Abstract
Long noncoding RNAs (lncRNAs) regulate gene expression. We investigated the role of lncRNAs in the inflammatory response to bacterial infection in the lungs. We identified the lncRNA MEG3 as a tissue-specific modulator of inflammatory responses during bacterial infection. Among the 10 transcript isoforms of MEG3, transcript 4 (referred to as MEG3–4) encodes the isoform with the lowest abundance in mouse lungs. Nonetheless, we found that MEG3–4 bound to the microRNA miR-138 in a competitive manner with mRNA encoding the proinflammatory cytokine interleukin-1β (IL-1β), thereby increasing IL-1β abundance and intensifying inflammatory responses to bacterial infection in alveolar macrophages and lung epithelial cells in culture and in lung tissue in mice. MEG3–4–mediated sponging of miR-138 in the cytoplasm increased the autocrine activity of IL-1β that subsequently induced a negative feedback mechanism mediated by nuclear factor κB that decreased MEG3–4 abundance and inflammatory cytokine production. This timely reduction in MEG3–4 abundance tempered proinflammatory responses in mice with pulmonary bacterial infection, preventing the progression to sepsis. Together, these findings reveal that MEG3–4 dynamically modulates pulmonary inflammatory responses through transcriptional regulation of immune response genes, extending the decoy and sponge mechanism associated with lncRNAs to antibacterial immunity, which affects both response and disease progression.
INTRODUCTION
Long noncoding RNAs (lncRNAs) function in various biological processes (1), including stem cell differentiation (2), cell fate determination (3), parental imprinting (4), tumorigenesis (5), and immune response (6). lncRNAs can facilitate or disrupt protein-protein interactions and directly bind to DNA and RNA to regulate gene expression and transcript abundance, respectively (7). In some cases, lncRNAs compete for microRNA (miRNA) binding, thereby acting as decoys to prevent transcript degradation. With such broad potential biochemically, lncRNAs have exceptionally diverse functions biologically. Emerging data show that the inflammatory response in both the adaptive and innate immune systems is particularly, and dynamically, regulated by lncRNAs. For example, the lncRNA Lethe is induced by proinflammatory cytokine production and functions as a negative feedback regulator to block DNA binding by the nuclear factor κB (NF-κB) subunit RelA, thereby suppressing inflammatory signaling (8). Another, the long intergenic noncoding RNA Cox2 coordinates the innate, antimicrobial immune response upon Toll-like receptor (TLR) activation by both promoting and repressing the expression of distinct classes of inflammatory genes, depending on its protein interactions (9). In addition, the lncRNA EPS (erythroid pro-survival, also known as Ttc39aos1) transcriptionally represses immune response genes (IRGs) in macrophages (10).
The lncRNA called maternally expressed gene 3 (MEG3) is encoded by an imprinted gene belonging to the DLK1-MEG3 locus located on chromosome 14q32.3 in humans (11). MEG3 lncRNA, a collective term for 10 distinct transcripts in mice, is expressed in many tissues, including the lung (12), liver (13), brain (14), and muscle (15), and is reportedly a tumor suppressor; MEG3 expression is lost in various primary tumors and tumor cell lines [including neuroblastomas (16), hepatocellular cancers (17), gliomas (18), and non–small cell lung cancer cells (12)] by various mechanisms, such as gene deletion, promoter hypermethylation, and hypermethylation of the intergenic differentially methylated region (19). Ectopic expression of MEG3 in glioma cells suppresses cell proliferation and promotes apoptosis in both p53-dependent and p53-independent mechanisms (20). However, it is not yet clear whether MEG3 has roles beyond tumor suppression. One study links MEG3 to infection by showing that its expression is reduced in macrophages upon mycobacterial infection, which facilitates elimination of the mycobacteria through autophagy (21). Hence, we speculate that MEG3 may play roles in modulating the immune response against pathogens.
Pseudomonas aeruginosa is an opportunistic Gram-negative bacterium that causes intractable infections (22). lncRNAs are differentially expressed in the bronchial epithelium of P. aeruginosa–infected cystic fibrosis (CF) patients relative to non-CF individuals (23), suggesting that lncRNAs may be involved in immunity to P. aeruginosa. Here, we defined lncRNA MEG3 as an important modulator that affects inflammatory responses upon P. aeruginosa infection in the lungs of mice. Our data demonstrated that MEG3 transcript 4 (MEG3–4) acts tissue-specifically as a natural decoy by binding to miRNA-138 (miR-138), thereby releasing interleukin-1β (IL-1β) to augment inflammatory responses.
RESULTS
MEG3–4 expression is tissue-specifically inhibited upon bacterial infection
We assessed the lncRNA expression profile of mouse lungs 24 hours after intranasal infection with the wild-type P. aeruginosa strain PAO1 using the mouse inflammatory response array-based RT2 lncRNA primer-profiling assay. The abundance of 73 lncRNAs was increased in the lungs from infected mice, whereas the abundance of three lncRNAs—MEG3, Rplp0, and C130021l20Rik—was significantly decreased, with MEG3 showing the greatest decrease (Fig. 1A and table S1). On the basis of this observation, we hypothesized that MEG3 is critically involved in pulmonary P. aeruginosa infection.
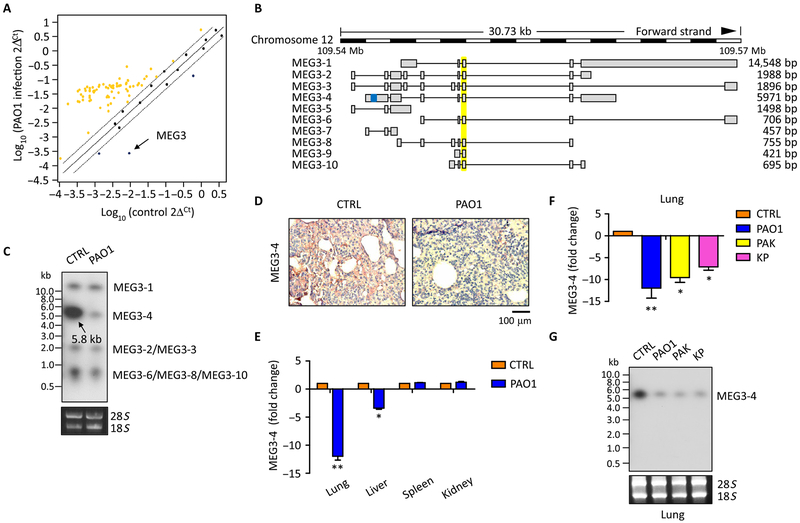
Fig. 1. MEG3–4 is in a decreased abundance in mouse lungs after bacterial infection.
(A) C57BL/6J mice were intranasally infected with 5 × 106 colony-forming units (CFU) of PAO1 for 24 hours. Lungs were lysed to assess the profiling of lncRNAs using the inflammatory response–based RT2 lncRNA primer array. Yellow dots indicate increased lncRNAs, and blue dots indicate decreased lncRNAs (more than twofold change and P ≤ 0.05 by Student’s t test). Data are from n = 2 biological replicates. (B) Schematic of MEG3 transcripts (http://vega.archive.ensembl.org/Mus_musculus/Transcript). Yellow, general MEG3 isoform probes; blue, specific MEG3–4 primers and probes. bp, base pair. (C) Northern blots of MEG3 transcripts in the lungs from control (CTRL) or PAO1-infected mice. 18S and 28S ribosomal RNAs (rRNAs) were used as loading controls. (D) In situ analysis of MEG3–4 abundance in the lungs from mice described in (C) with a digoxigenin (DIG)–labeled MEG3–4 probe (brown stain). Nuclei were counterstained by hematoxylin (blue). Scale bar, 100 μm. (E) qRT-PCR analysis of MEG3–4 expression in the lung, liver, spleen, and kidney from PAO1-infected mice. (F and G) qRT-PCR (F) and Northern blots (G) of MEG3–4 transcripts in the lungs from mice infected with PAO1, PAK, and KP. Data in (C), (D), and (G) are representative of data from three mice. Data in (E) and (F) are means ± SD from three mice (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01).
The mouse homolog MEG3 is a multiexon lncRNA that contains 10 alternatively spliced isoforms (Fig. 1B and fig. S1A), previously characterized by 5′-RACE (rapid-amplificantion of cDNA ends)–polymerase chain reaction (PCR) (http://vega.archive.ensembl.org/Mus_musculus/Transcript) (24). Using a general MEG3 probe (Fig. 1B), we detected four bands by Northern blotting, at about 15, 5.8, 2.0, and 0.8 kb, which correspond to the alternatively spliced isoforms of MEG3 (Fig. 1C). The strongest 5.8-kb signal represents MEG3–4 as the most significant responder in mouse lungs (Fig. 1C). Compared to MEG3–4, transcript of MEG3–1 shows much less abundance in lung tissue and cells, suggesting that MEG3–1 may play insignificant roles in bacterial infection. Ribosome profiling and quantitative real-time PCR (qRT-PCR) data confirmed that MEG3–4 is noncoding in mouse lungs (fig. S1, B to D) (25). Northern blotting and in situ analysis using a MEG3–4–specific probe (Fig. 1B, highlighted in blue) revealed that MEG3–4 is significantly repressed in mouse lungs after PAO1 infection (Fig. 1, D and G). qRT-PCR analysis using unique primers to target each of the MEG3 isoforms again showed that MEG3–4 in PAO1-infected mouse lungs was markedly and selectively repressed (fig. S1E).
qRT-PCR and Northern blotting data together showed that, after PAO1 infection, MEG3–4 was significantly reduced in mouse lungs and slightly reduced in livers but not altered in the spleen and kidney (Fig. 1, E and G, and fig. S1F), indicating that MEG3–4 plays tissue-restricted roles. Data also showed that MEG3–4 was the isoform with the lowest abundance in mouse lungs after infection by another P. aeruginosa strain, PAK, and another Gram-negative species, Klebsiella pneumoniae (KP) (Fig. 1, F and G). Northern blotting also showed that MEG3–4 was significantly repressed in PAO1-, PAK-, and KP-infected mouse lungs and less so in the liver, but not altered in the spleen and kidneys (Fig. 1G and fig. S1F). These findings reveal that the role of MEG3–4 may be tissue-restricted during infections by broad bacterial species.
