Abstract
RNA nanotechnology employs synthetically modified ribonucleic acid (RNA) to engineer highly stable nanostructures in one, two, and three dimensions for medical applications. Despite the tremendous advantages in RNA nanotechnology, unmodified RNA itself is fragile and prone to enzymatic degradation. In contrast to use traditionally modified RNA strands e.g. 2′-fluorine, 2′-amine, 2′-methyl, we studied the effect of RNA/DNA hybrid approach utilizing a computer-assisted RNA tetra-uracil (tetra-U) motif as a toolkit to address questions related to assembly efficiency, versatility, stability, and the production costs of hybrid RNA/DNA nanoparticles. The tetra-U RNA motif was implemented to construct four functional triangles using RNA, DNA and RNA/DNA mixtures, resulting in fine-tunable enzymatic and thermodynamic stabilities, immunostimulatory activity and RNAi capability. Moreover, the tetra-U toolkit has great potential in the fabrication of rectangular, pentagonal, and hexagonal NPs, representing the power of simplicity of RNA/DNA approach for RNA nanotechnology and nanomedicine community.
Keywords: RNA nanotechnology, Nucleic acid bionanotechnology, RNA 3D motifs, Hybrid nanoparticles
Natural RNA is a biopolymer that folds into complex and functional structures according to the principles of hierarchy. In living organisms, the structures of an RNA complex usually comprise different types of recurrent and modular 3D motifs that have been widely utilized to design self-assembling RNA NPs of diverse architectures and novel functionalities.1–4 The library of RNA motifs is populated by novel artificial sequences obtained by in vitro selection methods and having novel binding selectivity and enzymatic capability.5 The modularity of RNA building blocks and the catalytic features of these molecules together with programmability of its secondary structure with predicted thermodynamic parameters make RNA biopolymers highly attractive material for diverse applications in nanomedicine.
As a matter of fact, a large number of natural RNA motifs are suitable for nano-fabrication6: kissing hairpin loops,7 cognate hairpin loop/loop-receptor pairs,8 paranemic motifs,9 the right angle motif,10 kink-turns,11,12 C-loops,13 three-way14 and multi-helix junctions,15 protein binding motifs,16,17 just to name a few. Perhaps, RNA A-from double helix is the central building block as it provides highly regular supports of variable length for the regulation of other RNA modules arrangement in 3D space allowing creation of novel nucleic-acid-based nanoparticles.
Since the pioneering work on therapeutic RNA nanotechnology by Guo’s group,7 the development of novel nucleic acid-based NPs that possess desirable applications in nanomedicine is of prime importance for the future of RNA nanotechnology and therapeutics. The nucleic acid therapeutics community has begun to accept the concept that RNA displays higher thermodynamic stability than DNA. However, RNA is not only thermodynamically stable but also enzymatically in vivo and in vitro. 1 This is due the fact that RNAs used for therapeutic reagents are those containing chemical modifications such as 2′-fluorine, 2′-amine, 2′-methyl, or Locked Nucleic Acid. Recently, tremendous progress has been made towards that goal. For instance, this foundational approach was employed to generate in vitro self-assembling monomers, dimers, trimers,18,19 2D polygons such as triangles,20 squares,21–24 pentagons,25 hexameric nano-rings,26 heptamers,27 RNA supramolecular assemblies such as RNA filaments,28,29 RNA planar arrays,30,20 and 3D RNA nanoparticles (NPs).31,32 Importantly, RNA nanoparticles can be rationally engineered to fold into thermostable complexes with high assembly efficiency, increased resistance to nuclease digestion, yet possessing functional properties including binding to its ligand, producing siRNA for protein regulatory purposes. Extensive biophysical studies of RNA structural motifs have provided great experimental insight into some of the constraints that dominate RNA folding.33–37 By unraveling the sequence–structure relationship for RNA tertiary folds, researchers have begun to establish the toolkit for rationally designing and constructing more complex and sizeable RNA assemblies.38 The use of RNA nanostructures as delivery vehicles for potential therapeutic RNAs is still a novel direction that holds tremendous promise. Moreover, there is great potential in generating co-transcriptional RNA nanostructures able to serve as multi-purpose self-assembling scaffolds in cells.
Recently, different RNA 3D nano-cubes were modeled utilizing the 3D conformation of A-form helix and different numbers of single strand nucleotide sequence in the so-called helix-centric approach.39 Herein, we have taken the advantage of this approach and have computationally generated an artificial RNA module which utilizes three RNA helices connected via the four single-stranded uracils (ssUs) abbreviated as tetra-U helix linking motif (Figure 1). The overall building block resembles a three-way-junction (3WJ) type RNA secondary structure. However, in contrast to the reported 3D nanocubes, there is no evidence that this building block can be utilized for the construction of the 2D nano-objects e.g., polygons or rings. Both such RNA 2D architectures polygons and nano-rings have recently demonstrated tremendous potential in nanomedicine to be used as nanovehicles to carry out RNA functional moieties such as aptamers, proteins, riboswitches, ribozymes, or short interfering RNAs (siRNAs) to target and treat cancer cells.25,40
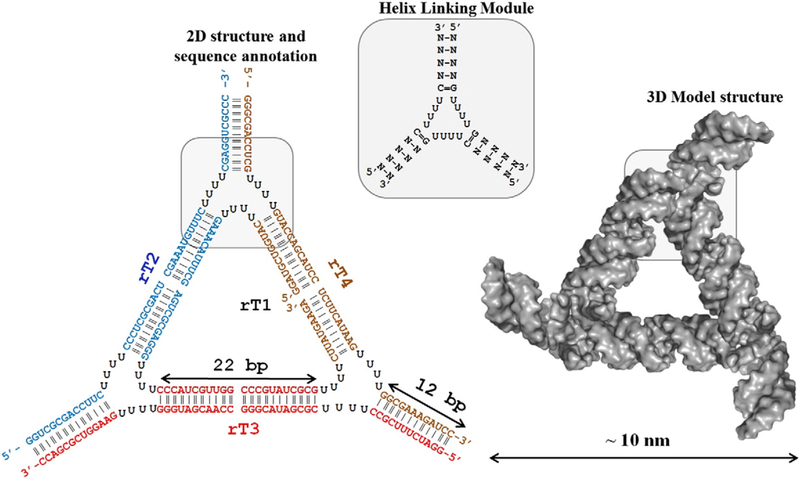
Figure 1.
