Abstract
Background
Utilization of quantitative proteomics data on the network level is still a challenge in proteomics data analysis. Currently existing models use sophisticated, sometimes hard to implement analysis techniques. Our aim was to generate a relatively simple strategy for quantitative proteomics data analysis in order to utilize as much of the data generated in a proteomics experiment as possible.
Results
In this study, we applied label-free proteomics, and generated a network model utilizing both qualitative, and quantitative data, in order to examine the early host response to Human Immunodeficiency Virus type 1 (HIV-1). A weighted network model was generated based on the amount of proteins measured by mass spectrometry, and analysis of weighted networks and functional sub-networks revealed upregulation of proteins involved in translation, transcription, and DNA condensation in the early phase of the viral life-cycle.
Conclusion
A relatively simple strategy for network analysis was created and applied to examine the effect of HIV-1 on host cellular proteome. We believe that our model may prove beneficial in creating algorithms, allowing for both quantitative and qualitative studies of proteome change in various biological and pathological processes by quantitative mass spectrometry.
Electronic supplementary material
The online version of this article (10.1186/s12859-019-2990-3) contains supplementary material, which is available to authorized users.
Keywords: Weighted network, Quantitative proteomics, Host response, HIV-1
Background
Utilization of state-of the art proteomics methods can generate thousands of data points, and extensive information on proteins present in the sample can be obtained. High-resolution shotgun proteomics can provide both qualitative and quantitative information about proteins, and can be applied in an unbiased way to study the complete proteome [1, 2]. Despite the high amount of data available, it is sometimes difficult to acquire relevant biological information, in which case sophisticated analytical methods and capable software are needed [3].
Network analysis is widely used in biological data analysis for examination of transcriptomic, proteomic or metabolomic datasets [4–6], and for analyzing interactions between various molecules [7, 8]. In the cellular environment, most of the proteins exert their biological function as part of a complex, or in the form of interactions with other proteins, therefore, application of protein-protein interaction (PPI) analysis methods is advantageous [9]. PPI networks can provide a new layer of information, allowing for the utilization of currently available data, in addition to possibly unravelling hidden biological phenomena [10, 11].
New concepts on network analysis are emerging helping the understanding of biological complexity [12], however, in most cases, only the presence or absence of the protein is considered, the available quantitative data can hardly be incorporated into the network analyses.
The replication cycle of human immunodeficiency virus-1 (HIV-1) is a complex, multi-step, and highly regulated process. The cycle typically begins with viral attachment to cell surface receptors, and ending with the production of infectious virions. Due to the multiple processes involved, the replication cycle has been classically divided into two distinct phases; the early and late phase. The early phase encompasses cell binding, fusion, internalization, uncoating, reverse transcription, as well as integration of the viral cDNA into the host genome. On the other hand, transcription of viral genome, export of viral RNA, assembly of virions at the plasma membrane, as well as budding and maturation of the released virions are parts of the late phase of the replication cycle [13, 14]. While late phase events are relatively well characterized, the precise mechanism and regulation of early phase steps remain poorly understood.
Genomics and proteomics studies were carried out to investigate how HIV-1 hijacks the host cellular machinery, avoiding being sensed by host immune responses. siRNA screens were implemented to study the cellular genes and proteins required for HIV-1 infection [9, 15, 16], HIV-1 protein – host protein protein-protein interaction networks were generated, and the data were deposited in HIV-1 Human Interaction Database [17].
In case of HIV infection, the network-based examinations have identified perturbed host cellular systems; such as the proteasome and transcriptional regulation, and have revealed that HIV-1 preferably interacts with highly connected and central cellular proteins [18–20].
In this study, we have generated the protein expression profiles of cells during early HIV-1 infection using protein mass spectrometry, and integrated the acquired data with knowledge-based protein-protein interaction network to understand how cellular network is perturbed by HIV.
Results
Our aim was to analyze the proteomic landscape of the early stage of HIV-1 based lentiviral vector transduction. 293 T cells were infected with VSV-G pseudotyped HIV-1 vector, and 0, 4 and 12 h post-infection, cell lysates were harvested. Label-free proteomics was applied to examine protein-level changes. Duplicate samples for three time points were collected (0, 4, and 12 h post-transduction) in case of virus transduced samples and in case of control, mock transduced samples. The collected 6 virus treated and 6 control samples were analyzed in duplicates, allowing for the measurement of two technical and two biological replicates for each time point.
The mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium [21] via the PRIDE partner repository with the dataset identifier PXD010436 and 10.6019/PXD010436.
Identified proteins (Additional file 1) were manually curated, and in the case of non-human or non-viral identifications, the sequences were verified. In many instances, they were mistakenly designated as non-human proteins, in which case it was corrected. In few instances, the non-human proteins could not be matched to any of the human or viral proteins, and consequently, these sequences were omitted from further analyses. The data for Rhodobacter capsulatus cytochrome c, bovine pancreatic trypsin inhibitor, bovine serum albumin and pig trypsin were kept to serve as reference for quantitative analyses, but were not used for further computations. The relative amount of proteins was computed based on spectral counting and in case of each protein the mean of the results of the four analyses corresponding to each condition was calculated (Additional file 2).
Statistical analysis
Firstly, a qualitative analysis was carried out to detect newly expressed or down-regulated proteins in the first 4 or 12 h after HIV-1 pseudovirion transduction. Only those proteins were considered for statistical analysis which could be quantified in at least 2 out of 4 replicates, and were not quantified in other conditions. HIST1H1E, HNRNPL, PRRC2A and TRIM28 were quantified only at H04, and there were no proteins quantified solely in H12 time point (Additional files 1, 2). HIST1H1E interacts with linker DNA between nucleosomes, and functions in DNA condensation, HNRNPL and TRIM28 play a role in translation and transcription, while PRRC2A plays a role in inflammatory processes.
Some of the proteins were quantified in all time points except H12. These include ALYREF, CCDC86, CSDA, COX5A, HN1, MYL6, PPIF, SEPT2, SRSF6, TCOF1, and TPM3 (Additional file 1). These proteins participate in RNA binding (ALYREF, CCDC86, SRSF6, TCOF1), DNA binding (CSDA), protein folding (PPIF), energy generation (COX5A), signalization (HN1) and cytoskeleton assembly (MYL6, SEPT2, TPM3).
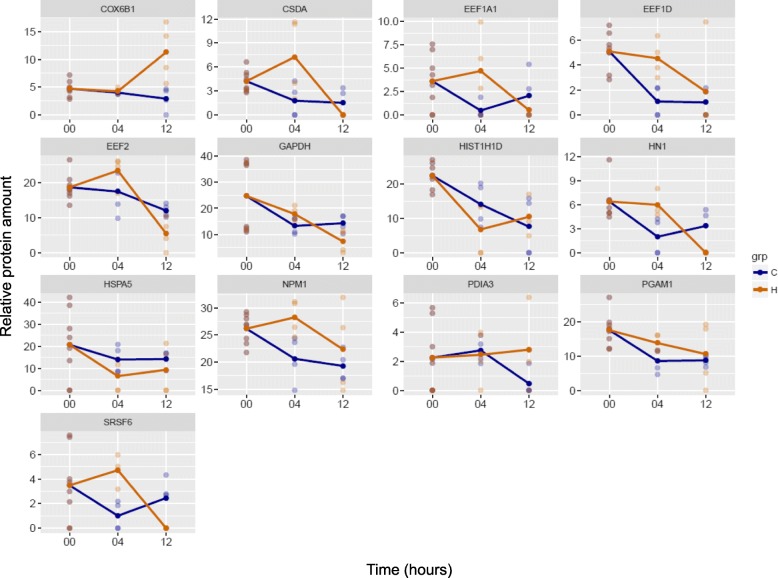
In order to examine changes in the amount of proteins, statistical analysis was carried out (Additional file 3). The amount of CSDA, EEF1A1, EEF1D, HN1, NPM1, PGAM1 and SRSF6 increased significantly, while that of HIST1H1D and HSPA5 significantly decreased in H04 (Fig. 1). It is interesting to note that after peaking in H04, CSDA, HN1 and SRSF6 were not quantified in H12. In H12, compared to C12, the amount of COX6B1 and PDIA3 increased, while that of EEF2 and GAPDH decreased significantly (Fig. 1). When the function of proteins showing statistically significant changes was examined, we observed an increase in the amount of proteins implicated in RNA binding in H04, and an overall decrease in their amount in H12.
Fig. 1.
