Abstract
In plant cells, the trans-Golgi network (TGN) is known to act as the early endocytic compartment, whereas RAB5-localizing multivesicular endosomes (MVEs) act as the later compartment. Land plants and certain green algal species possess plant-unique RAB5 homologs (ARA6/RABF1 in Arabidopsis thaliana) in addition to the orthologs of animal RAB5 (RHA1/RABF2a and ARA7/RABF2b in A. thaliana), and these two RAB5 members reside in substantially overlapping but different subpopulations of MVEs. Several studies indicate that the TGN and MVEs are closely related; however, the distribution of the two RAB5 groups in relation to the TGN remains elusive. Here, we quantitatively showed that ARA6 and ARA7 are closely associated with the TGN, and the subpopulation of ARA6 and ARA7 overlaps with the TGN in the root epidermal cells of A. thaliana.
Keywords: Arabidopsis thaliana, multivesicular endosomes, RAB5 members, SYP43, trans-Golgi network
Endocytosis is the process in which extracellular substances and cell surface proteins are taken up into cells by the invagination of the plasma membrane. Endocytosis plays pivotal roles in a wide variety of cellular functions, for example, the amplification or downregulation of signaling cascades by sequestrating receptor molecules, the maintenance of the intracellular environment by controlling the amount of transporters and channels on the plasma membrane, the uptake of extracellular nutrients or substances, and the establishment and maintenance of cell polarity (Hupalowska and Miaczynska 2012; Platta and Stenmark 2011). After endocytosis, the cargos are carried to compartments called early endosomes (EEs), and subsequently they are either returned to the plasma membrane by recycling pathways or directed to late endosomes before reaching the lysosome/vacuole for degradation (Huotari and Helenius 2011; Jovic et al. 2010). Thus, EEs function as the sorting center for the endocytic cargos.
In mammalian systems, a small GTPase, RAB5, localizes to the EEs. RAB5 acts as a molecular switch through cycling between the GTP-bound active state and the GDP-bound inactive state, and it mediates the tethering and fusion of EEs by recruiting various effector molecules (Grosshans et al. 2006; Stenmark 2009; Zerial and McBride 2001). RAB5 is conserved in a broad range of eukaryotic organisms, including plants. In addition to the orthologs of animal RAB5, land plants possess plant-unique RAB5 homologs. Arabidopsis thaliana, a well-studied model plant, possesses three RAB5 members: RHA1/RABF2a and ARA7/RABF2b, which have high similarity to mammalian RAB5, and a plant-unique RAB5, ARA6/RABF1 (Ueda et al. 2001). Several studies have demonstrated that plant RAB5 also participates in endosomal trafficking pathways. Confocal laser scanning fluorescence microscopy (CLSFM) revealed that endocytic indicators and/or cargos, such as FM4-64 (an endocytic tracer reagent), BOR1 (a boron transporter), FLS2 (a receptor-like kinase responsible for immune responses), and BRI1 (the brassinosteroid receptor) pass through RAB5-positive compartments during their endocytosis (Beck et al. 2012; Irani et al. 2012; Takano et al. 2005; Ueda et al. 2004). The overexpression of a dominant negative mutant of ARA7 was also shown to inhibit the endocytosis of endocytic cargos (Beck et al. 2012; Irani et al. 2012). Such dominant negative constructs also interfere in the trafficking of vacuolar cargos (Bolte et al. 2004; Kotzer et al. 2004; Sohn et al. 2003). Together with the genetic data (Ebine et al. 2011), these results indicate that conventional RAB5 plays critical roles in both the endocytic and vacuolar trafficking pathways.
These findings suggest the partially conserved function of conventional RAB5 among eukaryotic organisms; however, recent studies indicate that the organization of the post-Golgi organelles in plants exhibits distinctive features. Immunoelectron microscopy has revealed that all three RAB5 members are localized to spherical multivesiculated compartments (Haas et al. 2007), which is a prominent feature of the late endosome in animal cells (Hanson and Cashikar 2012; Huotari and Helenius 2011), in contrast to the early endosomal localization of RAB5 in animal cells. Furthermore, the trans-Golgi network (TGN), which functions in the sorting of cargo proteins in biosynthetic pathways in animal cells, is shown to act as the early endocytic compartment in plant cells; FM4-64 stains the TGN before reaching the RAB5-positive endosomes (Chow et al. 2008; Dettmer et al. 2006; Lam et al. 2007; Viotti et al. 2010). Thus, the TGN is often designated as TGN/EE.
