Abstract
Yersinia ruckeri is the causative agent of enteric redmouth disease in salmonids. In fish, the intestine represents an important site of nutrient uptake, host–pathogen interactions, and defense. The posterior intestine can be inflamed, reddened, and filled with an opaque, yellowish fluid during Y. ruckeri infection. Herein, we report an investigation on the proteome alteration in the posterior intestinal mucosa of rainbow trout (Oncorhynchus mykiss) after exposure to Y. ruckeri. The intestinal mucosal proteins were identified and quantified by a shotgun proteomic approach by applying data-independent quantification with sequential windowed acquisition of all theoretical mass spectra (SWATH). A total of 437 proteins were found to be differentially up- or downregulated in the posterior intestine. Gene ontology of upregulated proteins pointed to their involvement into exopeptidase, endopeptidase, and hydrolase activities, while the downregulated proteins were involved in lipid metabolism, actin binding, and translation processes. Additionally, upregulated proteins were predicted to be involved in lysosome, oxidative phosphorylation, and metabolic pathways, while downregulated proteins were implicated in focal adhesion, regulation of actin cytoskeleton, protein digestion and absorption pathways. This study showed that Y. ruckeri infection can alter protein abundance involved in serine-type carboxypeptidase, cysteine and aspartic-type endopeptidases, metallopeptidases, antioxidant defense, calcium ion binding, glycolytic and carbohydrate metabolic processes in the proteome of the intestinal mucosa of rainbow trout.
Electronic supplementary material
The online version of this article (10.1186/s13567-019-0673-8) contains supplementary material, which is available to authorized users.
Introduction
Enteric redmouth disease (ERM) is one of the major diseases of mainly salmonid fish. It was first described in the 1950s in rainbow trout (Oncorhynchus mykiss) in the Hagerman Valley of Idaho, USA. The disease is caused by Yersinia ruckeri, a Gram-negative rod shaped enterobacterium, and prevalent in Europe, North and South America, Australia, South Africa, the Middle East, and China [1, 2]. The signs of the disease include exophthalmia, subcutaneous hemorrhages, and splenomegaly [1]. In most cases, the intestine is inflamed, reddened, and filled with an opaque, yellowish fluid [1, 3]. In rainbow trout, up to 25% of infected fish could carry Y. ruckeri in their posterior intestines [4]. Y. ruckeri appeared to adhere or invade gills and gut tissue of rainbow trout [3, 5]. Y. ruckeri can also be seen in the lumen of the intestine, adhering to mucus and invading villi within 30 min after exposure and can be detected 7 days post-exposure (dpe) in the liver, spleen, brain, and heart [6].
The strains of Y. ruckeri have been classified on the basis of biotypes, serotypes (O1, O2, O3, and O4), and outer-membrane protein types [7]. The majority of epizootics in salmonids are caused by the motile serotype O1a [1]. There are two biotypes of Y. ruckeri; biotype 1 is positive for motility and lipase secretion, whereas biotype 2 is negative for both tests [8]. The outbreaks of ERM were recorded in biotype 1-vaccinated salmonids and the isolates associated with these outbreaks were identified as Y. ruckeri biotype 2 [9–12].
In fish, the gastrointestinal (GI) tract is a multifunctional organ, which serves a diverse range of functions from nutrient absorption to ionic and osmotic regulation and even air breathing [13–19]. Gut epithelial cells are protected by a mucus layer, which creates a physical and chemical barrier against an intruder and acts as an important mechanism of innate defense that maintains tissue homeostasis [20]. The GI tract is continuously challenged with food antigens as well as pathogens entering the body via feed and water intake, and acts as the first line of defense against pathogen attachment and invasion [20–22]. The posterior portion of the GI tract is a site where uptake of macromolecules and foreign antigens occurs, which leads to antigen uptake and processing as well as the initiation of a systemic immune response [20, 23]. The disease has been of a major concern in salmonids, which influences the physiological response of fish. Rainbow trout is highly susceptible to ERM [1]. The effect of natural herbal products on growth performance, hematological and biochemical values and resistance to Y. ruckeri infection was studied as a feed additives for rainbow trout [24, 25].
In ERM, hyperemia and hemorrhages throughout the intestinal mucosa in rainbow trout have been observed [1]. The intestinal response of rainbow trout has been mainly studied at the transcription level, especially regarding immune gene expression [26–29]. It is still largely unknown how the gut mucosal proteome of the rainbow trout responds to alternations in the luminal environment triggered by Y. ruckeri. Therefore, in the current study, we aimed to evaluate the possible effects of Y. ruckeri exposure on the modulation of the proteome profile in the posterior intestine of rainbow trout. In order to meet this aim, we applied a label-free shotgun proteomic approach.
Materials and methods
Collecting posterior intestine samples
The details of the experimental setup have been described in our previous study [30]. Briefly, specific pathogen free rainbow trout (15 ± 1 cm) were allocated to 9 aquaria, 18 fish per aquarium. In order to monitor the effect of Y. ruckeri exposure, the following three treatments were given in parallel—Treatment 1: an exposure with biotype 1 (CSF007–82), Treatment 2: an exposure with biotype 2 (A7959–11), and Treatment 3: a mock exposure to sterile tryptic soya broth. In both the test groups, the fish were challenged by bath exposing to 2 × 106 colony forming units (CFU) of Y. ruckeri biotype 1 (CSF007-82) and Y. ruckeri biotype 2 (A7959-11) strains for 2 h. The fish were maintained in a flow-through system supplied with UV-treated ground water at 19 ± 1 °C, monitored daily, and morbid, moribund, and dead fish were taken out immediately from the aquaria. The mortality of fish was only considered to be caused by Y. ruckeri if the bacteria were recovered from the head kidney and confirmed using the MONO-Yr kit (Bionor, Skien, Norway) or PCR [31]. No morbid and moribund fish were sampled for the study. The fish were maintained in starvation 48 h before the sampling. Nine fish from each group were sampled at 3, 9, and 28 dpe. The posterior intestine of each sample was dissected and placed in cold sterile PBS (Sigma-Aldrich, Neustadt, Germany). The intestine was opened along the mesenteric border and washed three times with cold sterile PBS containing a cocktail of mammalian protease inhibitors (Sigma-Aldrich) to remove digesta and fecal matter. Intestinal mucosa was scraped with a sterile large scalpel blade. Each sample was divided into two parts, one immediately snap-frozen in liquid nitrogen for proteomic analysis, and one fixed in RNALater (Sigma-Aldrich) for molecular analysis and stored at −80 °C.
Protein extraction
Equal amounts (30 mg) of nine individual intestinal mucosal samples from each group were pooled randomly as three pools of three samples each in order to minimize the effects of individual variation. It refers to N = 3 for the biological replicates per time point (3, 9 and 28 dpe) and per exposed group (control, Y. ruckeri biotype 1 and biotype 2). Each pool was ground using a sterilized mortar and pestle in the presence of liquid nitrogen to a fine powder, which was then mixed with 800 μL of precooled denaturing lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, and 1% DTT) containing mammalian protease inhibitor cocktail (Sigma-Aldrich). The samples were subjected to ultrasonic disruption for 10 cycles of 10 s pulse-on and 30 s pulse-off. The samples were then centrifuged at 12 000 rpm for 20 min at 4 °C to remove any cellular debris. The supernatant was collected. The protein content was measured using the Pierce 660 nm Protein Assay (Thermo Scientific, Vienna, Austria).
Protein digestion
The protein digestion was performed following a standard enhanced, filter-aided sample preparation protocol (FASP) [32]. Briefly, after washing, proteins were reduced and alkylated. On-filter digestion was performed with 1.2 µg Trypsin/Lys-C mix (Promega, Madison, USA) for 14 h at 37 °C. Digested peptides were recovered, dried, and redissolved in 0.1% aqueous trifluoroacetic acid. After desalting and sample cleanup by C18 ZipTips (Sigma-Aldrich), all samples were spiked with standardized indexed retention time reference peptides for facilitation of retention time alignment (iRT kit, Biognosys, Switzerland). For each liquid chromatography (LC)–mass spectrometer (MS) analysis, 3 µg of digested protein was loaded onto the LC column.
Microflow high-performance liquid chromatography electrospray-ionisation quadrupole time-of-flight tandem mass spectrometry (micro LC ESI QTOF MS/MS)
Micro LC ESI QTOF MS/MS was carried out as described earlier [30]. Briefly, peptides were separated on an Eksigent NanoLC 425 system using a microflow pump module (Sciex, Concord, Canada). The samples were pre-concentrated and desalted on a 5 mm YMC-Triart C18 precolumn using ultra-pure LC–MS grade water with 0.1% formic acid as a mobile phase and a flow rate of 10 μL/min. Desalted peptides were separated on a 15 cm YMC-Triart C18 column at a flow rate of 5 µL/min. Mobile phase A was ultra-pure water with 0.1% formic acid, whereas mobile phase B consisted of acetonitrile with 0.1% formic acid. The gradient started with 3% B and increased in two steps to 25% B (68 min) and 35% (73 min) followed by a washing step with 80% B. Total acquisition time was 87 min. For mass spectrometric analysis, LC was directly coupled via a DuoSpray ion source in electrospray mode to a high resolution quadrupole time-of-flight mass spectrometer (Triple TOF 5600+, Sciex). For information dependent data acquisition (IDA runs), the MS1 spectra were collected in a range of 400–1250 m/z for 250 ms. The 40 most intense precursors with charge state 2–4, which exceeded 150 counts per second, were selected for fragmentation. MS2 spectra were collected in the range of 200–1500 m/z for 50 ms. The precursor ions were dynamically excluded from reselection for 13 s. Based on the data-dependant acquisition spectra, an ion library was established for the next step of data-independent acquisition of SWATH spectra (Sequential Windowed Acquisition of All Theoretical Mass Spectra). Seventy-three variable windows were created in a mass range of 400–1250 Da depending on the precursor ion density. All precursors of each window were fragmented. MS2 spectra were acquired for 50 ms (SWATH runs).
