Abstract
Background
The increase in multidrug-resistant tuberculosis (MDR-TB) severely hampers tuberculosis prevention and control in China, a country with the second highest MDR-TB burden globally. The first nationwide drug-resistant tuberculosis surveillance program provides an opportunity to comprehensively investigate the epidemiological/drug-resistance characteristics, potential drug-resistance mutations, and effective population changes of Chinese MDR-TB.
Methods
We sequenced 357 MDR strains from 4600 representative tuberculosis-positive sputum samples collected during the survey (70 counties in 31 provinces). Drug-susceptibility testing was performed using 18 anti-tuberculosis drugs, representing the most comprehensive drug-resistance profile to date. We used 3 statistical and 1 machine-learning methods to identify drug-resistance genes/single-nucleotide polymorphisms (SNPs). We used Bayesian skyline analysis to investigate changes in effective population size.
Results
Epidemiological/drug-resistance characteristics showed different MDR profiles, co-resistance patterns, preferred drug combination/use, and recommended regimens among 7 Chinese administrative regions. These factors not only reflected the serious multidrug co-resistance and drug misuse but they were also potentially significant in facilitating the development of appropriate regimens for MDR-TB treatment in China. Further investigation identified 86 drug-resistance genes/intergenic regions/SNPs (58 new), providing potential targets for MDR-TB diagnosis and treatment. In addition, the effective population of Chinese MDR-TB displayed a strong expansion during 1993–2000, reflecting socioeconomic transition within the country. The phenomenon of expansion was restrained after 2000, likely attributable to the advances in diagnosis/treatment technologies and government support.
Conclusions
Our findings provide an important reference and improved understanding of MDR-TB in China, which are potentially significant in achieving the goal of precision medicine with respect to MDR-TB prevention and treatment.
Keywords: multidrug-resistant tuberculosis, whole-genome sequencing, drug resistance, epidemiological study, Bayesian skyline
We describe the epidemiological/drug-resistance characteristics, potential drug-resistance genes/intergenic regions/single-nucleotide polymorphisms, and effective population changes of multidrug-resistant (MDR) tuberculosis in China through comprehensive analysis of the epidemiological, drug-resistance, and whole-genome sequencing data of 357 MDR tuberculosis strains from the first Chinese national survey of drug-resistant tuberculosis.
According to the World Health Organization (WHO), tuberculosis has exceeded human immunodeficiency virus infection as the most lethal infectious disease globally for 3 consecutive years [1]. Moreover, the increase in drug-resistant tuberculosis seriously hampers its prevention and control. In 2016, there were approximately 600 000 new rifampicin-resistant (RR) tuberculosis cases, including approximately 490 000 multidrug-resistant tuberculosis (MDR-TB) cases [1]. Compared with treatment of drug-sensitive tuberculosis, treatment of MDR-TB requires more time (at least 2 years). In 2016, the median cost per MDR-TB person was estimated to be $9525, but the treatment success rate was only 54% [1]. China ranks second after India among all countries regarding MDR-TB burden, with approximately 73 000 new MDR/RR tuberculosis cases reported in 2016. MDR/RR tuberculosis occurred in 7.1% of new cases and 24% of previously treated cases, which is higher than those worldwide (4.1% and 19%, respectively) [1]. Furthermore, the cure rate for MDR-TB in China was only 41% (13% lower than the global experience) [1]. By analyzing epidemiological, drug-resistance, and genomic data, previous studies on MDR-TB have mainly focused on the epidemiological/drug-resistance characteristics [2–4], potential drug-resistance mutations [5–7], and evolution of MDR-TB [8, 9].
Although previous studies have revealed some features of MDR-TB in China, the first nationwide drug-resistant tuberculosis surveillance in China (covering 70 counties in 31 provinces) [10] provided a great opportunity for comprehensive investigation of the epidemiological/drug-resistance characteristics, drug-resistance genes/single-nucleotide polymorphisms (SNPs), and evolution of MDR-TB in China. Here, we performed whole-genome sequencing (WGS) for 357 MDR-TB strains from the survey. Drug-susceptibility testing (DST) was also conducted using 18 anti-tuberculosis drugs, representing the most comprehensive drug-resistance profile to date. Through the comprehensive analyses of epidemiological/drug-resistance phenotypic and genotypic data, we describe the epidemiological/drug-resistance characteristics, potential drug-resistance genes/intergenic regions (IGRs)/SNPs, and the effective population changes of MDR-TB in China.
METHODS
Sample Information and Sequencing
The first national survey of drug-resistant tuberculosis in China (2007–2008) included local/regional surveys (covering 70 counties in 31 provinces) by the Chinese Center for Disease Control and Prevention [10]. Cluster-randomized sampling was adopted to obtain representative samples of patients with tuberculosis in China [10]. The number of study participants was estimated at 3542 treatment naive patients and 1189 previously treated patients (Supplementary Table 1). Finally, 4600 representative sputum samples from smear-positive tuberculosis (3514 treatment naive and 1086 previously treated patients) cases were obtained. Written informed consent was also obtained from each patient in face-to-face interviews by trained clinicians.
