Abstract
Malignant peripheral nerve sheath tumor (MPNST) is an aggressive sarcoma with recurrent loss-of-function alterations in polycomb-repressive complex 2 (PRC2), a histone-modifying complex involved in transcriptional silencing. To understand the role of PRC2 loss in pathogenesis and identify therapeutic targets, we conducted parallel global epigenomic and proteomic analysis of archival formalin-fixed, paraffin-embedded (FFPE) human MPNST with and without PRC2 loss (MPNSTLOSS vs. MPNSTRET). Loss of PRC2 resulted in increased histone posttranslational modifications (PTM) associated with active transcription, most notably H3K27Ac and H3K36me2, whereas repressive H3K27 di- and trimethylation (H3K27me2/3) marks were globally lost without a compensatory gain in other repressive PTMs. Instead, DNA methylation globally increased in MPNSTLOSS. Epigenomic changes were associated with upregulation of proteins in growth pathways and reduction in IFN signaling and antigen presentation, suggesting a role for epigenomic changes in tumor progression and immune evasion, respectively. These changes also resulted in therapeutic vulnerabilities. Knockdown of NSD2, the methyltransferase responsible for H3K36me2, restored MHC expression and induced interferon pathway expression in a manner similar to PRC2 restoration. MPNSTLOSS were also highly sensitive to DNA methyltransferase and histone deacetylase (HDAC) inhibitors. Overall, these data suggest that global loss of PRC2-mediated repression renders MPNST differentially dependent on DNA methylation to maintain transcriptional integrity and makes them susceptible to therapeutics that promote aberrant transcription initiation.
Introduction
Malignant peripheral nerve sheath tumor (MPNST) is an aggressive sarcoma with a poor prognosis, particularly in the setting of metastatic disease, where tumors are poorly responsive to traditional chemotherapy (1). Thus, targeted therapeutics, rooted in the molecular biology of the tumor, could be highly useful. The molecular pathogenesis of MPNST has been elucidated, in part, due to its syndromic association with neurofibromatosis type I (NF1), which is the clinical setting of roughly one-half of MPNST cases. In NF1, patients harbor germline alterations resulting in the loss of one functional copy of the eponymous NF1 gene, a Ras-GTPase. Benign neurofibromas, which may be numerous, are associated with loss of heterozygosity for the remaining functional allele (2). Malignant progression is associated with additional molecular events, including loss of the CDKN2A tumor suppressor, and, in the majority of cases, loss of function alterations in polycomb-repressive complex 2 (PRC2) components (3, 4).
PRC2 is a key epigenetic regulator essential to transcriptional silencing of target genes (5). It is composed of a core complex containing the catalytic subunit, EZH1/2, along with SUZ12 and EED, which are necessary for histone methylation. Although the best characterized function of PRC2 is in trimethylation of histone H3K27 at promoters, PRC2 also mediates mono- and dimethylation, the roles of which are more poorly understood. K27me1 localizes to regions of active transcription (6). Like K27me3, K27me2 is associated with reduced levels of transcription, but its distribution is distinct. It is the most abundant form of methylation, present on more than half of total H3, and is deposited broadly across the genome (7). Recent work has established a role for K27me2 in the genome-wide suppression of inappropriate transcriptional activation (7, 8). This function underlies the ability of EZH2 inhibitors to induce inappropriate transcriptional activation of endogenous retroviral elements, and highlights the importance of mediators of transcriptional silencing in safeguarding the genome.
The crucial need to suppress inappropriate transcriptional initiation is underscored by the presence of multiple, partially redundant epigenetic repressive mechanisms. DNA methylation, a distinct epigenetic regulator that mediates transcriptional silencing can, in some instances, substitute for H3K27me3 in gene silencing (9). PRC2-mediated H3K27me3 may also spread into previously DNA-methylated regions upon loss of DNA methylation (9, 10). Likewise, in some circumstances, H3K27me3 can replace H3K9me3, a marker of constitutive.
To ensure that the appropriate transcriptional program is expressed in a particular cell type or at a particular stage in development, PRC2 engages in functional antagonism with epigenetic regulators that promote gene expression. The most notable polycomb antagonist is the trithorax group of proteins, which promotes active transcription and nucleosome remodeling (11). Imbalance between PRC2 and trithorax activity is found in numerous human cancers. Loss-of-function alterations in the SWI/SNF complex, a nucleosome remodeler in the trithorax family, promote oncogenesis through the inappropriate silencing of tumor suppressors by PRC2. Conversely, gain-of-function alterations in SWI/SNF lead to inappropriate activation of transcription through eviction of polycomb from chromatin (12). PRC2 is antagonistic to other regulators of chromatin as well, including NSD2 and CBP, which mediate H3K36 dimethylation (H3K36me2) and H3K27 acetylation (H3K27Ac), respectively (13, 14).
Prior work on PRC2 loss in MPNST identified a loss of H3K27me3 and a reciprocal gain in acetylation of H3K27Ac, which was shown to enhance Ras pathway output (3). Characterization of histone PTMs, however, was limited to Western blotting and IHC, and only dealt with these two modifications. No comprehensive investigation into the epigenetic consequences of PRC2 loss has been undertaken. Given the fundamental role of polycomb in transcriptional repression, and the complex interactions with other epigenetic regulators, we hypothesized that PRC2 loss would lead to widespread changes in the epigenetic landscape that might be therapeutically targetable. To test this, we conducted quantitative profiling of global histone posttranslational modifications (PTM) of tumors using archival formalin-fixed, paraffin-embedded (FFPE) tissue samples, in parallel with quantitative proteomics from the same samples.
Materials and Methods
Tissue samples
All tissue experiments were conducted on deidentified human tissue specimens taken as part of a diagnostic or therapeutic procedure independent from the research study and exempt from a separate consent process. The samples were obtained and analyzed in accordance with protocols approved by the Institutional Review Board at the University of Pennsylvania (Philadelphia, PA) and in accordance with ethical principles outlined in the Belmont Report. Autopsy liver samples were collected and flash-frozen in liquid nitrogen or fixed in formalin and processed according to standard institutional histopathology protocols. MPNST and precursor neurofibroma samples were identified through a database search. A subspecialty bone and soft tissue pathologist (J.B. Wojcik) reviewed selected slides from the initial diagnostic material to confirm the diagnoses for all tumors, and select blocks for histologic and tissue mass spectrometry (MS) analysis. Standard 5 μmol/L tissue sections were used for hematoxylin/eosin staining and IHC analysis in selected cases. For tissue MS analysis, 3-mm tissue cores were taken from representative areas of solid tumor for protein isolation, purification, and characterization.
IHC
IHC on FFPE tissue was done on a Leica Bond-IIITM instrument using the Bond Polymer Refine Detection System (Leica Micro-systems DS9800). Heat-induced epitope retrieval was done for 20 or 30 minutes with ER1 or ER2 solution (Leica Microsystems). Incubation with primary antibody was carried out at the clinically validated dilution for 15 minutes. A 15-minute incubation with the primary antibody was followed by detection of bound antibody using the Bond Polymer Refine Detection system (Leica, DS9800) according to the manufacturer’s protocol. The whole experiment was done at room temperature. Slides are washed three times between each step with bond wash buffer or water.
Antibody conditions:
H3K27me3, Cell Signaling Technologies, AB#9733, 1:200, ER2/30 minutes
H3K27me2, Abcam ab24684, 1:500, ER1/20 minutes
HLA-ABC, EMR8–5, Abcam, ab70328, 1:1200, ER1/20 minutes
HLA-DR, LN3, eBioscience, 14–9956-80, 1:1200, ER1/20 minutes
For interpretation of H3K27me3 IHC for the purpose of classification, only the tumors showing complete loss of nuclear reactivity with retained internal control staining in stromal cells were categorized as MPNSTLOSS. Any staining for H3K27me3 in tumor nuclei led to a classification as MPNSTRET.
Protein isolation from FFPE tissues
Tissue cores were cut into small pieces and resuspended in 10 × volume, based on dry weight of xylene and incubated at 37° C for 10 minutes with gentle agitation. Following centrifugation and removal of xylene, this process was repeated once with xylene, then, sequentially, with 100% ethanol, 95% ethanol, 85%, 70%, 50%, 20%, and then H2O. After the final incubation, the tissue was resuspended in 100 mmol/LTris HCl, pH 8.0, 100 mmol/L NaCl with 4-benzenesulfonyl fluoride hydrochloride (1 mmol/L), microcystin, and sodium butyrate (1 mmol/L) and homogenized with a mortar and pestle, then passed through 18-gauge needle 15×d and then a 21-gauge needle 15 ×. The lysate was sonicated three times for 30 seconds each at room temperature using a 50 watt benchtop sonicator (Fisher, FB50110) at the 20% power setting. One-tenth volume of 20% SDS was added to final concentration of 2%, and the sample was placed on a 95° C heated shaker for a total of 2 hours, with another sonication step at 1 hour. The lysate was centrifuged at 17,000–20,000 × g for 15 minutes. The concentration of solubilized protein was assessed with Pierce BCA Assay Kit (Thermo Fisher Scientific, 23225). For total protein isolation, 100 μg of total protein was precipitated using methanol and chloroform as published previously (15). The precipitated protein was resuspended in 40 μL of 6 mol/Lurea/2 mol/L thiourea in 50 mmol/L ammonium bicarbonate, pH 8.2 with protease inhibitors as above. The solution was diluted six times with 50-mm ammonium bicarbonate, reduced with 10-mm dithiothreitol for 1 hour at room temperature and alkylated with 20-mm iodoacetamide in the dark for 30 minutes at room temperature. Proteins were then digested with trypsin (Promega) at an enzyme-to-substrate ratio of approximately 1:50 for 16 hours at room temperature. After digestion, the samples were concentrated to a volume of approximately 100 μL by lyophilization. Dried samples were resuspended in trifluoroacetic acid (TFA) and desalted as described previously for histone peptides (16). Eluted samples were dried via lyophilization, and resuspended to a concentration of 1 μg/μLin 0.1% formic acid (FA) for MS analysis.
