Abstract
Background
Whole-genome sequencing (WGS) is a powerful method for revealing the diversity and complexity of the somatic mutation burden of tumours. Here, we investigated the utility of tumour and matched germline WGS for understanding aetiology and treatment opportunities for high-risk individuals with familial breast cancer.
Patients and methods
We carried out WGS on 78 paired germline and tumour DNA samples from individuals carrying pathogenic variants in BRCA1 (n = 26) or BRCA2 (n = 22) or from non-carriers (non-BRCA1/2; n = 30).
Results
Matched germline/tumour WGS and somatic mutational signature analysis revealed patients with unreported, dual pathogenic germline variants in cancer risk genes (BRCA1/BRCA2; BRCA1/MUTYH). The strategy identified that 100% of tumours from BRCA1 carriers and 91% of tumours from BRCA2 carriers exhibited biallelic inactivation of the respective gene, together with somatic mutational signatures suggestive of a functional deficiency in homologous recombination. A set of non-BRCA1/2 tumours also had somatic signatures indicative of BRCA-deficiency, including tumours with BRCA1 promoter methylation, and tumours from carriers of a PALB2 pathogenic germline variant and a BRCA2 variant of uncertain significance. A subset of 13 non-BRCA1/2 tumours from early onset cases were BRCA-proficient, yet displayed complex clustered structural rearrangements associated with the amplification of oncogenes and pathogenic germline variants in TP53, ATM and CHEK2.
Conclusions
Our study highlights the role that WGS of matched germline/tumour DNA and the somatic mutational signatures can play in the discovery of pathogenic germline variants and for providing supporting evidence for variant pathogenicity. WGS-derived signatures were more robust than germline status and other genomic predictors of homologous recombination deficiency, thus impacting the selection of platinum-based or PARP inhibitor therapy. In this first examination of non-BRCA1/2 tumours by WGS, we illustrate the considerable heterogeneity of these tumour genomes and highlight that complex genomic rearrangements may drive tumourigenesis in a subset of cases.
Keywords: familial breast cancers, BRCA1, BRCA2, whole-genome sequencing, mutation signatures
Key Message
We demonstrate the role for matched germline/tumour whole-genome sequencing and somatic mutational signatures to give insight into the aetiology of high-risk familial breast cancer. We illustrate how the approach can improve the diagnosis and management of individuals and their families by clarifying germline pathogenicity and enhancing the selection of platinum-based or PARP inhibitor therapy.
Introduction
Approximately 15%–20% of breast cancers (BCs) are associated with a strong family history of the disease. Pathogenic variants in BRCA1, BRCA2 or other moderate to highly penetrant susceptibility genes (e.g. TP53, ATM, CHEK2, PALB2 and PTEN) account for <50% of familial BC and thus for the majority of families the underlying genetic contribution to their cancer risk remains unknown. Approximately 10% of individuals may benefit from the identification of germline pathogenic variants using multigene panel sequencing; however, little insight is gained for a large number of individuals and there is an increasing identification of variants of uncertain significance (VUS) [1]. Germline testing is also impacting therapy, since carriers of germline pathogenic BRCA1/2 variants derive enhanced benefit from platinum-based chemotherapy or poly(ADP-ribose) polymerase inhibitors (PARPi) [2–7].
WGS detects a broad repertoire of somatic and/or germline alterations in an unbiased manner. The frequency and distribution of somatic mutations serve as an imprint, or signature, of mutational processes or exposures that contribute to tumour development [8–10]. Some somatic mutational signatures are strongly linked to pathogenic germline variants in risk genes that play functional roles in DNA repair; for instance, homologous recombination (HR; BRCA1, BRCA2), mismatch repair (MLH1, MSH2, MSH6) or base excision repair (BER; MUTYH) [7–9, 11]. In BC, 12 substitution and 6 structural rearrangement signatures were identified [9]. The combination of these signatures was used to develop HRDetect, a robust predictor of BRCA-deficiency and hence PARPi benefit [7].
Methods
We analysed 78 tumours from high-risk familial BC patients to investigate how WGS could impact the management of both risk and therapy for individuals and their families. Cases were carriers of a BRCA1 pathogenic germline variant (n = 26), a BRCA2 pathogenic germline variant (n = 22) or neither (non-BRCA1/2, n = 30) (supplementary Table S1, available at Annals of Oncology online). In order to characterise the somatic landscape of these cases, matched germline/tumour DNA underwent WGS using Illumina X-Ten sequencing to an average fold depth of 34× and 68×, respectively. WGS data were analysed to characterise somatic mutations [single nucleotide variants (SNVs), insertions-deletions, structural variants, copy number], mutational signatures and measures of HR-deficiency (HRDetect, HRD Index) (supplementary Table S2, available at Annals of Oncology online). This approach highlighted important mechanisms of genomic instability that underly familial BC.
Please refer to supplementary Material, available at Annals of Oncology online for details.
