Abstract
Researchers have realized that microRNAs (miRNAs) play significant roles in the pathogenesis of various diseases. Although many computational models have been proposed to predict the associations between miRNAs and diseases, prediction performance could still be improved. In this paper, we propose a novel self-weighted, multi-kernel, multi-label learning (SwMKML) method to predict disease-related miRNAs. SwMKML adaptively learns two optimal kernel matrices for both miRNAs and diseases from multiple kernels constructed from known miRNA-disease associations. Moreover, the miRNA-disease associations predicted from both spaces are updated simultaneously based on a multi-label framework. Compared with four state-of-the-art computational models, SwMKML achieved best results of 95.5%, 93.1%, and 84.1% in global leave-one-out cross-validation, 5-fold cross-validation, and overall prediction accuracy, respectively. A case study conducted on head and neck neoplasms further identified two potential prognostic biomarkers, hsa-mir-125b-1 and hsa-mir-125b-2, for the disease. SwMKML is freely available at Github, and we anticipate that it may become an effective tool for potential miRNA-disease association prediction.
Keywords: graph-based learning, multi-kernel learning, miRNA-disease association prediction
Introduction
MicroRNAs (miRNAs) are a class of evolutionarily conserved, non-coding, small-molecule RNAs that have the function of regulating gene expression at the post-transcriptional level.1 Recent studies have shown that miRNAs play crucial roles in various biological processes, such as cell growth and apoptosis, hemocyte differentiation, cardiac genesis, and late embryonic development.2, 3 Therefore, researchers have made great efforts to explore disease-related miRNAs by biological experiments to promote the understanding of the functional roles of miRNAs in the pathogenesis of human diseases and provide new clues for subsequent clinical treatment.4 Nevertheless, the experimental methods are usually costly and time-consuming, which hinders their applicability to large-scale prediction.5 Because of the relatively limited experimental data, recently, various studies regarding this topic have also been proposed to detect potential disease-related miRNAs based on computational biology methods.6, 7, 8
Existing computational models can be roughly divided into three categories: similarity-based approaches, network topology-based methods, and machine learning-based methods. Based on the assumption that functionally similar miRNAs are generally associated with phenotypically similar diseases, many similarity-based approaches have been developed.9, 10, 11, 12, 13, 14 For instance, Jiang et al.9 constructed a comprehensive human phenome-microRNAome network to prioritize the entire human microRNAome for diseases of interest. Chen et al.10 adopted global network similarity measures to infer potential disease-related miRNAs by implementing random walk with restart on the functional similarity network. Both Xuan et al.11 and Liu et al.12 constructed a bilayer heterogeneous network to effectively uncover miRNA-disease associations.
Another set of prediction methods utilized network topological characteristics and also achieved remarkable performance.15, 16, 17, 18, 19, 20 Zou et al.15 learned an integrated network similar to a social network composed of multiple heterogeneous networks to predict the potential associations between miRNAs and diseases. You et al.16 adopted a depth-first search algorithm to rank the associations between miRNAs and diseases in terms of their path length. Chen et al.19 used graphlet interaction to quantify the relationships between miRNAs and diseases. Qu et al.20 developed a novel KATZ model-based computational method through a reliable heterogeneous network by integrating multiple data sources. Although these methods have achieved great performance, their prediction performance could be easily affected by a change in network topology.
In addition, with the rapid development of artificial intelligence techniques, increasing numbers of computational models based on machine learning have also been designed to solve the prediction problem.21, 22, 23, 24, 25, 26, 27 Chen and Yan28 developed a regularized least-squares method to discover new disease-related miRNAs. Xiao et al.29 proposed a graph-regularized, non-negative matrix factorization framework to discover potential associations between miRNAs and diseases. Chen et al.30, 31 presented a novel model of inductive matrix completion for miRNA-disease association prediction. Zeng et al.32 proposed a structural perturbation method based on the metric of structural consistency to predict potential new associations.
Despite the tremendous efforts made to identify the possible associations between miRNAs and diseases, most computational methods still suffer from several limitations that affect their prediction accuracy and scalability. For instance, the similarity matrices constructed for miRNAs and diseases might be sub-optimal because of data incompleteness. Moreover, the prediction process in miRNA space is usually separated from that in disease space. To conquer the aforementioned limitations, we propose a novel method to predict potential disease-related miRNAs based on a self-weighted, multi-kernel, multi-label learning (SwMKML) framework. Specifically, our method first constructs a set of kernel matrices by fully taking advantage of known miRNA-disease associations. We then adaptively learn two optimal kernel matrices for both miRNAs and diseases from multiple kernels. Finally, the predicted miRNA-disease associations are updated synchronously according to a graph-based, multi-label learning framework. To illustrate the effectiveness of the proposed method, we apply several evaluation metrics to systematically measure prediction performance. The experimental results show that our method achieves favorable performance compared with several state-of-the-art methods. We further implement a case study of head and neck neoplasms to identify potential diagnostic biomarkers for the disease. In summary, our method demonstrates a superior ability to predict candidate disease-related miRNAs for future clinical trials.