Relative protein amounts showing statistically significant changes in HIV-1 treated samples compared to controls. The x axis shows the time-points of sample collection in hours, and the y axis shows the relative protein amounts. Blue color refers to control (C), and yellow color to the HIV-1 treated sample (H)
Network analysis
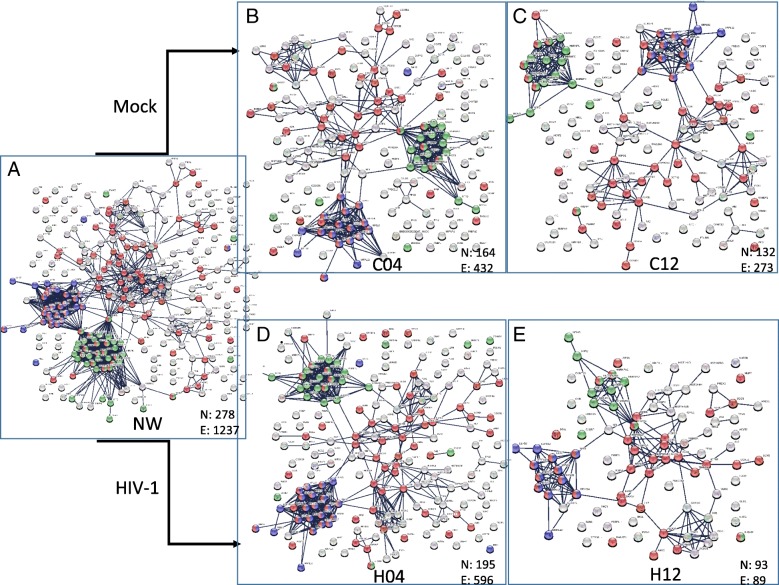
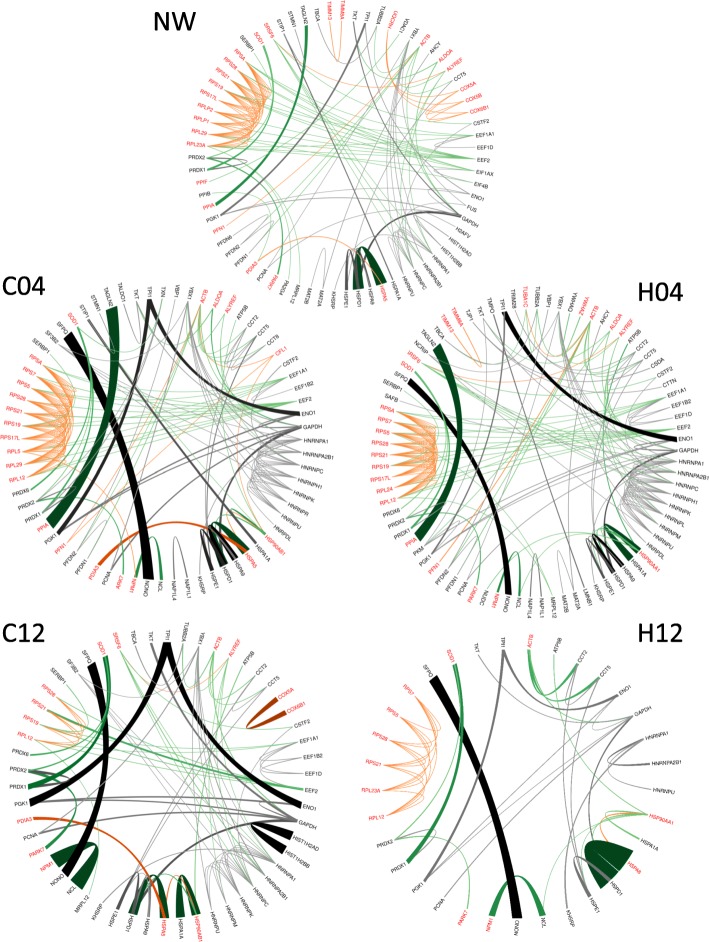
To broaden our insight, and to better understand the possible functional associations of protein changes upon HIV-1 pseudovirion transduction, we have searched for the available protein-protein interactions of the quantified proteins in our datasets. For evaluation of the interactions, the STRING database was used, which contains information on known and predicted, direct physical, and indirect functional protein-protein interactions [22]. Only interactions which were of high confidence (interaction score in STRING database > 0.95) were used. Initially, five binary interaction networks were generated: NW0 combined proteins from mock- and virion-treated cell lysates collected at 0 time-point, C04 and C12 networks contained proteins from the mock-treated cells collected 4 h and 12 h post-infection, respectively, and the H04 and H12 networks contained proteins from the HIV-1 transduced cells collected at 4 and 12 h time-points, respectively (Fig. 2). The number of nodes and the number of edges of the networks show a decreasing trend over time, with a marked shrinkage in H12.
Fig. 2.
Protein-protein interaction network of the proteins quantified in each condition. The PPI networks were generated by STRING (confidence 0.95) using the list of quantified proteins presented in Additional file 2 in case of each condition. The number of nodes (N) and the number of edges (E) according to STRING in case of each network is indicated. a. PPI network of proteins in the 0 h time point (NW), b. PPI network of proteins in the 4 h time point in control, mock-transduced cells (C04). c PPI network of proteins in the 12 h time point in control, mock-transduced cells (C12). d. PPI network of proteins in the 4 h time point in HIV vector-transduced cells (H04). e. PPI network of proteins in the 12 h time point in HIV vector- transduced cells (H12). Red dots represent proteins belonging to transport GO term, blue dots indicate proteins having a role in translation, while green dots indicate proteins with a role in RNA splicing according to the functional enrichment analysis provided by STRING
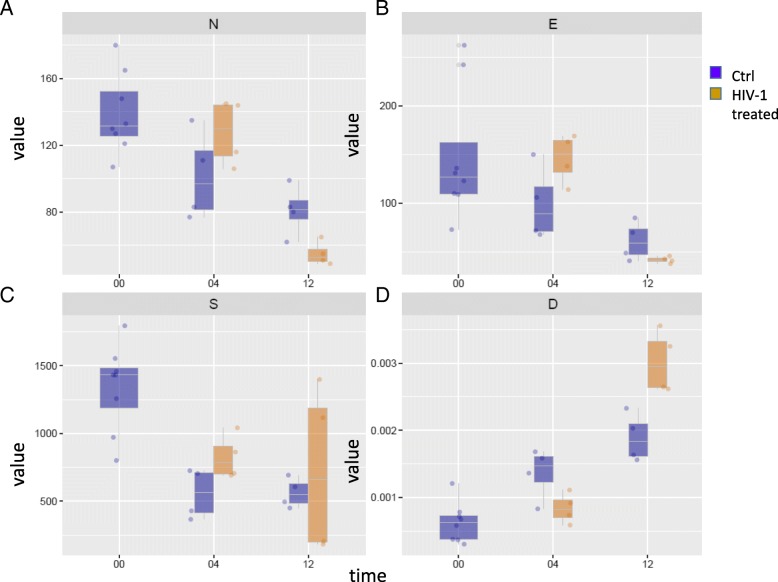
These binary networks provide information solely on the possibility of interaction between two proteins (Fig. 2, Fig. 3a, b), hence, in order to gain more realistic information, protein amounts measured by spectral counting were implemented into the network using a simple statistical model. In this way, binary edges were transformed into estimated protein pair’s interaction intensities in the sample, which is proportional to the amounts of proteins participating in the interaction, and inversely proportional to the number of interactions (Fig. 3c). The weighted networks were examined, and the number of nodes (N), edges (E), network strength (S), edge density (D) and functional and non-functional edge ratio (R) were calculated (Fig. 4).
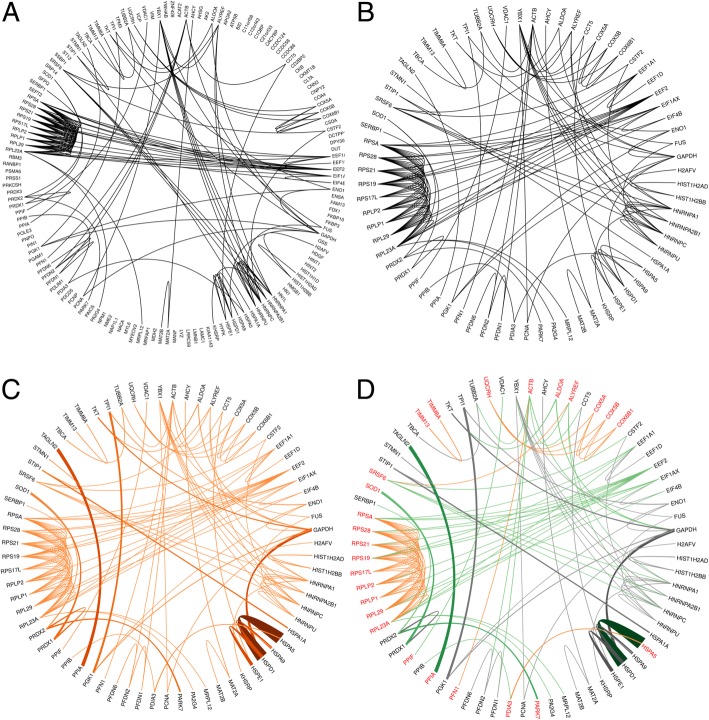
Fig. 3.
