Abstract
Bisphenol A (BPA) is a ubiquitous endocrine-disrupting chemical. Developmental exposure produces changes in behavior and gene expression in the brain. Here, we examined social recognition behaviors in mice from the third familial generation (F3) after exposure to gestational BPA. Second-generation mice were bred in one of four mating combinations to reveal whether characteristics in F3 were acquired via maternal or paternal exposures. After repeated habituation to the same mouse, offspring of dams from the BPA lineage failed to display increased investigation of a novel mouse. Genes involved in excitatory postsynaptic densities (PSDs) were examined in F3 brains using quantitative PCR. Differential expression of genes important for function and stability of PSDs were assessed at three developmental ages. Several related PSD genes―SH3 and multiple ankyrin repeat domains 1 (Shank1), Homer scaffolding protein 1c (Homer1c), DLG associated protein 1 (Gkap), and discs large MAGUK scaffold protein 4 (PSD95)―were differentially expressed in control- vs BPA-lineage brains. Using a second strain of F3 inbred mice exposed to BPA, we noted the same differences in Shank1 and PSD95 expression in C57BL/6J mice. In sum, transgenerational BPA exposure disrupted social interactions in mice and dysregulated normal expression of PSD genes during neural development. The fact that the same genetic effects were found in two different mouse strains and in several brain regions increased potential for translation. The genetic and functional relationship between PSD and abnormal neurobehavioral disorders is well established, and our data suggest that BPA may contribute in a transgenerational manner to neurodevelopmental diseases.
Bisphenol A (BPA) is a ubiquitous endocrine-disrupting compound (EDC) used to stabilize plastics and produce epoxy resins. It is present in a wide variety of daily use items, such as the ink on thermal receipts, plastic bottles, and linings in canned foods. Not surprisingly, when examined, detectable levels are present in the urine or blood of the majority of humans (1–4). In several strains of laboratory rodents, neonatal exposure to BPA has well-documented effects on behaviors, including anxiety, aggression, learning, and social interactions (5–10). Epidemiological studies of human populations report associations between exposure to environmental contaminants, including BPA, and increased likelihood of mild to more extreme neurobehavioral issues, including attention-deficit/hyperactivity disorder and autism (11–15). These data and others have increased public concerns about the potential dangers of BPA. In response, some manufacturers have replaced this component and are now using structurally similar but largely untested compounds. To date, animal studies using these replacement compounds have shown that their effects in first familial generation (F1) offspring are similar to or more severe than the effects of BPA (16–19).
BPA has documented transgenerational effects on social behavior, heart, and reproductive functions; thus, removing BPA from all products will not eliminate long-term effects (20–24). It is important to keep in mind that with gestational exposure to any substance, the offspring (F1) have experienced direct actions of the compound during development. When they mature and breed, the second familial generation (F2) is produced from germ cells that were likewise exposed to the substance during F1 gestation. We examined the third familial generation (F3), as these are the first offspring without any direct exposure to BPA. Effects in these animals are transgenerational and are expected to continue in subsequent generations.
The fundamental causes of mental health disorders are deficits and abnormalities in neural function, which are likely produced by a large number of mechanisms. BPA has been associated with alterations in spine postsynaptic densities (PSDs) (25–28) and synaptic plasticity (29). Effects of BPA may, at least in part, be caused by perturbation of ionotropic and/or metabotropic glutamate receptors (29–31). At the ultrastructural level, the combination of prenatal and postnatal BPA exposure produced thickening of the PSDs and increased the curvature of synapses in hippocampal pyramidal cells (32). In male mice ranging in age from postnatal day (PN) 14 to PN56, several PSD-related proteins were reduced by perinatal exposure to BPA (32, 33).