Data processing
The acquired raw data were processed with ProteinPilot Software version 5.0 (Sciex) for re-calibration and database searches as described by Kumar et al. [30]. The database contained entries of following taxonomies: Oncorhynchus mykiss (NCBI, Refseq: 71 285 entries), and Y. ruckeri (UniProt, taxonomy id 29486: 4493 entries) as well as cRAP (common Repository of Adventitious Proteins). The database search parameters applied were trypsin digestion, cysteine alkylation set to iodoacetamide, search effort set to rapid ID. The mass tolerance in MS mode was set by program default with 0.05 Da in MS and 0.1 Da in MSMS mode for the rapid recalibration search, and 0.0011 Da in MS and 0.01 Da in MSMS mode for the final search. False discovery rate (FDR) analysis was performed using the integrated tools in ProteinPilot with < 1% on peptide as well as on protein level.
For the quantification of proteins by SWATH, the identified proteins of a combined search of all 27 IDA runs identifying 3425 proteins served as a basis for the creation of the SWATH ion library with the MS/MS (ALL) with SWATH Acquisition MicroApp 2.0 in PeakView 2.2 (both Sciex) (dataset PRIDE PXD011087, IDA FASP intestine Control/CSF/7959). Shared peptides were excluded. So the resulting ion library (Additional file 1) contained a total of 3372 proteins, 25 469 peptides and 295 168 transitions to be quantified potentially. The key criteria for processing of the SWATH samples were to use only proteins with an FDR rate below 1%, furthermore up to 6 peptides per protein and up to 6 transitions per peptide were chosen, false discovery rate threshold was 5%, peptide confidence threshold 98%, XIC extraction window 5 min, XIC width 100 ppm and modified peptides were excluded. Retention time alignment based on iRT peptides (iRT kit, Biognosys) and processing of SWATH raw samples for calculation of raw peak areas were performed in PeakView 2.2. These were normalized in the software MarkerView 1.2.1 (Sciex) with the integrated tool based on the total area sums, which assumes that the abundance of most of the proteins within the label free quantification approach remains unchanged and only a few of them are differentially regulated.
Data analysis
All statistical analyses were performed in R programming language [33]. The protein abundance after retention time alignment and normalization on total area sums derived from MarkerView were first transformed into a logarithmic scale. Technical replicates compensating for instabilities during LC–MS acquisition (N = 2) were averaged for the mean values. Afterward, the differences in abundance of posterior intestinal proteins were assessed using one-way ANOVA for each protein comparing groups [(control vs. biotype 1 (CSF007-82) exposed vs. biotype 2 (A7959-11) exposed posterior intestine samples] for each time point (day 3, 9 and 28). The method of Benjamini and Hochberg [34] was applied to control the FDR. The differences between the biotypes-exposed and control posterior intestine samples were considered significant if FDR-adjusted p-values were smaller than the significance level of α = 0.05. In order to assess the significance of the pairwise comparisons, Tukey’s honest significant difference method was applied as a post hoc test. Protein level changes were considered differential if the adjusted p value was below α and the absolute fold change was at least two (fold change < −2 or > +2).
GO annotation
PANTHER classification system was used to categorize the Gene Ontology of all the differentially up- or downregulated proteins in biological process, cellular component, and protein class [35]. Molecular function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of differentially up- or downregulated proteins were further determined on the Cytoscape software (version 3.7.1) [36] plugin ClueGO (version 2.5.4) [37]. We tested the significance of GO term and KEGG pathway using Fisher’s exact test and FDR-correction at< 0.05 significant level.
Quantitative real time PCR
In order to validate the abundance of identified proteins, eight kinds of proteins (probable serine carboxypeptidase, cathepsin D, caspase 6, lysozyme C II, precerebellin, protein S100, and tubulin alpha) were chosen for qPCR analysis carried out on a CFX96 Touch Real-Time PCR detection system (Bio-Rad, Hercules, USA). The sequences of the primers used in this study are listed in Additional file 2. Total RNA was extracted from posterior intestinal mucosa samples using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and included an on-column DNase digestion step. cDNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad) with 500 ng total RNA according to the user’s manual.
All reactions were done in a final volume of 20 μL, which contained 4 μL of 1:10-fold diluted cDNA, 0.5 μL of each primer (10 pmol/μL), 10 μL of 2× SsoAdvanced™ Universal SYBR Green Supermix (Bio-Rad), and 5 μL of RNase-free water. After 5 min of cDNA denaturation at 95 °C, 38 cycles were performed at 95 °C for 30 s, 57 °C for 30 s and 72 °C for 30 s. Each qPCR was performed for all biological replicates. Relative gene expression was assessed by the CFX Manager Maestro Software in normalized expression mode (∆∆Cq), using elongation factor alpha 1 as a Ref. [38], and was compared to the control sample. Additionally, standard curve was generated for qPCR using plasmid containing Y. ruckeri 16S ribosomal RNA fragment [39]. Y. ruckeri load was measured in the exposed and unexposed control posterior intestine samples with a Y. ruckeri 16S rRNA specific primers [39]. The copy number of Y. ruckeri 16S rRNA was calculated for each sample. The differences between unexposed control and exposed group and between exposed groups were tested for significant differences using a t-test. The sequential differences within exposed groups were tested for using a one-way ANOVA and significant differences revealed with the Tukey’s post hoc test. The Pearson correlation coefficient (r) was calculated to estimate the relationship between qPCR and proteomic quantifications. For all statistical tests, a p-value of < 0.05 was regarded as significant and all the data were analyzed in IBM SPSS software version 24.
Results
Clinical signs and Y. ruckeri load
The Y. ruckeri-exposed fish were prepared as described in our previous study [30], where 30% fish showed severe clinical signs of the disease and the maximum morbidity and mortality were observed between 8 and 10 dpe. The fish exhibited external hemorrhages in the caudal and anal fins (Figure 1A) and internal signs, such as enlarged spleen and reddened intestine (Figure 1B), at 9 dpe. However, at 28 dpe, these organs were in normal form and did not show any clinical signs of the disease (Figure 1C). The Y. ruckeri 16S transcript increased and peaked in the fish exposed to both biotypes at 3 dpe and then decreased at 9 dpe and was almost undetected at 28 dpe (Figure 2). No Y. ruckeri was detected in unexposed control samples.
Figure 1.
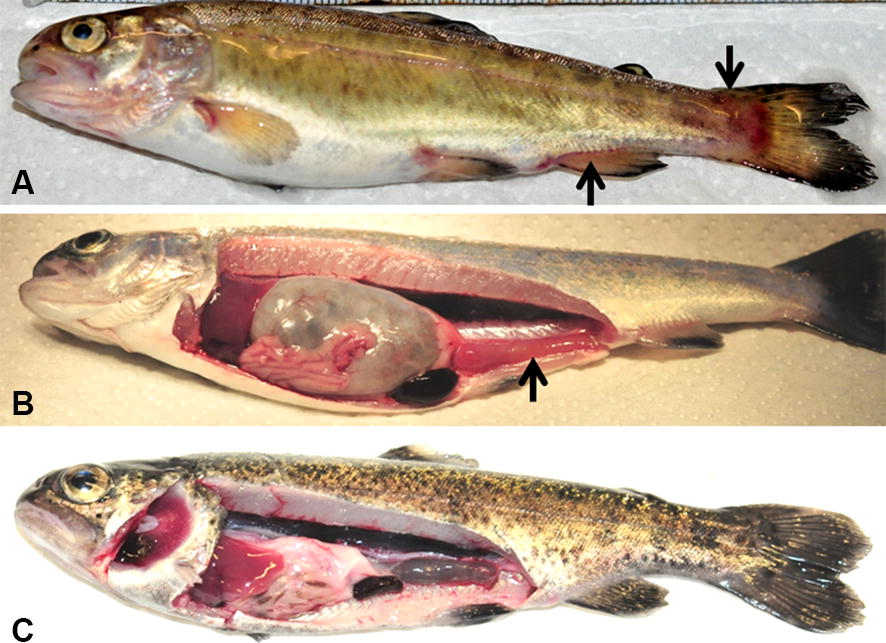
Rainbow trout showing clinical signs of enteric redmouth disease. A Hemorrhages in the caudal and anal fins (arrows). B Enlarged spleen and reddened intestine (arrow), and C rainbow trout at 28 days post-exposure.
