Abstract
Several studies have recently revealed the regulatory mechanisms underlying female germline stem cell (FGSC) differentiation, proliferation, and apoptosis, but other biological processes such as autophagy and its mechanism in FGSCs are largely unclear. The use of small chemical compounds may be a good approach to further investigate the process and mechanism of autophagy in FGSC development. In this study, we used ZCL-082, a derivative of benzoxaboroles, to treat FGSCs. Using a cell counting kit-8 (CCK8) and 5-ethynyl-2′-deoxyuridine (EdU) assays, we found that ZCL-082 could significantly reduce the viability, proliferation, and number of FGSCs in vitro. Moreover, western blotting revealed that the expression of light chain 3 beta 2 (LC3B-II) in FGSCs was significantly increased after treatment with ZCL-082 for 3 and 6 h. Meanwhile, the expression of sequestosome-1 (SQSTM1) was significantly decreased. These results suggested that ZCL-082 can induce autophagy of FGSCs in vitro. Regarding the molecular mechanism, ZCL-082 could significantly reduce the expression of growth arrest-specific 5 (GAS5) long non-coding RNA, which could directly bind to microRNA-21a (miR-21a) and negatively regulate each other in FGSCs. Knockdown of GAS5 induced the autophagy of FGSCs, while GAS5 overexpression inhibited the autophagy of FGSCs in vitro and rescued FGSC autophagy induced by ZCL-082. Additionally, overexpression of miR-21a significantly enhanced LC3B-II protein expression while significantly reducing the expression of programmed cell death protein 4 (PDCD4) and SQSTM1 protein in FGSCs compared with control cells. The inhibition of miR-21a significantly reduced the basal or ZCL-082-induced upregulated expression of LC3B-II, and it significantly enhanced the expression of PDCD4 while downregulating the basal or ZCL-082-induced expression of SQSTM1 in FGSCs. Furthermore, the overexpression of GAS5 enhanced the protein expression of PDCD4, but knockdown of GAS5 reduced the protein expression of PDCD4. Taken together, these results suggested that ZCL-082 induced autophagy through GAS5 functioning as a competing endogenous RNA (ceRNA) sponge for miR-21a in FGSCs. It also suggested that the GAS5/miR-21a axis may be a potential therapeutic target for premature ovarian failure in the clinic.
Introduction
Female germline stem cells (FGSCs) have been successfully isolated from postnatal mammal ovarian tissue,1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 enabling researchers to further understand human oogenesis, treat infertility, and delay menopause.15 This also provides a novel strategy for preserving fertility. While several studies recently revealed the regulatory mechanisms underlying FGSC differentiation,3, 16, 17, 18, 19, 20 proliferation,4, 21, 22, 23, 24 and apoptosis,21, 25 which are considered crucial for female fertility, other biological processes such as autophagy and its mechanism in FGSCs are largely unknown, and the use of small compounds may help us to understand them further.
Benzoxaboroles, which were synthesized and characterized by Torssell in 1957,26 have medical applications because 5-fluroben-zoxaborole (AN2690) can inhibit fungal protein synthesis by targeting leucyl-tRNA synthetase (LeuRS)27 and is a promising clinical treatment for onychomycosis.26 The wide application of benzoxaboroles in anti-viral,28 anti-bacterial,29 anti-inflammatory,30 anti-parasite,31 and anti-cancer32 treatments was shown in recent studies. For example, two benzoxaboroles, AN2728 and AN2898, underwent phase II clinical trials as treatments for atopic dermatitis and psoriasis, because of their anti-inflammatory characteristics.26 However, the cell biological function of benzoxaboroles was only studied in SKOV-3 cells, and the result has shown that ZCL-082, a derivative of benzoxaboroles, significantly inhibited the proliferation of human ovarian cancer SKOV-3 cells,32 suggesting that it probably induces cell autophagy or apoptosis. To date there have been no studies of the autophagy function of benzoxaboroles in cells.
Long non-coding RNAs (lncRNAs), which do not encode proteins, can recruit transcription factors and sponge microRNAs (miRNAs) to destabilize mRNA.33, 34, 35, 36 This type of RNA has been found to widely regulate the proliferation, differentiation, apoptosis, and autophagy of stem cells as competing endogenous RNAs (ceRNAs).37, 38, 39, 40 In our previous study, we determined that growth arrest-specific 5 (GAS5), a lncRNA, promoted proliferation and inhibited apoptosis of FGSCs in vitro;21 but, whether and how GAS5 executes other FGSC functions (e.g., autophagy or differentiation) are unclear.
miRNAs, short non-coding RNAs of 22–23 nt in size, can act as sponges for lncRNAs and silence the expression of coding or non-coding genes in animals and plants.33 Further, they are involved in cell signal transduction pathways and protein-protein networks by changing the expressions of proteins.33, 41 MiR-21a, which can be sponged by GAS5,42 has been reported to play an important regulatory role in cell apoptosis, differentiation, autophagy, and proliferation by suppressing the expression of coding genes, such as programmed cell death protein 4 (PDCD4).43, 44, 45, 46
In our study, we investigated how ZCL-082 affected the biological function of FGSCs in vitro, and we revealed that ZCL-082 induced the autophagy of FGSCs through the GAS5/miR-21a axis. The results suggested that ZCL-082 and the GAS5/miR-21a axis have critical roles in the autophagy of FGSCs. Our findings also provide new insights into reproductive disorders and the preservation of female fertility.
Results
ZCL-082 Reduced the Number, Viability, and Proliferation of FGSCs
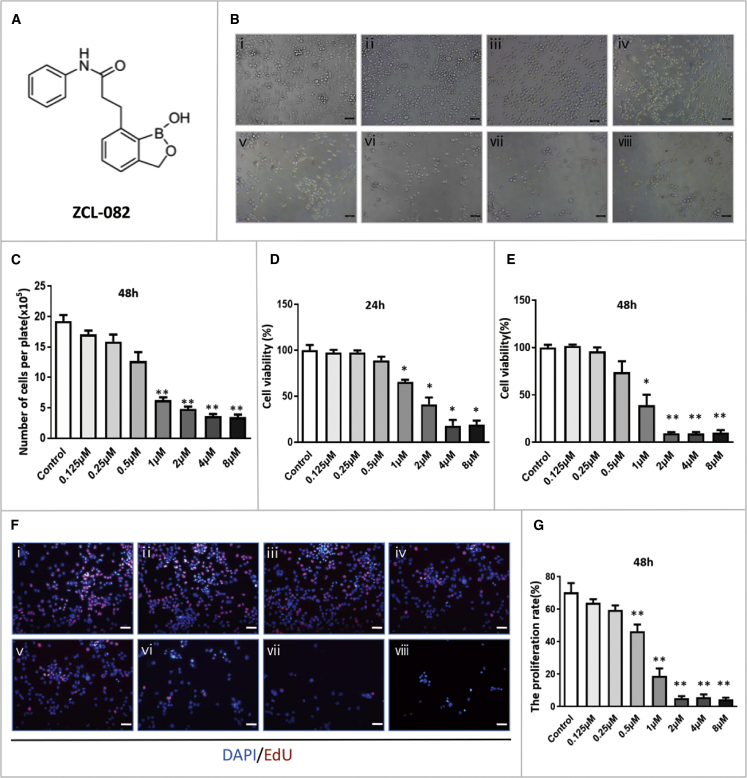
To detect whether ZCL-082 affects FGSC proliferation, the cells were treated with various concentrations of ZCL-082 (0.125, 0.25, 0.5, 1, 2, 4, and 8 μM), and FGSCs per well were counted using a hemocytometer. As shown in Figures 1B and 1C, the number of cells was significantly reduced by treatment with 1–8 μM ZCL-082 compared with the control (DMSO). Cell counting kit-8 (CCK8) and 5-ethynyl-2′-deoxyuridine (EdU) assays were used to detect cell viability and proliferation, respectively. As shown in Figures 1D and 1E, cell viability was significantly lower following treatment with 1, 2, 4, and 8 μM ZCL-082 for 24 or 48 h compared with the control. The results of the EdU assays showed that the cell proliferation rate was significantly lower in cells treated for 48 h with ZCL-082 at 0.5, 1, 2, 4, and 8 μM (Figure 1F). Taken together, these results indicated that ZCL-082 reduced the number, viability, and proliferation of FGSCs in vitro.