Results
Somatic landscape of familial BC
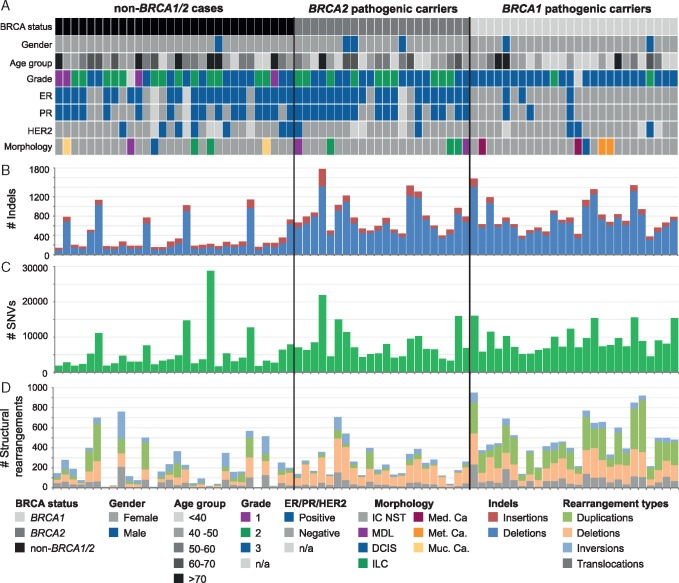
The somatic mutational landscape differed between tumours from BRCA1, BRCA2 and non-BRCA1/2 carriers (Figure 1; supplementary Figure S1, available at Annals of Oncology online). Seventy-nine out of the 93 previously identified BC driver genes [9] were mutated in at least one tumour; including a high frequency of mutations in TP53 (88%) in BRCA1-tumours and GATA3/PIK3CA (50%) in non-BRCA1/2-tumours (supplementary Figure S2, Tables S3 and S4, available at Annals of Oncology online). BRCA1- and BRCA2-tumours harboured more small deletions and SNVs compared with non-BRCA1/2 tumours; and BRCA1-tumours exhibited a higher number of structural rearrangements including duplications and translocations compared with BRCA2 and non-BRCA1/2 tumours (both P ≤ 0.001, Mann–Whitney U test).
Figure 1.
Somatic mutational landscape of 78 familial breast cancers grouped by BRCA status determined by original clinical diagnosis. (A) Clinical information for each sample includes: germline pathogenic variant status from clinical testing of BRCA1 and BRCA2 genes, gender, age at diagnosis, tumour morphological type, histological grade and biomarker status for estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). (B) The number of somatic indels per sample. (C) Number of somatic single nucleotide variants (SNVs). (D) Number and type of somatic structural rearrangements. IC NST, Invasive Carcinoma No Special Type; MDL, Mixed Ductal-Lobular Carcinoma; DCIS, Ductal Carcinoma In Situ; ILC, Invasive Lobular Carcinoma; Med. Ca., Medullary Carcinoma; Met. Ca., Metaplastic Carcinoma; Muc. Ca., Mucinous Carcinoma; #, number; n/a, not available.
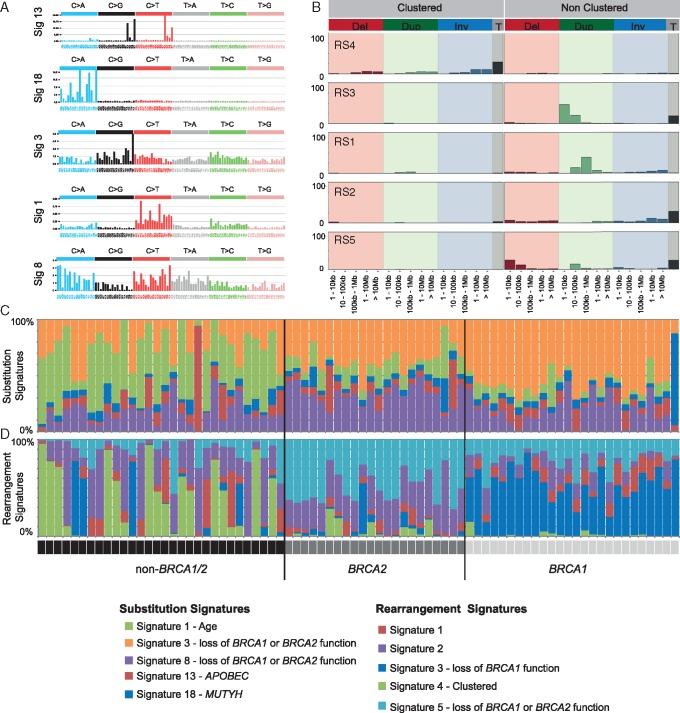
Five substitution and five rearrangement signatures [8, 9] were identified (Figure 2; supplementary Figure S3, available at Annals of Oncology online). BRCA1-tumours had the highest proportion of substitution signature 3 and rearrangement signature 3; while BRCA2-tumours had the highest burden of substitution signature 8 and rearrangement signature 5 (both P ≤ 0.001, Mann–Whitney U test).