After excluding 671 samples (culture failed to grow, 531; non-tuberculous mycobacteria, 140), we successfully obtained 3929 Mycobacterium tuberculosis isolates, which were preserved in the Beijing Bio-Bank of Clinical Resources on Tuberculosis (Beijing Chest Hospital, China). Through DST, we obtained 401 MDR isolates. Eliminating 28 samples that failed to regrow, 373 MDR strains were obtained for sequencing (Figure 1). Sixty-seven non-MDR control strains were also selected from the first national survey of drug-resistant tuberculosis in China according to distribution of MDR strains (Supplementary Table 2).
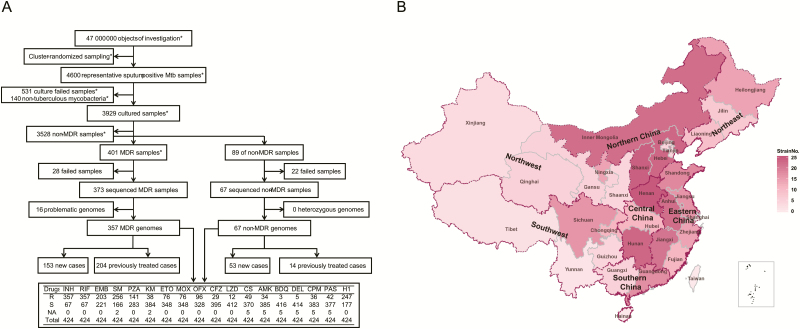
Figure 1.
Distribution of the 357 MDR isolates from the first national survey of drug-resistant tuberculosis in China. A, Screening process of the 357 MDR isolates. The drug-susceptible tests were performed using 18 anti-tuberculosis drugs. R indicates the number of resistance strains, S indicates the number of susceptible strains, and NA indicates the number of the strains with failed drug-susceptibility testing. Some screening procedures were completed as reported in the literature (denoted as *). B, Nationwide distribution of MDR isolates from the first national survey of drug-resistant tuberculosis in China. The shade of color indicates the number of MDR strains. Abbreviations: AMK, amikacin; BDQ, bedaquiline; CFZ, clofazimine; CPM, capreomycin; CS, cycloserine; DEL, delamanid; EMB, ethambutol; ETO, ethionamide; H1, high-dose isoniazid; INH, isoniazid; KM, kanamycin; LZD, linezolid; MDR, multidrug-resistant; MOX, moxifloxacin; Mtb, Mycobacterium tuberculosis; OFX, ofloxacin; PAS, para-aminosalicylic acid; PZA, pyrazinamide; RIF, rifampicin; SM, streptomycin.
The above strains were cultivated on Lowenstein-Jensen solid media. Genomic DNA samples were prepared and purified for sequencing using the MasterPure Complete DNA isolation kit (Epicentre, Madison, WI) following the standard protocols. DNA libraries were constructed using kits provided by Illumina according to the manufacturer’s instructions. Sequencing was performed on an Illumina HiSeq X-Ten sequencing platform. After excluding 16 problematic genomes, 357 MDR-TB genomes were used for subsequent bioinformatics analysis. For additional information, see the Supplementary Methods.
Drug Susceptibility Testing
The DST for 18 anti-tuberculosis drugs was performed with solid medium or an MGIT 960 system (BD Biosciences, Sparks, MD) using methods recommended by WHO or in other studies [11–15]. Bedaquiline, delamanid, and clofazimine were purchased from the Hanxiang Company (Shanghai, China); the other drugs were purchased from Sigma-Aldrich. DST methods are shown in Supplementary Table 3.
SNP Identification and Phylogenetic Analyses
Sequencing reads for the strains were aligned to the H37Rv reference genome (NC_000962) using SOAP2 [16] and the Burrows-Wheeler algorithm [17] (average coverage, >98%; average depth, 325×). SNPs were derived from high-quality unique mapping reads using SOAPsnp [16], SAMtools [18], and GATK [19]. Further filtration was performed based on the standards in the literature [20]. The 34 959 SNPs (excluding the SNPs within the known drug-resistance genes [21] and repeat regions [22]) in all strains were used to construct a phylogenetic tree using FastTree v2.1.9 [23]. Mycobacterium canettii CIPT 140010059 was used as an outlier (Supplementary Figure 1). Genotyping was implemented based on 62 SNP markers [24].
Identification of Potential Drug-resistance SNPs and Genes/IGRs
We used the following 4 identification methods: normal distribution analysis [6], Fisher exact test [11], PhyC test [7], and random forest model [25]. To ensure reliability, only the genes/IGRs/SNPs identified by at least 3 methods and with the proportion of drug-sensitive isolates less than 0.3 were considered as potentially drug-resistant candidates. The detailed protocols are provided in the Supplementary Methods.
