Abstract
Background
Circular RNAs (circRNAs) with miRNA response elements (MREs) could function as competing endogenous RNA (ceRNA) in regulating gene expression, thus playing vital roles in pathogenesis and progression of many diseases. However, the function of circRNAs in endometriosis remains unknown. This study was carried to profile the expression patterns of circRNAs in ovarian endometriosis.
Methods
High throughput RNA‐Seq was performed in six paired ectopic and eutopic endometrium tissues (ecEM vs. euEM), followed by quantitative real‐time polymerase chain reaction (qRT‐PCR) in 30 paired samples. Through bioinformatics prediction, we constructed a circRNA‐miRNA ‐mRNA network and elucidated circRNAs functions by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
Results
A total of 146 upregulated and 148 downregulated circRNAs were identified, binding with 2,495 MREs. The qRT‐PCR validation results of four upregulated circRNAs matched the RNA‐Seq data. The ceRNA network included 48 miRNAs and 296 mRNAs. Functional analysis revealed several important pathways such as MAPK signaling pathway, and PI3K‐AKT signaling pathway, which might be associated with the pathogenesis and development of endometriosis.
Conclusion
Our data suggested that circRNAs are differentially expressed in endometriosis, which might be candidate factors for pathogenesis of this disease and be considered as promising therapeutic targets in the future.
Keywords: circRNAs, endometriosis, miRNAs, mRNAs
1. INTRODUCTION
Endometriosis is characterized by the presence of endometrium‐like tissues (including glandular and stromal tissues) outside the uterine cavity, associated with dysmenorrhea, chronic pelvic pain, and infertility (Borghese, Zondervan, Abrao, Chapron, & Vaiman, 2017; Giudice, 2010; Shafrir et al., 2018; Tomassetti & D'Hooghe, 2018) . As a common estrogen‐dependent gynecologic disorder, it affects up to 10% women of reproductive age and up to 50% of women with pelvic pain and/or infertility (Giudice, 2010; Shafrir et al., 2018; Tomassetti & D'Hooghe, 2018). Although a benign condition, it shares malignant biological behaviors like tissue invasion, local spread, distant metastasis, and relapse, and seriously influences women's physical and mental health (Matias‐Guiu & Stewart, 2018). Unfortunately, up to date there are still no optimal treatments for endometriosis due to the complexity of the pathogenesis and diversity of symptoms. It is widely assumed that ectopic lesions arise through retrograde endometrial fragments during menstruation (Sampson, 1927). However, not all the women suffered endometriosis despite most women at reproductive age exhibit some degree of retrograde menstruation (Halme, Hammond, Hulka, Raj, & Talbert, 1984). It is postulated that affected women may have certain susceptible factors that contribute to the development and progression of this disease (Asghari, Valizadeh, Aghebati‐Maleki, Nouri, & Yousefi, 2018; Borghese et al., 2017; Ponandai‐Srinivasan et al., 2018).
With the development of high‐throughput sequencing, novel bioinformatics approaches and corresponding experimental validation, a broad spectrum of noncoding RNAs (ncRNAs), including miRNA, long ncRNA, and circRNA have been revealed and shown to play an important participation in various biological processes. Under certain conditions, those ncRNAs demonstrate abnormal expression pattern which may cause the occurrence or progression of multiple diseases (Ankasha, Shafiee, Wahab, Ali, & Mokhtar, 2018; Jain, Thakkar, Chhatai, Bhadrac, & Bhadra, 2017; Lekka & Hall, 2018; Zhang, Yang, & Xiao, 2018). Unlike traditional linear RNAs, circRNA is a distinct class of circular‐shaped RNA, where the 3′ and 5′ ends are covalently bond and form a closed continuous loop, thus is resistant to exonuclease digestion (Jeck & Sharpless, 2014). This confers stability on circRNAs and makes them very abundant in the cytoplasm of eukaryotic cells (Jeck et al., 2013). Their high abundance, stability, and evolutionary conservation between species suggest that circRNAs may have important regulatory roles (Greene et al., 2017; Jeck & Sharpless, 2014; Jeck et al., 2013; Zhang, Yang, et al., 2018). As competing endogenous RNA (ceRNA), circRNAs may sequester miRNAs with their own miRNA response elements (MREs) and function as miRNA “sponges,” which strongly suppress miRNA activity and result in increased levels of miRNA target genes, thus mediating cell differentiation, proliferation, apoptosis, and material metabolism etc. (Bossi & Figueroa‐Bossi, 2016; Greene et al., 2017). For example, a circRNA named ciRS‐7 or CDR1‐AS (OMIM#300898) contains more than 70 selectively conserved miRNA target sites, and regulate the initiation and progression of various malignancies in a miR‐7‐dependent manner (Hansen, Kjems, & Damgaard, 2013; Sang et al., 2018; Weng et al., 2017). Song and Li (2018) investigated circRNAs expression in osteosarcoma and screened 1,152 upregulated and 915 downregulated circRNAs using microarray. Further findings showed that upregulated hsa_circ_001564 in osteosarcoma tissues served as miR‐29c‐3p “sponge” to promote tumor progression. All abovementioned characteristics make circRNAs become important biological regulators for understanding the molecular mechanisms of diseases and identifying effective diagnostic biomarkers and therapeutic targets. However, there is little information available in literatures about the relationship between circRNAs and endometriosis.
In this study, we explored circRNA expression profiles using high throughput RNA‐Seq for six paired ecEM and euEM tissues and discovered 294 differentially expressed circRNAs (146 upregulated and 148 downregulated). Furthermore, nine up‐expressed circRNAs were validated by qRT‐PCR. We also performed a comprehensive bioinformatic analysis of the most four upregulated circRNAs (hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0008016, and hsa_circ_0077837) and discussed their functions in the pathogenesis and progression of endometriosis. This study may provide a new breakthrough point for etiology research and molecular targeted therapy of endometriosis.
2. PATIENTS AND METHODS
2.1. Ethical compliance
This study was approved by the Ethics Committee of Shengjing Hospital (Ethics No. 2018PS504K), and written informed consent was obtained from each patient before surgical procedures.
2.2. Clinical specimens acquisition
Cyst walls of ovarian endometriomas and matched eutopic endometrium samples from the same patient were collected from 30 women (20–44 years old) with a laparoscopic and histological diagnosis of rASRM (the Revised American Society for Reproductive Medicine classification system, 1997) stage III/IV endometriosis at Shengjing Hospital, China Medical University from February 1, 2017 to March 31, 2018. All patients had regular menstrual cycles (21–35 days) and none of them had received gonadotropin‐releasing hormone analogues or other hormonal medications for at least 6 months before surgery. All euEM samples were in the proliferative phase of menstrual cycle confirmed by histological diagnosis. Once removed from the body, tissue samples were frozen in liquid nitrogen immediately and then stored at −80℃ for subsequent experiments. We chose six pairs of ecEM and euEM for high throughput RNA‐Seq at random.
2.3. RNA isolation and quality control
Total RNA was isolated from about 100mg of tissue with TRIzol agent (TaKaRa, Japan) according to the manufacturer's protocol. RNA quantity and quality were measured using Nanophotometer®N50 (Implen, Germany; Supplementary Table S1). Only when the ratio of the absorbance at 260 nm and 280 nm was between 1.8 and 2.2, the total RNA sample was accepted. All RNA samples were stored at −80°C for further use.
2.4. RNA library preparation and circRNA sequencing
A total amount of about 5 μg RNA per sample was used as input material for the RNA sample preparation. First, ribosomal RNA was removed by Epicentre Ribozero™ rRNA Removal Kit (Epicentre, USA). Subsequently, the linear RNA was digested with 3 units of RNase R (Epicentre, USA) per µg of RNA. The sequencing libraries were generated by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations. First and second strand cDNA were synthesized and modified with adenylation of 3′ ends of DNA fragments. NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. Then 3 μl USER Enzyme (NEB, USA) was used with size‐selected (preferentially 150–200 bp in length), adaptor‐ligated cDNA before PCR. PCR was performed with Phusion High‐Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index‐coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3‐cBot‐HS (Illumia) according to the manufacturer's instructions. After clustering, the libraries were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired‐end reads were generated. The RNA‐Seq and data collection were performed by Novogene Co. LTD, Beijing, China.
