Abstract
Excessive reactive oxygen species (ROS) generated in mitochondria is known to be a causal event in diabetic cardiomyopathy. Recent studies suggest that microRNAs (miRNAs) are able to translocate to mitochondria to modulate mitochondrial activities, but the roles of such miRNAs in diabetic cardiomyopathy remain unclear. We observed a marked reduction of mitochondrial gene cytochrome-b (mt-Cytb) in the heart of db/db mice compared with controls. Downregulation of mt-Cytb by small interfering RNA (siRNA) recaptured some key features of diabetes, including elevated ROS production. Microarray revealed that none of the miRNAs were upregulated, but 14 miRNAs were downregulated in mitochondria of db/db heart. miR-92a-2-5p and let-7b-5p targeted mt-Cytb and positively modulated mt-Cytb expression. Re-expression of miR-92a-2-5p and let-7b-5p into cardiomyocytes led to reduced ROS production. Furthermore, recombinant adeno-associated virus (rAAV)-mediated delivery of miR-92a-2-5p, but not let-7b-5p, was sufficient to rescue cardiac diastolic dysfunction in db/db heart. Let-7b-5p not only upregulated mt-Cytb in mitochondria, but also downregulated insulin receptor substrate 1 in cytosol and finally lead to no efficiency for improvement of diastolic dysfunction in db/db mice. Our findings demonstrate that reduced mitochondrial miRNAs contribute to impaired mitochondrial gene expression and elevated ROS production. Re-expression of miR-92a-2-5p enhances mitochondrial translation and reduces ROS production and lipid deposition, which finally rescues diabetic cardiomyopathy.
Keywords: diabetic cardiomyopathy, mitochondrial miRNAs, diastolic dysfunction, mitochondrial Cytb
Introduction
Cardiovascular complications are the leading causes of diabetes-related morbidity and mortality.1, 2 In the near future, the cardiovascular complications will be able to account for 75% of deaths among the diabetes population.3, 4 Despite advances in developing anti-hyperglycemia therapies, the incidence of heart failure in diabetic patients was not significantly improved,5, 6 thus stressing the importance of understanding the diabetic cardiomyopathy pathogenesis in order to develop new intervention strategies.
MicroRNAs (miRNAs) are a class of small (∼22 nt) non-coding RNAs that negatively regulate gene expression at post-transcriptional levels.7 Interestingly, recent data suggest that miRNAs that may also regulate gene expression in a positive fashion.8 A recent report demonstrates that miR-1, a miRNA specifically induced during myogenesis, not only efficiently targets to mitochondria, but also directly enhances mitochondrial translation.9 Our previous study reveals a positive function of miR-21 in mitochondrial translation in the same way, which is sufficient to reduce blood pressure and alleviate cardiac hypertrophy in spontaneously hypertensive rats.10 These studies have raised a new regulatory pattern for miRNAs to function in mitochondrial reactive oxygen species (ROS) production.
Mitochondrial electron respiratory chain contains complex I (NADH:ubiquinone oxidoreductase), complex II (succinate dehydrogenase), complex III (cytochrome bc1 complex), and complex IV (cytochrome c oxidase).11 Complex I and complex III of the electron transport chain (ETC) are the major sites for ROS production.12 In fact, multiple studies suggest that complex III is more important than complex I in generating mitochondrial ROS (mt-ROS) in the heart.13, 14 Complex III contains 11 subunits, among which Cytb is the only mtDNA-encoded subunit of complex III.10 Emerging evidences suggest that mitochondrial dysfunction-induced ROS might be a central mediator of diabetic heart diseases. Therefore, the development of therapeutic approaches targeting mitochondrial biology holds promise for the management of heart failure in diabetic patients.15 However, the roles of mitochondrial miRNAs in diabetic cardiomyopathy remain unknown.
We hypothesized that altered expression of mitochondrial miRNAs might be causal to the initiation and/or progression of diabetic cardiomyopathy. In the present study, we identified that the downregulation of mtDNA-encoded Cytb in db/db mice heart appeared to directly contribute to the increase of mt-ROS. We found that miR-92a-2-5p, a key miRNA decreased in diabetic cardiomyopathy, was able to translocate into mitochondria to counteract mt-Cytb downregulation. Strikingly, exogenous miR-92a-2-5p delivered by recombinant adeno-associated virus (rAAV) preserved heart function in the db/db model, suggesting a new therapeutic strategy against diabetic cardiomyopathy.
Results
Downregulation of Mitochondrial Cytb Linked to Increased ROS in db/db Mouse Heart
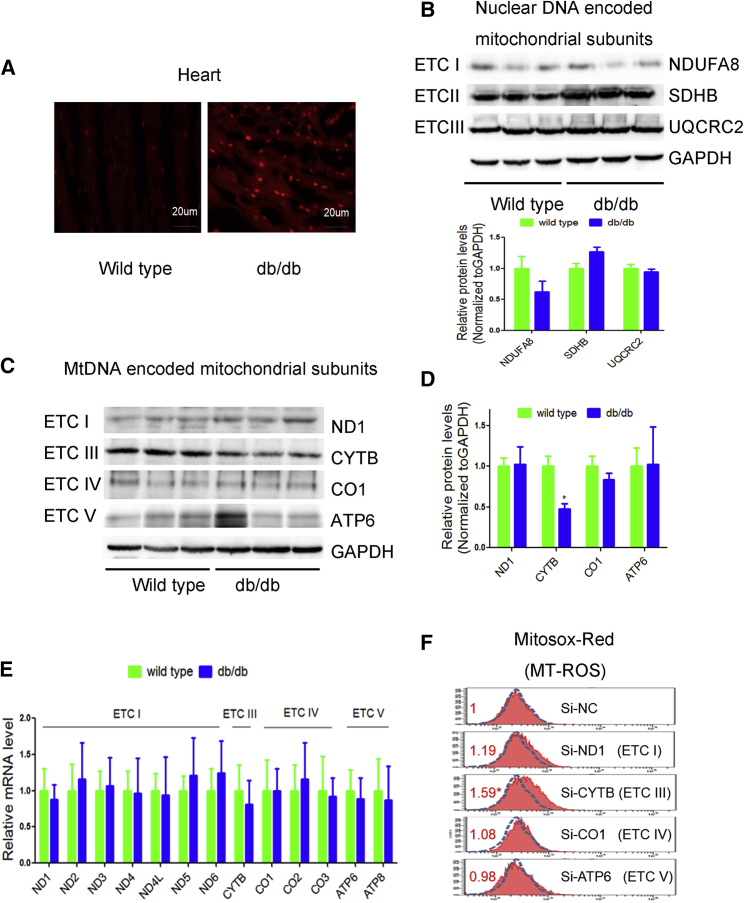
In db/db mouse heart, the amount of ROS was increased compared to controls (Figure 1A). As mitochondria are the main source of ROS, we next measured the expression of mitochondria-encoded subunits by western blotting and quantitative real-time PCR. We found that nuclear genome (nDNA)-encoded mitochondrial ETC subunits NDUFA8, SDHB, and UQCRC2 remained unchanged (Figure 1B). Interestingly, Cytb, the only mitochondrial gene-encoded subunit of ETC III, was significantly decreased in db/db hearts relative to controls. In contrast, other mtDNA-encoded subunits, such as ND1 (subunit of ETC I), COI (subunit of ETC IV), and ATP6 (subunit of ETC V), remained unaltered (Figures 1C and 1D). However, mRNA levels of mtDNA-encoded genes remained unchanged (Figure 1E). These data suggest a compromised coordination of nDNA and mtDNA-encoded ETC components in the heart of db/db mouse.
