Abstract
Poly(ADP-ribose) polymerase (PARP) enzymes are broadly involved in the cellular response to DNA damage. PARP-1 is the chief human PARP enzyme involved in the DNA damage response, acting as a first responder that detects DNA strand breaks, and contributes to repair pathway choice and the efficiency of repair through modulation of chromatin structure and through interaction with and modification of a multitude of DNA repair factors. This perspective summarizes our knowledge of PARP-1 involvement in DNA repair pathways, and highlights recent structural and functional data regarding the activation of PARP-1 upon detecting DNA damage, and the release and trapping of PARP-1 at sites of DNA damage.
Keywords: poly(ADP-ribose), PARP, DNA damage response
The PARP family of enzymes
The poly(ADP-ribose) polymerase (PARP) family of enzymes regulates virtually every aspect of human cell biology through the production of an ADP-ribose posttranslational modification of proteins using NAD+ [1,2]. The majority of the 17 family members create a mono(ADP-ribose) modification on the acceptor amino acids of target proteins, and a subset of family members extend the initial modification site to form poly(ADP-ribose), or PAR. The structures and activities of the catalytic domains of PARP family enzymes are related to each other and to the bacterial toxins that also use NAD+ to ADP-ribosylate proteins [3]; indeed, the PARP family members are also known as ADP-ribosyl transferase diphtheria toxin-like (ARTD) enzymes [4]. In contrast to the related catalytic domains, distinct regulatory domains of the 17 members of the PARP family of enzymes specify their cellular functions. Three human PARP enzymes, PARP-1, and PARP-2 and PARP-3, are regulated through interaction with DNA damage, and each of these DNA damage response PARPs are implicated in different aspects of the cellular response to DNA damage [5,6]. The Tankyrase enzymes (PARP-5a/PARP-5b) are perhaps best known for their contribution to genome stability through the regulation of telomeres and mitosis [7,8], and there is also growing evidence for other PARP family members involvement in the DNA damage response [6]. This Perspective will focus on PARP-1, for which there is the most advanced understanding of structure, mechanism, and biological function. Moreover, PARP-1 is the major source of poly(ADP-ribose) produced during the cellular response to DNA damage.
PARP-1 and the cellular response to DNA damage
PARP-1 plays an expansive and multifaceted role in the cellular response to DNA damage, with growing evidence for participation in multiple pathways of DNA damage repair and genome maintenance [5,6]. The currently appreciated roles within repair pathways can be summarized in three categories: (i) detection of DNA damage, (ii) poly(ADP-ribose) mediated recruitment of repair factors, and (in) poly(ADP-ribose) mediated regulation of biochemical activities. Early literature centered on PARP-1 involvement in single strand break repair (SSBR) and base excision repair (BER) [9–16], where critical events are the detection of single-stand break damage and the poly(ADP-ribose)-mediated recruitment of the scaffolding factor XRCC1 to DNA strand breaks. PARP-1 has now been implicated in the regulation of nucleotide excision repair (NER) [17,18], classical non-homologous end-joining (cNHEJ) [19–21], alternative non-homologous end-joining (aNHEJ) [22–28], microhomology-mediated end-joining (MMEJ) [29,30], homologous recombination (HR) [31,32], DNA mismatch repair (MMR) [33], and maintenance of replication fork stability [34–38]. The emerging details for the roles of PARP-1 in the various pathways has been the subject of two recent reviews [5,6]. Several representative roles for PARP-1 regulation of DNA repair are summarized in Table 1.
Table 1.
Examples of PARP-1 participation in DNA repair pathways
| Role of PARP-1/poly(ADP-ribose) | Repair factor | Pathway(s) |
|---|---|---|
| recruitment | XRCC1, scaffold protein | SSBR, BER |
| damage detection | DNA damage-binding protein 2 (DDB2) | NER |
| recruitment and activation | ALC1, chromatin remodeller | NER |
| recruitment and inhibition | RECQ1, helicase | replication fork reversal |
| recruitment | POLΘ, polymerase | aNHEJ |
| recruitment | MRE11, nuclease | HR, aNHEJ, replication fork stability |
| recruitment | CHD2, chromatin remodeller | cNHEJ |
| recruitment | BRCA1, scaffold protein | HR |
Note: refer to text for references linking these PARP-1/PAR functions to repair pathways.
An additional and perhaps more general role of PARP-1 in the response to DNA damage is the regulation of chromatin structure and composition. This role is carried out in part through the poly(ADP-ribose)-mediated recruitment and regulation of chromatin remodeling factors [39–42], but also through PARP-1 modification of histones [43]. Furthermore, poly(ADP-ribose) has been proposed to seed the formation of subnuclear structures through the principle of liquid demixing, a phase separation phenomenon that endows special physical properties for granting access to certain protein factors and excluding others [44]. Thus, in addition to specific roles in repair pathways, PARP-1 can be viewed as a general organizer of nuclear architecture [45].