Figure 2.
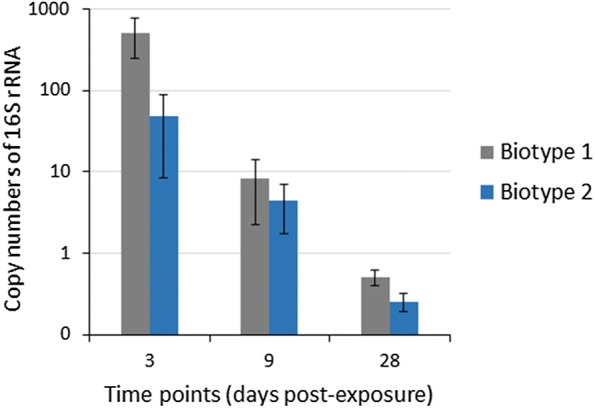
Yersinia ruckeri 16S rRNA copy numbers in the posterior intestine of rainbow trout. Quantitative real-time PCR shows the mean copy numbers of 16S rRNA gene in the exposed posterior intestine samples. There were significant differences of copy numbers of Y. ruckeri between time points (p value = 0.001), and between biotype 1 and biotype 2 (p value = 0.01). No Y. ruckeri was detected in unexposed control posterior intestine samples at any time point. Error bars indicate standard deviation (n = 9).
Protein quantification
A total of 3006 proteins in the posterior intestine of rainbow trout were identified at 1% FDR and minimum of two matching peptides (Additional file 3) over all 27 samples including controls as well as the exposed groups. In total, 1880 proteins with at least two unique peptides could be quantified. Sixty-two percent of the total 3006 proteins identified amongst all samples could be quantified, after the exclusion of shared and modified peptides and the exclusion of so-called one-peptide wonders. The statistical analysis (exposed versus control) revealed a total of 437 differentially abundant proteins in the posterior intestine at 3 dpe (Additional file 4). Of these, 205 proteins were upregulated and 232 proteins were downregulated in the posterior intestine at 3 dpe. We did not find statistically different changes in the protein abundance at 9 and 28 dpe. As shown in Table 1, upregulated proteins were related to serine -type carboxypeptidase, cysteine-type endopeptidase, aspartic-type endopeptidase, metallodipeptidase, metallopeptidase, hydrolase, peroxidase activity, signal transduction, and carbohydrate metabolic activities. On the other hand, the downregulated proteins were related to lipid metabolism, actin filament binding, stress response, and translation (Table 2).
Table 1.
List of upregulated posterior intestine proteins of rainbow trout in response to Yersinia ruckeri strains
| Accession Refseq | Protein name | Number of quantified peptides | Biological process | Intestine in response to Y. ruckeri strain | 3 dpe (fold change) | 9 dpe (fold change) | 28 dpe (fold change) |
|---|---|---|---|---|---|---|---|
| XP_021441860.1 | Probable serine carboxypeptidase CPVL | 6 | Serine-type carboxypeptidase activity | Biotype 1 | 3.6* | −1.7 | 1.4 |
| Biotype 2 | 4.9* | −1.3 | 1.4 | ||||
| XP_021462823.1 | Dipeptidase 1 | 6 | Metallodipeptidase activity | Biotype 1 | 2.1* | 1.3 | −1.0 |
| Biotype 2 | 2.7* | 1.5 | −1.2 | ||||
| XP_021442206.1 | Xaa-Pro dipeptidase | 6 | Metallopeptidase activity | Biotype 1 | 2.4* | −1.1 | 1.2 |
| Biotype 2 | 3.1* | 1.1 | 1.2 | ||||
| XP_021437213.1 | Dipeptidyl peptidase 1 | 5 | Cysteine-type endopeptidase activity | Biotype 1 | 1.8 | 1.2 | 1.9 |
| Biotype 2 | 2.3* | 1.3 | 1.5 | ||||
| XP_021460613.1 | Cathepsin D isoform X1 | 6 | Aspartic-type endopeptidase activity | Biotype 1 | 2.4* | −1.1 | 1.2 |
| Biotype 2 | 3.2* | 1.1 | 1.2 | ||||
| XP_021427100.1 | Pro-cathepsin H | 5 | Aminopeptidase activity | Biotype 1 | 4.3* | −2.0 | 1.3 |
| Biotype 2 | 6.3* | −1.3 | 1.0 | ||||
| XP_021430445.1 | Cathepsin L1 | 6 | Cysteine-type endopeptidase activity | Biotype 1 | 4.3* | −2.4 | 1.3 |
| Biotype 2 | 5.9* | −1.4 | 1.1 | ||||
| XP_021430112.1 | Acid phosphatase type 7 | 6 | Acid phosphatase activity | Biotype 1 | 2.5* | 1.1 | 1.8 |
| Biotype 2 | 4.4* | 1.7 | 1.3 | ||||
| XP_021457257.1 | Lysozyme C II | 5 | Hydrolase activity | Biotype 1 | 2.6* | 2.2* | 1.4 |
| Biotype 2 | 2.8* | 1.6 | 1.1 | ||||
| XP_021478264.1 | Macrophage mannose receptor 1 | 5 | Endocytosis | Biotype 1 | 3.2* | −1.1 | 1.3 |
| Biotype 2 | 4.7* | 1.4 | 1.2 | ||||
| XP_021446773.1 | Eosinophil peroxidase | 6 | Peroxidase activity | Biotype 1 | 3.2* | −1.2 | −1.1 |
| Biotype 2 | 1.6 | −1.4 | −1.2 | ||||
| XP_021424232.1 | Thioredoxin reductase 1 | 4 | Redox homeostasis | Biotype 1 | 1.7 | 1.1 | 1.1 |
| Biotype 2 | 2.2* | 1.0 | 1.1 | ||||
| XP_021412083.1 | Lysosomal alpha-mannosidase | 6 | Alpha-mannosidase activity | Biotype 1 | 2.8* | −1.8 | 1.4 |
| Biotype 2 | 4.8* | 1.1 | 1.3 | ||||
| NP_001154031.1 | Glutathione S-transferase A | 6 | Glutathione metabolic process | Biotype 1 | 1.7 | 1.1 | −1.0 |
| Biotype 2 | 2.0* | 1.2 | −1.0 | ||||
| XP_021460921.1 | Peroxisomal multifunctional enzyme type 2 | 6 | Oxidoreductase activity | Biotype 1 | 2.0* | 1.1 | −1.0 |
| Biotype 2 | 2.4* | 1.0 | −1.0 | ||||
| XP_021475660.1 | Endoplasmic reticulum aminopeptidase 1 | 4 | Aminopeptidase activity | Biotype 1 | 1.8 | 1.2 | 1.0 |
| Biotype 2 | 2.2* | 1.0 | −1.0 | ||||
| NP_001117743.1 | Caspase 6 precursor | 2 | Apoptotic process | Biotype 1 | 2.2* | −1.1 | 1.2 |
| Biotype 2 | 2.1* | −1.2 | −1.1 | ||||
| XP_021477916.1 | Interferon-induced GTP-binding protein Mx2 | 5 | GTPase activity | Biotype 1 | 2.0 | 1.3 | −1.1 |
| Biotype 2 | 2.4* | 1.3 | −1.3 | ||||
| XP_021468639.1 | Gamma-interferon-inducible lysosomal thiol reductase | 3 | Antigen processing and presentation | Biotype 1 | 3.0* | −1.9 | 1.4 |
| Biotype 2 | 4.4* | −1.1 | 1.3 | ||||
| XP_021466891.1 | Precerebellin-like protein | 5 | Response to lipopolysaccharide | Biotype 1 | 2.9* | 5.3* | 2.0 |
| Biotype 2 | 2.2 | 3.3* | 1.3 | ||||
| XP_021453615.1 | Calretinin-like | 6 | Calcium ion binding | Biotype 1 | 2.5* | 1.5 | 1.2 |
| Biotype 2 | 3.0* | 1.6 | 1.0 | ||||
| XP_021426220.1 | Regucalcin | 6 | Calcium ion binding | Biotype 1 | 2.1 | 1.0 | 1.4 |
| Biotype 2 | 3.1* | 1.2 | 1.1 | ||||
| XP_021480508.1 | Ras-related protein Rab-5C | 4 | GTPase activity | Biotype 1 | 1.7 | 1.0 | 1.3 |
| Biotype 2 | 2.4* | 1.1 | 1.0 | ||||
| XP_021477941.1 | Fructose-1,6-bisphosphatase 1 | 3 | Carbohydrate metabolic process | Biotype 1 | 3.8* | 1.4 | 1.0 |
| Biotype 2 | 2.2 | 1.0 | −1.2 | ||||
| XP_021434786.1 | Beta-hexosaminidase | 2 | Carbohydrate metabolic process | Biotype 1 | 2.4* | −1.5 | −1.0 |
| Biotype 2 | 2.5* | −1.1 | 1.2 | ||||
| XP_021467816.1 | Alpha-N-acetylgalactosaminidase | 6 | Carbohydrate metabolic process | Biotype 1 | 3.0* | −1.3 | 1.2 |
| Biotype 2 | 4.1* | 1.1 | 1.3 |
Fold change was statistically analyzed in the posterior intestine of rainbow trout exposed to Y. ruckeri biotype 1 (CSF007-82) and biotype 2 (A7959-11) versus control posterior intestine of rainbow trout samples (n = 3 per time point).