Another prominent characteristic of plant endosomal trafficking is the existence of ARA6. ARA6 exhibits a high overall similarity to conventional RAB5, but it lacks the C-terminal hyper-variable region and cysteine residues to be isoprenylated. Instead, ARA6 is N-myristoylated and palmitoylated at its N-terminus (Pereira-Leal and Seabra 2001; Ueda et al. 2001). Despite the partly overlapping localization with conventional RAB5 on multivesicular endosomes (MVEs; also referred to as prevacuolar compartments (PVCs)) (Ebine et al. 2011; Haas et al. 2007; Ueda et al. 2004), ARA6 is shown to act in a distinct endosomal trafficking pathway from conventional RAB5; ARA6 regulates the trafficking pathway from MVEs to the plasma membrane in A. thaliana (Ebine et al. 2011), while it is also proposed that ARA6 regulates vacuolar trafficking (Beck et al. 2012; Bolte et al. 2004), suggesting that ARA6 regulates multiple trafficking steps around the plant endosome.
Links between RAB5 and the TGN are also reported. A population of ARA7 was localized to the TGN by electron microscopy (Stierhof and El Kasmi 2010). It is also reported that some population of the TGN matures directly into the ARA7-positive MVEs (Scheuring et al. 2011). We also have reported the existence of a hybrid compartment between the TGN and MVEs in cells actively internalizing FLS2 upon flg22 treatment (Choi et al. 2013). ARA6 is also proposed to act in a recycling pathway from MVEs to the TGN (Bottanelli et al. 2012). However, detailed quantitative comparison of the subcellular distribution between the two RAB5 groups and the TGN has not yet been conducted. In this study, we analyzed the distribution of ARA6 and ARA7 and compared it with the TGN marker SYNTAXIN OF PLANT 43 (SYP43) using a macro constructed in the Metamorph software, which semi-automatically measures the distances from the center of a fluorescent signal to the center of the nearest fluorescent signal with the other color in an acquired image (Ito et al. 2012). Our result indicated that the RAB5-positive MVEs and the TGN are highly related compartments.
Materials and methods
Plant materials
To visualize the RAB5-positive compartment, TGN, and/or peroxisome in the same cells, we established transgenic plants expressing either GFP- or mRFP-tagged RAB5 (ARA6 and ARA7) under the regulation of their own regulatory elements (promoter, introns, terminator) (Ebine et al. 2011) and crossed them with transgenic plants expressing fluorescently tagged SYP43 (the TGN-marker) (Uemura et al. 2004; 2012) and/or GFP fused with peroxisomal targeting signal 1 (PTS1, (Mano et al. 2002; 2004)). Plants were plated on 1×Murashige Skoog (MS; Murashige and Skoog 1962) medium solidified with 0.3% gellan gum (Wako) and grown at 23°C under constant light.
Confocal microscopy
Root meristematic cells of 5-day-old seedlings were observed under the LSM710 confocal microscope (Carl Zeiss) with an oil immersion lens (×63, NA=1.40). Localization analysis was performed using a macro in the Metamorph software, as described previously (Boutté et al. 2006; Ito et al. 2012). In brief, the distances between two signals were measured and grouped into three categories: (i) colocalized: a distance between two centers that was below the resolution limit of the objective lens (0.24 µm in this study); (ii) associated: a distance less than the sum of the two radii of two signals (<1 µm in this study); and (iii) independent: a distance larger than the sum of the two radii of two punctate signals (>1 µm). We analyzed at least three images from at least four independent roots. The numbers of foci analyzed in this study are shown in Table 1.
Table 1. Number of fluorescent foci analyzed in this study.