Overview of theoretical 3D model and 2D structure of RNA triangular nanoparticle designed from RNA 4 × 4 × 4 Us HLM building unit.
In the current approach, the artificial module containing a bulge 4 × 4 × 4 uracils (Us) flexible region and three helical arms, is used as the proof-of-concept in the implementation of this building blocks containing unmodified RNA to fabricate triangular nano-scaffolds from both RNA and DNA strands, as well as their mixtures resulting in a hybrid DNA/RNA nanoparticle (Figure 1). Throughout the text, the 4 × 4 × 4 tetra U building-block will be referred to as “RNA 4Us Helical Linking Motif” or RNA 4Us HLM. We hypothesize that, depending on the nucleic acid inclusion within the NP, we can regulate (i) the thermodynamic stability of the complex, (ii) its resistance to nuclease degradation, (iii) its immunostimulatory activity, and (iv) simultaneous delivery of multiple siRNAs targeting specific genes. This work reevaluates the principles behind the fabrication of RNA nanoparticles by implementing the hybrid RNA/DNA nano-constructs, and will demonstrate significant economic advantages over the use of modified RNA bases. Furthermore, we have obtained preliminary data demonstrating versatility of the RNA 4Us HLM to construct other polygons including square, pentagon and hexagon. This project highlights the capacity of the artificially designed nucleic acid 2D nano-scaffolds to simultaneously deliver specific therapeutics targeting selective human tissues while controlling the formulation of the drugs.
Methods
The detailed method section can be found in Supporting Information.
Triangular nano-scaffolds self-assembly
All triangular complexes RNA, DNA and hybrids were assembled by mixing corresponding nucleic acid strands (1 μM) in TMS (50 mM TRIS pH = 8.0, 100 mM NaCl and 10 mM MgCl2) buffer by heating the mixture to 80 °C and slow cooling (over 1 h) to +4 °C.
Electrophoretic mobility shift assay
The assembled nucleic acid complexes were evaluated on native 7% PAGE. The gel run in 0.5× TBM buffer (45 mM Tris/borate pH = 8.2, 5 mM MgCl2) at room temperature with constant 80 V.
UV-melting experiments
Temperature-dependent absorption measurements were recorded at 260 nm on Agilent spectrophotometer (Agilent Inc.) equipped with a temperature Peltier controller.
Fetal bovine serum degradation assay
Four nucleic acid triangles were preassembled in TMS buffer at the 1 μM concentration prior to incubation with 2% (v/v) FBS (Sigma-Aldrich™) at 37 °C for various times ranging from 0 to 90 min.
AFM imaging and sample preparation
AFM images were acquired at room temperature in tapping mode using silicon probes on 5500 AFM system (Keysight Technologies). Images were processed by PicoView 1.2 software.
Transfection of human cell lines
To assess the delivery and intracellular activity of functionalized triangular nanoparticles, the human breast cancer cell line MDA-MB-231 (with or without GFP) was used. Lipofectamine 2000 was used for all transfections.
Fluorescent microscopy and flow cytometry
The silencing experiments were visualized using a UV 510 confocal microscope (Carl Zeiss, Oberkochen) and a Plan-Neofluar 40×/1.3 Oil lens. Flow Cytometry. BD Accuri C6 flow cytometer was used for all experiments. CellQuest or the CFlow Sampler software was used to retrieve the geometric mean fluorescence intensity (gMFI) and the standard error of the mean.
Immunotoxicity experiments
The human astrocyte-like cell line, U87 MG (ATCC HTB-14) and microglia-like cell line, hHμ, were used in all experiments.
Results
Rationale behind application and design of artificial RNA 4Us helical linking motif
The Protein Data Bank features myriad RNA multi-way junctions available for nanofabrication purposes. Most of them have been cataloged based on their authentic 3D geometries, e.g. 3WJ data base.41 Alternatively, they can be accessed using innovative search tools available under the Nucleic Acid Data Base.42 In general, natural RNA motifs have not been implemented in our studies due to the fact that most of the RNA structures have been folded to their 3D conformations under native conditions. Such folding often requires the use of specific mono- and divalent metal ions,43 proteins,23 a certain pH level,44 the presence of other short- and long-range RNA– RNA interactions etc. Ideally, these factors need to be considered prior the fabrication of de-novo RNA nano-constructs to preserve all canonical Watson–Crick and non-canonical (non-WC) interactions within the 3D geometry of the RNA motif. Indeed, any forces that disturb the native conformation of the motif will result in the instability of the overall complex and, consequently, in unfavorable thermodynamic parameters. For instance, the replacement of one of the RNA strands with a cognate DNA sequence during the nanoparticle fabrication disrupts the 3D geometry of the RNA motif, resulting in significant loss in thermodynamic stability.21
In the current approach, using 4 Us HLM, the main driving force for nanoparticle folding to the desired configuration is manifested by the formation of either A-form or B-form helices via a Watson–Crick (base-pair), referred to as helix-centric approach.39 The design mainly relies on the energy minima driven by the formation of the duplex including RNA/RNA, hybrid RNA/DNA and DNA/DNA. The assumption is that any 3D configuration that could potentially be formed by non-canonical interactions between uracils in 4 × 4 × 4 Us internal loop will acquire much higher ΔG value than formation of a helix, hence helix formation is always favorable and can be treated as rigid modules.