2.5. RNA‐seq data analysis
Clean reads were used for all the downstream analyses. Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using bowtie2 v2.2.8 and paired‐end clean reads were aligned to the reference genome using Bowtie (Langmead, Trapnell, Pop, & Salzberg, 2009). The circRNAs were detected and identified using find_circ (Memczak et al., 2013) and CIRI2 (Gao, Zhang, & Zhao, 2018). After quantile normalization of raw counts using Trans Per Million, differential expression analysis of two groups (matched ecEM vs. euEM) was performed using the DESeq R package (1.10.1) based on the negative binomial distribution. The resulting p values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate. CircRNA shaving fold change (FC, the ratio of the two groups’ averages) ≥2 and adjusted p‐values (Padj) ≤ 0.05 are selected as the significantly differentially expressed.
2.6. Construction of circRNA/miRNA interaction network
MiRNA target sites on circRNA were identified using miRNA target prediction software‐Novomagic (Novogene's homemade) based on TargetScan (Enright et al., 2003) and miRanda (John et al., 2004), and circRNA‐miRNA network was further constructed according to the seed match sequences and was drawn using Cytoscape 3.6.1.
2.7. qRT‐PCR
The circRNA was chosen for qRT‐PCR when it met all the following criteria: (a) upregulated in ecEM, (b) FC > 4.0, P adj < 0.01, (c) average readcount of each group >3, (d) sequence length between 200 and 1000 nt. According to these criteria, nine circRNAs (hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0002198, hsa_circ_0008016, novel_circ_0000153, hsa_circ_0077837, novel_circ_0020048, hsa_circ_0006856, and hsa_circ_0005777) were chosen. GAPDH (GeneBank Sequence #NM_002046.7) was used as a reference. During the qRT‐PCR validation stage, we first recruited 17 matched ecEM and euEM samples and screened out four most differentially expressed circRNAs, then we further expanded the sample number to 30 for validation. The expression level of circRNAs was evaluated by qRT‐PCR using one‐step SYBR® primescriptTM RT‐PCR kit (TaKaRa, Japan) on Applied Biosystems 7500 FAST (Applied Biosystems, USA) in a 20 µl reaction volume, including 11.2 µl 2× PCR master mix, 0.4 µl forward/reverse primers (10 µmol/L), 2 µl total RNA, and 6 µl RAase‐free H2O. The cycling program was initiated from 42°C for 5 min, 95°C for 10 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s, and last 95°C for 15 s, 60°C for 1min and 95°C for 15 s. All the PCR primers were designed and synthesized by Sangon Biotech (Shanghai, China, Table 1), and the specificity was verified by a single‐peak on the melting curve. Each sample had three individual technical replicates. The threshold cycle method (2−ΔΔCT) was used to calculate relative expression levels normalized to GAPDH levels.
Table 1.
Primers used in this paper
| circRNA ID | Primers | Amplicon length (bp) |
|---|---|---|
| hsa_circ_0003380 |
F:5′‐CACACAGTCAGAGAAACCGG‐3′ R:5′‐CTTCTTCGCAGCTCAGATTT‐3′ |
197 |
| hsa_circ_0020093 |
F:5′‐CTGGTCATGGGAAGTGTACAAC‐3′ R:5′‐ATCCCAGCTACATTCTGTAGCA‐3′ |
325 |
| hsa_circ_0002198 |
F:5′‐CCTGAAAGAGCCAAAGCTTG‐3′ R: 5′‐AAATCGGTGTAAGGTCACCATA‐3′ |
240 |
| hsa_circ_0008016 |
F: 5′‐CGTCCCGCAGCCAGTTGA‐3′ R: 5′‐TGGAGGTGCAGGACTCCGTG‐3′ |
219 |
| novel_circ_0000153 |
F:5′‐CTTGGACATGGTCAGCAGTATG‐3′ R:5′‐GCCTTCAATGAGCCAAGTACAT‐3′ |
224 |
| hsa_circ_0077837 |
F:5′‐CCTCTGGATCGGAAGACTGA‐3′ R:5′‐TGCCAAGGGACAAGTGTTATTT‐3′ |
241 |
| novel_circ_0020048 |
F:5′‐ATTTGATCGTCATGAAATCCAG‐3′ R:5′‐TCCCTTGAGGAAAATTAGGTCT‐3′ |
247 |
| hsa_circ_0006856 |
F:5′‐AGTCCTAGAACTTGCTTGTGCC‐3′ R:5′‐ATGTATTGCCTAGTTGGTCGAA‐3′ |
287 |
| hsa_circ_0002714 |
F:5′‐TCATTTGGATCAGTACCATCTG‐3′ R:5′‐TTTGATTGTTCGTATTGGCACT‐3′ |
273 |
| GAPDH |
F:5′‐CAGGAGGCATTGCTGATGAT‐3′ R:5′‐GAAGGCTGGGGCTCATTT‐3′ |
138 |
The circRNA ID is in circBase (http://circbase.org); GAPDH Genebank Reference Sequence: NM_002046.7
2.8. Construction of ceRNA network and enrichment analysis
We constructed a circRNA‐miRNA‐mRNA network based on ceRNA hypothesis using Cytoscape3.6.1 to investigate their interactions. With the known targeted miRNAs of each circRNA, we used the MiRWalk v3.0 (http://mirwalk.umm.uni-heidelberg.de/) with the strict screening conditions including three prediction algorithms (Targetscan,MiRDB and mirTarbase) to predict the downstream target genes. Finally, the ceRNA network was depicted using the validated four circRNAs and predicted miRNAs/mRNAs. GO and KEGG analyses further annotated the function and signaling pathways of these genes.
2.9. Statistical analysis
For comparisons of circRNAs expression levels between the ecEM and euEM group, the normal distribution of data was first tested, if satisfied, Student's t test (two‐tailed) was used; otherwise, nonparametric Mann–Whitney U‐test was used. The abovementioned statistical analyses were processed with GraphPad Prism7 (GraphPad Software, USA). A value of P < 0.05 was considered statistically significant.
3. RESULTS
3.1. CircRNA expression profiling
A total of 294 circRNAs were detected to be differentially expressed (FC ≥ 2.0 and P adj ≤ 0.05) in ecEM compared with matched euEM samples, with 146 upregulated and 148 downregulated. Hierarchical clustering analysis showed the overview of circRNA expression (Figure 1a), and the top 20 up‐ and downregulated circRNAs (Figure 1b). Table 2 listed the top 20 up‐ and downregulated circRNAs. The volcano and scatter plots displayed the variation in circRNA expression between ecEM and euEM groups (Figure 1c,d). The distribution of circRNAs in human chromosomes was depicted in the cluster‐shaped bar chart (Figure 2a). We investigated the general signatures of dysregulated circRNAs and found that the majority originated from the protein coding exons, and some were from introns and intergenic region (Figure 2b).
Figure 1.