Bayesian Skyline Analyses of MDR-TB
To evaluate the changes in effective population size since 1974 (when rifampicin was brought to the market in China), we performed Bayesian skyline analysis for the 357 MDR-TB strains based on the concatenated SNPs in the identified resistance genes/IGRs using BEAST v1.8.4 [26] (for details, see the Supplementary Methods).
Data Availability
Whole-genome sequencing raw data were deposited at Sequence Read Archive (SRP134826) and Genome Sequence Archive (CRA000786).
RESULTS
Epidemiological Characteristics of MDR-TB in China
The 357 MDR strains were screened from the 3929 M. tuberculosis isolates in the first national survey of drug-resistant tuberculosis (70 counties in 31 provinces). These data are still the most representative Chinese MDR strains to date (Figure 1 and Supplementary Table 4). Sixty-seven non-MDR control strains were also selected from the survey according to distribution of MDR strains (Supplementary Table 5).
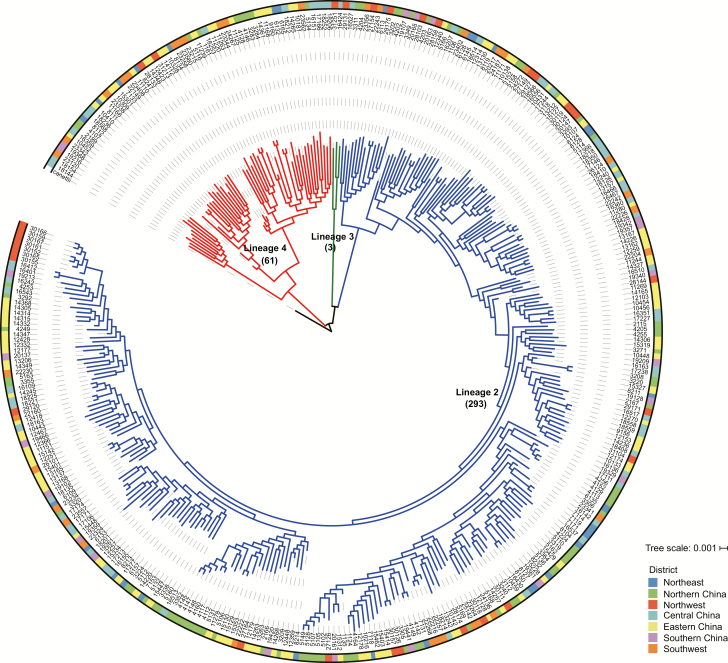
The corresponding patient information is shown in Table 1 and Supplementary Tables 4 and 5. As previously reported [10], the most prevalent MDR strains in China were lineage 2 Beijing strains (293, 82.1%; Figure 2); previously treated cases (204, 57.1%) with MDR exceeded new MDR cases (153, 42.9%); and the sex ratio (male-to-female) was approximately 2:1 (237/120). In addition, Chinese farmers (220, 61.6%) accounted for a higher percentage of MDR cases than others (137, 38.4%). The percentage of MDR isolates in Eastern China (104, 29.1%) and Northern China (77, 21.6%) ranked first and second among the 7 Chinese administrative regions.
Table 1.
Epidemiological and Drug-resistance Information for the 357 Multidrug-resistant Tuberculosis Isolates From the First National Survey of Drug-resistant Tuberculosis in China
| Characteristic | Patients With New Cases of Tuberculosis (n = 153) | Patients With Previously Treated Tuberculosis (n = 204) | Total (N = 357) |
|---|---|---|---|
| Drug resistance: first-line tuberculosis drugs | |||
| Isoniazid-Rifampicin | 153 (42.9%) | 204 (57.1%) | 357 (100.0%) |
| High-dose isoniazid | 99 (40.1%) | 148 (59.9%) | 247 (69.2%) |
| Streptomycin | 117 (45.7%) | 139 (54.3%) | 256 (71.7%) |
| Ethambutol | 83 (40.9%) | 120 (59.1%) | 203 (56.9%) |
| Pyrazinamide | 58 (42.7%) | 78 (57.4%) | 136 (38.1%) |
| Drug resistance: second-line tuberculosis drugs | |||
| Ofloxacin | 41 (42.7%) | 55 (57.3%) | 96 (26.9%) |
| Moxifloxacin | 18 (37.5%) | 30 (62.5%) | 48 (13.5%) |
| Ethionamide | 30 (39.5%) | 46 (60.5%) | 76 (21.3%) |
| Kanamycin | 22 (57.9%) | 16 (42.1%) | 38 (10.6%) |
| Linezolid | 5 (50.0%) | 5 (50.0%) | 10 (2.8%) |
| Clofazimine | 9 (36.0%) | 16 (64.0%) | 25 (7.0%) |
| Amikacin | 16 (47.1%) | 18 (52.9%) | 34 (9.5%) |
| Capreomycin | 15 (45.5%) | 18 (54.6%) | 33 (9.2%) |
| Cycloserine | 22 (44.9%) | 27 (55.1%) | 49 (13.7%) |
| Para-aminosalicylic acid | 22 (52.4%) | 20 (47.6%) | 42 (11.8%) |
| Bedaquiline | 1 (33.3%) | 2 (66.7%) | 3 (0.8%) |
| Delamanid | 2 (40.0%) | 3 (60.0%) | 5 (1.4%) |