MEG3–4 is regulated by a TLR4/NF-κB signaling pathway
Despite many cell types in the lung, alveolar epithelial cells and alveolar macrophages are critical resident populations with immune function. To investigate whether the decrease of MEG3–4 is cell type–dependent, we isolated mouse primary alveolar macrophages and alveolar type II epithelial cells (AECII). Data showed that expression of MEG3–4 in both primary alveolar macrophages and AECII cells was significantly decreased after PAO1 infection (Fig. 2A). Studies indicate that the localization of MEG3 is cell-specific (24). qRT-PCR revealed that in primary alveolar macrophages, about 62% of MEG3–4 was in nuclear fractions, whereas 38% was in cytosolic fractions (Fig. 2B), similar results observed in primary AECII cells (Fig. 2B). These data indicate that MEG3–4 plays potential roles in both alveolar macrophages and epithelial cells upon infection. We then chose mouse alveolar macrophages as model cells to further investigate molecular details of MEG3–4 regulation in innate immunity.
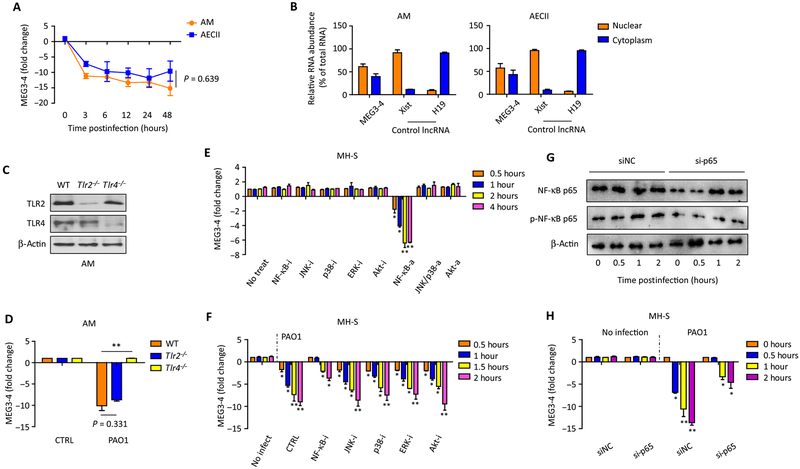
Fig. 2. MEG3–4 expression is regulated via a TLR4/NF-κB signaling pathway.
(A) Primary alveolar macrophages (AMs) and AECII were infected with PAO1 at a multiplicity of infection (MOI) of 20:1 for 1 hour, polymyxin B (100 μg/ml) was added, and cells were cultured for another 1 hour to kill bacteria outside of the cell membrane. Samples were collected at multiple time points over 48 hours, and the expression of MEG3–4 in AMs and AECII cells is time-dependent, as detected by qRT-PCR. (B) Nuclear and cytosolic expression of MEG3–4 in primary AMs and AECII cells was detected by qRT-PCR. lncRNA Xist and H19 were used as nuclear and cytoplasmic controls, respectively. (C) TLR2 and TLR4 expression in AMs from wild-type (WT), Tlr2−/−, and Tlr4−/− mice was measured by immunoblotting. (D) WT, Tlr2−/−, and Tlr4−/− mice (n = 3) were infected with 5 × 106 CFU of PAO1 per mouse for 24 hours. AMs were collected to assess MEG3–4 expression. (E and F) MH-S cells were pretreated with indicated signaling pathway activators (a) or inhibitors (i) for 4 hours and then infected for 2 hours with PAO1 at an MOI of 20:1. MEG3–4 expression before and after infection was analyzed by qRT-PCR. (G and H) MH-S cells were transfected with control siRNA [scrambled siRNA (siNC)] and NF-κB p65 siRNA (si-p65) for 48 hours, respectively, and then infected with PAO1 at 20:1 MOI for 2 hours. Expression and phosphorylation of NF-κB p65 were measured by immunoblotting, and MEG3–4 transcripts were detected by qRT-PCR. Data in (C) and (G) are representative of three independent mice or cell samples. Data in (A), (B), (D) to (F), and (H) are means ± SD for three independent mice or cell samples (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01). NS, no significant change.
TLR2 and TLR4 are important pattern recognition receptors for initial recognition of pathogenic bacteria (26). To determine whether TLR2 and TLR4 regulate the expression of MEG3–4, we isolated primary alveolar macrophages from Tlr2−/−, Tlr4−/−, and wild-type isogenic C57BL/6J mice. Having confirmed that TLR2 and TLR4 abundance had been ablated (Fig. 2C), we challenged these alveolar macrophages with PAO1. qRT-PCR assay revealed that reduction of MEG3–4 abundance upon PAO1 challenge was abolished in Tlr4−/− but not Tlr2−/− alveolar macrophages (Fig. 2D), suggesting that MEG3–4 inhibition was largely dependent on TLR4. We further transfected murine alveolar macrophage MH-S cells with small interfering RNA (siRNA) against TLR2 and TLR4 to inhibit their expression, alongside cells transfected with random-targeting siRNA as a negative control. Then, we infected TLR2- and TLR4-silenced cells with the Gram-positive bacterium Staphylococcus aureus. qRT-PCR analysis demonstrated that the suppression of MEG3–4 is mediated by specifically TLR4 (fig. S2).
To characterize signaling pathways involving MEG3–4, we pretreated MH-S cells with inflammatory inhibitors or activators (functionally validated by immunoblotting; figs. S3 and S4). Inhibitors did not alter MEG3–4 expression in uninfected cells, whereas activation of NF-κB decreased MEG3–4 expression (Fig. 2E). Upon PAO1 infection, inhibition of NF-κB in MH-S cells delayed the reduction of MEG3–4 expression (Fig. 2F). Because NF-κB modulators may alter many transcription factors and apoptosis proteins, we transfected MH-S cells with siRNA against the NF-κB subunit p65. Immunoblotting and quantitative PCR (qPCR) assays showed that the phosphorylation of NF-κB p65 was decreased (Fig. 2G and fig. S5A) and the reduction of MEG3–4 expression was delayed (Fig. 2H) in p65-silenced MH-S cells upon PAO1 infection. These results suggest that inhibition of MEG3–4 after PAO1 infection is mediated by a TLR4–NF-κB signaling pathway.
Overexpression of MEG3–4 exacerbates mouse susceptibility to PAO1 infection
Macrophages engineered to overexpress cytokines with subsequent airway delivery to the lung can efficiently magnify immune responses (27). We used this ex vivo approach to generate a mouse that harbors MH-S cells overexpressing MEG3–4 in the lungs. The MEG3–4 gene was amplified and cloned into the pcDNA3-EGFP (enhanced green fluorescent protein) vector to construct a plasmid expressing MEG3–4 (pWT-MEG3) that was further transfected into MH-S cells. Stable pWT-MEG3– or empty vector (EV)–transfected MH-S cells (fig. S6, A to C) were delivered intranasally into C57BL/6J mice to overexpress MEG3–4 in mouse lungs. qRT-PCR revealed that lung MEG3–4 expression in mice that received pWT-MEG3–transfected MH-S cells (hereafter called MEG3–4 mice) remained high 72 hours after infection compared to mice that received EV-transfected MH-S cells (hereafter called EV mice) and sham control mice (received no cells) (fig. S6, D and E).
The MEG3–4, EV, and sham control mice were then intranasally infected with PAO1 (each mouse received independently same amounts of transfected cells) to establish an acute pneumonia model. Kaplan-Meier survival curves showed that the MEG3–4 mice exhibited increased lethality (P = 0.0217, log-rank test; Fig. 3A) compared to EV mice, whereas survival between EV mice and sham control mice was not significantly different (Fig. 3A). To directly visualize the progression of infection, we also intranasally infected mice with PAO1 Xen-41 (an engineered bacterium emitting bioluminescence for imaging) to the MEG3–4 and EV mice. The MEG3–4–overexpressing mice displayed both a markedly broader distribution of bioluminescence in the thoracic cavity 4 hours after infection and continuous expansion compared to EV-transfection mice (Fig. 3, B and C), suggesting that MEG3–4 promotes bacterial dissemination in pneumonia models.
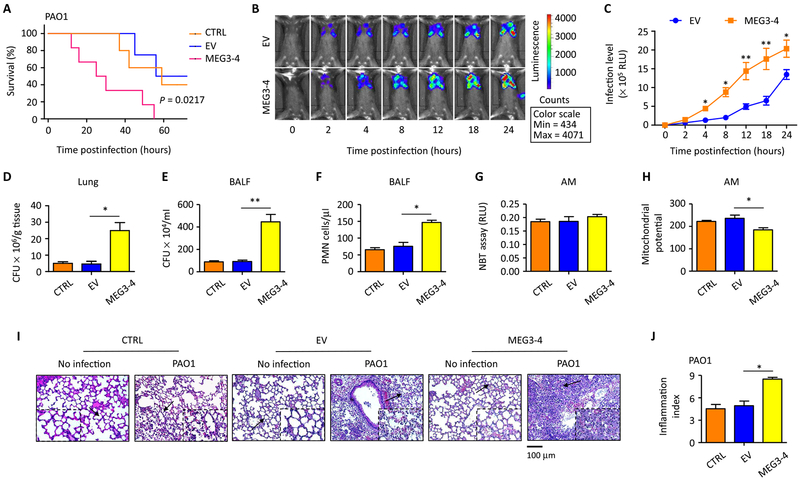
Fig. 3. MEG3–4 overexpression impairs host defense against P. aeruginosa.