Figure 1.
ZCL-082 Reduced the Number, Viability, and Proliferation of FGSCs
(A) Chemical structure of ZCL-082. (B) Morphology of FGSCs treated with various concentrations of ZCL-082 (i, control; ii, 0.125 μM; iii, 0.25 μM; iv, 0.5 μM; v, 1 μM; vi, 2 μM; vii, 4 μM; and viii, 8 μM). Scale bar, 50 μm. Control was DMSO. (C) Number of FGSCs per 35-mm plate at various concentrations of ZCL-082. Control was DMSO. (D and E) Fold change in cell viability under different concentrations was detected by CCK8 assay at 24 h (D) and 48 h (E). Control was DMSO. (F and G) Cell proliferation was detected by EdU assay at various concentrations of ZCL-082 (F) and their results were statistically significant (G) (i, control; ii, 0.125 μM; iii, 0.25 μM; iv, 0.5 μM; v, 1 μM; vi, 2 μM; vii, 4 μM; and viii, 8 μM). Control was DMSO. Scale bar, 50 μm. Values are presented as mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
ZCL-082 Induced FGSC Apoptosis and Autophagy In Vitro
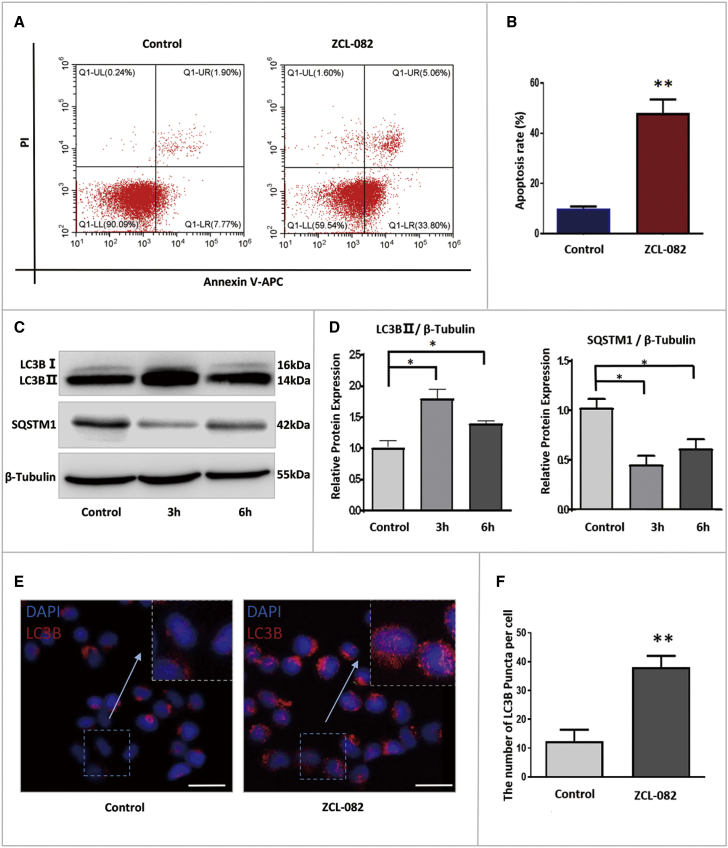
Next, we investigated why viability declined in FGSCs. The expression of SYCP3 and STRA8, differentiation markers of FGSCs, was determined by RT-PCR; no significant difference was found between the control and ZCL-082-treated cells for 48 h (Figure S1). The results suggested that ZCL-082 has no effect on FGSC differentiation. Next, we used flow cytometry to elucidate whether ZCL-082 induces the apoptosis of FGSCs. As shown in Figures 2A and 2B, ZCL-082 induced the apoptosis of FGSCs. Because of the close relationship between apoptosis and autophagy during cell death,47, 48 we determined whether ZCL-082 induced autophagy in FGSCs. Western blotting revealed that the protein expression of light chain 3 beta 2 (LC3B-II) in FGSCs was significantly increased after treatment with ZCL-082 for 3 and 6 h (Figures 2C and 2D). Meanwhile, sequestosome-1 (SQSTM1) protein expression was significantly decreased (Figures 2C and 2D). Immunofluorescence staining showed that the number of LC3B puncta per cell was significantly increased in treated cells (Figures 2E and 2F). Taken together, these results suggested that ZCL-082 could induce the autophagy of FGSCs in vitro.
Figure 2.