Figure 2.
Somatic mutational signatures in familial breast cancer. (A) Five substitution mutational signatures were identified and cosine similarity was used to compare the signatures to known signatures in COSMIC (Mutational Signatures v2 - March 2015; signatures were assigned based on highest similarity). (B) Five somatic rearrangement signatures were identified and cosine similarity was used to compare to rearrangement signatures previously reported in breast cancers [9]. Rearrangements were grouped as clustered in the genome or not, then grouped by type: deletion (Del), duplication (Dup), inversion (Inv), or translocations (T); and then by size (as indicated on the x-axis). (C) The proportion of each substitution signature present per tumour (see colour coding in legend): Tumours from BRCA1 carriers had a higher proportion of substitution signature 3 (orange); tumours from BRCA2 carriers had a higher proportion substitution signature 8 (purple); tumours from non-BRCA1/2 cases had heterogeneous patterns of signatures, but a high proportion of substitution signature 1 (previously associated with age; green). One tumour had a dominant signature 18 (MUTYH, blue) and one tumour had a prominent signature 13 (APOBEC, red). (D) The proportion of each rearrangement signature per tumour (see colour coding in legend): Tumours from BRCA1 carriers had a higher proportion of rearrangement signature 3 (blue), tumours from BRCA2 carriers had higher proportion of rearrangement signature 5 (light blue); tumours from non-BRCA1/2 cases had heterogeneous patterns of rearrangement signatures, but the highest proportion of rearrangement signatures 4 (green) and 2 (purple).
The mutation profile of non-BRCA1/2-tumours was diverse, suggesting a likely heterogeneous aetiology. Tumours had: (i) quiet genomes with few somatic mutations, (ii) a high SNV burden associated with APOBEC substitution signature [8], (iii) a high burden of rearrangement signature 4 (clustered rearrangements), or (iv) mutational signatures suggesting BRCA1/2-deficiency (Figures 1 and 2; supplementary Figures S1–S3 and Table S2, available at Annals of Oncology online).
Somatic mutational signatures to stratify tumours
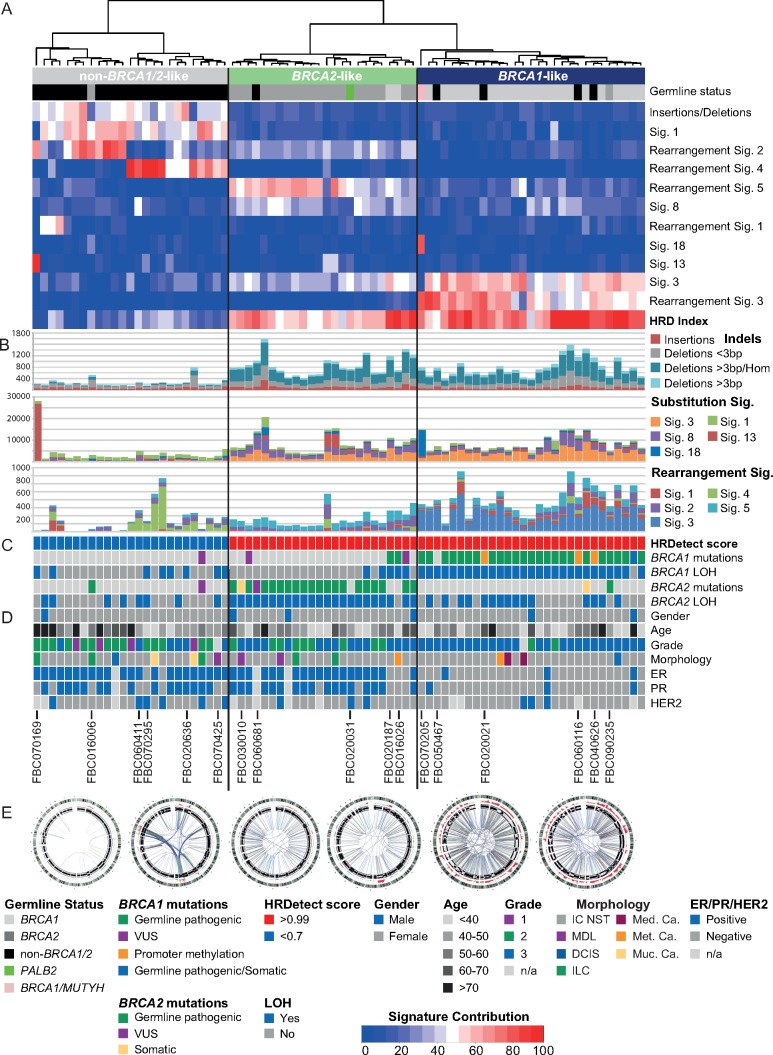
Unsupervised hierarchical clustering based on the contribution of multiple mutational signatures in each tumour stratified the cohort into three groups that broadly captured germline status, hence these groups were termed ‘BRCA1-like’, ‘BRCA2-like’ or ‘non-BRCA1/2-like’ (Figure 3). Ten tumours (13%) clustered away from their ‘original BRCA status’, including two BRCA1-, two BRCA2- and six non-BRCA1/2-tumours. All tumours stratified as ‘non-BRCA1/2-like’ were BRCA-proficient (HRDetect scores <0.7), while all ‘BRCA1-like’ and ‘BRCA2-like’ tumours were BRCA-deficient (Figure 3; supplementary Figure S4 and Table S2, available at Annals of Oncology online). We show that the combination of multiple mutational signatures or HRDetect [7] enabled better classification of the HR status in tumours than when using individual mutational signatures alone (Figure 3; supplementary Figure S4, available at Annals of Oncology online).