The single-stranded linkers using 4 uracils were chosen for several reasons. First, previous studies utilizing poly-Us as helical linkers were shown to be successful in the construction of functional RNA 3D nano-objects e.g. nanocube.32,39 Second, due to their small size, Us are particularly suitable for co-transcriptional chemical modification, increasing the resistance of the whole NP to ribonuclease activities. Third, computational studies using coarse-grained structure dynamic characterization of different length of Us have revealed that, upon increasing the number of the nucleotides, the overall structure becomes more flexible and, importantly, more stable.39 While previous studies have explored the effect of assembly efficiency only 0–3 Us to link three helices, here we utilize 4 Us with the main assumption that this will provide greater flexibility allowing for the assembly of higher order and more sophisticated polygons.
The edges of the triangle were formed by 22 bp nucleic acid duplexes. The sequences were generated randomly to have average ΔG = −37.0 ± 3.0 kcal/mol per double helix. The RNA 2D folding program mfold45 was extensively utilized to prevent misfolding of the sequences and formation of alternative stable conformations. Each edge possess two complete RNA turns resulting in a length of 6.4 nm (3.2 nm/11 bp). Each vertex of the triangle contains one helical turn (11 bp) to additionally stabilize the overall architecture. Moreover, to prevent the duplex from opening on the vertex, at least two GC/GC base pairs were included at the ends of triangle vertices. Based on the model structure, the triangle has equilateral geometry and is symmetric as well as planar. It is ~10 nm in size and provides three helical branches for the potential inclusion of three different (or same) RNA functional groups. Importantly, the regiospecificity of the functional groups can be easily regulated within the triangular nano-scaffold by design preferences. In addition, during folding, the core architecture is designed to remain in its original triangular shape, allowing for each functional group to fold to their authentic structures with specific functionality at each vertex. We expect that utilizing a “one-pot” assembly of different RNA strands will result in nanoparticle with addressable properties. For example, to make therapeutic RNA nanoparticles for medicinal purposes, one might include targeting (RNA aptamers), treatment (RNA interference by siRNA and miRNA) or detection modules (fluorophores) within one defined nanostructure to combat a disease.
Nucleic acid triangular nanoscaffolds assembly and characterization
For this study, we fabricated four RNA triangles that differ in their nucleic acid composition. The RNA NP is composed of RNA strands named rT1 (central strand), rT2, rT3, and rT4 (Figures 1 and 2, D structure). The DNA triangle has been constructed from DNA strands exclusively using dT1 (central strand) dT2, dT3, and dT4. The two hybrid triangles RNA/ DNA-center (or DNA-center) and DNA/RNA-center (or RNA-center) made from mixture of RNA and DNA as follows: RNA/DNA-center has dT1, rT2, rT3, rT4 the DNA/RNA-center hybrid is made of rT1, dT2, dT3, and dT4. Thus, by varying the central strand within RNA triangle or DNA triangle, we are aiming to obtain NPs possessing different physicochemical properties and selecting a triangle candidate for subsequent in vitro studies.
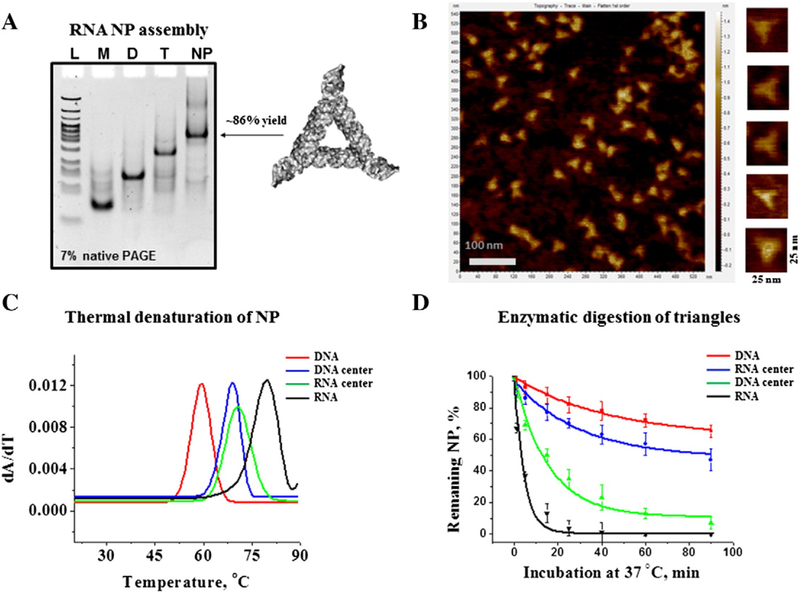
Figure 2.
RNA NP characterization. (A) Self-assembly properties of the triangle RNA were evaluated by 7% native PAGE in 0.5× TBM running buffer. Lane L is DNA ladder (Low Molecular Weight, New England Biolabs); Lanes M, D, and T are RNA monomer (rT2), dimer (rT2 and rT3), and trimer (rT2, rT3, and rT4) complexes, respectively. Lane NP (nanoparticle) contains fully assembled RNA triangle complex composed of all four RNA strands. The gel was stained in EtBr solution for the total RNA visualization. (B) RNA triangle AFM images acquired in air at ambient temperature. (C) Example of first derivatives of UV-melting curves of RNA, DNA and two hybrid RNA/DNA and DNA/RNA nanoparticles. The peaks correspond to Tm values of each individual NP. (D) Time dependent triangle NP (1 μM each) degradation profiles, obtained by incubation in 2% (v/v) fetal bovine serum at 37 °C demonstrating different decay rates. The data represent mean values of at least three independently repeated experiments.