(A) Mice that received MEG3–4–overexpressing MH-S cells (MEG3–4 mice, each mouse preinfected with independently transfected cells with pWT-MEG3), mice that received EV-expressing cells, and control mice that did not receive cells (n = 6) were intranasally challenged with PAO1 at 5 × 106 CFU per mouse and observed up to 72 hours. Kaplan-Meier survival curves were obtained (P = 0.0217 between MEG3–4 and EV mice, log-rank test, n = 2 biological replicates). (B and C) Whole-animal imaging of bioluminescence was obtained using the IVIS XRII system at different time points after MEG3–4 and EV mice (n = 6) were intranasally challenged with PAO1 Xen-41 at 5 × 106 CFU per mouse (n = 2 biological replicates). (D and E) Bacterial burdens in the lungs and BALF were determined 24 hours after PAO1 infection. (F) PMN percentages were evaluated in BALF versus total nuclear cells using HEMA-3 staining. (G) Superoxides in AMs were detected by nitroblue tetrazolium (NBT) assay. RLU, relative units. (H) Mitochondrial potential of AMs was measured by JC-1 fluorescence assay. (I) Lung injury and inflammation were assessed by hematoxylin and eosin (H&E) staining of paraffin-embedded sections (black arrows indicate the region of insets with tissue injury and inflammatory influx). Scale bar, 100 μm. (J) Inflammatory cell infiltration was determined in the lungs in (I) (10 random areas from 3 triplicate samples). Data in (C) are means ± SD for two independent mouse lungs in (B). Data in (I) show one representative of three independent mice. Data in (J) are means ± SD for three independent mouse lungs in (I). Data in (D) to (H) are means ± SD for three independent mice (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01).
Increasing MEG3–4 profoundly aggravates lung injury
To quantitatively measure bacterial dissemination, we evaluated bacterial burdens and found significantly increased CFU in the lung, liver, spleen, kidney, bronchoalveolar lavage fluid (BALF), and blood from MEG3–4 mice compared with samples from EV mice (Fig. 3, D and E, and fig. S6, F to I). Recruitment of polymorphonuclear neutrophils (PMNs) to the lung was increased in the BALF from MEG3–4 mice (Fig. 3F). Reactive oxygen species in primary alveolar macrophages from MEG3–4 and EV mice after PAO1 infection were not significantly different (Fig. 3G). In contrast, a JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3-tetracthylbenzimidazolylcarbocyanine iodide) fluorescence assay showed decreased mitochondrial membrane potential in alveolar macrophages of MEG3–4 mice (Fig. 3H). We identified clear histological alterations and inflammatory infiltration in lungs of MEG3–4 mice 24 hours after PAO1 infection (with an inflammation index of 1.8-fold over EV mouse lungs) (Fig. 3, I and J), which indicate severe lung injury in MEG3–4 mouse lungs. The data also confirmed that pcDNA3-EGFP vector alone did not alter immune responses in PAO1 infection (Fig. 3, D to J).
MEG3–4 specifically modulates IL-1β expression
MTT analysis showed that overexpression of MEG3–4 in MH-S cells markedly decreased cell survival after PAO1 infection (Fig. 4A), indicating that MEG3–4 may affect cell viability. Because excessive inflammatory responses cause cell death, we measured proinflammatory cytokine secretion in mouse BALF and lungs by enzyme-linked immunosorbent assay (ELISA), qRT-PCR, and immunoblotting. Data showed that abundance of tumor necrosis factor–α (TNF-α), IL-1β, and IL-6 was all significantly increased in BALF from PAO1-infected MEG3–4 mice compared to that from infected EV mice, and among these tested cytokines, IL-1β was the highest one (Fig. 4, B to D, and fig. S5B). Immunofluorescence microscopy also showed a more rapid induction of IL-1β expression after PAO1 infection in primary alveolar macrophages from MEG3–4 mice than in those from EV mice (Fig. 4E). These data suggest that MEG3–4 plays a specific role in IL-1β regulation.
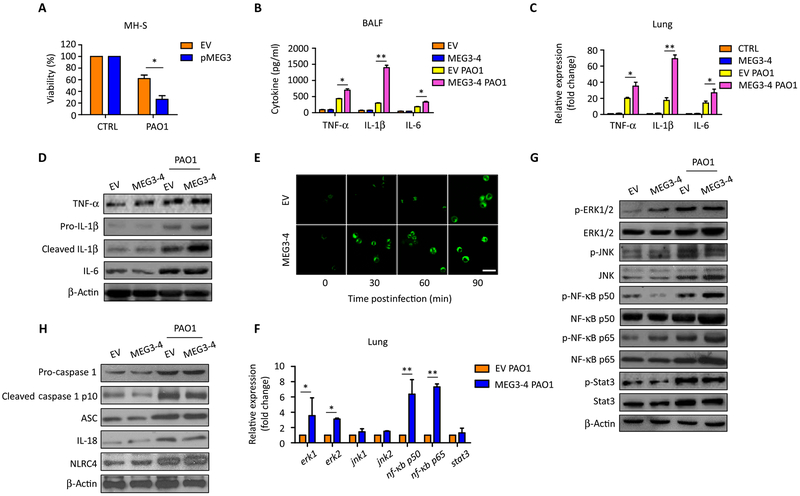
Fig. 4. MEG3–4 modulates IL-1β expression in mouse lungs.
(A) Plasmids pWT-MEG3 and empty pcDNA3-EGFP were transfected into MH-S cells with Lipofectamine 2000 for 24 hours. MEG3–4–overexpressing and control MH-S cells were infected with PAO1 at 20:1 MOI for 30 min, and cell viabilities were determined by MTT. (B) Mice that received MEG3–4–overexpressing MH-S cells (MEG3–4 mice) and mice that received EV-expressing cells (n = 3) were infected with PAO1 at 5 × 106 CFU per mouse for 24 hours. Cytokine expression in BALF was assessed by ELISA. (C and D) Cytokines in mouse lungs were detected by qRT-PCR and immunoblotting. (E) Normal AMs were transfected with pWT-MEG3 and empty pcDNA3-EGFP (100 ng) for 24 hours. Cells were challenged with PAO1 at an MOI of 20:1 for 0, 30, 60, and 90 min. Confocal laser scanning microscopy showed the production of IL-1β in AMs using immunochemistry staining. Scale bar, 50 μm. (F and G) Indicated transcription factors in the lungs were determined by qRT-PCR and immunoblotting. (H) Inflammasome factors in the lungs were determined by immunoblotting. Data in (D), (E), (G), and (H) are representative of three independent mice or cell samples. Data in (A) to (C) and (F) are means ± SD for three independent mice or cell samples (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01).
IL-1β activates NF-κB to drive the expression of other inflammatory factors, whereas IL-6 induces signal transducer and activator of transcription 3 (STAT3), and TNF-α activates NF-κB and c-Jun (28). qRT-PCR data revealed that the abundance of transcripts encoding extracellular signal–regulated kinase 1 (ERK1), ERK2, NF-κB p50, and NF-κB p65 was significantly increased by PAO1 infection in the lungs of MEG3–4 mice, whereas that of c-Jun N-terminal kinase (JNK1), JNK2, and STAT3 did not significantly change relative to each in the lungs from infected EV mice (Fig. 4F). Immunoblotting showed that total protein abundance of ERK1/2, NF-κB p50, and NF-κB p65 was highly increased in the lungs of PAO1-infected mice (Fig. 4G and fig. S5C). However, only the phosphorylation of p50 and p65 was significantly increased in PAO1-infected MEG3–4 mice compared to PAO1-infected EV mice (Fig. 4G and fig. S5C), indicating that MEG3–4 primarily modulates the IL-1β–NF-κB circuit instead of ERK1/2.
IL-1β is usually secreted as inactive precursors (pro-IL-1β), and inflammasomes then cleave pro-IL-1β to its mature form (cleaved IL-1β) (29). Immunoblotting analysis showed that the abundance of both pro-IL-1β and cleaved IL-1β were increased in the lungs of PAO1-infected MEG3–4 mice compared with EV mice (Fig. 4D and fig. S5B). However, none of the measured inflammasome factors, including caspase 1, IL-18, ASC (apoptosis-associated speck-like protein containing C-terminal), and NLRC4 (nod-like receptor family CARD domain containing 4), were significantly changed by overexpressing MEG3–4 (Fig. 4H and fig. S5D), suggesting that MEG3–4 may primarily affect the transcription or translation of IL-1β mRNA, rather than processing pro-IL-1β to its mature form.
We next examined morphologically inflammasome-mediated pyroptosis in macrophages (30). PAO1 infection induced membrane damage in MH-S cells, but it was not altered significantly by MEG3–4 overexpression (fig. S7A). Given that ASC expression is another marker for pyroptosis (31), we used immunostaining of ASC and found that ASC was increased in MH-S cells after PAO1 infection but was not further altered with MEG3–4 overexpression (fig. S7B). Immunoblotting data also showed increases in the abundance of caspase 1, IL-18, and ASC (pyroptosis-related factors) in the lungs of PAO1-infected mice (Fig. 4H). These data collectively demonstrate that overexpression of MEG3–4 did not significantly alter the course of pyroptosis.
MEG3–4 and IL-1β are targets of miR-138
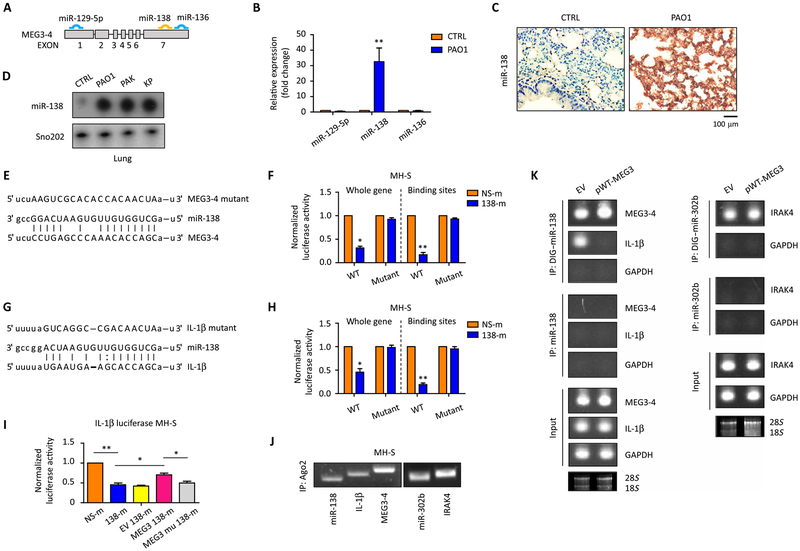
Most of the currently identified lncRNAs affect the expression of neighboring protein-coding genes in the genome (32); however, the location of the MEG3 gene (chromosome 12) is very far from the gene encoding IL-1β (chromosome 2). Because lncRNAs can act as sponges or decoys for miRNAs and prevent their binding from mRNA targets (33), we assumed that MEG3–4 may regulate IL-1β in this manner. Bioinformatics analysis indicated that three putative miRNAs (miR-129–5p, miR-138, and miR-136) may bind mouse MEG3–4 (Fig. 5A). However, our validation experiments identified miR-138 as the sole responder to MEG3–4 signaling using various bacteria-infected mouse models, as determined by qRT-PCR, immunostaining, and Northern blotting (Fig. 5, B to D, and fig. S1F).