Previously, we reported that F3 mice from a BPA lineage differed from controls in a social recognition task. BPA lineage mice displayed more investigatory behavior initially when paired with another mouse, and after investigation decreased over repeated exposures, they failed to display dishabituation when a novel mouse was presented (23). Here, we asked if these behavioral differences between F3 mice from BPA and control lineages could be traced to maternal and/or to paternal BPA exposure. We replicated our social recognition data, with additional groups of mice prepared by breeding a subset of BPA-lineage mice to controls (in both configurations of dams and sires) (Fig. 1A). Next, on the basis of candidate genes discovered in a prior RNA-sequencing data set from F3 brains (20), we tested the hypothesis that changes in PSD genes would differ between mice from control and BPA F3 lineages. We assessed the expression of four genes in which protein products form physical contacts in the PSD. SH3 and multiple ankyrin repeat domains 1 (SHANK1) is a master scaffolding protein in association with metabotropic glutamate receptors (mGluRs) that bridges to HOMER1 proteins by linking with PSD95 (also called DLG4) and GKAP (also called SAPAP). These proteins produce a functional link between N-methyl-d-aspartate and glutamate receptors in the PSD (34). Our results showed that maternal exposure to BPA has transgenerational effects on behavior, and we suggest that changes in genes and proteins in PSDs cause these alterations in male mice.
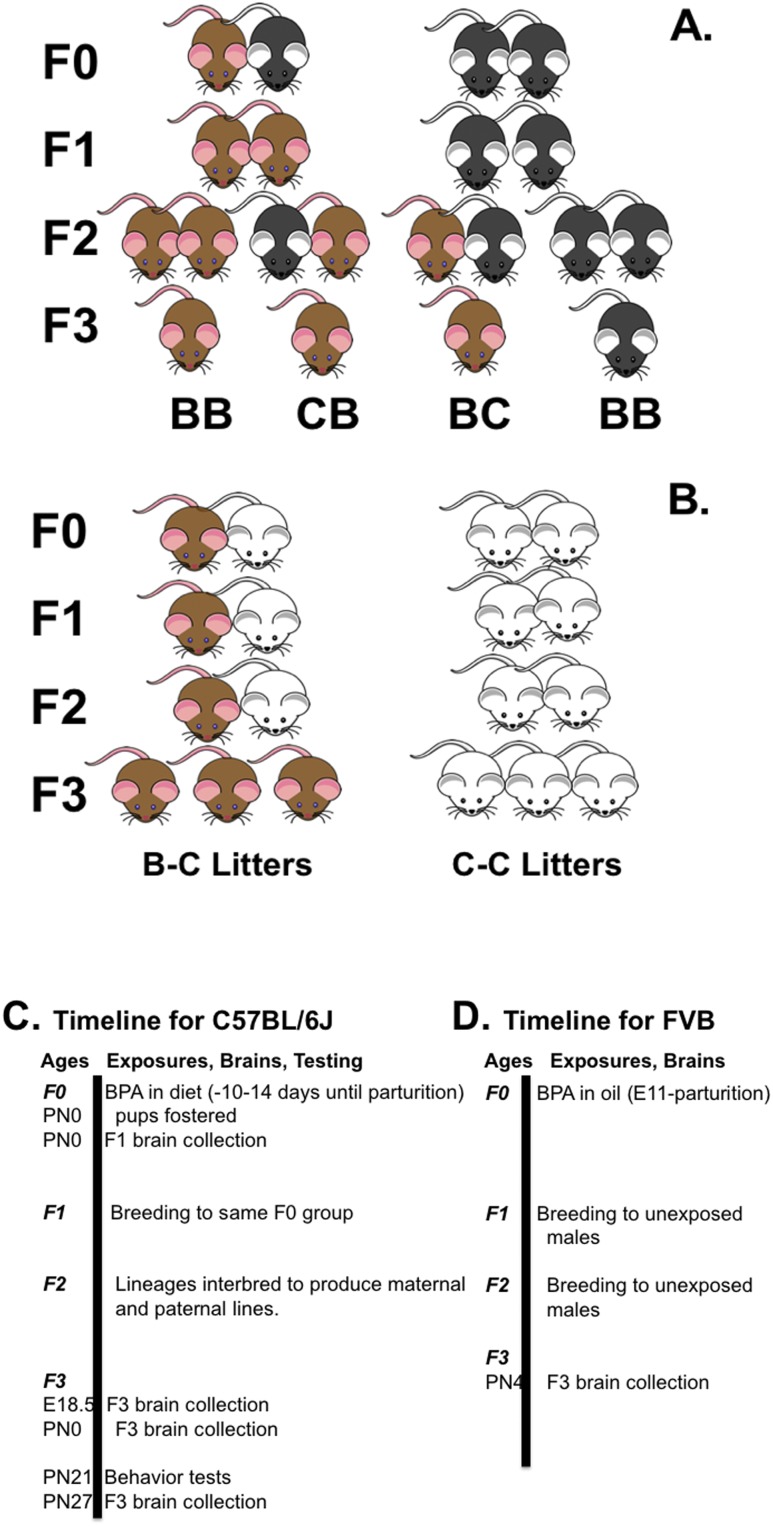
Figure 1.