Figure 1.
Downregulation of Mitochondrial Cytb Was Linked to Increased ROS in db/db Mouse Heart
(A) ROS was detected by DHE probe in frozen heart sections of db/db mice in comparison with wild-type controls. n = 3. (B–D) Western blot was performed to analyze the nuclear DNA encoded ETC subunits (B), mitochondrial DNA encoded ETC subunits (C), and statistical analysis (D) in the heart of db/db mice. (E) Quantitative real-time PCR was used to determine the mRNA levels of ETC subunits in the heart of db/db mice. (F) Mitochondrial ROS levels were detected by MitoSOX red in H9c2 cells transfected with si-ND1, si-Cytb, si-COI, or si-ATP6. n = 3; results from controls were set to 1; *p < 0.05 versus si-NC.
We then explored the use of specific small interfering RNAs (siRNAs) against mtDNA-encoded Cytb, ND1, COI, and ATP6 to determine their contributions to ROS production in H9c2 cells. We found that siRNA to cytochrome b (si-Cytb) specifically caused elevated ROS in comparison with si-ND1, si-COI, and si-ATP6 (Figure 1F), which is consistent with our previous study.10 These data indicate a critical role of Cytb in ROS generation. We therefore selected Cytb for further investigation.
Global Function of Cytb in Human Cardiomyocytes
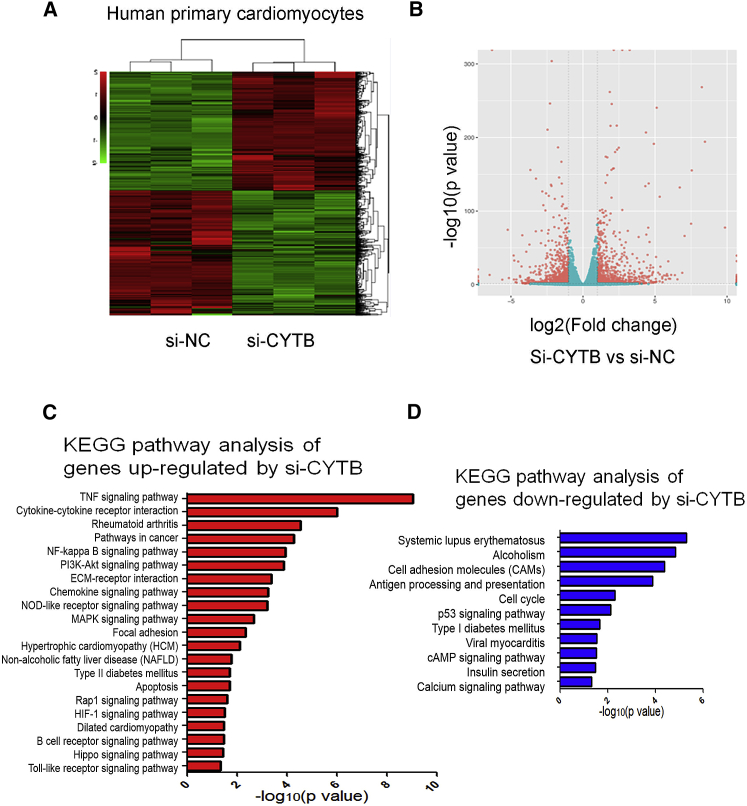
We further investigated the global roles of Cytb in cardiomyocytes by mRNA sequencing (mRNA-seq). Cytb knockdown led to dysregulation of hundreds of genes (Figures 2A and 2B; Data S1). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that tumor necrosis factor (TNF) signaling pathway, cytokine-cytokine receptor interaction pathway, and nuclear factor κB (NF-κB) signaling pathway were significantly upregulated in si-Cytb-treated cardiomyocytes (Figure 2C). Specifically, genes in the tumor necrosis factor (TNF) signaling pathway such as TNF, NF-κB, interleukin-1b (IL-1b), IL6, and chemokine (C-C motif) ligand 2 (Ccl2) were significantly upregulated in si-Cytb-treated cardiomyocytes (Figure S1). Meanwhile, cell cycle, the cyclic AMP (cAMP) signaling pathway, and the calcium signaling pathway were downregulated by si-Cytb (Figure 2D). Dysregulated pathways such as the TNF signaling pathway and NF-κB signaling pathway have been reported to be important regulators in the progression of diabetic cardiomyopathy.16, 17 These data suggest potential roles of Cytb in diabetic cardiomyopathy.
Figure 2.
Global Function of Cytb in Human Cardiomyocytes Was Analyzed by mRNA-seq
(A) Heatmap of the genes dysregulated in human primary cardiomyocytes (HCMs) treated with si-Cytb. n = 3. (B) Volcano plot of genes expressed in control and Cytb knockdown HCMs. (C) KEGG pathway analysis of genes upregulated in si-Cytb-treated HCMs. (D) KEGG pathway analysis of genes downregulated in si-Cytb treated HCMs.
miRNA Profile in Cytosol and Mitochondria of db/db Mouse Heart
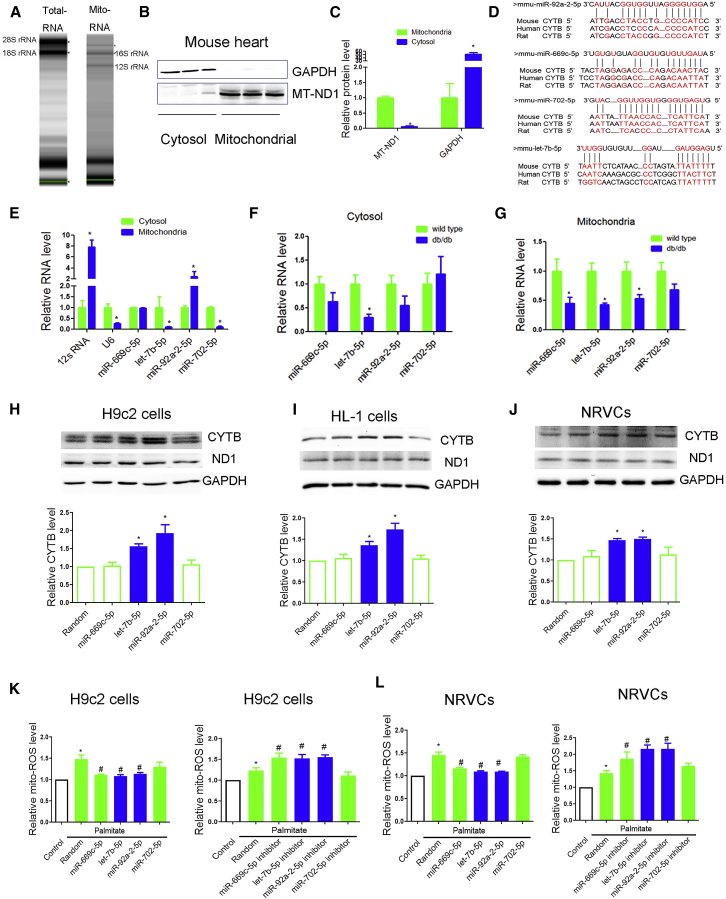
Because exogenous Cytb was not able to translocate into mitochondria, as evidenced by our own data (Figures S2A–S2C) and previous study,18 the clinical potential of exogenous Cytb overexpression was limited. Given recently reported roles of miRNAs in mitochondria, we tested whether Cytb downregulation was mediated by a specific miRNA in db/db mouse and whether miRNA-based therapy could modulate mitochondrial Cytb expression. We first performed a miRNA microarray on RNAs isolated from total lysis and mitochondria from db/db and control hearts, respectively. Total RNA isolated from heart were enriched with 18S and 28S rRNA, whereas the absence of 18S and 28S rRNA in the RNA isolated from mitochondria indicated no measurable cytosolic RNA contamination in the mitochondria preparation (Figure 3A). Moreover, western blotting further confirmed that there was no cytosolic protein contamination in the mitochondria preparation (Figures 3B and 3C).