* Statistically significant difference according to both ANOVA and post hoc Tukey’s HSD with FDR-adjusted p-value < 0.05 and fold change < −2 or > +2. (Full table is presented in Additional file 4).
Table 2.
List of downregulated posterior intestine proteins of rainbow trout in response to Yersinia ruckeri strains
| Accession Refseq | Protein name | Number of quantified peptides | Biological process | Intestine in response to Y. ruckeri strain | 3 dpe (fold change) | 9 dpe (fold change) | 28 dpe (fold change) |
|---|---|---|---|---|---|---|---|
| NP_001117719.1 | Apolipoprotein A-I-1 | 6 | Lipid metabolism | Biotype 1 | −1.3 | −1.1 | −1.6 |
| Biotype 2 | −2.2* | −1.2 | −1.1 | ||||
| NP_001154920.1 | Apolipoprotein A-II | 4 | Lipid metabolism | Biotype 1 | −2.1* | −1.0 | −1.7 |
| Biotype 2 | −4.0* | 1.1 | −1.3 | ||||
| XP_021447265.1 | Apolipoprotein C-I | 2 | Lipid metabolism | Biotype 1 | −1.7 | 1.1 | −3.1* |
| Biotype 2 | −4.6* | −1.3 | −2.7 | ||||
| XP_021422847.1 | Filamin-A | 6 | Actin filament binding | Biotype 1 | −3.2* | −2.8 | −1.5 |
| Biotype 2 | −10.4* | −2.8 | −1.4 | ||||
| XP_021438757.1 | Alpha-actinin-1 | 6 | Actin filament binding | Biotype 1 | −2.4* | −1.7 | −1.2 |
| Biotype 2 | −6.3* | −1.9 | 1.3 | ||||
| XP_021474949.1 | PDZ and LIM domain protein 3 | 2 | Alpha-actinin binding | Biotype 1 | −4.2* | −2.5* | −1.1 |
| Biotype 2 | −22.6* | −1.7 | 1.1 | ||||
| XP_021479779.1 | Dihydropyrimidinase-related protein 3 | 6 | Actin filament bundle assembly | Biotype 1 | −2.4* | −1.7 | −1.3 |
| Biotype 2 | −4.5* | −1.6 | −1.1 | ||||
| XP_021449109.1 | Tubulin alpha chain | 3 | Microtubule process | Biotype 1 | −2.1* | 1.2 | −1.8 |
| Biotype 2 | −4.9* | −1.2 | −1.6 | ||||
| XP_021451765.1 | Tubulin polymerization-promoting protein | 6 | Microtubule binding | Biotype 1 | −3.6* | −1.7 | −1.3 |
| Biotype 2 | −7.0* | −1.5 | −1.2 | ||||
| XP_021480500.1 | DnaJ homolog subfamily C member 7 | 2 | Chaperone binding | Biotype 1 | −1.9 | 1.1 | −1.2 |
| Biotype 2 | −3.2* | −1.4 | −1.0 | ||||
| XP_021478259.1 | Lysosome membrane protein 2 | 2 | Chaperone activity | Biotype 1 | −2.6* | 1.1 | −1.3 |
| Biotype 2 | −3.3* | −1.3 | −1.3 | ||||
| NP_001117706.1 | Heat shock 47 precursor | 6 | Stress response | Biotype 1 | −3.7* | −1.4 | −1.1 |
| Biotype 2 | −9.6* | −1.2 | 1.0 | ||||
| XP_021438340.1 | Heat shock cognate 70 kDa protein-like | 6 | Chaperone activity | Biotype 1 | −1.9 | −1.1 | −1.3 |
| Biotype 2 | −2.6* | −1.4 | −1.1 | ||||
| XP_021441996.1 | Programmed cell death protein 5 | 2 | DNA binding | Biotype 1 | −2.7* | 1.2 | −1.7 |
| Biotype 2 | −4.4* | −1.4 | −2.6* | ||||
| XP_021418468.1 | Annexin A13 | 6 | Calcium ion binding | Biotype 1 | −2.0* | −1.6 | −1.8 |
| Biotype 2 | −3.4* | −1.2 | −1.1 | ||||
| XP_021416101.1 | Protein S100 A13 | 2 | Calcium binding | Biotype 1 | −3.2* | −1.1 | −1.5 |
| Biotype 2 | −6.6* | −1.3 | −1.1 | ||||
| NP_001117701.1 | M-calpain | 6 | Calcium ion binding | Biotype 1 | −2.0* | −1.2 | −1.0 |
| Biotype 2 | −4.0* | −1.4 | 1.0 | ||||
| NP_001117963.1 | Calpain 2 catalytic subunit | 5 | Calcium ion binding | Biotype 1 | −1.8 | −1.1 | −1.1 |
| Biotype 2 | −2.6* | −1.0 | −1.0 | ||||
| XP_021419099.1 | l-Lactate dehydrogenase | 2 | Carboxylic acid metabolic process | Biotype 1 | −3.4* | −1.5 | −1.8 |
| Biotype 2 | −7.4* | −1.7 | 1.3 | ||||
| XP_021452473.1 | Glyceraldehyde-3-phosphate dehydrogenase | 4 | Glycolysis process | Biotype 1 | −1.8 | −1.2 | −1.3 |
| Biotype 2 | −2.9* | −1.1 | −1.1 | ||||
| XP_021447060.1 | Pyruvate dehydrogenase E1 | 4 | Glucose metabolic process | Biotype 1 | −2.1* | 1.2 | 1.0 |
| Biotype 2 | −1.7 | 1.1 | −1.0 | ||||
| XP_021440913.1 | Galectin-5-like isoform X2 | 2 | Carbohydrate binding | Biotype 1 | −2.3* | 1.0 | 1.1 |
| Biotype 2 | −3.9* | 1.4 | −1.2 | ||||
| XP_021476056.1 | Eukaryotic translation initiation factor 5A | 5 | Translation | Biotype 1 | −1.4 | 1.1 | −1.4 |
| Biotype 2 | −2.4* | −1.3 | −1.6 | ||||
| XP_021467048.1 | 40S ribosomal protein S12 | 5 | Translation | Biotype 1 | −1.1 | 1.4 | −1.2 |
| Biotype 2 | −2.2* | 1.2 | 1.3 | ||||
| XP_021456583.1 | 40S ribosomal protein S29 | 3 | Translation | Biotype 1 | −3.4* | 1.0 | −1.2 |
| Biotype 2 | −2.9* | −1.0 | −1.3 |
Fold change was statistically analyzed in the posterior intestine of rainbow trout exposed to Y. ruckeri biotype 1 (CSF007-82) and biotype 2 (A7959-11) versus control posterior intestine of rainbow trout samples (n = 3 per time point).
* Statistically significant difference according to both ANOVA and post hoc Tukey’s HSD with FDR-adjusted p-value < 0.05 and fold change < −2 or > +2. (Full table is presented in Additional file 4).
GO annotation
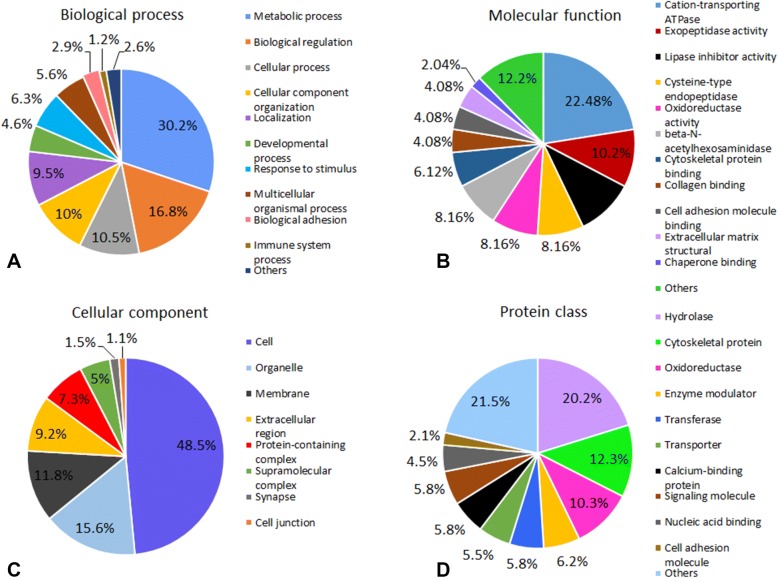
Within the classification of biological process, the differentially up- or downregulated posterior intestine proteins were mainly associated with metabolic process, biological regulation, cellular processes, and cellular component organization (Figure 3A). In terms of molecular function, the majorities of up- or downregulated posterior intestine proteins were involved in cation-transporting ATPase, exopeptidase, lipase inhibitor, cysteine-type endopeptidase, oxidoreductase, and beta-N-acetylhexosaminidase activities (Figure 3B). Most of the proteins were localized in the cell, organelle, membrane, and extracellular region (Figure 3C). Classification based on protein class resulted in the identification of ten major categories: hydrolase, cytoskeletal protein, oxidoreductase, enzyme modulator, transferase, transporter, calcium binding protein, signaling molecule, nucleic acid binding, and cell adhesion molecule (Figure 3D). Additionally, upregulated proteins were predicted to be involved in lysosome, oxidative phosphorylation, and metabolic pathways, while downregulated proteins were involved in focal adhesion, regulation of actin cytoskeleton, protein digestion and absorption pathways.