The RNA triangular NPs self-assemble from an equimolar mixture (1 μM each) of 4 nucleic acid strands under physiological condition using 1× TMS buffer with subsequent heating and cooling processes (annealing); all assembly experiments were monitored on 7% native PAGE (Figure 2, A, and Supplementary Figure S1). The gradually decreasing mobility of RNA species on the gel upon addition of corresponding strands indicated the formation of monomers (M), dimers (D), trimers (T) and fully assembled triangles (tetramers). In addition, to evaluate the participation of all four RNA strands within the tetramer complex we conducted radioactive labeling assays, where NPs were assembled using radiolabeled (gamma P-32 ATP) strands enabling the detection the signal of the labeled strand only. It was found that all RNA strands rT1, rT2, rT3, and rT4 contribute to the formation of the NP, as their individual activities have been detected on the gel corresponding to the RNA tetramer (Supplementary Figure S2).
The yield of the triangle nanoparticle folding was found to be different depending on the composition of the nucleic acid strands within the triangle. Table 1 summarizes the calculated percent yields for each individual NP. The NP formed by only RNA and only DNA strands resulted in almost the same assembly efficiencies of 85.9% and 88.4% respectfully. Triangles formed by hybrid strands have shown significantly different assembly yields: 90.6% DNA/RNA-center and 76.6% RNA/DNA-center (presumably because the affinity of three DNA strands to the central RNA is higher than three RNA strands to DNA). Nevertheless, the assembly experiment demonstrated that all NPs have a high efficiency of assembly in solution.
Table 1.
Physicochemical features of triangle NP.
| NP type | Assembly yield, % | Melting point (TM), °C | Decay constant (τ), min−1 |
|---|---|---|---|
| RNA | 85.9 ± 7.2 | 79.2 ± 0.2 | 4.6 ± 0.8 |
| RNA-center | 76.6 ± 3.3 | 71.0 ± 1.5 | 30.3 ± 8.2 |
| DNA-center | 90.6 ± 3.8 | 68.8 ± 1.2 | 15.7 ± 3.2 |
| DNA | 88.4 ± 6.9 | 59.5 ± 2.0 | 44.1 ± 9.2 |
Further evidence that the RNA complex folded into a triangular architecture was obtained from AFM experiments. The 50 nM gel-purified RNA nanoparticles were deposited on a freshly cleaved mica (Ted Pella) surface pre-treated with 1 mM NiCl2, air dried, and imaged on AFM 5500 (Keysight Technologies). Consistent with the theoretical model structures, the AFM images of RNA complexes revealed triangle shaped features (Figure 2, B). The measured average diameter of the triangles was found to be about 12 nm.
Thermal stabilities of triangular nanoscaffolds
Having confirmed the triangular nano-scaffold formation, we then addressed the question of the stability of the NPs in solution with respect to temperature. To achieve this goal, UV-absorbance of the nucleic acid NPs at 260 nm was monitored upon gradually increasing temperature, better known as UV-melting technique.46 Figure 2, C shows melting curves for RNA, DNA and the two hybrid triangles. The melting temperatures (TM) were obtained from the first derivative of resulting curves. All NPs displayed one strong transition and their corresponding TM values are summarized in Table 1. The order of stability decreases in the following direction from the most to the least stable: RNA, RNA/DNA, DNA/RNA and DNA. Accordingly, RNA is the most thermally stable NP (TM = 79.2 ± 0.2 °C) followed by two hybrid NPs. The thermal analysis of the hybrid triangles shows that these complexes have nearly the same TM values around 70 °C. The least stable NP is the triangle containing DNA strands only with the apparent TM = 59.5 °C. Interestingly, the transition temperatures vary by roughly 10 °C between RNA and hybrid NPs and between hybrid NPs and DNA triangle. The difference between the RNA-based triangle and the DNA triangle is significantly higher; the calculated ΔTM = 19.7 °C. Such differences between the thermal stabilities of the RNA NP and DNA NP can be attributed to the difference in thermodynamics between the RNA and DNA helices. In general, a higher melting temperature of RNA relative to the same sequence in DNA is observed under the same conditions.47 The dominant source of the higher energy for RNA is due to modestly better base-stacking energy in the A-form conformation of the helix compared to the B-form of the DNA. Therefore, by changing the ratio of RNA and DNA strands within NP, its thermodynamic stability can be precisely controlled, which demonstrates the advantage of this system for nanofabrication.
Chemical stabilities of triangular nanoscaffolds
The instability of RNA duplexes within the intracellular environment has posed major obstacles to the use of naked RNA strands for drug delivery purposes. To increase the life span of the RNA based nanovehicles in vivo, several methods have been introduced including modification RNAs with 2′-fluoro pyrimidines14,20,48, complexation RNA with proteins23, introduction DNA/RNA hybrid approach.21,49,50
Next, it was of particular interest to evaluate the behavior of the triangular NPs in presence of nucleases. The NPs were incubated in 2% FBS at 37 °C for various time points. Figure 2, D shows the fraction of remaining NPs with respect to incubation time. Supplementary Figures S3 and S4 show some raw data of the conducted experiment. The results suggest that the stability of the nanoparticles in FBS is highly dependent on the nucleic acid composition of the triangular complex. While the RNA NPs completely degraded after 1.5 h, the RNA‐center and DNA NPs were found to be stable (Figure S3). Although DNA‐center hybrid NP degrades much more rapidly than the RNA‐center hybrid NP, it is more stable overall than the RNA NP. To assess the NPs’ relative degradation rates, their corresponding decay constants were calculated assuming an exponential decay process. Table 1 summarizes the decay constants gleaned for the individual NPs. The data suggest that RNA triangle degrades ~10 times faster than its DNA counterpart, while the RNA‐center hybrid triangle is two times less stable than its DNA‐center counterpart. These results, taken together, suggest that the NPs’ resistance to blood serum can be increased by simply varying the ratio of DNA strand over RNA during the NP fabrication.