Protein isolation from cell culture
Cell pellets were resuspended in 5 × volume of 6 mol/L urea/2 mol/L thiourea in 50 mmol/L ammonium bicarbonate, pH 8.2 with protease inhibitors as above. The following steps were carried out as with FFPE protein. For decitabine and panobinostat treatment proteomics experiments, we constructed a data-dependent acquisition (DDA) library from high pH reverse-phase fractionation of the pooled peptides from all samples. We loaded the pooled peptides on a Harvard apparatus Micro SpinColumn (catalog no. 74–4601) under acidic conditions, and then washed with 0.1% TFA. Peptides were eluted in a step-wise gradient consisting of 12 separate elutions of increasing concentrations of MeCN (10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 35, 60%) in 100 mmol/L ammonium formate, pH 10.0. Samples from different portions of the gradient (1/7, 2/8, etc.) were pooled for a total of 6 samples. These peptides were lyophilized and desalted as above. Data-independent acquisition (DIA) and DDA library runs were resuspended in buffer A with 1:10 dilution of indexed retention time peptide standards (Biognosys, Ki-3002–1).
Histone isolation from FFPE
Fifty microgram of the total protein from FFPE purification was loaded in 2 adjacent wells of a 12.5% acrylamide SDS PAGE gel. Following the Coomassie dye staining, histone bands were excised from the gel, cut into approximately 2-mm cubes, rinsed with 5× volume of 100 mmol/L NH4HCO3, and placed on a shaker for 5 minutes. Buffer was removed and 100 μL of 100% acetonitrile (MeCN) was added to dehydrate the gel bands. MeCN was removed and 200 μL of a 1:3 propionic anhydride:MeCN mixture was added along with 200 μL of 100 mmol/L ammonium bicarbonate for 10 minutes on a shaker at room temperature. Excess liquid was removed and bands were rinsed with 100 mmol/L ammonium bicarbonate then dehydrated with MeCN as above. This procedure was repeated for three total rounds. Trypsin digestion was carried out by resuspending the gel pieces in 12.5 ng/μL trypsin in 100 mmol/L ammonium bicarbonate and incubating on ice for 45 minutes to allow the gel pieces to absorb the buffer. Additional buffer was then added to cover bands, and the sample was incubated for approximately 16 hours at room temperature. The eluted histones in the liquid were removed and saved. Bands were washed with 50 μL of ammonium bicarbonate for 10 minutes at room temperature on a shaker. The wash was removed and combined with the histones from the prior step. Two additional washes with 50% MeCN, 5% FA in water were added, with one 100% MeCN wash. This pooled liquid was lyophilized. Subsequent in solution propionylation and desalting was carried out as reported previously (16).
Histone isolation from cells
The procedure for histone isolation and preparation for MS analysis has been described elsewhere (16). Histone PTM quantification for PRC2 loss and PRC2-retained cell lines depicted in Fig. 3 were conducted using three replicates from four distinct MPNSTLOSS cell lines (T265, S462, 90–8TL, ST88–14) and six and four replicates, respectively, from an MPNSTRET cell line (STS26T) and a cell line derived from a benign nerve sheath tumor (HEI193). Histone PTMs depicted in Fig. 5 were measured on S462 following NSD2 knockdown and SUZ12 restoration. NSD2 knockdown was carried out using three replicates from two cell lines expressing two distinct NSD2 short hairpin RNA (shRNA) constructs, which were compared with three replicates from a nontarget control. Histones from SUZ12-restored lines were measured using three replicates each for control and SUZ12 vectors. Histone quantifications in Figs. 5G and 7G are provided for a single representative experiment, with each experiment repeated at least twice.
Data acquisition for histone PTMs with nanoLC/MS
Procedures for histone analysis have been described previously for both DDA (17) and DIA (18). Briefly, histone peptides derived from cells or FFPE were resuspended in 10 mL buffer A (0.1% FA in H2O). For cell-derived histones, peptides were resuspended at a concentration of 1 mg/mL in buffer A. Samples were analyzed by using a nanoLC/MS-MS setup. NanoLC was configured with a 75-μm internal diameter × 17-cm Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH) nano-column using an EASY-nLC nanoHPLC (Thermo Fisher Scientific), packed in-house. The high-performance liquid chromatography (HPLC) gradient was as follows: 2% to 28% solvent B (A = 0.1% FA; B = 95% MeCN, 0.1% FA) over 45 minutes, from 28% to 80% solvent B in 5 minutes, 80% B for 10 minutes at a flow-rate of 300 nL/minute. NanoLC was coupled to an Orbitrap Elite or Q-Exactive Mass Spectrometer (Thermo Fisher Scientific). For DDA acquisition, a full-scan MS spectrum (300–1,100 m/z) at a resolution of 60,000 (at 400 m/z) was followed by 10 DDAs of the 10 most abundant ions. The DIAMS-MS method consisted of a full-scan MS spectrum (300–1,100 m/z) at a resolution of 60,000 (at 400 m/z), followed by MS-MS of windows of 50 m/z at a resolution of 15,000. MS-MS data were collected in centroid mode.
Histone MS-MS data analysis
DIA data were searched using EpiProfile 2.0 (19). The peptide relative ratio was calculated using the total area under the extracted ion chromatograms of all peptides with the same amino acid sequence (including all of its modified forms) as 100%. For isobaric peptides, the relative ratio of two isobaric forms was estimated by averaging the ratio for each fragment ion with different mass between the two species. The quantifications of the H3 (27–40aa) peptides were independently validated using peak integration of MS1 parent ions using Skyline (20).
Bottom-up shotgun proteomic data acquisition
For FFPE proteomic samples and samples for the NSD2/SUZ12 cellular experiments, MS data acquisition was carried out as follows. Samples were analyzed by using a nanoLC/MS-MS setup. NanoLC was configured with a 75-μm internal diameter × 20-cm Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH) nano-column using an EASY-nLC nanoHPLC (Thermo Fisher Scientific). The HPLC gradient was 0%–30% solvent B (A = 0.1% FA; B = 95% MeCN, 0.1% FA) over 105 minutes followed by 5 minutes from 30% to 85% B and 10 minutes at isocratic 85% B.The flowrate was set to 300 nL/minute. NanoLC was coupled with an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific). Spray voltage was set at 2.6 kV and capillary temperature was set at 275°C. Full-scan MS spectrum (350–1,200 m/z) was performed in the Orbitrap with a resolution of 120,000 (at 200 m/z) with an automated gain control (AGC) target of 10e6. Fragmentation was performed with higher-energy collisional dissociation (HCD) with normalized collision energy of 27, an AGC target of 5 × 10e4, and a maximum injection time of 150 ms. Fragments were acquired in the Orbitrap with a resolution of 17,500 (at 200 m/z). The top 12 most intense ions above a threshold of 17,000 counts were selected for fragmentation. Only charge states 2 to 4 were included. MS-MS data were collected in centroid mode to optimize file size.
For panobinostat and decitabine treatment data, the same column setup was attached to a Dionex UltiMate 3000 LC system (Thermo Fisher Scientific). The HPLC buffers were as above. The HPLC conditions were 5% buffer B for 2 minutes, 5%–30% buffer B gradient over 72 minutes, 30%–45% buffer B gradient over 18 minutes, and isocratic 85% B for 8 minutes with a 10-minute reequilibration at 2% buffer B. The flowrate was set to 400 nL/minute. NanoLC was coupled with on a QE-HFX (Thermo Fisher Scientific). Spray voltage was set at 2.6 kV and capillary temperature was set at 275°C. Full-scan MS spectrum (350–1,200 m/z) was performed in the Orbitrap with a resolution of 120,000 (at 200 m/z) with an AGC target of 3e6 and a maximum injection time of 50 ms. Fragmentation was performed with HCD and normalized step-collision energy of 25.5, 27, and 30. For DDA, MS-MS spectra from the top 40 most abundant ions (1.3 m/z isolation window; 30-second dynamic exclusion) were collected with a resolution of 30,000 m/z (at 200 m/z), an AGC target of 1e5, and a maximum injection time of 50 ms. For DIA, MS-MS spectra were collected for 47 × 19.1 m/z windows covering the region from 350 to 1,200 m/z.
Proteomics data analysis
All DDA MS-MS spectra were processed through the MaxQuant program (21). Parameters for MS-MS database searching included the following: precursor mass tolerance 4.5 ppm; product mass tolerance 0.5 Da; enzyme trypsin; missed cleavages allowed 2; static modifications carbamidomethyl (C); variable modification oxidation (M); label-free quantification method iBAQ (for protein abundance estimation); match-between runs enabled; data-base used was Homo sapiens (UniProt, May 2015). Peptide-to-spectrum matches and protein FDR were filtered for <0.01. All DIA data were analyzed in Spectronaut v11.0.15038.4.29119 software (Biognosys AB) under default search and quantification parameters. DDA and DIA protein quantifications were log2-trans-formed and normalized by the average of the protein abundance within each sample. Protein abundances for FFPE samples were the average of normalized values from technical triplicates. Only the proteins identified by a minimum of two peptides were included for downstream analysis. Statistical differences were estimated using the t test (P < 0.05, two-tailed, either homo- or heteroscedastic according to the significance of the F test). For NSD2 knockdown and SUZ12 restoration experiments, log2 ratios of protein abundance from experimental/control are taken from average of three replicates. Pearson correlation and P value for correlation are shown. Heatmaps for Fig. 4A was generated with Perseus (22). Figures 4D and 7, and Supplementary Fig. S2 were generated in Qlucore Omics Explorer 3.3 software (Qlucore). Figure 4D contains all measured values for the listed protein groups. Data in Fig. 7 and Supplementary Fig. S2 were filtered for differential expression at a nominal P value of 0.05.
Cell culture
Patient-derived MPNST cell lines were generously provided by Karen Cichowski (Dana-Farber Cancer Center, Harvard Medical School, Cambridge, MA; 90–8TL, S462) and Jeffrey Field (Perelman School of Medicine, University of Pennsylvania; STS26T, ST88–14, T265, HEI193). Cell lines were not independently authenticated. All cell lines were maintained in DMEM (Corning10013CM) supplemented with 10% FBS (GE, HyClone, SH30071.03). All lines used in analysis tested negative for Mycoplasma contamination.