Fig. 5. MEG3–4 binds miR-138 to compete the binding to IL-1β.
(A) miRNA binding sites of MEG3–4 (www.microrna.org/). (B) Mice (n = 3) were intranasally infected with 5 × 106 CFU PAO1 for 24 hours, and the lungs were lysed to assess miR-129–5p, miR-138, and miR-136. (C) In situ analysis with a digoxigenin (DIG)–labeled miR-138 probe (brown) in PAO1-infected and control lungs. Nuclei were stained by hematoxylin (blue). Scale bar, 100 μm. (D) Northern blots of miR-138 in lungs after PAO1, PAK, and KP infection. (E) Duplex formations between MEG3–4 (bottom) and miR-138 (middle). Target mutagenesis sites (top) are indicated. (F) MEG3–4 WT and mutant cloned to downstream of the luciferase coding region in pGL3 and cotransfected in MH-S with 138-m. Luciferase activities were determined using Luciferase Reporter Assay. (G) IL-1β 3′UTRs contain miR-138 binding sequence. Predicted duplex formations between IL-1β 3′UTR (bottom) and miR-138 (middle). (H) IL-1β 3′UTR and its mutant cloned to pGL3 and cotransfected into MH-S cells with 138-m. Luciferase activities were determined by Luciferase Reporter Assay. (I) IL-1β 3′UTR and its mutant cloned to pGL3 and cotransfected into MH-S cells with 138-m and pWT-MEG3 or with 138-m and pMU-MEG3 [mutant (mu)]. Luciferase activities were determined by Luciferase Reporter Assay. Empty pcDNA3-EGFP vector was used as a control. (J) Binding of Ago2 with MEG3–4, miR-138, and IL-1β mRNA was detected by individual cross-linking immunoprecipitation (iCLIP) using anti-Ago2 antibody and qRT-PCR. (K) Plasmids pWT-MEG3 and empty pcDNA3-EGFP transfected into MH-S. Binding of miR-138 to MEG3–4 or IL-1β mRNA in MEG3–4–overexpressing and EV cells was detected by LAMP assay and endpoint gel PCR. miRNA alone without DIG labeling was used as a negative control, whereas these transcripts in total MH-S cell extracts (input) were used as a positive control. In (J) and (K), binding of miR-320b and IL-1 receptor–associated kinase 4 (IRAK4) was used as a positive control. In (K), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a negative control, and 18S and 28S rRNAs were used as loading controls. Data in (C), (D), (J), and (K) show one representative of three independent mice or cell samples. Data in (B), (F), and (H) are means ± SD for three independent mice or cell samples (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01). IP, immunoprecipitation.
After that identification, we inserted a MEG3–4 fragment complementary DNA (cDNA), containing wild-type miR-138 binding sites, and a MEG3–4 mutant cDNA (change purine to pyridine or change pyridine to purine) with mutated miR-138 binding sites (Fig. 5E) [as described previously (34, 35)] downstream of the luciferase gene in the plasmid pGL3 and transfected this plasmid together with miR-138 mimics (138-m) into MH-S cells. Luciferase expression was decreased by 70% in 138-m–transfected MH-S cells compared with nonspecific control mimic (NS-m)–transfected cells, an effect that was abolished by mutating miR-138 binding sites (Fig. 5F). Notably, bioinformatics analysis showed that the 3′ untranslated region (3′UTR) in the mRNA encoding IL-1β is another potential target of miR-138 (Fig. 5G). Similarly, luciferase expression was dampened by about 50% in 138-m–transfected MH-S cells compared to NS-m–transfected cells (Fig. 5H), suggesting that miR-138 binds to IL-1β 3′UTR. We have further validated these results by cloning only the miR-138 binding portion (and its mutant) of MEG3–4 and IL-1β into the pGL3 luciferase vector. Similar results were observed, indicating that the miR-138 binding site is necessary and sufficient for mediating MEG3–4 function as a miRNA “sponge” (Fig. 5, E to H). We then cotransfected the MEG3–4 overexpression constructs together with either wild-type or mutant miR-138 binding site and analyzed the reversal of the suppression of the IL-1β UTR containing a luciferase reporter. We found that in IL-1β UTR containing luciferase reporter cells, overexpression of wild-type MEG3–4 could reverse the suppression of luciferase expression, but overexpression of mutant MEG3–4 could not, confirming the specificity of the effects that MEG3–4 and IL-1β competitively bind with miR-138 (Fig. 5I).
Premature miRNAs (pre-miRNAs) are cleaved by ribonuclease III and transported into a miRNA-induced silencing complex (miRISC) to function as a mature miRNA (36). The core of miRISC is formed by the Argonaute (Ago) proteins capable of mediating endonucleolytic cleavage of the target mRNA, which requires complementarity between a miRNA and target site (36). To confirm the direct binding of miR-138 to MEG3–4 (and mRNA encoding IL-1β), we performed a pulldown assay on MH-S cell lysates using Ago2 antibody. qRT-PCR analysis readily detected miR-138, MEG3–4, and IL-1β mRNA in Ago2 immunoprecipitates (Fig. 5J), indicating that Ago2 mediates the MEG3–4/miR-138 interaction. In addition, we used the labeled miRNA pulldown (LAMP) assay (37) using DIG-labeled pre-miR138 oligonucleotides that were mixed with MH-S cell extracts. Labeled extracts were then pulled down with antibodies to DIG or with miR-138 oligonucleotides alone as a negative control. Endpoint PCR gel analysis detected both MEG3–4 and IL-1β mRNA in the miR-138 coimmunoprecipitated mRNAs (Fig. 5K). Binding of MEG3–4 to miR-138 increased with MEG3–4 overexpression in MH-S cells compared to control cells, whereas binding of IL-1β mRNA to miR-138 was almost abolished (Fig. 5K). These data collectively demonstrate that miR-138 can bind both MEG3–4 and IL-1β mRNA, indicating that MEG3–4 competitively binds with miR-138 to free IL-1β. We also excluded the possibility that MEG3 modulates inflammatory responses by directly producing miRNAs because inhibition of intragenic miRNAs miR-1906–1 and miR-770 (fig. S1A) did not alter cell viability and cytokine production during PAO1 infection (fig. S8).
MEG3–4 modulates the expression of IL-1β by competitively binding to miR-138
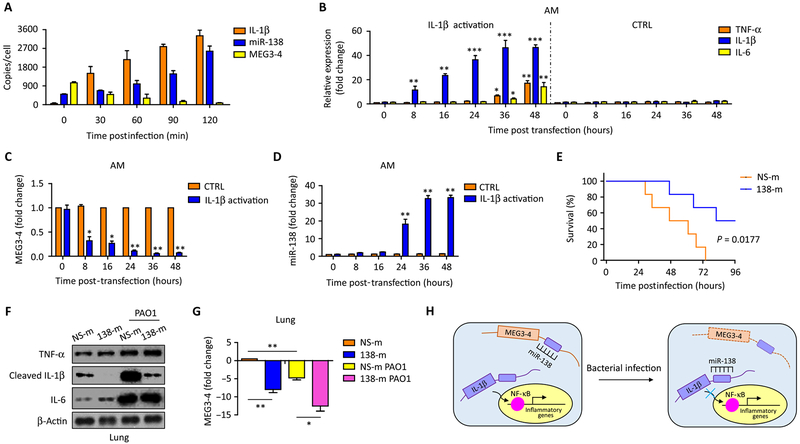
To elucidate how MEG3–4 regulates gene expression, we quantified exact copies of MEG3–4, miR-138, and IL-1β mRNA in cells by absolute qRT-PCR to determine their dynamic changes in PAO1 infection. We showed that PAO1 infection increased the abundance of mRNA encoding IL-1β from about 45 copies to 3300 copies per MH-S cell (Fig. 6A). Meanwhile, MEG3–4 was reduced from 1000 copies to 50 copies (Fig. 6A), whereas miR-138 was increased from 100 copies to 2500 copies per MH-S cell (Fig. 6A). These absolute qRT-PCR analyses showed that activation of IL-1β mRNA and shutdown of MEG3–4 occurred earlier than miR-138 induction after PAO1 infection.
Fig. 6. MEG3–4 modulates IL-1β and subsequent inflammatory responses by tightly modulating miR-138 expression.
(A) MH-S cells were infected with PAO1 at 20:1 for 2 hours. Copies of IL-1β mRNA, miR-138, and MEG3–4 in each MH-S cell were detected by absolute qRT-PCR. (B) Normal AMs (n = 3) transfected with IL-1β CRISPR-Cas activation plasmids or control plasmids (50 ng) or for 48 hours. TNF-α, IL-1β, and IL-6 were detected by qRT-PCR. (C and D) MEG3–4 and miR-138 in transfected AMs were measured by qRT-PCR. (E) Mice were intravenously injected with vehicle containing either NS-m or 138-m (50 μg per mouse) 24 and 48 hours (two time points in different animals) before PAO1 challenge. Kaplan-Meier survival curves of PAO1-infected NS-m– or 138-m–injected mice (n = 6, two independent experiments). Survival was determined up to 96 hours (P = 0.0177, log-rank test). (F) 138-m– and NS-m–injected mice (n = 3) were infected with PAO1 at 5 × 106 CFU per mouse for 24 hours. Expression of cytokines in mouse lungs was detected by immunoblotting. (G) qRT-PCR analysis of MEG3–4 expression in the lung from PAO1-infected NS-m– and 138-m–injected mice. (H) Proposed model for the role of lncRNA MEG3–4 in regulating IL-1β expression by competitively binding miR-138. Data in (A) to (D) are means ± SD for three independent mice or cell samples (Kruskal-Wallis test; *P ≤ 0.05 and **P ≤ 0.01). Error bars represent SD. Data in (F) and (G) are representative of three independent mice or cell samples.