Collective results obtained from thermal and enzymatic stability analysis suggest the following tendency: upon increasing the numbers of DNA strands within the triangle complex, thermal stability decreases and enzymatic stability increases. These data can be further implemented to design a triangular scaffold with finely tuned properties, as discussed below.
Immunostimulatory activities of triangular nano-scaffolds
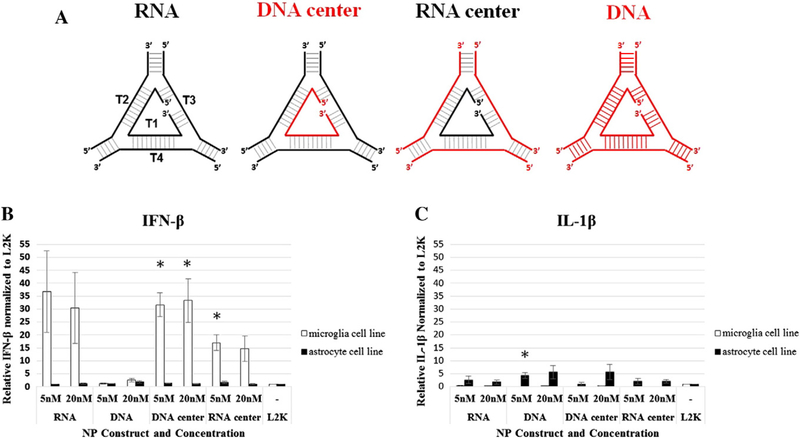
To determine the immunostimulatory activity of NPs, we assessed the release of inflammatory mediators from human astrocyte-like and microglia-like cells transfected with the triangle NPs. In the CNS, microglia and astrocytes are important for the development of protective immune responses and potentially damaging neuroinflammation. These cell types use pattern recognition receptors such as Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD)-like receptors (NLRs), and retinoic acid inducible gene (RIG)-like receptors (RLRs) to recognize pathogen-associated molecular patterns (PAMPs). In response to PAMPs, stimulated microglia and astrocytes release inflammatory and anti-viral mediators such as IL-6, IL-8, TNF-α, IL-1β, and IFN-β.51,53 The profile of inflammatory mediators released in response to NP transfection was dependent on nucleic acid composition (Figure 3, A) and cell type. Similar to recognition of viral RNA and DNA, we predicted NPs would be recognized by a combination of TLRs, NLRs, and RLRs resulting in an anti-viral response. In support of this hypothesis, we primarily observed an elevated release of IFN-β from human microglia-like cells. Interestingly, RNA/ DNA and DNA/RNA hybrid triangles significantly increased the release of IFN-β (Figure 3, B). The RNA triangle modestly increased release of IFN-β, although this was not statistically significant (Figure 3, B). In contrast, we observed no release of IFN-β in response to the DNA triangle suggesting the necessity of RNA to induce an IFN response (Figure 3, B). Additionally, in response to transfection with all NPs human astrocyte-like cells released modestly elevated levels of IL-1β (Figure 3, C). Only the DNA triangle caused a statistically significant elevation of IL-1β release. Regardless of NP nucleic acid composition or themicroglia cell type transfected, we observed no significant increase in the release of the inflammatory mediators IL-6 and IL-8 (Supplementary Figure S5) and no detectable release of TNF-α (data not shown). Together these data demonstrate that NPs are immunostimulatory and the composition of the NPs can alter the degree and profile of inflammatory mediators released. These data suggest the exciting potential of NPs to be eloquently designed to limit immune cell stimulation or serve as an adjuvant to elevate protective interferon responses and reduce damaging inflammatory response.
Figure 3.
Nanoparticle immunostimulatory activity. Human astrocyte-like and microglia-like cell lines were transfected with NPs at a final concentration of 5 nM and 20 nM. Cell supernatants were collected at 24 h post transfection and levels of IFN-β and IL-1β assessed by specific-capture ELISA. Differences in inflammatory mediator release between cells transfected with NPs and the L2K control were compared with Student’s two-tailed t test. Asterisks indicate a statistically significant differences (P value <0.05).
RNA/DNA-center hybrid nano-scaffold carrying siRNA demonstrate efficient gene silencing properties
To confirm that triangular NPs can be used as programmable nano-scaffolds for controllable cargo delivery, the RNA/ DNA-center triangles were functionalized with three siRNAs and fluorescent dyes. The formation of functionalized triangles was confirmed (Figure 4, A) and further tested in various human cells (Figure 4, B-E and Supporting Figure S7). As the proof of concept, green fluorescent protein (GFP), was chosen as target for siRNAs delivered with triangular NPs. To guarantee the intracellular release of siRNAs upon dicing, NPs were decorated with dicer substrate RNAs (DS RNAs)52 that have been used in multiple prior studies.40,54,55 Assembled functionalized NPs were further used for experiments with human breast cancer cells. In all cell culture experiments, the equivalent concentrations of free DS RNAs were used as a control.
