Abstract
Background
Pre-slaughter stress (PSS) impairs animal welfare and meat quality. Dark, firm and dry (DFD) are terms used to designate poor quality meats induced by PSS. Protein phosphorylation can be a potentially significant mechanism to explain rapid and multiple physiological and biochemical changes linked to PSS-dependent muscle-to-meat conversion. However, the role of reversible phosphorylation in the response to PSS is still little known. In this study, we report a comparative phosphoproteomic analysis of DFD and normal meats at 24 h post-mortem from the longissimus thoracis (LT) bovine muscle of male calves of the Rubia Gallega breed. For this purpose, two-dimensional gel electrophoresis (2-DE), in-gel multiplex identification of phosphoproteins with PRO-Q Diamond phosphoprotein-specific stain, tandem (MALDI-TOF/TOF) mass spectrometry (MS), novel quantitative phosphoproteomic statistics and bioinformatic tools were used.
Results
Noticeable and statistically significant differences in the extent of protein phosphorylation were detected between sample groups at the qualitative and quantitative levels. Overall phosphorylation rates across significantly changed phosphoproteins were about three times higher in DFD than in normal meat. Significantly changed phosphoproteins involved a variable number of isoforms of 13 myofibrillar and sarcoplasmic nonredundant proteins. However, fast skeletal myosin light chain 2 followed by troponin T, F-actin-capping and small heat shock proteins showed the greatest phosphorylation change, and therefore they were the most important phosphoproteins underlying LT muscle conversion to DFD meat in the Rubia Gallega breed.
Conclusions
This is the first study reporting global meat phosphoproteome changes in response to PSS. The results show that reversible phosphorylation is a relevant mechanism underlying PSS response and downstream effects on meat quality. This research opens up novel horizons to unravel the complex molecular puzzle underlying muscle-to-meat conversion in response to PSS.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5943-3) contains supplementary material, which is available to authorized users.
Keywords: Bos taurus - DFD meat - meat phosphoproteome, Beef quality, Meat tenderness, Pre-slaughter stress biomarkers, Post-mortem metabolism
Background
Phosphorylation is a ubiquitous protein post-translational modification that regulates a plethora of fundamental cell processes such as signal transduction pathways, cell cycle and apoptosis [1]. Reversible phosphorylation by the concerted action of a complex network of protein kinases and protein phosphatases plays a key regulatory role in the biochemical processes underlying muscle contraction and metabolism during the post-mortem muscle-to-meat conversion [2–5]. Post-mortem changes in the phosphorylation status of myofibrillar proteins and glycolytic enzymes in bovine, ovine and porcine muscles have been linked to differences in the meat quality traits of tenderness and color stability [2, 4, 6–8]. The available evidence suggests that reversible phosphorylation of proteins involved in muscle contraction and glycolysis can influence meat quality due to its post-mortem effects on pH decline and the development of rigor mortis [4, 9–11].
Stress is a key factor for animal welfare that influences meat quality traits [12, 13]. PSS can be elicited by multiple factors such as physical and psychological stressors linked to transport and handling activities from farms to abattoirs [14–17]. Physical and psychological stressors that trigger PSS response include but are not limited to environmental temperature, human presence, unfamiliar environments, mixing of animals from different social group, water and feed deprivation during transportation, loading and unloading practices and lairage in slaughter house [17–20]. The strength of PSS depends on the type, intensity and duration of stressors. PSS has been classified in acute or short stress when the duration of transport does not exceed a few days and chronic stress whether the transport lasts longer [17]. PSS can also be modulated by previous experiences and acquired learning, endogenous animal factors (e.g. genotype, age, sex) as well as by individual psychological and physiological state [21, 22]. Stress response to stressors is typically initiated through the activation of the autonomic nervous system (ANS) and the hypothalamic-pituitary-adrenal (HPA) axis mediated by catecholamines and glucocorticoids [21, 23, 24]. Neuroendocrine systems trigger a wide range of physiological and biochemical changes that affect the animal welfare and influence key processes when muscle turns into meat [12, 13, 25]. PSS can produce poor quality meats classified as DFD and pale, soft and exudative (PSE) meats [25]. They are therefore an excellent model to extent our knowledge about the biochemical processes underlying stress response and downstream effects on meat quality traits.
PSS can induce the ante-mortem depletion of glycogen reserves in bovine muscle which are used as source of energy to supply ATP for muscle contraction and relaxation [20, 26–28]. Glycogen-depleted muscle fibers in the immediate post-mortem period generate low amounts of lactic acid through anaerobic glycolysis which alters the normal process of meat acidification. In DFD meat, pH values at 12–48 h post-mortem are higher than 6.0, while in normal meat the corresponding pH falls to values of about 5.4–5.7 [25, 27]. Higher pH values can have a detrimental impact on the meat general appearance and many other determinants of meat quality. In comparison to normal meat, DFD condition is characterized by a darker color, a superficially drier and firmer texture, higher water-holding capacity (WHC) with little or no exudates, less protein denaturation, inferior taste, more susceptibility to microbial growth and a high potential of spoilage at an early meat aging [25–27, 29–35]. It must be highlighted, however, that DFD meat is usually tenderer than normal meat [20, 29, 32, 35]. Overall, it is a type of meat with lower consumer acceptability [36] that causes significant economic losses to the beef industry [20–25].
Proteomics has successfully contributed to unraveling the biochemical processes determining meat quality variations in response to varied stress inducers [37, 38]. To our knowledge, only a few recent studies have reported global proteome changes in DFD and PSE meat of cattle and broiler, respectively [35, 39, 40]. On the other hand, phosphoproteome studies have showed that two pre-slaughter stressors (i.e. transport and lairage) had no apparent effect on the global protein phosphorylation in lamb meat at 24 h post-mortem, as well as no differences in the phosphorylation levels of pork myofibrillar proteins between PSE and normal meats at early post-mortem time [41, 42]. In contrast, highly phosphorylated fast skeletal myosin regulatory light chain 2 (MYLPF) isoforms showed the most intense relative change across the proteome between DFD and normal meats from LT bovine muscle [35]. This finding suggests that reversible phosphorylation could be a significant mechanism in response to PSS. It is noteworthy that PSS triggers multiple physiological and biochemical changes in beef muscle in very short intervals of time. Accordingly, reversible phosphorylation has the potential to rapidly alter regulatory processes associated with muscle conversion into meat. In this regard, phosphoproteome changes have been reported in pigs with Halothane gene mutations and anomalous muscle energy metabolism that produce PSE meat [43].
This study aimed to assess for the first time the phosphoproteome differences between DFD and normal bovine meat. For this purpose, the phosphoproteome profiles of DFD and unaffected control meat from LT bovine muscle of the Rubia Gallega breed were compared using previously characterized meat samples at the proteome level. It will allow us to evaluate whether reversible phosphorylation plays a significant role in response to PSS with outstanding effects downstream on muscle-to-meat conversion processes.
Results
Phosphoproteome profiles of DFD and control meat samples by 2-DE
Figure 1 shows 2-DE representative proteomic profiles of DFD and control (non-DFD) meat samples at 24 h post-mortem derived from LT muscle on gels stained with phosphoprotein-specific Pro-Q Diamond stain and post-stained with non-specific SYPRO Ruby stain. PeppermintStick markers showed the specificity of Pro-Q Diamond for phosphoproteins under our experimental protocols. We found that DFD and normal meats exhibited markedly differentiated phosphoproteome profiles at the qualitative and quantitative levels. First of all, the percentage of Pro-Q-Diamond-stained reproducible spots was similar in DFD (14.6%, 46 out of 314 spots) and control (13.3%, 41 out of 308 spots) meat (P-value = 0.76, two-tailed Fisher’s exact test). Nevertheless, only 28 Pro-Q-Diamond-stained spots were shared between sample groups. Therefore, 18 and 13 spots were unique spots with phosphorylation signal only in DFD and control meats, respectively.
Fig. 1.
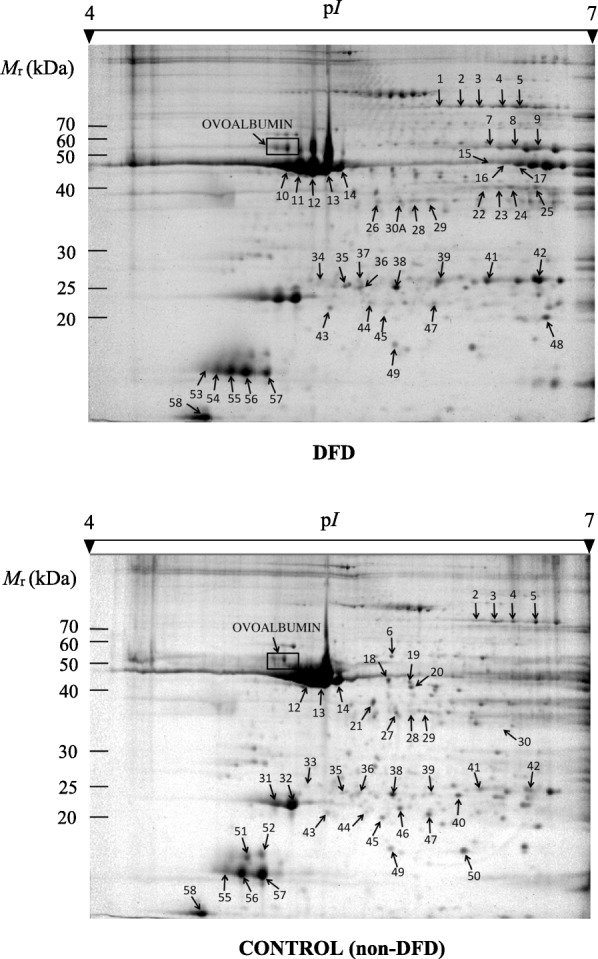
Representative 2-DE gel profiles of DFD (above) and control (below) meat samples from the LT bovine muscle stained with Pro-Q Diamond and subsequently with SYPRO Ruby. Phosphoprotein spots with statistically significant qualitative (presence/absence) and quantitative (changes in intensity) differential phosphorylation are marked and numbered. Numbered spots were excised from gels for phosphoprotein identification by MALDI-TOF and MALDI-TOF/TOF MS.
Quantitative estimates of phosphorylation levels over 2-DE spots in DFD and control samples were assessed by the phosphorylation rate (PR) statistic (Additional file 1: Table S1). In total, 54.2% (32 out of 59) of spots showed statistically significant differences in the PR mean value between sample groups by using 95% bootstrap CIs obtained by the bias-corrected percentile method and adjusted with the Bonferroni correction. Significantly changed phosphoprotein spots are shown in Table 1. Overall, phosphorylation levels were far higher in DFD meat than in control meat: PRDFD and PRC values across significant cases averaged (±SE, standard error) 0.33 ± 0.06 and 0.13 ± 0.03, respectively (P < 0.05, two-tailed Mann-Whitney test). It is also important to note that most (97%) significant cases were unique spots in either DFD or control meats.