| ARA6-GFPARA6-mRFP | ARA6-GFPmRFP-ARA7 | GFP-SYP43ARA6-mRFP | GFP-SYP43mRFP-ARA7 | GFP-PTS1ARA6-mRFP | GFP-PTS1mRFP-ARA7 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GFP | mRFP | GFP | mRFP | GFP | mRFP | GFP | mRFP | GFP | mRFP | GFP | mRFP | |
| Root 1 | 1107 | 1095 | 560 | 540 | 893 | 1205 | 730 | 721 | 669 | 1152 | 444 | 590 |
| Root 2 | 883 | 889 | 548 | 530 | 1033 | 1323 | 747 | 720 | 347 | 725 | 655 | 621 |
| Root 3 | 1809 | 1871 | 1326 | 1331 | 589 | 581 | 635 | 600 | 795 | 1044 | 1032 | 1088 |
| Root 4 | 1046 | 1096 | 1297 | 1358 | 471 | 447 | 1075 | 921 | 774 | 1306 | 444 | 295 |
| Root 5 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 666 | 576 | N.D. | N.D. | N.D. | N.D. |
N.D.: not determined
Results
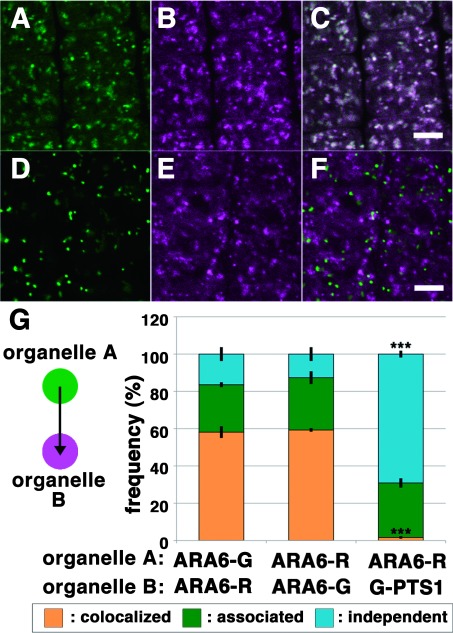
To validate the applicability of the quantification method we developed, we at first compared the localization of ARA6 tagged with distinct colors of fluorescent proteins. We acquired confocal images of transgenic plants expressing ARA6-GFP and ARA6-mRFP (Figure 1A to C) and automatically measured the distances from ARA6-GFP foci to the nearest ARA6-mRFP foci and the distances from ARA6-mRFP foci to the nearest ARA6-GFP foci. By our method, approximately 60% and 25–30% of ARA6-positive foci were classified as “colocalized” and “associated”, respectively. The fluorescent tags may make a small contribution to the subcellular localization of ARA6, as the frequency of the colocalization did not reach 100% (Figure 1G left and middle bars). We then compared the localization of ARA6-mRFP and GFP-PTS1, which labels the peroxisome (Mano et al. 2002; 2004). The transgenic plants expressing GFP-PTS1 and ARA6-mRFP were examined, finding little colocalization of these fluorescent markers (Figure 1D to F); 1.67±0.67% (mean±standard deviation) and 69.1±1.85% for “colocalized” and “independent”, respectively (Figure 1G).
Figure 1. Quantitative analysis of the localization patterns of ARA6-GFP/ARA6-mRFP and ARA6-RFP/GFP-PTS1 in root epidermal cells near meristematic regions of A. thaliana. (A–C) Comparison between subcellular localization of ARA6-GFP (A, green) and ARA6-mRFP (B, magenta). (C) Merged image of (A) and (B). Scale bar=5 µm. (D–F) Subcellular distribution of peroxisomes (GFP-PTS1) (D, green) and ARA6-mRFP (E), magenta. (F) Merged image of (D) and (E). Scale bar=5 µm. (G) Stack bar graph representing the relationships of ARA6-GFP/ARA6-mRFP and ARA6-mRFP/GFP-PTS1. The distances from the center of each ARA6-GFP spot to the center of the closest ARA6-mRFP spot (left bar) and the distances from the center of each ARA6-mRFP spot to the center of the closest ARA6-GFP spot (middle bar) were classified as described previously (Ito et al. 2012). The graph shows that there were no differences between the two measurements (p>0.5 by Student’s t-test). On the other hand, the frequency of colocalization was significantly lower for ARA6-mRFP/GFP-PTS1 (right bar) (p***<0.001 by Tukey’s test). Error bars indicate±standard deviation.