Results
Performance Evaluation
We compared the prediction performance of our method with four state-of-the-art computational models: L1-Norm, structural perturbation method for miRNA-disease association prediction (SPMMDA), path-based miRNA-disease association prediction (PBMDA), and extreme gradient boosting machine for miRNA-disease Association prediction (EGBMMDA). Specifically, L1-Norm is a graph-based, semi-supervised learning method that obtains sparse solutions for prioritizing disease-related miRNAs.33 SPMMDA uses structural consistency to estimate the link probability between miRNAs and diseases.32 PBMDA measures the association scores of miRNA-disease pairs by calculating the accumulative contributions from all paths.16 EGBMMDA utilizes an extreme gradient-boosting machine model for predicting miRNA-disease associations.31 Several different evaluation metrics were employed to comprehensively verify the performance of our method.
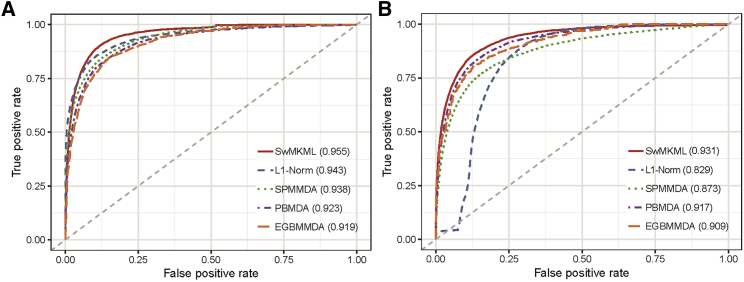
We first performed global leave-one-out cross-validation (LOOCV) and 5-fold cross-validation (CV) to evaluate our method based on the experimentally verified miRNA-disease association dataset from Human MicroRNA Disease Database (HMDD) v.2.0.34 In particular, global LOOCV considered each association as the test set and the rest as the training set to iteratively obtain a predicted ranking.35 For 5-fold CV, the entire miRNA-disease associations were randomly divided into five disjoint subsections, and then each part was selected as the test set, whereas the remaining parts were taken as the training set.36 To intuitively demonstrate prediction performance, the receiver operating characteristic (ROC) curve was drawn by plotting the true positive rate (TPR) against the false positive rate (FPR) at varying thresholds.37 Moreover, the area under the ROC curve (AUC) was calculated to quantitatively measure the performance of all methods.38 AUC = 1 means that the method achieves a perfect performance, whereas AUC = 0.5 indicates that the method has a random prediction performance. Figure 1 shows in detail the performance of our method compared with the other four methods in terms of global LOOCV and 5-fold CV. It can be observed that our method obtained the best performance within both frameworks.
Figure 1.
Performance Comparison of SwMKML with the Other Four Methods
(A and B) Comparisons in terms of (A) global LOOCV and (B) 5-fold CV.
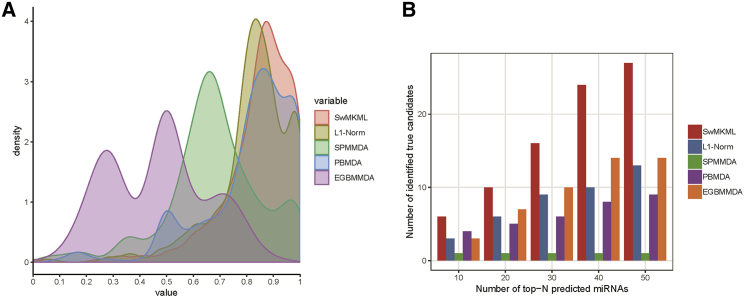
Next, a new evaluation metric, called leave-one-disease-out cross-validation (LODOCV) was adopted to assess the prediction power of our method in predicting diseases without known associated miRNAs. Specifically, for a given disease, LODOCV removed all miRNAs associated with this disease, and the predictions were carried out relying on the association information from other diseases. As shown in Figure 2A, our method also achieved the best performance among all methods. Furthermore, we also calculated the statistical significance of differences in performance obtained by our method and the other four methods (Table 1), and a Wilcoxon signed-rank test statistically confirmed the superiority of our method.
Figure 2.
Performance Comparison between SwMKML and the Other Methods under Two Different Evaluation Metrics
(A) Performance comparison of SwMKML with the other four methods in terms of LODOCV. (B) The number of predicted miRNAs that were confirmed in HMDD v.2.0.
Table 1.
Statistical Significance of Differences in Performance between SwMKML and the Other Four Methods in LODOCV
| L1-Norm | SPMMDA | PBMDA | EGBMMDA | |
|---|---|---|---|---|
| p Value | 3.99e−03 | 3.81e−37 | 4.01e−02 | 3.17e−87 |
We also selected four classical performance evaluation metrics—sensitivity (Sn), specificity (Sp), overall accuracy (Acc), and stability (Matthews correlation coefficient [MCC])—to objectively reflect the prediction performance of each method in a quantitative way.39, 40 The definitions of the four metrics are given as follows:41, 42
where and represent the total number of positive samples and negative samples investigated, respectively. is the number of positive samples incorrectly predicted to be negative, whereas is the number of negative samples incorrectly predicted to be positive. According to the definitions above, we obtained the values of the four metrics for each disease following the same process as that of LODOCV and calculated their average as the final results for each method. As shown in Table 2, SwMKML achieved the best performance under all evaluation metrics except Sn.
Table 2.