Table 1.
Significantly (P < 0.05) changed phosphoprotein spots between DFD and control (non DFD) meat samples of LT bovine muscle
| Spot no.a | PR DFD | PR C | ||
|---|---|---|---|---|
| Mean (±SE) | Adjusted 95% CIs (CL, CU)b | Mean (±SE) | Adjusted 95% CIs (CL, CU) | |
| 1 | 0.41 ± 0.10 | 0.292, 0.603 | 0 | N/A |
| 5 | 0.08 ± 0.03 | 0.051, 0.108 | 0.36 ± 0.12 | 0.169, 0.696 |
| 6 | 0 | N/A | 0.12 ± 0.06 | 0.060, 0.171 |
| 7 | 0.54 ± 0.21 | 0.206, 0.939 | 0 | N/A |
| 8 | 0.48 ± 0.08 | 0.281, 0.614 | 0 | N/A |
| 9 | 0.36 ± 0.10 | 0.153, 0.630 | 0 | N/A |
| 10 | 0.33 ± 0.06 | 0.205, 0.435 | 0 | N/A |
| 11 | 0.31 ± 0.03 | 0.238, 0.371 | 0 | N/A |
| 15 | 0.35 ± 0.16 | 0.154, 0.655 | 0 | N/A |
| 16 | 0.31 ± 0.04 | 0.237, 0.379 | 0 | N/A |
| 18 | 0 | N/A | 0.12 ± 0.04 | 0.067, 0.186 |
| 19 | 0 | N/A | 0.15 ± 0.05 | 0.041, 0.206 |
| 20 | 0 | N/A | 0.13 ± 0.02 | 0.105, 0.179 |
| 21 | 0 | N/A | 0.11 ± 0.05 | 0.031, 0.187 |
| 22 | 0.70 ± 0.12 | 0.458, 0.948 | 0 | N/A |
| 23 | 0.69 ± 0.08 | 0.505, 0.874 | 0 | N/A |
| 24 | 0.66 ± 0.10 | 0.452, 0.804 | 0 | N/A |
| 25 | 0.63 ± 0.06 | 0.447, 0.693 | 0 | N/A |
| 26 | 0.72 ± 0.19 | 0.531, 0.904 | 0 | N/A |
| 27 | 0 | N/A | 0.37 ± 0.19 | 0.175, 0.563 |
| 30 | 0 | N/A | 0.73 ± 0.19 | 0.358, 0.929 |
| 30A | 0.71 ± 0.15 | 0.402, 0.879 | 0 | N/A |
| 33 | 0 | N/A | 0.49 ± 0.24 | 0.033, 0.839 |
| 34 | 0.69 ± 0.25 | 0.192, 0.951 | 0 | N/A |
| 37 | 0.65 ± 0.19 | 0.467, 0.835 | 0 | N/A |
| 44 | 0 | N/A | 0.46 ± 0.20 | 0.087, 0.776 |
| 46 | 0 | N/A | 0.13 ± 0.03 | 0.073, 0,177 |
| 50 | 0 | N/A | 0.09 ± 0.05 | 0.011, 0.194 |
| 51 | 0 | N/A | 0.38 ± 0.14 | 0.211, 0.664 |
| 52 | 0 | N/A | 0.37 ± 0.19 | 0.020, 0.764 |
| 53 | 0.93 ± 0.05 | 0.831, 1.000 | 0 | N/A |
| 54 | 0.94 ± 0.04 | 0.875, 1.000 | 0 | N/A |
aGel position of assigned spots is shown in Fig. 1. bSimultaneous non-parametric bootstrap CIs (CL, lower bound; CU, upper bound) determined by the bias-corrected percentile method (10,000 replicates) and adjusted by the Bonferroni method. N/A, not applicable
Phosphoprotein identification by tandem MS
Phosphoprotein spots with significantly changed phosphorylation level in sample groups (i.e. 32 spots) were excised from 2-DE gels, processed for in-gel trypsin digestion and confidently (P < 0.05) identified by MALDI-TOF and MALDI-TOF/TOF MS. The resulting protein identifications are shown in Table 2 (see Additional file 1: Table S2 for further information). It can be seen that 13 nonredundant proteins with a variable number of isoforms (zero to seven) were identified. Protein identifications corresponded to myofibrillar proteins [actin (ACTA1), fast skeletal myosin regulatory light chain 2 (MYLPF), myosin regulatory light chain 2 (MYL2) and myosin, light chain 6B (MYL6B)]; muscle contraction regulation proteins [slow skeletal muscle troponin T (TNNT1) and fast skeletal muscle troponin T (TNNT3)]; actin polymerization protein [alpha-2 subunit of the F-actin-capping protein (CAPZA2)]; enzymes involved in glycogenolysis (phosphoglucomutase-1, PGM1), glycolysis (beta-enolase, ENO3) and interconversion between creatine and phosphocreatine (creatin kinase M-type, CKM); cytochrome b-c1 complex subunit (UQCRC1); and small heat shock proteins beta-1 (HSPB1) and beta-6 (HSPB6).
Table 2.
Identification of differentially (P < 0.05) phosphorylated 2-DE protein spots in DFD and control meat samples by MALDI-TOF and MALDI-TOF/TOF MS
| Spot no.a | Type of meat | Protein identityb | Abbrev. (isospot) | Accession | Mr (kDa) Obs./Expc | pI Obs./Expc |
Scored | Sequence coveragee |
|---|---|---|---|---|---|---|---|---|
| 1 | DFD | Phosphoglucomutase-1 | PGM1 (1) | PGM1_BOVIN | 66.2/61.8 | 5.90/6.36 | 415 | 45 |
| 5 | DFD | Unidentified | 69.9/61.8 | 6.98/6.36 | ||||
| Control | Phosphoglucomutase-1 | PGM1 (2) | PGM1_BOVIN | 69.9/61.8 | 6.98/6.36 | 552 | 41 | |
| 6 | Control | Cytochrome b-c1 complex subunit 1, mitocondrial | UQCRC1 | QCR1_BOVIN | 53.9/53.4 | 5.60/5.94 | 364 | 47 |
| 7 | DFD | Beta-enolase | ENO3 (1) | ENOB_BOVIN | 51.9/47.4 | 6.33/7.60 | 269 | 39 |
| 8 | DFD | Beta-enolase | ENO3 (2) | ENOB_BOVIN | 51.8/47.4 | 6.45/7.60 | 253 | 34 |
| 9 | DFD | Beta-enolase | ENO3 (3) | ENOB_BOVIN | 51.7/47.4 | 6.63/7.60 | 269 | 48 |
| 10 | DFD | Actin, alpha skeletal muscle | ACTA1 (1) | ACTS_BOVIN | 42.2/42.4 | 4.95/5.23 | 474 | 62 |
| 11 | DFD | Actin, alpha 1, skeletal muscle | ACTA1 (2) | A4IFM8_BOVIN | 41.6/42.4 | 4.99/5.23 | 439 | 51 |
| 15 | DFD | Creatin kinase M-type | CKM (1) | KCRM_BOVIN | 45.0/43.2 | 6.34/6.63 | 266 | 39 |
| 16 | DFD | Creatin kinase M-type | CKM (2) | KCRM_BOVIN | 44.1/43.1 | 6.42/6.63 | 369 | 49 |
| 18 | Control | Actin, alpha, skeletal muscle | ACTA1 (3) | ACTS_BOVIN | 45.3/42.4 | 5.58/5.23 | 519 | 43 |
| 19 | Control | Actin, alpha, skeletal muscle | ACTA1 (4) | ACTS_BOVIN | 44.5/42.4 | 5.78/5.23 | 453 | 39 |
| 20 | Control | Actin, alpha, skeletal muscle | ACTA1 (5) | ACTS_BOVIN | 40.0/42.4 | 5.80/5.23 | 423 | 33 |
| 21 | Control | Actin, alpha, skeletal muscle | ACTA1 (6) | ACTS_BOVIN | 36.5/42.4 | 5.45/5.23 | 484 | 53 |
| 22 | DFD | Troponin T, fast skeletal muscle | TNNT3 (1) | TNNT3_BOVIN | 37.8/32.1 | 6.31/5.99 | 66 | 8 |
| 23 | DFD | Troponin T fast skeletal muscle type | TNNT3 (2) | TNNT3_BOVIN | 37.8/32.1 | 6.38/5.59 | 62 | 13 |
| 24 | DFD | Troponin T, fast skeletal muscle | TNNT3 (3) | TNNT3_BOVIN | 37.8/32.1 | 6.44/5.99 | 145 | 13 |
| 25 | DFD | Troponin T, fast skeletal muscle | TNNT3 (4) | TNNT3_BOVIN | 37.8/32.1 | 6.62/5.99 | 60 | 13 |
| 26 | DFD | F-actin-capping protein subunit alpha-2 | CAPZA2 | CAZA2_BOVIN | 35.4/33.1 | 5.48/5.57 | 74 | 16 |
| 27 | Control | Actin, alpha skeletal muscle | ACTA1 (7) | ACTS_BOVIN | 35.8/42.4 | 5.54/5.23 | 300 | 23 |
| 30 | Control | Troponin T, slow skeletal muscle | TNNT1 (1) | TNNT1_BOVIN | 33.8/31.3 | 6.53/5.71 | 109 | 20 |
| 30A | DFD | Troponin T, slow skeletal muscle | TNNT1 (2) | TNNT1_BOVIN | 35.5/31.3 | 5.63/5.71 | 71 | 14 |
| 33 | Control | Heat shock protein beta-1 | HSPB1 (1) | HSPB1_BOVIN | 25.8/22.4 | 5.05/5.98 | 230 | 36 |
| 34 | DFD | Heat shock protein beta-1 | HSPB1 (2) | E1BEL7_BOVIN | 26.2/22.6 | 5.09/5.77 | 159 | 20 |
| 37 | DFD | Heat shock protein beta-1 | HSPB1 (3) | E1BEL7_BOVIN | 25.9/22.6 | 5.34/5.77 | 163 | 27 |