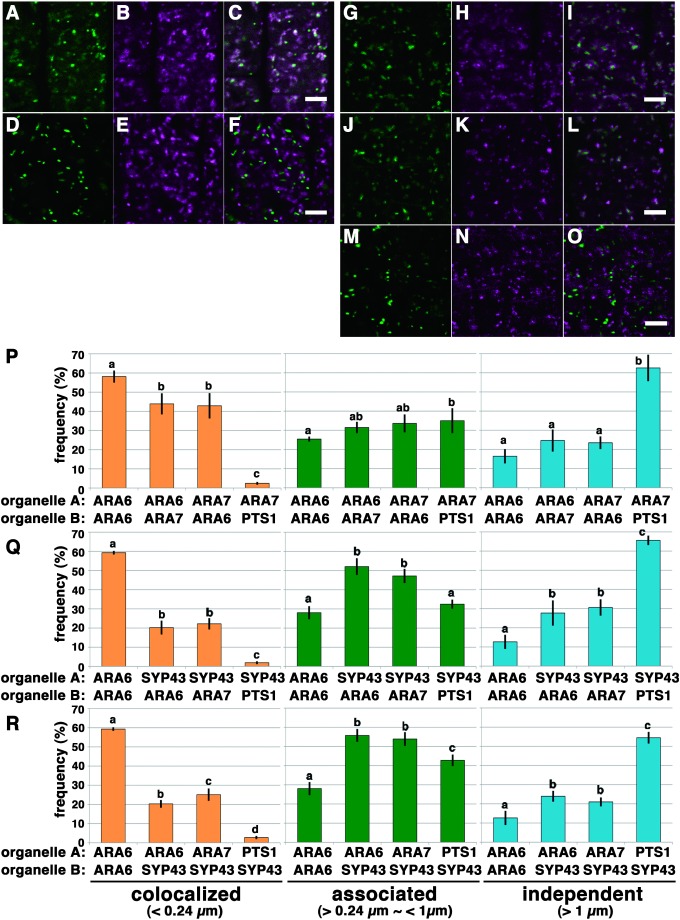
Next, we quantified the colocalization between ARA6 and ARA7 (Figure 2A–F and P) in transgenic plants expressing GFP-ARA6 and mRFP-ARA7. We measured the distances from GFP-positive foci to the nearest mRFP-positive foci, 43.9±5.52% of which was classified as colocalization. This ratio was significantly lower than the colocalization measured between ARA6-GFP and ARA6-mRFP (58.1±3.15%, p<0.05, Tukey’s test) (Figure 2P). The analysis of the distance from mRFP-ARA7 foci to GFP-ARA6 foci also resulted in a similar pattern (42.9±6.63%) (Figure 2P). These data were consistent with the distinct localization with considerable overlap between ARA6 and ARA7 that we reported previously (Ebine et al. 2011; Ueda et al. 2004).
Figure 2. Quantitative analysis of the localization pattern of two distinctive RAB5 endosomes and SYP43-localizing TGN in root epidermal cells near meristematic regions of A. thaliana. (A–C) Distribution of ARA6-GFP (A, green) and mRFP-ARA7 (B, magenta). (C) Merged image of (A) and (B). Scale bar=5 µm. (D–F) Distribution of peroxisomes (GFP-PTS1) (D, green) and mRFP-ARA7 (E, magenta). (F) Merged image of (D) and (E). Scale bar=5 µm. (G–I) Distribution of the TGN compartments labeled by GFP-SYP43 (G, green) and MVEs labeled by ARA6-mRFP (H, magenta). (I) Merged image of (G) and (H). Scale bars=5 µm. (J–L) Distribution of the TGN compartments labeled by GFP-SYP43 (J, green) and MVEs labeled by mRFP-ARA7 (K, magenta). (L) Merged image of (J) and (K). Scale bars=5 µm. (M–O) Distribution of the peroxisomes labeled by GFP-PTS1 (M, green) and the TGN compartments labeled by mRFP-SYP43 (N, magenta). (O) Merged image of (M) and (N). Scale bars=5 µm. (P) Bar graphs representing the localization relationships of ARA6/ARA7 and ARA7/PTS1. The frequency of colocalization for ARA6/ARA7 was significantly lower than for ARA6/ARA6, indicating that ARA6 and ARA7 only partially overlap with each other (p<0.05 by Tukey’s test). Localization frequency of ARA6/ARA6 shown in Figure 1G is represented here as the positive control. Error bars indicate±standard deviation. (Q) Bar graph representing the localization of the TGN in relation to ARA6, ARA7 and peroxisomes. The distances from the center of each SYP43 spot to the center of the closest marker spot (ARA6, ARA7 or PTS1) were classified. The graph indicates that the majority of the population of SYP43 foci was classified as “associated” with ARA6 or ARA7 foci (middle graph). A subpopulation of SYP43 was classified as “colocalized” with ARA6 or ARA7 (left graph) (p<0.05 by Tukey’s test). Localization frequency of ARA6/ARA6 shown in Figure 1G is represented here as the positive control. Error bars indicate±standard deviation. (R) Bar graph representing the localization of ARA6, ARA7 and peroxisomes in relation to the TGN. The distances from the center of each marker (either ARA6, ARA7 or PTS1) to the center of the closest SYP43 spot were classified (p<0.05 by Tukey’s test). Similarly to the result in Figure 2Q, the majority of the populations of ARA6 and ARA7 foci were classified as “associated” with SYP43 foci (middle graph). Subpopulations of ARA6 and ARA7 were classified as “colocalized” with SYP43 (left graph). The frequency of the colocalization of ARA7/SYP43 pairs was significantly higher than for ARA6/SYP43 pairs (p<0.05 by Tukey’s test). Localization frequency of ARA6/ARA6 shown in Figure 1G is represented here as the positive control. Error bars indicate±standard deviation.