Autophagy Was Induced by ZCL-082 in FGSCs
(A and B) Flow cytometry detected the apoptosis of FGSCs after being treated with ZCL-082. Apoptosis was detected by FACs with Annexin V-PI Apoptosis Kit (A) and statistics (B). Control was DMSO. (C and D) Differential expressions of LC3B-II and SQSTM1 after treatment with ZCL-082 for 3 and 6 h. (C) Western blot showing the relative expression levels of LC3B-II and SQSTM1. (D) Densitometry demonstrated differences in expression were statistically significant. Control was DMSO. (E and F) Immunofluorescence staining of LC3B-II after treatment with ZCL-082. (E) LC3B puncta (F) numbers were found to be statistically significant. Control was DMSO. Scale bar, 25 μm. Values are mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
The Expressions of Several Autophagy-Related lncRNAs Were Changed in FGSCs after Treatment with ZCL-082
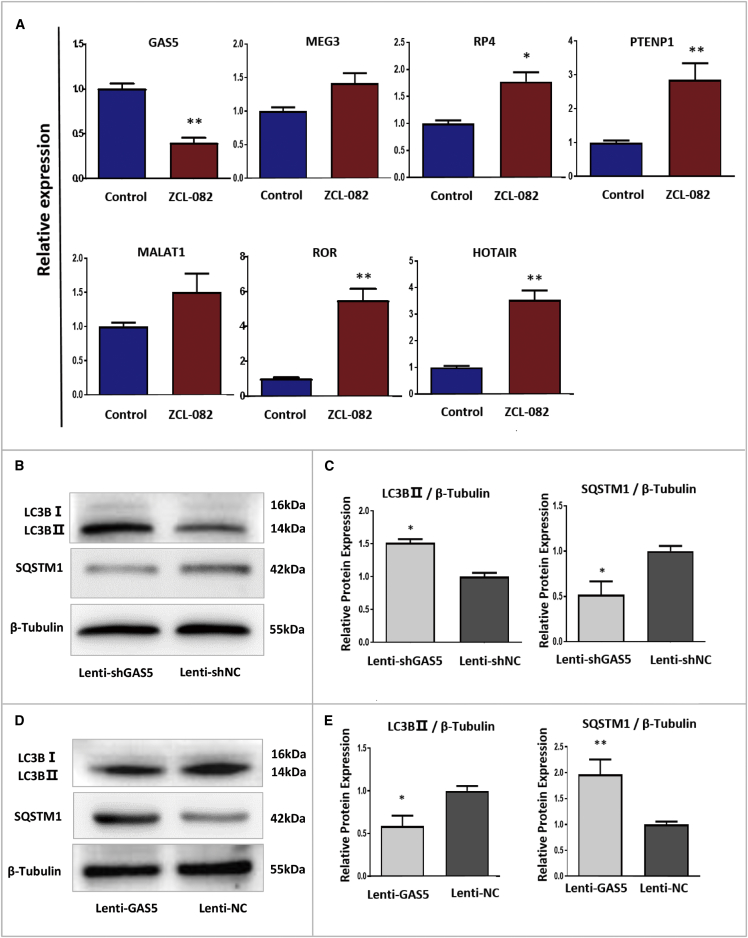
lncRNAs were reported to be involved in the process of cell autophagy in previous studies.49, 50 To discuss the ceRNA regulatory mechanism underlying FGSC autophagy induced by ZCL-082, we first selected lncRNAs related to autophagy, and we detected their expressions in FGSCs after ZCL-082 treatment. qRT-PCR showed that the expression of GAS5 was significantly decreased, while the expression levels of RP4, PTENP1, ROR, and HOTAIR were significantly increased (Figure 3A). Additionally, after treatment, the expressions of MEG3 and MALAT1 were not significantly changed (Figure 3A).
Figure 3.
Expression Changes of Several Autophagy-Related lncRNAs in FGSCs following ZCL-082 Treatment
(A) Fold changes in GAS5, RP4, PTENP1, ROR, HOTAIR, MEG3, and MALAT1 expressions after ZCL-082 treatment. Control was DMSO. (B–E) Differential expressions of LC3B-II and SQSTM1 after infection with Lenti-GAS5 and Lenti-shGAS5. Representative blots following infection with Lenti-GAS5 (B) and Lenti-shGAS5 (D) are shown. Relative protein expression levels of LC3B and SQSTM1 in Lenti-GAS5- (C) and Lenti-shGAS5-infected FGSCs (E) are shown. Lenti-shGAS5, lentivirus carrying GAS5-knockdown plasmid; Lenti-shNC, lentivirus carrying control plasmid; Lenti-GAS5, lentivirus carrying GAS5-overexpressing plasmid; Lenti-NC, lentivirus carrying control plasmid. Data are shown as mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
GAS5 Inhibited FGSC Autophagy In Vitro
In our previous study, although GAS5 could promote the proliferation of FGSCs and inhibit their apoptosis,21 the function of GAS5 in FGSC autophagy was not investigated. To define the function of GAS5 in FGSC autophagy, FGSCs were infected with GAS5-knockdown or -overexpressing lentiviral particles. Western blotting demonstrated that LC3B-II protein expression was significantly increased and that of SQSTM1 was significantly decreased compared with the control (Lenti-negative control of shRNA [shNC]), when cells were infected with GAS5-knockdown lentivirus (Figures 3B and 3C). Furthermore, LC3B-II protein expression was significantly decreased and that of SQSTM1 was significantly increased compared with the control (Lenti-negative control [NC]), after infection with GAS5-overexpressing lentivirus (Figures 3D and 3E). These results indicated that GAS5 could inhibit FGSC autophagy in vitro.
Overexpression of GAS5 Rescued FGSC Autophagy Induced by ZCL-082
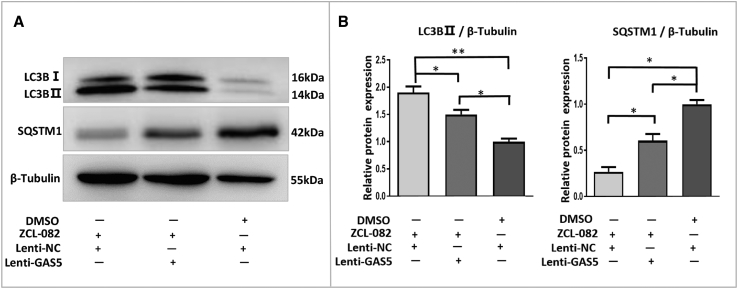
Based on the above results, we wondered whether overexpression of GAS5 protects FGSCs from ZCL-082-induced autophagy. We constructed three separate groups: treatment with ZCL-082 for 3 h in Lenti-GAS5-infected FGSCs (group ZCL-082 + GAS5), treatment with ZCL-082 for 3 h in Lenti-NC-infected FGSCs (group ZCL-082 + NC), and treatment with DMSO for 3 h in Lenti-NC-infected FGSCs (group DMSO + NC). After treatment, LC3B-II and SQSTM1 protein expressions in the different groups were detected by western blotting; LC3B-II expression in group ZCL-082 + GAS5 was significantly lower than that in group ZCL-082 + NC. Moreover, the expression of SQSTM1 in group ZCL-082 + GAS5 was significantly higher than that in group ZCL-082 + NC (Figures 4A and 4B). These results indicated that GAS5 overexpression could rescue the autophagy of FGSCs induced by ZCL-082.
Figure 4.
Autophagy in ZCL-082-Treated FGSCs Was Rescued by the Overexpression of GAS5
(A and B) After infection with Lenti-GAS5 or Lenti-NC, FGSCs were treated with ZCL-082 for 3 h. Protein expressions of LC3B-II and SQSTM1 were detected by western blotting (A), and the results were statistically significant (B). Lenti-shGAS5, lentivirus carrying GAS5-knockdown plasmid; Lenti-GAS5, lentivirus carrying GAS5-overexpressing plasmid; Lenti-NC, lentivirus carrying control. Data are presented as mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
GAS5 Regulated Autophagy by Sponging miR-21a
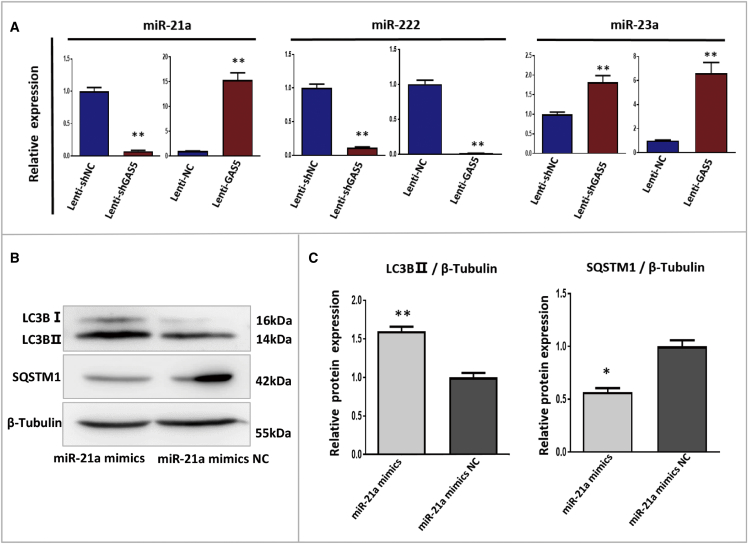
To further explore the ceRNA regulatory mechanism underlying FGSC autophagy induced by ZCL-082 through GAS5, we selected miRNAs (miR-21a, miR-222, and miR-23a) that were reported in previous studies to be sponged by GAS5.42, 51 We first detected the expressions of these miRNAs in Lenti-GAS5-infected FGSCs and Lenti-knockdown of GAS5 (shGAS5)-infected FGSCs. The qRT-PCR results showed that the expression of mir-21a was significantly decreased in Lenti-GAS5-infected FGSCs while it was significantly increased in Lenti-shGAS5-infected FGSCs compared with the control (Lenti-NC and Lenti-shNC, respectively) (Figure 5A). The qRT-PCR results also showed that, in both Lenti-GAS5- and Lenti-shGAS5-infected cells, miR-222 expression was significantly lower than in the control and miR-23a expression was significantly higher than in the control (Figure 5A). Combined with previous research,42 these results suggested that GAS5 could directly bind miR-21a to negatively regulate each other.