| 44 | Control | Myosin, light chain 6B, alkali, smooth muscle and non-muscle | MYL6B (1) | Q148H2_BOVIN | 20.9/23.5 | 5.43/5.40 | 109 | 26 |
| 46 | Control | Myosin, light chain 6B, alkali, smooth muscle and non-muscle | MYL6B (2) | Q148H2_BOVIN | 21.1/23.5 | 5.69/5.40 | 329 | 58 |
| 50 | Control | Heat shock protein beta-6 | HSPB6 | HSPB6_BOVIN | 18.7/17.5 | 6.28/5.95 | 146 | 39 |
| 51 | Control | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MYL2 (1) | MLRV_BOVIN | 18.1/18.9 | 4.86/4.86 | 399 | 65 |
| 52 | Control | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MYL2 (2) | F1ME15_BOVIN | 18.2/18.9 | 4.90/4.86 | 380 | 76 |
| 53 | DFD | Myosin regulatory light chain 2, fast skeletal muscle isoform | MYLPF (1) | MLRS_BOVIN | 17.7/19.1 | 4.74/4.91 | 363 | 61 |
| 54 | DFD | Myosin regulatory light chain 2, fast skeletal muscle isoform | MYLPF (2) | MLRS_BOVIN | 17.6/19.1 | 4.77/4.91 | 88 | 23 |
aGel position of assigned spots is shown in Fig. 1. bAll identified proteins were matched to Bos taurus proteins. cTheoretical (Th) isoelectric point (pI) and molecular mass (Mr) were obtained from UniProtKB/Swiss-Prot databases. Observed (Ob) pI and Mr were obtained from the spot position on the gel. dThe Mascot baseline statistically significant (P-value < 0.05) score was 56. ePercentage of coverage of the entire amino acid sequence by matched peptides
Quantitation of protein phosphorylation changes
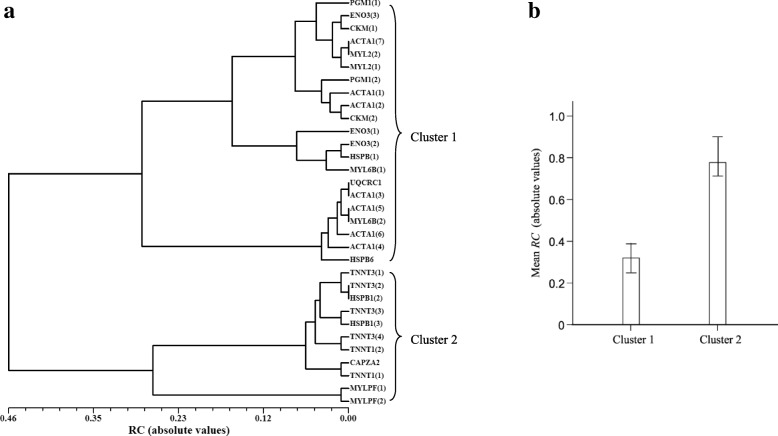
Quantitative changes of PR between DFD and control meats were measured by the fold change (FC) and relative change (RC) coefficients (Table 3). It can be seen that FC, commonly used for measuring changes in protein abundance, was not useful to quantify changes in the phosphorylation status. Thus, most FC-values over phosphoproteins were -∞ or + ∞ because of the high presence of unique phosphorylated protein spots in DFD or control meats. Unlike FC, the RC measure has shown to have advantageous statistical properties in a wide diversity of proteomic scenarios because it always ranges from − 1.0 and + 1.0 across both unshared and shared spots between sample groups [35, 44–46]. Applying RC, we found that MYLPF isoforms underwent the strongest quantitative change at the phosphorylation level (RCMYLPF1(1) = + 0.99; RCMYLPF1(2) = + 1.0); followed by TNNT1 (1–2), CAPZA2, TNNT3 (1–2) and HSPB1 (2) phosphoproteins with absolute RC values higher than 0.70. The UPGMA dendrogram based on pairwise mean differences in RC between sample groups (absolute values) distinguished two major phosphoprotein clusters (Fig. 2a). The phosphoprotein cluster including MYLPF (1–2), TNNT1 (1–2), TNNT3 (1–4), HSPB1 (2) and CAPZA2 exhibited statistically significant higher levels of phosphorylation than those of the other cluster (P < 0.05, 95% bootstrap CIs adjusted with the Bonferroni correction; Fig. 2b). The sign or direction of the change in phosphorylation levels between DFD and control meats was extremely variable across proteins identified in this study without any apparent function-dependent trend (Fig. 3).
Table 3.
Change in the PR of proteins between DFD and control meats measured by FC and RC coefficients
| Spot no.a | Protein (isospot) | FC | RC | Spot no. | Protein (isospot) | FC | RC |
|---|---|---|---|---|---|---|---|
| 1 | PGM1 (1) | +∞ | + 0.44 | 24 | TNNT3 (3) | +∞ | + 0.70 |
| 5 | PGM1 (2) | −4.50 | −0.30 | 25 | TNNT3 (4) | +∞ | + 0.67 |
| 6 | UQCRC1 | -∞ | −0.13 | 26 | CAPZA2 | +∞ | + 0.77 |
| 7 | ENO3 (1) | +∞ | + 0.57 | 27 | ACTA1 (7) | -∞ | −0.39 |
| 8 | ENO3 (2) | +∞ | + 0.51 | 30 | TNNT1 (1) | -∞ | −0.78 |
| 9 | ENO3 (3) | +∞ | + 0.38 | 30A | TNNT1 (2) | +∞ | + 0.76 |
| 10 | ACTA1 (1) | +∞ | + 0.35 | 33 | HSPB1 (1) | -∞ | −0.52 |
| 11 | ACTA1 (2) | +∞ | + 0.33 | 34 | HSPB1 (2) | +∞ | + 0.73 |
| 15 | CKM (1) | +∞ | + 0.37 | 37 | HSPB1 (3) | +∞ | + 0.69 |
| 16 | CKM (2) | +∞ | + 0.32 | 44 | MYL6B (1) | -∞ | −0.49 |
| 18 | ACTA1 (3) | -∞ | −0.13 | 46 | MYL6B (2) | -∞ | −0.14 |
| 19 | ACTA1 (4) | -∞ | −0.16 | 50 | HSPB6 | -∞ | −0.10 |
| 20 | ACTA1 (5) | -∞ | −0.14 | 51 | MYL2 (1) | -∞ | −0.40 |
| 21 | ACTA1 (6) | -∞ | −0.12 | 52 | MYL2 (2) | -∞ | −0.39 |
| 22 | TNNT3 (1) | +∞ | + 0.74 | 53 | MYLPF (1) | +∞ | + 0.99 |
| 23 | TNNT3 (2) | +∞ | + 0.73 | 54 | MYLPF (2) | +∞ | + 1.00 |
aGel position of assigned spots is shown in Fig. 1
Fig. 2.
Cluster analysis of phosphoproteins with significant (P-value < 0.05) differential phosphorylation level in DFD and control (non-DFD) meats based on RC-values. a UPGMA dendrogram constructed from the matrix of mean differences in RC between pairs of phosphoproteins (in absolute value) using NTSYS software. The two main clusters in the resulting dendrogram were denoted as clusters 1 and 2. b Mean values of RC (absolute values) for phosphoproteins of clusters 1 and 2 along with their 99% bootstrap CIs
Fig. 3.
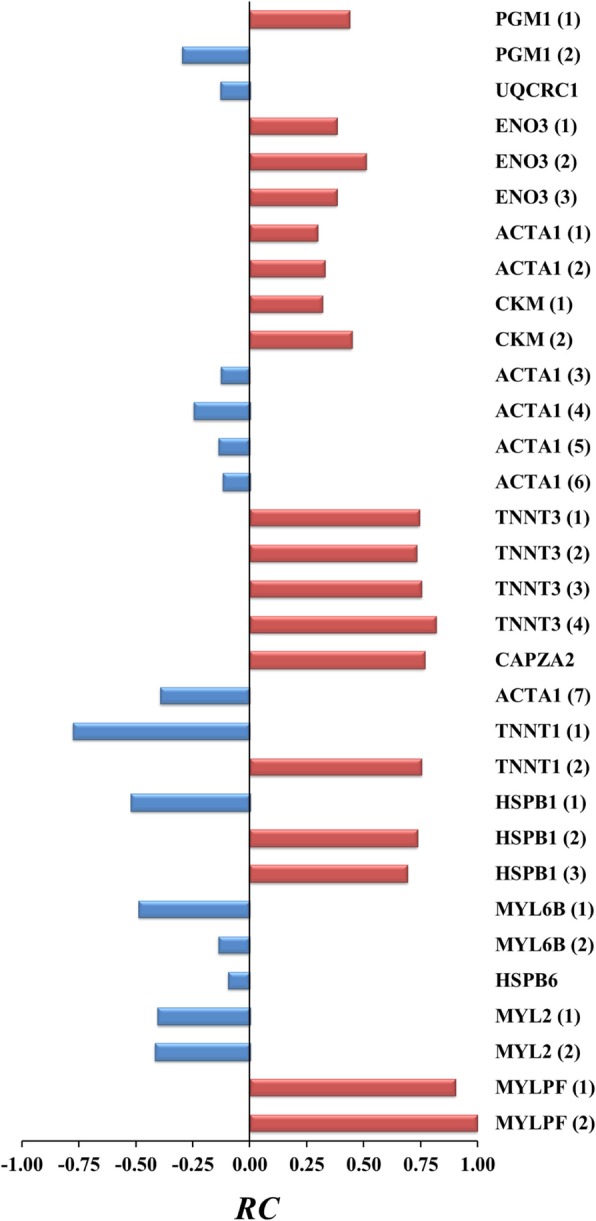
Quantitation of phosphorylation changes (P < 0.05) between phosphoproteins of DFD and control meat samples from the LT bovine muscle assessed by the RC coefficient. Phosphoproteins in DFD meat with higher (red) and lower (blue) phosphorylation levels than in control (or normal) meat are shown
Functional categorization of phosphoproteins from GO terms
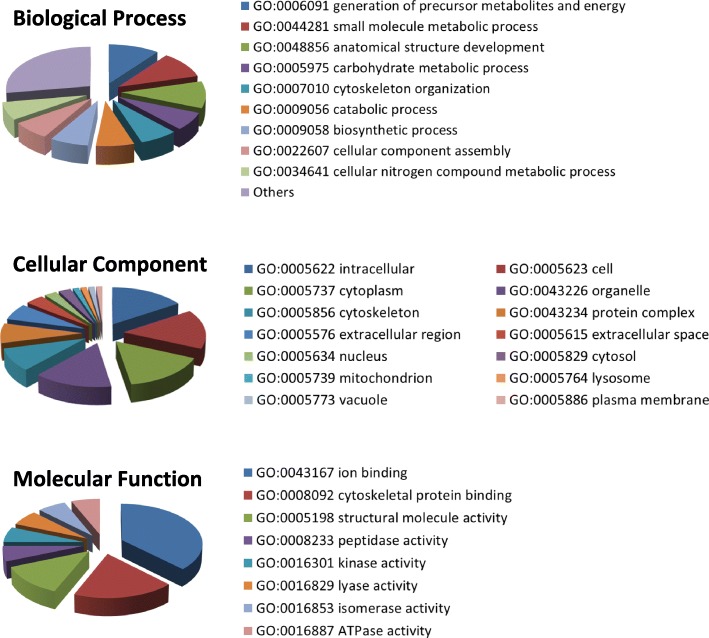
Analysis of broader or high level GO slim terms using the Slimmer tool of AmiGO software as well as fine-grained GO terms by means of QuickGo tool showed that the 13 differentially phosphorylated proteins participate in biological processes such as glycogen biosynthesis and skeletal muscle contraction and activities such as actin, tropomyosin and calcium ion binding. They can be found in different locations such as extracellular space, cell membrane and inside cells as a constituent component of myosin or troponin complex (Fig. 4; Additional file 1: Table S3). In addition, FatiGo enrichment analysis from GO, InterPro and KEGG database terms revealed that three InterPro terms [troponin (IPR001978), alpha crystallin/heat shock protein (IPR001436) and heat shock protein Hsp20 (IPR002068)] and three GO cellular component terms [actin cytoskeleton (GO: 0015629), myofibril (GO: 0030016) and contractile fiber (GO: 0043292)] were significantly (P < 0.05) overrepresented in the proteome of B. taurus (Additional file 1: Table S4).