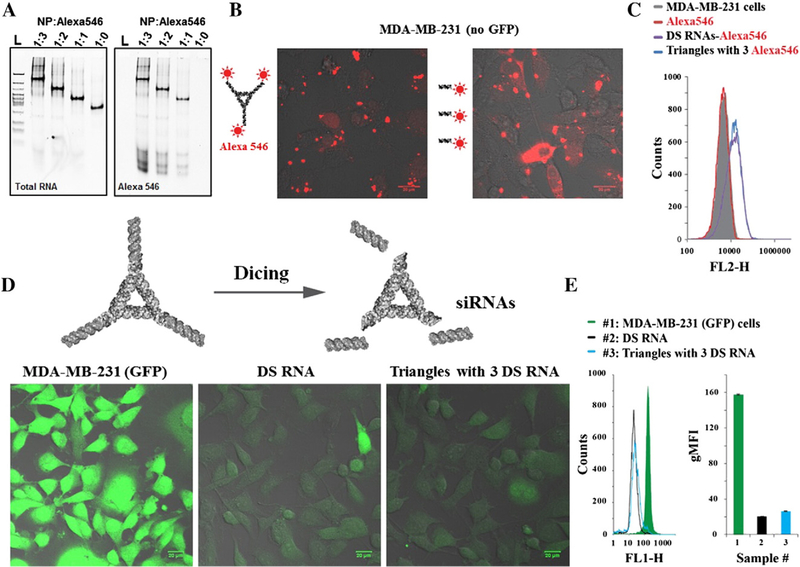
Figure 4.
Uptake and gene specific silencing experiments using human breast cancer cell line (MDA-MB-231) with and without GFP. (A) Native 7% PAGE experiment demonstrating sequential assembly of triangle NP with 1 μM, 2 μM, and 3 μM DNA-Alexa 546 oligodeoxynucleotide (ODN) per 1 μM NP, respectively. The gel was first scanned to detect signals from ODN-Alexa546 conjugates (left) then stained using EtBr for total nucleic acid imaging. Fluorescent microscopy (B) and flow cytometry (C) of non-fluorescent cells 24 h post-transfection with Alexa 546 labeled triangles (5 nM final) and DS RNAs (15 nM final) are shown. GFP expressing cells were analyzed three days post-transfection with functionalized triangle (20 nM final) and DS RNA (60 nM final) by fluorescent microscope (D) and flow cytometry (E). Error bars denote ±S.E.M.
To evaluate the relative cellular uptake efficiencies, human breast cancer cells were transfected with fluorescently labeled nanoparticles (Figure 4, B and C). The results analyzed by fluorescent microscopy and flow cytometry demonstrate that the transfection efficiencies for nanoparticles are comparable to the uptake of free fluorescently labeled DS RNAs tested at three-times higher concentration.
To confirm the successful dicer-assisted intracellular release of siRNAs and further activation of RNA interference, specific gene silencing experiments were carried out (Figure 4, D and E). As a proof of concept, GFP expressing breast cancer cells (MDA-MB-231/GFP) were transfected with NPs functionalized with three anti-GFP siRNAs and compared to free anti-GFP DS RNAs (Figure 4, C and D). The combined results show GFP silencing of ~90% with comparable efficiencies for triangles and DS RNA at nanomolar concentrations. However, at lower picomolar concentrations, significant GFP silencing was also observed (Supporting Figures S7, and S8 for negative controls). All the results clearly indicate that the RNA triangles can be used as scaffolds for controlled delivery of siRNAs and fluorescent probes to the diseased human cells.
Discussion
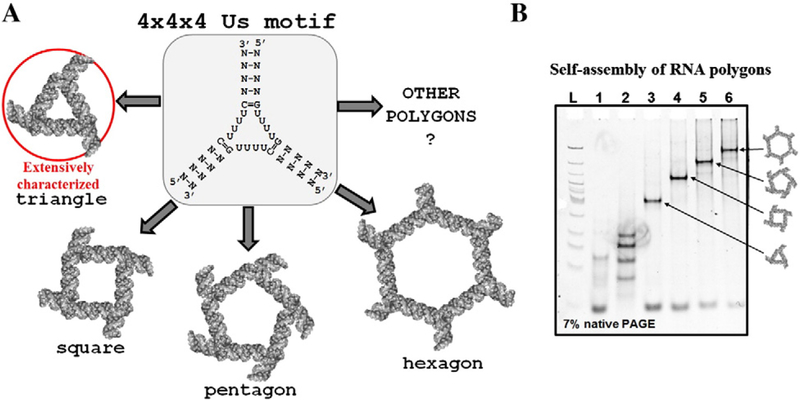
Precise control of RNA nanoparticle properties such as shape and size at nanometer levels is one of the most prominent goals of RNA nanotechnology. The progression in the field of RNA nanotechnology has begun achieving this goal as various elegant RNA nano-architectures using computer-aided 3D model software and RNA 2D folding programs have been recently reported.56–58 The novel multipurpose structural RNA 3D motifs are desirable to promote the art of nanofabrication to the more sophisticated level. We hypothesized that 4 × 4 × 4 tetra-U HLM can be further used to fabricate other nanoparticles, such as a square, pentagon, hexagon, heptagon, octagon etc. demonstrating its value as a toolkit in RNA nanotechnology (Figure 5, A).
Figure 5.
Versatile features of the 4 × 4 × 4 tetra-U linking motif. (A) Conceptual view of application of the 4 × 4 × 4 tetra-U HLM to fabricate 2D nucleic acid polygons. (B) Preliminary data obtained by gel shift assay suggest the formation of RNA stable complexes of tetragonal, pentagonal, and hexagonal shapes.