We then analyzed the distribution of ARA6, ARA7, and SYP43 by this qualification method (Figure 2G–O, Q and R). First, we analyzed the distances from GFP-SYP43-foci to ARA6-mRFP- or mRFP-ARA7-foci (Figure 2Q). More than half of the foci were classified as “associated”, which suggested that many of the TGN compartments are located in close proximity to ARA6- and/or ARA7-positive compartments. We also found that a subpopulation of the foci was classified as “colocalized”. This result may imply the existence of a hybrid compartment between the TGN and endosome bearing both SYP43 and RAB5 in root meristematic cells. We then analyzed the distances from ARA6 or ARA7-positive foci to the nearest GFP-SYP43-positive foci (Figure 2R). This result also gave a similar localization pattern; however, a significantly higher population of ARA7-foci was classified as “colocalized” with SYP43 compared with ARA6-positive foci (Tukey’s test p<0.05).
Discussion
In this study, we qualitatively depicted the localization relationship among two RAB5 groups and the TGN. ARA6 and ARA7 are considered to label functionally differentiated MVEs with overlap (Ebine et al. 2011; Ueda et al. 2004). Because FM4-64 reaches ARA6-localizing MVEs (hereafter referred to as “ARA6-MVEs”) at a different time point than ARA7-localizing MVEs (“ARA7-MVEs”) in the protoplast of A. thaliana suspension cultured cells (Ueda et al. 2004), ARA6 is considered to label “later” late endosomes in A. thaliana (Beck et al. 2012; Geldner and Jügens 2006). Pharmaceutical experiments also support the different natures of ARA6- and ARA7-MVEs. Both ARA6- and ARA7-MVEs are sensitive to brefeldin A (BFA), the inhibitor of ARF guanine nucleotide exchange factors, and form aggregates called “BFA bodies” upon BFA treatment (Ebine et al. 2011; Grebe et al. 2003; Jaillais et al. 2008). However, the effect of BFA on ARA7-MVEs is more prominent than on ARA6-MVEs (Beck et al. 2012; Ebine et al. 2011). It is also reported that the number of ARA6-MVEs, but not ARA7-MVEs, is severely affected by treatment with concanamycin A, a specific inhibitor of V-ATPase, in A. thaliana leaf cells (Beck et al. 2012). Here, we quantitatively confirmed that ARA6 and ARA7 localize to partially overlapping but different endosomal populations in cells of the meristematic zone of A. thaliana.
We also demonstrated that ARA6- and ARA7-positive compartments are well associated with SYP43-fluorescent foci, and subpopulations of the ARA6 and ARA7 signals overlap with SYP43. In tobacco leaf cells, RHA1 exclusively labels PVCs/MVEs and the late PVC, which is the intermediate compartment between the PVCs and vacuoles (Foresti et al. 2010). Meanwhile, ARA6 exhibits different localization patterns depending on its expression levels, as follows: ARA6 colocalized with RHA1 when expressed by a weak promoter, whereas TGN-localization of ARA6 was observed when it was expressed by a strong promoter (Bottanelli et al. 2012). However, our results did not provide clear evidence supporting the closer relationship between the TGN and ARA6 compared with ARA7, at least in our experimental system. We previously reported that a major population of intracellular clathrin localizes to the TGN, and clathrin is well associated with ARA6 compared to the conventional RAB5 (Ito et al. 2012). Given that ARA6 and ARA7 are equally associated with the TGN, it is likely that clathrin, which is independent of the TGN, is associated with ARA6. In animal and yeast systems, clathrin is shown to function as the platform for the formation of intraluminal vesicles (ILVs) in the multivesicularization of endosomes (Gruenberg and Stenmark 2004; Sachse et al. 2002). Clathrin plaque is also found on the MVEs in plant cells (Scheuring et al. 2011; Stierhof and El Kasmi 2010), although the function of clathrin in ILV formation remains elusive. Our results may indicate that, ARA7 is associated with the domain of the TGN where clathrin is less concentrated, whereas ARA6 is on the specific domain of the TGN enriched with clathrin.