Fig. 4.
Pie chart reporting the distribution of high level GO functional annotation terms (GO Slim) of the three different ontologies (biological process, molecular function and cellular component) for the 13 differentially phosphorylated proteins between DFD and control meats. GO slim terms were retrieved by mean of the Slimmer tool of AmiGO software
Phosphoprotein-phosphoprotein interaction networks
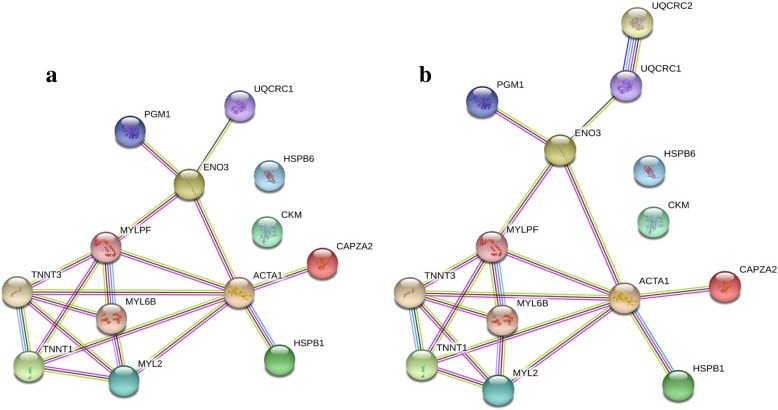
The network of known and predicted interactions of proteins with differential (P < 0.05) phosphorylation level in DFD and control meats according to STRING database searching is shown in Fig. 5. The interaction network map revealed several outstanding facts. All phosphoproteins were clustered into a single interaction network, with the exception of HSPB6 and CKM (Fig. 5a; settings: zero interactions to show in the first and second shell). Most phosphoprotein-phosphoprotein interactions involved muscle structural-contractile, muscle contraction regulation and actin polymerization functions. ACTA1 showed the largest number of interacting partners, which suggests that it could play a key role in response mechanisms to PSS. In addition, ACTA1 together with MYLPF were the only two nodes on the interaction map connecting structural-contractile muscle phosphoproteins with metabolism phosphoenzymes. It was found that no other protein interacts directly with the phosphoproteins identified in this study (Fig. 5b; settings: number of interaction to show, one in the first shell and none in the second shell). This result suggests that the comparative analysis of 2-DE-based phosphoproteome profiles between DFD and control meats was able to successfully identify the core of protein-protein interaction networks linked to PSS.
Fig. 5.
Graphs showing the interaction networks of differentially (P < 0.05) phosphorylated proteins in DFD and control meats, according to STRING confidence view. a) STRING network view only for differentially phosphorylated proteins identified in the present study (specific settings: number of interaction to show, zero in the first and the second shell). b) The same interaction networks adding other directly interacting proteins (specific settings: number of interaction to show, one in the first shell and zero in the second shell). The network nodes (circles) are phosphoproteins and the edges show known or predicted functional associations (threshold: 0.4, medium confidence interval). Colored lines between the phosphoproteins indicate the various types of interaction evidence (blue line: co-occurrence; light blue line: database evidence; black line: coexpression; green line: neighborhood evidence; purple line: experimental evidence; red line: fusion evidence; yellow line: text mining evidence)
Two small clusters of related phosphoproteins were obtained when a phosphoprotein-phosphoprotein co-expression specific network was retrieved from STRING database: a cluster of structural-contractile phosphoproteins (MYLPF and TNNT3) and other group of metabolism phosphoenzymes (PGM1, ENO3 and UQCRC1) (Additional file 1: Figure S1; settings: zero interactions to show in the first and second shell). It is noteworthy, however, that STRING database is based on pre-existing knowledge of protein-protein interactions completed in very diverse biological scenarios. By way of contrast, our observations provide previously unknown information on correlated changes at the phosphorylation level of proteins that occur specifically in response to PSS. Accordingly, we found more extensive correlations between proteins differentially phosphorylated in DFD and control meats than STRING database searching (Table 3, Fig. 2).
Discussion
Our observations revealed remarkable phosphoproteome changes induced by PSS. Global protein phosphorylation levels were found to be about three times higher in DFD meat than in normal meat and most differentially phosphorylated spots were only identified either in DFD or control samples. In addition, changes in the status of protein phosphorylation between DFD and normal meats were noticeably higher than those changes in protein abundance from the same meat samples previously reported by Franco et al. [35]. Thus, we identified a total of 32 protein spots with statistically significant differences in the phosphorylation status between DFD and normal meats, whereas only 10 protein spots showed statistically significant differential abundance over total proteome (P < 0.05, two-tailed Fisher’s exact test).
The huge majority of phosphorylation changes between sample groups involved a single interaction network according to STRING database, which included muscle contraction, glycolysis, actin polymerization, mitochondrial electron transport and stress response related phosphoproteins. This result suggests that reversible phosphorylation in a few interacting proteins can provoke rapid and extensive changes of meat proteome expected to occur in response to PSS. In addition, ACTA1 and MYLPF seem to play a pivotal role in the interaction network because they are the only two proteins mediating the interactions between structural-contractile muscle phosphoproteins and other interacting phosphoproteins. In particular, the ACTA1 protein binds to the largest number of interacting partners, which suggests that it might be a hub protein with a key role in PSS response. Most (71%) phosphorylated actin isoforms [i.e. ACTA1 (3–7)] were only identified in relatively less tender control meat. A number of studies have associated decreased beef tenderness with increasing ACTA1 phosphorylation [2, 5]. This relationship can be explained taking into account that ACTA1 phosphorylation prevents the activation of caspase 3, halting the pathway to apoptosis [2]. Li et al. [47] also reported that the phosphorylation of ACTA1 by protein kinase A prevents its degradation by μ-calpains.
The MYL2 isoforms [i.e. MYL2 (1–2), MYLPF (1–2)] showed opposite phosphorylation patterns in DFD and control meats. Thus, MYL2 (1–2) isoforms increased their phosphorylation levels in relatively less tender normal meat, which is in agreement with previous studies. Reversible phosphorylation of MYLPF regulates the myosin function and is accomplished by opposing activities of Ca2+/calmodulin-dependent skeletal muscle myosin light chain kinase and protein phosphatase type 1 [48–50]. In the post-mortem muscles, MYL2 phosphorylation by myosin light chain kinase is stimulated through sarcoplasmic reticulum Ca2+ release in a concentration-dependent manner [50, 51]. It has been reported that MYL2 phosphorylation occurs during rigor mortis formation and increases at 24 h post-mortem in bovine, porcine and ovine meats, which supports its possible involvement in the rigor mortis progress [4, 6, 7, 51]. In addition, the extent of MYLPF phosphorylation is proportionally related to increased skeletal muscle contraction force of fast-twitch fibers type IIb and tough meat [4, 52, 53]. However, two highly phosphorylated (PR > 90%) MYLPF (1–2) isoforms showed a contrary pattern to MYL2 (1–2) because they were identified only in more tender DFD meat samples. It suggests, therefore, that usual MYLPF phosphorylation pathways can be altered during meat ageing in response to PSS. In addition MYLPF (1–2) isoforms seem to be the most important protein-based meat biomarkers in PSS response. They achieved not only the strongest quantitative change in phosphorylation status between normal and DFD meats (RC-values > + 0.90), but also the most intense differential abundance at the global proteome level using the same control and DFD samples [35]. Other proteins involved in the regulation of muscle contraction (TNNT) and the polymerization of actin (CAPZ) were found to be differentially phosphorylated in DFD and normal meats. Numerous studies support the fact that TNNT is an important substrate of the proteolytic enzymes and the relationship between TNNT degradation and tenderization [54–57]. The phosphorylation of TNNT by cyclic AMP-dependent protein kinase C increases the rate of its proteolysis by μ-calpain probably due to phosphorylation-dependent dissociation of the troponin complex [58]. Huff-Lonergan et al. [59] showed that post-mortem μ-calpain-induced degradation was positively correlated with tenderness in meat samples from LT bovine muscle. Accordingly, most (83%) of the fast [i.e. TNNT3 (1–4)] and slow [i.e. TNNT1 (2)] TNNT skeletal isoforms were phosphorylated only in more tender DFD meat. CAPZA2 phosphorylation was detected only in DFD meat (RC = + 0.77). CAPZA2 is an alpha-2 subunit of the F-actin-capping protein that binds the barber end of actin filaments at Z-discs and blocks actin polymerization and depolymerization [60, 61]. It has been hypothesized that phosphorylation of actin capping protein subunits by protein kinase CK2 may affect the activity of the actin capping protein at the actin filaments [62].