Network generation pipeline. Representative network drawn by circlize, showing data for sample1 and interactions generated by STRING. a. Binary network containing all the identified proteins arranged in alphabetical order on the external ring of the circular plot. Thin black curves show the possible interactions generated by STRING. b. Binary network containing only the proteins with interactions. The isolated proteins (i.e. without any connection) were eliminated. Thin black curves show the possible interactions generated by STRING. c. Weighted network containing the interacting proteins. Orange lines represent interactions, the higher the intensity of the color and thickness of the line the higher the interaction strength. d. Weighted network with functional feature. A randomly selected GO function (GO:0044765) is used to illustrate the functional network. Red proteins are part of the functional sub-network, while black proteins are not, being considered as non-functional proteins. The weighted interactions are color-coded according to the protein-pair classification: functional – functional interactions are orange, non-functional – non-functional interactions are gray, and functional – non-functional interactions are green. The interaction strength is represented by the intensity of the color and thickness of the line: the higher the intensity of the color and the thickness of the line, the higher the interaction strength
Fig. 4.
Network parameters. a. Number of nodes (N), b. number of edges (E), c. network strength (S), d. strength or edge density (D) in case of networks observed in the examined conditions. The y axis show the mean value characteristic for each parameter, and the x axis indicates the time points. Blue color refers to control, while the yellow color to the HIV-1 treated conditions
The number of nodes decreased significantly in H12, indicating network shrinkage in H12, observed in the binary network (Fig. 4a). The number of edges and network strength did not change in a statistically significant manner (Fig. 4b, c), however, edge density decreased significantly in H04 while increasing significantly in H12 (Fig. 4d). These changes indicate the presence of a less interactive network in H04, and a smaller; yet more active, PPI network in H12 (Fig. 4a, d).
Next, we were eager to analyze the functionality of the networks, and hence, we generated functional sub-networks of proteins belonging to GO terms. All the Molecular Function, Biological Process and Cellular Component GO terms listed as enriched by STRING in C04, H04, C12 and H12; where at least 10 protein per GO function in any of the networks were present, were considered. To visualize network changes, the GO.0044765 term was chosen randomly (Fig. 3d), and the change of this sub-network was visualized in all time points (Fig. 5). Proteins present in a given GO term listed as enriched by STRING were considered as being part of the functional sub-network (f), whereas proteins not being part of the specific GO term, were considered as non-functional (n) proteins. Three types of interactions were analyzed: i) interactions between proteins belonging to functional sub-networks (f), ii) interactions between proteins not belonging to functional sub-network (n), and iii) interactions between functional and non-functional proteins (c – cross) (Fig. 3d). In order to better understand the changes, a statistical approach was applied, and the following network parameters were calculated: in case of each functional (f) network of proteins belonging to a specific GO term, the Nf, Ef, Sf, Df, and Rf, while for non-functional (n) proteins the Nn, En, Sn, Dn, and Rn network parameters were calculated. In case of interactions between the functional and non-functional proteins (c) the Ec, Sc, Dc, and Rc network parameters were calculated (Additional file 4).
Fig. 5.
Network changes visualized in case of a representative functional sub-network. The representative figure shows the changes in weighted networks in case of proteins belonging to randomly selected GO:0044765 GO term. NW represents the weighted PPI network of interacting proteins in the 0 h time point, C04 and C12 correspond to PPI networks of interacting proteins in the 4 h and 12 h time point, respectively, in control, mock-transduced cells. H04 and H12 represent the weighted PPI network of interacting proteins in the 4 h and 12 h time point, respectively, in HIV vector-transduced cells. Red proteins are part of the functional sub-network, while black proteins are non-functional proteins. The weighted interactions are color-coded according to the protein-pair classification: functional – functional interactions are orange, non-functional – non-functional interactions are gray, and functional – non-functional interactions are green. The interaction strength is represented by the intensity of the color and thickness of the line: the higher the intensity of the color and the thickness of the line, the higher the interaction strength
According to our hypothesis, those GO functions or functional sub-networks might be responsible for the changes induced by HIV-1, where the parameters in the functional network change significantly, whereas in the non-functional network, no statistically significant changes are shown. At the same time, those GO functions where the parameters in the functional network do not change in a statistically significant manner, yet do so in the non-functional sub-networks, are thought to not explain the changes related to HIV-1 transduction.
After statistical analysis and FDR correction of the results (Additional file 5), in case of some GO terms, statistically significant differences were observed. No significant difference in edge and strength values were observed in any of the functional sub-networks (Ef and Sf), and the number of nodes was significantly reduced in H12 only in the case of 5 functional sub-networks (Additional file 6). Considering edge density (D) and ratio (R), only those GO terms were further considered where (i) the significant difference was present only in the functional sub-network (Df and Rf, respectively) and (ii) where the significant difference was present both in the functional sub-network (Df and Rf) and in the cross network (Dc and Rc) (Additional file 6). According to our hypothesis, proteins belonging to the GO terms listed in Table 1 and Table 2, are responsible for the changes of cellular proteome observed in the H04 and H12 networks in response to HIV-1 transduction. In H04 sample, an increase in the node number (proteins present in the network) was observed, however, this increase was not significant. In the same time, a global decrease in interactivity; represented by the number of edges, was noticed. Proteins which might be responsible for this reduced interactivity belong to the RNA processing-related functions (splicing, RNA synthesis, RNA catabolism, translation, transcription), regulation of cell death, regulation of cellular response to stress, viral life cycle (viral gene expression, viral transcription, viral life cycle) and protein localization, and some very general GO terms; such as protein binding, cellular macromolecular biosynthetic process, purine nucleotide binding, organic substance transport, etc. (Table 1). In spite of the reduced global interactivity, some functional sub-networks; such as viral process, protein kinase binding, multi-organism process, de novo protein folding and protein complex subunit organization, show significantly increased interactivity (Table 1).
Table 1.
List of GO terms with significantly different changes in the functional sub-network in H04
| GO code | GO term | Significantly changed parameter | Direction of change in H04 |
|---|---|---|---|
| GO.0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | Df | decrease |
| GO.0000228 | nuclear chromosome | Df | decrease |
| GO.0000375 | RNA splicing, via transesterification reactions | Df | decrease |
| GO.0000398 | mRNA splicing, via spliceosome | Df | decrease |
| GO.0000956 | nuclear-transcribed mRNA catabolic process | Df | decrease |
| GO.0003735 | structural constituent of ribosome | Df | decrease |
| GO.0005198 | structural molecule activity | Df | decrease |
| GO.0005515 | protein binding | Df | decrease |
| GO.0005524 | ATP binding | Df, Dc | decrease |
| GO.0005622 | intracellular | Df | decrease |
| GO.0005737 | cytoplasm | Df | decrease |
| GO.0005739 | mitochondrion | Df | decrease |
| GO.0005840 | ribosome | Df | decrease |
| GO.0006351 | transcription, DNA-templated | Df, Dc | decrease |
| GO.0006366 | transcription from RNA polymerase II promoter | Df, Dc | decrease |
| GO.0006367 | transcription initiation from RNA polymerase II promoter | Df | decrease |
| GO.0006396 | termination of RNA polymerase II transcription | Df | decrease |
| GO.0006397 | mRNA processing | Df | decrease |
| GO.0006401 | RNA catabolic process | Df | decrease |
| GO.0006402 | mRNA catabolic process | Df | decrease |
| GO.0006412 | translation | Df | decrease |
| GO.0006413 | translational initiation | Df, Dc | decrease |
| GO.0006414 | translational elongation | Df, Dc | decrease |
| GO.0006415 | translational termination | Df | decrease |
| GO.0006417 | regulation of translation | Df, Dc | decrease |
| GO.0006518 | peptide metabolic process | Df | decrease |
| GO.0006605 | protein targeting | Df | decrease |
| GO.0006614 | SRP-dependent cotranslational protein targeting to membrane | Df | decrease |
| GO.0006732 | coenzyme metabolic process | Df, Dc | decrease |
| GO.0006886 | intracellular protein transport | Df | decrease |
| GO.0008104 | protein localization | Rf | decrease |
| GO.0008152 | metabolic process | Df | decrease |
| GO.0008380 | RNA splicing | Df | decrease |
| GO.0009056 | catabolic process | Df | decrease |
| GO.0009892 | negative regulation of metabolic process | Df | decrease |