Another important finding is that the subpopulations of ARA6, ARA7, and SYP43 overlap with each other. Electron microscopic analysis revealed that MVEs are frequently located in close proximity to the TGN in root tip cells, and this structure is referred to as the TGN/MVB complex (Kang et al. 2011). The overlapping distribution among the TGN and MVE markers could represent these distinctive but juxtaposed compartments that cannot be resolved by light microscopy. It is also possible that compartments labeled by all three markers are the intermediate compartments during maturation from the TGN to MVE (Choi et al. 2013; Scheuring et al. 2011). In the case of FLS2 endocytosis, such a hybrid compartment is observed only when the endocytosis of FLS2 is induced by the flg22 peptide (Choi et al. 2013). This finding implies that the MVEs, TGN, and their intermediate structure are dynamically reorganized when endocytosis is activated by environmental and/or developmental stimuli. Quantification methods such as the one employed in this study will facilitate the understanding of organelle dynamics during various physiological events, which will further unravel the dynamic interactions and functions of post-Golgi organelles in plant cells.
Acknowledgments
We thank Dr. T. Nakagawa for providing pGWB vectors and Dr. S. Mano for sharing GFP-PTS1 plants. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to E. I., A.N., and T.U.), JST, PRESTO, and a Grant-in-Aid for JSPS fellows (E.I., 2010649).
References
- Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S (2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Brown S, Satiat-Jeunemaitre B (2004) The N-myristoylated Rab-GTPase m-Rabmc is involved in post-Golgi trafficking events to the lytic vacuole in plant cells. J Cell Sci 117: 943–954 [DOI] [PubMed] [Google Scholar]
- Bottanelli F, Gershlick DC, Denecke J (2012) Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic 13: 338–354 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B (2006) The plasma membrane recycling pathway and cell polarity in plants: Studies on PIN proteins. J Cell Sci 119: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Choi SW, Tamaki T, Ebine K, Uemura T, Ueda T, Nakano A (2013) RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 25: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CM, Neto H, Foucart C, Moore I (2008) Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Foresti O, Gershlick DC, Bottanelli F, Hummel E, Hawes C, Denecke J (2010) A recycling-defective vacuolar sorting receptor reveals an intermediate compartment situated between prevacuoles and vacuoles in tobacco. Plant Cell 22: 3992–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Jügens G (2006) Endocytosis in signalling and development. Curr Opin Plant Biol 9: 589–594 [DOI] [PubMed] [Google Scholar]
- Grebe M, Xu J, Mobius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P (2006) Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA 103: 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H (2004) The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5: 317–323 [DOI] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martinez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS (2007) The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A (2012) Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28: 337–362 [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30: 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalowska A, Miaczynska M (2012) The new faces of endocytosis in signaling. Traffic 13: 9–18 [DOI] [PubMed] [Google Scholar]
- Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizon J, Hniliková J, Sísa M, Buyst D, Vilarrasa-Blasi J, Szatmári AM, et al. (2012) Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol 8: 583–589 [DOI] [PubMed] [Google Scholar]
- Ito E, Fujimoto M, Ebine K, Uemura T, Ueda T, Nakano A (2012) Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Gaude T (2008) Evidence for a sorting endosome in Arabidopsis root cells. Plant J 53: 237–247 [DOI] [PubMed] [Google Scholar]
- Jovic M, Sharma M, Rahajeng J, Caplan S (2010) The early endosome: A busy sorting station for proteins at the crossroads. Histol Histopathol 25: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L (2007) Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Hayashi M, Kato A, Kondo M, Nishimura M (2002) Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol 43: 331–341 [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M (2004) An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tabacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889–901 [DOI] [PubMed] [Google Scholar]
- Platta HW, Stenmark H (2011) Endocytosis and signaling. Curr Opin Cell Biol 23: 393–403 [DOI] [PubMed] [Google Scholar]
- Sachse M, Urbé S, Oorschot V, Strous GJ, Klumperman J (2002) Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell 13: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, et al. (2011) Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Stierhof YD, El Kasmi F (2010) Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur J Cell Biol 89: 285–297 [DOI] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, von Wiren N, Fujiwara T (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Kim H, Saito C, Ebine K, Ueda T, Schulze-Lefert P, Nakano A (2012) Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]