Comparison of the Proposed Method with the Four State-of-the-Art Methods in Terms of Acc, MCC, Sn, and Sp
| Method | Acc (%) | MCC | Sn (%) | Sp (%) |
|---|---|---|---|---|
| SwMKML | 84.10 | 0.3059 | 63.79 | 85.30 |
| L1-Norm | 83.34 | 0.3005 | 51.37 | 84.87 |
| SPMMDA | 82.45 | 0.2932 | 38.43 | 84.34 |
| PBMDA | 79.37 | 0.2613 | 65.00 | 79.78 |
| EGBMMDA | 54.87 | 0.1845 | 38.04 | 56.58 |
Finally, to demonstrate the prediction power of our method on real datasets, we implemented our method on the HMDD v.1.0 dataset and verified the prediction results based on the HMDD v.2.0 dataset. The older version of HMDD v.1.0 contained 1,616 association pairs involving 129 diseases and 280 miRNAs after filtering. Specifically, we compared the number of validated miRNA-disease pairs among the top 50 associations predicted by each method. As shown in Figure 2B, our method identified more validated associations than the other computational methods. Taken together, all results demonstrated the superiority and reliability of our method in predicting potential miRNAs associated with diseases.
Parameter Analysis
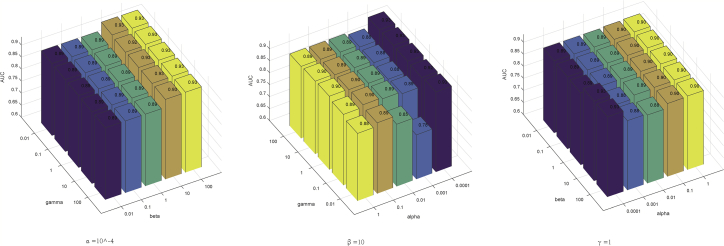
There were three trade-off parameters in our objective function. In this section, we varied their values to see their effects on the final prediction accuracy of 5-fold CV. Specifically, we tested the effects of two parameters each time by fixing the other parameter (Figure 3). We found that our method achieved the best performance when α = 10−4, β = 10, and γ = 1.
Figure 3.
Effects of the Three Parameters α, β, and γ on the Prediction Performance of SwMKML in 5-Fold CV
Convergence Analysis
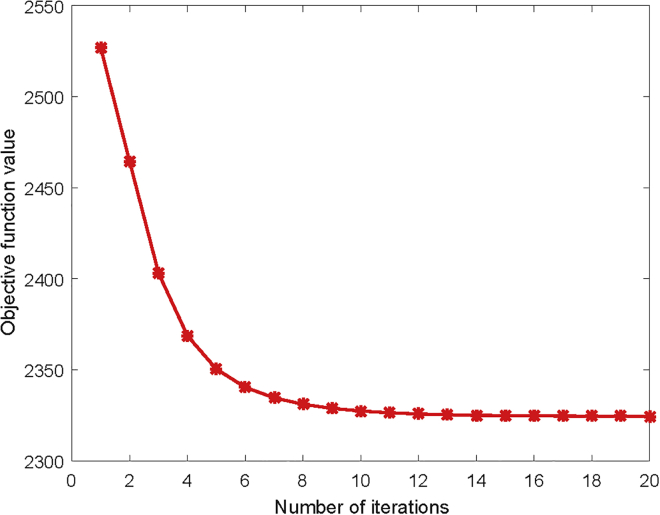
In this section, we verified the convergence of our method in practice based on 5-fold CV. As shown in Figure 4, our method quickly reached a steady state within 15 iterations, which clearly demonstrated that our method has a fast convergence speed. This characteristic ensures the extendibility of our method on large-scale datasets.
Figure 4.
The Variations of the Objective Function Value of SwMKML with Respect to the Number of Iterations
Case Study
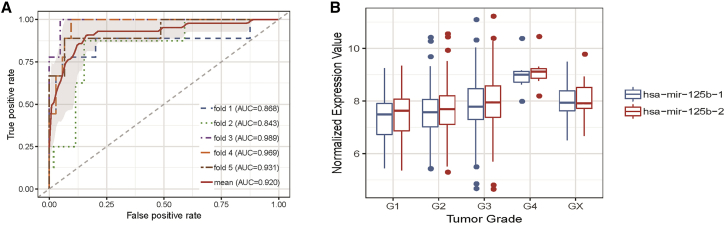
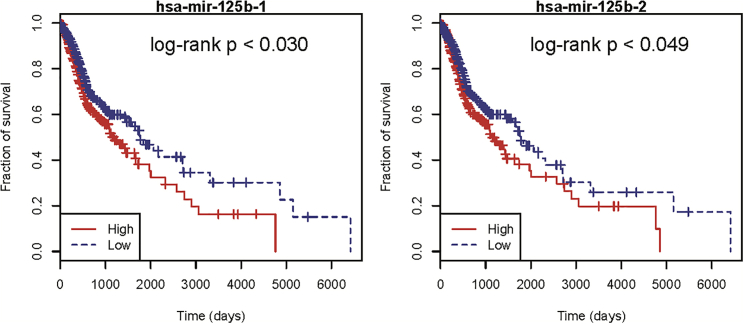
We conducted a case study analysis on head and neck neoplasms to further prove the reliability and prediction performance of our method. Head and neck squamous cell carcinoma (HNSC) is the sixth most common cause of cancer death worldwide, and the molecular mechanism of HNSC is not yet clear. In recent years, a handful of miRNAs were found to be differentially expressed in HNSC through clinical experiments, such as hsa-let-7a-1. The top10 miRNAs predicted to be related to HNSC by our method are listed in Table 3. Moreover, we downloaded miRNA expression data as well as clinical information of HNSC patients from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/repository)43 for further analysis. Concretely, the miRNA expression data contain 567 HNSC samples: 44 normal samples and 523 tumor samples. We first perform a 5-fold CV to assess the classification ability of the predicted miRNAs in differentiating the normal samples from tumor samples. As expected, these miRNAs achieved a mean classification accuracy of 0.92, indicating their strong classification power in HNSC (Figure 5A). We then carried out a differential expression analysis by using the R package edgeR.44 As a result, we found that 2 of the top 3 predicted miRNAs, hsa-mir-125b-1 and hsa-mir-125b-2, were significantly differentially expressed (false discovery rate [FDR] < 0.05 and log fold-change [|logFC|] > 1). Therefore, we further tested whether these two miRNAs were also significantly differentially expressed at different tumor stages by one-way ANOVA. Specifically, 5 pathological stages—G1, G2, G3, G4, and GX—were recorded in the clinical information, and the test results confirmed that their expression levels were indeed altered at varying stages (Figure 5B). Last, we carried out a Kaplan-Meier survival analysis to assess their potential prognostic role for HNSC by using the R package survival (Figure 6). Intriguingly, we found that patients with a lower expression level have a higher survival rate. In summary, our analysis indicated that the two miRNAs were closely related to HNSC and that they could serve as potential prognostic markers for clinical diagnosis.