| Sex | |||
| Male | 90 (38.0%) | 147 (62.0%) | 237 (66.4%) |
| Female | 63 (52.5%) | 57 (47.5%) | 120 (33.6%) |
| Age | |||
| Early youth | 8 (88.9%) | 1 (11.1%) | 9 (2.5%) |
| Youth | 65 (42.8%) | 87 (57.2%) | 152 (42.6%) |
| Middle | 60 (39.2%) | 93 (60.8%) | 153 (42.9%) |
| Elderly | 20 (46.5%) | 23 (53.5%) | 43 (12.0%) |
| District | |||
| Northeast | 11 (36.7%) | 19 (63.3%) | 30 (8.40%) |
| Northern China | 45 (58.4%) | 32 (41.6%) | 77 (21.57%) |
| Eastern China | 34 (32.7%) | 70 (67.3%) | 104 (29.13%) |
| Southern China | 10 (45.4%) | 12 (54.6%) | 22 (6.16%) |
| Central China | 15 (25.4%) | 44 (74.6%) | 59 (16.53%) |
| Northwest | 20 (64.5%) | 11 (35.5%) | 31 (8.68%) |
| Southwest | 18 (52.9%) | 16 (47.1%) | 34 (9.52%) |
| Ethnicity | |||
| Han | 131 (42.7%) | 176 (57.3%) | 307 (86.0%) |
| Other | 22 (44.0%) | 28 (56.0%) | 50 (14.0%) |
| Occupation | |||
| Chinese farmer | 94 (42.7%) | 126 (57.3%) | 220 (61.6%) |
| Other | 59 (43.1%) | 78 (56.9%) | 137 (38.4%) |
| Contact | |||
| Yes | 17 (32.1%) | 36 (67.9%) | 53 (14.9%) |
| No | 136 (44.9%) | 167 (55.1%) | 303 (84.9%) |
| Lineage | |||
| 2 (L2) | 130 (44.4%) | 163 (55.6%) | 293 (82.1%) |
| 3 (L3) | 3 (100.0%) | 0 (0.0%) | 3 (0.8%) |
| 4 (L4) | 21 (34.4%) | 40 (65.6%) | 61 (17.1%) |
Figure 2.
The evolutionary tree of 357 multidrug-resistant tuberculosis (MDR-TB) strains. A total of 357 MDR-TB strains included 293 L2 strains (blue), 3 L3 strains (green), and 61 L4 strains (red). Different colors on the outside ring indicate the strains from different administrative regions in China.
Drug-resistance Characteristics of MDR-TB in China
We performed the DSTs for 18 anti-tuberculosis drugs (Supplementary Table 3). The drug-resistance profiles are shown in Supplementary Table 6. We first calculated the resistance ratio of each anti-tuberculosis drug in MDR strains (Table 1, Supplementary Table 7, and Supplementary Figure 2). The results revealed that the resistance ratio of pyrazinamide (38.1%) was the lowest among the first-line drugs. Subsequent analysis of the resistance ratios for the other drugs showed that they were much lower than those for the first-line drugs in the following ranking order (from low to high): bedaquiline (0.8%) < delamanid (1.4%) < linezolid (2.8%) < clofazimine (7.0%) < capreomycin (9.2%) < amikacin (9.5%) < kanamycin (10.6%) < para-aminosalicylic acid (11.8%) < moxifloxacin (13.45%) < cycloserine (13.7%) < ethionamide (21.3%) < ofloxacin (26.9%).
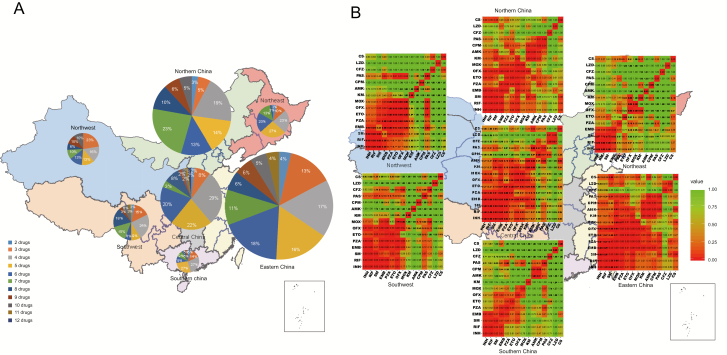
We further investigated the distribution of MDR-TB strains in China by calculating the proportion of MDR-TB strains with different drug-resistance profiles among the 7 administrative regions (Figure 3A). The results showed that more than half of MDR strains were resistant to at least 5 anti-tuberculosis drugs in each administrative region, especially in Northern (22%), Eastern (20%), Northwest (26%), and Southwest (29%) China, with more than 20% of MDR strains resistance to at least 8 drugs. This demonstrated severe drug resistance in China.
Figure 3.