Figure 3.
Stratification of tumours using unsupervised hierarchical clustering of the somatic mutational signatures. (A) Hierarchical clustering of the somatic genomic characteristics was based on the percentage contribution of each mutational signature per tumour (see colour coding at bottom), the ratio of insertion to deletions, and the HRD index [3, 4] using the Euclidean method for dissimilarity matrix and Ward.D2 for hierarchical clustering. Tumours were stratified into three groups based mainly on BRCA status (germline carriers of BRCA1 or BRCA2 or non-BRCA1/2) and so the three groups were termed ‘BRCA1-like’, ‘BRCA2-like’, or ‘non-BRCA1/2-like’. (B) Number of mutations in each tumour, including insertion and deletions, SNVs and structural rearrangements coloured by the number of mutations associated with the different mutational signatures. (C) Panel shows HRDetect scores [7] (the cut-off for HR-deficiency in the original study >0.7; all BRCA1- and BRCA2-like tumours had a score >0.99) and the presence of pathogenic germline variants and/or somatic alterations observed in BRCA1 or BRCA2. (D) Clinical information and tumour features according to Figure 1, together with tumour codes for cases discussed in text. (E) Examples of circos plots from each subgroup showing characteristic patterns of structural rearrangements. From the left: FBC050727 (non-BRCA1/2), FBC070474 (clustered rearrangements), FBC061542 (BRCA2 biallelic inactivation), FBC020031 (PALB2 biallelic inactivation), FBC060031 (BRCA1 biallelic inactivation), FBC060116 (BRCA1 promoter methylation/LOH). VUS, variant of unknown clinical significance; LOH, loss of heterozygosity; n/a, not available.
WGS characteristics of tumours with ‘BRCA1-like’ mutational signatures
Twenty-nine tumours showed ‘BRCA1-like’ mutational signatures; all had biallelic inactivation of BRCA1 and were BRCA-deficient according to HRDetect. Most tumours were grade-3 and triple-negative, but included two histological grade-2, four ER/PR-positive and three HER2-positive tumours (Figure 3).
Previously undetected carriers of dual germline pathogenic variants were discovered by WGS-derived mutational signatures. Case FBC090235 carried a BRCA2 germline pathogenic variant (c.574_575delAT; p.Met192ValfsX13) identified by germline testing. WGS revealed not only mono-allelic loss of BRCA2 but also a BRCA1 germline structural rearrangement (chr17: g.41230286_41236428dup) coupled with somatic loss of the wild-type allele (Figure 3; supplementary Figure S5, available at Annals of Oncology online). Case FBC070205 carried a BRCA1 germline pathogenic variant (c.5244_5245delAA; p.Lys1748fs) and somatic loss of the wild-type allele, and the tumour showed a high contribution of the somatic rearrangement signature 3. However, substitution signatures were dominated by C>A transversions in NpCpA or NpCpT contexts (Figures 2 and 3) previously associated with inactivation of the BER gene MUTYH [11]. We subsequently identified a MUTYH germline pathogenic variant (c.1556G>A; p.Arg519Gln) and somatic loss of the wild-type allele (supplementary Figure S6, available at Annals of Oncology online). The biallelic inactivation of both genes, together with evidence of their mutational signatures suggest that both genes contributed to tumourigenesis.
Four non-BRCA1/2-tumours harboured mutational signatures suggestive of BRCA1 loss-of-function (cases FBC020021, FBC040626, FBC060116, FBC050467) (Figure 3). Each tumour had somatic LOH of BRCA1 and three exhibited somatic BRCA1 promoter methylation (case FBC050467 had insufficient tumour DNA for methylation assessment). BRCA1 promoter methylation was tested in the blood of all four cases and other family members, but all were found to be unmethylated, suggesting that these tumours are likely driven by somatic biallelic inactivation of BRCA1 (supplementary Figure S7, available at Annals of Oncology online). We found no evidence of biallelic inactivation of 52 additional genes involved in the HR pathway [12] that could account for the HR defective signatures (supplementary Tables S3–S6, available at Annals of Oncology online).