PGM1, ENO3, UQCRC1 and CKM metabolism phosphoenzymes underwent phosphorylation changes between DFD and normal meats. PGM1, ENO3 and UQCRC1 are co-expressed phosphoenzymes that interact with MYLPF and ACTA1 phosphoproteins as shown by STRING database. PGM1 reversibly catalyzes the conversion of glucose 1-phosphate to glucose 6-phosphate in glycolysis and glycogenesis [5]. Phosphorylation of PGM1 significantly enhances its enzymatic activity in response to an increase of glycogenolysis during post-mortem metabolism [63, 64]. Anderson et al. [65] reported that more tender meats from longissimus dorsi bovine presented higher phosphorylated PGM1 isoforms than less tender meat samples. They hypothesized that PGM1 phosphorylation may alter the rate of conversion of glucose 1-phosphate to glucose 6-phosphate, inducing differences in the rate of pH decline. This suggests that energy demands caused by PSS response may provoke an increase of glycogenolysis, enhancing PGM1 phosphorylation in DFD meat. However, D’Alessandro et al. [2] proposed a phosphorylation-induced inhibition by preventing the kinase access to PGM1. On the other hand, all phosphorylated isoforms of ENO3 and CKM were only identified in DFD meat. In agreement with our results, higher phosphorylation levels of these enzymes were found to be positively correlated with pH increase in pork [18]. Β-enolase is a glycolytic enzyme that catalyses the conversion of 2-phospho-D-glycerate to phosphoenolpyruvate [66]. It has been shown that β-enolase phosphorylation increases phosphoenolpyruvate synthesis [66]. Accordingly, phosphorylation of ENO3 in DFD meat samples might be a response to the high energy demands induced by PSS. On the other hand, CKM catalyzes the interconversion of phosphocreatine and ADP to creatine and ATP [67]. Previous studies have shown that CKM is phosphorylated by AMP-activated protein kinase (AMPK) depending on phosphocreatine/creatine ratio and inhibits its activity [68, 69]. Ponticos et al. [68] proposed that long term muscle contraction under extreme situations reduces the phosphocreatine/creatine ratio and full activation of AMPK, ensuring that there will be sufficient ATP to sustain muscle contraction. In this line, increased muscle contraction caused by PSS could lead to increased CKM phosphorylation. Finally, UQCRC1 phosphorylation was only identified in control meat. UQCRC1 is a subunit of the cytochrome bc1 complex (or complex III) of the mitochondrial respiratory chain that transports the electrons from ubiquinol to cytochrome c [70]. The cleavage of cytochrome b-c1 by caspase 3 promotes mitochondrial disruption leading to increased cytochrome c release and apoptosis [71]. Previous studies have shown that protein phosphorylation can prevent protein cleavage by the onset of caspases and apoptosis [72–74]. Therefore, UQCRC1 phosphorylation may help understand that normal meat is less tender than DFD meat at 24 h post-mortem.
The small heat shock proteins (sHSP) HSPB1 and HSPB6 were found to be differentially phosphorylated in DFD and normal meats. HSPB1 (former name HSP27) is a phosphoprotein involved in stress response, actin stability and apoptotic signalling pathways [7, 75]. Herrera-Mendez et al. [76] have proposed that the over-abundance of HSPs at the time of programmed cellular death could have a protective function on structural proteins due to its anti-apoptotic role [76]. The over-abundance of HSPs could delay the apoptotic signaling pathway during meat aging with diverse actions such as hindering the activity of caspases and other intracellular proteolytic systems. On the contrary, decreased abundance of HSPB1 would favor the disorganization and degradation of actin which in turn would weaken the myofibrillar lattice leading to increased tenderness. Overall, the experimental evidence supports that HSPB1 concentration and actin degradation are indeed related to the mean tenderness bovine muscle [77–80]. However, the study of the type of relationship between HSPB1 abundance and tenderness provided conflicting results because the decreased abundance of HSPB1 was associated with either increased [79, 80] or decreased [77, 78] tenderness. It must be highlighted, however, that the anti-apoptotic role of HSPB1 not only depends on its concentration, but also of its phosphorylation status [75]. HSPB1 is capable of inhibiting actin polymerization at the unphosphorylated state [81], but the phosphorylation of HSPB1 following stress abolishes its actin polymerization-inhibiting activity contributing to the maintenance of actin and microfilament network stability [82, 83]. In the present study, we found that HSPB1 isoforms are unequally phosphorylated in meat samples which could contribute to explain their difference in tenderness. The effect of HSPB1 on meat tenderness could be reinforced by HSPB6 (former HSP20) phosphorylation. It has been shown that the phosphorylated isoform of HSPB6 interacts with universal adapter protein 14–3-3 inhibiting the interaction of phosphorylated cofilin with 14–3-3 that induces fragmentation and depolymerization of actin filaments [75].
Conclusions
The present study reports pronounced phosphoproteome changes in DFD beef. Phosphoproteins with differential phosphorylation status between DFD and normal meat were involved in structural-contractile, metabolism, electron transport chain, actin polymerization and stress response related functions. Most (97%) of these phosphoproteins were only detected in either DFD or unaffected meat samples. They could therefore be candidate biomarkers of DFD meat from the LT muscle of the Rubia Gallega breed. Phosphoprotein changes were consistently associated with expected tenderness variations according to previously reported proteomic studies. It is also noteworthy that most differentially changed phosphoproteins were clustered in a single protein-protein interaction network, which can help to understand extensive meat quality variations induced by PSS in a very short period of time. Further follow-up studies are clearly necessary to assess meat phosphoproteome changes linked to the wide range of exogenous and endogenous animal factors that modulate PSS effects, different muscles and breeds, as well as the identification of phosphosites and crosstalk between kinases and phosphatases. This huge challenge will probably require the application of multi-omics technologies.
Methods
Sample information
Phosphoproteomic analyses were performed from four biological replicates of DFD and control (non-DFD) meat samples of the LT muscle extracted from male calves of the Rubia Gallega breed (Spain), which were previously used to assess the response of total proteome to PSS [35]. Briefly, meat samples were taken according to the usual practices in the Spanish beef industry and European Union regulations (Council Directive 93/119/EEC). Animals with a mean age of 10 months were transported from family farms to the abattoir in a time not exceeding one hour, stunned with a captive bolt, slaughtered and dressed in an accredited abattoir (Lugo, Spain). Carcasses were chilled for 24 h in a refrigerated chamber at 2 °C and relative humidity of 98%. The LT muscle was excised from the left half of each carcass, 2.5 cm thick steaks were taken at the fifth rib, vacuum-packed and transported to the laboratory under chilled conditions. Meat quality parameters distinguishing DFD and control meat were evaluated: pH at 24 h post-mortem, color in the CIELAB space [lightness (L*), redness (a*) and yellowness (b*)], water holding capacity (WHC, cooking loss), Warner-Bratzler (WB, shear force) and textural profile analysis (TPA, hardness) tests [35]. Once DFD meat samples were identified according with quality parameter values, control samples from the same farm and slaughtered the same day than DFD samples were selected for the study. Quality parameters were evaluated in meat samples from a total of 76 male calves in order to eventually obtain four biological replicates of each type of meat. Therefore, the incidence of DFD defect was 5.3%. Statistically significant differences (P < 0.05, Mann-Whitney U test) between DFD and control meat samples were detected for all meat quality attributes analyzed (Additional file 1: Figure S2). It can be seen that DFD samples fulfil all typical meat-quality parameter scores of DFD condition [25]. Thus, ultimate pH values in DFD and control samples were always higher and lower than 6.0, respectively. In addition, DFD meats showed darker color in the CIELAB space, higher WHC, and lower shear force (WB-test) and hardness (TPA-test) cuts. A statistically significant correlation of pH-values with L* (P < 0.01, rs = − 0.98, n = 8, Spearman’s nonparametric coefficient of rank correlation), b* (P < 0.01, rs = − 0.91, n = 8), WB (P < 0.01, rs = − 0.93, n = 8), and TPA (P < 0.05, rs = − 0.73, n = 8) values was detected [35], in accordance with previous observations [32, 33]. Finally, meat samples were lyophilized separately under optimal conditions [84] and subsequently frozen at − 80 °C until the time of protein extraction.
Protein extraction and two-dimensional electrophoresis (2-DE)
Total protein extracts from lyophilized meat samples were obtained according to Franco et al. [35]. Extraction and protein purification were performed with the Clean-Up kit (GE Healthcare, Uppsala) from crude cell lysates obtained by ultrasonic disruption using a Branson digital sonifier (model S-250, Branson Ultrasonics, Danbury). Protein concentration in samples was assessed with an improved Bradford method using the CB-X protein assay kit (G-Biosciences, St. Louis) following the instructions of the manufacturer to remove interfering agents and use with a microplate reader. The bovine serum albumin (BSA) protein standard was used to determine protein concentration from calibration curves.
Proteins from lyophilized meat samples were separated by 2-DE as previously described by Franco et al. [35]. Briefly, 350 μg of each biological replicate were loaded onto an immobilized pH gradient (IPG) strip (24-cm long, pH 4–7 linear gradient, ReadyStrip IPG strips, Bio-Rad Laboratories, Hercules). First-dimension isoelectric focusing (IEF) of proteins in strip gel was performed using a PROTEAN IEF cell system (Bio-Rad Laboratories). The second dimension was run on 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using an Ettan DALTsix large vertical electrophoresis system (GE Healthcare).
Phosphoprotein and total protein gel staining
Pro-Q Diamond phosphoprotein stain (Thermo Fisher Scientific, Waltham) was used as a probe for multiplex in-gel detection of phosphorylated polypeptides following the procedure described in Agrawal and Thelen [85]. The PeppermintStick™ (Thermo Fisher Scientific) phosphoprotein molecular weight standards (phosphorylated proteins: ovalbumin/45.0 kDa and β-casein/23.6 kDa; unphosphorylated proteins: β-galactosidase/116.25 kDa, bovine serum albumin/66.2 kDa, avidin/18.0 kDa and lysozyme/14.4 kDa) were used as phosphorylation controls. Phosphorylated and unphosphorylated PeppermintStick protein markers were added to meat protein extracts prior to 2-DE. Pro-Q Diamond-stained gels were post-stained for total protein density with SYPRO Ruby protein gel fluorescent stain (Lonza, Rockland) following the manufacturer’s indications.
Image analysis
The 2-DE images from gels stained with Pro-Q Diamond and SYPRO Ruby fluorescent dyes were captured with the Gel Doc XR+ Imaging System (Bio-Rad Laboratories). Analysis of digitalized gel images was performed with PDQuest Advanced software v. 8.0.1 (Bio-Rad Laboratories). Protein volumes of detected and matched spots over biological replicates were measured following subtraction of background noise and total valid spot normalization. Automatic spot analysis by PDQuest software was manually validated. Only protein spots reproducibly detected in at the least two of four biological replicates were selected for image analyses. The observed isoelectric point (pI) value of protein spots was determined from their gel position relative to focused strips of linear pH gradient, whereas molecular mass markers ranging from 15 to 200 kDa (Fermentas, Ontario) were used to assess the observed molecular mass (Mr). Protein fragments were identified by comparing the Mr observed on 2-DE gels with the theoretical Mr of the full-length sequence and they were excluded from further analysis.