Drug-resistance profiles and multidrug co-resistance patterns of the multidrug-resistant tuberculosis (MDR-TB) strains among 7 administrative regions in China. A, The proportion of MDR-TB strains resistant to different numbers of antibiotics among the 7 administrative regions. Different colors in the circular sectors indicate different numbers of anti-tuberculosis drugs. The size of the circle indicates the number of MDR strains. B, Relationship matrices of the co-resistance/combination of drugs among 7 administrative regions. The P value of co-resistance for each pair of anti-tuberculosis drugs was obtained using Fisher exact test (shown in the cells). The color key indicates the degree of relationship (ie, frequency of co-resistance). Bedaquiline and delamanid were not included in the analysis due to the limited number of resistance strains (≤5). Abbreviations: AMK, amikacin; BDQ, bedaquiline; CFZ, clofazimine; CPM, capreomycin; CS, cycloserine; DEL, delamanid; EMB, ethambutol; ETO, ethionamide; H1, high-dose isoniazid; INH, isoniazid; KM, kanamycin; LZD, linezolid; MDR, multidrug-resistant; MOX, moxifloxacin; Mtb, Mycobacterium tuberculosis; OFX, ofloxacin; PAS, para-aminosalicylic acid; PZA, pyrazinamide; RIF, rifampicin; SM, streptomycin.
We then used the Fisher test to investigate the multidrug co-resistance patterns among the 7 Chinese administrative regions by constructing co-resistance relationship matrices for 15 anti-tuberculosis drugs (Figure 3B). In general, the 5 first-line anti-tuberculosis drugs showed a high frequency of co-resistance; the other anti-tuberculosis drugs displayed low frequency of co-resistance except for the 2 fluoroquinolones. Specifically, the MDR strains of each administrative region possessed their own characteristic co-resistance pattern. The MDR isolates in Northeast China displayed a high frequency of co-resistance to only 4 first-line drugs (isoniazid, rifampicin, streptomycin, and ethambutol) and low frequency of co-resistance to the other drugs. In contrast, the MDR isolates from Eastern China exhibited a high frequency of co-resistance to almost all anti-tuberculosis drugs except for linezolid and clofazimine. In addition, we also found a high frequency of co-resistance to 3 injectable anti-tuberculosis drugs in Northern, Central, and Southern China. The various multidrug co-resistance patterns might reflect different drug combinations/use preferences and other unmeasured variables in different regions.
Identification of Potential Drug-resistance SNPs and Genes/IGRs
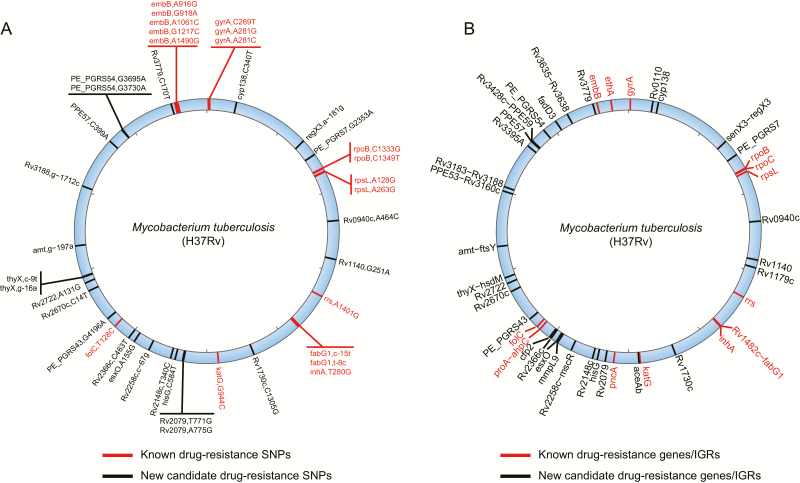
To investigate the relationship between the drug-resistance phenotypes and genotypes of the 357 MDR-TB strains, we sequenced them using the Illumina HiSeq sequencing platform (average coverage, 325×; Supplementary Table 8) and applied 4 methods (3 statistical and 1 machine-learning method) to identify the genetic drug-resistance determinants (Supplementary Figure 3 and Supplementary Table 9). A total of 42 SNPs and 44 genes/IGRs were found to correlate with drug resistance (Supplementary Tables 10–18) using at least 3 methods (see Supplementary Methods). Among them, 15 drug-resistance SNPs and 13 drug-resistance genes/IGRs had been experimentally validated [27], which cover most of the well-known drug-resistance SNPs and genes/IGRs (Figure 4 and Supplementary Tables 19 and 20), indicating the credibility of our screening strategy. Importantly, we identified 31 new drug-resistance genes/IGRs. Their functions were mainly associated with the known drug-resistance mechanisms of M. tuberculosis, such as cell wall synthesis and fatty acid metabolism (8 genes), secretion/transport (4 genes), DNA replication/repair (3 genes), oxidoreductase (1 gene), transcription/translation (1 IGR), and proline-glutamic acid (PE)/proline-proline-glutamic acid (PPE)-family genes (6 genes) (Supplementary Table 21). These genes/IGRs provide some potential molecular targets for the prevention and treatment of tuberculosis.