Figure 5.
ZCL-082 Induced Autophagy of FGSCs via GAS5 Sponging of miR-21a
(A) Relative expressions of miR-21a, miR-222, and miR-23a in Lenti-GAS5 and Lenti-shGAS5 FGSCs. (B and C) Protein expressions of LC3B-II and SQSTM1 were detected by western blotting (B) after transfection for 24 h with miR-21a mimics and NC in FGSCs. The results of differential blots (C) were statistically significant. miR-21a mimic-NC, negative control of miR-21a mimic. Data are mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
Next, FGSCs were transfected with miR-21a mimics, and the expressions of LC3B-II and SQSTM1 were detected by western blotting to determine whether the overexpression of miR-21a can induce FGSC autophagy. The results showed that miR-21a expression significantly enhanced the expression of LC3B-II protein (Figures 5B and 5C) while significantly reducing the expression of SQSTM1 protein in FGSCs compared with control cells (miR-21a mimic-NC) (Figures 5D and 5E). These results revealed that miR-21a could promote FGSC autophagy.
ZCL-082 Induced the Autophagy of FGSCs through miR-21a
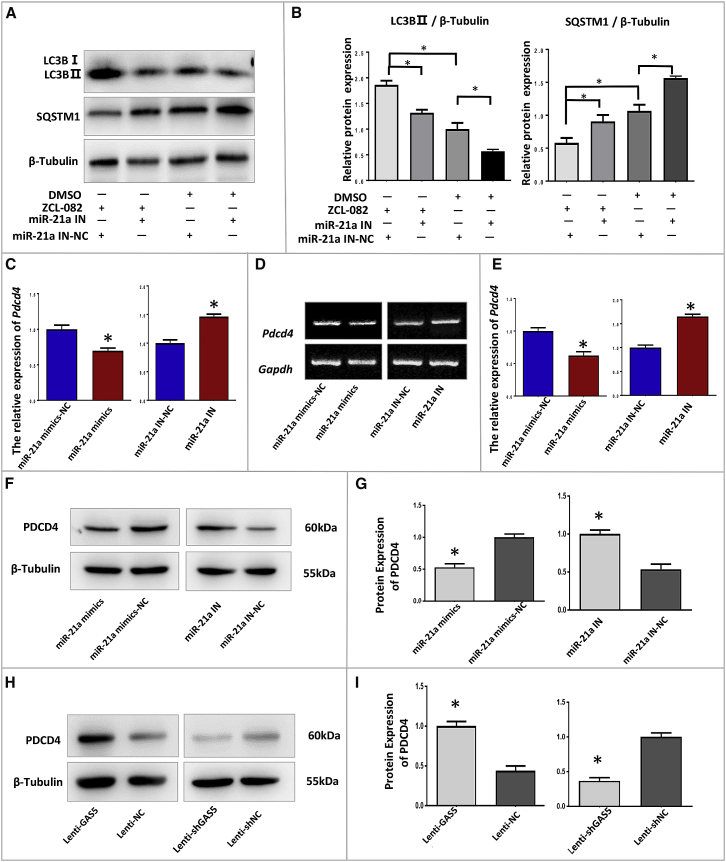
The abovementioned results revealed that ZCL-082 downregulated the expression of GAS5. Additionally, GAS5 could directly bind to miR-21a, which could promote FGSC autophagy. These conclusions suggest that ZCL-082-induced FGSC autophagy requires miR-21a. To test whether ZCL-082 induces the autophagy of FGSCs through miR-21a, FGSCs were transfected with a specific inhibitor of miR-21a. Western blotting showed that miR-21a inhibition significantly reduced basal and ZCL-082-induced LC3B-II protein expression while significantly enhancing the basal or ZCL-082-downregulated protein expression of SQSTM1 in FGSCs (Figures 6A and 6B). These data demonstrated that ZCL-082 induced the autophagy of FGSCs through miR-21a.
Figure 6.
ZCL-082 Induced Autophagy of FGSCs via miR-21a
(A and B) The expressions of LC3B-II and SQSTM1 in FGSCs were detected by western blotting. (A) After transfection for 24 h with miR-21a-specific inhibitor or NC, FGSCs were then treated with ZCL-082 or DMSO. The results of differential blots (B) were statistically significant. miR-21a IN, inhibitor of miR-21a; miR-21a IN-NC, negative control of inhibitor of miR-21a; miR-21a mimic-NC, negative control of miR-21a mimic. (C) PDCD4 expression was detected by qRT-PCR, and the results were statistically significant. (D and E) PDCD4 expression was detected by RT-PCR (D) and the results were statistically significant (E). (F and G) Protein expression of PDCD4 was detected after transfection for 24 h with miR-21a mimics and miR-21a IN in FGSCs (F). The results of differential blots (G) were statistically significant. (H and I) Protein expression of PDCD4 was detected by western blotting (H) after infection with Lenti-GAS5 and Lenti-shGAS5. The results were statistically significant (I). Lenti-shGAS5, lentivirus carrying GAS5-knockdown plasmid; Lenti-shNC, lentivirus carrying control plasmid; Lenti-GAS5, lentivirus carrying GAS5-overexpressing plasmid; Lenti-NC, lentivirus carrying control plasmid. Data are mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01.