| GO.0009987 | cellular process | Df | decrease |
| GO.0010468 | regulation of gene expression | Df | decrease |
| GO.0010556 | regulation of macromolecule biosynthetic process | Df | decrease |
| GO.0010608 | posttranscriptional regulation of gene expression | Df, Dc | decrease |
| GO.0010941 | regulation of cell death | Df | decrease |
| GO.0015031 | protein transport | Df | decrease |
| GO.0016032 | viral process | Df | increase |
| GO.0016043 | cellular component organization | Df | decrease |
| GO.0016070 | RNA metabolic process | Df, Dc | decrease |
| GO.0016071 | mRNA metabolic process | Df | decrease |
| GO.0016482 | cytoplasmic transport | Df | decrease |
| GO.0016604 | nuclear body | Df, Dc | decrease |
| GO.0017076 | purine nucleotide binding | Df, Dc | decrease |
| GO.0018130 | heterocycle biosynthetic process | Df, Dc | decrease |
| GO.0019058 | viral life cycle | Df | decrease |
| GO.0019080 | viral gene expression | Df | decrease |
| GO.0019083 | viral transcription | Df | decrease |
| GO.0019222 | regulation of metabolic process | Df | decrease |
| GO.0019438 | aromatic compound biosynthetic process | Df, Dc | decrease |
| GO.0019538 | aromatic compound biosynthetic process | Df | decrease |
| GO.0019899 | enzyme binding | Df | decrease |
| GO.0019900 | kinase binding | Df, Dc | increase |
| GO.0019901 | protein kinase binding | Df | increase |
| GO.0022607 | cellular component assembly | Df | decrease |
| GO.0022626 | cytosolic ribosome | Df | decrease |
| GO.0030554 | adenyl nucleotide binding | Df, Dc | decrease |
| GO.0031625 | ubiquitin protein ligase binding | Df, Dc | decrease |
| GO.0031974 | membrane-enclosed lumen | Df | decrease |
| GO.0032550 | purine ribonucleoside binding | Df, Dc | decrease |
| GO.0032553 | ribonucleotide binding | Df, Dc | decrease |
| GO.0032555 | purine ribonucleotide binding | Df, Dc | decrease |
| GO.0032774 | RNA biosynthetic process | Df, Dc | decrease |
| GO.0034248 | regulation of cellular amide metabolic process | Df | decrease |
| GO.0034613 | cellular protein localization | Rf | decrease |
| GO.0034622 | cellular macromolecular complex assembly | Df | decrease |
| GO.0034645 | cellular macromolecule biosynthetic process | Df | decrease |
| GO.0034654 | nucleobase-containing compound biosynthetic process | Df, Dc | decrease |
| GO.0034655 | nucleobase-containing compound catabolic process | Df | decrease |
| GO.0035639 | purine ribonucleoside triphosphate binding | Df, Dc | decrease |
| GO.0036094 | small molecule binding | Df, Dc | decrease |
| GO.0042981 | regulation of apoptotic process | Df | decrease |
| GO.0043065 | positive regulation of apoptotic process | Df | decrease |
| GO.0043066 | negative regulation of apoptotic process | Df, Dc | decrease |
| GO.0043226 | organelle | Df | decrease |
| GO.0043227 | membrane-bounded organelle | Df, Dc | decrease |
| GO.0043229 | intracellular organelle | Df | decrease |
| GO.0043231 | intracellular membrane-bounded organelle | Df | decrease |
| GO.0043233 | organelle lumen | Df | decrease |
| GO.0044085 | cellular component biogenesis | Df | decrease |
| GO.0044237 | cellular metabolic process | Df | decrease |
| GO.0044238 | primary metabolic process | Df | decrease |
| GO.0044248 | cellular catabolic process | Df | decrease |
| GO.0044249 | cellular biosynthetic process | Df | decrease |
| GO.0044260 | cellular macromolecule metabolic process | Df | decrease |
| GO.0044265 | cellular macromolecule catabolic process | Df | decrease |
| GO.0044267 | cellular protein metabolic process | Df | decrease |
| GO.0044271 | cellular nitrogen compound biosynthetic process | Df, Dc | decrease |
| GO.0044391 | ribosomal subunit | Df | decrease |
| GO.0044422 | organelle part | Df | decrease |
| GO.0044424 | intracellular part | Df | decrease |
| GO.0044444 | cytoplasmic part | Df | decrease |
| GO.0044446 | intracellular organelle part | Df | decrease |
| GO.0044451 | nucleoplasm part | Df, Dc | decrease |
| GO.0044454 | nuclear chromosome part | Df | decrease |
| GO.0044802 | single-organism membrane organization | Df, Dc | decrease |
| GO.0045184 | establishment of protein localization | Df | decrease |
| GO.0046907 | intracellular transport | Df, Dc | decrease |
| GO.0051084 | de novo posttranslational protein folding | Df | increase |
| GO.0051704 | multi-organism process | Df, Dc | increase |
| GO.0060255 | regulation of macromolecule metabolic process | Df | decrease |
| GO.0060548 | negative regulation of cell death | Df, Dc | decrease |
| GO.0065003 | macromolecular complex assembly | Df | decrease |
| GO.0070013 | intracellular organelle lumen | Df | decrease |
| GO.0070972 | protein localization to endoplasmic reticulum | Rf | decrease |
| GO.0071013 | catalytic step 2 spliceosome | Df | decrease |
| GO.0071702 | organic substance transport | Df, Dc | decrease |
| GO.0071704 | organic substance metabolic process | Df | decrease |
| GO.0071705 | nitrogen compound transport | Df, Dc | decrease |
| GO.0071822 | protein complex subunit organization | Df | increase |
| GO.0071840 | cellular component organization or biogenesis | Df | decrease |
| GO.0072594 | establishment of protein localization to organelle | Df | decrease |
| GO.0080090 | regulation of primary metabolic process | Df | decrease |
| GO.0080135 | regulation of cellular response to stress | Df, Dc | decrease |
| GO.0090304 | nucleic acid metabolic process | Df, Dc | decrease |
| GO.0097367 | carbohydrate derivative binding | Df, Dc | decrease |
| GO.1901362 | organic cyclic compound biosynthetic process | Df, Dc | decrease |
| GO.1901566 | organonitrogen compound biosynthetic process | Df | decrease |
| GO.1901575 | organic substance catabolic process | Df | decrease |
| GO.1901576 | organic substance biosynthetic process | Df | decrease |
| GO.1902580 | single-organism cellular localization | Df | decrease |
In case of each GO term where statistically significant changes were observed in network parameters in HIV-1 treated cells at 4 h time-point the GO identifier, the changed parameter and the direction of change is indicated
Table 2.
List of GO terms with significantly different changes in the functional sub-networks in H12
| GO code | GO term | Significantly changed parameter | Direction of change in H12 |
|---|---|---|---|
| GO.0005759 | mitochondrial matrix | Nf | decrease |
| GO.0003674 | molecular function | Df, Dc | increase |
| GO.0003723 | RNA binding | Rf, Rc | increase |
| GO.0005488 | binding | Df, Dc | increase |
| GO.0005615 | extracellular space | Rf, Rc | increase |
| GO.0005634 | nucleus | Df, Dc | increase |
| GO.0005654 | nucleoplasm | Df, Dc | increase |
| GO.0005739 | mitochondrion | Df | increase |
| GO.0005743 | mitochondrial inner membrane | Df, Dc | increase |
| GO.0005759 | mitochondrial matrix | Df | increase |
| GO.0005856 | cytoskeleton | Df, Dc | increase |
| GO.0006401 | RNA catabolic process | Df, Dc | increase |
| GO.0006402 | mRNA catabolic process | Df, Dc | increase |
| GO.0009058 | biosynthetic process | Rf, Rc | decrease |
| GO.0009987 | cellular process | Df, Dc | increase |
| GO.0010467 | gene expression | Rf, Rc | increase |
| GO.0016032 | viral process | Rf | increase |
| GO.0016604 | nuclear body | Nf | decrease |
| GO.0017076 | purine nucleotide binding | Df, Dc | increase |
| GO.0019058 | viral life cycle | Df, Dc | increase |
| GO.0022607 | cellular component assembly | Df | increase |
| GO.0030054 | cell junction | Df | increase |
| GO.0031966 | mitochondrial membrane | Df, Dc | increase |
| GO.0031981 | nuclear lumen | Df, Dc | increase |
| GO.0043066 | negative regulation of apoptotic process | Df | increase |
| GO.0043209 | myelin sheath | Df | increase |
| GO.0043232 | intracellular non-membrane-bounded organelle | Df | increase |
| GO.0043933 | macromolecular complex subunit organization | Rf, Rc | increase |
| GO.0044237 | cellular metabolic process | Df | increase |
| GO.0044265 | mitochondrial nucleoid | Df, Dc | increase |
| GO.0044428 | nuclear part | Df, Dc | increase |
| GO.0044430 | cytoskeletal part | Df, Dc | increase |
| GO.0044451 | nucleoplasm part | Nf | decrease |
| GO.0044712 | single-organism catabolic process | Nf | decrease |
| GO.0044765 | single-organism transport | Df, Dc | increase |
| GO.0044822 | poly(A) RNA binding | Rf | increase |
| GO.0051084 | de novo posttranslational protein folding | Rf, Rc | increase |
| GO.0051704 | multi-organism process | Rf, Rc | increase |
| GO.0060548 | negative regulation of cell death | Df | increase |
| GO.0071013 | catalytic step 2 spliceosome | Df | increase |
| GO.0071705 | nitrogen compound transport | Nf | decrease |
| GO.0071822 | protein complex subunit organization | Rf | increase |
| GO.1901575 | organic substance catabolic process | Df | increase |
In case of each GO term where statistically significant changes were observed in network parameters in HIV-1 treated cells at 12 h time-point the GO identifier, the changed parameter and the direction of change is indicated
In H12, a statistically significant reduction of the node numbers and shrinkage of the network; along with a significant increase in interactivity, was observed (Fig. 4). The proteins responsible for the increased interactivity (increased Df and Rf values) belong to RNA binding, RNA catabolic process, viral life cycle, viral process, negative regulation of cell death, de novo posttranslational protein folding, protein complex subunit organization, and cellular metabolic process, etc. (Table 2). The cell junction and the myelin sheet GO terms also appear in H12, however, when proteins belonging to these GO terms were examined, it was found that they are part of more general GO terms from the list; such as intracellular non-membrane-bounded organelle or nucleus, extracellular space, etc. In case of biosynthetic process functional sub-network (GO.0009058), a decrease in the Rf was observed.