PAO1 challenge often induces proinflammatory cytokines through complex signaling pathways. To determine whether the repression of MEG3–4 is responsible for IL-1β expression, we used IL-1β CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated protein) activation plasmids to generate MH-S cells that stably express IL-1β. At early time points after transfection (0 to 24 hours), IL-1β expression was enhanced in MH-S alveolar macrophages (Fig. 6B). This increased IL-1β led to subsequent induction of transcripts encoding NF-κB and production of downstream cytokines, such as TNF-α and IL-6 (at 36 and 48 hours; Fig. 6B). We observed that over the initial 24 hours, with increasing IL-1β in the cells (Fig. 6B), MEG3–4 abundance rapidly decreased (Fig. 6C), whereas miR-138 abundance gradually increased and continued to increase thereafter (Fig. 6D). These results demonstrate that the dynamics of MEG3–4 and miR-138 abundance correlated in a way that was consistent with down-regulating IL-1β during bacterial infection, but the actual modulation continues beyond that time course.
We then transfected 138-m into MH-S cells to induce miR-138 expression (fig. S9A). In miR-138–induced MH-S cells, IL-1β mRNA transcripts were significantly decreased after PAO1 infection compared to NS-m–transfected control cells (fig. S9B). Increased viability was also observed in miR-138–induced MH-S cells (fig. S9C). These findings attest that miR-138 regulates IL-1β expression and that over-expression of miR-138 promotes survival in MH-S cells.
We then performed an in vivo experiment to determine the physiological relevance of the mechanism. C57BL/6J mice were first intravenously injected with 138-m or NS-m and then challenged with PAO1. Notably, systemic delivery of 138-m significantly decreased mouse mortality after PAO1 infection (Fig. 6E). Bacterial burdens were also decreased in the lung and BALF of miR-138–injected mice (fig. S10, A and B). Infiltration of the lungs by PMNs was significantly decreased by assessment of the BALF from miR-138–injected mice (fig. S10C), but mitochondrial membrane potential in isolated primary alveolar macrophages was not significantly changed by miR-138 overexpression (fig. S10D).
We also measured critical cytokines in the BALF and lungs of 138-m– and NS-m–injected mice. Twenty-four hours after PAO1 infection, secretion of TNF-α, IL-1β, and IL-6 increased in the BALF from NS-m–injected mice (fig. S10E). Likewise, the secretion of TNF-α and IL-6 also increased in the BALF of 138-m–injected mice after PAO1 infection, but the secretion of IL-1β was significantly restrained (fig. S10E). Expression of TNF-α and IL-6 was induced similarly in the lungs from both 138-m–injected and NS-m–injected mice, whereas cleaved IL-1β was induced markedly less in 138-m–injected mice (Fig. 6F and fig. S5E). qRT-PCR analysis showed that reduction of MEG3–4 in 138-m mouse lungs decreased when IL-1β increased, suggesting that MEG3–4 responds to the abundance of IL-1β (Fig. 6G). Finally, we probed IL-1β expression in 138-m– and NS-m–transfected primary alveolar macrophages during PAO1 infection using immunofluorescence and found that IL-1β secretion was severely dampened in 138-m–transfected cells (fig. S10G). Collectively, these findings indicate that miR-138 decreases susceptibility to bacterial infection by repressing the expression of IL-1β.
Finally, we intravenously injected MEG3–4 wild-type and MEG3–4 mutant mice with 138-m and then challenged them with PAO1. Data showed that bacterial burdens were decreased in the lung and BALF of MEG3–4 wild-type/miR-138 mice but not in MEG3–4 mutant/miR-138 mice (fig. S11, A and B). Infiltration of PMN to the lung was significantly decreased in the BALF of MEG3–4 wild-type/miR-138 mice (fig. S11D), but mitochondrial membrane potential in isolated primary alveolar macrophages was not significantly changed in MEG3–4 wild-type/miR-138 mice (fig. S11C). These data revealed that the MEG3–4 expression phenotype can be partially reversed in vivo with 138-m.
In conclusion, we proposed a model for the role of lncRNA MEG3–4 in regulating IL-1β expression by competitively binding miR-138. Infection leads to excessive activation of IL-1β mRNA transcription in the lungs. miR-138 targets IL-1β mRNA and modulates its expression. In resting cells, miR-138 is sequestered by MEG3–4 for preventing the binding of IL-1β. Infection quickly shuts down MEG3–4 to release miR-138 to repress IL-1β and contain inflammation (Fig. 6H).
In addition, we noticed that sequences of mouse MEG3 are in low homology to human MEG3. To elucidate whether the mechanism also occurs in human cells, we performed bioinformatics analysis for human MEG3–4 and found that human MEG3–18 and human IL-1β mRNA share the complementary sites of human miR-330–5p (fig. S12A). We performed qRT-PCR in human primary alveolar macrophages and found that human MEG3–18 was significantly decreased in these cells after PAO1 infection (fig. S12C), suggesting that MEG3 may have similar functions in humans during bacterial infection. We also explored the role of MEG3–4 in carcinogenesis and found that its overexpression inhibits p53 expression in mouse B16 melanoma tumor cells, indicating that MEG3–4 may also act as a tumor suppressor (fig. S13), substantiating the concept that MEG3 plays multiple functions in mammalian cells.
DISCUSSION
Here, we report functional characterization of the conserved lncRNA MEG3, which is broadly expressed in mammalian cells and tissues. Our studies have yielded several important, unexpected findings. First, among its 10 diverse isoform transcripts, MEG3–4 is tissue-specifically inhibited in mouse lungs during pathogenic bacterial infection. Second, overexpression of MEG3–4 results in heightened inflammatory responses, severe lung injury, systemic dissemination, and increased mouse mortality, revealing an essential role for an lncRNA, in the regulation of inflammatory responses during bacterial infection. Third, MEG3–4 acts as a transcriptional controller of inflammation by modulating IRGs. Because of the miR-138 complementary 3′ region, MEG3–4 is able to sequester miR-138 in resting alveolar macrophages. During bacterial infection, repression of MEG3–4 frees more miR-138 in a temporal manner to regulate the expression of another target, IL-1β, and affects IL-1β–derived activation of NF-κB, thereby modulating expression of other proinflammatory cytokines (Fig. 6H). Elucidation of an MEG3–4/miR-138/IL-1β regulatory axis has expanded our understanding of lncRNA functions in regulation of inflammatory responses, providing additional evidence that suppression of lncRNA controls the expression of IRGs (10).
Our studies establish an intriguing mechanism for regulating the inflammatory cytokine, IL-1β, a critical mediator of host defense to microbial infections. IL-1β is a multifunctional cytokine that activates NF-κB and promotes production of proinflammatory cytokines, inducing fever and pyroptosis (38). Our findings that excessive MEG3–4 statistically significantly induces IL-1β expression and its related inflammatory responses reveal a previously unrecognized mechanism to IL-1β production. Current understanding of IL-1β production uses a two-step process requiring an initial signal to induce transcription of the 31-kDa preform and a second signal for posttranslational cleavage to the 17-kDa mature form, which depends on the activities of inflammasomes (29). Here, we report that MEG3–4 regulates IL-1β production (the most statistically significant responder) at the translational level rather than the posttranslational level and uncover that MEG3–4 is an inflammatory regulator of NF-κB activation in respiratory bacterial infections through negative feedback to TLR4. The miRNA feedback loop is a key modulator in inflammation. miR-24– and miR-629–governed hepatocyte nuclear factor 4α feedback is critical for liver tumorigenesis and inflammation (39). miR-146a constitutes a negative feedback loop in Streptococcus pneumoniae–induced macrophage activation (40). Previously, we reported that miR-302b regulates inflammatory responses via feedback to TLR/IRAK4 circuits (34). To date, only a few lncRNA feedback loops have been characterized for regulating inflammatory responses, and the underlying mechanisms remain highly elusive. Here, we delineate a model of IL-1β/MEG3–4 in host inflammatory responses: Bacterial infection is first sensed by TLR4 to activate NF-κB to initiate cytokine production. Activation of NF-κB and IL-1β also suppressed MEG3–4 expression to suppress inflammatory response by blocking IL-1β expression. This IL-1β/MEG3–4 loop provides a mechanism for balancing immune response during bacterial infection to protect host cells from damage.
Fundamentally, our discovery that MEG3–4 sequesters miR-138 contributes to an emerging concept that a subset of lncRNA functions as molecular decoys. lncRNA NORAD regulates genomic stability by sequestering Pumilio proteins (41), lncRNA Gadd7 interacts with TAR DNA binding protein 43 (TDP-43) and regulates Cdk6 mRNA decay (42). Besides inhibiting proteins by protein-lncRNA interactions, noncoding transcripts that sequester miRNAs, referred to competing endogenous RNAs (ceRNAs), have been identified to act as broad and robust regulators of gene expression (43). Relationships between MEG3 and miRNAs have been previously investigated in cancer models. miR-148a and miR-141 regulate MEG3 expression in gastric cancer by targeting DNA methyltransferase 1 (DNMT1) and E2F3 (44). miR-29 regulates hepatocellular MEG3 expression by targeting DNMT1 (45). However, the direct interaction between MEG3 and miRNAs and functional roles remain elusive. Several lines of evidence support MEG3’s function as a ceRNA of miR-138. First, alveolar macrophages express similar abundance of MEG3–4 in the nucleus and cytoplasm, enabling its interaction with miR-138. Second, in vitro luciferase and in vivo LAMP assays determined the direct binding of MEG3–4 and miR-138. Finally, both MEG3–4 and miR-138 were detected in Ago2 immunoprecipitates, indicating that this miRNA-mRNA binding protein mediates MEG3–4/miR-138 interaction.