Figure 3.
miRNA Profile Was Performed in Cytoplasmic and Mitochondrial Components of db/db Mouse Heart
(A) Gel electrophoresis was used to analyze the peak of mitochondrial RNA versus total heart. (B and C) Mitochondrial ND1 and GAPDH in mitochondria and cytosol analyzed by western blot (B) were evaluated for significance using Student’s t test (C). (D) Sequence alignment of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on the Cytb mRNA from different organisms. (E) Relative expression of selected miRNAs in mitochondria in comparison with cytosol were detected by quantitative real-time PCR. 100 ng of RNA from cytosol and mitochondria were reverse transcribed and used as template for quantitative real-time PCR. The expression of RNA in mitochondria was represented using a fold change relative to the value 1 of the cytosol. n = 3; *p < 0.05 versus cytosol. (F and G) Relative expression of selected miRNAs in cytosol (F) and mitochondria (G) of db/db mouse heart was detected by quantitative real-time PCR. Cytoplasmic RNAs were normalized to GAPDH mRNA and mitochondrial RNAs to 12 s rRNA. n = 8; *p < 0.05 versus wild-type. (H–J) Effects of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on mitochondrial subunits at the protein levels in H9c2 cardiomyocytes (H), HL-1 cardiomyocytes (I), and NRVCs (J) were detected by western blot. n = 3; *p < 0.05 versus random. (K and L) Effects of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on mitochondrial ROS under palmitate treatment were detected by MitoSOX red probe in H9c2 cardiomyocytes (K) and NRVCs (L). Cells were incubated with palmitate (0.25 mmol/L) for 24 h; n = 3; results from controls were set to 1; *p < 0.05 versus control, #p < 0.05 versus palmitate-random.
Total lysis and mitochondria samples from three control and three db/db hearts were used for miRNA profile detection. Global miRNA profile analysis revealed that 24 miRNAs were upregulated and 24 miRNAs were downregulated in the total lysis of db/db mouse heart (Table S1). However, no miRNAs were upregulated and 14 were downregulated in the mitochondria from db/db mouse heart (Table S2).
We have previously revealed a positive regulation of miRNAs in mitochondrial translation,10 so we hypothesize that Cytb downregulation might be mediated by specific mitochondrial miRNAs in db/db heart. We thus searched for potential miRNAs that may specifically target the Cytb transcript. The downregulated miRNAs (in mitochondria and total hearts) were chosen for miRNA-Cytb prediction. As a result, four miRNAs (miR-669c-5p, let-7b-5p, miR-92a-2-5p, and miR-702-5p) were predicted to target the Cytb transcript in conserved regions among human, rat, and mouse (Figure 3D). Quantitative real-time PCR validation further confirmed the downregulation of miR-669c-5p, let-7b-5p, and miR-92a-2-5p in mitochondria of db/db heart (Figures 3E–3G).
To further determine the potential effects of mitochondrial miRNAs on Cytb expression, we transfected miRNA mimic (miR-669c-5p, let-7b-5p, miR-92a-2-5p, and miR-702-5p) in cardiomyocytes, finding that let-7b-5p and miR-92a-2-5p increased Cytb expression in rat H9c2, murine HL-1, and neonatal rat ventricular cardiomyocytes (NRVCs) (Figures 3H–3J).
We then tested if overexpression of these miRNAs could protect cardiomyocytes against palmitate-induced mt-ROS outburst. We found that overexpression of miR-92a-2-5p or let-7b-5p decreased mt-ROS level and apoptosis in palmitate-treated cardiomyocytes (Figures 3K and 3L; Figure S2D). Consistently, inhibition of miR-92a-2-5p or let-7b-5p exacerbated palmitate-induced ROS production (Figures 3K and 3L). These data strongly suggest a protective role of miR-92a-2-5p and let-7b-5p in diabetic cardiomyopathy.
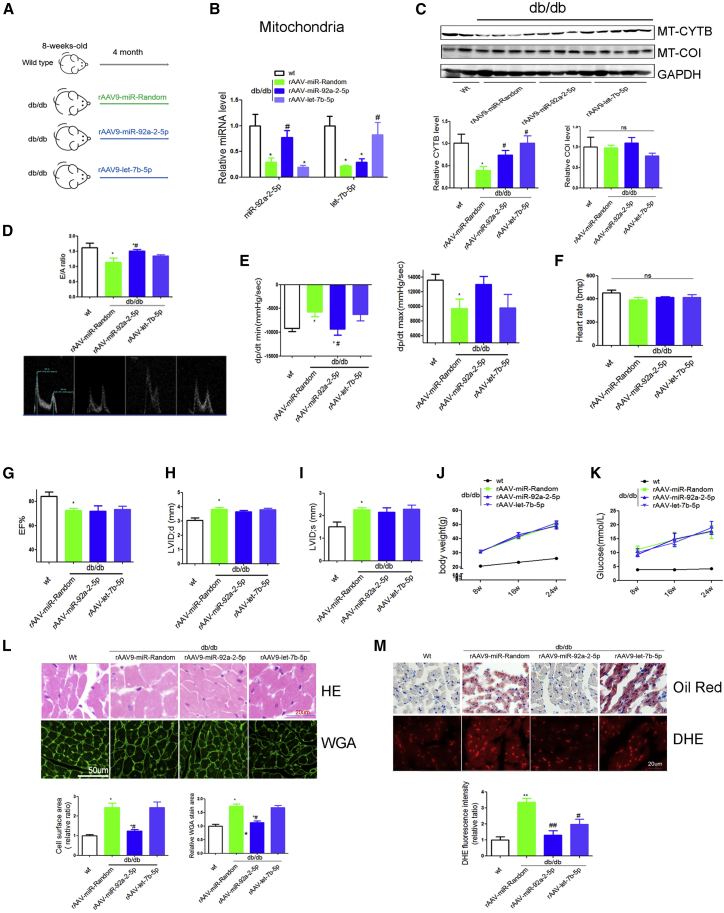
Overexpression of miR-92a-2-5p but Not let-7b-5p Alleviated Cardiac Dysfunction in db/db Mice
We next employed the db/db model to determine whether miR-92a-2-5p or let-7b-5p could alleviate diabetic cardiomyopathy in vivo. Eight-week-old male db/db mice were divided into three groups (n = 8 in each), each treated with rAAV-random, rAAV-miR-92a-2-5p, and rAAV-let-7b-5p for 4 months (Figure 4A). We found that rAAV-miR-92a-2-5p or rAAV-let-7b-5p treatment increased the levels of miR-92a-2-5p or let-7b-5p transcripts in mitochondria of db/db mouse heart as determined by quantitative real-time PCR (Figure 4B). In accordance, Cytb expression was significantly increased in rAAV-miR-92a-2-5p- and rAAV-let-7b-5p-treated db/db mouse heart (Figure 4C). Interestingly, rAAV-miR-92a-2-5p rescued cardiac diastolic dysfunction such as mitral peak velocity of early filling to mitral peak velocity of late filling (E/A ratio) and peak instantaneous rate of LV pressure decline (dp/dt min) in db/db mice (Figures 4D and 4E). However, rAAV-let-7b-5p had no effect on cardiac systolic or diastolic dysfunction in db/db mice (Figures 4D and 4E). rAAV-miR-92a-2-5p or rAAV-let-7b-5p treatments both had no effect on echocardiography markers such as left ventricle ejection fraction value, left ventricular internal dimension-systoles (LVIDs) value, body weight, or blood glucose levels among various db/db groups (Figures 4F–4K). Consistently, rAAV-miR-92a-2-5p significantly decreased the section area of myocardial cells, heart fat deposition, and ROS in db/db mice (Figures 4L–4M). In normal wild-type mice, overexpression of miR-let-7b-5p and miR-92a-2-5p had no influence on cardiac function (Figure S2E).