Mass spectrometry (MS) analysis
Protein identification was performed by MALDI–TOF and MALDI-TOF/TOF MS following the procedure described in Franco et al. [35]. In-gel proteolytic digestion of selected protein spots was performed with modified porcine trypsin (Promega, Madison) as described previously [86]. Tryptic peptides were concentrated in a SpeedVac (Thermo Fisher Scientific) and stored at − 20 °C. Dried peptide samples were dissolved in 4 μl 0.5% formic acid and subsequently mixed with an equal volume (0.5 μL) of matrix solution: 3 mg of cyano-4-hydroxycinnamic acid (CHCA) dissolved in 1 mL of 50% acetonitrile (ACN) and acidified with 0.1% trifluoroacetic acid (TFA). The resulting mixture was deposited onto a 384 Opti-TOF MALDI target plate (Applied Biosystems, Foster City) by applying the “thin layer” procedure [87]. MS data were acquired with a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems). Mass spectra of each sample were obtained in positive-ion reflector mode with an Nd:YAG laser operating at 355 nm, an average accumulation of 1000 laser shots and at least three trypsin autolysis peaks for internal calibration. Fragmentation of selected precursor ions was detected with a relative resolution of 300 (FWHM) and metastable suppression. Mass data were analyzed automatically using the 4000 Series Explorer Software v. 3.5 (Applied Biosystems). Combined search of peptide mass-fingerprinting (PMF) and MSMS fragment-ion spectra against the B. taurus UniProtKB/Swiss-Prot databases was performed with GPS Explorer Software v. 3.6 using Mascot software v. 2.1 (Matrix Science, Boston). Mascot search parameters were: one missed cleavage site allowed, precursor mass tolerance of 30 ppm, fragment mass tolerance of 0.35 Da, carbamidomethyl cysteine (CAM) as fixed modification and oxidized methionine as variable modification. All identifications and spectra were manually checked. Protein identification was validated with at least four matched peptides and statistically significant (P-value < 0.05) Mascot probability scores.
Statistical analysis
The phosphorylation rate (PR) for each protein spot was calculated by the ratio PR = P/T, where P and T are the volumes of the same spot on 2-DE gels stained with Pro-Q Diamond and post-stained with SYPRO Ruby, respectively [4, 6]. Quantitative changes of PR between DFD and control meat samples over protein spots were estimated by the fold change (FC) and relative change (RC) coefficients [35, 44]. The coefficient FC was computed for each spot by FC = PRDFD/PRC, where PRDFD and PRC are the mean values of PR across replicate gels in DFD and control meats, respectively. FC-values less than one were represented as their negative reciprocal. Therefore, FC ranges from - ∞ to + ∞ and takes a value of + 1.0 when there is no PR change. The RC coefficient, previously used for measuring changes in protein abundance between treatments [35], was adapted to estimate changes in the status of protein phosphorylation and calculated for each spot by RC = DPR/|DPRmax|, where DPR is the difference in PR between the two types of samples (i.e. PRDFD - PRC) and DPRmax is the maximum observed value of DPR over spots in the study. The RC coefficient has the advantage that it always ranges between − 1.0 and + 1.0 and achieves a value of zero when there is no PR change.
Non-parametric bootstrap confidence intervals (CIs) for mean values of PR across four biological replicates were obtained by the bias-corrected percentile method as previously shown [35, 44]. For each observed mean of PR, 20000 bootstrap samples of size N = 4 were drawn with replacement using the random number generator of Schrage [88]. The 95% bootstrap CIs were computed from bootstrapped empirical distribution of 20000 mean values after bias correction by the percentile method using the proportion of bootstrap mean replications with a mean value lower than the observed value of the mean and the normal distribution [89]. The usual experimental type I error rate of α = 0.05 was controlled for multiple statistical comparisons on the same data using the very conservative Bonferroni adjustment. Descriptive statistics and conventional statistical tests (Mann-Whitney, Spearman’s correlation, etc.) were performed using the IBM SPSS Statistics 20 (SPSS, Chicago) software package.
Phosphoproteins were grouped into clusters by the unweighted pair-group method with arithmetic averaging (UPGMA) from the matrix of pairwise mean differences in RC (absolute values). The UPGMA dendrogram was generated using NTSYSpc v. 2.1 software (Applied Biostatistics, Setauket).
Bioinformatic analysis
Functional classification of phosphoproteins grouped into biological process, molecular function and cellular component categories was carried out using high level Gene Ontology (GO) slim terms retrieved from the pre-existing GO slim generic subset (GO Consortium) by means of the Slimmer tool of AmiGO software [90]. Fine-grained information for each phosphoprotein using GO and all associated electronic and manual GO annotations provided by the GO Consortium annotation groups was retrieved by means of web-based QuickGO tool [91]. GO term enrichment analysis to find over-representation of functional annotations was performed using the FatiGO software available within the set of functional analysis tools of Babelomics 4.0 [92]. Statistically significant overrepresented functional annotations of the genes of interest with respect to the rest of the genome of B. taurus were determined in different databases (GO categories, KEGG and InterPro) using two-tailed Fisher’s exact tests. Adjusted P-values for multiple comparisons were calculated using the false discovery rate (FDR).
Map of known and predicted interaction networks for the proteins with statistically significant differences at the phosphorylation level in DFD and control samples was obtained by using the STRING v10.5 software [93, 94]. In basic settings, the “max. number of interactors to show (1st shell)” was set to zero and 1 to obtain interaction networks only between the proteins identified in our study and other proteins directly associated, respectively.
Additional file
Table S1. Differences in the phosphorylation rate (PR) over 2-DE spots between DFD and control (non-DFD) meat samples from the LT bovine muscle. Table S2. Differentially phosphorylated polypeptides in DFD and control (non-DFD) meat samples from the LT bovine muscle of the Rubia Gallega breed identified by MALDI-TOF and MALDI-TOF/TOF MS. Table S3. List of GO identifiers and terms (biological process, molecular function and cellular component) obtained by the QuickGo tool for differentially phosphorylated proteins in DFD and control meats samples from the LT bovine muscle. Table S4. Significantly (P < 0.05) overrepresented ontologies (study vs. rest of the bovine genome) in DFD and normal meats from the LT bovine muscle, after enrichment analysis by means of the FatiGo software. Figure S1. Phosphoprotein-phosphoprotein interaction network by means of String v10.5 considering exclusively co-expression of differentially phosphorylated proteins in DFD and control bovine meat samples. The network nodes (circles) are phosphoproteins and the edges represent co-expression associations. Threshold: 0.4. Number of interactions to show: none (1st and 2nd shell). Figure S2. Mean values of meat quality parameters (i.e. pH, color, water holding capacity and textural parameters measurements) over four biological replicates of DFD and control meat samples (LT bovine muscle). *Statistically significant (P < 0.05) differences between mean values of control and DFD meat quality parameters. (PDF 1115 kb)
Acknowledgements
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions to improve the quality of the article.
Abbreviations
- 2-DE
Two-dimensional electrophoresis
- ACTA1
Actin
- CAPZA2
F-actin-capping protein subunit alpha-2
- CKM
Creatin kinase M-type
- DFD
Dark, firm and dry
- ENO3
Beta-enolase
- GO
Gene Ontology
- HSPB1
Heat shock protein beta-1
- HSPB6
Heat shock protein beta-6
- LT
Longissimus thoracis
- MALDI-TOF
Matrix-assisted laser desorption/ionization time-of-flight
- Mr
Relative molecular mass
- MS
Mass spectrometry
- MYL2
Myosin regulatory light chain 2
- MYL6B
Myosin, light chain 6B
- MYLPF
Fast skeletal myosin regulatory light chain 2
- PGM1
Phosphoglucomutase-1
- pI
Isoelectric point
- PR
Phosphorylation rate
- PRC
Phosphorylation rate in control meat
- PRDFD
Phosphorylation rate in DFD meat
- PSE
Pale, soft and exudative
- PSS
Pre-slaughter stress
- RC
Relative change
- SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- TNNT1
Slow skeletal muscle troponin T
- TNNT3
Fast skeletal muscle troponin T
- UQCRC1
Cytochrome b-c1 complex subunit 1
Authors’ contributions
CZ conceived and designed the experiments. AM, ML-P, DF, SB and RR-V performed the experiments. AM, ML-P, DF and CZ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
Mass spectrometry analysis, writing of the manuscript and article-processing charges were supported by grant RTA 2014–00034-C04 from the Instituto Nacional de Investigación y Tecnología Agraria (INIA, Spain). Meat samples were obtained by a project FEADER 2010–04 (Consellería de Medio Rural of Xunta de Galicia, Spain). The funding bodies had no involvement in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this article and its supplementary information files.
Ethics approval and consent to participate
Ethic approval was not required. Meat samples were obtained from an accredited abattoir (Lugo, Spain) following the usual practices in the Spanish beef industry and European Union regulations (Council Directive 93/119/EEC). Farmers from “Indicación Geográfica Protegida Ternera Gallega” (Santiago de Compostela, Spain) provided the animals for this research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ariadna Mato, Email: ariadna.matomontero@gmail.com.
Raquel Rodríguez-Vázquez, Email: raquel.rodriguez.vazquez@usc.es.
María López-Pedrouso, Email: mariadolores.lopez@usc.es.
Susana Bravo, Email: sbbravo@gmail.com.
Daniel Franco, Email: danielfranco@ceteca.net.
Carlos Zapata, Email: c.zapata@usc.es.