Cells with defects in homologous recombination DSB repair due to BRCA1/2 loss are exquisitely sensitive to chemical inhibition of PARP activity [46,47]. PARP inhibition and HR deficiency exemplify the concept of synthetic lethality, in which two deficiencies that are not individually lethal are rendered lethal in combination. PARP inhibitors are the first example of synthetic lethality that has shown success in the clinic, and they have heralded the potential of targeting DNA damage repair for the treatment of cancer [48,49]. Given PARP-1 connections to multiple repair pathways, there have been several different proposals to explain the sensitivity of HR-deficient cells to PARPi-mediated deficiencies in repair pathways [5]: (i) deficiency in SSBR: elevated number of unrepaired single strand breaks that are converted to a DSB end after encountering a replication fork, (ii) deficiency in aNHEJ: eliminates another pathway of DSB repair in addition to HR, and (iii) deficiency in stability of replication forks: further reduces the capacity to cope with unstable replication forks. There is a growing appreciation for the importance of PARP-1 contribution to repair pathways outside of SSBR/BER, and this shift in understanding has also shifted the focus to alternate explanations for the underlying mechanism of PARPi sensitivity. A Perspective from Alan D’Andrea in this issue further explores the mechanisms of PARP inhibitor sensitivity and resistance.
PARP-1 detecting and signaling DNA damage
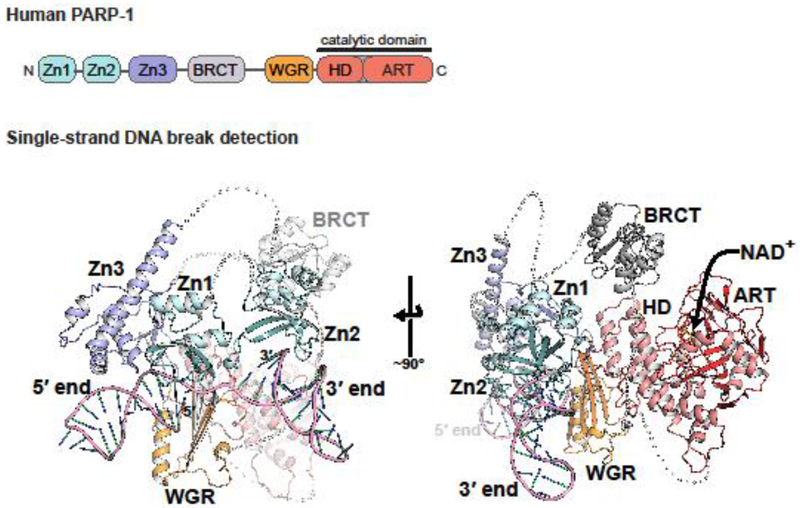
The structural analysis of PARP-1 has provided the most keen insights into how PARPs recognize DNA damage and into how DNA damage detection allosterically leads to robust production of poly(ADP-ribose) [50–57]. PARP-1 has a multi-domain architecture composed of six domains (see Fig. 1 schematic). Although the structure of full-length PARP-1 has yet to be determined, a composite model based on X-ray and NMR structures of fragments and biophysical and biochemical analysis of full-length protein provides a rather complete view of PARP-1 in the act of detecting DNA damage and being primed for poly(ADP-ribose) production (Fig. 1; see legend for model details).
Figure 1. PARP-1 detecting DNA damage and primed for poly(ADP-ribose) production.
Human PARP-1 domain names and organization, and a structural model for PARP-1 interaction with single-strand break DNA damage. PARP-1 binding to the ends of DNA strand break damage is achieved through coordinated action of two zinc finger domains, Zn1 and Zn2, located at the N-terminus of the protein. The Zn1 domain binds to the side of the DNA break with a terminal 5’ strand (5’ end), and the Zn2 domain binds to the side of the DNA break with a terminal 3’ strand (3’ end). A structurally unrelated third zinc binding domain, Zn3, and the WGR (Trp-Gly-Arg) domain also make contacts with DNA. The catalytic domain (CAT) is composed of an ADP-ribosyl transferase (ART) fold and a helical subdomain (HD). The linker residues (each residue represented by an individual sphere) and the BRCT (BRCA1 C-terminus) domain were manually positioned based on linker lengths and the domain termini. The NAD+ binding site is noted. The combined crystallographic and NMR model is shown in two orientations that are related by an approximately 90-degree rotation about a vertical axis in the plane of the page. The left view highlights the structure of the DNA break and PARP-1/DNA contacts, with the CAT domain and the BRCT in the background. The right view highlights the HD contacts with the domains that are assembled on DNA.