Figure 4.
Overexpression of miR-92a-2-5p but Not let-7b-5p Alleviated Cardiac Dysfunction in db/db Mice
(A) Overview of the experimental setup for rAAV-miR-92a-2-5p and rAAV-let-7b-5p treatment (n = 8). (B) Mitochondrial miR-92a-2-5p and let-7b-5p levels were determined by quantitative real-time PCR in various groups. (C) Cytb protein in the heart of db/db mice were detected by western blot. (D–I) Comparison of echocardiographic and hemodynamic measurements including E/A ratio (D), dp/dt (E), heart rate (F), EF value (G), LVIDd (H), and LVIDs (I) in rAAV-miR-92a-2-5p- and rAAV-let-7b-5p-treated db/db mice were shown. (J and K) Body weight (J) and blood glucose (K) in various groups were analyzed every 4 weeks. (L and M) H&E and WGA (L) and oil red and DHE staining (M) of myocardial tissue were detected in db/db mice. *p < 0.05 versus wild-type, #p < 0.05 versus db/db-rAAV-random.
These data demonstrate that miR-92a-2-5p, but not let-7b-5p, rescued cardiac diastolic dysfunction in db/db mice.
let-7b-5p Decreased IRS1 Expression in Cytosol
miR-92a-2-5p and let-7b-5p both increased Cytb and lowered ROS level in db/db mice. However, only miR-92a-2-5p exacerbated cardiac diastolic dysfunction and lipid deposition in db/db heart. We next tried to reveal the mechanisms underlying the different effects of miR-92a-2-5p and let-7b-5p in db/db mice.
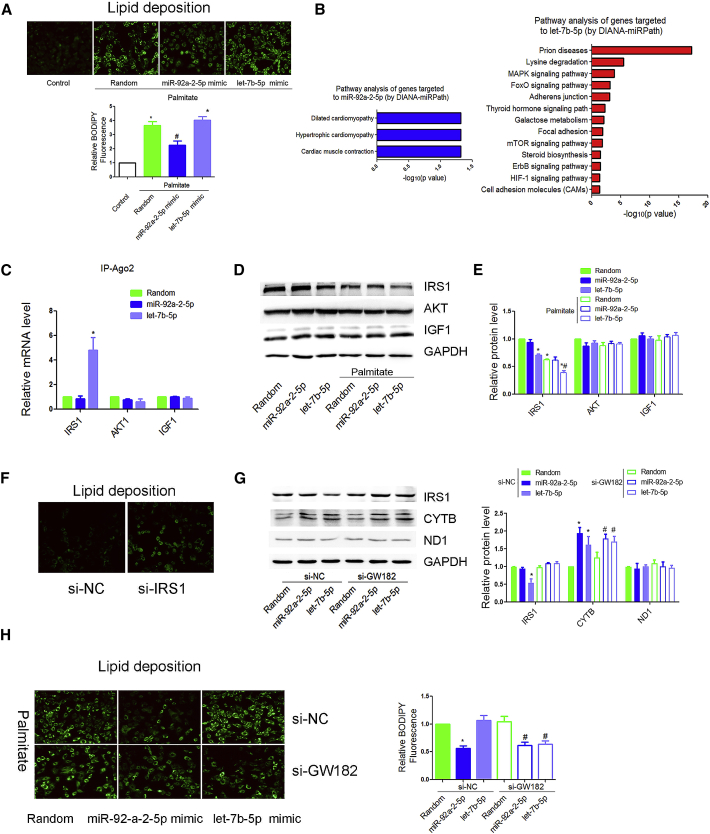
Consistent with in vivo studies, overexpression of miR-92a-2-5p, but not let-7b-5p, decreased palmitate-induced lipid deposition in H9c2 cardiomyocytes (Figure 5A). We then tested whether miR-92a-2-5p or let-7b-5p could regulate lipid metabolism by targeting non-mitochondrial genes. Pathway analysis of genes targeted by let-7b-5p reveal that mitogen-activated protein kinase (MAPK) signaling-related genes such as IRS1, AKT1, and IGF1 were potentially regulated by let-7b-5p (Figure 5B). We then performed RNA co-immunoprecipitation with anti-Ago2, finding that Ago2 showed increased association with the IRS1 mRNA after let-7b-5p transfection (Figure 5C). Western blotting further confirmed the downregulation of let-7b-5p on IRS1 expression in cardiomyocytes (Figures 5D and 5E). Interestingly, downregulation of IRS1 significantly increased lipid deposition in cardiomyocytes (Figure 5F). These data suggest that let-7b-5p might increase lipid deposition in cardiomyocytes via cytoplasmic targets such as IRS1.
Figure 5.
let-7b-5p Decreased IRS1 Expression in Cytosol
(A) Lipid deposition in miR-92a-2-5p- and let-7b-5p-treated H9c2 cardiomyocytes was detected by BODIPY probe. n = 3; *p < 0.05 versus control. (B) Pathway analysis was used to explore the function of genes potentially targeted to miR-92a-2-5p and let-7b-5p by DIANA-miRPath database. (C) mRNA in association with Ago2 in H9c2 cells was determined by quantitative real-time PCR. n = 3; results from control were set to 1. (D and E) Effects of transfected miR-92a-2-5p and let-7b-5p on IRS1, AKT, and IGF1 at the protein levels were detected by western blot (D) and were repeated three times independently (E). *p < 0.05 versus control. (F) Lipid deposition in si-IRS1-treated H9c2 cardiomyocytes was detected by BODIPY probe. n = 3. (G) Effects of miR-92a-2-5p and let-7b-5p on Cytb and IRS1 protein expression were detected by BODIPY probe with the miRNA machinery selectively inactivated by knocking down GW182 in the cytoplasm; n = 3; results from si-NC-random were set to 1. (H) Effects of miR-92a-2-5p and let-7b-5p on lipid deposition were detected by BODIPY probe with the miRNA machinery selectively inactivated by knocking down GW182 in the cytoplasm. Cells were incubated with palmitate (0.25 mmol/L) for 24 h. n = 3; *p < 0.05 versus si-NC-random; #p < 0.05 versus si-GW182-random.