Figure 3.
Classification of differentially up or down-regulated posterior intestine proteins of rainbow trout in response to Yersinia ruckeri at 3 dpe. Proteins were classified by gene ontology terms for biological processes, molecular functions, cellular components and protein classes using PANTHER and ClueGO tools. A Biological process, B molecular function, C cellular component, and D protein class.
Validation of differentially abundant proteins
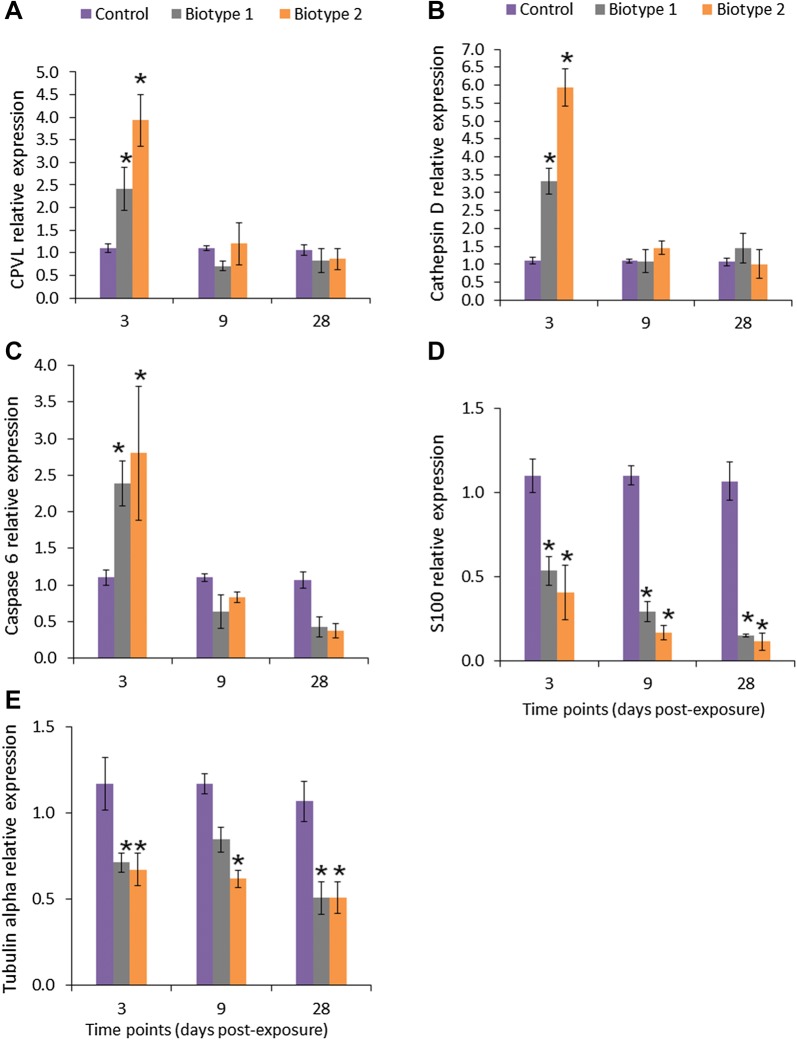
The upregulation of six proteins (probable serine carboxypeptidase, cathepsin D, caspase 6, lysozyme C II, and precerebellin-like protein) and downregulation of two proteins (protein S100 and tubulin alpha) in the posterior intestine of the fish exposed to both biotypes were confirmed at the mRNA level by qPCR analysis. The transcript levels of these candidate genes were significantly (p < 0.05) either increased or decreased compared to the control posterior intestine samples (Figure 4). The results of qPCR were consistent with those obtained from the proteomic results mainly at 3 dpe (Figure 5), which confirmed reliability of our proteomic data (r = 0.406 or 0.877). Nevertheless, the transcript expression slightly varied in parallel to the corresponding protein abundance perhaps suggesting additional post-transcriptional regulation.
Figure 4.
Relative expression levels of probable serine carboxypeptidase, cathepsin D, caspase-6, protein S100 and tubulin alpha in the posterior intestine of rainbow trout. Quantitative real-time PCR shows mean relative expression profiles of each selected gene in the posterior intestine of rainbow trout in response to Y. ruckeri biotype 1 (CSF007-82) and biotype 2 (A7959-11) at different time points. Relative gene expression changes in each gene were determined in the exposed and control posterior intestine samples by the CFX Manager Maestro Software in normalized expression mode (∆∆Cq), using elongation factor alpha 1 as a reference at each time point. A Probable serine carboxypeptidase CPVL, B cathepsin D, C caspase 6, D protein S100, and E tubulin alpha. Stars indicate statistically significant differences in the gene expression compared to the control group. Error bars indicate standard deviation (n = 9).
Figure 5.
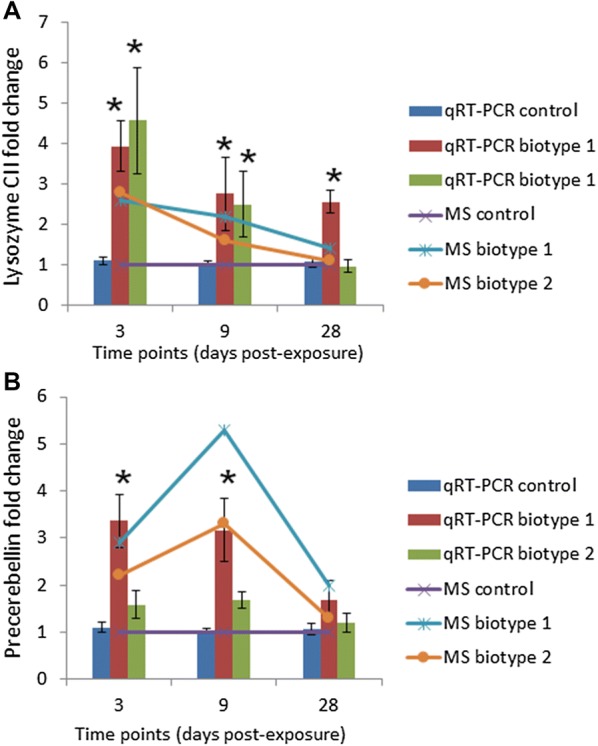
Comparison of transcriptional analysis and MS-based proteomic results of lysozyme C II and precerebellin. The qRT-PCR and the MS-based proteomic results are presented by the bar and line charts, respectively. Stars indicate statistically significant differences in the fold change compared to the control group. Error bars indicate standard deviation (n = 9). A Lysozyme C II, and B precerebellin-like protein.
Discussion
The gut immune system of teleost fish deserves special attention in the biology of the fish intestine. This provides important insights into the mechanisms induced by the progression of diseases [14]. In the case of common carp (Cyprinus carpio), the response of intestinal mucosa against Aeromonas hydrophila was investigated at the proteome level [40], which improved the understanding of defense mechanisms of carp intestinal mucosa and associated molecular mechanisms. In rainbow trout, the intestinal response was monitored at transcription levels against pathogens like Y. ruckeri, Enteromyxum leei, Edwardsiella ictaluri, and Vibrio harveyi [26–29]. In the current study, the elements of the gut mucosal responses were examined by proteome profiling of the posterior intestine of rainbow trout following experimental exposure to Y. ruckeri. Additionally, proteins were significantly up- or downregulated in the posterior intestine mainly at 3 dpe, which correlated with increasing bacterial transcripts found in the posterior intestine at 3 dpe (Figure 2). This suggests that Y. ruckeri might affect the gut and modulate the intestinal proteome of fish whenever a high load of bacteria is present in the fish intestine.
In total, 30% mortality was found in rainbow trout following the exposure to Y. ruckeri (2 × 106 CFU) [30]. This mortality rate is in contrast with the previous studies [41, 42], where mortality in rainbow trout was observed to be 20% and 66%, respectively, at lower (5 × 105 CFU) and higher (1.8 × 109 CFU) levels of the challenge dose of Y. ruckeri. These differences in the mortality data may be due to variations between the bacterial strains, challenge doses, fish size, and laboratory conditions. At 28 dpe, the intestine of rainbow trout exposed to Y. ruckeri strains was in normal form and also transcripts of Y. ruckeri 16S were almost undetectable in the posterior intestine. It seems that rainbow trout slowly cleared the Y. ruckeri during the analyzed period.