Breeding crosses used to produce mice. (A) The C57BL/6J mating paradigm is shown. Brown mice represent direct or historic BPA exposure. Black mice represent controls (C57BL/6J mice have black coats). (B) The FVB cross is shown. Again, brown mice represent BPA exposure, and white mice are controls (FVB mice have white coats). By convention, dams are shown on the left. F3 is the first generation capable of displaying transgenerational effects. (C and D) Timelines are presented for BPA treatment to F0 dams, brain collection, and testing. (C) The C57BL/6J studies are shown. (D) The FVB protocol is illustrated. BB, bisphenol A lineage pair; BC, dam from BPA lineage and sire from control lineage; CB, dam from control lineage and sire from bisphenol A lineage; E, embryonic day; F0, parental generation.
Methods
Animals: C57BL/6J mice
Most of the experiments were conducted with mice that were progeny of C57BL/6J mice purchased from Jackson Laboratory (Bar Harbor, ME). Adult females were randomly assigned to one of two diets, either a phytoestrogen-free chow (TD95092; Harlan Teklad, Madison WI) or the same chow supplemented with 5 mg of BPA per kilogram of diet (TD09386; Harlan Teklad). We have estimated that this dose in food results in an average daily consumption during pregnancy of 20 μg of BPA (22). Female mice were placed on diets 10 days before pairing with a male. Pairs remained together for up to 2 weeks. All mice consumed food and water ad libitum; dams continued on their diets throughout gestation. Lights were on a 12:12 light/dark cycle (lights off at 1200 hours). During the dosing period, mice were observed daily for abnormal behavior and signs of toxicity.
As in our previous studies (22, 23, 35), all F1 offspring (produced by seven control and six BPA-consuming dams) were fostered within 24 hours of birth to dams on the control diet. Four littermates (two of each sex) were fostered, and dams retained two of their own pups, which were not used in our studies. At weaning (PN21), mice were placed on standard chow (7912; Harlan Teklad) containing phytoestrogens and were group housed (three to five per cage) by litter and sex. Adult F1 males and females (nonsiblings) were mated (10 control pairs and seven BPA-exposed pairs) to produce F2 mice. As adults, F2 mice were paired in one of four combinations. Either mice from control lineages were paired (CC; n = 10 pairs), or mice from BPA lineages were paired (BB; n = 10 pairs) as we did in the past. To determine whether the effects of BPA are inherited from the dam or the sire, we also made pairs with either a dam from the BPA lineage and a sire from the control lineage (BC; n = 10 pairs) or the reverse (CB; n = 13 pairs) (Fig. 1A and 1C). As juveniles, a subset of the F3 mice was tested for behavior. No more than one mouse of each sex per litter was used for each experiment (CC: n = 14 males, 15 females; CB: n = 20 males, 16 females; BC: n = 14 males, 13 females; BB: n = 13 males, 17 females). All animal procedures were approved by the North Carolina State University Institutional Animal Care and Use Committee and were in accordance with National Institutes of Health and National Academy of Sciences guidelines.