Figure 4.
Genomic locations of the 42 drug-resistance SNPs and 44 drug-resistance genes/IGRs. A, Genomic locations of the 42 drug-resistance SNPs. B, Genomic locations of the 44 drug-resistance genes/IGRs. The red lines on the blue circle indicate the known drug-resistance SNPs/ genes/IGRs; the black lines indicate the new candidates found in this study. Abbreviations: IGR, intergenic region; SNP, single-nucleotide polymorphism.
We also validated our screening strategy by analyzing the nonsynonymous to synonymous substitutions (Ka/Ks) ratios of 33 drug-resistance genes (excluding 11 drug-resistance IGRs) that were subject to a higher selection pressure. We observed that these genes were under positive selection with Ka/Ks >1 in the MDR strains relative to the ratio of approximately 0.4 for other genes (Supplementary Figure 4). These findings are consistent with those from a previous study [6] and further support the feasibility and accuracy of our screening strategy in identifying drug-resistance genes.
Compensatory Mutation
Previous studies have shown that some particular mutations of rpoA and rpoC compensate for fitness costs in rifampicin-resistant strains with rpoB mutations [28]. Here, we identified 38 potential compensatory mutations in rpoA and rpoC [20, 28], 16 of which have not been reported previously. Thirty-seven compensatory mutations are associated with RpoB S450L, including 15 novel mutations. A “hot-spot” compensatory mutation region in rpoC has also been discovered (Supplementary Figure 5) as previous reported [29]. Incidentally, we found 2 pairs of potential compensatory mutations in RpoA and RpoC that co-occurred with RpoB S450L (RpoA E274G and RpoC E1033A; RpoA P250L and RpoC L527V; Supplementary Table 22).
Changes in Effective Population Size of Chinese MDR-TB
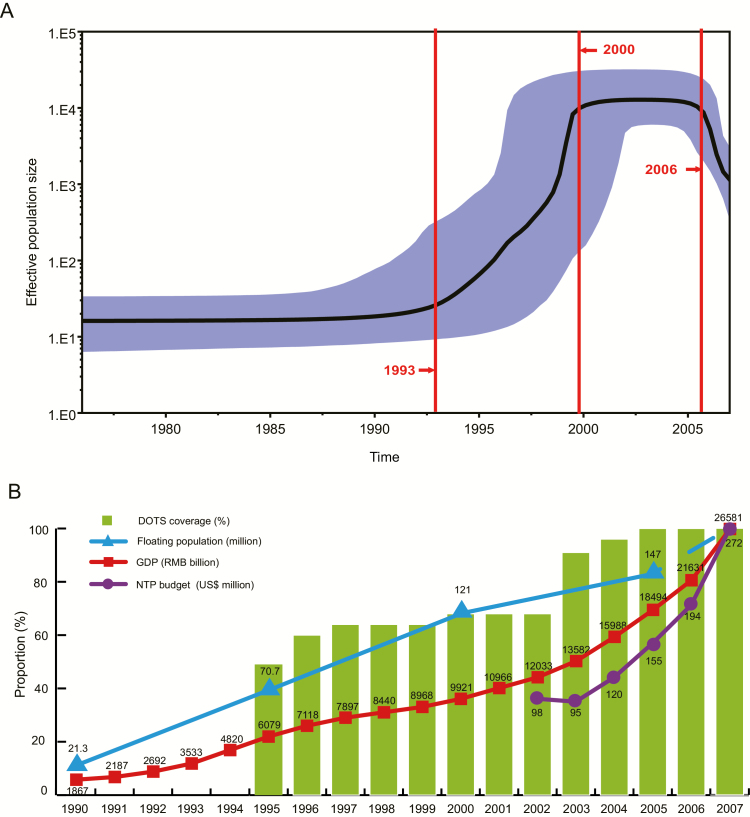
To investigate changes in the effective population size of Chinese MDR-TB strains since 1974 (Figure 5A), we constructed a Bayesian skyline plot based on the SNPs in the 44 drug-resistance genes/IGRs. A strong population expansion was detected during 1993–2000, while this expansion was restrained with unchanged population size from 2000 to 2006. There was even a significant decline after 2006.
Figure 5.
Bayesian skyline plot showing the changes in effective population size of Chinese multidrug-resistant tuberculosis (MDR-TB) strains. A, Bayesian skyline plot based on 357 MDR-TB strains shows the effective population size of Chinese MDR-TB since 1974. The shadowed area indicates 95% probability density intervals for the estimated population sizes. B, Schematic diagram showing the socioeconomic factors that might influence the population size changes of Chinese MDR-TB strains during 1990–2007. The green bar indicates the DOTS coverage (%) in China. The blue triangle indicates the number of floating population (million). The red square indicates the gross domestic product of China (RMB billion). The purple circle indicates the national tuberculosis program budget in China ($US million). Abbreviations: DOTS, Directly Observed Treatment, Short Course; GDP, gross domestic product; NTP, national tuberculosis program; RMB, Renminbi.