There are several key aspects of the PARP-1 structural model. First, the domains are organized through interaction with DNA damage, and the domains that contact DNA directly (Znl, Zn2, Zn3, and WGR) have mutually compatible binding sites. This aspect is particularly important for a single-strand break that presents two DNA ends, the key feature recognized by PARP-1 zinc fingers Zn1 and Zn2 [50]. A composite NMR/x-ray structure of the Zn1-Zn2 fragment of PARP-1 bound to a model single-strand DNA break illustrated that the zinc fingers are partitioned to specific ends of the DNA structure (Znl to the 5’ end, Zn2 to the 3’ end), rather than competing for the same DNA end or forming a mixed population [50]. This specific arrangement presents the Zn1 to form essential contacts with the WGR domain, which binds near the 5’ end of the DNA. The Zn1 domain has particular sequence features that allow it to communicate with the WGR domain and which are not present in the Zn2 domain [52,58]. It is also noteworthy that the extended linker region connecting the Zn1 and Zn2 domains is likely to allow the Zn1-Zn2 fragment to accommodate greater distances between the two ends of the DNA, for example on a single-strand break with a multi-nucleotide gap. The precise composition of the DNA structure could thus influence the efficiency with which the PARP-1 domains are assembled, and could thereby regulate the catalytic output (e.g. single-strand break versus stalled replication fork structure).
A second feature of the model is that PARP-1 binds to DNA damage as a monomer, thus indicating an intramolecular mechanism of activation. This aspect of the model is supported by both structural data and biophysical analysis [50,58–60]. The organization of domains positions a heavily modified region of PARP-1, the linker residues following the BRCT fold [61], in close proximity to the catalytic domain. The structural model thus can explain PARP-1 preference for automodification, and biochemical analysis using PARP-1 variants of different size and using defined DNA substrates has indicated a preference for in cis modification [50]. Recent cell-based studies using different tagged versions of PARP-1 have also supported the intramolecular activation mechanism of PARP-1 [62].
A third important aspect of the model concerns the mechanism of allosteric activation. A structural distortion in the regulatory helical domain (HD) of PARP-1 was observed in the crystal structure of the essential Zn1, Zn3, WGR, and CAT domains in complex with a DNA double strand break [58]. Along with supporting protein mutagenesis and biochemical analysis, it was expected that a conformational change in the HD, promoted through contacts with the Zn1, Zn3, and WGR domains, was the underlying trigger for an increase in poly(ADP-ribose) production [53]. However, the mechanism was not clear. An analysis of PARP-1 structural dynamics in the absence and presence of DNA strand break damage using hydrogen-deuterium exchange with mass spectrometry (HXMS) indicated that the DNA damage-dependent structural distortion of the HD is actually much more severe than what was captured in the crystal structure, leading to the unfolding of specific HD regions that are held in a helical conformation in the absence of DNA [51]. Deletion of the HD domains from PARP-1, PARP-2, and PARP-3 leads to constitutively active PARP enzymes, thus indicating that the HD is an autoinhibitory domain for each of the DNA damage-dependent PARPs [51]. A recent study using a non-hydrolyzable NAD+ analog has provided the final important step of the mechanism, indicating that the folded conformation of the HD acts to block NAD+ access to the PARP-1 catalytic domain; HD unfolding allows unrestricted access to NAD+ [54].
The full potential of PARP-1 catalysis of poly(ADP-ribose) production is remarkably robust. Indeed the HD-deleted, constitutively active forms of PARP-1 produced for biochemical and structural analysis require special considerations for production in E. coli, including the presence of PARP inhibitors and sortase-mediated joining of PARP fragments [51,63]. The HD provides a mechanism to keep PARP-1 inactive by restricting access to the key requirement for poly(ADP-ribose) synthesis: NAD+. Interaction with DNA damage acts to distort the structure of the HD, increasing the frequency of the active site being in the “open” state. Thus, the HD can be viewed as a throttle for the PARP-1 active site, regulating access to the NAD+ fuel.
PARP-2 and PARP-3 also exhibit HD autoinhibition [51], and are predicted to form similar WGR contacts with the HD [64]. Their WGR contacts with the DNA are likely to differ somewhat from what has been observed with the WGR of PARP-1, based on the fact that PARP-2 and PARP-3 are most potently activated by DNA single strand breaks and DNA phosphorylated on the 5’ terminus [64–66]. Thus, the nature of the damaged DNA structure (i.e. gapped or nicked single-strand break) can influence the catalytic output. A more complete understanding of how the DNA structure influences PARP-2 and PARP-3 allosteric activation will require structural information for DNA-bound enzymes.