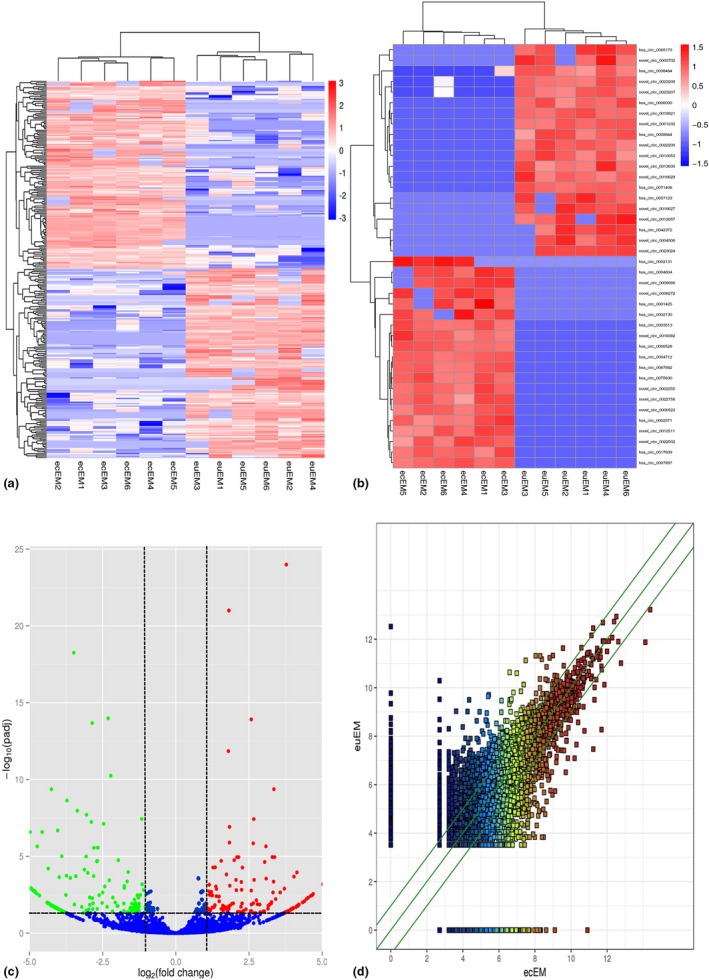
CircRNA expression patterns in ecEM relative to those in euEM. (a) Hierarchical cluster analysis of all differentially expressed circRNAs. (b) Hierarchical cluster analysis of the top 20 up‐ and downregulated circRNAs. Each column represents a sample and each row represents a circRNA. The color scale indicates relative expression, upregulation (red), and downregulation (blue). (c) Volcano plots demonstrate differential expression between two different conditions. Red points indicated upregulated while the green points indicated downregulated expression. (d) Scatter plots show the difference in the expression of circRNAs between ecEM and euEM. The values plotted on X and Y axes are the averaged normalized signal values of each group (log2 scaled). FC ≥ 2 and P adj ≤ 0.05 were regarded as the differentially expressed circRNAs
Table 2.
The top 20 upregulated and downregulated circular RNAs ranked by fold changes
| circRNA ID | chromosome | Spliced length | circRNA type | Source gene | Fold change | Padj value |
|---|---|---|---|---|---|---|
| The top 20 upregulated circRNAs | ||||||
| novel_circ_0012511 | chr2 | 318 | Exonic | SCN7A | 63.2108364 | 5.424E‐06 |
| hsa_circ_0067682 | chr3 | 668 | Exonic | PLOD2 | 47.9546895 | 4.596E‐05 |
| hsa_circ_0006528 | chr5 | 496 | Exonic | PRELID2 | 41.0326379 | 0.0001469 |
| hsa_circ_0007897 | chr3 | 457 | Exonic | BOC | 40.5828984 | 0.0001564 |
| hsa_circ_0002571 | chr14 | 14,092 | Exonic | / | 36.8808500 | 0.0003091 |
| novel_circ_0000523 | chr10 | 34 | Exonic | MKX | 36.3227115 | 0.0003238 |
| novel_circ_0022502 | chr9 | 371 | Exonic | NTRK2 | 34.9858588 | 0.0004293 |
| hsa_circ_0004712 | chr6 | 545 | Exonic | PDE7B | 34.3609972 | 0.0004443 |
| novel_circ_0002255 | chr12 | 534 | Exonic | NR1H4 | 32.6090386 | 0.000641 |
| novel_circ_0009666 | chr1 | 292 | Intergenic | / | 25.8143255 | 0.0027854 |
| hsa_circ_0001425 | chr4 | 1,127 | intronic | HERC6 | 25.3620595 | 0.0032733 |
| hsa_circ_0004804 | chr9 | 327 | Exonic | PBX3 | 24.4032265 | 0.0036856 |
| novel_circ_0018382 | chr6 | 298 | Exonic | LAMA2 | 22.9114999 | 0.0048829 |
| novel_circ_004 | chrX | 437 | Exonic | ARHGAP6 | 22.1586794 | 0.0058839 |
| novel_circ_0006272 | chr16 | 168 | Exonic | / | 21.0084818 | 0.008401 |
| hsa_circ_0003513 | chr1 | 463 | Exonic | GCLM | 20.1470960 | 0.0098272 |
| hsa_circ_0017639 | chr10 | 536 | Exonic | SFMBT2 | 18.4660066 | 0.013729 |
| hsa_circ_0002130 | chr19 | 195 | Exonic | C3 | 18.0960134 | 0.016081 |
| hsa_circ_0002131 | chr8 | 511 | Exonic | BNIP3L | 18.0496630 | 0.016694 |
| hsa_circ_0075830 | chr6 | 523 | Exonic | / | 17.9175317 | 0.015469 |
| The top 20 downregulated circRNAs | ||||||
| novel_circ_0018621 | chr6 | 314 | Exonic | ESR1 | 120.1172882 | 8.594E‐09 |
| novel_circ_0023209 | chrX | 371 | Intergenic | / | 74.2438696 | 9.04E‐13 |
| hsa_circ_0008844 | chr1 | 299 | Exonic | MFSD2A | 64.7990160 | 5.172E‐06 |
| novel_circ_0022200 | chr9 | 269 | Exonic | / | 55.1771869 | 1.773E‐05 |
| hsa_circ_0006030 | chr12 | 392 | Exonic | TMEM120B | 48.1211762 | 5.376E‐05 |
| novel_circ_0023207 | chrX | 320 | Intergenic | / | 44.5946891 | 2.861E‐09 |
| hsa_circ_0085173 | chr8 | 394 | Exonic | GRHL2 | 42.2623182 | 0.0001586 |
| novel_circ_0001232 | chr11 | 423 | Exonic | PGR | 39.0567500 | 0.000236 |
| novel_circ_0004306 | chr14 | 245 | Exonic | NPAS3 | 35.7061332 | 0.0004703 |
| novel_circ_0010653 | chr1 | 196 | Exonic | FYB2 | 35.4201889 | 0.0004526 |
| novel_circ_0018629 | chr6 | 475 | Exonic | ESR1 | 35.1974742 | 0.0004703 |
| hsa_circ_0038484 | chr16 | 220 | Exonic | VWA3A | 31.5266506 | 2.621E‐07 |
| novel_circ_0013630 | chr2 | 108 | Exonic | EPCAM | 30.7092208 | 0.0011808 |
| novel_circ_0012657 | chr2 | 95 | Intergenic | / | 30.2822396 | 0.0013858 |
| novel_circ_0023024 | chrX | 234 | Exonic | / | 30.2465775 | 0.001332 |
| hsa_circ_0057123 | chr2 | 363 | Exonic | CDCA7 | 29.6631520 | 0.0015094 |
| novel_circ_0003732 | chr13 | 327 | Exonic | / | 29.1050986 | 0.0017015 |
| novel_circ_0018627 | chr6 | 336 | Exonic | ESR1 | 29.0486659 | 0.0016979 |
| hsa_circ_0071408 | chr4 | 463 | Exonic | PALLD | 28.2073651 | 0.0018081 |
| hsa_circ_004237082 | chr17 | 320 | Exonic | SLC47A1 | 27.9175416 | 0.0021405 |
Figure 2.