DISCUSSION
Our findings elucidate the epidemiological/drug-resistance characteristics, some potential drug-resistance genes/SNPs, and effective population changes of MDR tuberculosis in China. We did this by analyzing the epidemiological, drug-resistance phenotypic, and genotypic data of 357 MDR-TB strains from the first national survey of drug-resistant tuberculosis. Here, we evaluated data with important recommendations for precision medicine and future research, as discussed below.
The resistance ratios of the anti-tuberculosis drugs also provide important reference to regimens for treatment of MDR-TB. As for the 2 fluoroquinolones we tested, moxifloxacin showed a lower resistance ratio (13.45%) than ofloxacin (26.9%). According to the longer MDR-TB regimens included in WHO evidence-based guidelines, moxifloxacin is also one of the prioritized medicines based on the latest evidence that refer to the balance of effectiveness to safety [30]. Importantly, bedaquiline (0.8%), delamanid (1.4%), and linezolid (2.8%) exhibited the lowest drug-resistance ratios. At present, bedaquiline and linezolid are considered prioritized medicines in the WHO guidelines for treatment of MDR-TB [30]. Bedaquiline is strongly recommended because of its effectiveness and low toxicity [31]. Linezolid also shows effectiveness but has limitations due to its high toxicity [30]. Although delamanid exhibited a low drug-resistance ratio, it did not belong to the prioritized group of medicines for treatment of MDR tuberculosis due to its short time to market and uncertain safety and effectiveness beyond 6 months [30]. These 3 drugs are rarely used in China currently and may constitute the last line of defense against MDR-TB; therefore, their prescription should be cautiously administered by clinicians.
Importantly, the 357 nationally representative MDR strains offered an opportunity to investigate the changes in MDR-TB population size in China since 1974, reflecting some realities during socioeconomic transition. The strong population expansion of Chinese MDR-TB strains during 1993–2000 might be predominantly associated with rapid economic growth [32] and incomplete tuberculosis surveillance and control systems during that period of socioeconomic transition. Rapid economic growth in the 1990s was accompanied by increased population movements from 21.3 million (1990) to 121 million (2000) (Figure 5B) [32], contributing to significant MDR-TB transmission and expansion. Additionally, reduced patient compliance with tuberculosis treatment, antibiotic abuse, and sharp increases in the number of drug addicts and human immunodeficiency virus–infected individuals are also likely to have contributed to the transmission and spread of Chinese MDR-TB [33, 34]. Furthermore, the coverage of the DOTS course for tuberculosis was only 68% in 2000 [35] (Figure 5B).
The increase in the size of the Chinese MDR-TB population from 1993–2000 was restrained, while it remained unchanged from 2000 to 2006, and there was even had a significant decline after 2006. The proportion of MDR-TB in all tuberculosis cases also showed a sharp decrease from 2000 (10.7%) to 2010 (6.8%) [36, 37]. This favorable change in combating tuberculosis was mainly ascribed to the advances in new diagnostic techniques and anti-tuberculosis drugs, as well as increased government support for MDR-TB patients [1]. The Chinese government introduced a policy for free diagnostic tests and drug use for tuberculosis patients in 2004, which effectively reduced the economic burden on patients and improved their treatment compliance [38]. The national tuberculosis program budget was also increased from $98 million (2002) to $272 million (2007) [35] (Figure 5B). The coverage of DOTS also reached 100% in 2005 [35]. Importantly, the government has provided an additional budget for MDR-TB since 2006, including subsidies for second-line drugs [35]. The WHO published guidelines for management of drug-resistant tuberculosis in 2006 (version 1), which also provide important guidance and advice for the prevention and control of MDR-TB in China [39].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. F. C., H. H., and S. X. conceived the study. G. W., G. J., G. M., S. W., Z. Z., and W. J. designed and performed the experiments. N. D., T. Y., C. L., X. J., and J. Z. performed the bioinformatics analyses. N. D., T. Y., C. L., Y. Z., and F. C. prepared the manuscript. All authors revised and approved the final manuscript.
Financial support. This work was supported by the National Natural Science Foundation of China (31601047, 31770870, 81672065, 81703632); National Science and Technology Major Project (2017ZX10201301-004-002, 2017ZX09304009-004, 2017ZX10302301-003-004); Beijing Natural Science Foundation (7172050); Beijing Municipal Science & Technology Commission (Z171100001017065); Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20181602); and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201809).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global tuberculosis report 2017 Available at: http://www.who.int/tb/publications/global_report/en/ Accessed 31 December 2017.