WGS characteristics of tumours with ‘BRCA2-like’ mutational signatures
Twenty-four tumours contained ‘BRCA2-like’ signatures and all were BRCA1/2-deficient according to HRDetect. This group included 20 of the 22 BRCA2-tumours, each with biallelic inactivation of BRCA2; 19 ER/PR-positive tumours, 9 grade-2 tumours, and 2 invasive lobular carcinomas (Figure 3).
WGS of non-BRCA1/2 case FBC020031 confirmed the previously identified PALB2 nonsense variant (rs180177132; c.3113G>A p.Trp1038*) and detected somatic loss of the wild-type allele. The tumour was ‘BRCA2-like’ with a high burden of substitution signature 3 and rearrangement signature 5 supporting previous studies linking loss of PALB2 function with BRCA-deficient signatures [7, 10, 13].
Non-BRCA1/2 case FBC060681 harboured a BRCA2 VUS (c.7828G>A p.Val2610Met). The tumour presented somatic loss of the wild-type allele, as well as ‘BRCA2-like’ mutational signatures. The variant was not reported in gnomAD or in the 560 BC genomes cohort [9] and is described by ClinVar (Variation ID: 135816) as ‘Class 3 Uncertain significance’ (supplementary Table S5, available at Annals of Oncology online). The variant is in a highly conserved amino acid and is predicted to create a de novo donor splice-site in exon 17, although no experimental evidence supported this [14]. Protein modelling of the variant predicted a damaging effect on protein structure and function (supplementary Figure S8, available at Annals of Oncology online). No evidence of biallelic inactivation in 52 HR-related genes [12] were identified in this case (supplementary Tables S3–S6, available at Annals of Oncology online).
WGS characteristics of tumours with ‘non-BRCA1/2-like’ mutational signatures
Twenty-five tumours were classified as ‘non-BRCA1/2-like’ and included 24 tumours from non-BRCA1/2 cases and a BRCA2-tumour (FBC016006, NM_000059.3, c.1310_1313delAAGA) that lacked somatic inactivation of the BRCA2 wild-type allele (supplementary Figure S9, available at Annals of Oncology online). Twenty-two tumours were ER/PR-positive; six were HER2-positive; and all were considered BRCA-proficient (Figure 3). Case FBC070169 had a strong APOBEC substitution signature accounting for 94% of the somatic mutations, yet there was no evidence of the APOBEC3A or APOBEC3B germline variants previously associated with this signature [15, 16].
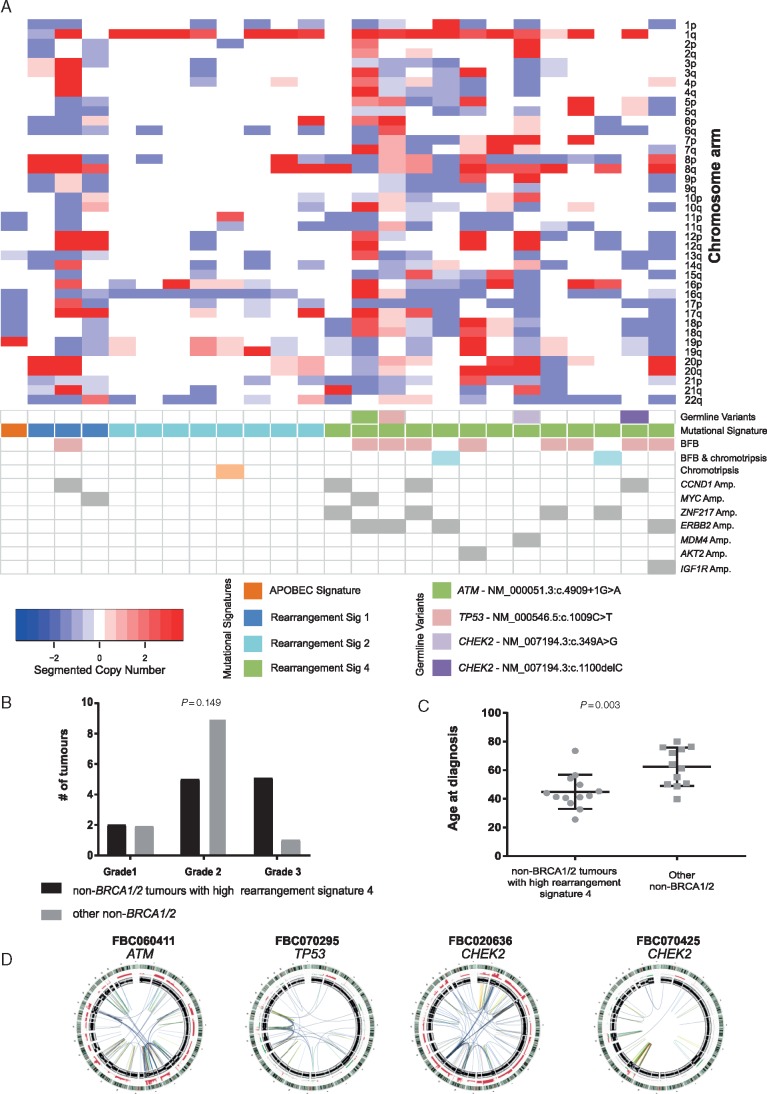
Unsupervised hierarchical clustering stratified the ‘non-BRCA1/2-like’ tumours into two groups based on the contributions of rearrangement signatures 1, 2 and 4 (Figures 3 and 4; supplementary Figure S10, available at Annals of Oncology online). Twelve tumours had either relatively quiet genomes dominated by rearrangement signature 2 (n = 9; with recurrent gain of 1q and/or loss of 16q) or rearrangement signature 1 (n = 3). The remaining thirteen tumours were younger at diagnosis (median age 44 versus 62 years) and displayed a high contribution of rearrangement signature 4, involving complex rearrangements clustered to one or a few chromosomes. These clustered events coincided with amplifications of known BC oncogenes (e.g. ZNF217, ERBB2, CCND1, MYC) [9], and in 8/13 tumours, the patterns of rearrangements suggested evidence for Breakage–Fusion–Bridge (BFB) cycles; two additional tumours had events resembling chromothripsis and BFB (Figure 4). We observed that ‘non-BRCA1/2-like’ tumours had shorter telomeres than BRCA-deficient tumours (supplementary Figure S11, available at Annals of Oncology online) raising the possibility that dicentric chromosome formation due to telomeric erosion could lead to BFB and/or chromothripsis [17, 18]. We were unable to identify germline or somatic variants in candidate genes associated with chromosomal segregation or telomere maintenance that could potentially lead to this pattern of rearrangements. In four cases, however, we identified rare germline variants in TP53 (NM_000546.5: c.1009C>T), ATM (NM_000051.3: c.4909+1G>A) and CHEK2 (NM_007194.3: c.349A>G; NM_007194.3: c.1100delC), together with somatic loss of the wild-type allele, implying functional loss of the relevant protein (Figure 4).
