine-specific miRNAs (Supplementary Table S11) was evaluated by qPCR. The primer sequences were obtained from the miRBase database (www.mirbase.org). Bta-miR-99b and -425-5p, consistently detected among the groups according to the NormFinder platform18,52,53, were used to generate the relative levels. The PCR mix was prepared using SYBR Green® (Qiagen, Hilden, Germany). A 384-well plate was used for PCR and the reaction was performed on QuantStudioTM 6 Flex (Thermo Fisher Scientific) with the following cycle conditions: an initial incubation of 95 °C/15 minutes and 45 cycles of 94 °C/15 seconds, 55 °C/30 seconds and 70 °C/30 seconds. The melting-curve was used to confirm the amplification of a single product.
In vitro embryo production
HF (n = 3) and LF (n = 3) selected batches were used to produce embryos. In vitro embryo production was performed in triplicate for each bull to give a total of nine HF and LF replicates. Parthenogenetic embryos and mature oocytes were produced in all replicates. Incubator conditions for oocytes, sperm cells and embryos were maintained at 38.5 °C during all steps under 5% CO2, air (20% oxygen) with saturating humidity. For embryo production, bovine ovaries were collected from a local slaughterhouse and transported at 35 °C in thermal bottles filled with sterile saline solution 0.9%. In the laboratory, ovarian follicles (3 to 6 mm) were aspirated with 18 G needles. The evaluation of cumulus–oocyte complexes (COCs) was performed under a stereomicroscope (SMZ 745T model, Nikon, Tokio, Japan). Then, COCs were washed in tissue culture medium 199 (TCM 199, Gibco, Thermo Scientific) supplemented with 10% extracellular vesicle-depleted foetal bovine serum (EV-free FBS; extracellular vesicle-depletion was performed by ultracentrifugation at 120,000 g for 16 hours), 22 µg/mL sodium pyruvate and 50 µg/mL gentamicin. Only grade I and grade II COCs54 were selected and washed in maturation medium [TCM 199 supplemented with 26 mM sodium bicarbonate, 10% EV-free FBS, 22 µg/mL sodium pyruvate, 50 µg/mL gentamicin, 0.5 µg/mL follicle-stimulating hormone (FSH; FolltropinTM, Bioniche Animal Health, Belleville, Canada) and 5 U/mL human chorionic gonadotropin (hCG; VetecorTM, Hertape Calier, London, England)]. A total of 20 to 25 COCs were placed in 90 µL of maturation medium covered with mineral oil for 22 hours. After a period of in vitro maturation (IVM), oocytes were divided into three groups: parthenogenetic embryos (control, without sperm cells) and in vitro fertilized embryos with sperm from high and low fertility bulls; also, pools of five mature oocytes were collected from each replicate. For parthenogenetic embryos, presumptive mature oocytes were completely denuded with 0.2% hyaluronidase in PBS and were evaluated for extrusion of the first polar body. Only mature oocytes (oocytes that present extrusion of first polar body) were submitted to parthenogenetic activation (at 26 hours post IVM) by incubation in 5 µM ionomycin in HEPES-buffered TCM-199 for 5 minutes and 2 mM 6-dimethylaminopurine (6-DMAP) diluted in modified synthetic oviduct fluid (SOF) for 3 hours55. For in vitro fertilization (IVF), presumptive mature oocytes were incubated for 18 hours in IVF medium composed of Tyrode’s lactate stock supplemented with 50 µg/mL gentamicin, 22 µg/mL sodium pyruvate, 40 µg/mL PHE (2 mM D-penicillamine, 1 mM hypotaurine and 245 µM epinephrine), 5.5 IU/mL heparin and 6 mg/mL bovine serum albumin (BSA) and sperm cells from frozen–thawed semen from each bull with either high or low fertility. Each batch was processed on a Percoll gradient (45% and 90%) to obtain sperm cells at a concentration of 1 × 106 sperm/mL. Presumptive zygotes from each group were divided into drops for evaluation and collection of zygotes, two-cell embryos and blastocysts. The presumptive zygotes were completely denuded using a pipette and were cultured in synthetic oviductal fluid with amino acids, sodium citrate and inositol (SOF) supplemented with 5 mg/mL BSA, 22 µg/mL sodium pyruvate, 50 µg/mL gentamicin and 2.5% EV-free FBS. Zygotes were evaluated and collected 14 to 16 hours post activation (hpa) or 16 to 18 hours post insemination (hpi; second polar body extrusion evaluation); two-cell embryos were evaluated and collected 20 to 22 hpa or 28 to 30 hpi. Cleavage rates were assessed on day-4 of development and blastocysts were evaluated and collected on day-7. Samples were either stored at −80 °C for miRNA and mRNA analyses, or fixed in paraformaldehyde 4% in PBS with 0.1% PVP (PBS + PVP) for imaging analysis.