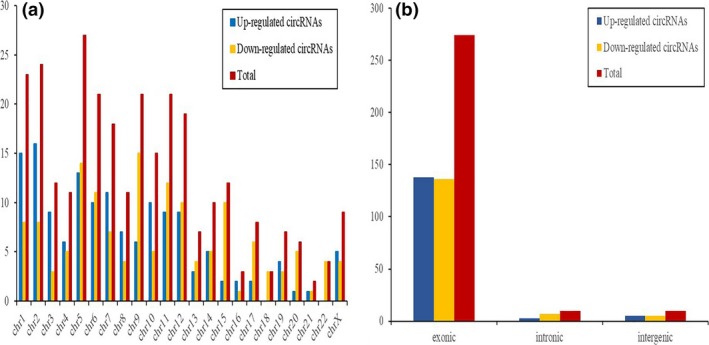
Characteristics of differentially expressed circular RNAs between ecEM and euEM. (a) The cluster shaped bar chart demonstrates chromosome distribution of differentially expressed circRNAs. (b) The histogram demonstrates the circRNA transcription origination
3.2. Construction of the circRNA/miRNA interaction network
To explore the function of circRNAs, specific binding patterns between circRNAs and miRNAs were bioinformatically predicted using Novomagic software (Novogene's homemade) based on TargetScan and miRanda according to the complementary miRNA matching sequence. A total of 2,495 MREs were formulated to bind with the 294 differentially expressed circRNAs. The circRNA/miRNA interaction network of the top 20 up‐and downregulated circRNAs was drawn by Cytoscape3.6.1 (Figure 3).
Figure 3.
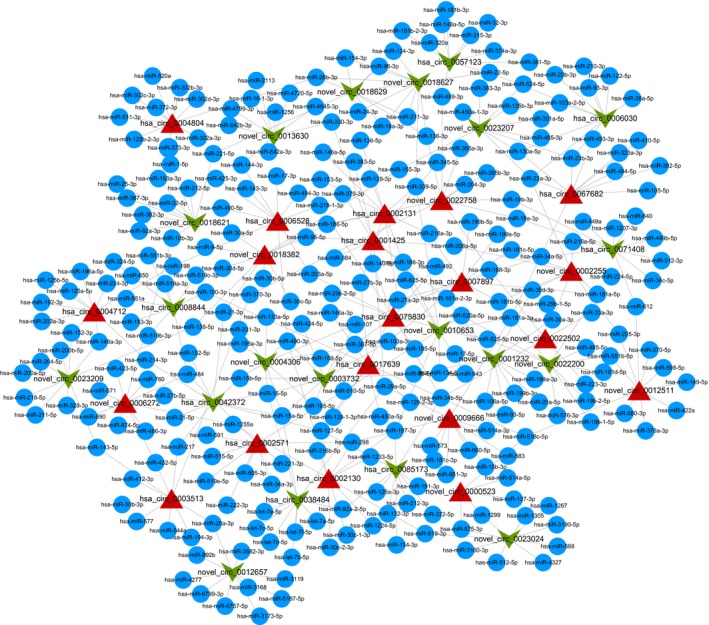
circRNA/miRNA network analysis. The network comprising the top 20 upregulated (red triangles) and downregulated circRNAs (green inverted arrows) and their target miRNAs (only top 10 included) are presented
3.3. QRT‐PCR validation for the selected circRNAs
To validate the circRNA sequencing results, qRT‐PCR was used to detect nine upregulated circRNAs which were chosen according to the criteria above mentioned. Five of nine circRNAs (hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0002198, hsa_circ_0008016, hsa_circ_0077837) were significantly overexpressed in 17 ecEM samples, consistent with RNA‐seq data, while novel_circ_0000153 expression level displayed no statistically significant difference between the two groups (p > 0.05; Figure 4a). Meanwhile, novel_circ_0020048, hsa_circ_0006856, and hsa_circ_0005777 could not be amplified by qPCR (data not displayed). We further expanded the number of sample to 30 and compared the expression levels of hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0002198, hsa_circ_0008016, and hsa_circ_0077837 in ecEM and euEM groups. The data confirmed that hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0008016, and hsa_circ_0077837 were all significantly overexpressed in the ecEM group (p < 0.001; Figure 4b).
Figure 4.
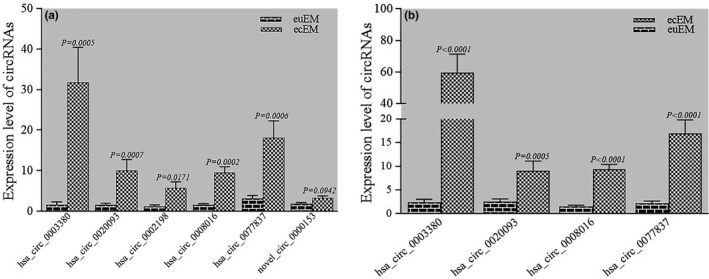
Validation results for the selected circRNAs. (a) The expression levels of the selected circRNAs were analyzed in 17 paired ecEM and euEM tissues by qPCR; (b) Further validation of the four most differentially expressed circRNAs in total 30 paired ecEM and euEM tissues (p < 0.05 considered significant)
3.4. Construction of ceRNA network
ceRNA network of the four differentially expressed circRNAs hsa_circ_0003380, hsa_circ_0020093, hsa_circ_0008016, and hsa_circ_0077837 in ecEM was constructed based on the ceRNA theory that circRNAs may share a common binding site of MRE with mRNAs so as to regulate each other (Figure 5). The ceRNA network contained 48 miRNAs and 296 mRNAs.
Figure 5.
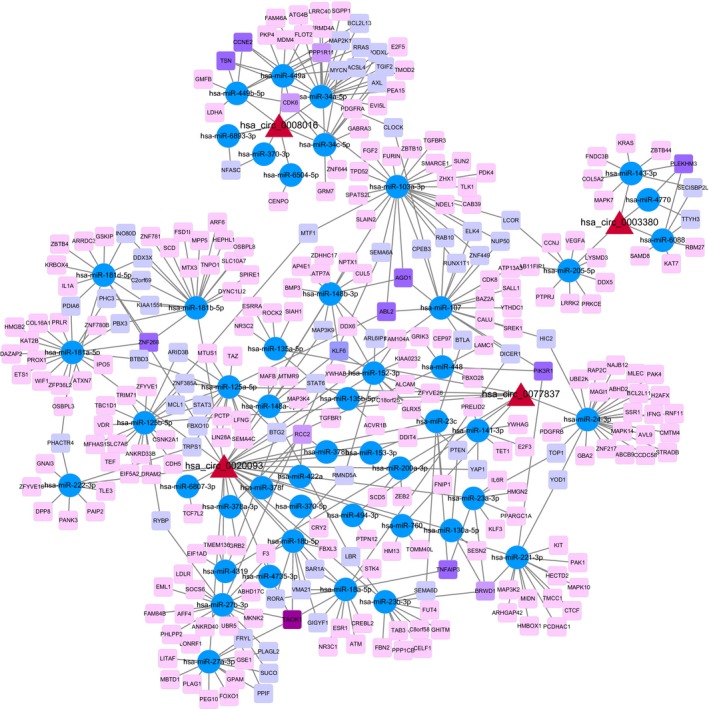
CircRNAs‐miRNAs‐mRNAs network. The ceRNA network was constructed with the four differentially expressed circRNAs. The network consists of 48 miRNAs and 296 mRNAs. The pink to purple square node represents a protein‐coding gene (the deeper color means the mRNA is targeted by more miRNAs), blue round node represents miRNA and red triangle represents a circRNA
3.5. Enrichment analysis of circular RNA‐targeted genes
GO and KEGG analysis was applied to annotate the function of circRNA‐targeted genes predicted in the ceRNA analysis. We totally obtained 57 GO terms and the data indicated that these target genes were mainly involved in the biological processes of extrinsic apoptotic signaling pathway, regulation of protein serine/threonine kinase activity, positive regulation of cellular component movement and so on (Figure 6a). Additionally, KEGG analysis revealed 41 signaling pathways such as MAPK signaling pathway, pathways in cancer, FOXO signaling pathway, RAS signaling pathway, and PI3K‐AKT signaling pathway (Figure 6b).
Figure 6.