Table 3.
Top 10 miRNAs related to HNSC based on our method.
| miRNA | p Value | logFC | FDR |
|---|---|---|---|
| hsa-mir-125b-1 | 2.59e−16 | −1.001610441 | 5.23e−15 |
| hsa-let-7a-1 | 2.50e−08 | −0.606930394 | 2.13e−07 |
| hsa-mir-125b-2 | 1.15e−17 | −1.061612358 | 2.70e−16 |
| hsa-let-7a-3 | 2.64e−08 | −0.60496417 | 2.23e−07 |
| hsa-let-7a-2 | 2.23e−08 | −0.609628999 | 1.92e−07 |
| hsa-let-7b | 0.000766178 | −0.367610987 | 3.24e−03 |
| hsa-let-7e | 0.606511339 | −0.068722721 | 9.64e−01 |
| hsa-mir-1-1 | 9.77e−27 | −3.369305544 | 4.08e−25 |
| hsa-mir-221 | 0.045288057 | 0.252113845 | 1.16e−01 |
| hsa-mir-145 | 8.05e−06 | −0.563207754 | 4.80e−05 |
The first column represents the miRNA names predicted by SwMKML. The second column represents the p value of the significance of differential expression for each miRNA. The third column represents the log2 fold change. The fourth column represents the adjusted p value of the differential analysis.
Figure 5.
Analysis for the Top 10 Predicted miRNAs
(A) Classification accuracy of the top 10 predicted miRNAs under 5-fold CV. (B) The expression level of has-mir-125b-1 and hsa-mir-125b-2 at different tumor stages.
Figure 6.
Kaplan-Meier Survival Analysis for hsa-mir-125b-1 and hsa-mir-125b-2, Identified as Prognostic Biomarkers in HNSC
As observed, patients with a lower expression level have a higher survival rate.
Discussion
It has been found that miRNAs play increasingly important roles in physiological processes and even complex human diseases. Researchers have attempted to make miRNAs valuable biomarkers for disease prevention, diagnosis, and treatment. Because of the inefficiency and high cost of experimental methods, many computational models have been developed to make effective predictions, such as graph-based methods, network topology-based methods, and the most widely used machine learning-based methods. In this paper, we propose a novel SwMKML method to predict potential miRNA-disease associations based on a miRNA functional similarity matrix, disease semantic similarity matrix, Gaussian interaction profile kernel similarity matrix, and association matrix between miRNAs and diseases. Specifically, our method learned an optimal kernel matrix adaptively from multiple kernel matrices for both miRNAs and diseases, respectively. We also propose a unified optimization process to update the predicted miRNA-disease association synchronously according to a graph-based, multi-label learning framework. As a result, comparative experiments conducted using our method and several state-of-the-art methods confirmed the superior performance and practicability of the proposed method. Last, the case study of head and neck neoplasms further validated the prediction ability of our method, and two miRNAs, hsa-mir-125b-1 and hsa-mir-125b-2, were identified as potential prognostic markers for HNSC.
The main reasons for the success of our model are 3-fold. First, the kernel matrices learned for both miRNAs and diseases during the optimization process were optimal kernels instead of a simple linear combination of base kernels. Moreover, the set of Gaussian kernels constructed with varying bandwidth parameters better characterized the known miRNA-disease associations from multiple views. Notably, our method is highly scalable because it only requires the miRNA-disease associations to fulfill the prediction task. Last but not least, the predictions of the miRNA-disease associations from both optimization spaces were unified by leveraging the multi-label learning framework. Nevertheless, there were also some limitations in our model. Because our method is a multi-kernel learning method, the given miRNA similarity matrix as well as disease similarity matrix has to be kernelized in advance, and different kernelization strategies might lead to different results. It remains a challenging task to balance the three trade-off parameters involved in our objective function to reach a global optimum.