Embryo mRNA and miRNA levels
Prior to being snap frozen, the zona pellucida of zygotes and two-cell embryos was removed by enzymatic digestion with pronase 0.5% (Protease from Streptomyces griseus). Nine pools of five embryos and three pools of five oocytes were snap frozen in PBS + PVP and used to evaluate miRNA and mRNA levels. RNA extraction and purification were performed using miRNeasy Mini Kit® (Qiagen, Hilden, Germany) according to the manufacturer’s instructions; concentration and purity were determined by NanoDrop™ 1000 (Thermo Scientific, Massachusetts, USA) spectrophotometer. For miRNA analyses, cDNA was generated using the miScript RT Kit® (Qiagen, Hilden, Germany) as previously described. Only miRNAs differentially abundant between sperm cells from HF and LF bulls were evaluated by qPCR. Bta-miR-99b and RNU43 snoRNA were consistently detected among the groups and were used to evaluate the relative levels of these miRNAs. The qPCR reaction was performed under the same conditions detailed before. For mRNA analyses, cDNA was generated using High-Capacity Kit® (Applied Biosystem, Foster City, California, USA) according to the manufacturer’s instructions. Next, the levels of miRNA target genes that were different in sperm cells and zygotes were analysed by qPCR. Target genes were determined by first evaluating the homology between bovine miRNAs and human miRNAs using the miRBase platform (http://www.mirbase.org/). Strongly validated target genes of human miRNAs were then researched on the miRTarBase platform (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). Conservation of the 3′ untranslated region (UTR) of the target gene was also investigated using the TargetScanHuman platform (http://www.targetscan.org/vert_72/). Target gene primer sequences (Supplementary Table S12) were designed based on GenBank and sequence specificity was verified by Nucleotide Blast of the NCBI platform (https://blast.ncbi.nlm.nih.gov/). β-Actin was used as a housekeeping gene. qPCR was performed with 8.5 ng of RNA per reaction following the manufacturer’s instructions for the Power Sybr® kit (Applied Biosystem, Foster City, California, USA). The PCR reaction was performed on a QuantStudioTM 6 Flex (Thermo Fisher Scientific) with the following cycle conditions: an initial incubation of 95 °C/10 minutes and 40 cycles of 95 °C/15 seconds and 60 °C/60 seconds. The melting-curve was used to confirm the amplification of a single product.
Blastocyst qualitative analysis
On day-7 following IVF or activation, blastocysts were evaluated according to stage (early blastocyst, blastocyst, expanded and hatched). Blastocysts were also collected and fixed for immunofluorescence proliferation evaluation: eight blastocysts from the HF group and 10 blastocysts from the LF group were fixed for 12 minutes in 4% PFA in PBS + PVP. Next, blastocysts were permeabilized with 1% Triton X-100 for 20 minutes and blocked with 5% BSA and 22 mg/mL glycine for 1 hour at room temperature. They were then incubated overnight at 4 °C with a rabbit anti-Ki-67 (1:100, 5500134, Sigma-Aldrich) primary antibody. Primary antibody was detected with anti-rabbit IgG AlexaFluor 488 (1:500, A11008, Thermo Fisher Scientific). Finally, the embryos were stained with 10 µL/mL Hoechst/15 minutes. Blastocyst slides were mounted with Prolong Antifade® (Thermo Fisher Scientific). The images were captured by confocal microscopy SP5 (Leica, Wetzlar, Germany) using argon (488 nm) and diode (405 nm) laser under 400× magnification. A negative control was performed for each set of analyses. Blastocyst diameter, number of cells stained with Hoechst 33342 and stained with anti-Ki-67 were analysed using Z-project on ImageJ® (NIH, https://imagej.nih.gov/ij/). The percentage of proliferation was determined by calculating the number of cells stained with Ki-67 divided by the total of cells stained with Hoechst. The cell number was determined by counting Hoechst-stained cells. The cell division number (n) was calculated using the formula: Log2(n) = cell number.