Previous studies showed that PDCD4 is a downstream target of miR-21a in other cell types and is closely related to autophagy.46, 52, 53 To confirm that PDCD4 is a downstream target of miR-21a in FGSCs, the cells were transfected with miR-21a mimic and specific inhibitor of miR-21a. The western blot showed that the transfection of miR-21a showed that the transfection of miR-21a mimic significantly reduced the expression of PDCD4 compared with control miR-21a (mimic-NC). Conversely, the inhibition of miR-21a significantly enhanced the expression of PDCD4 compared with the control (miR-21a inhibitor-negative control [IN-NC]) (Figures 6C, 6D, 6F, and 6G). Furthermore, we detected the protein expression of PDCD4 in Lenti-GAS5-infected FGSCs and Lenti-shGAS5-infected FGSCs. The results of western blot showed that the expression of PDCD4 was significantly decreased in Lenti-shGAS5-infected FGSCs while it was significantly increased in Lenti-GAS5-infected FGSCs compared with the control (Lenti-NC and Lenti-shNC, respectively) (Figures 6H and 6I). These data showed that PDCD4 was a downstream target of miR-21a in FGSCs and GAS5 was able to regulate PDCD4.
Discussion
Since a previous study reported that AN2690 targets LeuRS to inhibit fungal infections,27 benzoxaboroles and their derivatives have garnered attention in recent years. With their low toxicity and physicochemical properties, benzoxaboroles are considered a promising anti-infective therapy.26, 29 Our present study investigated for the first time the roles of ZCL-082 in regulating the proliferation, apoptosis, and autophagy of FGSCs in vitro. Moreover, this study revealed that ZCL-082 induced the autophagy of FGSCs through GAS5 acting as a ceRNA sponge for miR-21a. These results may provide new insights for drug discovery and clinical treatment.
Premature ovarian failure (POF) is defined as menopause among women under 40 years of age who present with amenorrhea, hypergonadotropic hypogonadism, infertility, and other complicated disorders in the clinic.54 Genetic predisposition, enzymatic disease, autoimmune disease, and environmental factors are reported to lead to POF. Moreover, non-physiological inhibition of autophagy leads to POF.55 The clinical application of FGSCs is considered a novel strategy to preserve fertility in women.2, 3 In addition, as one of the hallmarks of aging, autophagy affects the number and function of stem cells.56 Studies have shown that mesenchymal stem cells and hematopoietic stem cells lose their regenerative capacity when they reach an advanced age.57, 58 In the aged stem cell population, autophagy is defective.56, 57, 58 Some studies have shown that stem cells required autophagy to clear away cellular waste.56 Many experiments have also confirmed the important role of autophagy in both cell survival and cell death.57, 58 The function of stem cells requires correct autophagic activity, and pharmacological recovery of autophagy can act as a novel potential strategy to maintain stem cell activity for regenerative medicine and aging.56
A few studies have shown that autophagy is a cell survival mechanism to maintain the development of female germ cells prior to establishing primordial follicle pools in the ovary.59, 60 As stem cells in female germ cells, FGSCs have the capacity for self-renewal and differentiation to oocytes. It is significant for the treatment of POF to study the molecular mechanism of FGSC autophagy. In this study, we used compound ZCL-082 to induce FGSC autophagy, and we found that the GAS5/miR-21a axis was involved in the molecular mechanism of FGSC autophagy. Our results show that GAS5 and miR-21a are potential targets for the treatment of POF, by activating the appropriate autophagy level of FGSCs to protect cell survival or inhibiting excessive autophagy of FGSCs to prevent cell autophagic death.
GAS5 recruits transcription factors during embryonic development for the regulation of cell growth, differentiation, and development.61, 62 Additionally, GAS5 was reported to be a tumor suppressor in several cancer cells.63, 64 Previous work in our laboratory showed that GAS5 promoted FGSC proliferation and inhibited FGSC apoptosis in vitro.21
Recently, several studies showed that lncRNAs may function as ceRNA sponges of miRNAs.65, 66 As a well-known onco-microRNA, miR-21a was sponged up by GAS5 to induce the death of breast tumor cells in vitro and in vivo.42, 67 In this study, we revealed that GAS5 acts as a ceRNA by sponging miR-21a to inhibit autophagy in FGSCs.
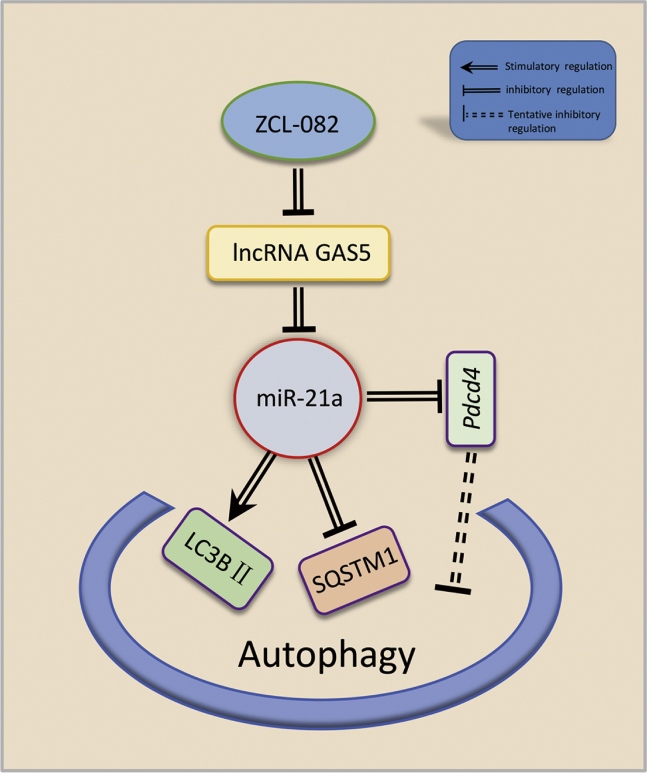
In conclusion, autophagy in female germ cells is rarely studied, but our study revealed for the first time that the AN2690 analog ZCL-082 induces lower viability and autophagy of FGSCs through GAS5. In addition, we revealed a novel autophagic regulatory mechanism for FGSCs treated with ZCL-082 in vitro (Figure 7). We identified that ZCL-082 induced autophagy through the function of GAS5 as a ceRNA sponge for mir-21a in FGSCs. Our study also suggested that the GAS5/miR-21a axis may be a potential therapeutic target for POF in the clinic. In further studies, we will focus on the effect of ZCL-082 on ovarian function in vivo and also the GAS5/miR-21a axis as a drug target for clinical application.
Figure 7.
Proposed Model of GAS5/miR-21a Axis in the Regulation of ZCL-082-Induced Autophagy
ZCL-082 induces autophagy in FGSCs by regulating GAS5, which acts as a ceRNA sponge for miR-21a, thus modulating the expression of PDCD4.
Materials and Methods
Animals
The 5-day-old mice were purchased from SLRC Laboratory (Shanghai, China). All animal procedures were approved by the Institutional Animal Care and Use Committee of Shanghai. The procedures were also performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals.
Structure of Chemical Compound
ZCL-082 is a derivative of benzoxaboroles and its structure is shown in Figure 1.
FGSC Culture
We established the FGSC line in a previous study.4 In brief, the cells were cultured in minimum essential medium alpha (MEM-alpha; Invitrogen Life Sciences, MA, USA), supplemented with 10% fetal bovine serum (FBS), 10 ng/mL mouse leukemia inhibitory factor (mLIF; Santa Cruz Biotechnology, CA, USA), 10 ng/mL mouse epidermal growth factor (mEGF; PeproTech, NJ, USA), 40 ng/mL mouse glial cell line-derived neurotrophic factor (mGDNF; R&D Systems, Minneapolis, MN, USA), 10 ng/mL mouse basic fibroblast growth factor (mbFGF; BD Biosciences, Franklin Lakes, NJ, USA), 10 mg/mL penicillin (Amresco, Lardner, PA, USA), 30 mg/mL pyruvate (Amresco, Lardner, PA, USA), β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), 1 mM non-essential amino acids (Invitrogen Life Sciences, MA, USA), and 2 mM L-glutamine (Amresco, Lardner, PA, USA).