Materials and Methods
Human miRNA-Disease Associations
The human miRNA-disease associations dataset used in this paper was downloaded from HMDD v.2.0, which includes 6,088 experimentally verified associations between 328 diseases and 550 miRNAs.45 For simplicity, we use to represent the known miRNA-disease association matrix. If miRNA mi is related to disease dj, then the entry Y(mi,dj) is 1 and 0 otherwise. Furthermore, the variables nm and nd represent the number of miRNAs and diseases in the dataset, respectively.
Disease Semantic Similarity
The development of the Mesh database provides great convenience for studying the relationship among diseases.46 Concretely, the relationship between different diseases in the database can be represented by a directed acyclic graph (DAG).47 A disease D can be represented as DAG(D) = (D,T(D),E(D)), where T(D) represents both D and its ancestor nodes, and E(D) represents all direct edges from parent nodes to child nodes. The contribution value of disease d to the semantic value of disease D can be formed as follows:
| (Equation 1) |
where Δ = 0.5 is the semantic contribution factor. For disease D, the contribution value to itself can be set to 1. From the representation of DAG mentioned above, we can finally conclude the semantic value of disease D as
| (Equation 2) |
Therefore, the semantic similarity between disease di and disease dj can be calculated as follows:
| (Equation 3) |
According to Equation 3, we obtained the disease semantic similarity matrix.
miRNA Functional Similarity
Wang et al.48 introduced a novel method to calculate miRNA functional similarity in terms of the associated disease terms. Here we directly downloaded the miRNA functional similarity score for the 550 miRNAs from http://www.cuilab.cn/files/images/cuilab/misim.zip.45 We use to denote the obtained similarity matrix for miRNAs, and (AM)ij measures the closeness between mi and mj.
Gaussian Interaction Profile Kernel Similarity
Based on the current miRNA-disease interaction prediction problem, we prefer the Gaussian kernel approach, which can construct a kernel matrix from the miRNA-disease interaction profiles. Gaussian interaction profile kernel similarity is the most popular method, and it has already been confirmed as an effective method for measuring similarities. For a given miRNA i or disease j, y(mi) or y(dj) is the interaction profile for the i-th row or the j-th column of the miRNA-disease association matrix. Therefore, the Gaussian interaction profile kernel similarity is defined as follows for both miRNA mi and disease dj:
| (Equation4) |
| (Equation 5) |
where γd and γm are determined by the following transformation:
| (Equation 6) |
| (Equation 7) |
where and are the kernel bandwidth. We denote and (i,j = 1,2,…,7) for the KGIP,m, and KGIP,d for both the miRNA space and disease space.
Kernelization
Because our method is based on multi-kernel learning, we first need to make the given miRNA similarity matrix AM as well as the disease similarity matrix AD positive semi-definite. As we know, a real symmetric matrix S could be decomposed into S = UΛUT, where U is an orthogonal matrix, and Λ is a diagonal matrix of real eigenvalues with Λ = diag(λ1,λ2,…,λn). Previous studies have considered different spectrum modifications to make S positive semi-definite, such as spectral shift, flip, and clip. Here we adopted spectrum shift because it only strengthens the self-similarities and does not change the similarity between any two different samples:49
| (Equation 8) |
where λmin(S) is the minimum eigenvalue of S. According to Equation 8, we converted AM and AD into the corresponding kernel matrices.
SwMKML
To fully understand the rationale behind our model, we first briefly introduced the single-kernel learning (SKL) framework on which SwMKML is based. In general, the SKL could be formulated as50
| (Equation 9) |
where K represents the kernel matrix constructed from the input data, and S is the similarity matrix that will be learned from K. L = D-S is the Laplacian matrix, and D is the diagonal degree matrix, with its i-th diagonal element defined as . In particular, F could be the class indicator matrix or label matrix, depending on whether this framework is applied to unsupervised or semi-supervised problems.51, 52 Therefore, we can obtain the multi-kernel learning framework by extending Equation 9 as follows:
| (Equation 10) |
where Hi (i = 1,…,l) is one of the input kernel matrices. Specifically, the kernel weight parameter wi is defined as
| (Equation 11) |
Although wi is dependent on K, we could update its value alternatively after obtaining K. As a result, the weight assignment for each kernel matrix is totally self-weighted. According to Equation 10, we could obtain the optimization function in miRNA space by substituting the variables in Equation 10 with matrices constructed in miRNA space:
| (Equation 12) |
where is the Laplacian matrix, and the degree matrix is defined as a diagonal matrix whose i-th diagonal element is . Similarly, we define the objective function in the disease space as follows:
| (Equation 13) |
The definition of variables in the disease space is equivalent to that in the miRNA space. Finally, instead of simply combining these two objective functions with equal weights, we integrate them into one overall optimization formulation in terms of the graph-based, multi-label learning framework:
| (Equation 14) |
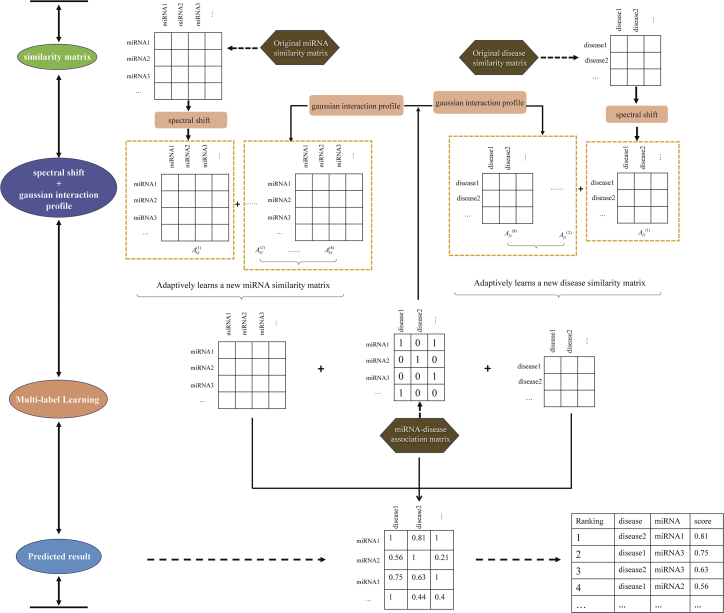
An overall workflow of the SwMKML method to predict the disease-related miRNAs is shown in Figure 7.