Vectors
NSD2 shRNA lentiviral vectors were generously provided by Irfan Asangani (Perelman School of Medicine, University of Pennsylvania). The vectors and their uses were described previously (23). NSD2 knockdown was confirmed via Western blot analysis. pCMVHA SUZ12 was a gift from Kristian Helin (Memorial Sloan Kettering Cancer Center, New York, NY; Addgene plasmid # 24232) and has been described previously (24). NSD2KD cells for MPNSTLOSS lines were harvested 10 days after transduction. SUZ12-transfected and control cell lines were harvested 7 days after transfection. The K36M expression vector was a gift from David Allis (The Rockefeller University, New York, NY). This vector contains the previously described H3.3K36M sequence (25) subcloned into a pTetOne doxycycline-inducible expression system (Takara Bio). Transduced cells were selected with 1 week of growth in 1.5 mg/mLpuromycin. Doxycyline induction (100 ng/mL) was carried out for 7 days prior to the start of the cell growth experiment and continued through the duration. Histones were isolated from cells following 10 days of induction.
Cell growth and viability assays
For cell growth, 2,000 cells per well were seeded in a 96-well plate. After 24 hours, cell density was measured using RealTime-Glo MT Cell Viability Assay (Promega). Cells grew for an additional 10 days with media changes on days 3, 6, and 8. Cell density was then measured again, and the values normalized to the initial readings. Each cell line was measured in triplicate. For decitabine treatment, 4,000 cells were plated and initial cell measurements were carried out in the same fashion. Cells were treated with decitabine or DMSO vehicle control each day for 3 days with media changes daily. After 3 days, the cells were transitioned to media without drug, and allowed to grow for additional 7 days, with media changed every 2 days. Cell density was measured at the end of this period again, and readings were normalized to the initial, predrug reading to plot log2 cell density. Each treatment was carried out in triplicate. For panobinostat treatment, approximately 10,000 cells were seeded in a 96-well plate. After 24 hours, cells were treated with panobinostat or DMSO control. Cell density measurements were carried out 72 hours after treatment in the same manner as above. Cell growth response curves were plotted using Graphpad Prism (growth assays, panobinostat) or Excel (decitabine). Panobinostat response was fit using nonlinear regression of variable slope (four parameters) and least squares (ordinary) fitting. The P value of this comparison is stated in plots. Error bars in all graphs are ± SD.
Western blot analysis
Western blotting was carried out according to standard protocols with the following antibodies:
Primary: GAPDH (Sigma, G8795); SUZ12 (Cell Signaling Technology, 3737), 1:2,000 dilution; MMSET/WHSC1 (Active Motif, 39878), 1:2,000 dilution. Secondary: anti-mouse (Jackson ImmunoResearch, 115-035-003) 1:10,000 dilution; anti-rabbit (Pierce, 31463), 1:10,000 dilution.
Bands were visualized using ECL Prime detection reagent (GE Healthcare, RPN2232).
IP-MS
Standard immunoprecipitation (IP) was carried out using a custom rabbit polyclonal antibody raised against EZH2 (described in ref. 26) or an isotype control antibody to assess background (anti-rabbit, above). This was incubated with 200 μg of total lysate from control or SUZ12 add back cell lines. Antibody complexes were isolated using DynaMag protein G beads (Thermo Fisher Scientific, 10003D). On-bead derivatization and digestion were followed by C18 stage tipping, LC/MS-MS data acquisition, and downstream analysis as with the DDA protocol described above for the FFPE tumor proteome runs.
qRT-PCR
Total RNA extraction from cells and tissue was performed with the RNeasy mini Kit according to manufacturer’s protocol (74104, Qiagen). RNA was transcribed to cDNA using random hexamers as primers and Superscript III Reverse Transcriptase Fist-strand Synthesis SuperMix (Invitrogen at#18080400). Expression analysis was performed using a 2 × SyBr Green PCR Mix (Applied Biosystems, catalog no. 4367659) on a 7900HT Fast Real-Time PCR machine (Thermo Fisher Scientific). Expression levels were calculated using the 2−ΔΔCt method with RPLP0 as a housekeeping control. Primer sequences for the specified genes were based on previously published primers, with sequences included in Table 1 (27).
Table 1.
Primer sequences for the specified genes based on previously published primers, with sequences
| Forward | Reverse | |
|---|---|---|
| RPLPO | CAGACAGACACTGGCAACA | ACATCTCCCCCTTCTCCTT |
| ERVL | ATATCCTGCCTGGATGGGGT | GAGCTTCTTAGTCCTCCTGTGT |
| MER21C | GGAGCTTCCTGATTGGCAGA | ATGTAGGGTGGCAAGCACTG |
| MER4D | CCCTAAAGAGGCAGGACACC | TCAAGCAATCGTCAACCAGA |
| MER57B1 | CCTCCTGAGCCAGAGTAGGT | ACCAGTCTGGCTGTTTCTGT |
| MLT1B | TGCCTGTCTCCAAACACAGT | TACGGGCTGAGCTTGAGTTG |
| MLT1C49 | TATTGCCGTACTGTGGGCTG | TGGAACAGAGCCCTTCCTTG |
| MLT1C627 | TGTGTCCTCCCCCTTCTCTT | GCCTGTGGATGTGCCCTTAT |
Methylation array analysis
We obtained raw idat files from the authors of a previously published methylation analysis of human nerve sheath tumor samples conducted using Infinium 450k Methylation Arrays (Illumina; ref. 28). Sample annotations containing the IHC staining pattern for H3K27me3 and the diagnosis were used to subdivide the tumors into cohorts of MPNST with loss of H3K27me3 (19 tumors) and with retained H3K27me3 (7 samples). We conducted global and differential methylation analysis using default settings in the R package RNBeads (Bioconductor release 3.7; ref. 29). P values on the site level were computed using the limma method. Hierarchical linear models from the limma package were employed and fitted using an empirical Bayes approach on the derived M values. For the assessment of differential methylation across all sites, mean β values for MPNSTLOSS and MPNSTRET were numerically ordered and plotted against the position in the list/total number of sites (cumulative distribution). For the assessment of differential methylation at promoters, CpG islands, and genes, we compared mean β values for every position between MPNSTLOSS/MPNSTRET. P values are from unpaired, two-tailed t tests. Boxplots parameters follow the Tukey method.
Whole-genome bisulfite sequencing
Whole-genome sequencing libraries were generated from 700 to 1,000 ng of genomic DNA spiked with 0.1% (w/w) unmethylated λ DNA (Promega) previously fragmented to 300–400 bp peak sizes using the Covaris Focused-ultrasonicator E210. Fragment size was controlled on a Bioanalyzer DNA 1000 Chip (Agilent) and the KAPA High-throughput Library Preparation Kit (KAPA Biosystems) was applied. End repair of the generated double-stranded DNA (dsDNA) with 3′ or 5′ overhangs, adenylation of 3′ ends, adaptor ligation, and clean-up steps was carried out asper KAPA Biosystems’ recommendations. The cleaned-up ligation product was then analyzed on a Bioanalyzer High-sensitivity DNA Chip (Agilent) and quantified by PicoGreen (Life Technologies). Samples were then bisulfite converted using the Epitect Fast DNA Bisulfite Kit (Qiagen) according to the manufacturer’s protocol. Bisulfite-converted DNA was quantified using OliGreen (Life Technologies) and, based on quantity, amplified by nine to 12 cycles of PCR using the Kapa Hifi Uracil + DNA Polymerase (KAPA Biosystems), according to the manufacturer’s protocol. The amplified libraries were purified using Ampure XP Beads (Beckman Coulter), validated on Bioanalyzer High-sensitivity DNA Chips, and quantified by PicoGreen. Sequencing of the whole-genome bisulfite sequencing (WGBS) libraries was performed on the Illumina HiSeqX system using 150-bp paired-end sequencing.
Analysis of WGBS data
Raw reads were aligned to human genome build (UCSC hg19) using Burrows-Wheeler Aligner (BWA; version 0.6.1; ref. 30) after converting the reference genome to bisulfite mode. Low-quality sequences at the 3′ ends were trimmed. For the overlapping paired-end reads, we clipped the 3′ end of one of them to avoid double counting on both forward and reverse strands. After alignment, we filtered duplicated or poorly mapping reads (>2% mismatches or aberrant insert size). To call methylation of individual CpGs, we used Samtools (version 0.1.18; ref. 31) mpileup. We filtered the CpGs with less than 5× coverage, which were overlapping with SNPs from Single Nucleotide Polymorphism Database (dbSNP; build 137) or were located within the ENCODEDACblacklisted regions or Duke-excluded regions (32). For visualization in Integrative Genomics Viewer (IGV), the coordinates of individual CpGs were artificially extended to the midpoint between their neighbors, as described previously (33). To compare DNA methylation of MPNSTLOSS and MPNSTRET cell lines, we selected CpG in autosomal chromosomes and computed the mean β-value in genomic compartments such as CpG islands and intergenic and exonic regions of high (top expression quartile) and low or unexpressed genes (bottom quartile).
RNA sequencing
Total RNA was extracted from cell pellets using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to instructions from the manufacturer. Two replicates of independent culture samples were obtained for each cell line. Library preparation was performed with rRNA depletion according to instructions from the manufacturer (Epicentre) to achieve greater coverage of mRNA and other long noncoding transcripts. Paired-end sequencing (125 bp) was performed on the Illumina HiSeq 4000 platform.
Analysis of RNA sequencing data
Raw sequencing reads were aligned using STAR (34) version 2.5.3a to the reference genomes mentioned above. Gene-level read counts were obtained from featureCounts (35) v1.5.3 using the RefSeq gene annotation and normalized using the Reads per Kilobase per Million mapped metric for each gene to quantify relative expression levels.