Cells were cultured in SIM-6-thiogunaniaoualiain cells (STOs) as a feeder line, and cells were passaged every 5 days. Cells were infected with lentivirus carrying GAS5 knockdown plasmid, GAS5 overexpression plasmid, or control plasmid, as previously described.21 Cells were also transfected with inhibitor or mimic of miR-21a (Ribobio, Guangzhou, China) using Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions.
CCK8 Assay
Cells were seeded at a density of 5,000 cells/well in a 96-well plate and cultured overnight. After treatment with ZCL-082 for 24 and 48 h, 100 μL CCK8, diluted 1:100 in cell culture medium, was added per well and incubated for 1 h at 37°C before the optical density (OD) value was detected at 450 nm, using a microplate spectrophotometer (Bio-Tek Instruments, Thermo Fisher Scientific, Winooski, VT, USA).
EdU Assay
When the cells had reached 80% confluency, they were treated with ZCL-082 for 48 h and incubated with 50 mM EdU for 2 h. Then the cells were fixed in 4% paraformaldehyde and stained with Apollo Dye Solution and Hoechst 33342. Images were obtained with a Leica microscope (Leica, Wetzlar, Germany), and the number of EdU-positive cells was counted.
RT-PCR and Real-Time qRT-PCR
Trizol reagent (QIAGEN, Hilden, Germany) was used to extract total RNA. Approximately 1,000 ng RNA was reverse transcribed to cDNA using a reverse transcription kit (Takara, Tokyo, Japan). Then we used an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) to detect the expression levels of mRNA, lncRNA, and miRNA. The PCR reaction conditions were as follows: 94°C for 10 min; followed by 94°C for 5 s, 58°C for 30 s, and 72°C for 10 s for 35 cycles; and then 72°C for 10 min. All experiments were repeated three times. Expression was analyzed by applying the 2−ΔΔCt method. Primers are listed in Table S1.
Western Blotting
Cells were lysed in radio immunoprecipitation assay (RIPA) buffer (Yeasen, Shanghai, China), and protein was collected by centrifugation (4°C, 12,000 × g, 10 min). Protein concentrations were determined by bicinchoninic acid (BCA) protein assay kit (Yeasen, Shanghai, China). Proteins were separated by 15% SDS-PAGE and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked with 5% non-fat milk in Tris-buffered saline Tween 20 (TBST) for 2 h with gentle shaking at room temperature, then incubated overnight with the following primary antibodies: anti-LC3B (1:1,000; Abcam, Cambridge, MA, USA), anti-SQSTM1 (1:1,000; Abcam, Cambridge, MA, USA), anti-PDCD4 (1:1,000; Abcam, Cambridge, MA, USA), and anti-β-tubulin (1:6,000; Abcam, Cambridge, MA, USA). The membranes were washed three times with TBST for 10 min and incubated for 1 h with secondary antibodies (1:2,000, Proteintech, Rosemont, IL, USA) at room temperature. Protein bands were scanned using a Tanon 4600SF (Tanon, Shanghai, China). The density of protein bands was quantified with Image-Pro Plus 6.0 software, and the ratio of target protein to β-tubulin, which reflects the changes in expression levels, was calculated.
Cell Immunofluorescence Assay
The cells were fixed in 4% paraformaldehyde and washed twice with 3% BSA. Then cells were treated with 0.5% Triton X-100 for 15 min at room temperature and washed twice with 3% BSA. Using 10% goat serum in BSA, cells were blocked for 1 h at room temperature. Cells were incubated with an anti-LC3B antibody in BSA (1:150; Abcam, Cambridge, MA, USA) overnight at 4°C. After incubation, cells were washed twice with PBS and incubated with fluorescent secondary antibodies in PBS at room temperature for 1 h (1:250; Proteintech, Wuhan, China). Then the cells were washed twice with PBS and stained with DAPI at room temperature for 5 min. Finally, the cells were washed twice with PBS, and images were obtained with a Leica XP8 fluorescence confocal microscope (Leica, Wetzlar, Germany).
Apoptosis Analysis
FGSCs were treated with ZCL-082 for 48 h. Then annexin V-propidium iodide (PI) (Thermo Fisher Scientific, MA, USA) was used to detect apoptotic cells. In brief, cells were collected and washed with 1× binding buffer and incubated with annexin V and PI for 20 min at room temperature in the dark. Finally, apoptotic cells were detected by flow cytometry (FACSCalibur, BD Biosciences, NJ, USA).
Statistical Analyses
All experiments were replicated at least three times. The data are presented as mean ± SEM. Student’s t test was performed with SPSS software; p values <0.05 were considered statistically significant.
Author Contributions
J.W. initiated and designed the entire project and J.W. and H.Z. supervised the entire project. B.L. and X.H. conducted the experiments, analysis, and interpretation of the data. B.L. drafted the article. Y.Y., M.Z., J.Z., Y.W., and X.P. did data analysis. All authors revised the article critically for important intellectual content and approved the final version to be published.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFA0505200 and 2017YFA0504201), the National Natural Science Foundation of China (81720108017 and 81573264), and the National Major Scientific Instruments and Equipment Development Project, National Nature Science Foundation of China (61827814).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.012.
Contributor Information
Huchen Zhou, Email: hczhou@sjtu.edu.cn.
Ji Wu, Email: jiwu@sjtu.edu.cn.