Figure 7.
Integrated Flow Chart of SwMKML to Predict Disease-Related miRNAs
Optimization
We divide the problem in Equation 14 into three subproblems with regard to miRNA space and disease space, respectively. We then develop an iterative algorithm to solve these problems alternatively.
Update SM. By fixing the other variables, the optimization for SM from Equation 14 can be derived as
| (Equation 15) |
Note that the problem (Equation 15) is independent for different i; thus, we can solve the problem separately for each i. Based on , we can equivalently solve the following problem for each i individually:
| (Equation 16) |
where with . By setting its first derivative with respect to (SM)i to zero, we can obtain
| (Equation17) |
Update KM. By fixing the other variables, Equation 14 can be rewritten as
| (Equation 18) |
By differentiating Equation 18 with respect to KM, we could obtain:
| (Equation 19) |
After we obtained KM, we could update the weight value for each view as follows:
| (Equation 20) |
Because the optimization in disease space is the same as that in miRNA space, we could derive the formulas to optimize SD, KD and as follows:
| (Equation 21) |
| (Equation 22) |
| (Equation 23) |
where with its j-th element defined as.
Update F. Equation 14 is transformed into the following formula by fixing the other four variables:
| (Equation 24) |
By differentiating Equation 24 with respect to F and setting it to zero, we could obtain the following formula:
| (Equation 25) |
Obviously, Equation 25 is a Sylvester equation and can be easily solved.53 The overall procedure for solving Equation 14 is summarized in Box 1. The dataset used in this paper as well as the source code of SwMKML is available at https://github.com/JiaMuL/SwMKML.
Box 1. Algorithm to Solve Equation 14.
Input: miRNA similarity matrices of n views {,, …, }, disease similarity matrices of m views {, , …, }, known association matrix , the parameters α, β and γ.
Output: Predicted association matrix F.
1. Initialize the weights of each view for both miRNAs and diseases with , .
2. Repeat:
3. Repeat:
4. Update SM by solving problem (17).
5. Update KM by solving problem (19).
6. Update SD by solving problem (21).
7. Update KD by solving problem (22).
8. Update F by solving problem (25).
9. Until convergence
10. Update , according to Equation (20) and Equation (23).
11. Until convergence
12. Return SM, KM, SD, KD, F
Author Contributions
C.L. and Z.P. conceived the study. C.L., Z.P., and J.L. developed the algorithm and analyzed the results. C.L. and Z.P. wrote this paper. H.Z., G.L., Q.X., and P.D. supervised this study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank the anonymous reviewers for their valuable suggestions. This work was supported by the National Natural Science Foundation of China (61602283, U1836216, 61772322, and 61873089) and the Key Research and Development Foundation of Shandong Province (2017GGX10117 and 2017CXGC0703).
Contributor Information
Huaxiang Zhang, Email: huaxzhang@hotmail.com.
Cheng Liang, Email: alcs417@sdnu.edu.cn.