Figure 4.
Whole-genome DNA copy number profile of non-BRCA1/2-like tumours. (A) Chromosome arm level copy number data [gains (red) and losses (blue)] across the genome in non-BRCA1/2-like tumours stratified according to rearrangement signatures 1, 2, and 4. Tumours are plotted in the same order as Figure 3. Tumours are identified which: harbour germline pathogenic variants in risk genes ATM, TP53 or CHEK2; show evidence of Break–Fusion–Bridge and/or chromothripsis; and harbour amplification of various oncogenes. (B) The distribution of histological grade was not significantly different (χ2). (C) Non-BRCA1/2-like tumours with a high proportion of rearrangement signature 4 (clustered rearrangements) were diagnosed at a significantly younger age to non-BRCA1/2 tumours with other genome characteristics (Mann -Whitney U test, two-tailed). (D) Circos plots are shown for the cases with germline variants in ATM, TP53 and CHEK2.
Discussion
This study represents the largest cohort of familial BC cases examined by WGS, and the first report from high-risk, non-BRCA1/2 families. The findings demonstrate the impact that combined germline and somatic WGS can offer as a companion diagnostic in clarifying cancer risk in individuals and in aiding decisions regarding treatment. We confirm the importance of biallelic inactivation of germline risk genes to drive the accumulation of specific patterns of somatic mutational signatures. Loss of function of BRCA1, BRCA2 and PALB2 yield a high mutation burden and specific signatures inferring defective HR [7, 9, 10, 13]. Despite the role of TP53, ATM and CHEK2 in DNA-damage signalling and double-strand breaks detection, these tumours do not show evidence of BRCA-deficient signatures, as previously described [9, 13, 19], but instead they harbour highly complex clustered chromosomal rearrangements.
WGS-derived signatures identified two individuals as carriers of dual germline pathogenic variants (BRCA1/BRCA2 and BRCA1/MUTYH), which were previously unreported. Mutation signatures indicated that biallelic inactivation of BRCA1 rather than BRCA2 was driving tumorigenesis in the BRCA1/BRCA2 case. Clinical BRCA1 and BRCA2 testing (in 2004) identified a pathogenic BRCA2 germline variant (c.574_575delAT; Met192ValfsX13), thus further testing was not pursued. During this study, a family member diagnosed with BC was found not to carry this variant, and more extensive testing revealed the BRCA1 germline rearrangement. Segregation of disease in the BRCA1/BRCA2 family was incomplete until the BRCA1 rearrangement was considered. Co-occurring germline pathogenic variants in BRCA1 and BRCA2 are rare (0.3%, 93/32,295 cases) and carriers are more likely to be diagnosed with BC than BRCA1 or BRCA2 only carriers [20]. Little is known about the incidence and risk profile for BRCA1/MUTYH carriers, and biospecimens from other family members were unavailable, so it was unclear how the variants segregate with disease. However, it is clear that biallelic inactivation of both genes contributed to tumorigenesis. These cases exemplify the importance of thorough germline testing and the power of WGS as a single test to (i) screen entire genes for different types of variants (SNV and rearrangements); and (ii) derive somatic mutational signatures for both variant discovery and delineating the aetiology of disease.