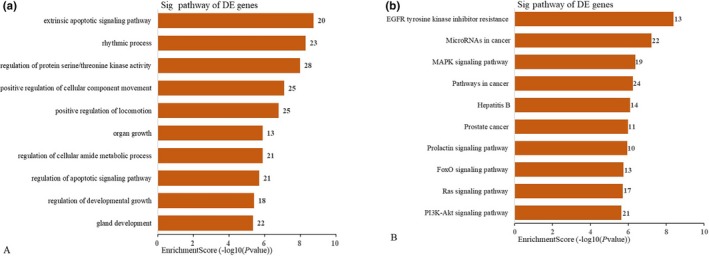
Functional ananlysis for the four validated circRNAs gathering genes. The vertical axis shows the annotated functions of the target genes. The horizontal axes show the enrichment score (−log10 transformed p‐value) and the gene number of each cluster, respectively. Only the top 10 significantly enriched clusters are included. (a) GO analysis. (b) KEGG pathway analysis
4. DISCUSSION
Recently, a growing body of evidence indicates that ncRNAs play crucial regulatory roles in the pathogenesis and development of endometriosis, including miRNAs (Agrawal et al., 2018; Nothnick, 2017) and long ncRNAs (Cui et al., 2018; Wang et al., 2016). For example, miR‐29c (Long, Wan, La, Gong, & Cai., 2015) and miR‐200b (Eggers et al., 2016) were reported as downregulated in endometriosis and affecting endometrial cell proliferation, apoptosis, and invasion. LncRNA LINC00261 downregulation inhibited endometriosis cell growth and migration (Sha et al., 2017). Ghazal et al. (2015) proposed that perturbation of H19/Let‐7/IGF1R regulatory pathway might represent a potential mechanism for endometriosis‐associated infertility. However, to the best of our knowledge, studies on the identification and functional characterization of circRNAs in endometriosis are limited.
In this study, we investigated circRNA expression profiles in ecEM compared with euEM of endometriosis patients and identified 294 differentially expressed circRNAs, 146 upregulated and 148 downregulated. Most of those dysregulated circRNAs originate from exons or introns which are in accordance with previous researches (Shen et al., 2018; Xu et al., 2018a, 2018b; Zhang, Ren, Xiao, Xia, & Fang, 2018). It has been reported that both exonic and intronic circRNAs may exert potential functions in gene regulation (Greene et al., 2017). Xu et al. (2018a) revealed for the first time the circRNA expression patterns between women with and without endometriosis and found that the increased level of circ_0004712 and circ_0002198 could help identify the patients with endometriosis. Zhang, Ren, et al. (2018) assessed the expression profiles of circRNAs in endometriosis by microarray analysis and detected 2,237 circRNAs differentially expressed among ectopic endometrium (EC) (endometriotic lesions), paired eutopic endometrium (EU), and normal endometrium from women free of endometriosis, and qRT‐PCR validation suggested that downregulated circ_103470 and circ_101102 matched the microarray results. In our study, four of nine upregulated circRNAs (hsa_circ_0003380, hsa_circ_0020093,hsa_circ_0008016 and hsa_circ_0077837) were confirmed by qRT‐PCR, which was consistent with sequence assays. Hsa_circ_0003380 is derived from the ZFPM2 gene. ZFPM2 is known as an inhibitor of PI3K/AKT pathway (Xiao et al., 2014). Previous studies have demonstrated that activating PI3K/AKT pathway lead to cellular events including proliferation, invasion, and angiogenesis in developing endometrial lesions (Cao et al., 2017; Diao et al., 2017). Hsa_circ_0020093 is encoded by ATRNL1 which is a highly conserved transmembrane protein involved in cell adhesion and signaling events (Stark et al., 2010). EP B41L2 gene is the parental gene of hsa_circ_0077837 that encodes 4.1G protein, which is a membrane skeletal protein that can also serve as an adapter to mediate cell adhesion, migration, and apoptosis in tumorgenesis (Bosanquet, Ye, Harding, & Jiang, 2014). At the initial stages of endometriosis, the attachment of retrograde endometrial tissues onto the pelvic mesothelium is a critical step and cell adhesion molecules are taken as key factors in this process (Kuessel et al., 2017; Tsai et al., 2018). FGFR1 is the source gene of hsa_circ_0008016. Zhao et al. (2015) found that FGFR1 was overexpressed in ectopic endometrium of endometriosis and correlated with dysmenorrhea severity and recurrence of this disease. Considering the various functions of those parent genes, we speculated that dysregulation of these circRNAs might play a role in the development and progression of endometriosis.
CircRNAs can function as miRNA sponge through the ceRNA network. Based on the ceRNA hypothesis, circRNAs can affect the miRNA activity through sequestration, thereby increasing or decreasing miRNA target gene expression (Bossi & Figueroa‐Bossi, 2016; Greene et al., 2017). We constructed the ceRNA crosstalk network to further explore the role of dysregulated circRNAs in endometriosis. The four validated upregulated circRNAs (hsa_circ_0003380, hsa_circ_0020093,hsa_circ_0008016, and hsa_circ_0077837) were found to target 48 miRNAs, of which 26 miRNAs have been reported in previous studies that might be associated with endometriosis (Agrawal et al., 2018; Long et al., 2015; Nothnick, 2017). Ohlsson‐Teague and coworkers (2009) first assessed the miRNA expression profiles in paired eutopic and ectopic endometrial tissues in 2009 and identified 14 upregulated and 8 downregulated miRNAs specific to endometriotic lesion by microarray analysis. For example, miR‐143 was reported as upregulated in eutopic endometrial tissues (Ohlsson‐Teague et al., 2009). Another study in vitro showed that expression of miR‐27b‐3p was significantly higher in human endometrial stromal cells from patients with endometriosis than that of controls, which was related to fibrosis modulation (Kim et al., 2017). miR‐34a‐5p was significantly downregulated in patients with endometriosis (Ma, Huang, & Chen, 2017). Additionally, several studies have published that miRNAs as putative biomarkers of endometriosis in biofluids. Seral miR‐125b‐5p levels, combined with miR‐451a and miR‐3613‐5p distinguished subjects with endometriosis from those without (Cosar et al., 2016). Expressions of miR‐200a‐3p and miR‐141‐3p were also downregulated in plasma samples of endometriosis patients (Rekker et al., 2015).
MiRNAs can regulate a wide array of biological processes through post‐transcriptionally repressing downstream mRNAs expression. Through bioinformatic prediction, there are 296 mRNAs targeted by the 48 miRNAs. In our study, bioinformatics analysis presented that E2F transcription factor 3 (E2F3), one target of miR‐141‐3p, is oncogenic in tumorigenesis and corresponds with poor prognosis in various cancers (Gao et al., 2017). Considering that endometriosis shares many characteristics with malignant diseases, we hypothesized that the miR‐141‐3p/E2F3 axis might play a role in the pathogenesis of endometriosis. Previous studies demonstrated that the miR‐200 family members (miR‐200a‐3p and miR‐141‐3p) regulated a cohort of target genes such as ZEB1 and E‐cadherin involved in epithelial‐mesenchymal transition (EMT) and metastasis (Bhardwaj et al., 2017; Xiong et al., 2016), which are important events in the initiation and progression of endometriosis. Thus, we inferred that hsa_circ_0020093/miR‐141‐3p/miR‐200a‐3p/ ZEB1 axis might regulate EMT in endometriosis. Angiogenesis is one of the important pathophysiological conditions in endometriosis and VEGFA was ever reported to be upregulated in endometriotic tissues (Ma et al., 2017). By constructing the ceRNA network, we supposed that hsa_circ_0003380/miR‐205/ VEGFA axis might modulate the angiogenic process in endometriosis. CDK6, predicted as a common target gene of miR‐103a‐3p, miR‐34c‐5p, miR‐449a, and miR‐449b‐5p, mainly serves as an oncogenic kinase that regulates cell cycle and was found upregulated in endometriosis (Wang, Li, Yang, Liu, & Wang, 2015). Genetic studies have demonstrated that endometriotic lesions commonly have mutations in genes, particularly the TP53, KRAS, PTEN, and ARID1A genes, which suggest a direct transition from a subset of endometriotic lesions to invasive neoplasms (Matias‐Guiu & Stewart, 2018; Ponandai‐Srinivasan et al., 2018). Lin et al. (2017) identified that disruption of hypoxia‐induced YAP1 signaling by siRNA knockdown or inhibitor abolished critical biological processes in endometriosis development like steroidogenesis, angiogenesis, inflammation, migration, innervation, and cell proliferation.