References
- 1.Trzybulska D., Vergadi E., Tsatsanis C. miRNA and Other Non-Coding RNAs as Promising Diagnostic Markers. EJIFCC. 2018;29:221–226. [PMC free article] [PubMed] [Google Scholar]
- 2.Song Y.S., Joo H.W., Park I.H., Shen G.Y., Lee Y., Shin J.H., Kim H., Kim K.S. Bone marrow mesenchymal stem cell-derived vascular endothelial growth factor attenuates cardiac apoptosis via regulation of cardiac miRNA-23a and miRNA-92a in a rat model of myocardial infarction. PLoS ONE. 2017;12:e0179972. doi: 10.1371/journal.pone.0179972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Xie D., Zhao Q., You Z.H. MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2019;20:515–539. doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]
- 4.Hiddingh L., Raktoe R.S., Jeuken J., Hulleman E., Noske D.P., Kaspers G.J., Vandertop W.P., Wesseling P., Wurdinger T. Identification of temozolomide resistance factors in glioblastoma via integrative miRNA/mRNA regulatory network analysis. Sci. Rep. 2014;4:5260. doi: 10.1038/srep05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pian C., Zhang G., Gao L., Fan X., Li F. miR+Pathway: the integration and visualization of miRNA and KEGG pathways. Brief. Bioinform. 2019 doi: 10.1093/bib/bby128. Published online January 16, 2019. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Yan C.C., Zhang X., You Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D., Li G., Zuo Y. Function determinants of TET proteins: the arrangements of sequence motifs with specific codes. Brief. Bioinform. 2018 doi: 10.1093/bib/bby053. Published online June 26, 2018. [DOI] [PubMed] [Google Scholar]
- 8.Qin L., Liu Y., Li M., Pu X., Guo Y. The landscape of miRNA-related ceRNA networks for marking different renal cell carcinoma subtypes. Brief. Bioinform. 2018 doi: 10.1093/bib/bby101. Published online November 16, 2018. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Q., Hao Y., Wang G., Juan L., Zhang T., Teng M., Liu Y., Wang Y. Prioritization of disease microRNAs through a human phenome-microRNAome network. BMC Syst. Biol. 2010;4(Suppl 1):S2. doi: 10.1186/1752-0509-4-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Liu M.-X., Yan G.-Y. RWRMDA: predicting novel human microRNA-disease associations. Mol. Biosyst. 2012;8:2792–2798. doi: 10.1039/c2mb25180a. [DOI] [PubMed] [Google Scholar]
- 11.Xuan P., Han K., Guo Y., Li J., Li X., Zhong Y., Zhang Z., Ding J. Prediction of potential disease-associated microRNAs based on random walk. Bioinformatics. 2015;31:1805–1815. doi: 10.1093/bioinformatics/btv039. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Zeng X., He Z., Zou Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017;14:905–915. doi: 10.1109/TCBB.2016.2550432. [DOI] [PubMed] [Google Scholar]
- 13.Shi H., Xu J., Zhang G., Xu L., Li C., Wang L., Zhao Z., Jiang W., Guo Z., Li X. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst. Biol. 2013;7:101. doi: 10.1186/1752-0509-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Yan C.C., Zhang X., You Z.-H., Huang Y.-A., Yan G.-Y. HGIMDA: Heterogeneous graph inference for miRNA-disease association prediction. Oncotarget. 2016;7:65257–65269. doi: 10.18632/oncotarget.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Q., Li J., Hong Q., Lin Z., Wu Y., Shi H., Ju Y. Prediction of microRNA-disease associations based on social network analysis methods. Biomed Res. Int. 2015;2015:810514. doi: 10.1155/2015/810514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You Z.-H., Huang Z.-A., Zhu Z., Yan G.-Y., Li Z.-W., Wen Z., Chen X. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput. Biol. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D., Li A., Feng H., Wang M. NTSMDA: prediction of miRNA-disease associations by integrating network topological similarity. Mol. Biosyst. 2016;12:2224–2232. doi: 10.1039/c6mb00049e. [DOI] [PubMed] [Google Scholar]
- 18.Li G., Luo J., Xiao Q., Liang C., Ding P., Cao B. Predicting microRNA-disease associations using network topological similarity based on deepwalk. IEEE Access. 2017;5:24032–24039. [Google Scholar]
- 19.Chen X., Guan N.N., Li J.Q., Yan G.Y. GIMDA: Graphlet interaction-based MiRNA-disease association prediction. J. Cell. Mol. Med. 2018;22:1548–1561. doi: 10.1111/jcmm.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu Y., Zhang H.X., Liang C., Dong X. KATZMDA: Prediction of miRNA-Disease Associations Based on KATZ Model. IEEE Access. 2018;6:3943–3950. [Google Scholar]
- 21.Agarwal V., Subtelny A.O., Thiru P., Ulitsky I., Bartel D.P. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 2018;19:152. doi: 10.1186/s13059-018-1504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo Y.-C., Peng Y., Liu L., Chen W., Yang L., Fan G.-L. Predicting peroxidase subcellular location by hybridizing different descriptors of Chou’ pseudo amino acid patterns. Anal. Biochem. 2014;458:14–19. doi: 10.1016/j.ab.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 23.Zuo Y., Lv Y., Wei Z., Yang L., Li G., Fan G. iDPF-PseRAAAC: a web-server for identifying the defensin peptide family and subfamily using pseudo reduced amino acid alphabet composition. PLoS ONE. 2015;10:e0145541. doi: 10.1371/journal.pone.0145541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo Y., Li Y., Chen Y., Li G., Yan Z., Yang L. PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition. Bioinformatics. 2017;33:122–124. doi: 10.1093/bioinformatics/btw564. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Wang C.-C., Yin J., You Z.-H. Novel human miRNA-disease association inference based on random forest. Mol. Ther. Nucleic Acids. 2018;13:568–579. doi: 10.1016/j.omtn.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W., Chen Y., Li D. Drug-target interaction prediction through label propagation with linear neighborhood information. Molecules. 2017;22:2056. doi: 10.3390/molecules22122056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., Qu Q., Zhang Y., Wang W. The linear neighborhood propagation method for predicting long non-coding RNA–protein interactions. Neurocomputing. 2018;273:526–534. [Google Scholar]