Discussion
Genome-wide RNA interference-based screens were carried out to evaluate more than 20,000 human gene products to determine their alteration in HIV infection [23, 24]. A previous study showed an overall downregulation of cellular genes encoding for nuclear proteins, and genes involved in DNA replication and protein synthesis in the early stages of the early phase of viral infection [25], in a pattern that was confirmed by our analysis (Table 1). Upregulation of cellular genes was only found to occur at a later time point, peaking at 22 h post-infection, additionally, analysis on T cells showed that the most profound changes in cellular proteome appear 24 h after infection, at time points related to the late phase of infection [26].
It was found that up to 300 host cellular genes were involved in the life cycle of HIV-1, and while the identity of the genes was divergent among different studies, they were found more or less to belong to similar pathways [27, 28]. Network analysis is widely used in the examination of protein-protein interactions, providing information regarding protein changes on a different level, giving a more ample view of the alterations and perturbations of the biological systems as a result of a particular treatment. During analysis of PPIs, the presence or absence of a protein is evaluated, and the interactions, in light of existing evidence (ex. experimental data, literature search, computational methods), are displayed [29, 30]. STRING is a widely used, constantly updated, and expanding database of PPIs [22], used for the examination of verified, or potential interactions among proteins of interests. These networks are rich in information on protein clusters and functions based on Gene Ontology (GO), however, enrichment of GO terms does not handle protein amounts, therefore, reflecting theoretical, rather than actual parameters. Meanwhile, the use of highly accurate mass spectrometry techniques provide analytical data that is wealthy in quantity as well as quality. There were few attempts made to introduce the quantitative data into the network analysis [31, 32]. In order to implement quantitative data into the PPI networks, instead of the widely used binary networks, a weighted network often utilized in information science [33] was used in this study. Taking into account the protein amount reflected by the normalized total spectra, instead of the probabilistic assumption [32], we choose a simple statistical model. In our model, the protein pair’s interaction is proportional to their amount in the sample, and inversely proportional to the number of possible interactions listed in the PPI network generated by STRING for proteins present in the sample. After including the interaction density values as network edge weights; calculated by our method, we could determine a sort of weighed network parameters for the statistical investigation of network alterations.
In our study, we aimed at characterizing the cellular proteome changes in the early stage of HIV-1 infection, within the 0–12 h time interval. Generation of weighted networks, and analysis of functional sub-networks revealed that the dynamics of protein level changes in sub-networks is different in HIV-1 transduced samples 12 h post-infection. Expectedly, in the very early stages of infection, proteins involved in translation, transcription and DNA condensation were upregulated, notably HIST1H1E, HNRNPL, PRRC2A and TRIM28. Some other proteins; such as ALYREF, CCDC86, CSDA, COX5A, HN1, MYL6, PPIF, SEPT2, SRSF6, TCOF1, and TPM3, prominently associated with RNA binding, cytoskeleton assembly, and signaling were quantified in all time points except H12.
Examining the binary networks, two protein clusters could be observed. One comprising proteins having a role in translation and ribosome biogenesis, and the other containing proteins from the hnRNP family with a role in RNA splicing (Fig. 2). The functional sub-network containing the ribosome component proteins did not show a statistically significant change, and with this, we can demonstrate on protein level the same findings observed by Kleinman et.al. at gene level, who could not observe statistically significant difference in case of genes having a role in ribosome biogenesis at 12 h time point [34]. Regarding the other cluster containing mainly hnRNP proteins, we could not observe a statistically significant change in network parameters among the different time points. However, literature data show that host RNA splicing is altered upon HIV-1 infection, and the level of class A/B and H of hnRNP proteins changes; initially decreased 6–12 days post infection, thereafter increased [35]. At the same time, it was shown that some proteins of this cluster; such as HNRNPH1, HNRNPU and SRSF6, are so called HIV-1 dependency factors [36] and are required by HIV-1. These data are derived from later time-points, as most of the experiments do not examine such early events at 4 h or 12 h post infection.
Considering the results of the analyses, based on the weighted networks, we could identify increased cellular metabolic processes comprising increased RNA binding and catabolism, cellular component assembly, along with increased viral process and inhibition of apoptosis (increased negative regulation of apoptotic process). RNA binding was shown to be increased upon RNA virus infection; Garcia-Moreno et al. observed an increased activity of RNA-binding proteins upon sindbis virus (SINV) infection at 18 h time point [37]. At the same time, they observed an increased binding of RNA binding proteins to viral RNAs. This implies a massive downregulation of the host mRNAs 18 h post infection, involving mainly the housekeeping genes [37]. In case of HIV-1 infection, global siRNA studies indicate that a statistically significant portion of the host factors participate in mRNA transport [18].
Cells infected with HIV-1 usually die by apoptosis, hence prevention of apoptosis might help maintain the viral reservoir in the host [18, 38]. It was shown that a fraction of infected immune cells survive, highlighting the importance of escaping from apoptosis in the development of viral reservoirs [38]. A mixed pattern of upregulation and downregulation of genes involved in antiviral defense and cell death signaling were observed by Mohammadi et al. at early time points [24]. Inhibition of apoptosis increases the virus production in HIV-1 infected cells [39], and modulation of this system might be a good possibility for a therapeutic intervention [40].
Based on our data on the weighted networks, HSPA8 shows an increased interactivity in H12 datasets (Fig. 5a). HSPA8 and other members of the Hsp70 family play a key role during viral infection either as receptors for the virus, as chaperons aiding the protein folding, or as transporters between organelles [18, 41, 42].
Hijacking of the host system by HIV-1 is a complex phenomenon with early and late events. In the early phases of the viral infection, the virus utilizes cellular RNA and protein production machinery for its replication. It was observed that by 15 h post infection, all viral transcripts were produced by the cells, and 18 h after infection, the virus budding commences [24]. Chang et al.;. using next generation sequencing, observed a considerable viral mRNA level in infected cells 12 h post-infection [43]. In this sense, examining the host response 48 h [15, 44] or 6 days post-infection [45] cannot provide us with information on the very early events. Observations made by Kleinmann et al. analyzing the dataset generated by Chang et al., show that at 12 h post infection, the gene expression profiles are similar to the mock samples, and clear distinctions could only be made after 24 h, highlighting the necessity of more sensitive methods for the examination of early events of HIV-1 infection.
It is challenging to properly compare our results to those presented in the scientific literature, since the commonly used starting time point examined is 48 h post infection, in case of HIV-1. However, considering the findings presented by different groups; either on HIV-1 or other RNA virus infections, our findings are in good agreement with previous studies analyzing transcriptomic and proteomic changes upon virus infection in these very early time points. The use of non-primary HIV-1 cell targets; such as HEK, and pseudotyped virions, and the application of data-dependent sampling [46], may indeed limit interpretation of the results. The utilization of other cell types and data acquisition methods with higher reproducibility; such as parallel reaction monitoring [2] or data independent acquisition [47], might give more accurate input data. In spite of the above limitations, we believe that this model of proteomic data evaluation serves as a good starting point for further development of algorithms implementing not only qualitative, but also quantitative data generated in a given proteomic experiment, and that such a combination will undoubtedly aid in the understanding and deciphering of complex biological phenomena.
Conclusion
A weighted network model facilitating the use of both qualitative and quantitative data, acquired in a label-free proteomics experiment was generated and applied to examine the early host response to HIV-1. Upregulation of proteins involved in translation, transcription and DNA condensation in the early phase of the viral life-cycle could be observed, highlighting the utility of our weighted PPI network data analysis approach. More studies are required to further demonstrate the utility of this new data-driven weighted network based analysis, and it should be noted that the current model has a serious limitation. The strength of different protein-protein interactions in the edge weight calculation; due to the lack of information, is not yet included. However, the applied weight-model can easily be extended to use this type of information as soon as any public database becomes available. We hope that this approach can open new ways for creating algorithms, allowing for both quantitative and qualitative studies of proteome change in various biological and pathological processes by quantitative mass spectrometry.
Methods
Production of viral particles
Viral particles were produced with some modifications of a previously utilized protocol [48]. Briefly, recombinant viruses were produced by transient transfection of 293 T cells (ATCC® CRL3216™) using pWOX-CMV-GFP (transfer vector plasmid), pMDLg/pRRE (packaging plasmid), pRSV.rev (Rev-coding plasmid), and pMD. G (VSV-G envelope protein-coding plasmid). Vectors were a kind gift from D. Trono (University of Geneva Medical School, Geneva, Switzerland) [49], and were subsequently modified by our research group [48]. Salmon sperm DNA (Sigma-Aldrich) was also added. Media containing virus particles was concentrated by Ultracel-100 K Amicon Ultra Centrifugal Filter (Millipore), and stored in − 70 °C. Quantity of pseudovirions produced was assessed by measurement of reverse transcriptase (RT) activity using a colorimetric kit (Sigma-Aldrich, Roche).