Analysis of microarray data
GSE52777 were imported from Gene Expression Omnibus (GEO) and analyzed using Qlucore Omics Explorer 3.3 software (Qlucore). Supplementary Fig. S2 depicts differentially expressed genes from the combined IFN signature gene set described above, between 90–8TL control cells and 90–8TL cells with SUZ12 over expression, filtered at a nominal P value of 0.05.
Gene ontology and gene set enrichment analysis
Gene ontology (GO) analysis was conducted using the GOrilla server (cbl-gorilla.cs.technion.ac.il/). All GO analyses relied on the ranked list of differentially expressed proteins (log2 ratio of protein abundance) or differential DNA methylation (log2 ratio of mean β values) for MPNSTLOSS versus MPNSTRET or treatment versus control. The lone exception to this was the NSD2/SUZ12 experiment, where GO was conducted on a ranked list based on the sum of the ranks in the individual experimental manipulations (i.e., rank NSD2KD+ rank SUZ12ADD). Each ranked list was queried using the single list function (i.e., no separate background list of genes). Gene set enrichment analysis (GSEA) (36) was carried out using GSEA v 3.0 with the GSEA preranked tool using the same ranked lists as for GO analysis. Gene sets are described at MSigDB. For PRC2 targets, BENPORATH_PRC2_TARGETS is derived from ref. 37. The YAP-TAZ JQ1 responsive signature was described in ref. 38. The collection of HALLMARK gene sets and KEGG gene sets were searched. The IFN/MHC genes used for the heatmap in 7C represents the union of HALLMARK_INTERFER-ON_ALPHA, HALLMARK_INTERFERON_GAMMA, and KEG-G_ANTIGEN_PROCESSING_AND_PRESENTATION gene sets from MSigDB. Nominal P values are shown.
Data availability
All raw data files for proteomics experiments are available on https://chorusproject.org at the project no.1539.
RNA sequencing (RNA-seq) and WGBS data are available at the following URL: https://datahub-7woy4ejm.udes.genap.ca/.
Results
Global analysis of histone PTMs reveals extensive changes associated with PRC2 loss
The general workflow for histone and proteomic analysis from FFPE human tissue samples is depicted in Fig. 1. To identify MPNST with PRC2 loss, we screened MPNST cases from our pathology archives using IHC for H3K27me3 as a surrogate for polycomb mutation status, because previous work suggested that this method is more sensitive than targeted sequencing approaches, which may miss complex structural alterations (4). On the basis of this screening, we selected nine MPNST for further analysis, including five with a global H3K27me3 loss (MPNSTLOSS) and four with retained PRC2 function (MPNSTRET). Prior to conducting the analysis on human MPNsT samples, we validated histone and proteome extraction procedures using fresh paired and fixed human autopsy liver specimens. Measurements of proteins and histone PTMs quantified from fresh and FFPE tissues showed excellent agreement (Supplementary Fig. S1).
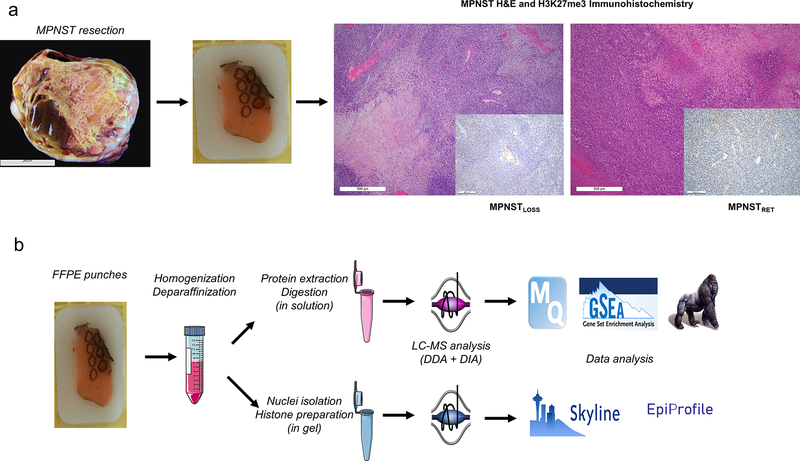
Figure 1.
Workflow for parallel proteomic and histone analysis of MPNSTLOSS and MPNSTRET. A, MPNST are resected (gross image of representative tumor at right) and processed according to standard pathology procedures. Representative sections from tumor blocks from pathology archives were reviewed on hematoxylin and eosin (H&E) histologic sections to identify the regions of interest. IHC (inset) on adjacent sections allowed for separation of MPNST tumors into MPNSTLOSS (left) and MPNSTRET (right) on the basis of H3K27me3 staining. MPNSTLOSS were defined by global loss of H3K27me3 nuclear staining, with retained internal control staining in nontumor tissue (blood vessel, center of IHC inset). MPNSTRET were defined by any degree of retained staining in tumor nuclei. B, From blocks identified in A, tissue was isolated from representative cores of FFPE tissues, and total protein was extracted. A portion of the total protein was processed in solution for MS and proteomic analysis using standard processing and analysis software (top), and a portion was sampled for SDS-PAGE, from which the histone bands were excised for in-gel digestion, derivatization, and histone PTM analysis using distinct analysis software (bottom).
Principal component analysis of the global histone PTM profiles separated tumors with PRC2 loss from those with retained activity along the first component, with one exception (Fig. 2A). Interestingly, this case was scored as retained by IHC, but exhibited mosaic staining, with patchy, variable reactivity for H3K27me3. In three cases, a histologically identifiable precursor benign neurofibroma was present adjacent to a high-grade MPNST with PRC2 loss. The precursor lesions were analyzed in parallel with the remainder of the tumors, and segregated with one of the cases with MPNSTRET, consistent with prior data showing retention of H3K27me3 in these tumors. They were included with MPNSTRET as PRC2RET for all pairwise comparisons.
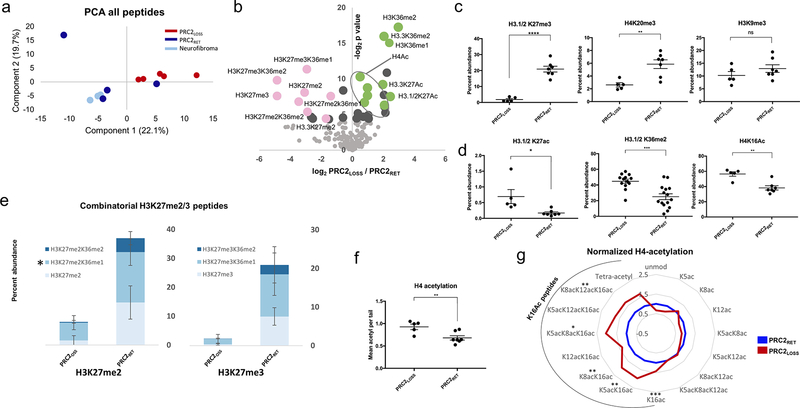
Figure 2.
Global analysis of histone PTMs reveals extensive changes associated with PRC2 loss. A, Principal component analysis (PCA) of global histone PTMs in MPNST with PRC2 loss (PRC2LOSS; 5 replicates; red), MPNST with retained PRC2 function (PRC2RET; 4 replicates; blue), and benign neurofibromas (3 replicates; light blue). B, Volcano plot illustrating differential histone PTMs in all samples as a function of PRC2 status. The log2-fold change in abundance (PRC2LOSS/PRC2RET) is plotted along the x-axis and the −log2 P value is plotted on the y-axis (P = 0.05 corresponds to 4.32). Individual modifications are highlighted with labels. Pink dots, peptides with PRC2-mediated modifications; green dots, modifications typically associated with active transcription. Quantitative comparison of individual histone PTMs between PRC2LOSS and PRC2RET samples, marks associated with transcriptional repression (C) or transcriptional activation (D). E, Stacked bar plots depicting combinatorial modifications for the peptides containing H3K27me2 (left) and H3K27me3 (right). Darker shades of blue indicate higher levels of coexisting methylation at H3K36. PRC2RET and PRC2LOSS samples are depicted with separate stacked bars. For the 27–40 peptide, the quantification of K27me2/K36me1 also includes K36me3 (*), because these peptides were not reliably distinguished in this dataset. Bar and dot plot of total H4K16Ac levels (F) and radar plot depicting all histone H4 acetylation states (G), normalized to levels in PRC2RET. The black arc highlights peptides containing H4K16Ac, PRC2LOSS (red), and PRC2RET (blue). For all panels, error bars are ± SD. ns, no significant change, two-tailed t test; *, P < 0.05;**, P < 0.01; ***·, P < 0.0001.
Overall, repressive histone modifications, most notably those containing H3K27me2 or H3K27me3, were markedly reduced, whereas modifications associated with active transcription were increased (Fig. 2B). Consistent with prior IHC data, there was a global loss of H3K27me3. This was true of both H3.1/2 and H3.3 variants (Fig. 2B; Supplementary Fig. S2A). There was also a significant reduction in H4K20me3, another marker associated with transcriptional silencing (39). Levels of H3K9me3, a modification associated with constitutive silencing in heterochromatin, were not significantly changed from baseline, indicating that this modification is unlikely to functionally replace the silencing activity of H3K27 methylation in the absence of polycomb activity (Fig. 2C). Modifications associated with active transcription displayed the opposite trend. H3K27Ac was significantly increased on both H3.1/2 and H3.3 variants (Fig. 2D; Supplementary Fig. S2B). Similarly, H3K36me2 levels were elevated following PRC2 loss, as was H4K16Ac, a modification that promotes open chromatin by decreasing internucleosome interactions (Fig. 2D; refs. 40, 41).
MS analysis allows for the simultaneous profiling of tandem modifications on the same histone tail. Histone H3 has a tryptic peptide containing modification sites at both K27 and K36. This allowed us to see that not only were K27me2/3 levels globally reduced, but this reduction included histone tails bearing H3K27me2/3 in combination with methylation at H3K36 (Fig. 2E). Because K36 methylation is typically associated with active transcription, these histone tails exhibited “bivalence” with respect to K27 and K36 methylation. In MPNSTLOSS, this bivalence is resolved in favor of unopposed K36 methylation. MS also allowed us to identify changes in acetylation present on histone H4, which contains multiple acetylation sites (K5, K8, K12, K16). There was a shift toward increased acetylation of H4, which is associated with greater transcriptional activity (42). This was entirely driven by increases in K16Ac-containing histone tails (Fig. 2F).