Supplemental Information
References
- 1.Zou K., Yuan Z., Yang Z., Luo H., Sun K., Zhou L., Xiang J., Shi L., Yu Q., Zhang Y. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat. Cell Biol. 2009;11:631–636. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 2.White Y.A., Woods D.C., Takai Y., Ishihara O., Seki H., Tilly J.L. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat. Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding X., Liu G., Xu B., Wu C., Hui N., Ni X., Wang J., Du M., Teng X., Wu J. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci. Rep. 2016;6:28218. doi: 10.1038/srep28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C., Wu J. Production of offspring from a germline stem cell line derived from prepubertal ovaries of germline reporter mice. Mol. Hum. Reprod. 2016;22:457–464. doi: 10.1093/molehr/gaw030. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L., Wang L., Kang J.X., Xie W., Li X., Wu C., Xu B., Wu J. Production of fat-1 transgenic rats using a post-natal female germline stem cell line. Mol. Hum. Reprod. 2014;20:271–281. doi: 10.1093/molehr/gat081. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Yang Z., Yang Y., Wang S., Shi L., Xie W., Sun K., Zou K., Wang L., Xiong J. Production of transgenic mice by random recombination of targeted genes in female germline stem cells. J. Mol. Cell Biol. 2011;3:132–141. doi: 10.1093/jmcb/mjq043. [DOI] [PubMed] [Google Scholar]
- 7.Xiong J., Lu Z., Wu M., Zhang J., Cheng J., Luo A., Shen W., Fang L., Zhou S., Wang S. Intraovarian Transplantation of Female Germline Stem Cells Rescue Ovarian Function in Chemotherapy-Injured Ovaries. PLoS ONE. 2015;10:e0139824. doi: 10.1371/journal.pone.0139824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E.S., Tilly J.L. Use of DEAD-box polypeptide-4 (Ddx4) gene promoter-driven fluorescent reporter mice to identify mitotically active germ cells in post-natal mouse ovaries. Mol. Hum. Reprod. 2015;21:58–65. doi: 10.1093/molehr/gau071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Z., Wu M., Zhang J., Xiong J., Cheng J., Shen W., Luo A., Fang L., Wang S. Improvement in Isolation and Identification of Mouse Oogonial Stem Cells. Stem Cells Int. 2016;2016:2749461. doi: 10.1155/2016/2749461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khosravi-Farsani S., Amidi F., Habibi Roudkenar M., Sobhani A. Isolation and enrichment of mouse female germ line stem cells. Cell J. 2015;16:406–415. doi: 10.22074/cellj.2015.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou K., Hou L., Sun K., Xie W., Wu J. Improved efficiency of female germline stem cell purification using fragilis-based magnetic bead sorting. Stem Cells Dev. 2011;20:2197–2204. doi: 10.1089/scd.2011.0091. [DOI] [PubMed] [Google Scholar]
- 12.Zheng W., Zhou J., Song S., Kong W., Xia W., Chen L., Zeng T. Dipeptidyl-Peptidase 4 Inhibitor Sitagliptin Ameliorates Hepatic Insulin Resistance by Modulating Inflammation and Autophagy in ob/ob Mice. Int. J. Endocrinol. 2018;2018:8309723. doi: 10.1155/2018/8309723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo K., Li C.H., Wang X.Y., He D.J., Zheng P. Germ stem cells are active in postnatal mouse ovary under physiological conditions. Mol. Hum. Reprod. 2016;22:316–328. doi: 10.1093/molehr/gaw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Z., Sun M., Liang X., Li J., Zhou F., Zhong Z., Zheng Y. The Controversy, Challenges, and Potential Benefits of Putative Female Germline Stem Cells Research in Mammals. Stem Cells Int. 2016;2016:1728278. doi: 10.1155/2016/1728278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silvestris E., D’Oronzo S., Cafforio P., D’Amato G., Loverro G. Perspective in infertility: the ovarian stem cells. J. Ovarian Res. 2015;8:55. doi: 10.1186/s13048-015-0184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou K., Wang J., Bi H., Zhang Y., Tian X., Tian N., Ma W., Wu J. Comparison of different in vitro differentiation conditions for murine female germline stem cells. Cell Prolif. 2019;52:e12530. doi: 10.1111/cpr.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park E.S., Woods D.C., Tilly J.L. Bone morphogenetic protein 4 promotes mammalian oogonial stem cell differentiation via Smad1/5/8 signaling. Fertil. Steril. 2013;100:1468–1475. doi: 10.1016/j.fertnstert.2013.07.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imudia A.N., Wang N., Tanaka Y., White Y.A., Woods D.C., Tilly J.L. Comparative gene expression profiling of adult mouse ovary-derived oogonial stem cells supports a distinct cellular identity. Fertil. Steril. 2013;100:1451–1458. doi: 10.1016/j.fertnstert.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacchiarotti J., Maki C., Ramos T., Marh J., Howerton K., Wong J., Pham J., Anorve S., Chow Y.C., Izadyar F. Differentiation potential of germ line stem cells derived from the postnatal mouse ovary. Differentiation. 2010;79:159–170. doi: 10.1016/j.diff.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X.L., Wu J., Wang J., Shen T., Li H., Lu J., Gu Y., Kang Y., Wong C.H., Ngan C.Y. Integrative epigenomic analysis reveals unique epigenetic signatures involved in unipotency of mouse female germline stem cells. Genome Biol. 2016;17:162. doi: 10.1186/s13059-016-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Gong X., Tian G.G., Hou C., Zhu X., Pei X., Wang Y., Wu J. Long noncoding RNA growth arrest-specific 5 promotes proliferation and survival of female germline stem cells in vitro. Gene. 2018;653:14–21. doi: 10.1016/j.gene.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y., Wu J., Yang W., Xia C., Shi X., Li H., Sun J., Shao Z., Wu J., Zhao X. STAT3 is required for proliferation and exhibits a cell type-specific binding preference in mouse female germline stem cells. Mol. Omics. 2018;14:95–102. doi: 10.1039/c7mo00084g. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Yang Y., Xia Q., Song H., Wei R., Wang J., Zou K. Cadherin 22 participates in the self-renewal of mouse female germ line stem cells via interaction with JAK2 and β-catenin. Cell. Mol. Life Sci. 2018;75:1241–1253. doi: 10.1007/s00018-017-2689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Wang M., Chen Y., Wang W., Wu J., Yu C., Zheng Y., Pan Z. Role of the Hedgehog Signaling Pathway in Regulating the Behavior of Germline Stem Cells. Stem Cells Int. 2017;2017:5714608. doi: 10.1155/2017/5714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X., Tian G.G., Yu B., Yang Y., Wu J. Effects of bisphenol A on ovarian follicular development and female germline stem cells. Arch. Toxicol. 2018;92:1581–1591. doi: 10.1007/s00204-018-2167-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Zhu M., Lin Y., Zhou H. The synthesis of benzoxaboroles and their applications in medicinal chemistry. Sci. China Chem. 2013;56:1372–1381. [Google Scholar]