References
- 1.Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther. 1999;82:111–121. doi: 10.1016/S0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 2.D’Alessandro A, Marrocco C, Rinalducci S, Mirasole C, Failla S, Zolla L. Chianina beef tenderness investigated through integrated omics. J Proteome. 2012;75:4381–4398. doi: 10.1016/j.jprot.2012.03.052. [DOI] [PubMed] [Google Scholar]
- 3.D’Alessandro A, Rinalducci S, Marrocco C, Zolla V, Napolitano F, Zolla L. Love me tender: an omics window on the bovine meat tenderness network. J Proteome. 2012;75:4360–4380. doi: 10.1016/j.jprot.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Li X, Ni N, Liu Y, Chen L, Wang Z, et al. Phosphorylation of myofibrillar proteins in post-mortem ovine muscle with different tenderness. J Sci Food Agric. 2016;96:1474–1483. doi: 10.1002/jsfa.7244. [DOI] [PubMed] [Google Scholar]
- 5.de Souza Rodrigues RT, Luiz Chizzotti M, Elber Vital C, Bacarat-Pereira MC, Barros E, Costa Busato K, et al. Differences in beef quality between Angus (Bos taurus taurus) and Nellore (Bos taurus indicus) cattle through a proteomic and phosphoproteomic approach. PLoS One. 2017;12:e0170294. doi: 10.1371/journal.pone.0170294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Larsen MR, Lametsch R. Changes in phosphorylation of myofibrillar proteins during postmortem development of porcine muscle. Food Chem. 2012;134:1999–2006. doi: 10.1016/j.foodchem.2012.03.132. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, Larsen MR, Palmisano G, Dai J, Lametsch R. Quantitative phosphoproteomic analysis of porcine muscle within 24 h postmortem. J Proteome. 2014;106:125–139. doi: 10.1016/j.jprot.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Wei Y, Li X, Quek SY, Zhao J, Zhong H, et al. Quantitative phosphoproteomic analysis of caprine muscle with high and low meat quality. Meat Sci. 2018;141:103–111. doi: 10.1016/j.meatsci.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Huang H, Larsen MR, Karlsson AH, Pomponio L, Costa LN, Lametsch R. Gel-based phosphoproteomics analysis of sarcoplasmic proteins in postmortem porcine muscle with pH decline rate and time differences. Proteomics. 2011;11:4063–4076. doi: 10.1002/pmic.201100173. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Li X, Xin J, Li Z, Li G, Zhang Y, et al. Effects of protein phosphorylation on color stability of ground meat. Food Chem. 2017;219:304–310. doi: 10.1016/j.foodchem.2016.09.151. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Li M, Li X, Xin J, Wang Y, Shen QW, et al. Quantitative phosphoproteomic analysis among muscles of different color stability using tandem mass tag labeling. Food Chem. 2018;249:8–15. doi: 10.1016/j.foodchem.2017.12.047. [DOI] [PubMed] [Google Scholar]
- 12.Gregory NG. Animal welfare and meat science. Wallingford: CABI Publishing; 1998. [DOI] [PubMed] [Google Scholar]
- 13.Moberg GP, Mench JA. The biology of animal stress: basic principles and implications for animal welfare. Wallingford: CABI Publishing; 2000. [Google Scholar]
- 14.Grandin T. Assessment of stress during handling and transport. J Animal Sci. 1997;75:249–275. doi: 10.2527/1997.751249x. [DOI] [PubMed] [Google Scholar]
- 15.Grandin T. Auditing animal welfare at slaughter plants. Meat Sci. 2010;86:56–65. doi: 10.1016/j.meatsci.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Rushen J, de Passillé AM, von Keyserlingk AG, Wary DM. The welfare of cattle. Dordrecht: Springer; 2008. [Google Scholar]
- 17.Costa LN. Short-term stress: the case of transport and slaughter. Ital J Anim Sci. 2009;8(sup1):241–252. doi: 10.4081/ijas.2009.s1.241. [DOI] [Google Scholar]
- 18.Li X, Fang T, Zong M, Shi X, Xu X, Dai C, et al. Phosphoproteome changes of myofibrillar proteins at early post-mortem time in relation to pork quality as affected by season. J Agric Food Chem. 2015;63:10287–10294. doi: 10.1021/acs.jafc.5b03997. [DOI] [PubMed] [Google Scholar]
- 19.Terlouw EMC, Arnould C, Auperin B, Berri C, Bihan-Duval E, Deiss V, et al. Pre-slaughter conditions, animal stress and welfare: current status and possible future research. Animal. 2008;2:1501–1517. doi: 10.1017/S1751731108002723. [DOI] [PubMed] [Google Scholar]
- 20.Ponnampalam EN, Hopkins DL, Bruce H, Li D, Baldi G, Bekhit AED. Causes and contributing factors to “dark cutting” meat: current trends and future directions: a review. Compr Rev Food Sci F. 2017;16(3):400–430. doi: 10.1111/1541-4337.12258. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson DM, Warner RD. Have we underestimated the impact of pre-slaughter stress on meat quality in ruminants? Meat Sci. 2008;80:12–19. doi: 10.1016/j.meatsci.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Miranda-de la Lama GC, Villarroel M, María GA. Livestock transport from the perspective of the pre-slaughter logistic chain: a review. Meat Sci. 2014;98:9–20. doi: 10.1016/j.meatsci.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Moberg GP. Biological response to stress: implications for animal welfare. In: Moberg GP, Mench JA, editors. The biology of animal stress-basic principles and implications for animal welfare. Oxon: CABI publishing; 2001. pp. 1–22. [Google Scholar]
- 24.Greenberg N, Carr JA, Summers CH. Causes and consequences of stress. Integ Comp Biol. 2002;42:508–516. doi: 10.1093/icb/42.3.508. [DOI] [PubMed] [Google Scholar]
- 25.Adzitey F, Nurul H. Pale soft exudative (PSE) and dark firm dry (DFD) meats: causes and measures to reduce these incidences – a mini review. Int Food Res J. 2011;18:11–20. [Google Scholar]
- 26.Warriss PD. Meat science: An introductory text. 1st ed. New York: CABI Publishing; 2000. [DOI] [PubMed]
- 27.Maltin C, Balcerzak R, Tilley R, Delday M. Determinants of meat quality: tenderness. Proc Nutr Soc. 2003;62:337–347. doi: 10.1079/PNS2003248. [DOI] [PubMed] [Google Scholar]
- 28.Muchenje V, Dzama K, Chimonyo M, Strydom PE, Hugo A, Raats JG. Some biochemical aspects pertaining to beef eating quality and consumer health: a review. Food Chem. 2009;112:279–289. doi: 10.1016/j.foodchem.2008.05.103. [DOI] [Google Scholar]
- 29.Dransfield E. Eating quality of DFD beef. Curr T Vet. 1981;10:344–361. [Google Scholar]
- 30.O’Halloran GR, Troy DJ, Buckley DJ. The relationship between early post-mortem pH and the tenderization of beef muscles. Meat Sci. 1997;2:239–251. doi: 10.1016/S0309-1740(96)00074-5. [DOI] [PubMed] [Google Scholar]
- 31.Purchas RW, Aungsupakorn R. Further investigations into the relationship between ultimate pH and tenderness for beef samples from bulls and steers. Meat Sci. 1993;34:163–178. doi: 10.1016/0309-1740(93)90025-D. [DOI] [PubMed] [Google Scholar]
- 32.Silva JA, Patarata L, Martins C. Influence of ultimate pH on bovine meat tenderness during ageing. Meat Sci. 1999;52:453–459. doi: 10.1016/S0309-1740(99)00029-7. [DOI] [PubMed] [Google Scholar]
- 33.Abril M, Campo MM, Önenç A, Sañudo C, Albertí P, Negueruela AI. Beef colour evolution as a function of ultimate pH. Meat Sci. 2001;58:69–78. doi: 10.1016/S0309-1740(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 34.Wulf DM, Emnett RS, Leheska JM, Moeller SJ. Relationships among glycolytic potential, dark cutting (dark, firm, and dry) beef, and cooked beef palatability. J Animal Sci. 2002;80:1895–1903. doi: 10.2527/2002.8071895x. [DOI] [PubMed] [Google Scholar]
- 35.Franco D, Mato A, Salgado FJ, López-Pedrouso M, Carrera M, Bravo S, et al. Tackling proteome changes in the longissimus thoracis bovine muscle in response to pre-slaughter stress. J Proteome. 2015;122:73–85. doi: 10.1016/j.jprot.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Viljoen HF, De Kock HL, Webb EC. Consumer acceptability of dark, firm and dry (DFD) and normal pH beef steaks. Meat Sci. 2002;61:181–185. doi: 10.1016/S0309-1740(01)00183-8. [DOI] [PubMed] [Google Scholar]
- 37.Ouali A, Gagaoua M, Boudida Y, Becila S, Boudjellal A, Herrera-Mendez CH, et al. Biomarkers of meat tenderness: present knowledge and perspectives in regards to our current understanding of the mechanisms involved. Meat Sci. 2013;95:854–870. doi: 10.1016/j.meatsci.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Marco-Ramell A., de Almeida A. M., Cristobal S., Rodrigues P., Roncada P., Bassols A. Proteomics and the search for welfare and stress biomarkers in animal production in the one-health context. Molecular BioSystems. 2016;12(7):2024–2035. doi: 10.1039/C5MB00788G. [DOI] [PubMed] [Google Scholar]
- 39.Desai MA, Jackson V, Zhai W, Suman SP, Nair MN, Beach CM, et al. Proteome basis of pale, soft, and exudative-like (PSE-like) broiler breast (Pectoralis major) meat. Poult Sci. 2016;95:2696–2706. doi: 10.3382/ps/pew213. [DOI] [PubMed] [Google Scholar]
- 40.Fuente-Garcia Claudia, Aldai Noelia, Sentandreu Enrique, Oliván Mamen, García-Torres Susana, Franco Daniel, Zapata Carlos, Sentandreu Miguel Angel. Search for proteomic biomarkers related to bovine pre-slaughter stress using liquid isoelectric focusing (OFFGEL) and mass spectrometry. Journal of Proteomics. 2019;198:59–65. doi: 10.1016/j.jprot.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Xia A-Q, Chen L-J, Du M-T, Chen L, Kan N, et al. Effects of lairage after transport on post-mortem muscle glycolysis, protein phosphorylation and lamb meat quality. J Integr Agr. 2018;17:2336–2344. doi: 10.1016/S2095-3119(18)61922-7. [DOI] [Google Scholar]
- 42.Li X, Fang T, Zong M, Shi X, Xu X, Dai C, et al. Phosphorproteome changes of myofibrillar proteins at early post-mortem time in relation to pork quality as affected by season. J Agr Food Chem. 2015;63(47):10287–10294. doi: 10.1021/acs.jafc.5b03997. [DOI] [PubMed] [Google Scholar]
- 43.Huang H, Scheffler TL, Gerrard DE, Larsen MR, Lametsch R. Quantitative proteomics and phosphoproteomics analysis revealed different regulatory mechanisms of halothane and Rendement Napole genes in porcine muscle metabolism. J Proteome Res. 2018;17(8):2834–2849. doi: 10.1021/acs.jproteome.8b00294. [DOI] [PubMed] [Google Scholar]
- 44.Franco D, Mato A, Salgado FJ, López-Pedrouso M, Carrera M, Bravo S, et al. Quantification of proteome changes in bovine muscle from two-dimensional electrophoresis data. Data Brief. 2015;4:100–104. doi: 10.1016/j.dib.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López-Pedrouso M, Pérez-Santaescolática C, Franco D, Fulladosa E, Carballo J, Zapata C, et al. Comparative proteomic profiling of myofibrillar proteins in dry-cured ham with different proteolysis indices and adhesiveness. Food Chem. 2018;244:238–245. doi: 10.1016/j.foodchem.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 46.López-Pedrouso M., Pérez-Santaescolástica C., Franco D., Carballo J., Garcia-Perez José V., Benedito J., Zapata C., Lorenzo J.M. Proteomic footprint of ultrasound intensification on sliced dry-cured ham subjected to mild thermal conditions. Journal of Proteomics. 2019;193:123–130. doi: 10.1016/j.jprot.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Li X, Gao X, Shen QW, Du M, Zhang D. Phosphorylation prevents in vitro myofibrillar proteins degradation by μ-calpain. Food Chem. 2017;218:455–462. doi: 10.1016/j.foodchem.2016.09.048. [DOI] [PubMed] [Google Scholar]