Transduction and sample collection
293 T cells in T-25 cell culture flasks were either mock-treated or transduced at 50% confluency with 5 ng RT equivalent of the HIV-based pseudovirions, in the presence of 4 μg/ml polybrene (Sigma-Aldrich), in 1 ml total volume, and incubated at 37 C°. After 0, 4, and 12 h, cells were trypsinized for 10 min, then washed tree times with ice-cold PBS to remove non-fused pseudovirion particles. The final pellet was suspended in 4 ml lysis buffer (150 mM sodium chloride, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and 50 mM Tris) pH 8.0, supplemented with cOmplete protease inhibitor cocktail (Sigma-Aldrich), incubated for 30 min at room temperature, centrifuged, and the supernatant was mixed with 24 ml cold (− 20 C°) acetone and stored at − 20 C° overnight.
Mass spectrometry analysis
The cleared cell lysates were acetone-precipitated with six volumes of cold acetone overnight. The precipitates were re-dissolved in 25 mM ammonium bicarbonate (Sigma-Aldrich) and digested in-solution with trypsin [50]. The tryptic fragments were used for replicate LC-MS/MS analyses at University of Arizona in Tucson, AZ, USA.
500 ng per 5 μL injected protein lysate spiked with 300 fmol of Rhodobacter capsulatus cytochrome c T33 V mutant, was analyzed using a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with an Advion nanomate ESI source (Advion), after Omix (Agilent Technologies) C18 sample clean-up according to the manufacturer’s instructions. Peptides were eluted from a C18 precolumn (100-μm × 2 cm, Thermo Fisher Scientific) onto an analytical column (75-μm × 10 cm, C18, Thermo Fisher Scientific) using a 165 min gradient of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid). The flow rate was 500 nl/minute. Data-dependent analysis (DDA) was performed by the Xcalibur v 2.1.0 software [51] using a survey mass scan at 60,000 resolution in the Orbitrap analyzer scanning mass/charge 350–1600, followed by collision-induced dissociation tandem mass spectrometry (MS/MS) at 35 normalized collision energy of the 14 most intense ions in the linear ion trap analyzer. Precursor ions were selected by the monoisotopic precursor selection setting with selection or rejection of ions held to a +/− 10 ppm window. Singly charged ions were excluded from MS/MS. Dynamic exclusion was set to place any selected m/z on an exclusion list for 45 s after a single MS/MS. Tandem mass spectra were searched against the UniprotKB/Swiss-Prot release available on December 12, 2014 without species restriction. At the time of the search, this database contained 459,734 entries. All MS/MS spectra were searched using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific) considering fully tryptic peptides with up to 2 missed cleavage sites. Variable modifications considered during the search included methionine oxidation (15.995 Da), and cysteine carbamidomethylation (57.021 Da). The parent ion mass tolerance was 10 ppm, while the fragment tolerance was 0.8 Da. Proteins were identified at 99% confidence with XCorr score cut-offs [52] as determined by a reversed database search. The protein and peptide identification results were validated with Scaffold v4.4.6. (Proteome Software Inc.) [1]. Peptide identifications were accepted if they had greater than 89% probability to achieve an FDR less than 0.1% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they had greater than 99% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm [53]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Protein quantification was done based on spectral counting; the quantitative values were generated by the Scaffold program based on the normalized total spectra. In case of protein clusters, each peptide was used only once for quantification for the first human protein in the cluster, as listed by Scaffold. All quantitative data were used for statistical analyses; none of the data points were removed.
Statistical analysis of proteomics data
For both statistical and network analysis, we used in-house developed R-software based on STRING [54–57], circlize (https://jokergoo.github.io/circlize_book/book/), MASS [55], lsmeans [56], matrixStats [57], reshape2 [58] and ggplot2 [59] packages. Assuming that data from technical repetitions are often characterized by Poisson distribution [60], and the large variances of biological replicas can be modelled by negative binomial distribution [61], we used modified general linear models to describe group-level differences in measured protein data in the 4 and 12 h time points. For each protein; after fitting negative binomial generalized linear model [55], we performed a post-hoc analysis [62] to characterize time-dependent mean differences by z score, and corrected p values for multiple comparisons.
Network analysis
Gene names of the identified human proteins were subjected to STRING database [22] and five PPI networks were generated. The NW0 combined proteins from mock- and HIV-1 plasmid-treated cell lysates collected at 0 time-point, the C04 and C12 networks contained proteins from the mock cells collected 4 and 12 h post-infection, respectively, while the H04 and H12 networks contained proteins from the HIV-1 treated cells collected at 4 and 12 h time-points, respectively. Very high confidence interactions (interaction score > 0.95) in between the query proteins were used for the generation of each binary network. In these networks, the nodes were the proteins and the edges indicated the interactions between proteins as they were present in STRING. For network generation, the SRING R-package and the STRING database was applied, and the 0.95 combined score value to generate the binary networks Bt,s (B0, B4h,C, B4h,H, B12h,C, B12h,H) corresponding to the protein sets. In these networks, the binary edges indicated only the possibility of the interactions, taking no notice of the quantity.
To estimate the real interaction density, binary networks (Bt,s) generated by STRING were further modified, and the amount of proteins measured by spectral counting was used to add wij weights to the edges. In this way, the existence of edges provides information on the existence of interaction, and the strength of protein pair’s interactions were estimated by this edge-weight model:
| 1 |
where wij represents the interaction density between protein Pi and Pj; ni, nj means the quantity while ki, kj denote the degree (the number of edges) of Pi and Pj in the given Bt,s binary network.
In this calculation, we used the measured data (ni, nj), which enabled us to alter the theoretical binary PPI network into a realistic, sample related interaction network, in which the weights of the edges are in direct proportion to the quantities and in inverse proportion to all interaction possibilities of the connected proteins in the given sample.
Because we can consider the ni as the number of molecules of the protein Pi, the ni/ki ratio represents the number of Pi molecules involved in one interaction of Pi, and thus, the interaction density between Pi and Pj can be described by the product of ni/ki and nj/kj. It should be mentioned that the used edge-weight model in the absence of a strong interactor protein may overestimate the effect of other weak interactor proteins, also, interaction strength data cannot be achieved in a classical quantitative proteomics experiment, and currently are unavailable in publicly accessible databases.
Functional subnetwork construction
In order to investigate the PPI networks of the proteins belonging to GO (geneontology.org/) terms, we marked in each Wt,s the nodes by a function flag, which indicated whether or not the protein belongs to a given f-function; in our case, to a GO term. The so-called functional enrichment according to GO terms was done by STRING, using default settings and the Molecular Function, Biological Process and Cellular Component GO terms listed as enriched by STRING in C04, H04, C12 and H12, where at least 10 protein per GO function in any of the networks was present, were considered. This procedure defined a sort of Wf t,s functional networks, and divided them into two disjunctive sub-networks (F f t,s functional, belonging to the GO term and NF f t,s non-functional not being part of the respective GO term), containing the functional and the non-functional nodes, respectively. Because of this separation, the edges (i.e. the interactions) were also classified into three classes: functional edges between the functional nodes, non-functional edges between non-functional nodes and cross-edges in between functional and non-functional nodes, depending on the f-markers of the connected proteins.
Examination of the global characteristics of the evaluated PPI networks
Any undirected weighted PPI network W(N,E) consists of two sets: N nodes and E edges. Each of the links (interactions) is defined by a couple of nodes (proteins) Pi and Pj, and its value is wij. Since the direction of interaction cannot be ordered, the connectivity matrix became symmetric: wij = wji.
Number of nodes (N) and edges (E)
N, Nf and Nn denotes the number of nodes (i.e. proteins) in the whole network and the functional and non-functional sub-networks, respectively, with the following relation:
| 2 |
E denotes the number of edges (i.e. interactions) in the whole network. Ef and En are the number of edges within the functional and the non-functional sub-networks, respectively. The number of cross-edges (Ec) shows the connected proteins between the functional and the non-functional sub-networks. The edge numbers follow the next relation:
| 3 |
Network strength and averaged node strength (S)
We defined the network strength S as the total sum of the weights of edges:
| 4 |
In the functional networks we can calculate strength of whole network (S), and the functional (Sf) and non-functional sub-networks (Sn), as well. The sum of cross connection edges can be calculate as follows:
| 5 |
Edge-weight density or strength density (D)
the edge-weight density measures how the weighted network is saturated by strong edges:
| 6 |
In the functional networks we can measure the edge-weight density of the whole network (D) and the functional (Df) and non-functional sub-networks (Dn), as well.
Edge-weight ratio (R): using the network strength we can define the edge-weight ratio parameter for the two sub-networks:
| 7 |
and the non-functional relative edge-weight density:
| 8 |
Since the distribution of network parameters was not Gaussian or negative binomial, we used Wilcoxson tests [63] to characterize the group-related differences at the 4 and 12 h time points. The evaluated p-values were corrected for multiple comparisons by false discovery rate methods [64].