To further dissect the role of those target genes contained in the ceRNA network, we also performed GO and KEGG analysis and obtained 57 functional terms and 41 important signaling pathways, most of which were consistent with the current knowledge on endometriosis. For example, the most significant GO term of biological process is extrinsic apoptotic signaling pathway (GO:0097191) which enriched 22 differentially expressed genes such as BCL2L11, YAP1, PTEN, and PIK3R1. Once mutated, these abnormal molecules can influence the cell survival through cascading activation of kinase signaling pathways and may cause malignant ovarian tumors (Matias‐Guiu & Stewart, 2018; Ponandai‐Srinivasan et al., 2018). Song et al. (2016) highlighted that dysregulation of HIPPO/YAP1 pathway components or amplification of its downstream effector YAP1 resulted in increased endometrial stromal cell proliferation and decreased cell apoptosis. Through KEGG analysis, we found that target genes participated in several signal pathways which were well‐researched in many diseases, such as MAPK signaling pathway, pathways in cancer, FOXO signaling pathway, RAS signaling pathway, and PI3K‐AKT signaling pathway. Xu et al. (2018b) constructed a circRNA‐miRNA‐mRNA network based on five identified differentially expressed circRNAs and eight mRNAs between ovarian ectopic and paired eutopic endometria, and revealed the primary roles of cancer‐related, purine metabolism, glycerophospholipid metabolism, and thyroid hormone signaling pathways in endometriosis pathogenesis. As outlined in many articles, the establishment and progression of endometriosis requires a number of signal pathways regulating cooperatively, like the canonical IKKβ/NF‐κB pathway, the MAPK pathways, and the PI3K/AKT/mTOR pathway, which also have been taken as viable targets for endometriosis treatment (McKinnon, Kocbek, Nirgianakis, Bersinger, & Mueller, 2016). Genome‐wide association study uncovered top pathways significantly associated with endometriosis and stage A disease included several MAPK‐related pathways (Uimari et al., 2017). Endometriosis lesions exist in a unique microenvironment and demonstrate a series of malignant biological behaviors. Pathways in cancer may also involve in pathogenesis and progression of endometriosis. Cao et al. (2017) first revealed a dose‐dependent inhibition effect of ginsenoside Rg3 on the growth of ectopic endometrium in endometriosis rats through blocking VEGFR‐2‐mediated PI3K/AKT/mTOR signaling pathway. Increased activation of RAS/RAF/MAPK and RhoA/ROCKII signaling pathways in primary eutopic endometrial stromal cells of patients with endometriosis promoted cell proliferation and migration (Yotova et al., 2011). Cross‐talk between FOXO signaling pathway and other pathways like PI3K‐AKT, MAPK, β‐CATENIN/WNT involves in cell differentiation, proliferation, and apoptosis which may provide a new avenue for the prevention and treatment of diseases (Parker et al., 2012; Xing et al., 2018). Certainly, the role of FOXO signaling pathway in endometriosis deserves further exploration. Future studies are expected to clearly define the functions of those signaling pathways in endometriosis pathophysiology.
In conclusion, our study revealed that the circRNA expression profiles in ovarian endometriosis and obtained 294 differentially expressed circRNAs between paired ecEM and euEM. The ceRNA network and functional analysis of potential target genes highlighted the relationship between dysregulated circRNAs and ovarian endometriosis, which may provide a new perspective on researches of endometriosis. However, there were several limitations existing in this study. First, the sample size was small and only rASRM stage III/IV cases were included. Second, the tissue samples were only derived from ectopic endometrioma wall and eutopic endometrium, neither peritoneal lesions nor deep infiltrated endometriosis, thus further validation involving in larger cohorts of patients with early stages as well as other types of endometriosis is warranted. Third, the tissue RNA isolation relied on the removal of ribosomal RNA by rRNA Removal Kit and a digest of free RNA by RNase R, and this approach had certain limitations as some other RNA types except circular RNAs could have been preserved which might interfered with the final data, therefore new RNA purification methods are expected. And then validation of the RNA‐seq assays by qRT‐PCR using specific primers is essential. Fourth, the molecular mechanism and function of the present circRNAs should be tested in vivo and vitro experiments further. Our group has started to study the overall effect and in‐depth mechanism of hsa_circ_0003380 and hsa_circ_0020093 in endometriosis, which may be promising biomarkers and therapeutic targets of endometriosis.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
Supporting information
Wang D, Luo Y, Wang G, Yang Q. Circular RNA expression profiles and bioinformatics analysis in ovarian endometriosis. Mol Genet Genomic Med. 2019;7:e756 10.1002/mgg3.756
Funding information
This project was supported by grants from the Outstanding Scientific Fund of Shengjing Hospital (No.201704), the Public Research Foundation of Liaoning Province (No. 20170017), and the Support Program for Youth Backbone of China Medical University (No. QGZD2018062).
REFERENCES
- Agrawal, S. , Tapmeier, T. , Rahmioglu, N. , Kirtley, S. , Zondervan, K. , & Becker, C. (2018). The miRNA mirage, how close are we to finding a non‐Invasive diagnostic biomarker in endometriosis? A systematic review. International Journal of Molecular Sciences, 19(2), pii: E599 10.3390/ijms19020599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankasha, S. J. , Shafiee, M. N. , Wahab, N. A. , Ali, R. A. R. , & Mokhtar, N. M. (2018). Post‐transcriptional regulation of microRNAs in cancer, from prediction to validation. Oncology Reviews, 12(1), 344 10.4081/oncol.2018.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghari, S. , Valizadeh, A. , Aghebati‐Maleki, L. , Nouri, M. , & Yousefi, M. (2018). Endometriosis, perspective, lights, and shadows of etiology. Biomedicine & Pharmacotherapy, 106, 163–174. 10.1016/j.biopha.2018.06.109 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, M. , Sen, S. , Chosdol, K. , Sharma, A. , Pushker, N. , Kashyap, S. … Bajaj, M. S. (2017). miRNA‐200c and miRNA‐141 as potential prognostic biomarkers and regulators of epithelial‐mesenchymal transition in eyelid sebaceous gland carcinoma. British Journal of Ophthalmology, 101(4), 536–542. 10.1136/bjophthalmol-2016-309460 [DOI] [PubMed] [Google Scholar]
- Borghese, B. , Zondervan, K. T. , Abrao, M. S. , Chapron, C. , & Vaiman, D. (2017). Recent insights on the genetics and epigenetics of endometriosis. Clinical Genetics, 91(2), 254–264. 10.1111/cge.12897 [DOI] [PubMed] [Google Scholar]
- Bosanquet, D. C. , Ye, L. , Harding, K. G. , & Jiang, W. G. (2014). FERM family proteins and their importance in cellular movements and wound healing (review). International Journal of Molecular Medicine, 34(1), 3–12. 10.3892/ijmm.2014.1775 [DOI] [PubMed] [Google Scholar]