To rigorously rule out the effects of let-7b-5p via cytoplasmic targets, we performed a diagnostic analysis on the differential requirement for the Ago2 cofactor GW182, which is required for miRNA effects in the cytoplasm, but not in the mitochondria.9 We have previously confirmed the knockdown effect of siRNA on GW182 in H9c2 cardiomyocytes.10 As expected, GW182 knockdown prevented let-7b-5p-mediated translational repression of its cytoplasmic target IRS1 (Figure 5G). However, GW182 RNAi-treated H9c2 cells continued to show an enhanced Cytb translation by the let-7b-5p mimic (Figure 5G). Interestingly, after ruling out the indirect effects via cytoplasmic targets, let-7b-5p mimic decreased lipid content in GW182 RNAi-treated cells (Figure 5H). MiR-92a-2-5p continued to decrease lipid deposition in GW182 RNAi-treated cardiomyocytes (Figure 5H).
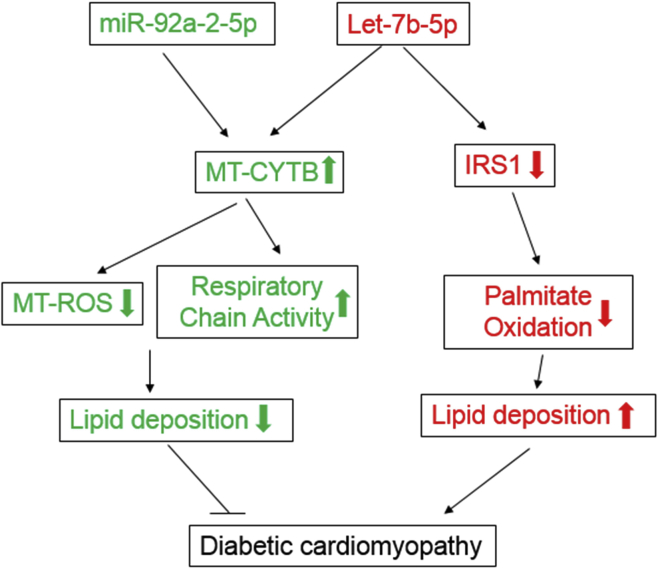
Together, these data strongly indicate that miR-92a-2-5p decreased lipid content in cardiomyocytes via the mitochondrial pathway, while let-7b-5p both increased and decreased lipid deposition by cytosol IRS1 and mitochondrial Cytb, respectively (Figure 6; see further in Discussion).
Figure 6.
A Model Was Provided to Illustrate the Role of miR-92a-2-5p and let-7b-5p in Diabetic Cardiomyopathy
Roles of miR-92a-2-5p and let-7b-5p in diabetic cardiomyopathy. miR-92a-2-5p and let-7b-5p were downregulated in mitochondria of db/db mice. Re-expression of miR-92a-2-5p increased mitochondrial Cytb expression, decreased mitochondrial ROS production, and eventually protected against lipid deposition and cardiac dysfunction in db/db mice. Let-7b-5p showed a similar regulation on Cytb as miR-92a-2-5p in the mitochondria; however, let-7b-5p also negatively regulated IRS1 and increased palmitate deposition, leading to unimproved cardiac function in db/db mice.
Discussion
In the present study, we observed a downregulation of the mitochondrial genome-encoded Cytb in db/db mouse, which appeared to directly contribute to the increase of mt-ROS. We found that miR-92a-2-5p and let-7b-5p were able to translocate into the mitochondria to counteract mt-Cytb downregulation. We showed that the delivery of exogenous miR-92a-2-5p by rAAV9 completely rescued cardiac diastolic dysfunction in the db/db model, suggesting a new therapeutic strategy against diabetic cardiomyopathy. However, let-7b-5p not only upregulated mitochondrial Cytb, but also decreased IRS1 expression in cytosol, which lead to unalleviated lipid deposition and cardiac dysfunction in db/db mice. Our study illustrated the complex roles of miRNAs in subcellular fractions such as mitochondria and cytosol, which may lead to new insights into the mechanisms and treatment strategies for diabetic cardiomyopathy in the future.
In the cardiovascular system, complex III was suggested to be the major site for ROS production.14, 19 Consistently, we found that siRNA-mediated silencing of Cytb, which is the only mtDNA-encoded subunit of complex III, caused elevated ROS. Mutations in Cytb have been suggested to be responsible for decreased complex III activity and increased ROS production and associated with cardiac hypertrophy and heart failure.20, 21 In fact, mitochondrial cardiomyopathy has been regarded as a distinct clinical entity in patients who have underlying genetic defects involving mitochondrial respiratory chain. For clinical practice, Cytb restoration might be beneficial for developing therapeutics against mitochondria-related diseases such as diabetic cardiomyopathy. However, we found that exogenous Cytb (Figures S2A and S2B) could not be efficiently imported into mitochondria by pcDNA3.1 or rAAV vector. Consistent with this, a previous study also demonstrated the low efficiency of transporting exogenous Cytb into mitochondria, even when a mitochondrial targeting sequence was inserted.18 In sharp contrast, multiple specific and non-specific miRNAs were able to enter purified mitochondria efficiently,9 which might serve as an alternative choice for upregulating mitochondrial gene expression.
miRNAs are in the spotlight as post-transcriptional regulators of gene expression.22 They usually act as negative regulators of protein translation by affecting mRNA stability in the cytoplasm.23 Recently, the roles of miRNAs in subcellular fractions such as nucleus and mitochondria were revealed. The majority of cellular miRNAs were present in both the nucleus and the cytoplasm. In the nucleus, miRNAs acted to regulate the stability of nuclear transcripts and induce epigenetic alterations that either silence or activate transcription at specific gene promoters.24 miRNAs were also enriched in or associated with mitochondria in various species.25, 26 Functionally, a recent report demonstrates that miR-1, a miRNA that specifically induces during myogenesis, is not only efficiently targeted to mitochondria, but also is able to enhance mitochondrial translation directly.9 Our previous study also found that miR-21 was able to translocate into the mitochondria to counteract mitochondrial gene downregulation.10 Moreover, our previous work also did not show evidence for liver and kidney toxicity of rAAV9, which implied that the rAAV9 was an ideal vector for miRNA delivery. Whether rAAV was inserted into the mitochondrial genome along with miRNAs in our system remains to be investigated. The future studies should minimize the potential off-target effects of rAAV-mediated mitochondrial-miRNA delivery.