Additional files
List of identified proteins. (XLSX 1147 kb)
List of quantified proteins. The gene name according to UniProt in case of quantified proteins is given, and for each protein, the mean amount of four replicates is presented for each time point except NW, where the mean amount of 8 replicates is given. (XLSX 23 kb)
Statistical analysis of protein quantities. The gene name according to UniProt, the p value and z score for the 4 h and 12 h time points are listed in case of each protein. The lists are presented in ascending order of the p values. (XLSX 24 kb)
Network parameters calculated for functional sub-networks. The y axis show the mean value characteristic for each parameter, and the x axis indicates the time points. Blue color refers to the control, while the yellow color to the HIV-1 treated conditions. N refers to the number of nodes, E to the number of edges, S show network strength, D represents the edge density and R the edge ratio. The f refers to the functional sub-network, the n to the non-functional subnetwork containing the proteins not present in the functional sub-network, while the c refers to the interactions between the functional and the non-functional sub-networks. (PDF 9676 kb)
Statistical analysis of network parameters. The FDR-corrected p value and z score for the 4 h and 12 h time points, respectively, in case of each network parameter calculated (N, Nf, Nn, E, Ef, En, Ec, S, Sf, Sn, Sc, D, Df, Dn, Dc, Rf, Rn, Rc) for each GO function presented in Additional file 4. (XLSX 287 kb)
List of GO terms with network parameters that were significantly changed in the functional sub-networks. (XLSX 16 kb)
Acknowledgements
Not applicable.
Abbreviations
- cDNA
Complementary DNA
- D
Saturation of the PPI interaction density in the sample
- Dc
Saturation of the PPI interaction density between the functional and non-functional proteins in the sample
- Df
Saturation of the PPI interaction density of the functional proteins in the sample
- DMEM
Dulbecco’s modified Eagle’s medium
- Dn
Saturation of the PPI interaction density of the nonfunctional proteins in the sample
- DNA
Deoxyribonucleic acid
- E
Number of interactions in the network generated from STRING with combined score of 0.95
- Ec
Number of interactions between the functional and non-functional proteins
- Ef
Number of interactions between the functional proteins
- En
Number of interactions between the non-functional proteins
- FBS
Fetal bovine serum
- GO
Gene Ontology
- HEK
Human embryonic kidney cells
- HIV
Human immunodeficiency virus
- MIPS
Monoisotopic precursor selection
- N
Number of proteins in the network
- Nf
Number of proteins in the functional sub-network
- Nn
Number of proteins in the non-functional sub-network
- PBS
Phosphate buffered saline
- PPI
Protein-protein interaction
- ppm
Parts per million
- R
Relative PPI density, edge-weight ratio
- Rc
Relative cross-functional PPI density, edge-weight ratio between the functional and non-functional PPI networks
- Rf
Relative functional PPI density, edge-weight ratio between in the functional PPI network
- Rn
Relative non-functional PPI density, edge-weight ratio in the non-functional PPI network
- RNA
Ribonucleic acid
- RT
Reverse transcriptase
- S
PPI density of the sample
- Sc
PPI density of the cross-functional proteins in the sample
- Sf
PPI density of the functional proteins in the sample
- siRNA
Small interfering RNA
- Sn
PPI density of the non-functional proteins in the sample
- VSV
Vesicular stomatitis virus
Authors’ contributions
JT conceptualized the study, designed the HIV-1 transduction experiments, reviewed the manuscript and provided the resources for the study. FT produced the viruses, carried out the cell transductions, prepared the samples for mass spectrometry and curated the mass spectrometry data. EC designed the data analysis strategy, analyzed the mass spectrometry, statistical and network analysis data, prepared the tables and wrote the manuscript. GT performed the mass spectrometry experiment and helped with the manuscript preparation. ME designed and performed the statistical analysis and network analysis, assisted in manuscript writing and prepared the Figs. MM participated in manuscript preparation, critically reviewed the manuscript, performed the English editing and helped with the evaluation of HIV-1 transduction results. All authors read and approved the final manuscript.
Funding
This work was supported by the Hungarian Scientific Research Fund (NKFI-6, 125238) to JT, by the Higher Education Institutional Excellence Programme of the Ministry of Human Capacities in Hungary, within the framework of the Biotechnology thematic programme of the University of Debrecen, GINOP-2.3.3-15-2016-00020, and partially by Janos Bolyai Research Scholarship of the Hungarian Academy of Sciences. Mass spectrometry data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant ES06694 to the SWEHSC, NIH/NCI grant CA023074 to the UA Cancer Center and by the BIO5 Institute of the University of Arizona. The Thermo Fisher LTQ Orbitrap Velos mass spectrometer was provided by grant 1S10 RR028868–01 from NIH/NCRR. Funding bodies did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The mass spectrometry datasets generated during the current study were deposited to the ProteomeXchange database and are available via the PRIDE repository with the dataset identifier PXD010436 and 10.6019/PXD010436. All data analyzed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
Non applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Miklós Emri and József Tőzsér contributed equally to this work.
Contributor Information
Éva Csősz, Phone: +36-52-416432, Email: cseva@med.unideb.hu.
József Tőzsér, Email: tozser@med.unideb.hu.
References
- 1.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28(7):710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 3.Codrea MC, Nahnsen S. Platforms and pipelines for proteomics data analysis and management. Modern Proteomics - Sample Preparation, Analysis and Practical Applications. 2016;919:203–215. doi: 10.1007/978-3-319-41448-5_9. [DOI] [PubMed] [Google Scholar]
- 4.Kentaro Kawata AH, Yugi K, Kubota H, Sano T, Fujii M, Tomizawa Y, Kokaji T, Tanaka KY, Uda S, Yutaka S, Matsumoto M, Nakayama KI, Saitoh K, Kato K, Ueno A, Ohishi M, Hirayama A, Kuroda S. Trans-omic Analysis Reveals Selective Responses to Induced and Basal Insulin across Signaling, Transcriptional, and Metabolic Networks. iScience. 2018;7:1–18. doi: 10.1016/j.isci.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koberlin MS, Snijder B, Heinz LX, Baumann CL, Fauster A, Vladimer GI, Gavin AC, Superti-Furga G. A conserved circular network of Coregulated lipids modulates innate immune responses. Cell. 2015;162(1):170–183. doi: 10.1016/j.cell.2015.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Li YP, Li YX, Zhu XH, Du XG, Zhou M, Li WB, Deng HY. Effect of regulatory network of exosomes and microRNAs on neurodegenerative diseases. Chin Med J. 2018;131(18):2216–2225. doi: 10.4103/0366-6999.240817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szilagyi A, Nussinov R, Csermely P. Allo-network drugs: extension of the allosteric drug concept to protein- protein interaction and signaling networks. Curr Top Med Chem. 2013;13(1):64–77. doi: 10.2174/1568026611313010007. [DOI] [PubMed] [Google Scholar]
- 9.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481(7381):365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csermely P, Sandhu KS, Hazai E, Hoksza Z, Kiss HJ, Miozzo F, Veres DV, Piazza F, Nussinov R. Disordered proteins and network disorder in network descriptions of protein structure, dynamics and function: hypotheses and a comprehensive review. Curr Protein Pept Sci. 2012;13(1):19–33. doi: 10.2174/138920312799277992. [DOI] [PubMed] [Google Scholar]
- 11.Dai LY, Zhao TY, Bisteau X, Sun WD, Prabhu N, Lim YT, Sobota RM, Kaldis P, Nordlund P. Modulation of Protein-Interaction States through the Cell Cycle. Cell. 2018;173(6):1481. doi: 10.1016/j.cell.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 12.Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchhoff Frank. Encyclopedia of AIDS. New York, NY: Springer New York; 2013. HIV Life Cycle: Overview; pp. 1–9. [Google Scholar]
- 14.Lehmann-Che J, Saib A. Early stages of HIV replication: how to hijack cellular functions for a successful infection. AIDS Rev. 2004;6(4):199–207. [PubMed] [Google Scholar]
- 15.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 16.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacPherson JI, Dickerson JE, Pinney JW, Robertson DL. Patterns of HIV-1 protein interaction identify perturbed host-cellular subsystems. PLoS Comput Biol. 2010;6(7):e1000863. doi: 10.1371/journal.pcbi.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickerson JE, Pinney JW, Robertson DL. The biological context of HIV-1 host interactions reveals subtle insights into a system hijack. BMC Syst Biol. 2010;4:80. doi: 10.1186/1752-0509-4-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinney JW, Dickerson JE, Fu W, Sanders-Beer BE, Ptak RG, Robertson DL. HIV-host interactions: a map of viral perturbation of the host system. Aids. 2009;23(5):549–554. doi: 10.1097/QAD.0b013e328325a495. [DOI] [PubMed] [Google Scholar]