Regulation of the PARP-1 complex with DNA damage
The presented structural model represents the expected state of PARP-1 immediately following DNA damage detection and just prior to the burst in production of poly(ADP-ribose). The DNA damage activated complex is dependent on a quite elaborate network of protein-DNA and protein-protein contacts, likely providing points of regulation through posttranslational modifications (including ADP-ribose) and through the binding of partner proteins. As an example, the protein HPF1 (Histone PARylation Factor 1) was recently identified as a binding partner of PARP-1 that regulates catalytic output [67]. The mechanistic details for how this is accomplished are still unknown, but HPF1 could potentially act by adjusting the organization of PARP-1 domains, and thereby controlling the catalytic output. Timeless is another binding partner of PARP-1, for which a complex of the Timeless PARP-1 binding domain has been determined in complex with the PARP-1 catalytic domain [68]. There is no apparent affect of this interaction on PARP-1 catalytic output. In this case, the repair factor Timeless might simply use the PARP-1 protein as an anchoring site to promote homologous recombination [68]. PARP-1 is an abundant nuclear protein and an ever-present chromatin-associated factor. As such, there are likely to be more repair factors with interfaces designed to engage or associate with PARP-1, similar to the transcription-related proteins that are influenced by PARP-1 [69].
Another mechanism of regulating PARP-1 assembly on DNA is automodification with poly(ADP-ribose). Automodification eventually leads to PARP-1 release from DNA and a concomitant drop in catalytic output. The balance between the cellular level of PARP-1 automodification that can contribute to repair factor recruitment, and the level of automodification that can lead to PARP-1 release from cellular sites of DNA damage is not clear. One challenge to investigating the function and mechanism of PARP-1 automodification is the sheer number of automodification sites that have been identified, which is likely to reflect some level of redundancy and the importance of PARP-1 automodification to the DNA damage response. However, the key sites required for regulation of PARP-1 have not been definitively established.
Interestingly, a recent study from the Van Houten and Wilson groups analyzed the mobility of unmodified and ADP-ribose-modified forms of PARP-1 on DNA tight ropes containing abasic site DNA damage using single molecule imaging and quantum dot-labeled PARP-1 [59]. Unmodified PARP-1 was stably associated with the DNA damage sites, whereas an automodified form of PARP-1 exhibited constrained motion around the DNA damage site, suggesting that there could also be an intermediate state of modification that regulates PARP-1 function, rather than just an on/off switch for interaction with DNA.
PARP inhibitors compete for the NAD+ binding site of PARP-1, thus preventing poly(ADP-ribose) production and its contribution to the recruitment and regulation of repair factors, and also its contribution to PARP-1 release from DNA damage. Pommier and colleagues have noted that certain PARP inhibitors are more efficient at “trapping” PARP-1 and PARP-2 in a tight complex with chromatin, and that the inhibitors with the higher trapping potential are more efficiently able to kill HR-deficient cells [70,71]. The differences in inhibitor trapping ability were proposed to arise from potential differences in their ability to invoke reverse allostery, in which the mode of inhibitor binding might influence PARP-1 structure in a way that would propagate to the DNA binding domains. However, there has been no evidence that this actually occurs with clinically used PARP inhibitors [72]. A recent study using a non-hydrolyzable NAD+ analog showed clear evidence of reverse allostery, where the presence of the NAD+ analog invoked a 10-fold increase in PARP-1 affinity for single-strand break damage and a resistance for PARP-1 to release from DNA in a competition experiment [54]. Moreover, there was a change in the dynamics of the DNA binding domains of PARP-1 in response to analog binding as measured by hydrogen/deuterium exchange mass spectrometry [54]. The study indicates the potential for substrate NAD+ to influence PARP-1 stability and residence time on DNA damage, and also indicates the design principles that should lead to inhibitors that can invoke reverse allostery and potentially lead to enhanced trapping.
Emerging topic: ADP-ribose modification of serine residues
PARP enzymes modify a variety of amino acid side chains with ADP-ribose. Glutamate and aspartate residues have been most frequently reported as the key residue type for modification by PARP-1. However, recent studies have discovered the ADP-ribose modification of serine residues mediated by PARP-1 [73]. Interestingly, HPF1 (histone PARylation factor 1) is essential for ADP-ribose-serine modification [74], in effect changing the specificity of PARP-1 from glutamate-directed modification to serine-directed modification. HPF1 also influences the target of PARP-1 catalysis toward histone modification, rather than almost exclusively being directed toward PARP-1 automodification [67]. The mechanism of HPF1 influence on PARP-1 catalytic output and the target amino acid specificity switch are not known. It is also not known whether HPF1 exerts its influence over other PARP enzymes, besides just PARP-1 and PARP-2. The enzyme ADP-ribosylhydrolase 3 (ARH3) reverses the ADP-ribose modification of serine residues [75,76], and the ADP-ribose-serine modification has recently been reported to be the major type of ADP-ribose modification in response to DNA damage [77]. A specificity switch is an attractive mechanism to create a different pattern of ADP-ribose modification in stress conditions versus normal physiological conditions.