In terms of the mechanism of miRNA-enhanced mitochondrial translation, three critical requirements for converting miRNA-dependent translational repression to activation have been proposed: (1) lack of the cap at the 5′ end, (2) lack of a typical poly(A) tail at the 3′ end, and (3) detachment of GW182 from an Ago protein.27 Interestingly, all mitochondrial transcripts fulfill these requirements, because mt-mRNAs have no cap nor typical long poly(A) tail. Moreover, GW182 was detected only in the cytoplasm, but not mitochondria.9 Consistent with this, we also found that GW182 knockdown prevented let-7b-5p-mediated translational repression of its cytoplasmic target IRS1. Under these conditions, GW182 RNAi-treated cardiomyocytes continued to show enhanced Cytb translation by miR-92a-2-5p and let-7b-5p treatments. Based on these results, the rearranged miRNA machinery, coupled with the prokaryotic characteristics of the mitochondrial transcripts, might be responsible for translational enhancement in the mitochondria.
Interestingly, miR-92a-2-5p rescued diastolic dysfunction in db/db mice, independently of blood glucose and body weight, which suggests a direct role of miR-92a-2-5p in heart. Diabetic cardiomyopathy in human is characterized by diastolic dysfunction, which may precede the development of systolic dysfunction.28 Myocardial steatosis (lipid accumulation) is reported to be an independent predictor of diastolic dysfunction in type 2 diabetes.29 We observed an increased cardiac lipid accumulation in diabetic mice, which was rescued by miR-92a-2-5p. Meanwhile, there was no difference for fibrosis among various groups (Figure S2F). These data suggest that miR-92a-2-5p might protect against diastolic dysfunction through decreased cardiac lipid accumulation, but not fibrosis. However, miR-92a-2-5p had no effect on systolic dysfunction in db/db mice. It is possible that decreased miR-92a-2-5p is only part of the mechanisms underlying diabetic cardiomyopathy, as overexpression of miR-92a-2-5p alone was insufficient to rescue systolic dysfunction. In addition, rAAV-miR-92a-2-5p may require extended periods of time to protect against diabetic systolic dysfunction.
miR-92 (miR-92a-3p) has been extensively studied in various diseases,30, 31, 32 but the role of miR-92a-2-5p was largely unknown. In the present study, we found that miR-92a-2-5p was downregulated in mitochondria of db/db mice heart, and re-expression of miR-92a-2-5p increased mitochondrial gene Cytb expression. Meanwhile, miR-92b-5p, another miRNA in the miR-92 family, was also decreased in mitochondria of db/db mouse heart (Figure S3A). However, these miRNAs in the miR-92 family, such as miR-92b-5p, miR-92a-1-5p, and miR-92a-3p, do not target Cytb (Figure S3B). Nevertheless, these miRNAs might still play crucial roles in the progression of diabetic cardiomyopathy, which needs further investigation.
In terms of let-7b-5p, it is interesting that let-7b-5p acts differently in mitochondria and cytosol. In fact, even in the cytoplasmic fraction, a single miRNA could target multiple genes; some of the targets may be protective, while others may be destructive in the same disease. For example, miR-494 targeted both pro-apoptotic genes including PTEN and CAMK2D, as well as anti-apoptotic genes such as FGFR-2 and LIF.33 As such, the global effects of a specific miRNA would be the network results of all its target genes. It is also the case for let-7b-5p in our study—that let-7b-5p promoted lipid deposition and suppressed lipid deposition via cytosol and mitochondrial gene, respectively. Our study revealed a complicated and sophisticated regulation of miRNAs in subcellular organelles during lipid deposition.
In summary, our findings reveal a positive function of miR-92a-2-5p in mitochondrial translation, and its cardiac delivery to animals is sufficient to rescue diastolic dysfunction in db/db mice. These observations provide a theoretical basis for developing miRNA-based therapeutics against diabetic cardiomyopathy.
Materials and Methods
Western Blotting
Cell lysate was resolved by SDS-PAGE, transferred to nitrocellulose membrane, and blocked with 5% non-fat dry milk in Tris-buffered saline and Tween-20 (TBS-T). The membrane was incubated with primary antibody overnight at 4°C, followed by peroxidase-conjugated secondary antibody for 1–2 h, and finally developed with the enhanced chemiluminescence (ECL) system (Beyotime Institute of Biotechnology, Nanjing, China). Antibodies used in the present study were listed in Table S3.
Quantification of ROS Production
Dihydroethidium (DHE; Invitrogen, Carlsbad, CA, USA) was applied to frozen heart sections at 40 μM for 30 min. Fluorescence intensity was measured under a Nikon DXM1200 fluorescence microscope and analyzed with the Image-Pro software (Media Cybernetics, Bethesda, MD, USA).
Mt-ROS was measured with MitoSOX red (Invitrogen, Carlsbad, CA, USA) in H9c2 cells as described.10 In brief, cells were washed three times with PBS. MitoSOX red (5 μM) was added to the media and incubated for 30 min at 37°C in the dark. mt-ROS was quantified by flow cytometry (BD Biosciences, CA, USA) with 510 nm excitation/580 nm emission filters.
mRNA Sequencing (mRNA-Seq)
RNA-seq and data analyses were performed by Personal Biotechnology (Shanghai, China). The threshold value for significance was a fold change > 2 with a p value of p < 0.05.
Mitochondria Isolation
Mitochondria were isolated using a MACS mitochondria extraction kit from Miltenyi Biotec (Bergisch Gladbach, Germany) as described.26 In brief, cells were lysed and mitochondria were magnetically captured with anti-TOM22 antibody-coated beads, and eluted mitochondria were collected by centrifugation at 13,000 × g for 2 min at 4°C.
miRNA Profile
RNAs were isolated from total hearts and mitochondria of three controls and three db/db mice, respectively. RNA quality and quantity were assessed by Agilent 2200 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
RNA samples were labeled and hybridized onto the surface of a 1 × 12k mouse miRNA microarray (RiboArray miDETECT mouse array, miRbase v21.0; RiboBio, Guangzhou, China). Scanning was performed with the GenePix 4000B microarray scanner (Molecular Devices). Scanned images were then imported into GenePix Pro 6.0 software for grid alignment and data extraction.34 p < 0.05 was considered significant.
miRNA Target Prediction
The miRBase (http://www.mirbase.org/) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html)35 websites were used for miRNA-(mito)RNA prediction. A minimum free energy of hybridization lower than −20 kcal/mol is sufficient for formation of a miRNA/mRNA complex,36, 37 and was used as a cut-off value.
For miRNA-mRNA (cytosol) prediction, miRPath was utilized to predict miRNA-mRNA targeting provided by experimental validation data derived from DIANA-TarBase v6.0.38, 39
Cell Culture and Transfection
The H9c2 cells and 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA). Human primary cardiomyocytes (HCMs) used in this experiment are from a single fetal donor, not pooled (ScienCell research laboratories; lot# 18387). HCM cells were maintained in cardiac myocyte medium (CMM; ScienCell research laboratories), according to the instruction manual. CMM consists of basal medium, FBS, cardiac myocyte growth supplement (CMGS), and penicillin/streptomycin solution. Murine HL-1 cardiomyocytes were cultured in Claycomb medium supplemented 100 μM norepinephrine stock (consisting of 10 mM norephinephrine [Sigma, the Netherlands] dissolved in 0.3 mM l-ascorbic acid [Sigma]), 4 mM l-glutamine (Gibco, the Netherlands), and 10% FBS (Life Technologies, Gaithersburg, MD, USA) as described.40 miRNA mimics (50 nM), inhibitors (50 nM), siRNAs (50 nM; Smart Silencer is a mixture of three siRNAs and three antisense oligonucleotides), and random small RNA controls (50 nM) were transfected into cells by Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). All of them were designed and synthesized by Riobio (Guangzhou, China), and sequences were listed in Table S4.