We found upregulation of exopeptidase and endopeptidase [probable serine carboxypeptidase (3.4- to 4.7-fold), dipeptidase I (2.1- to 2.7-fold), Xaa-Pro dipeptidase (2.4- to 3.1-fold), dipeptidyl peptidase 1 (2.3-fold), cathepsin D (2.4- to 3.2-fold), pro-cathepsin H (4.3- to 6.3-fold) and cathepsin L1 (4.3- to 5.9-fold)] in the posterior intestine of rainbow trout exposed to Y. ruckeri. These exo- and endopeptidases participate in several physiological and cellular processes in the guts of animals [43]. The expression of these peptidase (dipeptidase I and cathepsin D) genes was observed in the gut of channel catfish (Ictalurus punctatus) and grass carp (Ctenopharyngodon idella) following Edwardsiella ictaluri and Aeromonas hydrophila challenges [44, 45], thereby suggesting that they participate in various degradation functions and digestive processes and are closely involved in the immune response of rainbow trout. Overall, our study suggests that proteolysis and peptide hydrolysis activities are upregulated in the GI tract during the bacterial infection, indicating that these identified peptidases might play some role in host defense.
Furthermore, we found upregulation of phagocytosis proteins such as lysozyme C II, macrophage mannose receptor 1, eosinophil peroxidase, and thioredoxin reductase 1 in the posterior intestine of rainbow trout. These phagocytosis proteins were also upregulated in head kidney and spleen of rainbow trout exposed to Y. ruckeri strains [30]. Lysozymes are important defense proteins of the innate immune system of fish against bacterial pathogens [24, 46]. The upregulation of lysozyme C II (3.3-fold) in the posterior intestine, suggests that, besides its role in defense against bacterial pathogens, this protein may also play a significant role in the digestion during the infection in fish. In addition, the upregulation of macrophage mannose receptor 1 (3.2- to 4.7-fold) and thioredoxin reductase 1 (2.2-fold) in the posterior intestine of rainbow trout exposed to Y. ruckeri showed that the abundance of phagocytosis in the gut of rainbow trout results in enhanced intestinal defense against Y. ruckeri. Finally, these results suggest that Y. ruckeri triggers intestinal inflammation which enhances growth in the intestinal lumen and supports the invasion of the intestinal epithelium and mucosal macrophages. This macrophagic process promotes phagocytic activity in the gut to eliminate the bacteria and plays an important role in maintaining gut homeostasis and immune response [20].
In addition, caspase 6 and precerebellin-like protein were upregulated in the posterior intestine. Caspases are a family of proteases engaged in various important biological processes and play a significant role in the execution phase of the apoptotic death cascade [47]. Caspase-6 was found to be involved in apoptosis and immune response in puffer fish (Takifugu obscurus) against Aeromonas hydrophila [48]. The upregulation of caspase-6 (2.2-fold) in the gut mucosa of rainbow trout suggests that caspase-6 plays a role in the immune response and cell apoptosis against Y. ruckeri infection. However, another apoptosis related protein, programmed cell death protein 5 (PDCD5), was downregulated (−2.7- to 4.4-fold) in the posterior intestine as well. This indicates that PDCD5 may have an important role in the pathogenesis and development of the disease. However, the mechanisms underlying its apoptotic function are largely unknown in the fish. Precerebellin-like protein is a part of the acute phase response and has been characterized in rainbow trout [49]. The expression of precerebellin-like protein gene was upregulated (sevenfold) in the liver of rainbow trout in response to Y. ruckeri at 3 dpe [50]. This suggests that acute phase proteins act as a defensive agent against Y. ruckeri on the gut mucosal surface of rainbow trout. In support of this, the expression of some acute phase proteins such as serum amyloid protein and hepcidin was observed to be significantly increased in rainbow trout after Y. ruckeri challenge [50].
Evidently, calcium is engaged in signal transduction by acting as a second messenger [51]. In our study, we found two calcium binding proteins (calretinin and regucalcin) and seven signal transduction proteins (Ras-related protein Rab-1B, Rab-5C, Rab7-like, Rab-32-like, Rab-25-like, Rab7-like and ORAB-1) being upregulated. Calretinin is a calcium-binding protein found broadly distributed in the central nervous system and regucalcin is crucial for the regulation of Ca2+ ion homeostasis [52, 53]. The distribution of calretinin immunoreactivity in the developing olfactory system of the rainbow trout was observed by using an immunocytochemistry technique [54], and the expression of regucalcin gene was upregulated in rainbow trout at different temperatures and Aeromonas salmonicida challenges [55]. It could be speculated that calretinin and regucalcin execute their functions indirectly by controlling Ca2+ homeostasis in infected rainbow trout. Additionally, four calcium binding proteins [annexin A-13 (−4.1-fold), protein S100 (−6.6-fold), M-calpain (−4.0-fold), and calpain 2 catalytic subunit (−2.6-fold)] were downregulated. Annexins are a member of a multigene family of Ca2+ and phospholipid binding proteins [56]. S100 proteins take part in many cellular processes such as modulation of protein kinases and signal transduction pathways, maintenance of cell shape, regulation of calcium homoeostasis [57] and interaction with the parasite, Tetracapsuloides bryosalmonae [58]. However, calpains are calcium regulated proteases involved in many cellular functions and have been characterized in rainbow trout [59]. In conclusion, the presented results establish the significance of calcium binding proteins and their downregulation in the gut of rainbow trout in response to Y. ruckeri.
Yersinia ruckeri causes profound alterations in the nutritional and metabolic status of the fish [18, 60]. The interaction of a pathogen with intestinal mucosa leads to a variety of physiological responses aimed at adjusting to the new condition and triggers different processes in the gut epithelial cells [20]. Thus, we found upregulation of anabolic and catabolic proteins [fructose-1,6-bisphosphatase 1 (3.8-fold), beta-hexosaminidase (2.6-fold), alpha-N-acetylgalactosaminidase (4.1-fold), and long-chain specific acyl-CoA dehydrogenase (2.6-fold)], transport proteins [epididymal secretory protein E1 (3.9-fold), v-type proton ATPase subunit a (2.2-fold), and AP-1 complex subunit mu-2 (3.1-fold)]. The up-regulation of fructose-1,6-bisphosphatase and epididymal secretory protein E1 was observed in the spleen of rainbow trout in response to Y. ruckeri [30]. This suggests that these proteins are involved in the gut’s immune response and might result in physiological adaptations that contribute to extended longevity of fish during bacterial infection. Additionally, we found some downregulated proteins in the posterior intestine that are involved in gluconeogenesis [l-lactate dehydrogenase (−7.4-fold)], lipid transport [apolipoprotein A-I-1 (−2.2-fold), apolipoprotein A-II (−4.0-fold), and apolipoprotein C-I (−4.6-fold)], and translation [eukaryotic translation initiation factor 5A (−2.4-fold), 40S ribosomal protein S29 (−3.4-fold), and 40S ribosomal protein S12 (−2.2-fold)]. This suggests that fish might adjust their metabolism to channel the energy to the process of defense by decreasing glycogenesis, lipid metabolism, and translation, and increasing carbohydrate and protein catabolism during yersiniosis.
In conclusion, this study provides the first evidence illustrating the proteomic alteration of the intestinal mucosa of rainbow trout in response to Y. ruckeri, suggesting Y. ruckeri exerts a profound impact on posterior intestine mainly at 3 dpe. Intestinal endopeptidase, exopeptidase, and the proteins involved in antioxidant defense processes were upregulated upon Y. ruckeri exposure, while those involved in lipid metabolism, actin filament, and translation processes were downregulated. It is expected that this new information will exemplify further how Y. ruckeri influences the intestinal mucosa proteome of fish. The predicted lysosomal, metabolic, and focal adhesion pathways might be useful in understanding the gut defense mechanisms of rainbow trout and further research work in this direction. Utilization of these results may improve approaches for selection of disease-resistant rainbow trout broodstock and evaluation of prevention opportunities. Finally, the relationship among the intestinal bacterial pathogen and host protein responses will be explored to improve nutrient uptake, fish performance, and vaccine efficiency. Further studies are needed to address the functions of these intestinal proteins in immune response and protection against bacterial infection for both disease control and improved performance.
Additional files
Additional file 1. Ion library. Text file based on IDA runs (independent data acquisition) of the intestine samples used for data interpretation of SWATH acquisitions obtained by PeakView 2.2 software (Sciex).
Additional file 2. List of quantitative real-time PCR primers for confirmation of expression data. PCR primers specific to the selected genes were designed using NCBI Primer BLAST software.
Additional file 3. The details of total identified posterior intestine proteins of rainbow trout. The number of proteins was identified at false discovery rate of 1% and minimum two peptides.
Additional file 4.List of up or down-regulated posterior intestine proteins of rainbow trout in response to Yersinia ruckeri strains at 3 dpe. Fold change was statistically analyzed in the posterior intestine of rainbow trout exposed to Y. ruckeri biotype 1 (CSF007-82) and biotype 2 (A7959–11) versus control posterior intestine rainbow trout samples (n = 3 per time point). * denotes statistically significant difference according to both ANOVA and post hoc Tukey’s HSD with FDR-adjusted p-value < 0.05 and fold change < −2 or > +2.
Acknowledgements
This research was supported using resources of the VetCore Facility (Proteomics) of the University of Veterinary Medicine Vienna. We are thankful to Dr. Maike Ahrens for helping in R statistical analysis. Yersinia ruckeri strain CSF007-82 was kindly provided by Dr. Timothy J. Welch, National Center for Cool and Cold Water Aquaculture, Kearneysville, USA.