Emerging topic: ADP-ribose modification of DNA
It has recently been reported that human PARP-1, PARP2 and PARP3 can modify DNA with ADP-ribose [78–81]. The ADP-ribose modification of DNA bases occurs through the action of certain toxins [82]. In contrast, the recent reports indicate modification of the 5’phosphoiylated end of a DNA strand in the context of duplex DNA. 5’phosphorylated DNA is the same structure that maximally activates PARP2 and PARP3 through a WGR contact with the 5’ terminus [64]. Thus, it is unclear whether the same 5’ phosphate feature serves as both a binding site and a point of modification. The biochemical analysis indeed has required DNA structures bearing at least two 5’ phosphoiylated breaks. Despite the current uncertainties in the reaction mechanism, the biochemical activity appears to be robust in vitro, and there is evidence that the reaction takes place in cellular extracts [79]. The biological relevance of the modification is still unclear. Whether the ADP-ribose modified DNA represents a bona fide signaling modification or repair pathway intermediate has yet to be determined. Alternatively, the modification could represent an unwanted side reaction that occurs in the context of excessive DNA damage and PARP over-activation, akin to the abortive DNA ligation reaction that leaves an AMP nucleotide on the 5’ phosphate group of DNA, which can be reversed by the enzyme aprataxin [83,84]. ADP-ribosylhydrolase 3 (ARH3) is able to reverse the ADP-ribose modification of DNA [81], suggesting a pathway to reverse either a signaling event or an unwanted side reaction. DNA modification could represent an interesting extension of the use and prevalence of the ADP-ribose modification.
Conclusion
PARP-1 has long been appreciated as important for DNA repair [85], but the role of PARP-1 has been fairly enigmatic and difficult to clearly define and categorize. The understanding of PARP-1 contribution to DNA damage repair continues to grow in more specific and mechanistic ways. Collectively, we can start to appreciate that PARP-1 and the other DNA response PARPs have a pervasive influence on the nuclear environment in which repair processes take place, and we gain an even greater appreciation for the mechanisms by which the ADP-ribose modification can influence the cellular response to DNA damage
Acknowledgements
Research on DNA damage response PARP enzymes in the Pascal laboratory is supported by the CIHR (BMA342854) and the NCI (CA92584).
Abbreviations:
- PAR
poly(ADP-ribose)
- PARP
PAR polymerase
- PARylation
poly(ADP-ribosylation)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The author claims no conflict of interest.
References
- [1].Gibson BA, Kraus WL, New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs, Nat. Rev. Mol. Cell Biol. 13 (2012) 411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- [2].Hassa PO, Hottiger MO, The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases., Front. Biosci. 13 (2008) 3046–82. http://www.ncbi.nlm.nih.gov/pubmed/17981777 (accessed March 22, 2018). [DOI] [PubMed] [Google Scholar]
- [3].Karlberg T, Langelier M-F, Pascal JM, Schüler H, Structural biology of the writers, readers, and erasers in mono- and poly(ADP-ribose) mediated signaling., Mol. Aspects Med. 34 (2013) 1088–108. doi: 10.1016/j.mam.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F, Toward a unified nomenclature for mammalian ADP-ribosyltransferases, Trends Biochem. Sci. 35 (2010) 208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- [5].Ray Chaudhuri A, Nussenzweig A, The multifaceted roles of PARP1 in DNA repair and chromatin remodelling, Nat. Rev. Mol. Cell Biol. 18 (2017) 610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martin-Hernandez K, Rodriguez-Vargas J-M, Schreiber V, Dantzer F, Expanding functions of ADP-ribosylation in the maintenance of genome integrity, Semin. Cell Dev. Biol. 63 (2017) 92–101. doi: 10.1016/j.semcdb.2016.09.009. [DOI] [PubMed] [Google Scholar]
- [7].Smith S, de Lange T, Tankyrase promotes telomere elongation in human cells, 2000. doi: 10.1016/S0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- [8].Chang P, Coughlin M, Mitchison TJ, Tankyrase-1 polymerization of poly ( ADP-ribose ) is required for spindle structure and function, 7 (2005) 1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- [9].Leppard JB, Dong Z, Mackey ZB, Tomkinson AE, Physical and functional interaction between DNA ligase Illalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair., Mol. Cell. Biol. 23 (2003) 5919–27. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fisher AEO, Hochegger H, Takeda S, Caldecott KW, Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase., Mol. Cell. Biol. 27 (2007) 5597–605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW, A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage., Nucleic Acids Res. 31 (2003) 5526–33. http://www.ncbi.nlm.nih.gov/pubmed/14500814 (accessed March 23, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G, XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage., Mol. Cell. Biol. 18 (1998) 3563–71. http://www.ncbi.nlm.nih.gov/pubmed/9584196 (accessed March 23, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dantzer F, de La Rubia G, Ménissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V, Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1., Biochemistry. 39 (2000) 7559–69. http://www.ncbi.nlm.nih.gov/pubmed/10858306 (accessed March 23, 2018). [DOI] [PubMed] [Google Scholar]