- Bossi, L. , & Figueroa‐Bossi, N. (2016). Competing endogenous RNAs, a target‐centric view of small RNA regulation in bacteria. Nature Reviews Microbiology, 14(12), 775–784. 10.1038/nrmicro.2016.129 [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Ye, Q. , Zhuang, M. , Xie, S. , Zhong, R. , Cui, J. , … Cao, L. (2017). Ginsenoside Rg3 inhibits angiogenesis in a rat model of endometriosis through the VEGFR‐2‐mediated PI3K/Akt/mTOR signaling pathway. PLoS ONE, 12(11), e0186520 10.1371/journal.pone.0186520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosar, E. , Mamillapalli, R. , Ersoy, G. S. , Cho, S. , Seifer, B. , & Taylor, H. S. (2016). Serum microRNAs as diagnostic markers of endometriosis, a comprehensive array‐based analysis. Fertility and Sterility, 106(2), 402–409. 10.1016/j.fertnstert.2016.04.013 [DOI] [PubMed] [Google Scholar]
- Cui, D. , Ma, J. , Liu, Y. , Lin, K. , Jiang, X. , Qu, Y. , … Xu, K. (2018). Analysis of long non‐coding RNA expression profiles using RNA sequencing in ovarian endometriosis. Gene, 673, 140–148. 10.1016/j.gene.2018.06.046 [DOI] [PubMed] [Google Scholar]
- Diao, R. , Wei, W. , Zhao, J. , Tian, F. , Cai, X. , & Duan, Y. G. (2017). CCL19/CCR11 contributes to the pathogenesis of endometriosis via PI3K/Akt pathway by regulating the proliferation and invasion of ESCs. American Journal of Reproductive Immunology, 78(5), e12744– 10.1111/aji.12744 [DOI] [PubMed] [Google Scholar]
- Eggers, J. C. , Martino, V. , Reinbold, R. , Schäfer, S. D. , Kiesel, L. , Starzinski‐Powitz, A. , … Götte, M. (2016). microRNA miR‐200b affects proliferation, invasiveness and stemness of endometriotic cells by targeting ZEB1, ZEB2 and KLF4. Reproductive Biomedicine Online, 32(4), 434–445. 10.1016/j.rbmo.2015.12.013 [DOI] [PubMed] [Google Scholar]
- Enright, A. J. , John, B. , Gaul, U. , Tuschl, T. , Sander, C. , & Marks, D. S. (2003). MicroRNA targets in drosophila. Genome Biology, 5(1), R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Feng, B. , Lu, L. , Lu, L. , Han, S. , Chu, X. , … Wang, R. (2017). MiRNAs and E2F3: A complex network of reciprocal regulations in human cancers. Oncotarget, 8(36), 60624–60639. 10.18632/oncotarget.17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Zhang, J. , & Zhao, F. (2018). Circular RNA identification based on multiple seed matching. Briefings in Bioinformatics, 19(5), 803–810. 10.1093/bib/bbx014 [DOI] [PubMed] [Google Scholar]
- Ghazal, S. , McKinnon, B. , Zhou, J. , Mueller, M. , Men, Y. I. , Yang, L. , … Taylor, H. S. (2015). H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Molecular Medicine, 7(8), 996–1003. 10.15252/emmm.201505245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice, L. C. (2010). Clinical practice: Endometriosis. New England Journal of Medicine, 362(25), 2389–2398. 10.1056/NEJMcp1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, J. , Baird, A. M. , Brady, L. , Lim, M. , Gray, S. G. , McDermott, R. , & Finn, S. P. (2017). Circular RNAs, biogenesis, function and role in human diseases. Frontiers in Molecular Biosciences, 4, 38 10.3389/fmolb.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme, J. , Hammond, M. G. , Hulka, J. F. , Raj, S. G. , & Talbert, L. M. (1984). Retrograde menstruation in healthy women and in patients with endometriosis. Obstetrics and Gynecology, 64(2), 151–154. [PubMed] [Google Scholar]
- Hansen, T. B. , Kjems, J. , & Damgaard, C. K. (2013). Circular RNA and miR‐7 in cancer. Cancer Research, 73(18), 5609–5612. 10.1158/0008-5472 [DOI] [PubMed] [Google Scholar]
- Jain, S. , Thakkar, N. , Chhatai, J. , Bhadrac, M. P. , & Bhadra, U. (2017). Long non‐coding RNA, functional agent for disease traits. RNA Biology, 14(5), 522–535. 10.1080/15476286.2016.1172756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck, W. R. , & Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nature Biotechnology, 32(5), 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck, W. R. , Sorrentino, J. A. , Wang, K. , Slevin, M. K. , Burd, C. E. , Liu, J. , … Sharpless, N. E. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, B. , Enright, A. J. , Aravin, A. , Tuschl, T. , Sander, C. , & Marks, D. S. (2004). Human microRNA targets. PLoS Biology, 2(11), e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. K. , Lee, S. K. , Park, J. H. , Lee, J. H. , Yun, B. H. , Park, J. H. , … Choi, Y. S. (2017). Ginsenoside Rg3 decreases fibrotic and invasive nature of endometriosis by modulating miRNA‐27b, in vitro and in vivo studies. Scientific Reports, 7(1), 17670 10.1038/s41598-017-17956-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuessel, L. , Wenzl, R. , Proestling, K. , Balendran, S. , Pateisky, P. , Yotova, G. , … Husslein, H. (2017). Soluble VCAM‐1/soluble ICAM‐1 ratio is a promising biomarker for diagnosing endometriosis. Human Reproduction, 32(4), 770–779. 10.1093/humrep/dex028 [DOI] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. , & Salzberg, S. L. (2009). Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biology, 10(3), R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekka, E. , & Hall, J. (2018). Noncoding RNAs in disease. FEBS Letters, 5(92), 2884–2900. 10.1002/1873-3468.13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. C. , Lee, H. C. , Hou, P. C. , Fu, J. L. , Wu, M. H. , & Tsai, S. J. (2017). Targeting hypoxia‐mediated YAP1 nuclear translocation ameliorates pathogenesis of endometriosis without compromising maternal fertility. The Journal of Pathology, 242(4), 476–487. 10.1002/path.4922 [DOI] [PubMed] [Google Scholar]
- Long, M. , Wan, X. , La, X. , Gong, X. , & Cai, X. (2015). miR‐29c is downregulated in the ectopic endometrium and exerts its effects on endometrial cell proliferation, apoptosis and invasion by targeting c‐Jun. International Journal of Molecular Medicine, 35(4), 1119–1125. 10.3892/ijmm.2015.2082 [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Huang, Y. X. , & Chen, Y. Y. (2017). miRNA‐34a‐5p downregulation of VEGFA in endometrial stem cells contributes to the pathogenesis of endometriosis. Molecular Medicine Reports, 16(6), 8259–8264. 10.3892/mmr.2017.7677 [DOI] [PubMed] [Google Scholar]
- Matias‐Guiu, X. , & Stewart, C. J. R. (2018). Endometriosis‐associated ovarian neoplasia. Pathology, 50(2), 190–204. 10.1016/j.pathol.2017.10.006 [DOI] [PubMed] [Google Scholar]
- McKinnon, B. D. , Kocbek, V. , Nirgianakis, K. , Bersinger, N. A. , & Mueller, M. D. (2016). Kinase signalling pathways in endometriosis, potential targets for non‐hormonal therapeutics. Human Reproduction Update, 22(3), 382–403. 10.1093/humupd/dmv060 [DOI] [PubMed] [Google Scholar]
- Memczak, S. , Jens, M. , Elefsinioti, A. , Torti, F. , Krueger, J. , Rybak, A. , & Rajewsky, N. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Nothnick, W. B. (2017). MicroRNAs and endometriosis, distinguishing drivers from passengers in disease pathogenesis. Seminars in Reproductive Medicine, 35(2), 173–180. 10.1055/s-0037-1599089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson Teague, E. M. , Van der Hoek, K. H. , Van der Hoek, M. B. , Perry, N. , Wagaarachchi, P. , Robertson, S. A. , … Hull, L. M. (2009). MicroRNA‐regulated pathways associated with endometriosis. Molecular Endocrinology, 23(2), 265–275. 10.1210/me.2008-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J. A. , Vazquez‐Manrique, R. P. , Tourette, C. , Farina, F. , Offner, N. , Mukhopadhyay, A. , … Neri, C. (2012). Integration of β‐catenin, sirtuin, and FOXO signaling protects from mutant huntingtin toxicity. Journal of Neuroscience, 32(36), 12630–12640. 10.1523/JNEUROSCI.0277-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponandai‐Srinivasan, S. , Andersson, K. L. , Nister, M. , Saare, M. , Hassan, H. A. , Varghese, S. J. , … Lalitkumar, P. G. L. (2018). Aberrant expression of genes associated with stemness and cancer in endometria and endometrioma in a subset of women with endometriosis. Human Reproduction, 33(10), 1924–1938. 10.1093/humrep/dey241 [DOI] [PubMed] [Google Scholar]