Animals: Friend virus B mice
We validated and extended our findings by including a second inbred strain, FVB (Charles River), which was produced in a different manner. All mice received food containing phytoestrogens (Rodent Diet 8604; Harlan Teklad)) and water ad libitum. Using an out-crossed design, female FVB mice were mated with males at 12 weeks of age. Pregnancy was confirmed by the appearance of a vaginal sperm plug, and at that time, females were separated from the males and individually caged. At embryonic day (E) 9, each pregnant female was randomly assigned to a treatment group (six to nine for each condition). From E11 to birth, dams were orally dosed once per day in the early part of the lights-on phase with tocopherol-stripped corn oil or oil with one of three BPA doses (0.5, 20, or 50 µg/kg/d). The solution was administered into the corner of the mouth, as described in previous studies (24, 36). During the dosing period, mice were observed for abnormal behavior and signs of toxicity. F1 females were used to generate F2 females, and F2 females were used to generate F3 offspring. In these experiments, control and exposed F1 and F2 females (at least three per conditions, each from a different litter) were mated with fertility-confirmed, non‒BPA-exposed males to produce the next generation. Thus, any differences between BPA and control lineages had to be attributed to maternal inheritance. Using the terminology introduced previously, F3 mice were in either the CC or the BC group (Fig. 1B and 1D). No more than one mouse of each sex per litter was used for each experiment. All animal procedures were approved by the University of Illinois Institutional Animal Care and Use Committee and were in accordance with National Institutes of Health and National Academy of Sciences guidelines.
Social recognition tests
Between PN21 and PN23, juvenile F3 C57BL/6J mice were tested for social recognition according to previously described methods (23, 37, 38). Mice habituated to test boxes for at least 30 minutes. During the habituation period, an empty “cylinder” was placed in the cage. All trials were scored live and conducted in the lighted portion of the 12-hour light/-hour dark cycle. Nine investigation trials, 1 minute each, were separated by 9 minutes. During the first eight trials, the same stimulus C57BL/6J ovariectomized (OVX) mouse was placed into the test box under the cylinder. On the ninth trial, a novel OVX mouse was used. The total amount of time each young test mouse spent investigating the cylinder and/or the stimulus female was recorded. Investigation was defined as contact with the head, tail, or body of the stimulus mouse at a distance of 1 cm or less or directly touching the bars of the cylinder. Test boxes were cleaned with 70% ethanol and wiped dry between tests. All behavioral tests were scored by observers who were blinded to the animal lineages.
Brain collection
Pups and embryos were euthanized by cooling on ice, followed by cervical dislocation. Brains collected from F3 CC and BB litters on E18.5 and PN0 were dissected into regions of interest. Here, we present data from the hypothalami for quantitative polymerase chain reaction (qPCR) and from the amygdala for Western blotting (only from E18.5 brains). From CC and BC pairs of F3 FVB mice, we used whole brains collected on PN4. We also examined gene expression in whole brains from newborn F1 C57BL/6J male pups. The oldest (F3, PN27) mice were euthanized with CO2 and cervical dislocation. Brains were rapidly dissected and frozen. At a later time, brains were sectioned in a cryostat and mounted onto glass plus slides. Coronal thick (120-μM) sections were punched (21 gauge) to collect the bilateral lateral septum. We collected this region because it is a major terminal field for vasopressin fibers, and this neuropeptide is one of the essential regulators of social recognition behavior (39, 40). For this experiment, CC, CB, BC, and BB juveniles were examined. For the gene expression and protein experiments, no more than one mouse of each sex per litter was used for each experiment.