We demonstrate that transcription and function of MEG3 isoforms are tissue-dependent, and suppression of MEG3 plays a general part in pulmonary host defense to bacterial infection. Mouse MEG3 is differentially expressed during embryonic development through alternative splicing (46), and 12 different human MEG3 transcripts were detected in human gonadotroph–derived pituitary adenomas and most human tumor cell lines (47). However, little is known about how these lncRNA isoforms are selectively edited and transcribed. Only one study suggested that mouse MEG3 alternative splicing was perhaps from promoter switching (46). In addition, our data and others’ studies suggest a general role of MEG3 against both bacterial and viral infections because down-regulation of MEG3–4 was observed in mouse lungs with broad strains or species and MEG3 has been reported to be significantly reduced in human T cells infected with human T cell leukemia virus type 1 (48).
In summary, multiple molecular and cellular approaches demonstrate that the precise regulation of lncRNA MEG3–4 expression in mouse lungs is essential to regulate inflammatory responses by modulating the transcription of the gene of IL-1β. MEG3–4 modulates activation of NF-κB and affects the transcription of downstream genes chiefly in alveolar macrophages. MEG3–4 exhibits lung-restricted expression patterns and isoform-specific functions in bacterial immunity both in vitro and in vivo. These findings uncover that MEG3–4, through a sponge effect, sequesters miR-138 to modulate IL-1β expression, particularly in the early to middle phases of infection to affect levels of inflammatory cytokines via the NF-κB axis. IL-1β, a critical proinflammatory cytokine itself and a regulator of NF-κB, precisely modulates transcription of inflammatory factors, thus reaching a well-balanced immune response. The insights obtained indicate that lncRNA MEG3 and associated pathways may serve as potential therapeutic targets to treat bacterial infection and thereby advancing our understanding of the roles of lncRNAs in immunity.
MATERIALS AND METHODS
Mice
C57BL/6J mice (6 to 8 weeks) were obtained from the Jackson Laboratory, and Tlr2−/− and Tlr4−/− mice were provided by J. Sharma (University of North Dakota). Mice were maintained in the animal facility at the University of North Dakota. All animal studies were approved by the University of North Dakota Institutional Animal Care and Use Committee (IACUC) and performed in accordance with the animal care and institutional guidelines (IACUC approval no. 1204–4). The animal experimental procedures, including treatment, care, and endpoint choice, followed the “Animal Research: Reporting In Vivo Experiments” guidelines. Animal experiments were performed with randomization. We used six mice (8 weeks old, both male and female randomly grouped) for survival assays, it will give an effective power of better than 0.9 (when α is 0.1). Other infection experiments used at least three mice (8 weeks old, both sex for C57BL/6J, Tlr2−/−, and Tlr4−/− mice) for getting better statistic results.
Primary cells and cell lines
Mouse primary AECII cells were isolated as previously described (49), whereas primary alveolar macrophages were isolated by BAL concurrent to AECII isolation, as described (22). AECII cells were grown in Dulbecco’s modified Eagle’s medium (Life Technologies) and alveolar macrophages in RPMI 1640 (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and incubated on culture plate for 1 hour at 37°C/5% CO2 incubator to allow attachment of macrophages. Nonadherent cells were removed by washing with normal saline. Murine MH-S alveolar macrophages were obtained from American Type Culture Collection and cultured following the manufacturer’s instructions (49). This cell line was authenticated by phenotypic analysis of critical traits of the cells, and they were tested for mycoplasma. Human primary alveolar macrophages from BALF samples were obtained through assistance by A. Bansal at Altru Hospital. All experiments with human alveolar macrophages were approved by the University of North Dakota Institutional Review Board Committee (IRB-200908–036, exempt 5) and have been conducted according to the principles expressed in the Declaration of Helsinki.
Bacterial infection
The wild-type P. aeruginosa strain PAO1 was provided by S. Lory (Harvard University). PAK was obtained from G. Pier (Harvard University). PAO1 Xen-41 was obtained from PerkinElmer-Caliper. KP (ATCC43816, serotype II) was provided by V. Miller (University of North Carolina). S. aureus (CPCC 160001) was obtained from W. Wei (Nanjing University). Bacteria were grown for about 16 hours in lysogeny broth (LB) at 37°C with 220-rpm shaking. The bacteria were pelleted by centrifugation at 5000g. Various mammalian cells were then cultured in antibiotic-free medium and infected by bacteria in an MOI of 20:1 bacteria/cell ratio. Mice were anesthetized with ketamine (45 mg/kg) and intranasally infected with 5 × 106 CFU of PAO1 in 50 μl of phosphate-buffered saline (PBS) (six mice per group) (22). Mice were monitored for symptoms and euthanized when they were moribund.
RNA isolation and qRT-PCR
Total RNA was extracted using TRIzol (Life Technologies) according to the manufacturer’s instructions. For lncRNA assay, a total of 50 ng of DNA-free RNA was subjected to first-strand cDNA synthesis using the High-Capacity RNA-to-cDNA kit (Life Technologies). For miRNA assay, cDNA is synthesized using the miScript II RT kit (Qiagen). For mRNA assay, cDNA is synthesized using the SuperScript III First-Strand Synthesis System (Life Technologies). The qRT-PCR assay was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) and gene-specific primers (table S2; synthesized by Integrated DNA Technologies) in a CFX Connect Real-Time PCR Detection System (Bio-Rad). The separate well 2−ΔΔCt cycle threshold method was used to determine the relative quantitative expression of individual lncRNAs, miRNAs, and mRNAs. lncRNAs and mRNAs were expressed as the fold difference to GAPDH, whereas miRNAs were expressed as the fold change to Sno202, and all these genes showed fold change compared to indicated reference group in each individual experiment (34).
lncRNA primer-profiling assay
Total RNA was extracted, and first-strand cDNA was synthesized using the High-Capacity RNA-to-cDNA kit. A PCR primer assay was performed using iTaq Universal SYBR Green Supermix and gene-specific primers that attached to the bottom of the mouse inflammatory response and autoimmunity array-based RT2 lncRNA primer-profiling panel (Qiagen) in the CFX Connect Real-Time PCR Detection System. PCR primer assay data were analyzed on Qiagen analysis website (www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/), and scatter plot result was the output.
Polyribosome analysis
Polyribosome analysis was performed as previously described (25). Briefly, cell suspension was prepared from 0.1 mg of mouse lung by using mechanical trituration method and treated with cycloheximide (CHX) (0.1 mg/ml) for three times. Cells were then lysed in 400 μl of polyribosome extraction buffer [15 mM tris-HCl (pH 7.4), 15 mM MgCl2, 0.3 M NaCl, CHX (0.1 mg/ml), heparin (1 mg/ml), and 1% Triton X-100). Then, the cell lysate was centrifuged, and the supernatant was loaded onto a sucrose gradient (10 to 50%) in Beckman 9/16 × 3–3/4 mm polyclear centrifuge tubes. Fractions were collected after the centrifugation of 35,000 rpm at 4°C for 190 min. Two microliters of fractions was used to measure the absorbance at 254 nm to obtain the polyribosome profile, which was used to identify the polyribosome fractions and nonpolyribosome fractions. RNA of each fraction was then extracted with 8 M guanidine HCl and purified by lithium chloride precipitation and RNeasy Mini kit (Qiagen). For qPCR, relative MEG3 abundance was calculated with ΔCt method and normalized with total MEG3 in all the fractions.
In situ hybridization analysis
Lungs of three independent mice were fixed in 10% formalin (Sigma-Aldrich) for 24 hours and then embedded in paraffin using a routine histologic procedure, and 4-μm sections were cut. The DIG-labeled MEG3 (only specific targeting MEG3–4 exon 1, blue; Fig. 1B) or miR-138–targeting probes were synthesized by Integrated DNA Technologies (table S2). In situ hybridization (ISH) analysis was performed following a standard ISH protocol in Singer Lab (www.einstein.yu.edu/labs/robert-singer/protocols/). A mouse monoclonal antibody against DIG was obtained from Abcam. MEG3 and miR-138 in mouse lungs were detected using EXPOSE Mouse and Rabbit Specific HRP/DAB Detection IHC kit (Abcam) and observed by Nikon Eclipse 80i (Upright) microscopy (Nikon Instruments Inc.).
Northern blotting
Total RNA was extracted using TRIzol according to the manufacturer’s instructions. 32P-ATP–labeled MEG3 (a general probe targeting MEG3–1/MEG3–2/MEG3–3/MEG3–4/MEG3–6/MEG3–8/MEG3–9/MEG3–10, yellow in Fig. 1B, and a probe specific for MEG3–4, same with ISH analysis) and miR-138 and Sno202 probes were synthesized by Integrated DNA Technologies (table S2). Northern blotting was performed according to a published protocol, as previously described (34). MEG3 transcripts and miR-138 were visualized by exposure to x-ray film (Thermo Fisher Scientific) at −80°C overnight.
Nuclear and cytoplasmic fraction preparation
A total of 5 × 106 MH-S cells scraped from culture flasks were washed twice with cold PBS and then transferred into a prechilled microcentrifuge tube. Cells were gently resuspended in 500 μl of hypotonic buffer [20 mM tris-HCl (pH 7.4), 10 mM NaCl, and 3 mM MgCl2] by pipetting for 20 times and then incubated on ice for 15 min. Detergent (10% NP-40; 25 μl) was added and vortexed for 10 s at the highest setting. Cell lysate were centrifuged for 10 min at 3000 rpm at 4°C. The supernatant (cytoplasmic fraction) was removed, and the nuclear pellet was resuspended in 50 μl of complete cell extraction buffer [10 mM tris-HCl (pH 7.4), 2 mM Na3VO4, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 1 mM EGTA, 0.1% SDS, 1 mM NaF, 0.5% deoxycholate, and 20 mM Na4P2O7] for 30 min on ice with vortexing at 10-min intervals. RNA of each fraction was then extracted with 8 M guanidine HCl and purified by lithium chloride precipitation and RNeasy Mini kit.
Plasmid construction
Wild-type MEG3–4 or its mutant gene was amplified from MH-S cDNA with specific primers (table S2) by PCR and cloned into the BamH I and Hind III sites of the pcDNA3-EGFP vector (#13031, Addgene). MEG3–4, IL-1β, and their relative mutants were also amplified from MH-S cDNA and cloned into the Fse I and Xba I sites of pGL3 vector in the 3′ end of a reporter luciferase gene (Promega). miR-138 coding gene fragment (500 base pairs) was amplified from MH-S cDNA and cloned into pMD19-T vector. Constructed plasmids were electroporated into DH5α using an Electroporator 2510 system (settings: 25 μF, 200 ohms, 2.5 kV; Eppendorf). Transformants were selected and maintained in an LB medium containing ampicillin (100 μg/ml; Sigma-Aldrich). All of the nucleases, polymerases, and ligases used in molecular cloning were bought from New England Biolabs Inc.