- 2. Manson AL , Abeel T , Galagan JE , et al. Mycobacterium tuberculosis whole genome sequences from Southern India suggest novel resistance mechanisms and the need for region-specific diagnostics. Clin Infect Dis 2017; 64:1494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoa NB , Sy DN , Nhung NV , Tiemersma EW , Borgdorff MW , Cobelens FG. National survey of tuberculosis prevalence in Viet Nam. Bull World Health Organ 2010; 88:273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mao TE , Okada K , Yamada N , et al. Cross-sectional studies of tuberculosis prevalence in Cambodia between 2002 and 2011. Bull World Health Organ 2014; 92:573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coll F , Phelan J , Hill-Cawthorne GA , et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat Genet 2018; 50:307–16. [DOI] [PubMed] [Google Scholar]
- 6. Zhang H , Li D , Zhao L , et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 2013; 45:1255–60. [DOI] [PubMed] [Google Scholar]
- 7. Farhat MR , Shapiro BJ , Kieser KJ , et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 2013; 45:1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merker M , Blin C , Mona S , et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 2015; 47:242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comas I , Coscolla M , Luo T , et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 2013; 45:1176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y , Xu S , Wang L , et al. National survey of drug-resistant tuberculosis in China. N Engl J Med 2012; 366:2161–70. [DOI] [PubMed] [Google Scholar]
- 11. Desjardins CA , Cohen KA , Munsamy V , et al. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat Genet 2016; 48:544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. World Health Organization. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: World Health Organization, 2014. [PubMed] [Google Scholar]
- 13. van Ingen J , Simons S , de Zwaan R , et al. Comparative study on genotypic and phenotypic second-line drug resistance testing of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 2010; 48:2749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torrea G , Coeck N , Desmaretz C , et al. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 2015; 70:2300–5. [DOI] [PubMed] [Google Scholar]
- 15. Schena E , Nedialkova L , Borroni E , et al. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC™ MGIT™ 960 system. J Antimicrob Chemother 2016; 71:1532–9. [DOI] [PubMed] [Google Scholar]
- 16. Li R , Yu C , Li Y , et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 2009; 25:1966–7. [DOI] [PubMed] [Google Scholar]
- 17. Li H , Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H , Handsaker B , Wysoker A , et al. ; 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKenna A , Hanna M , Banks E , et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casali N , Nikolayevskyy V , Balabanova Y , et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 2014; 46:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sandgren A , Strong M , Muthukrishnan P , Weiner BK , Church GM , Murray MB. Tuberculosis drug resistance mutation database. PLoS Med 2009; 6:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tarailo-Graovac M , Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009; Chapter 4: Unit 4 10. [DOI] [PubMed] [Google Scholar]
- 23. Price MN , Dehal PS , Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009; 26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coll F , McNerney R , Guerra-Assunção JA , et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 2014; 5:4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liaw A , Wiener M. Classification and regression by randomForest. R News 2002; 2:18–22. [Google Scholar]
- 26. Drummond AJ , Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Müller B , Borrell S , Rose G , Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet 2013; 29:160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Comas I , Borrell S , Roetzer A , et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 2011; 44:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gygli SM , Borrell S , Trauner A , Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev 2017; 41:354–73. [DOI] [PubMed] [Google Scholar]
- 30. World Health Organization. Rapid communication: key changes to treatment of multidrug- and rifampicin-resistant tuberculosis (MDR/RR-TB) Available at: http://www.who.int/tb/publications/2018/rapid_communications_MDR/en/ Accessed 02 October 2018.
- 31. Word Health Organization. Frequently asked questions on bedaquiline Available at: http://www.who.int/tb/challenges/mdr/bedaquilinefaqs/en/ Accessed 02 October 2018.
- 32. National Statistics Bureau of the People’s Republic of China. The China statistical yearbook Available at: http://www.stats.gov.cn/tjsj/ndsj/ Accessed 10 March 2018.
- 33. Bi P , Tong S , Parton KA. Family self-medication and antibiotics abuse for children and juveniles in a Chinese city. Soc Sci Med 2000; 50:1445–50. [DOI] [PubMed] [Google Scholar]
- 34. Shao Y. AIDS epidemic at age 25 and control efforts in China. Retrovirology 2006; 3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. World Health Organization. Global tuberculosis report 2008. Available at: http://www.who.int/tb/publications/global_report/archive/en/ Accessed 31 December 2017.
- 36. National Technic Steering Group of the Epidemiological Sampling Survey for Tuberculosis , Duanmu H.Report on fourth national epidemiological sampling survey of tuberculosis. Zhonghua Jie He He Hu Xi Za Zhi 2002; 25:3–7. [PubMed] [Google Scholar]
- 37. Technical Guidance Group of the Fifth National TB Epidemiological Survey; The Office of the Fifth National TB Epidemiological Survey. The fifth national tuberculosis epidemiological survey in 2010. Chin J Antituberculosis. Chin J Antituberc 2012; 34:485–508. [Google Scholar]
- 38. National Health Commission of the People’s Republic of China. Health service reform and development report Available at: http://www.gov.cn/ztzl/2005–10/20/content_80720.htm Accessed 10 March 2018.
- 39. World Health Organization. Guidelines for the programmatic management of drug-resistant tuberculosis, 2011 update Available at: http://whqlibdoc.who.int/publications/2011/9789241501583_eng.pdf Accessed 10 October 2017. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing raw data were deposited at Sequence Read Archive (SRP134826) and Genome Sequence Archive (CRA000786).