- [14].Trucco C, Oliver FJ, de Murcia G, Ménissier-de Murcia J, DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines., Nucleic Acids Res. 26 (1998) 2644–9. http://www.ncbi.nlm.nih.gov/pubmed/9592149 (accessed March 23, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH, Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair., J. Biol. Chem. 276 (2001) 25541–8. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- [16].Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH, DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis., J. Biol. Chem. 276 (2001) 32411–4. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- [17].Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, Sugasawa K, Thoma N, Vermeulen W, Vrieling H, Mullenders L, PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1., J. Cell Biol. 199 (2012) 235–49. doi: 10.1083/jcb.201112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, Shah GM, Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair, Proc. Natl. Acad. Sci. 110 (2013) 1658–1663. doi: 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, van Attikum H, PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining, Mol. Cell. 61 (2016) 547–562. doi: 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR, Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase., J. Biol. Chem. 273 (1998) 14461–7. http://www.ncbi.nlm.nih.gov/pubmed/9603959 (accessed March 23, 2018). [DOI] [PubMed] [Google Scholar]
- [21].Spagnolo L, Barbeau J, Curtin NJ, Morris EP, Pearl LH, Visualization of a DNA-PK/PARP1 complex, Nucleic Acids Res. 40 (2012) 4168–4177. doi: 10.1093/nar/gkr1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cheng Q, Barboule N, Frit P, Gomez D, 0. Bombarde, B. Couderc, G.-S. Ren, B. Salles, P. Calsou, Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks, Nucleic Acids Res. 39 (2011) 9605–9619. doi: 10.1093/nar/gkr656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA, Ku Regulates the Non-Homologous End Joining Pathway Choice of DNA Double-Strand Break Repair in Human Somatic Cells, PLoS Genet 6 (2010) el000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mansour WY, Rhein T, Dahm-Daphi J, The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies, Nucleic Acids Res. 38 (2010) 6065–6077. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G, PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways., Nucleic Acids Res. 34 (2006) 6170–82. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sfeir A, de Lange T, Removal of Shelterin Reveals the Telomere End-Protection Problem, Science (80-,). 336 (2012) 593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MIR, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD, Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair, Nature. 518 (2015) 258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A, Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination., Nature. 518 (2015) 254–7. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sfeir A, Symington LS, Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway?, Trends Biochem. Sci. 40 (2015) 701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dutta A, Eckelmann B, Adhikari S, Ahmed KM, Sengupta S, Pandey A, Hegde PM, Tsai M-S, Tainer JA, Weinfeld M, Hegde ML, Mitra S, Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex, Nucleic Acids Res. 45 (2016) gkw1262. doi: 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G, Takeda S, Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells., EMBO J. 25 (2006) 1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hu Y, Petit SA, Ficarro SB, Toomire KJ, Xie A, Lim E, Cao SA, Park E, Eck MJ, Scully R, Brown M, Marto JA, Livingston DM, PARP1-Driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination-Mediated DNA Repair, Cancer Discov. 4 (2014) 1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu Y, Kadyrov FA, Modrich P, PARP-1 enhances the mismatch-dependence of 5’-directed excision in human mismatch repair in vitro, DNA Repair (Amst). 10 (2011) 1145–1153. doi: 10.1016/j.dnarep.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M, Topoisomerase I poisoning results in PARP-mediated replication fork reversal, Nat. Struct. Mol. Biol. 19 (2012) 417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- [35].Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Lopes M, Vindigni A, Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition., Nat. Struct. Mol. Biol. 20 (2013) 347–54. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bryant HE, Petermann E, Schultz N, Jemth A-S, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T, PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination, EMBO J. 28 (2009) 2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang Y-G, Cortes U, Patnaik S, Jasin M, Wang Z-Q, Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks, Oncogene. 23 (2004) 3872–3882. doi: 10.1038/sj.one.1207491. [DOI] [PubMed] [Google Scholar]