For primary cardiomyocyte NRVC isolation, hearts were removed from newborn rats (0–3 days) and cut into pieces. The tissues were then incubated in a balanced salt solution containing 0.2% collagenase type 2 (Invitrogen) for 5 min at 37°C. The digestion buffer was replaced six times, at which point the tissues were completely digested. The collected primary cells were passed through a cell strainer (200 mesh) and then seeded onto Petri dishes and incubated for 90 min. The supernatant (cardiomyocytes) was collected and plated in DMEM supplemented with 10% FBS, and the adherent cells (cardiac fibroblasts) were cultured under the same conditions as above. Primary cells were confirmed by immunofluorescence staining with antibodies directed against cardiomyocyte-specific marker α2-actinin (ACTN2, Sigma) and fibroblast-specific antigen prolyl-4-hydroxylase (P4HB, Acris), as previously described.41
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated using TRIzol and reverse transcribed with SuperScript III first strand synthesis kit (Life Technologies, Carlsbad, CA, USA). Real-time PCR assays were performed with the SYBR select master mix (Life Technologies, Carlsbad, CA, USA) on a 7900HT FAST real-time PCR system (Life Technologies, Carlsbad, CA, USA). All reactions were performed in triplicate. The data were analyzed as described.41 Primers used in the present study were listed in Table S5.
Fatty Acid Stain
Washed cells were incubated in PBS containing 10 nM BODIPY (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene; Molecular Probes, Eugene, OR, USA) for 15 min at 37°C. BODIPY lipid probes was used as a stain for neutral lipids and a tracer for oil and other non-polar lipids. After washing twice with ice-cold PBS, cells were analyzed by fluorescence microscopy.
Construction and Preparation of rAAVs
rAAV vectors (type 9) were used to express miR-92a-2-5p, let-7b-5p, and the appropriate controls in heart tissues in vivo. The rAAV-9 system was a kind gift from Dr. Xiao Xiao (University of North Carolina at Chapel Hill). Three plasmids used to co-transfect HEK293 cells were purified as we described previously.34, 42
Animals
The animal study was approved by the Institutional Animal Research Committee of Tongji Medical College. For in vivo experiments, male db/db mice generated in C57BLKS/JNju background and control wild-type mice (Model Animal Research Center of Nanjing University) were used. All experiments were performed in accordance with the ARRIVE guidelines and NIH guidelines for animal welfare. All animals were maintained under a 12 hr light/12 hr dark photoperiod, with free access to water and food. We randomly divided the db/db mice into three groups (n = 8 in each group): rAAV-miR-random, rAAV-miR-92a-2-5p, and rAAV-let-7b-5p. The mice were injected with the corresponding rAAVs via tail vein at 8 weeks of age.
Measurement of Cardiac Echocardiography
Upon anesthetization, echocardiography was performed with a 30-MHz high-frequency scanhead (VisualSonics Vevo770, VisualSonics) as described previously.43 To monitor LV catheterization, a catheter manometer (Millar 1.4F, SPR 835, Millar Instruments) was inserted via the right carotid artery into the left ventricle. After stabilization, data were continuously recorded. Cardiac function parameters were calculated with the PVAN software (Millar Instruments, Houston, TX, USA) as described.43
Histology Staining
Formalin-fixed hearts were embedded in paraffin and sectioned into 4-mm slices. The morphology, fibrosis, and lipid deposition were detected by H&E, wheat germ agglutinin (WGA), Sirius red, and oil red staining, respectively. Tissue sections were visualized by microscope and measured by mage-Pro Plus version 6.0 (Media Cybernetics, Bethesda, MD, USA).
RNA Immunoprecipitation
Lysed cell extracts were immunoprecipitated with anti-Ago2 antibody (Abnova, Taiwan, China) or immunoglobulin G (IgG) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) using protein G Sepharose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA), as described.10 After eluted from the beads, bound RNA were extracted with TRIzol and quantified by quantitative real-time PCR.
Statistical Analysis
The data were expressed as mean values ± SEM (n noted in specific figure legends). Differences between groups were evaluated for significance using Student’s t test of unpaired data or one-way ANOVA with Bonferroni post-test. p < 0.05 was considered statistically significant.
Author Contributions
H.L., J.F., and B.D. designed and performed the experiments, analyzed the data, and wrote the manuscript; C.C., X.N., Z.Y., Y.Z., and X.Z. participated in performing the experiments; and D.W.W. designed the experiments and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank colleagues in D.W.W’s group for various technical help and stimulating discussion during the course of this investigation. D.W.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. We also appreciate Dr. Xiaoquan Rao for English editing of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (nos. 31800973, 81822002, 81790624, 81630010, 91439203, and 31771264) and the National Precision Medicine Project (no. 2017YFC0909400). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.013.
Contributor Information
Huaping Li, Email: lhp@tjh.tjmu.edu.cn.
Dao Wen Wang, Email: dwwang@tjh.tjmu.edu.cn.