Previously, the design and construction of RNA polygons have been undertaken using naturally occurring RNA 3WJ from pRNA with 60° angles that served20,59 as corners of the polygons. A 60° junction is ideal for triangular nanostructures, but not for squares, or pentagons. As the angle of the junctions increases with the number of sides, an artificial stretch is exerted on the native conformation of the RNA 3WJ motif. This results in decreasing stabilities when progressing from a triangle conformation to a square or pentagon. The formation of hexagonal RNA nanostructure has not been achieved25 because of the tension applied to stretch native 60° angle to 120° is too high leading to distortion of the overall complex. In the current approach, the artificial 4 × 4 × 4 uracils (Us) flexible module can be beneficial as there are little, if any, structural hindrance observed while stretching the intra-helical angles. The design principle of RNA polygons using RNA 3WJ motifs is detailed elsewhere.56 Following this strategy, the transition from a triangle NP to a square, pentagon, and even a hexagon has been achieved using the 4 × 4 × 4 tetra-U HLM. The assembled RNA complexes corresponding to polygons have been analyzed by mobility shift assays (Figure 5, B). The formation of homogeneous and distinct RNA bands on native PAGE suggests that RNA strands corresponding to triangle, square, pentagon, and hexagon fold into stable and compact RNA complexes.
So far, we have obtained only these preliminary data demonstrating versatility of the 4 × 4 × 4 tetra-U HLM. The current project is under investigation and there are more details to be addressed before we can draw a conclusion as to whether the artificial module is universal for nanofabrication purposes.
Importantly, enzymatic instability hampers the application of nano-scaffolds assembled from pure RNA strands. This is due to the structural nature of ribonucleic acid: specifically, the hydroxyl group at the 2′-C position makes it susceptible to hydrolysis by nucleases, as well as by divalent metal ions. Thus, a variety of techniques to chemically modify ribose rings have been developed to improve enzymatic resistance and elevate its efficiency in drug delivery.60 However, this makes the production costs of such modified RNA nanoparticles expensive. Hybrid RNA/DNA nanovehicles, on the other hand, are less thermodynamically stable than pure RNA, yet they remain resistant to nucleases in the blood stream for a longer period of time.55 In addition, the production economy of RNA/DNA nanoparticles is significantly favorable. With the current solid-state synthesis approach, DNA strands are available from many vendors for less than ~$0.50/base. It’s worth noting that, while RNA/DNA are native molecules to cells, the chemically modified nucleic acids, specifically those at ribose rings, are alien. Unfortunately, very little to no examination has been conducted on the toxicity aspects of modified RNA nanovehicles.
To conclude, we have demonstrated the design and construction of fine-tunable triangular nanoarchitectures using unmodified de-novo 4 × 4 × 4 tetra-U helix linking motif. The highly versatile properties of the tetra-U HLM were also demonstrated by fabrication of equilateral RNA/DNA polygons such as a rectangle, a pentagon, and a hexagon with a great potential for extension the polygonal structures. The modulation of RNA and DNA strand composition makes it possible to engineer economically advantageous triangular nano-scaffolds with fine-tunable thermodynamic and enzymatic stability as well as immunostimulatory activity that can be used for effective delivery of fluorescent probes and gene-silencing agents to cancer cells. The employment of the hybrid RNA and DNA strands to fabricate functional nanoparticles holds tremendous potential to advance medicine. The technique shown here for a simple design to precisely tune physicochemical properties adds a new angle to exploit RNA/DNA hybrid nanoparticles in a clinical setting. Further work is currently under investigation and rapidly developing with a goal to implement modified synthetic RNA strands and investigate properties of the resulting nanoparticles in vitro and in vivo.
Supplementary Material
Acknowledgments
Funding: Emily Satterwhite was supported by a summer undergraduate research fellowship from the Charlotte Research Scholars program. The work was supported by Department of Chemistry BSU start-up funds and Indiana Academy of Science grant # G9000602A to Emil Khisamutdinov. Also, this work was supported by Department of Chemistry UNCC start-up funds and UNCCS Faculty Research Grant to Kirill Afonin and Ian Marriott. This research has been funded in part with the Federal funds of Dr. Ian Marriott, National Institutes of Health NS050325. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of Interests: The authors declare no competing financial interest.
Associated content
Please refer to supplementary information for detailed description of gene silencing, immunotoxicity experiments as well as RNA and DNA sequences used in this project. This material is available free of charge via the Internet at http://pubs.acs.org.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nano.2016.12.018.