Several lines of evidence support that the BRCA2 VUS (c.7828G>A p.Val2610Met) contributed to tumorigenesis: (i) somatic biallelic inactivation of BRCA2 (VUS and LOH); (ii) the tumour was ‘BRCA2-like’ according to the pattern of mutation signatures and was BRCA-deficient according to HRDetect; (iii) no variants were identified in other HR pathway genes that could explain this BRCA-deficiency; and (iv) protein modelling predicted a negative impact on protein function. This rare variant has only been described once in the literature [14], evaluating the same individual as reported here. Further investigation of the pathogenicity of this rare p.Val2610Met variant is warranted.
Most familial BC fall under the umbrella term of non-BRCA1/2, involving germline variants in other moderate- to highly penetrant genes, or where the underlying genetic cause is unknown. Non-BRCA1/2 tumours exhibit considerable morphological [21], molecular [22] and genomic heterogeneity. WGS provided evidence regarding the drivers of tumorigenesis in many cases, including germline carriers of variants in BRCA2 (VUS), PALB2, CHEK2, ATM and TP53; BRCA1 promoter methylation, APOBEC mutagenesis and oncogene amplification.
The subgroup of ‘non-BRCA1/2-like’ tumours harbouring complex patterns of clustered structural rearrangements was early onset cancers with an ER-positive and/or HER2-positive phenotype. The complex rearrangements were associated with amplification of various oncogenes and showed evidence of BFB and chromothripsis as a potential mechanism driving tumorigenesis [18]. Four cases within this group harboured germline pathogenic variants in ATM [23], CHEK2 [24] or TP53 [25], all with somatic LOH. Chromothripsis and BFB have been reported in numerous tumour types, and intriguingly, germline variants in ATM and TP53 were associated with such complex structural rearrangements in acute lymphoblastic leukaemia [26] and medulloblastoma [27]. Whilst these patterns of rearrangements have been reported in BC [28, 29], they have not been associated with a germline predisposition. Interestingly, >60% of BC in germline TP53 carriers are ERBB2/HER2 amplified [30] and complex structural rearrangements are suggested to drive amplification in such cancers [29].
Better predicting response to therapy is critical to advance oncology. Investigators have used various means to improve the utility of DNA-damaging chemotherapies and PARPi, for instance: the germline status of BRCA1 or BRCA2; somatic LOH of these genes; a triple-negative tumour phenotype; or patterns of somatic mutations [2–7]. We confirm the utility of WGS and HRDetect [7] to enhance the stratification of tumours as being BRCA-proficient versus deficient compared with individual genomic parameters (e.g. HRD score and substitution signature 3). Using this approach, all BRCA1-tumours, 91% of BRCA2-tumours and the PALB2 tumour had bialleic inactivation of said gene and strong evidence of HR-deficiency. These cases would be predicted to be good candidates for platinum-based chemotherapy or PARPi, and included 12 tumours that would not otherwise fall into this recommendation of treatment (i.e. histological grade-2, ER/PR-positive [31], or invasive lobular carcinoma). In contrast to a recent report, only two tumours (2/22, 9% versus 46% [5]) from BRCA2 carriers did not exhibit biallelic inactivation of the gene; one tumour was BRCA-deficient due to the biallelic inactivation of BRCA1, whereas the other was BRCA-proficient, and hence this latter patient would be unlikely to benefit from such therapies.
In summary, matched germline/tumour WGS can improve the identification of the underlying genetic cause of disease over BRCA1 and BRCA2 germline screening alone and that this will likely improve the clinical management of individuals and potentially their families. Furthermore, WGS yields the most robust assessment of BRCA-deficiency and can also identify oncogenic drivers in BRCA-proficient tumours, which collectively may enhance the selection of targeted therapies for patients.
Supplementary Material
Acknowledgements
We wish to thank Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study [which have received funding from the National Health and Medical Research Council (NHMRC, Australia), the National Breast Cancer Foundation (NBCF) Australia, Cancer Australia, and the National Institute of Health (USA)] for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by a grant from the NBCF Australia, and previously by the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. We also thank patients and staff who contributed to the Australian Breast Cancer Tissue Bank and the Brisbane Breast Bank. We acknowledge the support of Metro North Hospital and Health Services in the collection of the Clinical Subject Data and Clinical Subject Materials. We acknowledge the large body of work that has been published in this field that cannot be directly referenced herein.