- [38].Ronson GE, Piberger AL, Higgs MR, Olsen AL, Stewart GS, McHugh PJ, Petermann E, Lakin ND, PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation, Nat. Commun. 9 (2018) 746. doi: 10.1038/s41467-018-03159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ, Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1., Science. 325 (2009) 1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC, Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler., Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 13770–4. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, Kozlowski M, Bultmann S, Ladurner AG, Timinszky G, Huet S, The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage., Mol. Biol. Cell. 27 (2016) 3791–3799. doi: 10.1091/mbc.E16-05-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Singh HR, Nardozza AP, Möller IR, Knobloch G, V Kistemaker HA, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, Huet S, Mueller-Planitz F, V Filippov D, Timinszky G, Rand KD, Ladurner AG, A Poly-ADP-Ribose Trigger Releases the Auto-Inhibition of a Chromatin Remodeling Oncogene., Mol. Cell. 68 (2017) 860–871. e7. doi: 10.1016/j.molcel.2017.11.019. [DOI] [PubMed] [Google Scholar]
- [43].Strickfaden H, McDonald D, Kruhlak MJ, Haince J-F, Th’ng JPH, Rouleau M, Ishibashi T, Coriy GN, Ausio J, Underhill DA, Poirier GG, Hendzel MJ, Poly(ADP-ribosyl)ation-dependent Transient Chromatin Decondensation and Histone Displacement following Laser Microirradiation., J. Biol. Chem. 291 (2016) 1789–802. doi: 10.1074/jbc.M115.694992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, Lukas J, Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose)., Nat. Commun. 6 (2015) 8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Leung AKL, Poly(ADP-ribose): An organizer of cellular architecture, J. Cell Biol. 205 (2014) 613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NMB, Jackson SP, Smith GCM, Ashworth A, Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy., Nature. 434 (2005) 917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- [47].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T, Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase., Nature. 434 (2005) 913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- [48].Feng FY, de Bono JS, Rubin MA, Knudsen KE, Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function., Mol. Cell. 58 (2015) 925–34. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG, PARP inhibition: PARP1 and beyond., Nat. Rev. Cancer. 10 (2010) 293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Eustermann S, Wu W-F, Langelier M-F, Yang J-C, Easton LE, Riccio AA, Pascal JM, Neuhaus D, Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1., Mol. Cell. 60 (2015) 742–54. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dawicki-McKenna JM, Langelier M-F, DeNizio JE, Riccio AA, Cao CD, Karch KR, McCauley M, Steffen JD, Black BE, Pascal JM, PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain., Mol. Cell. 60 (2015) 755–68. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Langelier M-F, Planck JL, Roy S, Pascal JM, Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity., J. Biol. Chem. 286 (2011) 10690–701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Langelier M-F, Pascal JM, PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis., Curr. Opin. Struct. Biol. 23 (2013) 134–43. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Langelier M-F, Zandarashvili L, Aguiar PM, Black BE, Pascal JM, NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains, Nat. Commun. 9 (2018) 844. doi: 10.1038/s41467-018-03234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Langelier M-F, Servent KM, Rogers EE, Pascal JM, A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation., J. Biol. Chem. 283 (2008) 4105–14. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- [56].Eustermann S, Videler H, Yang J-C, Cole PT, Gruszka D, Veprintsev D, Neuhaus D, The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger., J. Mol. Biol. 407 (2011) 149–70. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lilyestrom W, van der Woerd MJ, Clark N, Luger K, Structural and biophysical studies of human PARP-1 in complex with damaged DNA., J. Mol. Biol. 395 (2010) 983–94. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Langelier M-F, Planck JL, Roy S, Pascal JM, Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1., Science. 336 (2012) 728–32. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu L, Kong M, Gassman NR, Freudenthal BD, Prasad R, Zhen S, Watkins SC, Wilson SH, Van Houten B, PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1, Nucleic Acids Res. 45 (2017) 12834–12847. doi: 10.1093/nar/gkxl047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mansoorabadi SO, Wu M, Tao Z, Gao P, Pingali SV, Guo L, Liu H, Conformational activation of poly(ADP-ribose) polymerase-1 upon DNA binding revealed by small-angle X-ray scattering., Biochemistry. 53 (2014) 1779–88. doi: 10.1021/bi401439n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gagné J-P, Ethier C, Defoy D, Bourassa S, Langelier M-F, Riccio AA, Pascal JM, Moon KM, Foster LJ, Ning Z, Figeys D, Droit A, Poirier GG, Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs., DNA Repair (Amst). 30 (2015) 68–79. doi: 10.1016/j.dnarep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- [62].Steffen JD, McCauley MM, Pascal JM, Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage., Nucleic Acids Res. 44 (2016) 9771–9783. doi: 10.1093/nar/gkw710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Langelier M-F, Steffen JD, Riccio AA, McCauley M, Pascal JM, Purification of DNA Damage-Dependent PARPs from E. cob for Structural and Biochemical Analysis., Methods Mol. Biol. 1608 (2017) 431–444. doi: 10.1007/978-1-4939-6993-7_27. [DOI] [PubMed] [Google Scholar]