Quantitative RT-PCR
We examined mRNA expression of SH3 and multiple ankyrin repeat domains 1 (Shank1), Homer scaffolding protein 1- c (Homer1c), DLG associated protein 1 (Gkap), and discs large MAGUK scaffold protein 4 (Dlg4, also called PSD95). We present the ages, groups, sexes, brain regions, and numbers per group in Table 1. Primers for specific targets were designed to amplify all known variants. Each set of primers was initially tested for efficiency (between 95% and 105%) and specificity using melting curve analysis to verify that one amplicon was represented by a single peak (Table 2). For data evaluation, the comparative ΔΔCt method was used. Each sample was analyzed in triplicate. Expression levels of target genes were normalized to endogenous control β-2-microglobulin or 60S ribosomal protein L19. A calibrator sample was run on each plate to adjust for plate-to-plate variation. Samples with Ct values >35 cycles, as well as outliers identified as samples with values above (or below) the 1.5-fold of the interquartile range from the third (or the first) quartile, were excluded from the analyses.
Table 1.
Description of Groups Used for qPCR
| Age | Generation | Sex-Group (Maximum Number per Group) | Region | Strain |
|---|---|---|---|---|
| E18.5 | F3 | CCM (8), BBM (8), CCF (9), BBM (8) | Hypothalamus | C57BL/6 |
| PN0 | F3 | CCM (7), BBM (6), CCF (6), BBF (8) | Hypothalamus | C57BL/6 |
| PN0 | F1 | CCM (4), BCM (6) | Whole brain | C57BL/6 |
| PN4 | F3 | CCM-0 (5), BCM-0.5 (5), BCM-20 (5), BCM-50 (5), | Whole brain | FVB |
| CCF-0 (6), BCF-0.5 (5), BCF-20 (5), BCF-50 (5) | ||||
| PN27 | F3 | CCM (6), CBM (6), BCM (6), BBM (6), CCF (5), CBF (6), BCF (6), BBF (6) | Lateral septum | C57BL/6 |
Experimental groups used for qPCR. In the FVB study, numbers following group designations are the doses (in micrograms) of BPA (per kilogram of body weight) consumed by F0 dams per day.
Abbreviations: BB, both parents from BPA line; BC, dam BPA line/sire control line; CB, dam control line/sire BPA line; CC, both parents from control line; F; female; M, male.
Table 2.
Primers for qPCR
| Target | Primer Sequence | Amplicon Size, bp | Targeted mRNA Variants |
|---|---|---|---|
| Shank1 | F: 5′-AGCTCCTGGACACCTACGTT-3′ | 168 | XM_006540868.3 |
| R: 5′-GATCTCGAAGTCCCCCACTG-3′ | XM_006540865.3 | ||
| XM_006540866.3 | |||
| R: 5′-TGTTTGGAGCCTGTGTTCTTTG-3′ | XM_006540867.3 | ||
| R: 5′-CCTGGAGGATCCTGAGTGGA-3′ | XM_006540868.3 | ||
| XM_006540869.2 | |||
| XM_006540870.2 | |||
| XM_006540872.1 | |||
| XM_006540874.1 | |||
| XM_006540871.2 | |||
| XM_006540875.3 | |||
| XM_006540876.3 | |||
| Homer1c | F: 5′-GAGAAGTCGCAGGAGAAGATG-3′ | 249 | NM_147176.3 |
| R: 5′-TTGCTGAACTAGCATGAGAGAG-3′ | |||
| Gkap1 | F: 5′-AAGGTGGCTGCAAGAAGAGA-3′ | 136 | NM_177639.6 |
| R: 5′-CACTTACGGCCCTCAGGTAG-3′ | |||
| Psd95 | F: 5′-GCCCCAGGATATGTGAACGG-3′ | 204 | XR_0017779872.1 |
| R: 5′-GATGCTGTCGTTGACCCTGA-3′ | |||
| B2M | F: 5′-GGCTCACACTGAATTCACCCCCAC-3′ | 104 | NM_009735.3 |
| R: 5′-ACATGTCTCGATCCCAGTAGACGGT-3′ | |||
| Rpl19 | F: 5′-GAAGGTCAAAGGGAATGTGTTCA-3′ | 74 | NM_009078.2 |
| R: 5′- CCTTGTCTGCCTTCAGCTTGT-3′ | NM_001159483.1 |
Primer sequences used for qPCR and variant coverage.
Abbreviations: B2M, β-2-microglobulin; F, forward primer; R, reverse primer; Rpl19, 60S ribosomal protein L19.