- 21.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou HL, Xu M, Huang Q, Gates AT, Zhang XHD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Arhel N, Kirchhoff F. Host proteins involved in HIV infection: new therapeutic targets. Bba-Mol Basis Dis. 2010;1802(3):313–321. doi: 10.1016/j.bbadis.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Mohammadi P, Desfarges S, Bartha I, Joos B, Zangger N, Munoz M, Gunthard HF, Beerenwinkel N, Telenti A, Ciuffi A. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9(1):e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemeth J, Vongrad V, Metzner KJ, Strouvelle VP, Weber R, Pedrioli P, Aebersold R, Gunthard HF, Collins B. In vivo and in vitro proteome analysis of human immunodeficiency virus (HIV)-1-infected, human CD4(+) T cells. Mol Cell Proteomics. 2017;16(4):S108–S123. doi: 10.1074/mcp.M116.065235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135(3):417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung ML, Houzet L, Yedavalli VSRK, Jeang KT. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284(29):19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307(5710):724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- 30.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celaj A, Schlecht U, Smith JD, Xu W, Suresh S, Miranda M, Aparicio AM, Proctor M, Davis RW, Roth FP, et al. Quantitative analysis of protein interaction network dynamics in yeast. Mol Syst Biol. 2017;13(7):934. doi: 10.15252/msb.20177532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Complex networks: structure and dynamics. Phys Rep. 2006;424(4–5):175–308. [Google Scholar]
- 34.Kleinman Claudia L., Doria Margherita, Orecchini Elisa, Giuliani Erica, Galardi Silvia, De Jay Nicolas, Michienzi Alessandro. HIV-1 Infection Causes a Down-Regulation of Genes Involved in Ribosome Biogenesis. PLoS ONE. 2014;9(12):e113908. doi: 10.1371/journal.pone.0113908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowling Dinushka, Nasr-Esfahani Somayeh, Tan Chun H, O'Brien Kate, Howard Jane L, Jans David A, Purcell Damian FJ, Stoltzfus C Martin, Sonza Secondo. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008;5(1):18. doi: 10.1186/1742-4690-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sertznig Helene, Hillebrand Frank, Erkelenz Steffen, Schaal Heiner, Widera Marek. Behind the scenes of HIV-1 replication: Alternative splicing as the dependency factor on the quiet. Virology. 2018;516:176–188. doi: 10.1016/j.virol.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Moreno Manuel, Noerenberg Marko, Ni Shuai, Järvelin Aino I., González-Almela Esther, Lenz Caroline E., Bach-Pages Marcel, Cox Victoria, Avolio Rosario, Davis Thomas, Hester Svenja, Sohier Thibault J.M., Li Bingnan, Heikel Gregory, Michlewski Gracjan, Sanz Miguel A., Carrasco Luis, Ricci Emiliano P., Pelechano Vicent, Davis Ilan, Fischer Bernd, Mohammed Shabaz, Castello Alfredo. System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Molecular Cell. 2019;74(1):196-211.e11. doi: 10.1016/j.molcel.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lum Julian, Badley Andrew. Resistance to Apoptosis: Mechanism for the Development of HIV Reservoirs. Current HIV Research. 2003;1(3):261–274. doi: 10.2174/1570162033485203. [DOI] [PubMed] [Google Scholar]
- 39.Antoni BA, Sabbatini P, Rabson AB, White E. Inhibition of apoptosis in human immunodeficiency virus-infected cells enhances virus production and facilitates persistent infection. J Virol. 1995;69(4):2384–392. [DOI] [PMC free article] [PubMed]
- 40.Badley AD, Sainski A, Wightman F, Lewin SR. Altering cell death pathways as an approach to cure HIV infection. Cell death & disease. 2013;4:e718. [DOI] [PMC free article] [PubMed]
- 41.Stricher François, Macri Christophe, Ruff Marc, Muller Sylviane. HSPA8/HSC70 chaperone protein. Autophagy. 2013;9(12):1937–1954. doi: 10.4161/auto.26448. [DOI] [PubMed] [Google Scholar]
- 42.Sherman Michael P., Greene Warner C. Slipping through the door: HIV entry into the nucleus. Microbes and Infection. 2002;4(1):67–73. doi: 10.1016/s1286-4579(01)01511-8. [DOI] [PubMed] [Google Scholar]
- 43.Chang ST, Sova P, Peng X, Weiss J, Law GL, Palermo RE, Katze MG. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. mBio. 2011;2(5). [DOI] [PMC free article] [PubMed]
- 44.Rato Sylvie, Rausell Antonio, Muñoz Miguel, Telenti Amalio, Ciuffi Angela. Single-cell analysis identifies cellular markers of the HIV permissive cell. PLOS Pathogens. 2017;13(10):e1006678. doi: 10.1371/journal.ppat.1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao S, Amorim R, Niu M, Breton Y, Tremblay MJ, Mouland AJ. Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocyte-derived macrophages. Retrovirology. 2019;16(1):3. [DOI] [PMC free article] [PubMed]
- 46.Tabb David L., Vega-Montoto Lorenzo, Rudnick Paul A., Variyath Asokan Mulayath, Ham Amy-Joan L., Bunk David M., Kilpatrick Lisa E., Billheimer Dean D., Blackman Ronald K., Cardasis Helene L., Carr Steven A., Clauser Karl R., Jaffe Jacob D., Kowalski Kevin A., Neubert Thomas A., Regnier Fred E., Schilling Birgit, Tegeler Tony J., Wang Mu, Wang Pei, Whiteaker Jeffrey R., Zimmerman Lisa J., Fisher Susan J., Gibson Bradford W., Kinsinger Christopher R., Mesri Mehdi, Rodriguez Henry, Stein Stephen E., Tempst Paul, Paulovich Amanda G., Liebler Daniel C., Spiegelman Cliff. Repeatability and Reproducibility in Proteomic Identifications by Liquid Chromatography−Tandem Mass Spectrometry. Journal of Proteome Research. 2010;9(2):761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heaven Michael R., Funk Adam J., Cobbs Archie L., Haffey Wendy D., Norris Jeremy L., McCullumsmith Robert E., Greis Kenneth D. Systematic evaluation of data-independent acquisition for sensitive and reproducible proteomics-a prototype design for a single injection assay. Journal of Mass Spectrometry. 2015;51(1):1–11. doi: 10.1002/jms.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miklossy G, Tozser J, Kadas J, Ishima R, Louis JM, Bagossi P. Novel macromolecular inhibitors of human immunodeficiency virus-1 protease. Protein engineering, design & selection : PEDS. 2008;21(7):453–461. doi: 10.1093/protein/gzn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Csosz E, Markus B, Darula Z, Medzihradszky KF, Nemes J, Szabo E, Tozser J, Kiss C, Marton I. Salivary proteome profiling of oral squamous cell carcinoma in a Hungarian population. FEBS open bio. 2018;8(4):556–569. doi: 10.1002/2211-5463.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, 3rd, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2(9):1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4(1):53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 53.Nesvizhskii Alexey I., Keller Andrew, Kolker Eugene, Aebersold Ruedi. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Analytical Chemistry. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 54.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.W.N. Venables BDR . Modern applied statistics with S. New York: Springer-Verlag; 2002. [Google Scholar]
- 56.Lenth RV. Least-squares means: the R package lsmeans. J Stat Softw. 2016;69(1):1–33. [Google Scholar]
- 57.matrixStats: Functions that Apply to Rows and Columns of Matrices (and to Vectors). R package version 0.52.2 [https://github.com/HenrikBengtsson/matrixStats].
- 58.Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21(12):1–20. [Google Scholar]
- 59.Ginestet C. ggplot2: elegant graphics for data analysis. J R Stat Soc a Stat. 2011;174:245. [Google Scholar]
- 60.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:3. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Searle SR, Speed FM, Milliken GA. Population marginal means in the linear-model - an alternative to least-squares means. Am Stat. 1980;34(4):216–221. [Google Scholar]
- 63.MHaDA W. Nonparametric statistical methods. New York: John Wiley & Sons; 1999. [Google Scholar]
- 64.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of identified proteins. (XLSX 1147 kb)
List of quantified proteins. The gene name according to UniProt in case of quantified proteins is given, and for each protein, the mean amount of four replicates is presented for each time point except NW, where the mean amount of 8 replicates is given. (XLSX 23 kb)
Statistical analysis of protein quantities. The gene name according to UniProt, the p value and z score for the 4 h and 12 h time points are listed in case of each protein. The lists are presented in ascending order of the p values. (XLSX 24 kb)
Network parameters calculated for functional sub-networks. The y axis show the mean value characteristic for each parameter, and the x axis indicates the time points. Blue color refers to the control, while the yellow color to the HIV-1 treated conditions. N refers to the number of nodes, E to the number of edges, S show network strength, D represents the edge density and R the edge ratio. The f refers to the functional sub-network, the n to the non-functional subnetwork containing the proteins not present in the functional sub-network, while the c refers to the interactions between the functional and the non-functional sub-networks. (PDF 9676 kb)
Statistical analysis of network parameters. The FDR-corrected p value and z score for the 4 h and 12 h time points, respectively, in case of each network parameter calculated (N, Nf, Nn, E, Ef, En, Ec, S, Sf, Sn, Sc, D, Df, Dn, Dc, Rf, Rn, Rc) for each GO function presented in Additional file 4. (XLSX 287 kb)
List of GO terms with network parameters that were significantly changed in the functional sub-networks. (XLSX 16 kb)
Data Availability Statement
The mass spectrometry datasets generated during the current study were deposited to the ProteomeXchange database and are available via the PRIDE repository with the dataset identifier PXD010436 and 10.6019/PXD010436. All data analyzed during this study are included in this published article [and its supplementary information files].