- Rekker, K. , Saare, M. , Roost, A. M. , Kaart, T. , Sõritsa, D. , Karro, H. , … Peters, M. (2015). Circulating miR‐200‐family micro‐RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertility and Sterility, 104(4), 938–946.e2. 10.1016/j.fertnstert.2015.06.029 [DOI] [PubMed] [Google Scholar]
- Sampson, J. A. (1927). Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. American Journal of Pathology, 3(2), 93–110.43. [PMC free article] [PubMed] [Google Scholar]
- Sang, M. , Meng, L. , Sang, Y. , Liu, S. , Ding, P. , Ju, Y. , … Shan, B. (2018). Circular RNA ciRS‐7 accelerates ESCC progression through acting as a miR‐876‐5p sponge to enhance MAGE‐A family expression. Cancer Letters, 426, 37–46. 10.1016/j.canlet.2018.03.049 [DOI] [PubMed] [Google Scholar]
- Sha, L. X. , Huang, L. X. , Luo, X. S. , Bao, J. , Gao, L. , Pan, Q. … Wang, H. (2017). Long non‐coding RNA LINC00261 inhibits cell growth and migration in endometriosis. Journal of Obstetrics and Gynaecology Research, 43(10), 1563–1569. 10.1111/jog.13427 [DOI] [PubMed] [Google Scholar]
- Shafrir, A. L. , Farland, L. V. , Shah, D. K. , Harris, H. R. , Kvaskoff, M. , Zondervan, K. , & Missmer, S. A. (2018). Risk for and consequences of endometriosis, a critical epidemiologic review. Best Practice & Research Clinical Obstetrics & Gynaecology, 51, 1–15. 10.1016/j.bpobgyn.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Shen, L. , Zhang, Y. , Zhou, W. , Peng, Z. , Hong, X. , & Zhang, Y. (2018). Circular RNA expression in ovarian endometriosis. Epigenomics, 10(5), 559–572. 10.2217/epi-2017-0079 [DOI] [PubMed] [Google Scholar]
- Song, Y. , Fu, J. , Zhou, M. , Xiao, L. , Feng, X. , Chen, H. , & Huang, W. (2016). Activated Hippo/Yes‐associated protein pathway promotes cell proliferation and anti‐apoptosis in endometrial stromal cells of endometriosis. Journal of Clinical Endocrinology and Metabolism, 101(4), 1552–1561. 10.1210/jc.2016-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. Z. , & Li, J. F. (2018). Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochemical and Biophysical Research Communications, 495(3), 2369–2375. 10.1016/j.bbrc.2017.12.050 [DOI] [PubMed] [Google Scholar]
- Stark, Z. , Bruno, D. L. , Mountford, H. , Lockhart, P. J. , & Amor, D. J. (2010). De novo 325 kb microdeletion in chromosome band 10q25.3 including ATRNL1 in a boy with cognitive impairment, autism and dysmorphic features. European Journal of Medical Genetics, 53(5), 337–339. 10.1016/j.ejmg.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Tomassetti, C. , & D'Hooghe, T. (2018). Endometriosis and infertility, Insights into the causal link and management strategies. Best Practice & Research Clinical Obstetrics & Gynaecology, 51, 25–33. 10.1016/j.bpobgyn.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Tsai, H. W. , Huang, M. T. , Wang, P. H. , Huang, B. S. , Chen, Y. J. , & Hsieh, S. L. (2018). Decoy receptor 3 promotes cell adhesion and enhances endometriosis development. The Journal of Pathology, 244(2), 189–202. 10.1002/path.5000 [DOI] [PubMed] [Google Scholar]
- Uimari, O. , Rahmioglu, N. , Nyholt, D. R. , Vincent, K. , Missmer, S. A. , Becker, C. , … Zondervan, K. T. (2017). Genome‐wide genetic analyses highlight mitogen‐activated protein kinase (MAPK) signaling in the pathogenesis of endometriosis. Human Reproduction, 32(4), 780–793. 10.1093/humrep/dex024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. T. , Sun, Y. M. , Huang, W. , He, B. , Zhao, Y. N. , & Chen, Y. Q. (2016). Genome‐wide long non‐coding RNA analysis identified circulating LncRNAs as novel non‐invasive diagnostic biomarkers for gynecological disease. Scientific Reports, 6, 23343 10.1038/srep23343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Li, Y. , Yang, Z. , Liu, K. , & Wang, D. (2015). Genome‐wide microarray analysis of long non‐coding RNAs in eutopic secretory endometrium with endometriosis. Cellular Physiology and Biochemistry, 37(6), 2231–2245. 10.1159/000438579 [DOI] [PubMed] [Google Scholar]
- Weng, W. , Wei, Q. , Toden, S. , Yoshida, K. , Nagasaka, T. , Fujiwara, T. , … Goel, A. (2017). Circular RNA ciRS‐7‐a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Journal of Clinical Cancer Research, 23(14), 3918–3928. 10.1158/1078-0432.CCR-16-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. T. , Wang, J. , Chen, Y. W. , Zhou, K. , Wen, J. , Wang, Y. , … Cai, W. (2014). Up‐regulation of miR‐200b in biliary atresia patients accelerates proliferation and migration of hepatic stellate cells by activating PI3K/Akt signaling. Cellular Signalling, 26(5), 925–932. 10.1016/j.cellsig.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Xing, Y. Q. , Li, A. , Yang, Y. , Li, X. X. , Zhang, L. N. , & Guo, H. C. (2018). The regulation of FOXO1 and its role in disease progression. Life Sciences, 193, 124–131. 10.1016/j.lfs.2017.11.030 [DOI] [PubMed] [Google Scholar]
- Xiong, Y. , Liu, Y. , Xiong, W. , Zhang, L. , Liu, H. , Du, Y. , & Li, N. (2016). Hypoxia‐inducible factor 1α‐induced epithelial‐mesenchymal transition of endometrial epithelial cells may contribute to the development of endometriosis. Human Reproduction, 31(6), 1327–1338. 10.1093/humrep/dew081 [DOI] [PubMed] [Google Scholar]
- Xu, X.‐X. , Jia, S.‐Z. , Dai, Y. , Zhang, J.‐J. , Li, X.‐Y. , Shi, J.‐H. , … Lang, J.‐H. (2018a). Identification of circular RNAs as a novel biomarker for ovarian endometriosis. Chinese Medical Journal, 131(5), 559–566. 10.4103/0366-6999.226070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Jia, S.‐Z. , Dai, Y. I. , Zhang, J.‐J. , Li, X. , Shi, J. , … Lang, J. (2018b). The relationship of circular RNAs with ovarian endometriosis. Reproductive Sciences, 25, 1292–1300. 10.1177/1933719118759439 [DOI] [PubMed] [Google Scholar]
- Yotova, I. Y. , Quan, P. , Leditznig, N. , Beer, U. , Wenzl, R. , & Tschugguel, W. (2011). Abnormal activation of Ras/Raf/MAPK and RhoA/ROCKII signaling pathways in eutopic endometrial stromal cells of patients with endometriosis. Human Reproduction, 26(4), 885–897. 10.1093/humrep/der010 [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Ren, C. , Xiao, Y. , Xia, X. , & Fang, X. (2018). Expression profile analysis of circular RNAs in ovarian endometriosis by microarray and bioinformatics. Medical Science Monitor, 24, 9240–9250. 10.12659/MSM.913885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. R. , Yang, T. T. , & Xiao, J. J. (2018). Circular RNAs, promising biomarkers for human diseases. EBioMedicine, 34, 267–274. 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Yang, H. , Xuan, Y. U. , Luo, Z. , Lin, Q. , Zhao, J. , … Zhao, X. (2015). Increased expression of fibroblast growth factor receptor1 in endometriosis and its correlation with endometriosis‐related dysmenorrhea and recurrence. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 184, 117–124. 10.1016/j.ejogrb.2014.11.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