MPNST cell lines validate the changes observed in human tumors and allow for more precise quantification of changes in H3 PTMs
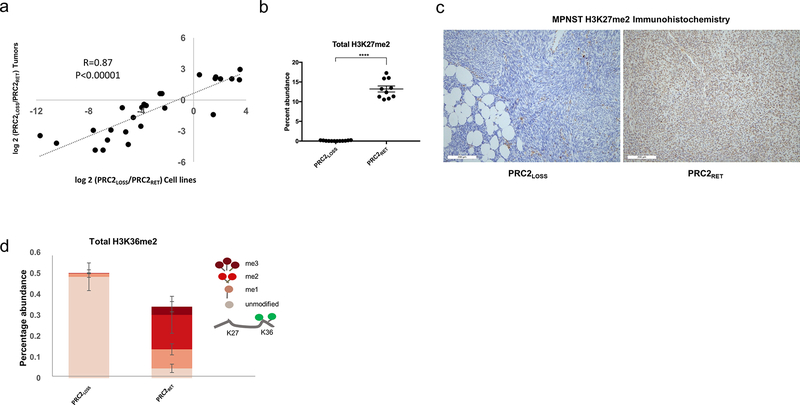
The DDA strategy utilized on the tumor histone samples was unable to reliably discriminate between the isobaric and coeluting H3K27me2/H3K36me1 and H3K36me3. Consequently, we could not accurately determine the degree of H3K27me2 loss using our tumor data. To more precisely quantify global changes resulting from PRC2 loss in MPNST, and to validate that MPNST cell lines adequately recapitulate the changes seen in human tumors, we conducted MS analysis of histones from four MPNSTLOSS cell lines and compared them with histones from an MPNSTRET cell line and a line derived from benign nerve sheath cells.
The consequences of PRC2 loss in MPNST lines were in excellent agreement with those seen in the tumors, with cell lines showing an increased magnitude of changes, likely reflecting reduced biological variability of cell lines compared with tumor samples (Fig. 3A). Because the cell line data was gathered using a DIA, it was possible to reliably distinguish between H3K27me2/H3K36me1 and H3K36me3. This allowed us to confirm complete, global loss of H3K27me2 in PRC2LOSS cell lines (Fig. 3B). We validated H3K27me2 loss on MPNST tumors using IHC for H3K27me2 (Fig. 3C). The DIA approach also enabled a more complete assessment of the combinatorial modifications on the histone H3 K27–40 peptide. As with the tumors, there was an increase in global H3K36me2 levels, and a loss of “bivalent” K27-K36 methylations. In line with retained PRC2 function, the majority of K36me2 is found on histone tails containing either an H3K27me2 or H3K27me3 modification, and in PRC2LOSS tumors, nearly all H3K36me2 is present on histone tails containing no modification at K27 (Fig. 3D).
Figure 3.
MPNST cell lines validate the changes observed in human tumors and allow for more precise quantification of changes is H3 PTMs. A, Correlation between changes in histone PTMs as a function of PRC2 status in cell lines (x-axis) and human tumors (y-axis) for peptides with significant changes in either comparison. P values for Pearson correlation are shown. B, Quantitative comparison of H3K27me2 levels between PRC2LOSS (12 replicates) and PRC2RET (10 replicates) samples. Two-tailed t test; ****, P< 0.0001. Using DIA acquisition, H3K27me2/1 and H3K36me3 could be distinguished, enabling more precise quantification of H3K27me2 levels. Error bars, ± SD. C, IHC for histone H3K27me2 in representative MPNST cases: PRC2LOSS (left) and PRC2RET (right). D, Quantitative comparison of H3K36me2 levels between PRC2LOSS (left) and PRC2RET (right) samples. The peptide quantifications are represented as stacked bar plots, with each level of methylation colored according to the schematic depicted at right, where higher levels of methylation at K27 lead to progressively darker red colors. Unpaired t test, P < 0.0001 for pairwise peptide comparisons at the same level of methylation. Error bars, ± SD.
Taken together, the tumor and cell line data demonstrate that PRC2LOSS tumors are characterized by the loss of H3K27me3 and the even more broadly distributed H3K27me2 repressive mark, and a global shift toward markers of open, active chromatin without a compensatory gain in other markers of silent chromatin (H3K9me3, H4K20me3). This includes an increase in multiple histone H3 and H4 acetyl marks including the canonical enhancer mark, H3K27Ac, and loss of “bivalent” H3K27/K36 methylations, which are resolved in favor of elevated levels of unopposed H3K36me2.
Global proteomic analysis reveals PRC2 loss enhances tumor growth pathways and promotes immune evasion
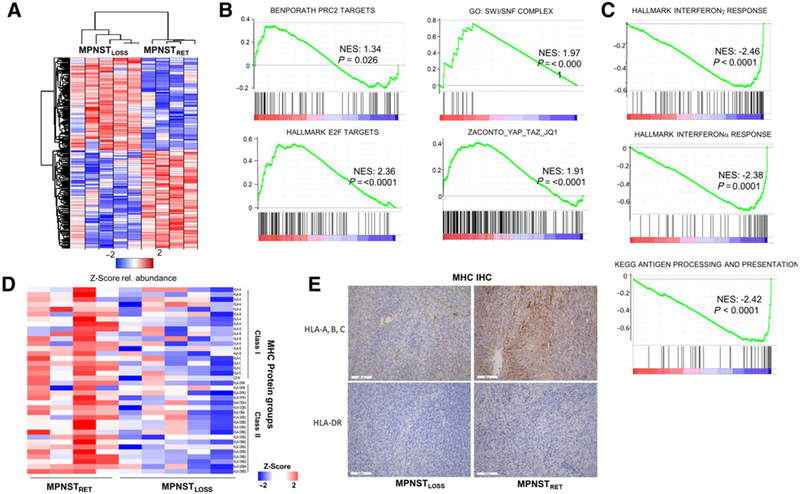
In parallel with our analysis of the global histone PTM landscape of MPNsT, we conducted label-free proteomics analysis to assess the consequences of PRC2 loss at the level of the proteome, strictly in the MPNST samples. Across the entire dataset, total 5,632 proteins were quantified by at least two peptides (Supplementary Table S1). Hierarchical clustering of the differentially expressed proteins (P < 0.05) between MPNSTLOSS and MPNSTRET revealed distinct sets of proteins that were both upregulated and, surprisingly given the role of PRC2 as a transcriptional repressor, downregulated (Fig. 4A). Similar to prior studies, we saw the upregulation of polycomb targets following PRC2 loss (4), consistent with derepression of these genes (Fig. 4B). Prior work identified an upregulation of Ras transcriptional program (3). In the proteome dataset, we did not observe a specific activation of the Ras pathway. Rather, we observed a generalized increase in proteins associated with growth and division, for example, E2F targets and other markers of cell-cycle progression (Fig. 4B; Supplementary Table S2). We also observed an increase in proteins associated with nucleosome remodeling and transcriptional activation. This included increased levels of all 11 of the detected proteins in the SWI/SNF complex (Fig. 4B), and upregulation of a recently characterized YAP_TAZ target gene set linked to transcriptional addiction and sensitivity to JQ1 (38), consistent both with the upregulation of the Yap pathway in MPNST (43) and the sensitivity of MPNSTLOSS to JQ1 (3). These data, coupled with the histone PTM data, suggest that a global shift toward more open, active chromatin enhances growth and division pathways.
Figure 4.
Global proteomic analysis reveals PRC2 loss enhances tumor growth pathways and promotes immune evasion. A, Heatmap depicting Z-score for proteomic data, for proteins with P < 0.05 between conditions. B and C, GSEA for selected annotated gene sets run using the GSEA preranked tool, with all identified proteins ordered by differential expression between MPNSTLOSS versus MPNSTRET. Proteins showing increased expression in MPNSTLOSS have positive enrichment values and are shown in B. Those with decreased expression in MPNSTLOSS have negative enrichment values and are shown in C. NES, normalized enrichment score. D, Heatmap depicting Z-scores for all MHC protein groups identified across proteomic dataset, ordered with MPNSTRET (left) and MPNSTLOSS (right). Class I MHC proteins are at the top and class I are at the bottom. E, IHC staining for MHC class I (top) and MHC class II (bottom) from representative sections of MPNSTLOSS (left) and MPNSTLOSS (right). In MPNSTLOSS, HLA-A, B, C staining is largely confined to blood vessels, whereas in MPNSTRET, membranous staining is visible in tumor cells as well. MHC class II, represented with HLA-DR staining, is essentially absent in MPNSTLOSS, with only rare, positive-staining antigen-presenting cells. In MPNSTRET, these cells are more numerous. The IHC findings agree with the quantitative values seen across the dataset in the MS analysis.
Perhaps, more surprisingly, several pathways were strongly downregulated in MPNST with PRC2 loss (Fig. 4C; Supplementary Table S3). GSEA revealed significant downregulation of IFN signaling and antigen presentation following PRC2 loss. The relative reduction of MHC expression was uniform across the proteomic dataset, and included both MHC class I as well as MHC class II, presumably representing both decreased MHC I expression by the tumor cells as well as a decreased infiltration of MPNSTLOSS tumors by antigen-presenting cells (Fig. 4D). We confirmed the MHC downregulation observed in the proteomic data by staining of whole-mount tissue sections from MPNSTLOSS and MPNSTRET with antibodies detecting MHC class I and class II expression (Fig. 4E). MHC I expression was reduced in MPNSTLOSS tumor cells, which also showed decreased levels of infiltrating inflammatory cells expressing MHC II.