Transfection of siRNA, plasmid, activators, and inhibitors
NF-κB p65 (sc-29411), TLR2 (sc-40257), TLR4 (sc-40261), siNCs (sc-37007), IL-1β CRISPR-Cas9 activation (sc-421097-ACT), and control plasmids (sc-437275) were obtained from Santa Cruz Biotechnology. Signaling pathway activators and inhibitors were all obtained from Sigma-Aldrich. The 138-m (C-310415–07) and negative NS-m (CN-001000-01-05) were purchased from GE Dharmacon, whereas miR-770 (MCM03174) (770-m) and miR-1906–1 mimics (MCM01312) (1906–1-m) were obtained from Applied Biological Materials. MH-S cells were transfected with siRNAs (5 nM) for 48 hours, constructed and control plasmids, IL-1β CRISPR activation and control plasmids (100 ng), and miRNA and control mimics (50 ng) using Lipofectamine 2000 (Life Technologies) for 24 hours before bacterial infection. MH-S cells were treated with indicated signaling pathway activator or inhibitor, including NF-κB inhibitor (SN50), JNK inhibitor (SP600125), p38 inhibitor (SB203580), ERK inhibitor (FR180204), Akt inhibitor (GSK690693), NF-κB activator (betulinic acid), JNK/p38 activator (anisomycin), and Akt activator (SC79), at 10 nM, respectively, for 4 hours before and together with bacteria during infection.
Mouse models
pWT-MEG3, pMU-MEG3, or empty pcDNA3-EGFP vector was transfected MH-S cells and then treated with G418 (1 mg/ml) to select stable and homogeneous cell population to express MEG3–4 or EV from single colonies, as evaluated through the EGFP marker. For ex vivo cell transfer model, mice were intranasally instilled with the stable MH-S cells that were transfected with pWT-MEG3, pMU-MEG3, or empty pcDNA3-EGFP vector (1 × 106 cells per mouse in 50 μl of PBS) for 24 hours before bacterial infection, as described previously, with modification (27). For miRNA transfection model, mice were intravenously administered with vehicle (in vivo-jetPEI, Polyplus-transfection), NS-m, or 138-m (50 μg per mouse) twice (at 24 and 48 hours) before bacterial infection, as described previously, with modification (34).
Immunoblotting
Mouse monoclonal antibodies against β-actin (sc-47778), TLR4 (sc-293072), p-ERK (sc-7383), ERK1/2 (sc-514302), p-JNK (sc-6254), JNK (sc-7345), p-NF-κB p50 (sc-33022), NF-κB p50 (sc-166588), NF-κB p65 (sc-8008), p-Stat3 (sc-8059), pro-caspase I (sc-56036), MAPK p38 (sc-7972), and p53 (sc-377567); goat polyclonal antibodies against cleaved IL-1β (sc-23460), IL-6 (sc-1265), TNF-α (sc-1349), and NLRC4 (sc-49395); rabbit polyclonal antibodies against TLR2 (sc-10739), Stat3 (sc-482), p-NF-κB p65 (sc-33020), pro-IL-1β (sc-7884), cleaved caspase I (sc-514), ASC (sc-22514), and Akt1 (sc-8312); and chicken polyclonal antibodies against IL-18 (sc-7954-Y) were obtained from Santa Cruz Biotechnology. The samples derived from cells and lung homogenates were lysed in radioimmunoprecipitation assay buffer, separated by electrophoresis on 12% SDS–polyacrylamide gel electrophoresis gels, and transferred to nitrocellulose transfer membranes (GE Amersham Biosciences). Proteins were detected by Western blotting using primary antibodies at a concentration of 1/200 (Santa Cruz Biotechnology) and were incubated overnight. Specific interaction with the primary antibodies was detected using corresponding secondary antibodies conjugated to horseradish peroxidase (HRP) (Santa Cruz Biotechnology) and detected using enhanced chemiluminesence reagents (Santa Cruz Biotechnology). Gel bands were quantified by Quantity One software (Bio-Rad), and error bar represents three independent immunoblotting assays. Phosphorylated and total protein levels were determined and quantified by three independent successive immunoblotting membranes (22).
In vivo imaging
At various time points after infection, P. aeruginosa Xen-41–infected mice were imaged with an IVIS XRII system (PerkinElmer-Caliper), following the user guides provided by PerkinElmer-Caliper (50).
Bacterial burden assay
Alveolar macrophages from BALF and tissues of the lung, spleen, liver, and kidney were homogenized with PBS and spread on LB dishes to enumerate bacteria. The dishes were cultured in a 37°C incubator overnight, and colonies were counted. Triplicates were performed for each sample and control (50).
NBT assay
This assay is based on the color change of NBT dye upon reduction by released superoxide. Alveolar macrophages from BALF were grown on a 96-well plate in serum-containing medium at 37°C for 4 hours, and NBT dye (1 μg/ml; Sigma-Aldrich) was added to each well. Cells were incubated for an additional hour or until color developed. The dye is yellow and gives a blue-colored formazan product upon reduction by superoxide. The reduction was terminated by adding 100 μl of stop solution (10% dimethyl sulfoxide and 10% SDS in 50 mM Hepes buffer). The plate was kept at room temperature overnight for complete dissolution of formazan, and absorbance at 560 nm was recorded using a multiscan plate reader to quantify the concentration of super-oxide anion (49). Triplicates were performed for each sample and control.
Mitochondrial assay
JC-1 Mitochondrial Membrane Potential Assay kit (Cayman Chemical) was used for this assay, following the manufacturer’s instructions. The cytofluorimetric, lipophilic cationic dye, JC-1, can selectively enter into mitochondria and reversibly change color from green to red as the membrane potential increases. Alveolar macrophages were treated as above, and an equal amount of dye was added. After 30-min incubation, fluorescence was measured using a fluorometer, a 560-nm excitation and 595-nm emission filter for detecting healthy cells, and a 485-nm excitation and 535-nm emission filter for detecting dead cells (50). Triplicates were performed for each sample and control.
MTT cell proliferation assay
This assay measures the color change in MTT (Sigma-Aldrich) upon regulation by enzymes to assess the viability of cells. Cells were treated as above, and an equal amount of dye was added. After 1-hour incubation, the reaction was stopped by stop solution and left at room temperature overnight for complete dissolution of formazan. Absorbance at 560 nm was recorded using a multiscan plate reader to quantify the concentration (50). Triplicates were performed for each sample and control.
Histological analysis
Lung tissues of three independent mice were fixed in 10% formalin (Sigma-Aldrich) for 24 hours and then embedded in paraffin using a routine histologic procedure. Four-micrometer sections were cut, stained by standard H&E, and examined for differences in morphology after infection. The degree of cellular infiltration was scored using previously described methods. The index was calculated by multiplying severity by extent in 10 random areas, with a maximum possible score of 9 (22).
Inflammatory cytokine profiling
Cytokine concentrations of TNF-α, IL-6, and IL-1β were measured by ELISA kits (eBioscience Co.) in samples of BALF collected at the indicated times after infection. BALF was collected, and 100 μl of aliquots of samples was added to the coated microtiter wells. The cytokine concentrations were determined with corresponding detection HRP-conjugated antibodies. The values were read at 450 nm (22).
Immunostaining
Alveolar macrophages isolated from linc-MEG3–4–expressing mice after 24 hours after PAO1 infection and from EV control mice. Cells were individually incubated with both primary anti–IL-1β antibody (or ASC antibody) and then the second fluorescein isothiocyanate–conjugated antibodies, as described. Colocalization was observed under an LSM 510 Meta Confocal Microscope (Carl Zeiss Micro-Imaging) (34).
Luciferase assay
MH-S cells were transfected with a series of pGL3-based plasmids, with or without 138-m together. Twenty-four hours after transfection, cell lysate was subjected to luciferase activity analysis by the Luciferase Reporter Assay System (Promega), following the user guides provided by the manufacturer (34).
Ago iCLIP
iCLIP was performed on MH-S cells lysates using Pierce Classic Magnetic IP kit (Thermo Fisher Scientific) with a monoclonal Ago antibody, 2A8 (SAB4800048, Sigma-Aldrich), to pull down Ago2, according to the manufacturer’s instructions. Ago2-associated RNA was cross-linked with Ago before immunoprecipitation by ultraviolet (UV) light (150 mJ/cm2) using a Stratalinker 1800 UV Cross-linker (Agilent Technologies). Total RNA was isolated from the precipitate, and qPCR was performed (51).
LAMP assay
LAMP assay was performed following the previous study (37). Briefly, cell extracts were incubated with 70 μg of DIG-labeled pre-miRNA at 4°C for 30 min, the total volume was adjusted to 1 ml with binding buffer [25 mM tris-HCl (pH 7.4), 60 mM KCl, 2.5 mM EDTA, 0.2% Triton X-100, and 80 U of ribosomal RNA synthesis inhibitor], and the mixture was incubated at 37°C for 60 min. The sample was transferred to a tube containing 20 μl of anti-DIG agarose beads and rotated slowly overnight at 4°C. After the mixture had been spun down at 15,600g for 30 min at 4°C, it was washed with washing buffer [20 mM tris-HCl (pH 7.4), 350 mM KCl, and 0.02% NP-40] and spun again for 15 min. The sample was heated to 95°C for 15 min, and a clear lysate was obtained after being spun down at 15,600g for 10 min at 4°C. The purified RNA was collected when deoxyribonuclease I (20 U) was added and incubated at 37°C for 30 min before the phenol/chloroform extraction was performed. miRNA alone without DIG labeling was used as a negative control, whereas these transcripts in total MH-S cell extracts (input) were examined as an untreated control.
Absolute qPCR
Absolute copy numbers of mRNA, lncRNA, and miRNA were measured as previously described (52). Plasmid pWT-MEG3 was used for constructing MEG3–4 standard curve, whereas pGL3–IL-1β was used for IL-1β mRNA and pMD19-T–miR-138 for miR-138. The RNA copy numbers were calculated for each sample from the standard curve by the instrument software using the Ct value.