- 27.Rock F.L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y.K., Hernandez V., Akama T., Baker S.J. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 28.Li X., Zhang S., Zhang Y.K., Liu Y., Ding C.Z., Zhou Y., Plattner J.J., Baker S.J., Bu W., Liu L. Synthesis and SAR of acyclic HCV NS3 protease inhibitors with novel P4-benzoxaborole moieties. Bioorg. Med. Chem. Lett. 2011;21:2048–2054. doi: 10.1016/j.bmcl.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Xia Y., Cao K., Zhou Y., Alley M.R., Rock F., Mohan M., Meewan M., Baker S.J., Lux S., Ding C.Z. Synthesis and SAR of novel benzoxaboroles as a new class of β-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2011;21:2533–2536. doi: 10.1016/j.bmcl.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 30.Akama T., Baker S.J., Zhang Y.K., Hernandez V., Zhou H., Sanders V., Freund Y., Kimura R., Maples K.R., Plattner J.J. Discovery and structure-activity study of a novel benzoxaborole anti-inflammatory agent (AN2728) for the potential topical treatment of psoriasis and atopic dermatitis. Bioorg. Med. Chem. Lett. 2009;19:2129–2132. doi: 10.1016/j.bmcl.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Qiao Z., Wang Q., Zhang F., Wang Z., Bowling T., Nare B., Jacobs R.T., Zhang J., Ding D., Liu Y., Zhou H. Chalcone-benzoxaborole hybrid molecules as potent antitrypanosomal agents. J. Med. Chem. 2012;55:3553–3557. doi: 10.1021/jm2012408. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Yang F., Qiao Z., Zhu M., Zhou H. Chalcone-benzoxaborole hybrids as novel anticancer agents. Bioorg. Med. Chem. Lett. 2016;26:5797–5801. doi: 10.1016/j.bmcl.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Costa F.F. Non-coding RNAs: Meet thy masters. BioEssays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 34.Au P.C., Zhu Q.H., Dennis E.S., Wang M.B. Long non-coding RNA-mediated mechanisms independent of the RNAi pathway in animals and plants. RNA Biol. 2011;8:404–414. doi: 10.4161/rna.8.3.14382. [DOI] [PubMed] [Google Scholar]
- 35.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Pal D., Rao M.R.S. Long Noncoding RNAs in Pluripotency of Stem Cells and Cell Fate Specification. Adv. Exp. Med. Biol. 2017;1008:223–252. doi: 10.1007/978-981-10-5203-3_8. [DOI] [PubMed] [Google Scholar]
- 38.Li W.D., Zhou D.M., Sun L.L., Xiao L., Liu Z., Zhou M., Wang W.B., Li X.Q. LncRNA WTAPP1 Promotes Migration and Angiogenesis of Endothelial Progenitor Cells via MMP1 Through MicroRNA 3120 and Akt/PI3K/Autophagy Pathways. Stem Cells. 2018;36:1863–1874. doi: 10.1002/stem.2904. [DOI] [PubMed] [Google Scholar]
- 39.Lu W., Han L., Su L., Zhao J., Zhang Y., Zhang S., Zhao B., Miao J. A 3'UTR-associated RNA, FLJ11812 maintains stemness of human embryonic stem cells by targeting miR-4459. Stem Cells Dev. 2015;24:1133–1140. doi: 10.1089/scd.2014.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao S., Wang P., Hua Y., Xi H., Meng Z., Liu T., Chen Z., Liu L. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget. 2016;7:1608–1618. doi: 10.18632/oncotarget.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Zhu Z., Watabe K., Zhang X., Bai C., Xu M., Wu F., Mo Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 44.Bera A., Das F., Ghosh-Choudhury N., Kasinath B.S., Abboud H.E., Choudhury G.G. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKβ-mTORC1 axis to regulate renal cancer cell invasion. Exp. Cell Res. 2014;328:99–117. doi: 10.1016/j.yexcr.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Zhou L., Li Y., Lin S., Tomuleasa C. MicroRNA-21 stimulates gastric cancer growth and invasion by inhibiting the tumor suppressor effects of programmed cell death protein 4 and phosphatase and tensin homolog. J. BUON. 2014;19:228–236. [PubMed] [Google Scholar]
- 46.Liu X., Luo F., Ling M., Lu L., Shi L., Lu X., Xu H., Chen C., Yang Q., Xue J. MicroRNA-21 activation of ERK signaling via PTEN is involved in arsenite-induced autophagy in human hepatic L-02 cells. Toxicol. Lett. 2016;252:1–10. doi: 10.1016/j.toxlet.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Nikoletopoulou V., Markaki M., Palikaras K., Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta. 2013;1833:3448–3459. doi: 10.1016/j.bbamcr.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Booth L.A., Tavallai S., Hamed H.A., Cruickshanks N., Dent P. The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell. Signal. 2014;26:549–555. doi: 10.1016/j.cellsig.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu M.L., Zhang Q., Yuan X., Jin L., Wang L.L., Fang T.T., Wang W.B. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World J. Gastroenterol. 2018;24:1004–1012. doi: 10.3748/wjg.v24.i9.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol. Res. 2018;129:151–155. doi: 10.1016/j.phrs.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Gu J., Wang Y., Wang X., Zhou D., Wang X., Zhou M., He Z. Effect of the LncRNA GAS5-MiR-23a-ATG3 Axis in Regulating Autophagy in Patients with Breast Cancer. Cell. Physiol. Biochem. 2018;48:194–207. doi: 10.1159/000491718. [DOI] [PubMed] [Google Scholar]
- 52.Gu H., Liu Z., Li Y., Xie Y., Yao J., Zhu Y., Xu J., Dai Q., Zhong C., Zhu H. Serum-Derived Extracellular Vesicles Protect Against Acute Myocardial Infarction by Regulating miR-21/PDCD4 Signaling Pathway. Front. Physiol. 2018;9:348. doi: 10.3389/fphys.2018.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S., Gao G., Wu F., Liu D., Zhao H., Ke J., Liu Y., Li F., Li J., Chen Z. Programmed cell death protein 4 deficiency suppresses foam cell formation by activating autophagy in advanced glycation end-product low-density lipoprotein-induced macrophages. J. Cell. Biochem. 2019;120:7689–7700. doi: 10.1002/jcb.28043. [DOI] [PubMed] [Google Scholar]
- 54.Jankowska K. Premature ovarian failure. Prz. Menopauzalny. 2017;16:51–56. doi: 10.5114/pm.2017.68592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witkin S.S., Kanninen T.T., Sisti G. The Role of Hsp70 in the Regulation of Autophagy in Gametogenesis, Pregnancy, and Parturition. Adv. Anat. Embryol. Cell Biol. 2017;222:117–127. doi: 10.1007/978-3-319-51409-3_6. [DOI] [PubMed] [Google Scholar]
- 56.Revuelta M., Matheu A. Autophagy in stem cell aging. Aging Cell. 2017;16:912–915. doi: 10.1111/acel.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.García-Prat L., Muñoz-Cánoves P., Martinez-Vicente M. Dysfunctional autophagy is a driver of muscle stem cell functional decline with aging. Autophagy. 2016;12:612–613. doi: 10.1080/15548627.2016.1143211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho T.T., Warr M.R., Adelman E.R., Lansinger O.M., Flach J., Verovskaya E.V., Figueroa M.E., Passegué E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gawriluk T.R., Hale A.N., Flaws J.A., Dillon C.P., Green D.R., Rucker E.B., 3rd Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction. 2011;141:759–765. doi: 10.1530/REP-10-0489. [DOI] [PubMed] [Google Scholar]
- 60.Rodrigues P., Limback D., McGinnis L.K., Plancha C.E., Albertini D.F. Multiple mechanisms of germ cell loss in the perinatal mouse ovary. Reproduction. 2009;137:709–720. doi: 10.1530/REP-08-0203. [DOI] [PubMed] [Google Scholar]
- 61.Fleming J.V., Fontanier N., Harries D.N., Rees W.D. The growth arrest genes gas5, gas6, and CHOP-10 (gadd153) are expressed in the mouse preimplantation embryo. Mol. Reprod. Dev. 1997;48:310–316. doi: 10.1002/(SICI)1098-2795(199711)48:3<310::AID-MRD2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 62.Coccia E.M., Cicala C., Charlesworth A., Ciccarelli C., Rossi G.B., Philipson L., Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell. Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 64.Shi X., Sun M., Liu H., Yao Y., Kong R., Chen F., Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol. Carcinog. 2015;54(Suppl 1):E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 65.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J., Ahn C., Chun C.H., Jin E.J. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J. Orthop. Res. 2014;32:1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 67.Li W., Zhai L., Wang H., Liu C., Zhang J., Chen W., Wei Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–27786. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.