Together, the proteomic data suggest that PRC2 loss contributes to two key hallmarks of cancer. It amplifies growth and division pathways and promotes immune evasion through the down-regulation of IFN signature genes and MHC expression.
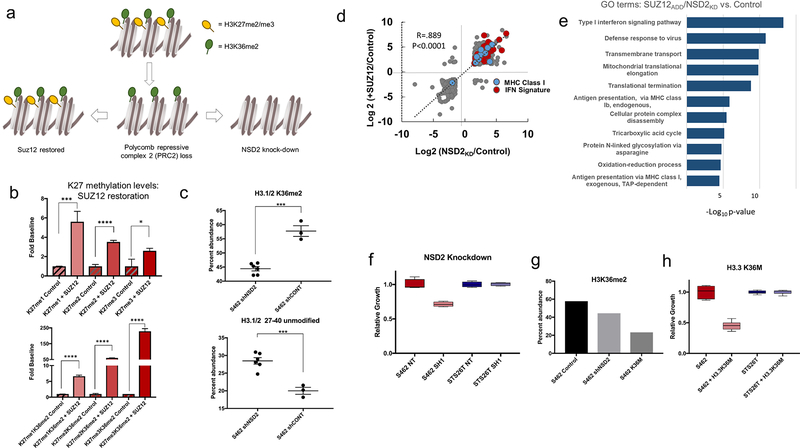
NSD2 knockdown and PRC2 restoration restore IFN pathway protein expression and antigen presentation
To directly link the changes in histone PTMs to the observed proteomic changes in human tumors, and to assess whether histone PTMs could be directly targeted as a therapeutic strategy, we restored PRC2 function in MPNSTLOSS cell lines by adding back SUZ12 and depleted NSD2 in the MPNSTRET lines (Fig. 5A). H3K36 methylation was chosen as a target for several reasons. It was among the most robustly upregulated modifications in MPNSTLOSS, and, as was noted previously, it was no longer opposed by H3K27 methylation present in tandem on the same histone tail. In addition, prior work identified NSD2 as a potentiator of Ras-driven malignancies (44). Finally, prior sequencing studies of MPNST identified putative deleterious alterations or reduced expression of KDM2A or KDM2B, the lysine demethy-lases responsible for the removal of methylation from K36 (45). This suggests that imbalance between K27 methylation and K36 methylation may be selected for in MPNST.
Figure 5.
NSD2 knockdown and PRC2 restoration restore IFN pathway protein expression and antigen presentation. A, Experimental design. In MPNSTRET, most H3K36me2 (green circles) is present in tandem with H3K27me2/3 (yellow circles). PRC2 loss increases unopposed K36me2 (bottom row, center). SUZ12 restores bivalence (bottom row, left) and NSD2 knockdown increases unmethylated peptides. B, H3K27 methylation states in control (3 replicates; striped) and SUZ12-expressing cells (+SUZ12; 3 replicates; solid) are shown, with mono-, di-, and trimethylation from left to right. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Top, total H3K27 methylation values; bottom, tandem modifications. Error bars, ± SD. C, H3K36me2 levels (top) and unmodified peptide levels (bottom) following NSD2 shRNA knockdown (shNSD2; 6 replicates) or control shRNA (shCONT; 3 replicates). ***, P < 0.001. Error bars, ± SD. D, Correlation between log2 ratios of protein abundance following NSD2 knockdown (x-axis) versus SUZ12 restoration (y-axis) for proteins with a log2-fold change >1.5 or <1.5. MHC I (blue) and IFN pathway proteins (red) are highlighted. E, GO enrichment: −log10 P values for GO term enrichment for GO process terms based on combined ranked list of protein abundance changes following NSD2 knockdown and SUZ12 restoration. F, Cell growth following NSD2 knockdown (light shade; 3 replicates) compared with control (dark shade; 3 replicates) in MPNSTLOSS (left, red) and MPNSTRET cell lines (right, blue). G, Levels of H3K36me2 from a single representative experiment in control (left, black), NSD2 knockdown (shNSD2; middle, light gray), and K36M-expressing (right, dark gray) cells. H, Cell growth following K36M induction (light shade; 3 replicates) relative to control values (dark shade; 3 replicates) in MPNSTLOSS (left, red) and MPNSTRET cell lines (right, blue).
We confirmed successful restoration of SUZ12 expression by Western blotting (Supplementary Fig. S3A) and demonstrated, by IP of EZH2 followed by MS, that SUZ12 reestablished the core PRC2 complex (Supplementary Fig. S3B). The accessory subunits RBBP4 and RBBP7 were only identified in immunoprecipitates from SUZ12-expressing cell lines, consistent with PRC2 complex structural data (46).
Short-term restoration of PRC2 activity significantly increased the levels of all three methylation states at K27 relative to baseline, and began to restore “bivalence” to K36me2 peptides (Fig. 5B). NSD2 knockdown, confirmed by Western blotting (Supplementary Fig. S3C), significantly decreased H3K36me2 levels, and resulted in a reciprocal gain in unmodified forms of the H3 K27–40 peptide (Fig. 5C). At the proteome level, these two manipulations had similar effects. Both restored the IFN pathway expression and led to an increase in MHC class I expression (Fig. 5D). By GO analysis, the pathways downregulated in MPNSTLOSS tumor samples were among the most robustly upregulated following targeting of histone PTMs (Fig. 5E; Supplementary Table S4).
Because IFN pathway genes may be induced as an artifact of introduction of exogenous DNA or expression of shRNA, it is important to note that neither the control vector with a short N-terminal SUZ12 peptide insert nor the nontarget shRNA induced IFN pathway genes. Moreover, analysis of the transcriptional data from a previous study using distinct vectors likewise showed robust IFN pathway upregulation following SUZ12 restoration as compared with control vectors (Supplementary Fig. S4A), and the same shRNA constructs, used in a prior study, did not induce an IFN response in prostate cancer lines (Supplementary Fig. S4B).
Prior work demonstrated that restoration of SUZ12 decreased cell growth (3). To determine whether targeting H3K36 dimethylation would similarly slow proliferation, we compared the effects of NSD2 knockdown in an MPNSTLOSS line with NSD2 knockdown in MPNSTRET. NSD2 knockdown decreased cell proliferation in MPNSTLOSS, but had no effect in MPNSTRET (Fig. 5F). To confirm that the observed decrease in cell growth was due to H3K36me2 loss, we transduced the same MPNSTLOSS line with an inducible vector expressing an H3.3K36M mutant, originally identified in chondroblastoma, which markedly reduces global levels of K36me2 (25). Induction was associated with an even more marked reduction in K36me2 levels (Fig. 5G) and an even greater reduction in cell growth (Fig. 5H).
Together, these results suggest that global changes in histone PTMs can be targeted to slow cell growth and restore immune surveillance through the activation of cell-intrinsic IFN signaling and MHC expression.
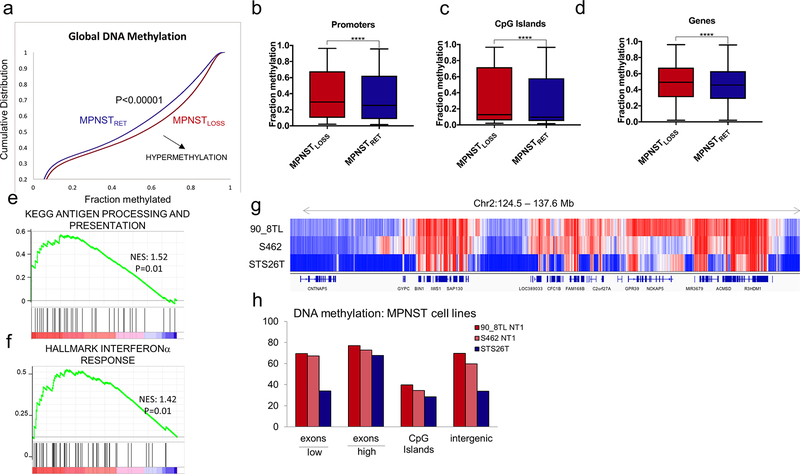
MPNSTLOSS exhibit global DNA hypermethylation
It was somewhat paradoxical that loss of a transcriptional repressor led to decreased expression of so many proteins. We reasoned that perhaps a countervailing epigenetic mechanism contributed to the observed changes in protein expression. As noted previously, no repressive histone modifications showed a compensatory increase. Thus, we turned to DNA methylation. We took advantage of a previously published analysis of DNA methylation arrays in nerve sheath tumors (28). Critically, the MPNSTs within this dataset were also annotated for H3K27me3 status by IHC, enabling a direct comparison between MPNSTLOSS and MPNSTRET.
MPNSTLOSS and MPNSTRET had distinction methylation profiles. MPNSTLOSS exhibited increased levels of DNA methylation genome-wide (Fig. 6A). This was not confined to any particular region. Rather, methylation levels were elevated at promoters, in CpG islands, and within gene bodies (Fig. 6B–D). To determine whether promoter hypermethylation was observed in MPNSTLOSS at the same gene sets that exhibited decreased protein expression, we queried the rank-ordered list of differentially methylated promoters against the antigen presentation and IFN signature gene sets upregulated in the proteome data. Both antigen presentation and IFNα gene sets exhibited significant enrichment. Although the IFNγ gene set did exhibit hypermethylation, it did not meet the threshold for significant enrichment above levels seen across all genes (P = 0.12).
Figure 6.
MPNSTLOSS exhibit global DNA hypermethylation. A, Methylation array data (62) for MPNST tumors analyzed as a function of H3K27me3 status (loss, red; retained, blue). Cumulative distribution function for methylation B-values across all probes in the dataset. Shift of the curve toward the lower right indicates hypermethylation in MPNSTLOSS (P< 0.00001). B-D, Tukey boxplot of methylation B-values for probes within the specified genomic regions for MPNSTLOSS versus MPNSTRET. ****, P< 0.0001, two-tailed t test, all comparisons. E and F, GSEA for rank-ordered list of differentially methylated promoters (log2 MPNSTLOSS/MPNSTRET) queried against antigen presentation (E) and IFN response (F) gene sets previously shown to have decreased protein abundance in MPNSTLOSS. Nominal P values are shown. G and H, WGBS DNA methylation comparison of MPNSTLOSS and MPNSTRET cell lines. G, Visualization of DNA methylation of a representative genomic region containing expressed and unexpressed genes and intergenic regions illustrating genome-wide trends summarized in H. The methylation differences between MPNSTLOSS (90_8TL and S462) and MPNSTRET (STS26T) are least prominent in the expressed genes (designated as “high” with the bar below the x-axis) and CpG islands, and highest within silenced genes (designated as “low” with the bar below the x-axis) and intergenic regions, where global loss of K27me3 leads to replacement by DNA methylation.