Data availability: The WGS data have been deposited in the European Genome-phenome Archive (EGA) repository under the accession code 90 (EGAS00001003305).
Funding
The work was funded by NHMRC Australia project grants (APP1080985 and APP1028742) and supported by NHMRC Program grants (APP1017028 and APP1113867). ALD is supported by an Australian Government Research Training Program (RTP) Scholarship. KN is supported by Keith Boden fellowship. NW and ABS are supported by NHMRC senior research fellowships (APP1139071 and APP1061779).
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Couch FJ, Shimelis H, Hu C. et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol 2017; 3(9): 1190–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watkins JA, Irshad S, Grigoriadis A, Tutt AN.. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 2014; 16(3): 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Telli ML, Hellyer J, Audeh W. et al. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res Treat 2018; 168(3): 625–630. [DOI] [PubMed] [Google Scholar]
- 4. Marquard AM, Eklund AC, Joshi T. et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res 2015; 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maxwell KN, Wubbenhorst B, Wenz BM. et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun 2017; 8(1): 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riaz N, Blecua P, Lim RS. et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun 2017; 8(1): 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies H, Glodzik D, Morganella S. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017; 23(4): 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Signatures of mutational processes in human cancer. Nature 2013; 500(7463): 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nik-Zainal S, Davies H, Staaf J. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016; 534(7605): 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waddell N, Pajic M, Patch AM. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518(7540): 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scarpa A, Chang DK, Nones K. et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017; 543(7643): 65–71. [DOI] [PubMed] [Google Scholar]
- 12. Pearl LH, Schierz AC, Ward SE. et al. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 2015; 15(3): 166–180. [DOI] [PubMed] [Google Scholar]
- 13. Polak P, Kim J, Braunstein LZ. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017; 49(10): 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiley PJ, Parsons MT, Leary J. et al. Multifactorial likelihood assessment of BRCA1 and BRCA2 missense variants confirms that BRCA1: c.122A>G(p.His41Arg) is a pathogenic mutation. PLoS One 2014; 9(1): e86836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nik-Zainal S, Wedge DC, Alexandrov LB. et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet 2014; 46(5): 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Middlebrooks CD, Banday AR, Matsuda K. et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet 2016; 48(11): 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crasta K, Ganem NJ, Dagher R. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012; 482(7383): 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glodzik D, Purdie C, Rye IH. et al. Mutational mechanisms of amplifications revealed by analysis of clustered rearrangements in breast cancers. Ann Oncol 2018; 29(11): 2223–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weigelt B, Bi R, Kumar R. et al. The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J Natl Cancer Inst 2018; 110(9): 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rebbeck TR, Friebel TM, Mitra N. et al. Inheritance of deleterious mutations at both BRCA1 and BRCA2 in an international sample of 32, 295 women. Breast Cancer Res 2016; 18(1): 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lakhani SR, Gusterson BA, Jacquemier J. et al. The pathology of familial breast cancer: histological features of cancers in families not attributable to mutations in BRCA1 or BRCA2. Clin Cancer Res 2000; 6(3): 782–789. [PubMed] [Google Scholar]
- 22. Waddell N, Arnold J, Cocciardi S. et al. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res Treat 2010; 123(3): 661–677. [DOI] [PubMed] [Google Scholar]
- 23. Sandoval N, Platzer M, Rosenthal A. et al. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum Mol Genet 1999; 8(1): 69–79. [DOI] [PubMed] [Google Scholar]
- 24. Schmidt MK, Hogervorst F, van Hien R. et al. Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers. J Clin Oncol 2016; 34(23): 2750–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bougeard G, Renaux-Petel M, Flaman JM. et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 2015; 33(21): 2345–2352. [DOI] [PubMed] [Google Scholar]
- 26. Ratnaparkhe M, Hlevnjak M, Kolb T. et al. Genomic profiling of acute lymphoblastic leukemia in ataxia telangiectasia patients reveals tight link between ATM mutations and chromothripsis. Leukemia 2017; 31(10): 2048–2056. [DOI] [PubMed] [Google Scholar]
- 27. Rausch T, Jones DT, Zapatka M. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012; 148(1–2): 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stephens PJ, Greenman CD, Fu B. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011; 144(1): 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferrari A, Vincent-Salomon A, Pivot X. et al. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nat Commun 2016; 7: 12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson JR, Bateman AC, Hanson H. et al. A novel HER2-positive breast cancer phenotype arising from germline TP53 mutations. J Med Genet 2010; 47(11): 771–774. [DOI] [PubMed] [Google Scholar]
- 31. Lips EH, Debipersad RD, Scheerman CE. et al. BRCA1-mutated estrogen receptor-positive breast cancer shows BRCAness, suggesting sensitivity to drugs targeting homologous recombination deficiency. Clin Cancer Res 2017; 23(5): 1236–1241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.