References
- 1.Guo PX. The emerging field of RNA nanotechnology. Nat Nanotechnol 2010;5:833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Lee T, Dziubla T, Pi FM, Guo SJ, Xu J, et al. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 2015;10:631–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabow WW, Jaeger L. RNA self-assembly and RNA nanotechnology. Acc Chem Res 2014;47:1871–80. [DOI] [PubMed] [Google Scholar]
- 4.Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, et al. In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc Chem Res 2014;47:1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. Invitro selection of RNA molecules that bind specific ligands. Nature 1990;346:818–22. [DOI] [PubMed] [Google Scholar]
- 6.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 2006;16:279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 1998;2:149–55. [DOI] [PubMed] [Google Scholar]
- 8.Novikova IV, Hassan BH, Mirzoyan MG, Leontis NB. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res 2011;39:2903–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J Am Chem Soc 2008;130:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabow WW, Zhuang Z, Swank ZN, Shea JE, Jaeger L. The right angle (RA) motif: a prevalent ribosomal RNA structural pattern found in group I introns. J Mol Biol 2012;424:54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Lilley DM. The kink turn, a key architectural element in RNA structure. J Mol Biol 2016;428:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J 2001;20:4214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonin KA, Leontis NB. Generating new specific RNA interaction interfaces using C-loops. J Am Chem Soc 2006;128:16131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol 2011;6:658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA 2006;12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, et al. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol 2011;6:116–20. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Inoue T. RNA and RNP as new molecular parts in synthetic biology. J Biotechnol 2007;132:1–7. [DOI] [PubMed] [Google Scholar]
- 18.Chen CP, Sheng ST, Shao ZF, Guo PX. A dimer as a building block in assembling RNA — a hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem 2000;275:17510–6. [DOI] [PubMed] [Google Scholar]
- 19.Shu D, Huang LP, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J Nanosci Nanotechnol 2003;3:295–302. [DOI] [PubMed] [Google Scholar]
- 20.Khisamutdinov EF, Jasinski DL, Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 2014;8:4771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasinski DL, Khisamutdinov EF, Lyubchenko YL, Guo P. Physico-chemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS Nano 2014;8:7620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett 2009;9:1270–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno H, Inoue T. Designed regular tetragon-shaped RNA-protein complexes with ribosomal protein L1 for bionanotechnology and synthetic biology. ACS Nano 2015;9:4950–6. [DOI] [PubMed] [Google Scholar]
- 24.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. S A 2011;108:6405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J, Guo P. Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res 2014;42:9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 2011;11:878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu Y, Shu D, Haque F, Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat Protoc 2013;8:1635–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res 2006;34:1381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 2004;4:1717–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, et al. Building programmable jigsaw puzzles with RNA. Science 2004;306:2068–72. [DOI] [PubMed] [Google Scholar]
- 31.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat Chem 2010;2:772–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 2010;5:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiore JL, Nesbitt DJ. An RNA folding motif: GNRA tetraloop-receptor interactions. Q Rev Biophys 2013;46:223–64. [DOI] [PubMed] [Google Scholar]
- 34.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res 2003;31:5544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X, Huang L, Lilley DM, Harbury PB, Herschlag D. The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol 2016;12:146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binzel DW, Khisamutdinov E, Vieweger M, Ortega J, Li J, Guo P. Mechanism of three-component collision to produce ultrastable pRNA three-way junction of Phi29 DNA-packaging motor by kinetic assessment. RNA 2016. [DOI] [PMC free article] [PubMed]
- 37.Binzel DW, Khisamutdinov EF, Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 2014;53:2221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halman JR, Satterwhite E, Roark B, Chandler M, Viard M, Ivanina A, et al. Nucleic Acid Res 2017, 10.1093/nar/gkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonin KA, Kasprzak W, Bindewald E, Puppala PS, Diehl AR, Hall KT, et al. Computational and experimental characterization of RNA cubic nanoscaffolds. Methods 2014;67:256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, et al. Multifunctional RNA nanoparticles. Nano Lett 2014;14:5662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res 2008;36:D392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coimbatore Narayanan B, Westbrook J, Ghosh S, Petrov AI, Sweeney B, Zirbel CL, et al. The nucleic acid database: new features and capabilities. Nucleic Acids Res 2014;42:D114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, et al. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 2013;19:1226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goh GB, Knight JL, Brooks III CL. pH-dependent dynamics of complex RNA macromolecules. J Chem Theory Comput 2013;9:935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31:3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philippe Dumas EE, Disdier F, Walter P. UV Melting Studies with RNA. In: Roland AB, Hartmann K, Schön A, Westhof E, editors. Handbook of RNA Biochemistry: second, completely revised and enlarged edition Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2014. [Google Scholar]
- 47.Conte MR, Conn GL, Brown T, Lane AN. Conformational properties and thermodynamics of the RNA duplex r(CGCAAAUUUGCG)2: comparison with the DNA analogue d(CGCAAATTTGCG)2. Nucleic Acids Res 1997;25:2627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haque F, Shu D, Shu Y, Shlyakhtenko LS, Rychahou PG, Evers BM, et al. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 2012;7:245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, et al. Activation of different split functionalities on re-association of RNA–DNA hybrids. Nat Nanotechnol 2013;8:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoerter JA, Krishnan V, Lionberger TA, Walter NG. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PloS One 2011;6:e20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furr SR, Marriott I. Viral CNS infections: role of glial pattern recognition receptors in neuroinflammation. Front Microbiol 2012;3:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by dicer processing of short substrate RNAs. Nucleic Acids Res 2005;33:4140–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Mesa Y, Jay TR, Checkley MA, Luttge B, Dobrowolski C, Valadkhan S, et al. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol 2016, 10.1007/s13365-016-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, et al. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 2011;6:2022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, Hofmann J, et al. Triggering of RNA interference with RNA–RNA, RNA– DNA, and DNA–RNA nanoparticles. ACS Nano 2015;9:251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khisamutdinov EF, Bui MN, Jasinski D, Zhao Z, Cui Z, Guo P. Simple method for constructing RNA triangle, square, pentagon by tuning interior RNA 3WJ angle from 60 degrees to 90 degrees or 108 degrees. Methods Mol Biol 2015;1316:181–93. [DOI] [PubMed] [Google Scholar]
- 57.Parlea L, Bindewald E, Sharan R, Bartlett N, Moriarty D, Oliver J, et al. Ring catalog: a resource for designing self-assembling RNA nanostructures. Methods 2016. [DOI] [PMC free article] [PubMed]
- 58.Sharma A, Haque F, Pi F, Shlyakhtenko LS, Evers BM, Guo P. Controllable self-assembly of RNA dendrimers. Nanomedicine 2016;12:835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bindewald E, Afonin K, Jaeger L, Shapiro BA. Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano 2011;5:9542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bramsen JB, Kjems J. Chemical modification of small interfering RNA. Methods Mol Biol 2011;721:77–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.