To validate both the tumor methylation array data and gain greater resolution on differential methylation between MPNSTLOSS and MPNSTRET, we conducted whole-genome bisulfite sequencing (WGBS) on two separate MPNSTLOSS cell lines (S462 and 90–8TL) and an MPNSTRET line (STS26T). The global methylation trends were consistent with those seen in the tumors (Fig. 6F and G). Relative to MPNSTRET, MPNSTLOSS showed global hypermethylation, encompassing both coding and noncoding regions. Comparison between WGBS and RNA-seq data demonstrated that the methylation differences were most pronounced in silenced genes and intergenic regions—the portions of the genome typically associated with K27me2/3. Together with the histone data, the changes in DNA methylation suggest that as polycomb retreats across the genome, it is replaced by broadly increased DNA methylation, both at gene promoters and intergenic regions.
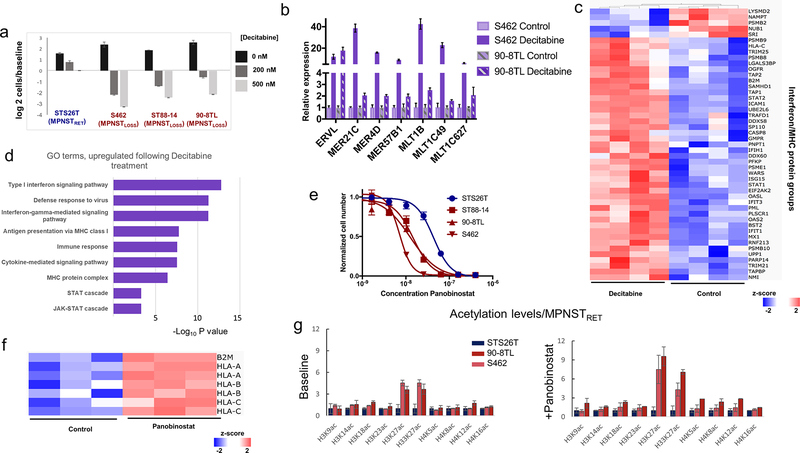
MPNSTLOSS are highly sensitive to DNA methyltransferase and histone deacetylase inhibition
Epigenetic repression safeguards against aberrant transcription of repetitive sequences including endogenous retroviral elements (ERV). Given the complete loss of polycomb-associated H3K27me2/3 across broad regions of the genome and the absence of compensatory histone-mediated repressive marks, we hypothesized that MPNSTLOSS would be disproportionately reliant on the observed increases in DNA methylation for the suppression of ERVs, and thus, sensitive to DNA methyltransferase inhibitors (DNMTi). To test this, we compared the sensitivity of MPNSTLOSS and MPNSTRET cell lines to decitabine treatment. MPNSTLOSS cell lines exhibited increased sensitivity to decitabine relative to MPNSTRET. Decitabine treatment not only halted cell growth, but led to marked levels of cell death (Fig. 7A). The induction of cell death was associated with increased expression of endogenous retroviral elements and IFN signature genes, consistent with the previously characterized mechanism of action of these drugs (Fig. 7B; refs. 27, 47). Proteomic analysis of decitabine-treated MPNSTLOSS lines verified the strong upregulation of IFN pathways (Fig. 7C and D; Supplementary Table S5). Decitabine also restored MHC class I expression in these cell lines demonstrating that, as with perturbations of histone PTMs, DNA methyltransferase inhibition restored the pathways most robustly downregulated in human tumors following PRC2 loss.
Figure 7.
MPNSTLOSS are highly sensitive to DNA methyltransferase and HDAC inhibition. A, Log2 plots of cell numbers normalized to time 0 control, following decitabine treatment for MPNSTRET (blue labels) and MPNSTLOSS lines (red labels). Error bars, ± SD for 3 replicates. B, mRNA expression levels measured by RT-PCR for endogenous retroviral elements (x-axis) in two MPNST cell lines. Values calculated using the 2−ΔΔCt method with three independent replicates. Error bars, ± SD. C, Heatmap depicting Z-scores for combined IFN signature protein groups and antigen processing/presentation (listed along y-axis) in MPNST cell lines ordered by treatment (decitabine, left; control, right), with hierarchical clustering of samples. Proteins depicted in heatmap were those exhibiting differential expression between treatment and control. Two-tailed t test, P < 0.05. D, GO enrichment: −log10 P values for GO term enrichment for GO process terms based on ranked list of protein abundance changes (treatment vs. control). E, Dose-response curves for MPNST cell lines treated with panobinostat. MPNSTRET (blue) are compared with MPNSTLOSS (red). Three replicates for each concentration. F, Heatmap depicting Z-scores for MHC I protein groups (listed along y-axis) in MPNSTLOSS cell lines grouped by treatment with panobinostat (right) or control (left). G, Acetylation levels for the designated H2, H3, and H4 sites, normalized to the levels in STS26T, an MPNSTRET cell line. Left, baseline levels; right, levels following treatment with sub IC50 concentrations of panobinostat. Blue bars, MPNSTRET cell lines; red/pink bars, MPNSTLOSS cell lines. Error bars, SD for 3 replicates.
Both DNMTi and histone deacetylase (HDAC) inhibitors (HDACi) function by promoting transcriptional instability; DNMTi through derepression and HDACi through activation associated with increased histone acetylation (48). As MPSNTLOSS were shown to be differentially sensitive to DNMTi-mediated derepression, we hypothesized that they might be differentially sensitive to HDAC due to higher baseline levels of activating histone acetyl marks and, thus, a propensity toward transcriptional instability.
MPNSTLOSS cell lines were indeed significantly more sensitive to the HDACi panobinostat (Fig. 7E). Moreover, as with DNMTi, panobinostat markedly increased the expression of MHC I (Fig. 7F). Because HDACi broadly increase acetylation levels across all sites, it is difficult to assess which acetyl marks are most likely to account for the differential sensitivity of MPMNSTLOSS lines to panobinostat. Nonetheless, it is possible to look at which acetylation sites differ most between MPNSTLOSS and MPNSTRET, both at baseline and following a low-dose treatment with panobinostat (sub IC50). As compared with MPNSTRET, MPNSTLOSS lines show the greatest level of differential acetylation at histone H3K27, both at baseline and following panobinostat treatment (Fig. 7G). The marked differential increase in H3K27Ac following panobinostat treatment is most manifested in the H3.3 variant of histone H3, which is enriched at the sites of active transcription and ERV (49).
In conjunction with the DNMTi results, these findings indicate that, as a consequence of PRC2 loss, MPNST are sensitized to therapeutics that promote transcriptional instability. This likely reflects a decreased capacity to counteract aberrant transcriptional activation genome-wide due to the loss of a major transcriptional suppressor.
Discussion
Alterations in epigenetic regulatory complexes are common place in human cancers (50). Although they promote oncogenesis, they may also lead to selective vulnerabilities, often to chemical or molecular perturbations of other epigenetic regulatory complexes. The selective vulnerabilities identified here converge on the central role of repressive epigenetic modifications in safeguarding the genome from inappropriate transcriptional activation.
Histone modifications and DNA methylation coordinate across the genome to suppress aberrant transcription. H4K20me3, H3K27me3, and H3K9me3 have all been implicated in silencing of repeat elements (39, 51, 52). Targeted disruption of these silencing mechanisms with DNMTi, HDACi, as well as pharmacologic or genetic targeting of histone methyltransferases, all function, at least in part, by inducing transcriptional disarray (27, 47, 48, 52). The selective vulnerability of MPNSTLOSS to these agents is likely a function of synthetic lethality due to a preexisting “hit” in a major silencing mechanism, PRC2. In this setting, the global loss of H3K27me2, which we demonstrate here, may be even more important than H3K27me3, given its significantly broader distribution, greater abundance, and greater association with intergenic and repeat regions (7). Although it was not investigated here, it would be interesting to see whether MPNSTLOSS are sensitive to the disruption of other mediators of transcriptional silencing, such as SETDB1.
Further work will be needed to establish the extent to which the drug susceptibilities in MPNST are present in other tumors. There is intriguing data in pediatric gliomas with lysine K27M alterations that suggests the epigenetic vulnerabilities shown here are shared in that system. In this group of pediatric gliomas, expression of a K27M-mutant version of the histone variant, H3.3, is associated with globally reduced K27me3 levels and, in contrast to MPNST, DNA hypomethylation (53). Recent data indicates that the K27M mutation leads to markedly increased levels of K27Ac, which is not deposited at enhancer elements, but rather, is broadly distributed across intergenic regions promoting transcriptional instability and increased sensitivity to the combination of DNMTi and HDACi (Krug and colleagues, in review). Although prior work has shown that HDACi promotes transcriptional instability, this was attributed to increases in the acetylation of H2AK9, H3K14, and H3K23 (48). Our data, like those in K27M glioma models, suggests a role for K27Ac in HDACi sensitivity, because none of the other acetylation states differed dramatically between the highly sensitive MPNSTLOSS lines and the less sensitive MPNSTRET, either at baseline or following HDACi treatment. This would be in keeping with the known interplay between K27me3 and K27Ac, which was recently shown to influence sensitivity to EZH2 inhibitors (54). It would be interesting to see whether this histone signature (K27me3 low, K27Ac high), associated with sensitivity to drugs targeting epigenetic repression in K27M gliomas and MPNSTLOSS, is more generally true for other tumor types. If it is, then the baseline levels of H3K27me3 or H3K27Ac on H3.1/2 or H3.3 might serve as biomarkers for susceptibility to these agents.