Embryo kinetic evaluation
Early embryonic kinetics of HF (n = 3) and LF (n = 3) selected batches were evaluated by subjecting semen samples for in vitro embryo production as previously described with an IVF duration of 8 hours. Embryos were evaluated with regard to the two-cell and four-cell rates 28 to 30 hpi and 40 to 42 hpi, respectively.
Experiment 2: Higher level of miR-216b is associated with polyspermic zygotes
Semen samples and experimental design
For this experiment, in order to select semen samples that present the greatest difference in miR-216b relative levels, frozen–thawed semen from two bulls (one semen sample from one HF bull and one semen sample from one LF bull) were selected based on the results of experiment 1. For validation of polyspermy induction, the HF and LF semen samples selected were pooled and tested to give a total of seven replicates. Afterwards, a total of five HF and LF semen replicates were individually evaluated to determine the relative levels of miR-216b in polyspermic embryos.
Validation of polyspermy induction and polyspermy evaluation
To verify polyspermy induction rates, a sperm concentration of 1 × 106 sperm/mL per IVF drop was considered the control and a concentration of 8 × 106 sperm/mL was considered polyspermy induced. Both concentrations were tested in seven replicates using pools of the selected semen batches from HF and LF bulls. IVM and IVF were performed as described before, with the exception that the sperm concentration in the IVF drop varied according to the group. Second polar body-positive embryos were collected and fixed 12 hpi. Embryo fixation was performed with 4% PFA in PBS + PVP and embryos were stained with 10 µL/mL Hoechst/15 minutes. Slides with embryos were mounted with Prolong Antifade® and the number of pronuclei was evaluated by epifluorescence microscopy (Nikon, model 80i, Tokyo, Japan) at 600× magnification using UV-2E/C filter (excitation 340–380 nm emission 435–485 nm).
MiR-216b relative level in polyspermic embryos
HF and LF semen samples selected according to miR-216b relative level were used to perform five replicates of IVF with adjusted sperm concentrations for control (1 × 106 sperm/mL) and polyspermy induction (8 × 106 sperm/mL). Embryos were collected 16 to 18 hpi and stored at −80 °C to evaluate the abundance of miR-216b. Five pools of 15 embryos were subjected to RNA extraction and the relative levels of miR-216b were verified by qPCR following the protocols previously described.
Statistical analysis
Statistical Analysis System (SAS Institute Inc, 9.3) software was used for all statistical analysis. Shapiro-Wilk was performed to evaluate the normality of data. When necessary, data were transformed or outliers were removed. The significance level was P < 0.05 except for miRNAs in sperm cells where P ≤ 0.10. Analysis of variance (ANOVA) using the mixed procedure was performed to evaluate sperm characteristics, the miRNA and mRNA relative levels and blastocyst quality features. The Tukey test was used when the parthenogenetic and/or mature oocyte groups were added. The Chi-Square frequency test was performed to evaluate embryo development and polyspermy rates. Analysis of variance (ANOVA) using the mixed procedure respecting the factorial design was performed to evaluate the relative level of miR-216b in polyspermic embryos (Experiment 2). For molecular analysis, ΔCT was considered for statistical analysis and relative level data were presented as 2−ΔCT as shown in the graphics, tables and Venn Diagram (http://bioinfogp.cnb.csic.es/tools/venny/). Exclusively miRNA detection was based on the miRNA detection in at least two semen samples. Non-detection was considered when it appeared in only one or none of the samples.
Supplementary information
Acknowledgements
The present study was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (M.B.R.A. was granted by FAPESP 2015/09154-6 and 2016/23243-4; E.C.C.C. was granted by FAPESP 2016/05395-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank A.F.C. Andrade and M.A. Torres of Laboratory of Andrology and Technology of Swine Embryos of School of Veterinary Medicine and Animal Science, Universidade de São Paulo for technical assistance during flow cytometry analysis; Olhos D’Água slaughterhouse for providing ovaries; and Alta Genetics for providing semen samples and field fertility data of the bulls used on the present study.
Author Contributions
M.B.R.A., R.P.A., T.H.C.D.B., J.C.S., F.P. and E.C.C.C. designed the study; M.B.R.A. and S.A.F.R. performed semen analyses; M.B.R.A. and T.H.C.D.B. performed embryo production, collection of the samples and immunofluorescence analysis; M.B.R.A. performed molecular and images analyses; M.B.R.A., J.C.S., F.P. and E.C.C.C. analysed the data; M.F.S.F. assessed the bulls’ fertility data; M.B.R.A. wrote the manuscript; M.B.R.A, T.H.C.D.B., C.B., F.V.M., J.C.S., F.P. and E.C.C.C. discussed the results. All authors reviewed the manuscript.