Abbreviations
- ERM
enteric redmouth disease
- FDR
false discovery rate
- IDA
information dependent data acquisition
- LC–MS
liquid chromatography–mass spectrometry
- Micro LC ESI QTOF MS/MS
microflow high-performance liquid chromatography electrospray-ionisation quadrupole time-of-flight tandem mass spectrometry
- PCA
principal component analysis
- SWATH
sequential window acquisition of all theoretical spectra
- TOF
triple quadrupole time of flight
Authors’ contributions
GK and MEL conceptualized and designed the study. GK performed the experiment and wrote the manuscript. KH, GK, and ERF carried out LC–MS/MS. ERF and MEL reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Austrian Science Fund (FWF) Project No. P 27489-B22 to GK.
Availability of data and materials
Shotgun proteomics data generated during the current study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository [61] with the dataset identifiers PXD011087.
Ethics approval and consent to participate and to publication
This study was approved by the institutional ethics committee of the University of Veterinary Medicine Vienna and the national authority, according to §26 of the Austrian Law for Animal Experiments, Tierversuchsgesetz 2012 under approval number BMWFW-68.205/0041-WF/V/3b/2015.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gokhlesh Kumar, Email: Gokhlesh.Kumar@vetmeduni.ac.at.
Karin Hummel, Email: Karin.Hummel@vetmeduni.ac.at.
Ebrahim Razzazi-Fazeli, Email: Ebrahim.Razzazi@vetmeduni.ac.at.
Mansour El-Matbouli, Email: Mansour.El-Matbouli@vetmeduni.ac.at.
References
- 1.Horne MT, Barnes AC. Enteric redmouth disease (Yersinia ruckeri) In: Woo PTK, Bruno DW, editors. Fish diseases and disorders. Viral, bacterial and fungal infections. Wallingford: CABI Publishing; 1999. pp. 445–477. [Google Scholar]
- 2.Kumar G, Menanteau-Ledouble S, Saleh M, El-Matbouli M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet Res. 2015;46:103. doi: 10.1186/s13567-015-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zorriehzahra MJ, Adel M, Torabi Delshad S. Enteric redmouth disease: past, present and future: a review. Iran J Fish Sci. 2017;16:1135–1156. [Google Scholar]
- 4.Busch RA, Lingg AJ. Establishment of an asymptomatic carrier state infection of enteric redmouth disease in rainbow trout (Salmo gairdneri) J Fish Res Board Can. 1975;32:2429–2432. doi: 10.1139/f75-279. [DOI] [Google Scholar]
- 5.Tobback E, Hermans K, Decostere A, Van den Broeck W, Haesebrouck F, Chiers K. Interactions of virulent and avirulent Yersinia ruckeri strains with isolated gill arches and intestinal explants of rainbow trout Oncorhynchus mykiss. Dis Aquat Org. 2010;90:175–179. doi: 10.3354/dao02230. [DOI] [PubMed] [Google Scholar]
- 6.Ohtani M, Villumsen KR, Strøm HK, Raida MK. 3D Visualization of the initial Yersinia ruckeri infection route in rainbow trout (Oncorhynchus mykiss) by optical projection tomography. PLoS One. 2014;9:e89672. doi: 10.1371/journal.pone.0089672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies RL. Clonal analysis of Yersinia ruckeri based on biotypes, serotypes and outer membrane protein types. J Fish Dis. 1991;14:221–228. doi: 10.1111/j.1365-2761.1991.tb00591.x. [DOI] [Google Scholar]
- 8.Romalde JL, Margariños B, Barja JL, Toranzo AE. Antigenic and molecular characterization of Yersinia ruckeri. Proposal for a new intraspecies classification. Syst Appl Microbiol. 1993;16:411–419. doi: 10.1016/S0723-2020(11)80274-2. [DOI] [Google Scholar]
- 9.Austin DA, Robertson PAW, Austin B. Recovery of a new biogroup of Yersinia ruckeri from diseased rainbow trout (Oncorhynchus mykiss, Walbaum) Syst Appl Microbiol. 2003;26:127–131. doi: 10.1078/072320203322337416. [DOI] [PubMed] [Google Scholar]
- 10.Fouz B, Zarza C, Amaro C. First description of non-motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiss (Walbaum), cultured in Spain. J Fish Dis. 2006;29:339–346. doi: 10.1111/j.1365-2761.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 11.Arias CR, Olivares-Fuster O, Hayden K, Shoemaker CA, Grizzle JM, Klesius PH. First report of Yersinia ruckeri biotype 2 in the U.S.A. J Aquat Anim Health. 2007;19:35–40. doi: 10.1577/H06-011.1. [DOI] [PubMed] [Google Scholar]
- 12.Strom-Bestor M, Mustamaki N, Heinikainen S, Hirvela-Koski V, Verner-Jeffreys D, Wiklund T. Introduction of Yersinia ruckeri biotype 2 into Finnish fish farms. Aquaculture. 2010;308:1–5. doi: 10.1016/j.aquaculture.2010.08.009. [DOI] [Google Scholar]
- 13.Axelsson M, Fritsche R. Effects of exercise, hypoxia and feeding on the gastrointestinal blood flow in the Atlantic cod Gadus morhua. J Exp Biol. 1991;158:181–198. doi: 10.1242/jeb.158.1.181. [DOI] [PubMed] [Google Scholar]
- 14.Buddington RK, Krogdahl A, Bakke-Mckellep AM. The intestines of carnivorous fish: structure and functions and the relations with diet. Acta Physiol Scand Suppl. 1997;638:67–80. [PubMed] [Google Scholar]
- 15.Seth H, Axelsson M, Farrell AP. The circulation and metabolism of the gastrointestinal tract. In: Grosell M, Farrell AP, Brauner CJ, editors. Fish physiology: the multifunctional gut of fish. Amsterdam: The Multifunctional Gut of Fish Academic Press; 2011. pp. 351–393. [Google Scholar]
- 16.Wulff T, Petersen J, Nørrelykke MR, Jessen F, Nielsen HH. Proteome analysis of pyloric ceca: a methodology for fish feed development? J Agric Food Chem. 2012;60:8457–8464. doi: 10.1021/jf3016943. [DOI] [PubMed] [Google Scholar]
- 17.Baumgarner BL, Bharadwaj AS, Inerowicz D, Goodman AS, Brown PB. Proteomic analysis of rainbow trout (Oncorhynchus mykiss) intestinal epithelia: physiological acclimation to short-term starvation. Comp Biochem Physiol Part D Genomics Proteomics. 2013;8:58–64. doi: 10.1016/j.cbd.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Ingerslev HC, Strube ML, Lv Jørgensen, Dalsgaard I, Boye M, Madsen L. Diet type dictates the gut microbiota and the immune response against Yersinia ruckeri in rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2014;40:624–633. doi: 10.1016/j.fsi.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Na-Phatthalung P, Teles M, Voravuthikunchai SP, Tort L, Fierro-Castro C. Immune-related gene expression and physiological responses in rainbow trout (Oncorhynchus mykiss) after intraperitoneal administration of Rhodomyrtus tomentosa leaf extract: a potent phytoimmunostimulant. Fish Shellfish Immunol. 2018;77:429–437. doi: 10.1016/j.fsi.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Salinas I, Parra D. Fish mucosal immunity: intestine. In: Beck B, Peatman E, editors. Mucosal health in aquaculture. San Diego: Academic Press; 2015. pp. 135–170. [Google Scholar]
- 21.Tobback E, Decostere A, Hermans K, Ryckaert J, Duchateau L, Haesebrouck F, Chiers K. Route of entry and tissue distribution of Yersinia ruckeri in experimentally infected rainbow trout Oncorhynchus mykiss. Dis Aquat Org. 2009;84:219–228. doi: 10.3354/dao02057. [DOI] [PubMed] [Google Scholar]
- 22.Wilson JM, Castro LFC. Morphological diversity of the gastrointestinal tract in fishes. In: Grosell MA, Farrell P, Brauner CJ, editors. The multifunctional gut of fish. Amsterdam: Academic Press; 2011. pp. 1–155. [Google Scholar]
- 23.Martin SA, Dehler CE, Król E. Transcriptomic responses in the fish intestine. Dev Comp Immunol. 2016;64:103–117. doi: 10.1016/j.dci.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Acar Ü, Parrino V, Kesbiç OS, Lo Paro G, Saoca C, Abbate F, Yılmaz S, Fazio F. Effects of different levels of pomegranate seed oil on some blood parameters and disease resistance against Yersinia ruckeri in rainbow trout. Front Physiol. 2018;9:596. doi: 10.3389/fphys.2018.00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrino V, Kesbiç OS, Acar Ü, Fazio F. Hot pepper (Capsicum sp.) oil and its effects on growth performance and blood parameters in rainbow trout (Oncorhynchus mykiss) Nat Prod Res. 2019 doi: 10.1080/14786419.2018.1550769. [DOI] [PubMed] [Google Scholar]