Our human tumor data and cellular experiments strongly implicate PRC2 loss in immune evasion, and point to multiple pathways to restore immune surveillance. Although PRC2 restoration is not therapeutically feasible, NSD2 knockdown and both DNMTi and HDACi promoted significant increases in MHC I expression. If replicated in vivo, these agents might help restore immune surveillance and render these tumors susceptible to immune-modulating agents. Indeed, therapeutics aimed at making cold tumors hot are highly desirable, as the recovery of MHC class I expression is a critical precursor to successful immunotherapy (55). It is important to note here that the role of epigenetic regulators in immune evasion is likely to be complex and system-dependent. Although, in MPNST, PRC2 loss may promote immune evasion, recent work in other tumor types suggest that, in other contexts, increased polycomb activity is associated with a suppression of immune pathways (56).
It is unclear whether the epigenetic effects on immune surveillance and IFN signaling are direct or indirect. Although we did find increased DNA methylation at the promoters of IFN-related genes and MHC I, changes in DNA methylation were not strongly enriched at these genes. We suspect that another contribution may be coming from the global increases in growth-promoting oncogenic pathways. Evasion of IFN signaling and MHC I down-regulation have been linked to upregulation of the Ras pathway, which downregulates IFN responses by preventing STAT2 activation and IRF1 expression (57, 58). This may explain the association of K36me2, typically considered an activating mark, with IFN silencing, because it is a potentiator of Ras signaling (44). Myc transcriptional activity has also been associated with IFN suppression (59). Thus, it is possible that the decreased IFN gene expression seen following PRC2 loss is driven by amplification of oncogenic pathways. This phenomenon has been described across numerous malignancies, and, aside from the examples cited above, remains poorly understood (60). Irrespective of the mechanism, we demonstrate that epigenetic modifications are prime targets to both unleash intrinsic IFN signaling and restore MHC I expression.
Finally, this work illustrates the utility of translational approaches to combined proteomic and epigenetic analyses, which can be undertaken using archival pathology materials. Only one other laboratory has published analyses of histone PTMs carried out in FFPE (61). Looking at a narrower set of H3 and H4 histone peptides, the authors did not see significant effects of fixation and paraffin-embedding on histone PTMs, and were able to detect differences in histone H3 modifications in different subtypes of breast cancer. To the best of our knowledge, this is the first study to investigate histone PTMs and global proteomes in parallel on the same tissue specimens. We demonstrate that the measured effects of PRC2 loss on histone PTMs in human tissues can be reliably quantified in tissue stored for over a decade. A similar approach could be applied to characterize global epigenetic changes in other cancer types or subtypes. In future, we think it will be even more important for researchers to avail themselves ofarchival materials, as the movement toward minimally invasive diagnostic techniques and neoadjuvant therapy will greatly reduce the amount of viable fresh or frozen tumor tissue available for translational research.
Supplementary Material
Significance:
Global profiling of histone PTMs and protein expression in archival human MPNST illustrates how PRC2 loss promotes oncogenesis but renders tumors vulnerable to pharmacologic modulation of transcription.
Acknowledgments
This research was supported by US NIH grants (R01-GM110174, R01-AI118891, and CA196539 to B.A. Garcia; P01CA196539 to J. Majewski and B.A. Garcia; 5K12CA076931 to J.B. Wojcik; and T32GM008275 and TL1TR001880 to D.M. Marchione). B.A. Garcia is also supported by a Robert Arceci Scholar award from the Leukemia and Lymphoma Society. The authors would like to acknowledge Dr. David Reuss for providing raw DNA methylation array data from his previously published work (28). They would also like to acknowledge Drs. Paul J. Zhang and Kumarasen Cooper in the Department of Pathology at the Hospital of the University of Pennsylvania for their support and helpful discussions regarding MPNST histopathology.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Kroep JR, Ouali M, Gelderblom H, Le Cesne A, Dekker TJA, Van Glabbeke M, et al. First-line chemotherapy for malignant peripheral nerve sheath tumor (MPNST) versus other histological soft tissue sarcoma subtypes and as a prognostic factor for MPNST: an EORTC soft tissue and bone sarcoma group study. Ann Oncol 2011;22:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenbarth I, Beyer K, Krone W, Assum G. Toward a survey of somatic mutation of the NF1 gene in benign neurofibromas of patients with neurofibromatosis type 1. Am J Hum Genet 2000;66: 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014;514:247–51. [DOI] [PubMed] [Google Scholar]
- 4.Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 2014;46:1227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006;125:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner LA, Schulz VP, Maksimova Y, Wong C, Gallagher PG. Patterns of histone H3 lysine 27 monomethylation and erythroid cell type-specific gene expression. J Biol Chem 2011;286:39457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 2014;53:49–62. [DOI] [PubMed] [Google Scholar]
- 8.Lee H-G, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res 2015;25:1170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet 2008;4:e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkman AB, Gu H, Bartels SJJ, Zhang Y, Matarese F, Simmer F, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 2012; 22:1128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016;352:aad9780–0. [DOI] [PubMed] [Google Scholar]
- 12.Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SMG, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 2016;49:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009;136:3131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 2011;286: 7983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessel D, FlüKigge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 1984;138:141–3. [DOI] [PubMed] [Google Scholar]
- 16.Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp 2016;111: 54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karch KR, Zee BM, Garcia BA. High resolution is not a strict requirement for characterization and quantification of histone post-translational modifications. J Proteome Res 2014;13:6152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidoli S, Simithy J, Karch KR, Kulej K, Garcia BA. Low resolution data-independent acquisition in an LTQ-orbitrap allows for simplified and fully untargeted analysis of histone modifications. Anal Chem 2015;87: 11448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Z-F, Sidoli S, Marchione DM, Simithy J, Janssen KA, Szurgot MR, et al. EpiProfile 2.0: a computational platform for processing Epi-proteomics mass spectrometry data. J Proteome Res 2018;17:2533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 22.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Meth 2016;13:731–40. [DOI] [PubMed] [Google Scholar]
- 23.Asangani IA, Ateeq B, Cao Q, Dodson L, Pandhi M, Kunju LP, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell 2013;49:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004;23:4061–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016;352:844–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, McKenzie NJ, Warneford-Thomson R, Gail EH, Flanigan SF, Owen BM, et al. RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat Struct Mol Biol 2019;26:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Röhrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol 2016;131:877–87. [DOI] [PubMed] [Google Scholar]
- 29.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Meth 2014;11: 1138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hovestadt V, Jones DTW, Picelli S, Wang W, Kool M, Northcott PA, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014;510:537–41. [DOI] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008;40:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med 2018;24:1599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 2013;41:2797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang R, Erler J, Langowski J. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys J 2017;112:450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006;311:844–7. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008;40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu LMN, Deng Y, Wang J, Zhao C, Wang J, Rao R, et al. Programming of Schwann cells by Lats1/2-TAZ/YAP signaling drives malignant peripheral nerve sheath tumorigenesis. Cancer Cell 2018;33:292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Carpizo V, Sarmentero J, Han B, Graña O, Ruiz-Llorente S, Pisano DG, et al. NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation. Sci Rep 2016;6:32952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sohier P, Luscan A, Lloyd A, Ashelford K, Laurendeau I, Briand-Suleau A, et al. Confirmation of mutation landscape of NF1-associated malignant peripheral nerve sheath tumors. Genes Chromosom Cancer 2017;56: 421–6. [DOI] [PubMed] [Google Scholar]
- 46.Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018;359:940–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162: 974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet 2017;49: 1052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elsöasser SJ, Noh K-M, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015;522:240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gnad F, Doll S, Manning G, Arnott D, Zhang Z. Bioinformatics analysis of thousands of TCGA tumors to determine the involvement of epigenetic regulators in human cancer. BMC Genomics 2015;16:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishak CA, Marshall AE, Passos DT, White CR, Kim SJ, Cecchini MJ, et al. An RB-EZH2 complex mediates silencing of repetitive DNA sequences. Mol Cell 2016;64:1074–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato M, Takemoto K, Shinkai Y. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat Commun 2018; 9:1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013;24:660–72. [DOI] [PubMed] [Google Scholar]
- 54.Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X, et al. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-aberrant solid tumors. Cell 2018;175:186–199.e19. [DOI] [PubMed] [Google Scholar]
- 55.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol 2016;39:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015;527:249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christian SL, Collier TW, Zu D, Licursi M, Hough CM, Hirasawa K. Activated Ras/MEK inhibits the antiviral response of alpha interferon by reducing STAT2 levels. J Virol 2009;83:6717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AbuSara N, Razavi S, Derwish L, Komatsu Y, Licursi M, Hirasawa K. Restoration of IRF1-dependent anticancer effects by MEK inhibition in human cancer cells. Cancer Lett 2015;357:575–81. [DOI] [PubMed] [Google Scholar]
- 59.Schlee M, Hölzel M, Bernard S, Mailhammer R, Schuhmacher M, Reschke J, et al. C-myc activation impairs the NF-κB and the interferon response: implications for the pathogenesis of Burkitt’s lymphoma. Int J Cancer 2007;120:1387–95. [DOI] [PubMed] [Google Scholar]
- 60.Lin C-F, Lin C-M, Lee K-Y, Wu S-Y, Feng P-H, Chen K-Y, et al. Escape from IFN-γ-dependent immunosurveillance in tumorigenesis. J Biomed Sci 2017;24:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noberini R, Uggetti A, Pruneri G, Minucci S, Bonaldi T. Pathology tissue-quantitative mass spectrometry analysis to profile histone post-translational modification patterns in patient samples. Mol Cell Proteomics 2016; 15:866–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roöhrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol 2016;131:877–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data files for proteomics experiments are available on https://chorusproject.org at the project no.1539.
RNA sequencing (RNA-seq) and WGBS data are available at the following URL: https://datahub-7woy4ejm.udes.genap.ca/.