Statistical analysis
Experiments were conducted in triplicate and repeated at least three times (except for lncRNA primer-profiling assay, performed twice). The differences in outcomes of mice are presented as percent or amount changes compared with the control after treatment. Differences between two groups were compared by Kruskal-Wallis test using SPSS Statistics software with means ± SD, whereas mice survival rates were calculated using Kaplan-Meier curves (22).
Supplementary Material
www.sciencesignaling.org/cgi/content/full/11/536/eaao2387/DC1
Fig. S1. Characterization of lncRNA MEG3.
Fig. S2. MEG3–4 inhibition signaling in alveolar macrophage cells is TLR4-specific during S. aureus infection.
Fig. S3. Immunoblotting validation of signaling factors in inhibitor- or activator-treated MH-S cells.
Fig. S4. Dissection of signaling molecules in PAO1-infected MH-S cells.
Fig. S5. Densitometric quantification of the immunoblotting data.
Fig. S6. Restoration of the phenotype in a MEG3–4–overexpressing model.
Fig. S7. Imaging of pyroptosis in MH-S cells.
Fig. S8. Functional analysis of miRNAs generated by MEG3–4.
Fig. S9. miR-138 regulates IL-1β expression and cell viability in alveolar macrophages.
Fig. S10. miR-138 enhances host defense against P. aeruginosa by repressing IL-1β expression in mouse lungs.
Fig. S11. MEG3–4 overexpression phenotypes in mice were reversed by treatment with 138-m.
Fig. S12. Analysis of MEG3 function and expression in human alveolar macrophages.
Fig. S13. MEG3–4 overexpression inhibits p53 expression in mouse B16 melanoma tumor cells.
Table S1. lncRNA expression in response to PAO1 infection.
Table S2. Primers used in this study.
Acknowledgments:
We thank S. Nechaev, C. Brissette, and M. Nilles of our department for reading the manuscript and S. Abrahamson and S. Adkins (University of North Dakota Core Facilities, supported by NIH grants INBRE P20GM103442, COBRE P30GM103329, and COBRE P20GM113123) for the help with imaging and flow cytometry.
Funding: This work was supported by the NIH (AI109317–01A1, AI101973–01, and AI097532–01A1) and the National Natural Science Foundation of China (81703569).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Yoon J-H, Abdelmohsen K, Gorospe M, Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol 425, 3723–3730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loewer S, Cabili MN, Guttman M, Loh Y-H, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL, Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet 42, 1113–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM, A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. U.S.A. 103, 5781–5786 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleutels F, Zwart R, Barlow DP, The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE, A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113, 2526–2534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imamura K, Akimitsu N, Long non-coding RNAs involved in immune responses. Front. Immunol 5, 573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald KA, Caffrey DR, Long noncoding RNAs in innate and adaptive immunity. Curr. Opin. Immunol 26, 140–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY, A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2, e00762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA, A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA, A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F, Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5, 211–220 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Lu KH, Li W, Liu X.-h., Sun M, Zhang M.-l., Wu W.-q., Xie W.-p., Hou Y.-y., Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13, 461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Wu Y.-t., Huang C, Meng X-M, Ma T.-t., Wu B-M, Xu F.-y., Zhang L, Lv X-W, Li J, Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim. Biophys. Acta 1842, 2204–2215 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A, Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology 151, 2443–2452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oczkowicz M, Piestrzyska-Kajtoch A, Piórkowska K, Rejduch B, Rózycki M, Expression of DLK1 and MEG3 genes in porcine tissues during postnatal development. Genet. Mol. Biol 33, 790–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astuti D, Latif F, Wagner K, Gentle D, Cooper WN, Catchpoole D, Grundy R, Ferguson-Smith AC, Maher ER, Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: Analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br. J. Cancer 92, 1574–1580 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T, MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30, 4750–4756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Ren Z, Sun P, Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem 113, 1868–1874 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A, Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70, 2350–2358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A, Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem 282, 24731–24742 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Pawar K, Hanisch C, Palma Vera SE, Einspanier R, Sharbati S, Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci. Rep 6, 19416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Fang L, Tan S, Yu M, Li X, He S, Wei Y, Li G, Jiang J, Wu M, Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 26, 1273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKiernan PJ, Molloy K, Cryan SA, McElvaney NG, Greene CM, Long noncoding RNA are aberrantly expressed in vivo in the cystic fibrosis bronchial epithelium. Int. J. Biochem. Cell Biol. 52, 184–191 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A, The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn 212, 214–228 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Hu G, Lou Z, Gupta M, The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLOS ONE 9, e107016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salcedo SP, Alexopoulou L, Investigating TLR signaling responses in murine dendritic cells upon bacterial infection. Methods Mol. Biol 1197, 209–225 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Wu M, Hussain S, He Y-H, Pasula R, Smith PA, Martin II WJ, Genetically engineered macrophages expressing IFN-γ restore alveolar immune function in scid mice. Proc. Natl. Acad. Sci. U.S.A. 98, 14589–14594 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Han S, Sun Y, An IL6-STAT3 loop mediates resistance to PI3K inhibitors by inducing epithelial-mesenchymal transition and cancer stem cell expansion in human breast cancer cells. Biochem. Biophys. Res. Commun 453, 582–587 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Netea MG, Simon A, van de Veerdonk F, Kullberg B-J, Van der Meer JWM, Joosten LAB, IL-1β processing in host defense: Beyond the inflammasomes. PLOS Pathog. 6, e1000661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbé K, Saleh M, Cell death in the host response to infection. Cell Death Differ. 15, 1339–1349 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Fernandes-Alnemri T, Wu J, Yu J-W, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES, The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14, 1590–1604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogu GK, Vizánb P, Stantone LW, Beatob M, Di Croceb L, Marti-Renoma MA, Chromatin and RNA maps reveal regulatory long noncoding RNAs in mouse. Mol. Cell. Biol 36, 809–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I, A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Li X, Ye Y, Zhao K, Zhuang Y, Li Y, Wei Y, Wu M, MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nat. Commun 5, 3619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, He S, Li R, Zhou X, Zhang S, Yu M, Ye Y, Wang Y, Huang C, Wu M, Pseudomonas aeruginosa infection augments inflammation through miR-301b repression of c-Myb-mediated immune activation and infiltration. Nat. Microbiol 1, 16132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD, Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat. Struct. Mol. Biol 16, 1259–1266 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Hsu R-J, Tsai H-J, Performing the Labeled microRNA pull-down (LAMP) assay system: An experimental approach for high-throughput identification of microRNA-target mRNAs. Methods Mol. Biol 764, 241–247 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Dinarello CA, Biologic basis for interleukin-1 in disease. Blood 87, 2095–2147 (1996). [PubMed] [Google Scholar]
- 39.Gougelet A, Colnot S, MicroRNA-feedback loop as a key modulator of liver tumorigenesis and inflammation. World J. Gastroenterol 19, 440–444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griss K, Bertrams W, Sittka-Stark A, Seidel K, Stielow C, Hippenstiel S, Suttorp N, Eberhardt M, Wilhelm J, Vera J, Schmeck B, MicroRNAs constitute a negative feedback loop in Streptococcus pneumoniae–induced macrophage activation. J. Infect. Dis 214, 288–299 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Lee S, Kopp F, Chang T-C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT, Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Li D, Zhang W, Guo M, Zhan Q, Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 31, 4415–4427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP, A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Ji G, Ke X, Gu H, Jin W, Zhang G, MiR-141 inhibits gastric cancer proliferation by interacting with long noncoding RNA MEG3 and down-regulating E2F3 expression. Dig. Dis. Sci 60, 3271–3282 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, Zhao W, Jin S, MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med. Oncol 31, 879 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Croteau S, Charron M-C, Latham KE, Naumova AK, Alternative splicing and imprinting control of the Meg3/Gtl2-Dlk1 locus in mouse embryos. Mamm. Genome 14, 231–241 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A, Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 151, 939–947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJM, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C, MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun 6, 7743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kannan S, Huang H, Seeger D, Audet A, Chen Y, Huang C, Gao H, Li S, Wu M, Alveolar epithelial type II cells activate alveolar macrophages and mitigate P. aeruginosa infection. PLOS ONE 4, e4891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li R, Tan S, Yu M, Jundt MC, Zhang S, Wu M, Annexin A2 regulates autophagy in Pseudomonas aeruginosa infection through the Akt1–mTOR–ULK1/2 signaling pathway. J. Immunol 195, 3901–3911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi SW, Zang JB, Mele A, Darnell RB, Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460, 479–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Formisano-Tréziny C, de San Feliciano M, Gabert J, Development of plasmid calibrators for absolute quantification of miRNAs by using real-time qPCR. J. Mol. Diagn 14, 314–321 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencesignaling.org/cgi/content/full/11/536/eaao2387/DC1
Fig. S1. Characterization of lncRNA MEG3.
Fig. S2. MEG3–4 inhibition signaling in alveolar macrophage cells is TLR4-specific during S. aureus infection.
Fig. S3. Immunoblotting validation of signaling factors in inhibitor- or activator-treated MH-S cells.
Fig. S4. Dissection of signaling molecules in PAO1-infected MH-S cells.
Fig. S5. Densitometric quantification of the immunoblotting data.
Fig. S6. Restoration of the phenotype in a MEG3–4–overexpressing model.
Fig. S7. Imaging of pyroptosis in MH-S cells.
Fig. S8. Functional analysis of miRNAs generated by MEG3–4.
Fig. S9. miR-138 regulates IL-1β expression and cell viability in alveolar macrophages.
Fig. S10. miR-138 enhances host defense against P. aeruginosa by repressing IL-1β expression in mouse lungs.
Fig. S11. MEG3–4 overexpression phenotypes in mice were reversed by treatment with 138-m.
Fig. S12. Analysis of MEG3 function and expression in human alveolar macrophages.
Fig. S13. MEG3–4 overexpression inhibits p53 expression in mouse B16 melanoma tumor cells.
Table S1. lncRNA expression in response to PAO1 infection.
Table S2. Primers used in this study.