- [64].Langelier M-F, Riccio AA, Pascal JM, PARP-2 and PARP-3 are selectively activated by 5’ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1., Nucleic Acids Res. 42 (2014) 7762–75. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Grundy GJ, Polo LM, Zeng Z, Rulten SL, Hoch NC, Paomephan P, Xu Y, Sweet SM, Thorne AW, Oliver AW, Matthews SJ, Pearl LH, Caldecott KW, PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2BGlu2, Nat. Commun. 7 (2016) 12404. doi: 10.1038/ncomms12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Obaji E, Haikarainen T, Lehtiö L, Characterization of the DNA dependent activation of human ARTD2/PARP2., Sci. Rep. 6 (2016) 34487. doi: 10.1038/srep34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gibbs-Seymour I, Fontana P, Rack JGM, Ahel I, HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity., Mol. Cell. 62 (2016) 432–442. doi: 10.1016/j.molcel.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Xie S, Mortusewicz O, Ma HT, Herr P, Poon RYC, Helleday T, Qian C, Qian C, Timeless Interacts with PARP-1 to Promote Homologous Recombination Repair, Mol. Cell. 60 (2015) 163–176. doi: 10.1016/j.molcel.2015.07.031. [DOI] [PubMed] [Google Scholar]
- [69].Ryu KW, Kim D-S, Kraus WL, New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases., Chem. Rev. 115 (2015) 2453–81. doi: 10.102l/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y, Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors., Cancer Res. 72 (2012) 5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pommier Y, O’Connor MJ, de Bono J, Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action., Sci. Transl. Med. 8 (2016) 362psl7. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- [72].Hopkins TA, Shi Y, Rodriguez LE, Solomon LR, Donawho CK, DiGiammarino EL, Panchal SC, Wilsbacher JL, Gao W, Olson AM, Stolarik DF, Osterling DJ, Johnson EF, Maag D, Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors, Mol. Cancer Res. 13 (2015) 1465–1477. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- [73].Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, Matic I, Serine is a new target residue for endogenous ADP-ribosylation on histones, Nat. Chem. Biol. 12 (2016) 998–1000. doi: 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, Matic I, Serine ADP-Ribosylation Depends on HPF1., Mol. Cell. 65 (2017) 932–940. e6. doi: 10.1016/j.molcel.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, Ahel I, Serine ADP-ribosylation reversal by the hydrolase ARH3., Elife. 6 (2017). doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Abplanalp J, Leutert M, Frugier E, Nowak K, Feurer R, Kato J, Kistemaker HVA, Filippov DV, Moss J, Caflisch A, Hottiger MO, Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase, Nat. Commun. 8 (2017) 2055. doi: 10.1038/s41467-017-02253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I, Serine is the major residue for ADP-ribosylation upon DNA damage., Elife. 7 (2018). doi: 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, Matkarimov BT, Gasparutto D, Saparbaev MK, Lavrik OI, Ishchenko AA, Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro., Nucleic Acids Res. 44 (2016) 9279–9295. doi: 10.1093/nar/gkw675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, Biard D, Gasparutto D, Lavrik OI, Ishchenko AA, Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation., Nucleic Acids Res. (2018). doi: 10.1093/nar/gkxl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Belousova EA, Ishchenko AA, Lavrik OI, Dna is a New Target of Parp3., Sci. Rep. 8 (2018) 4176. doi: 10.1038/s41598-018-22673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Munnur D, Ahel I, Reversible mono-ADP-ribosylation of DNA breaks, FEBS J. 284 (2017) 4002–4016. doi: 10.1111/febs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Jankevicius G, Ariza A, Ahel M, Ahel I, The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA., Mol. Cell. 64 (2016) 1109–1116. doi: 10.1016/j.molcel.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC, The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates., Nature. 443 (2006) 713–6. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- [84].Tumbale P, Appel CD, Kraehenbuehl R, Robertson PD, Williams JS, Krahn J, Ahel I, Williams RS, Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease., Nat. Struct. Mol. Biol. 18 (2011) 1189–95. doi: 10.1038/nsmb.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Satoh MS, Lindahl T, Role of poly(ADP-ribose) formation in DNA repair, Nature. 356 (1992) 356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]