- 26.Davey GC, Calduch-Giner JA, Houeix B, Talbot A, Sitja-Bobadilla A, Prunet P, Perez-Sanchez J, Cairns MT. Molecular profiling of the gilthead sea bream (Sparus aurata L.) response to chronic exposure to the myxosporean parasite Enteromyxum leei. Mol Immunol. 2011;48:2102–2112. doi: 10.1016/j.molimm.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Evenhuis JP, Cleveland BM. Modulation of rainbow trout (Oncorhynchus mykiss) intestinal immune gene expression following bacterial challenge. Fish Shellfish Immunol. 2012;146:8–17. doi: 10.1016/j.vetimm.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Zhang Y, Wang R, Lu J, Nandi S, Mohanty S, Terhune J, Liu Z, Peatman E. RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immunol. 2012;32:816–827. doi: 10.1016/j.fsi.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Xia JH, Liu P, Liu F, Lin G, Sun F, Tu R, Yue GH. Analysis of stress responsive transcriptome in the intestine of Asian seabass (Lates calcarifer) using RNA-seq. DNA Res. 2013;20:449. doi: 10.1093/dnares/dst022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar G, Hummel K, Noebauer K, Razzazi-Fazeli E, El-Matbouli M. Proteome analysis reveals a role of rainbow trout lymphoid organs during Yersinia ruckeri infection process. Sci Rep. 2018;8:13998. doi: 10.1038/s41598-018-31982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibello A, Blanco MM, Moreno MA, Cutuli MT, Domenech A, Domínguez L, Fernández-Garayzábal JF. Development of a PCR assay for detection of Yersinia ruckeri in tissues of inoculated and naturally infected trout. Appl Environ Microbiol. 1999;65:346–350. doi: 10.1128/aem.65.1.346-350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erde J, Loo RR, Loo J. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13:1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team R (2015) A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. https://www.R-project.org/ Accessed 15 Mar 2019
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 35.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-into decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar G, Abd-Elfattah A, El-Matbouli M. Differential modulation of host genes in the kidney of brown trout Salmo trutta during sporogenesis of Tetracapsuloides bryosalmonae (Myxozoa) Vet Res. 2014;45:101. doi: 10.1186/s13567-014-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raida MK, Buchmann K. Development of adaptive immunity in rainbow trout, Oncorhynchus mykiss (Walbaum) surviving an infection with Yersinia ruckeri. Fish Shellfish Immunol. 2008;5:533–541. doi: 10.1016/j.fsi.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Di G, Li H, Zhang C, Zhao Y, Zhou C, Naeem S, Li L, Kong X. Label-free proteomic analysis of intestinal mucosa proteins in common carp (Cyprinus carpio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2017;66:11–25. doi: 10.1016/j.fsi.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 41.Wiens GD, Vallejo RL. Temporal and pathogen-load dependent changes in rainbow trout (Oncorhynchus mykiss) immune response traits following challenge with biotype 2 Yersinia ruckeri. Fish Shellfish Immunol. 2010;29:639–647. doi: 10.1016/j.fsi.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Raida MK, Holten-Andersen L, Buchmann K. Association between Yersinia ruckeri infection, cytokine expression and survival in rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2011;30:1257–1264. doi: 10.1016/j.fsi.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 43.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng T, Zhang H, Liu H, Zhou Z, Niu D, Wong L, Kucuktas H, Liu X, Peatman E, Liu Z. Molecular characterization and expression analysis of the channel catfish cathepsin D genes. Fish Shellfish Immunol. 2011;31:164–169. doi: 10.1016/j.fsi.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Dong ZD, Zhang J, Ji XS, Zhou FN, Fu Y, Chen W, Zeng YQ, Li TM, Wang H. Molecular cloning, characterization and expression of cathepsin D from grass carp (Ctenopharyngodon idella) Fish Shellfish Immunol. 2012;33:1207–1214. doi: 10.1016/j.fsi.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Grinde B, Jollès J, Jollès P. Purification and characterization of two lysozymes from rainbow trout (Salmo gairdneri) Eur J Biochem. 1988;173:269–273. doi: 10.1111/j.1432-1033.1988.tb13994.x. [DOI] [PubMed] [Google Scholar]
- 47.Laing KJ, Holland J, Bonilla S, Cunningham C, Secombes CJ. Cloning and sequencing of caspase 6 in rainbow trout, Oncorhynchus mykiss, and analysis of its expression under conditions known to induce apoptosis. Dev Comp Immunol. 2001;25:303–312. doi: 10.1016/S0145-305X(00)00061-6. [DOI] [PubMed] [Google Scholar]
- 48.Fu S, Ding M, Yang Y, Kong J, Li Y, Guo Z, Wang A, Ye J. Molecular cloning, characterization and expression analysis of caspase-6 in puffer fish (Takifugu obscurus) Aquaculture. 2018;490:311–320. doi: 10.1016/j.aquaculture.2018.03.008. [DOI] [Google Scholar]
- 49.Gerwick L, Reynolds WS, Bayne CJ. A precerebellin-like protein is part of the acute phase response in rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol. 2000;24:597–607. doi: 10.1016/S0145-305X(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 50.Raida MK, Buchmann K. Innate immune response in rainbow trout (Oncorhynchus mykiss) against primary and secondary infections with Yersinia ruckeri O1. Dev Comp Immunol. 2009;33:35–45. doi: 10.1016/j.dci.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Machaca K. Ca2+ signaling, genes and the cell cycle. Cell Calcium. 2010;48:243–250. doi: 10.1016/j.ceca.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi H, Yamaguchi M. Increase of (Ca(2+)–Mg2+)-ATPase activity in hepatic plasma membranes of rats administered orally calcium: the endogenous role of regucalcin. Mol Cell Biochem. 1995;144:1–6. doi: 10.1007/BF00926733. [DOI] [PubMed] [Google Scholar]
- 53.Castro A, Becerra M, Manso MJ, Anadón R. Calretinin immunoreactivity in the brain of the zebrafish, Danio rerio: distribution and comparison with some neuropeptides and neurotransmitter-synthesizing enzymes. II. Midbrain, hindbrain, and rostral spinal cord. J Comp Neurol. 2006;494:792–814. doi: 10.1002/cne.20843. [DOI] [PubMed] [Google Scholar]
- 54.Porteros A, Arévalo R, Weruaga E, Crespo C, Briñón JG, Alonso JR, Aijon J. Calretinin immunoreactivity in the developing olfactory system of the rainbow trout. Brain Res Dev Brain Res. 1997;100:101–109. doi: 10.1016/S0165-3806(97)00037-0. [DOI] [PubMed] [Google Scholar]
- 55.Verleih M, Rebl A, Köllner B, Korytář T, Anders E, Wimmers K, Goldammer T. Comparative molecular characterization of the regucalcin (RGN) gene in rainbow trout (Oncorhynchus mykiss) and maraena whitefish (Coregonus marena) Mol Biol Rep. 2012;39:4291–4300. doi: 10.1007/s11033-011-1216-1. [DOI] [PubMed] [Google Scholar]
- 56.Farber SA, De Rose RA, Olson ES, Halpern ME. The zebrafish annexin gene family. Genome Res. 2003;13:1082–1096. doi: 10.1101/gr.479603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar G, Gotesman M, El-Matbouli M. Interaction of Tetracapsuloides bryosalmonae, the causative agent of proliferative kidney disease, with host proteins in the kidney of Salmo trutta. Parasitol Res. 2015;114:1721–1727. doi: 10.1007/s00436-015-4357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salem M, Nath J, Rexroad CE, Killefer J, Yao J. Identification and molecular characterization of the rainbow trout calpains (Capn1 and Capn2): their expression in muscle wasting during starvation. Comp Biochem Physiol B: Biochem Mol Biol. 2005;140:63–71. doi: 10.1016/j.cbpc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Skov J, Kania PW, Holten-Andersen L, Fouz B, Buchmann K. Immunomodulatory effects of dietary β-1,3-glucan from Euglena gracilis in rainbow trout (Oncorhynchus mykiss) immersion vaccinated against Yersinia ruckeri. Fish Shellfish Immunol. 2012;33:111–120. doi: 10.1016/j.fsi.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 61.Vizcaíno JA, Csordas A, del Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Ion library. Text file based on IDA runs (independent data acquisition) of the intestine samples used for data interpretation of SWATH acquisitions obtained by PeakView 2.2 software (Sciex).
Additional file 2. List of quantitative real-time PCR primers for confirmation of expression data. PCR primers specific to the selected genes were designed using NCBI Primer BLAST software.
Additional file 3. The details of total identified posterior intestine proteins of rainbow trout. The number of proteins was identified at false discovery rate of 1% and minimum two peptides.
Additional file 4.List of up or down-regulated posterior intestine proteins of rainbow trout in response to Yersinia ruckeri strains at 3 dpe. Fold change was statistically analyzed in the posterior intestine of rainbow trout exposed to Y. ruckeri biotype 1 (CSF007-82) and biotype 2 (A7959–11) versus control posterior intestine rainbow trout samples (n = 3 per time point). * denotes statistically significant difference according to both ANOVA and post hoc Tukey’s HSD with FDR-adjusted p-value < 0.05 and fold change < −2 or > +2.
Data Availability Statement
Shotgun proteomics data generated during the current study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository [61] with the dataset identifiers PXD011087.