Data Availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
M.F.S.F. is employed by Alta Genetics; M.B.R.A., R.P.A., T.H.C.D.B., S.A.F.R., C.B., F.V.M., J.C.S., F.P. and E.C.C.C. have nothing to declare.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46775-8.
References
- 1.Vincent P, et al. Bovine semen quality control in artificial insemination centers. Anim. Reprod. 2012;9:153–165. [Google Scholar]
- 2.Krawetz SA. Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 3.Zhang BR, Larsson B, Lundeheim N, Ha MGH. Prediction of bull fertility by combined in vitro assessments of frozen-thawed semen from young dairy bulls entering an AI-programme. Int. J. Androl. 1999;22:253–260. doi: 10.1046/j.1365-2605.1999.00178.x. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira BM, et al. Fertility and uterine hemodynamic in cows after artificial insemination with semen assessed by fluorescent probes. Theriogenology. 2014;82:767–772. doi: 10.1016/j.theriogenology.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira LZ, et al. Assessment of field fertility and several in vitro sperm characteristics following the use of different Angus sires in a timed-AI program with suckled Nelore cows. Livest. Sci. 2012;146:38–46. doi: 10.1016/j.livsci.2012.02.018. [DOI] [Google Scholar]
- 6.Gillan L, Kroetsch T, Chis Maxwell WM, Evans G. Assessment of in vitro sperm characteristics in relation to fertility in dairy bulls. Anim. Reprod. Sci. 2008;103:201–214. doi: 10.1016/j.anireprosci.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Amann RP, Hammerstedt RH. In vitro evaluation of sperm quality: an opinion. J. Androl. 1993;14:397–406. [PubMed] [Google Scholar]
- 8.Sellem E, et al. Use of combinations of in vitro quality assessments to predict fertility of bovine semen. Theriogenology. 2015;84:1447–1454. doi: 10.1016/j.theriogenology.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 9.Saunders, C. M. et al. PLCζ: a sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. 3544, 3533–3544 (2002). [DOI] [PubMed]
- 10.Liu W, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotaja N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014;101:1552–1562. doi: 10.1016/j.fertnstert.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Conine CC, Sun F, Song L, Rivera-Pérez JA, Rando OJ. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell. 2018;46:470–480. doi: 10.1016/j.devcel.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J. Androl. 2005;26:70–74. [PubMed] [Google Scholar]
- 16.Krawetz SA, et al. A survey of small RNAs in human sperm. Hum. Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-halima M, et al. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013;99:1249–1255. doi: 10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 18.Fagerlind M, Stalhammar H, Olsson B, Klinga-Levan K. Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reprod. Domest. Anim. 2015;50:587–594. doi: 10.1111/rda.12531. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, et al. Sperm-borne miR-449b influences cleavage, epigenetic reprogramming and apoptosis of SCNT embryos in bovine. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan S, et al. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 2016;143:635–647. doi: 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RS, Van Lingen B, Papaioannou VE, Spiegelman BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 22.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koera K, et al. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- 24.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira LZ, et al. Assessment of in vitro sperm characteristics and their importance in the prediction of conception rate in a bovine timed-AI program. Anim. Reprod. Sci. 2013;137:145–155. doi: 10.1016/j.anireprosci.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Tartaglione CM, Ritta MN. Prognostic value of spermatological parameters as predictors of in vitro fertility of frozen-thawed bull semen. Theriogenology. 2004;62:1245–1252. doi: 10.1016/j.theriogenology.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Kovac JR, Pastuszak AW, Lamb DJ. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil. Steril. 2013;99:998–1007. doi: 10.1016/j.fertnstert.2013.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero Y, et al. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jerczynski O, et al. Role of Dicer1-dependent factors in the paracrine regulation of epididymal gene expression. PLoS One. 2016;11:e0163876. doi: 10.1371/journal.pone.0163876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Björkgren I, et al. Dicer1 ablation in the mouse epididymis causes dedifferentiation of the epithelium and imbalance in sex steroid signaling. PLoS One. 2012;7:e38457. doi: 10.1371/journal.pone.0038457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sendler E, et al. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41:4104–4117. doi: 10.1093/nar/gkt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:2603. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 33.Govindaraju A, et al. Dynamics of microRNAs in bull spermatozoa. Reprod. Biol. Endocrinol. 2012;10:1–10. doi: 10.1186/1477-7827-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salas-Huetos A, et al. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil. Steril. 2015;104:591–601. doi: 10.1016/j.fertnstert.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Zanatta GM, et al. Effect of the delay at TAI in Nelore cows (Bos indicus) using low fertility bulls semen. Anim. Reprod. 2017;14:660. [Google Scholar]
- 36.He S, et al. MiR-216b inhibits cell proliferation by targeting FOXM1 in cervical cancer cells and is associated with better prognosis. BMC Cancer. 2017;17:1–12. doi: 10.1186/s12885-016-3022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. MicroRNA-216b reduces growth, migration and invasion of pancreatic ductal adenocarcinoma cells by directly targeting ρ-associated coiled-coil containing protein kinase 1. Oncol. Lett. 2018;15:6745–6751. doi: 10.3892/ol.2018.8109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, et al. MiR-216b inhibits pancreatic cancer cell progression and promotes apoptosis by down-regulating KRAS. Arch. Med. Sci. 2018;14:1321–1332. doi: 10.5114/aoms.2018.72564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferino A, et al. MicroRNA therapeutics: design of single-stranded miR-216b mimics to target KRAS in pancreatic cancer cells. RNA Biol. 2018;15:1273–1285. doi: 10.1080/15476286.2018.1526536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng M, et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J. Cell Sci. 2011;124:2997–3005. doi: 10.1242/jcs.085050. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, et al. miR-216b Post-transcriptionally downregulates oncogene KRAS and inhibits cell proliferation and invasion in clear cell renal cell carcinoma. Cell. Physiol. Biochem. 2018;49:1755–1765. doi: 10.1159/000493621. [DOI] [PubMed] [Google Scholar]
- 43.Kong X, et al. The relationship between cell number, division behavior and developmental potential of cleavage stage human embryos: a time-lapse study. PLoS One. 2016;11:e0153697. doi: 10.1371/journal.pone.0153697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wydooghe E, et al. Autocrine embryotropins revisited: how do embryos communicate with each other in vitro when cultured in groups? Biol. Rev. 2017;92:505–520. doi: 10.1111/brv.12241. [DOI] [PubMed] [Google Scholar]
- 45.Sponchiado M, et al. Pre-hatching embryo-dependent and -independent programming of endometrial function in cattle. PLoS One. 2017;12:e0175954. doi: 10.1371/journal.pone.0175954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blom E. The ultrastructure of some characteristic sperm defects and a proposal for a new classification of the bull spermiogram. Nord. Vet. Med. 1973;25:383–391. [PubMed] [Google Scholar]
- 47.Celeghini ECC, de Arruda RP, de Andrade AFC, Nascimento J, Raphael CF. Practical techniques for bovine sperm simultaneous fluorimetric assessment of plasma, acrosomal and mitochondrial membranes. Reprod. Domest. Anim. 2007;42:479–488. doi: 10.1111/j.1439-0531.2006.00810.x. [DOI] [PubMed] [Google Scholar]
- 48.Alves MBR, et al. An efficient technique to detect sperm reactive oxygen species: the CellRox Deep Red® fluorescent probe. Biochem. Physiol. Open Access. 2015;4:1–5. [Google Scholar]
- 49.Li Y, Trush MA. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 1998;253:295–299. doi: 10.1006/bbrc.1998.9729. [DOI] [PubMed] [Google Scholar]
- 50.Villani P, et al. Sperm DNA fragmentation induced by DNAse I and hydrogen peroxide: an in vitro comparative study among different mammalian species. Reproduction. 2010;140:445–452. doi: 10.1530/REP-10-0176. [DOI] [PubMed] [Google Scholar]
- 51.Simões R, et al. Influence of bovine sperm DNA fragmentation and oxidative stress on early embryo in vitro development outcome. Reproduction. 2013;146:433–441. doi: 10.1530/REP-13-0123. [DOI] [PubMed] [Google Scholar]
- 52.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 53.Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64.1–R64.10. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Bem THC, et al. The Influence of morphology, follicle size and Bcl-2 and bax transcripts on the developmental competence of bovine Oocytes. Reprod. Domest. Anim. 2014;49:576–583. doi: 10.1111/rda.12325. [DOI] [PubMed] [Google Scholar]
- 55.De Bem THC, et al. Low levels of exosomal-miRNAs in maternal blood are associated with early pregnancy loss in cloned cattle. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.