- 48.Sweeney H, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol Cell Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 49.Takashima MD. Phosphorylation of myosin regulatory light chain by myosin light chain kinase, and muscle contraction. Circ J. 2009;73:208–213. doi: 10.1253/circj.CJ-08-1041. [DOI] [PubMed] [Google Scholar]
- 50.Stull JT, Kamm KE, Vandenboom R. Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch Biochem Biophys. 2011;510:120–128. doi: 10.1016/j.abb.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muroya S, Ohnishi-Kameyama M, Oe M, Nakajima I, Shibata M, Chikuni K. Double phosphorylation of the myosin regulatory light chain during rigor mortis of bovine longissimus muscle. J Agric Food Chem. 2007;55:3998–4004. doi: 10.1021/jf063200o. [DOI] [PubMed] [Google Scholar]
- 52.Ryder JW, Lau KS, Kamm KE, Stull JT. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J Biol Chem. 2007;282:20447–20454. doi: 10.1074/jbc.M702927200. [DOI] [PubMed] [Google Scholar]
- 53.Lana A, Zolla L. Proteolysis in meat tenderization from the point of view of each single protein: a proteomic perspective. J Proteome. 2016;47:85–97. doi: 10.1016/j.jprot.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Purchas RW, Yan X, Hartley DG. The influence of period of ageing on the relationship between ultimate pH and shear values of beef M. longissimus thoracis. Meat Sci. 1999;51:135–141. doi: 10.1016/S0309-1740(98)00111-9. [DOI] [PubMed] [Google Scholar]
- 55.van Laack RLJM, Stevens SG, Stalder KJ. The influence of ultimate pH and intramuscular fat content on pork tenderness and tenderization. J Anim Sci. 2001;79:392–397. doi: 10.2527/2001.792392x. [DOI] [PubMed] [Google Scholar]
- 56.Iwanowska A, Iwanska E, Grzes B, Mikolajczak B, Pospiech E, Rosochacki, et al. Changes in proteins and tenderness of meat from young bulls of four breeds at three ages over 10 days of could storage. Anim Sci Paper Rep. 2010;28:13–25. [Google Scholar]
- 57.Sun X, Chen KJ, Berg EP, Newman DJ, Schwartz CA, Keller WI, et al. Prediction of troponin-T degradation using color image texture features in 10 d aged beef longissimus steaks. Meat Sci. 2014;96:837–842. doi: 10.1016/j.meatsci.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 58.Di Lisa F, De Tullio R, Salamino F, Barbato R, Melloni E, Siliprandi N, et al. Specific degradation of troponin T and I by μ-calpain and its modulation by substrate phosphorylation. Biochem J. 1995;308:57–61. doi: 10.1042/bj3080057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huff-Lonergan E, Mitsuhashi T, Beekman DD, Parrish FC, Jr, Olson DG, Robson RM. Proteolysis of specific muscle structural proteins by μ-calpain at low pH and temperature is similar to degradation in postmortem bovine muscle. J Animal Sci. 1996;74:993–1008. doi: 10.2527/1996.745993x. [DOI] [PubMed] [Google Scholar]
- 60.Caldwell JE, Heiss SG, Mermall V, Cooper JA. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry. 1989;28:8506–8514. doi: 10.1021/bi00447a036. [DOI] [PubMed] [Google Scholar]
- 61.Dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 62.Canton DA, Olsten MEK, Kim K, Doherty-Kirby A, Lajoie G, Cooper JA, et al. The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol Cell Biol. 2005;25:3519–3534. doi: 10.1128/MCB.25.9.3519-3534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gururaj A, Barnes CJ, Vadlamudi RK, Kumar R. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene. 2004;23:8118. doi: 10.1038/sj.onc.1207969. [DOI] [PubMed] [Google Scholar]
- 64.Longo V, Lana A, Bottero MT, Zolla L. Apoptosis in muscle-to-meat aging process: the omic witness. J Proteome. 2015;125:29–40. doi: 10.1016/j.jprot.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 65.Anderson MJ, Lonergan SM, Huff-Lonergan E. Differences in phosphorylation of phosphoglucomutase 1 in beef steaks from the longissimus dorsi with high or low star probe values. Meat Sci. 2014;96:379–384. doi: 10.1016/j.meatsci.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 66.Nettelblad FA, Engström L. The kinetic effects of in vitro phosphorylation of rabbit muscle enolase by protein kinase C. FEBS Lett. 1987;214:249–252. doi: 10.1016/0014-5793(87)80064-9. [DOI] [PubMed] [Google Scholar]
- 67.Cruzen SM, Pearce SC, Baumgard LH, Gabler NK, Huff-Lonergan E, Lonergan SM. Proteomic changes to the sarcoplasmic fraction of predominantly red or white muscle following acute heat stress. J Proteome. 2015;128:141–153. doi: 10.1016/j.jprot.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 68.Ponticos M, Lu QL, Morgan JE, Hardie DG, Partridge TA, Carling D. Dual regulation of the AM-activated protein kinase provides a novel mechanism for the control of creatine kinase in skeletal muscle. EMBO J. 1998;17:1688–1699. doi: 10.1093/emboj/17.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin G, Liu Y, MacLeod KM. Regulation of muscle creatine kinase by phosphorylation in normal and diabetic hearts. Cell Mol Life Sci. 2009;66:135. doi: 10.1007/s00018-008-8575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y, Li M, Wang X, Jin H, Liu S, Xu J, et al. Caspase cleavage of cytochrome c1 disrupts mitochondrial function and enhances cytochrome c release. Cell Res. 2012;22:127. doi: 10.1038/cr.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, et al. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/S1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 73.Fluhrer R, Friedlein A, Haass C, Walter J. Phosphorylation of presenilin 1 at the caspase recognition site regulates its proteolytic processing and the progression of apoptosis. J Biol Chem. 2004;279:1585–1593. doi: 10.1074/jbc.M306653200. [DOI] [PubMed] [Google Scholar]
- 74.Schweigreiter R, Stasyk T, Contarini I, Frauscher S, Oertle T, Klimaschewski L, et al. Phosphorylation-regulated cleavage of the reticulon protein Nogo-B by caspase-7 at a noncanonical recognition site. Proteomics. 2007;7:4457–4467. doi: 10.1002/pmic.200700499. [DOI] [PubMed] [Google Scholar]
- 75.Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potential of small heat shock proteins. Physiol Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- 76.Herrera-Mendez C, Becila S, Boudjellal A, Ouali A. Meat ageing: reconsideration of the current concept. Trends Food Sci Technol. 2006;17:394–405. doi: 10.1016/j.tifs.2006.01.011. [DOI] [Google Scholar]
- 77.Kim NK, Cho S, Lee SH, Park HR, Lee CS, Cho YM, et al. Proteins in longissimus muscle of Korean native cattle and their relationship to meat quality. Meat Sci. 2008;80:1068–1073. doi: 10.1016/j.meatsci.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 78.Morzel M, Terlow C, Chambon C, Micol D, Picard B. Muscle proteome and meat eating qualities of Longissimus thoracis of “blonde d’Aquitaine” young bulls: a central role of HSP27 isoforms. Meat Sci. 2008;78:297–304. doi: 10.1016/j.meatsci.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 79.Guillermin N, Cassar-Malek I, Hocquette JF, Jurie C, Micol D, Listrat A, et al. INRA Prod. Anim. 2009;22:331–334. [Google Scholar]
- 80.Laville E, Sayd T, Morzel M, Blinet S, Chambon C, Lepetit J, et al. Proteome changes during meat aging in tough and tender beef suggest the importance of apoptosis and protein solubility for beef aging and tenderization. J Agric Food Chem. 2009;57(22):10755–10764. doi: 10.1021/jf901949r. [DOI] [PubMed] [Google Scholar]
- 81.Miron T, Vancompernolle K, Vandekerckhove J, Wilchek M, Geiger B. A 25-kD inhibitor of actin polimerization is a low molecular mass heat shock protein. J Cell Biol. 1991;114:255–261. doi: 10.1083/jcb.114.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benndorf R, Hayess K, Ryazantsev S, Wieske M, Behlke J, Lutsch G. Phosphorylation and supramolecular organization of murine small heat shock protein HSP25 abolish its actin polymerization-inhibiting activity. J Biol Chem. 1994;269:20780–20784. [PubMed] [Google Scholar]
- 83.Guay J, Lambert H, Gringas-Breton G, Lavoie JN, Huot J, Landry J. Regulation of actin filament dynamics by p38 kinase-mediated phosphorylation of heat shock protein 27. J Cell Sci. 1997;110:357–368. doi: 10.1242/jcs.110.3.357. [DOI] [PubMed] [Google Scholar]
- 84.Carpentier SC, Dens K, Van den Houwe I, Swennen R, Panis B. Lyophilization, a practical way to store and transport tissues prior to protein extraction for 2-DE analysis? Pract Proteomics. 2007;1:64–69. doi: 10.1002/pmic.200700529. [DOI] [PubMed] [Google Scholar]
- 85.Agrawal GK, Thelen JJ. Development of a simplified, economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics. 2005;5:4684–4688. doi: 10.1002/pmic.200500021. [DOI] [PubMed] [Google Scholar]
- 86.Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- 87.Vorm O, Roepstorff P, Mann N. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. doi: 10.1021/ac00091a044. [DOI] [Google Scholar]
- 88.Schrage L. A more portable Fortran random number generator. ACM Transactions Mathematical Software. 1979;5:132–138. doi: 10.1145/355826.355828. [DOI] [Google Scholar]
- 89.Efron B. The jackknife, the bootstrap and other resampling plans. CBMS-NSF regional conference series in applied mathematics no. 38. 1st ed. Philadelphia: Society for Industrial and Applied Mathematics; 1982. [Google Scholar]
- 90.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R. QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;22:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein networks integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differences in the phosphorylation rate (PR) over 2-DE spots between DFD and control (non-DFD) meat samples from the LT bovine muscle. Table S2. Differentially phosphorylated polypeptides in DFD and control (non-DFD) meat samples from the LT bovine muscle of the Rubia Gallega breed identified by MALDI-TOF and MALDI-TOF/TOF MS. Table S3. List of GO identifiers and terms (biological process, molecular function and cellular component) obtained by the QuickGo tool for differentially phosphorylated proteins in DFD and control meats samples from the LT bovine muscle. Table S4. Significantly (P < 0.05) overrepresented ontologies (study vs. rest of the bovine genome) in DFD and normal meats from the LT bovine muscle, after enrichment analysis by means of the FatiGo software. Figure S1. Phosphoprotein-phosphoprotein interaction network by means of String v10.5 considering exclusively co-expression of differentially phosphorylated proteins in DFD and control bovine meat samples. The network nodes (circles) are phosphoproteins and the edges represent co-expression associations. Threshold: 0.4. Number of interactions to show: none (1st and 2nd shell). Figure S2. Mean values of meat quality parameters (i.e. pH, color, water holding capacity and textural parameters measurements) over four biological replicates of DFD and control meat samples (LT bovine muscle). *Statistically significant (P < 0.05) differences between mean values of control and DFD meat quality parameters. (PDF 1115 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files.