- 28.Chen X., Yan G.-Y. Semi-supervised learning for potential human microRNA-disease associations inference. Sci. Rep. 2014;4:5501. doi: 10.1038/srep05501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Q., Luo J., Liang C., Cai J., Ding P. A graph regularized non-negative matrix factorization method for identifying microRNA-disease associations. Bioinformatics. 2017;34:239–248. doi: 10.1093/bioinformatics/btx545. [DOI] [PubMed] [Google Scholar]
- 30.Chen X., Wang L., Qu J., Guan N.-N., Li J.-Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 31.Chen X., Huang L., Xie D., Zhao Q. EGBMMDA: extreme gradient boosting machine for MiRNA-disease association prediction. Cell Death Dis. 2018;9:3. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X., Liu L., Lü L., Zou Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics. 2018;34:2425–2432. doi: 10.1093/bioinformatics/bty112. [DOI] [PubMed] [Google Scholar]
- 33.Liang C., Yu S.P., Wong K.C., Luo J.W. A novel semi-supervised model for miRNA-disease association prediction based on 1-norm graph. J. Transl. Med. 2018;16:357. doi: 10.1186/s12967-018-1741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S.P., Liang C., Xiao Q., Li G.H., Ding P.J., Luo J.W. GLNMDA: a novel method for miRNA-disease association prediction based on global linear neighborhoods. RNA Biol. 2018;15:1215–1227. doi: 10.1080/15476286.2018.1521210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bo L., Wang L., Jiao L. Feature scaling for kernel fisher discriminant analysis using leave-one-out cross validation. Neural Comput. 2006;18:961–978. doi: 10.1162/089976606775774642. [DOI] [PubMed] [Google Scholar]
- 36.Isaksson A., Wallman M., Goransson H., Gustafsson M.G. Cross-validation and bootstrapping are unreliable in small sample classification. Pattern Recognit. Lett. 2008;29:1960–1965. [Google Scholar]
- 37.Zhang H.X., Cao L.L., Gao S. A locality correlation preserving support vector machine. Pattern Recognit. 2014;47:3168–3178. [Google Scholar]
- 38.Hand D.J., Till R.J. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach. Learn. 2001;45:171–186. [Google Scholar]
- 39.Liu B., Li K., Huang D.S., Chou K.C. iEnhancer-EL: identifying enhancers and their strength with ensemble learning approach. Bioinformatics. 2018;34:3835–3842. doi: 10.1093/bioinformatics/bty458. [DOI] [PubMed] [Google Scholar]
- 40.Long C.S., Li W., Liang P.F., Liu S., Zuo Y.C. Transcriptome Comparisons of Multi-Species Identify Differential Genome Activation of Mammals Embryogenesis. IEEE Access. 2019;7:7794–7802. [Google Scholar]
- 41.Zuo Y.C., Su W.X., Zhang S.H., Wang S.S., Wu C.Y., Yang L., Li G.P. Discrimination of membrane transporter protein types using K-nearest neighbor method derived from the similarity distance of total diversity measure. Mol. Biosyst. 2015;11:950–957. doi: 10.1039/c4mb00681j. [DOI] [PubMed] [Google Scholar]
- 42.Zuo Y.C., Chen W., Fan G.L., Li Q.Z. A similarity distance of diversity measure for discriminating mesophilic and thermophilic proteins. Amino Acids. 2013;44:573–580. doi: 10.1007/s00726-012-1374-z. [DOI] [PubMed] [Google Scholar]
- 43.Katsoulakis E., Oh J.H., Leeman J.E., Yu Y., Tsai C.J., McBride S., Katabi N., Apte A., Deasy J.O., Lee N. Identifying Biological Subtypes of Head and Neck Squamous Cell Carcinoma (HNSCC) From Contrast Enhanced CT Scans Using Radiomic and the Cancer Genome Atlas (TCGA) Int. J. Radiat. Oncol. 2018;102:S60–S61. [Google Scholar]
- 44.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Qiu C., Tu J., Geng B., Yang J., Jiang T., Cui Q. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mottaz A., Yip Y.L., Ruch P., Veuthey A.L. Mapping proteins to disease terminologies: from UniProt to MeSH. BMC Bioinformatics. 2008;9(Suppl 5):S3. doi: 10.1186/1471-2105-9-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akinkugbe A.A., Sharma S., Ohrbach R., Slade G.D., Poole C. Directed Acyclic Graphs for Oral Disease Research. J. Dent. Res. 2016;95:853–859. doi: 10.1177/0022034516639920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D., Wang J., Lu M., Song F., Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26:1644–1650. doi: 10.1093/bioinformatics/btq241. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Gupta M.R., Recht B. Learning kernels from indefinite similarities. Proceedings of the 26th Annual International Conference on Machine Learning. 2009;2009:145–152. [Google Scholar]
- 50.Kang Z., Lu X., Yi J., Xu Z. Self-weighted multiple kernel learning for graph-based clustering and semi-supervised classification. Proceedings of the 27th International Joint Conference on Artificial Intelligence. 2018;2018:2312–2318. [Google Scholar]
- 51.Zhu L., Shen J.L., Xie L., Cheng Z.Y. Unsupervised Topic Hypergraph Hashing for Efficient Mobile Image Retrieval. IEEE Trans. Cybern. 2017;47:3941–3954. doi: 10.1109/TCYB.2016.2591068. [DOI] [PubMed] [Google Scholar]
- 52.Zhu L., Huang Z., Li Z., Xie L., Shen H.T. Exploring Auxiliary Context: Discrete Semantic Transfer Hashing for Scalable Image Retrieval. IEEE Trans. Neural Netw. Learn. Syst. 2018;29:5264–5276. doi: 10.1109/TNNLS.2018.2797248. [DOI] [PubMed] [Google Scholar]
- 53.Zha Z.J., Mei T., Wang J., Wang Z., Hua X.S. Graph-based semi-supervised learning with multiple labels. J. Vis. Commun. Image Represent. 2009;20:97–103. [Google Scholar]