Supplemental Information
References
- 1.Matheus A.S., Tannus L.R., Cobas R.A., Palma C.C., Negrato C.A., Gomes M.B. Impact of diabetes on cardiovascular disease: an update. Int. J. Hypertens. 2013;2013:653789. doi: 10.1155/2013/653789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marso S.P., Bain S.C., Consoli A., Eliaschewitz F.G., Jódar E., Leiter L.A., Lingvay I., Rosenstock J., Seufert J., Warren M.L., SUSTAIN-6 Investigators Semaglutide and Cardiovascular Outcomes in Patients with Type 2 Diabetes. N. Engl. J. Med. 2016;375:1834–1844. doi: 10.1056/NEJMoa1607141. [DOI] [PubMed] [Google Scholar]
- 3.King H., Aubert R.E., Herman W.H. Global burden of diabetes, 1995-2025: prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–1431. doi: 10.2337/diacare.21.9.1414. [DOI] [PubMed] [Google Scholar]
- 4.Falcão-Pires I., Leite-Moreira A.F. Diabetic cardiomyopathy: understanding the molecular and cellular basis to progress in diagnosis and treatment. Heart Fail. Rev. 2012;17:325–344. doi: 10.1007/s10741-011-9257-z. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert R.E., Krum H. Heart failure in diabetes: effects of anti-hyperglycaemic drug therapy. Lancet. 2015;385:2107–2117. doi: 10.1016/S0140-6736(14)61402-1. [DOI] [PubMed] [Google Scholar]
- 6.Duckworth W., Abraira C., Moritz T., Reda D., Emanuele N., Reaven P.D., Zieve F.J., Marks J., Davis S.N., Hayward R., VADT Investigators Glucose control and vascular complications in veterans with type 2 diabetes. N. Engl. J. Med. 2009;360:129–139. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 7.Thomson D.W., Bracken C.P., Goodall G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasudevan S., Steitz J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Zhang X., Wang F., Zhou L., Yin Z., Fan J., Nie X., Wang P., Fu X.D., Chen C., Wang D.W. MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation. 2016;134:734–751. doi: 10.1161/CIRCULATIONAHA.116.023926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archakov A.I. [Molecular organization and function of the electron transport chains of liver endoplasmic reticulum membranes] Usp. Sovrem. Biol. 1971;71:163–183. [PubMed] [Google Scholar]
- 12.Murphy E., Steenbergen C. Preconditioning: the mitochondrial connection. Annu. Rev. Physiol. 2007;69:51–67. doi: 10.1146/annurev.physiol.69.031905.163645. [DOI] [PubMed] [Google Scholar]
- 13.Demin O.V., Kholodenko B.N., Skulachev V.P. A model of O2.-generation in the complex III of the electron transport chain. Mol. Cell. Biochem. 1998;184:21–33. [PubMed] [Google Scholar]
- 14.Chen Q., Vazquez E.J., Moghaddas S., Hoppel C.L., Lesnefsky E.J. Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 15.Schilling J.D. The mitochondria in diabetic heart failure: from pathogenesis to therapeutic promise. Antioxid. Redox Signal. 2015;22:1515–1526. doi: 10.1089/ars.2015.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas C.M., Yong Q.C., Rosa R.M., Seqqat R., Gopal S., Casarini D.E., Jones W.K., Gupta S., Baker K.M., Kumar R. Cardiac-specific suppression of NF-κB signaling prevents diabetic cardiomyopathy via inhibition of the renin-angiotensin system. Am. J. Physiol. Heart Circ. Physiol. 2014;307:H1036–H1045. doi: 10.1152/ajpheart.00340.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzo O., Picatoste B., Ares-Carrasco S., Ramírez E., Egido J., Tuñón J. Potential role of nuclear factor κB in diabetic cardiomyopathy. Mediators Inflamm. 2011;2011:652097. doi: 10.1155/2011/652097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oca-Cossio J., Kenyon L., Hao H., Moraes C.T. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics. 2003;165:707–720. doi: 10.1093/genetics/165.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St-Pierre J., Buckingham J.A., Roebuck S.J., Brand M.D. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 20.Valnot I., Kassis J., Chretien D., de Lonlay P., Parfait B., Munnich A., Kachaner J., Rustin P., Rötig A. A mitochondrial cytochrome b mutation but no mutations of nuclearly encoded subunits in ubiquinol cytochrome c reductase (complex III) deficiency. Hum. Genet. 1999;104:460–466. doi: 10.1007/s004390050988. [DOI] [PubMed] [Google Scholar]
- 21.Marin-Garcia J., Hu Y., Ananthakrishnan R., Pierpont M.E., Pierpont G.L., Goldenthal M.J. A point mutation in the cytb gene of cardiac mtDNA associated with complex III deficiency in ischemic cardiomyopathy. Biochem. Mol. Biol. Int. 1996;40:487–495. doi: 10.1080/15216549600201053. [DOI] [PubMed] [Google Scholar]
- 22.Barwari T., Joshi A., Mayr M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016;68:2577–2584. doi: 10.1016/j.jacc.2016.09.945. [DOI] [PubMed] [Google Scholar]
- 23.Wojciechowska A., Braniewska A., Kozar-Kamińska K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017;26:865–874. doi: 10.17219/acem/62915. [DOI] [PubMed] [Google Scholar]
- 24.Roberts T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S., Ferlito M., Kent O.A., Fox-Talbot K., Wang R., Liu D., Raghavachari N., Yang Y., Wheelan S.J., Murphy E., Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrey E., Saint-Auret G., Bonnamy B., Damas D., Boyer O., Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 28.Bell D.S. Diabetic cardiomyopathy. Diabetes Care. 2003;26:2949–2951. doi: 10.2337/diacare.26.10.2949. [DOI] [PubMed] [Google Scholar]
- 29.Rijzewijk L.J., van der Meer R.W., Smit J.W.A., Diamant M., Bax J.J., Hammer S., Romijn J.A., de Roos A., Lamb H.J. Myocardial steatosis is an independent predictor of diastolic dysfunction in type 2 diabetes mellitus. J. Am. Coll. Cardiol. 2008;52:1793–1799. doi: 10.1016/j.jacc.2008.07.062. [DOI] [PubMed] [Google Scholar]
- 30.Al-Nakhle H., Burns P.A., Cummings M., Hanby A.M., Hughes T.A., Satheesha S., Shaaban A.M., Smith L., Speirs V. Estrogen Receptor β1 Expression Is Regulated by miR-92 in Breast Cancer. Cancer Res. 2010;69:4139. doi: 10.1158/0008-5472.CAN-09-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith L., Baxter E., Hanby A., Hughes T., Millican-Slater R., Verghese E., Speirs V. Loss of miR-92 expression in breast epithelial cells is associated with cancer progression. Mol. Cancer Res. 2013;11(10 Suppl.):B127. [Google Scholar]
- 32.Cardin R., Piciocchi M., Sinigaglia A., Lavezzo E., Bortolami M., Kotsafti A., Cillo U., Zanus G., Mescoli C., Rugge M., Farinati F. Oxidative DNA damage correlates with cell immortalization and mir-92 expression in hepatocellular carcinoma. BMC Cancer. 2012;12:177. doi: 10.1186/1471-2407-12-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdellatif M. Differential expression of microRNAs in different disease states. Circ. Res. 2012;110:638–650. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J., Yao K., Wang Q., Guo J., Shi H., Ma L., Liu H., Gao W., Zou Y., Ge J. Ischemic Postconditioning-Regulated miR-499 Protects the Rat Heart Against Ischemia/Reperfusion Injury by Inhibiting Apoptosis through PDCD4. Cell. Physiol. Biochem. 2016;39:2364–2380. doi: 10.1159/000452506. [DOI] [PubMed] [Google Scholar]
- 35.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng L., Chen Y., Wang Y., Yu L.R., Knox B., Chen J., Shi T., Chen S., Ren Z., Guo L. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem. Pharmacol. 2017;140:139–149. doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D., Tolleson W.H., Knox B., Jin Y., Guo L., Guo Y., Kadlubar S.A., Ning B. Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem. Pharmacol. 2015;98:671–680. doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., Hatzigeorgiou A.G. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claycomb W.C., Lanson N.A., Jr., Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., Izzo N.J., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Fan J., Yin Z., Wang F., Chen C., Wang D.W. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan M., Chen C., Gong W., Yin Z., Zhou L., Chaugai S., Wang D.W. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc. Res. 2015;105:340–352. doi: 10.1093/cvr/cvu254. [DOI] [PubMed] [Google Scholar]
- 43.Chen C., Yang S., Li H., Yin Z., Fan J., Zhao Y., Gong W., Yan M., Wang D.W. Mir30c Is Involved in Diabetic Cardiomyopathy through Regulation of Cardiac Autophagy via BECN1. Mol. Ther. Nucleic Acids. 2017;7:127–139. doi: 10.1016/j.omtn.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.