Western blots
We examined protein expression of SHANK1 and MGLUR5 in E18.5 amygdala from F3 males (n = 5 to 7 per group) and females (n = 6 to 7 per group) from the CC and BB groups using Western blotting techniques (41). Briefly, protein was homogenized in radioimmunoprecipitation assay buffer and quantified using a BCA assay (Pierce, Rockford, IL). Total protein (20 μg) was electrophoresed on Tris-acetate gels and transferred to polyvinylidene difluoride membranes; blots were incubated with SHANK1 [1:250 rabbit; NB300-167, lot 6162; Novus Biologicals, Littleton, CO (42)], MGLUR5 [1:5000 rabbit; ab76316, lot GR3241176-4; Abcam, Cambridge, MA (43)], or β-actin [1:10,000 mouse; ab8226, lot 085M4754V; Abcam (44)] and visualized using horseradish peroxidase antibodies and enhanced chemiluminescence (Thermo Fisher Scientific, Waltham, MA). Images were quantified using ImageJ (V2.0). Data are reported as mean ± SEM of band intensity.
Statistics
All data were analyzed using the Number Cruncher Statistical System v.11 (Kaysville, UT). To analyze social recognition, we used two-way repeated-measures ANOVA, with maternal diet history and paternal diet history as main factors and trials as the repeated measure. Data from trials 1 to 8 (habituation) and trial 9 (dishabituation) were subjected to two-way repeated-measures ANOVA, separately for each sex. Data from the qPCR and Western blots were analyzed within each age. We used two-way ANOVA when the two factors were diet lineage and sex. The PN27 brains were analyzed with three-way ANOVA using paternal diet history, maternal diet history, and sex as main factors. After ANOVA, significant main effects or interactions were further assessed with Fisher’s exact post hoc tests, which adjust significance levels to account for multiple comparisons. Data on gene expression in F1 male brains were analyzed with Student t tests.
Results
Social recognition
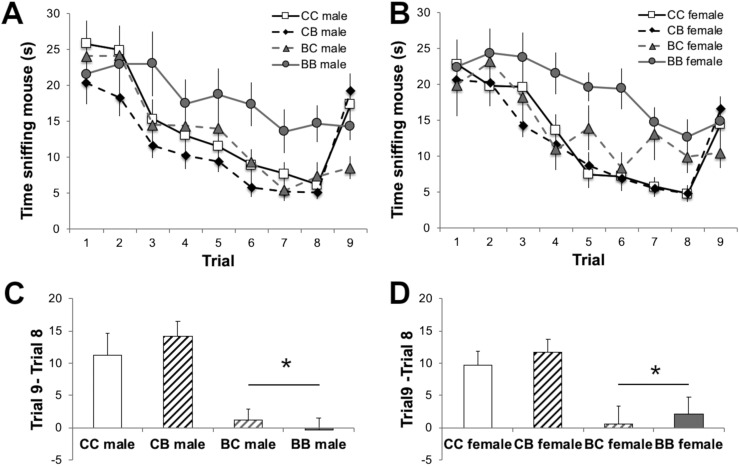
Our results show that gestational BPA exposure had transgenerational effects on social recognition behavior. Moreover, these effects can be attributed to the parent’s EDC lineage. During habituation trials when the same stimulus animal was presented on each trial (1–8), a main effect of parental lineage was noted [males: F(3, 399) = 4.27; P < 0.01; females: F(3, 399) = 6.33; P < 0.001]. Social investigation of an OVX adult by juveniles was greatest in male and female mice with both parents from BPA lineages (BB). Thus, BB mice were more active and took longer to habituate than other mice from different combinations of parental lineages (Fig. 2A and 2B). To examine dishabituation, we subtracted the time of investigation in trial 8 (with the familiar stimulus mouse) from the time of investigation in trial 9 (with a novel mouse) (Fig. 2C and 2D). In both males and females, a significant main effect of maternal diet lineage was found [males: F(1, 57) = 23.66; P < 0.001; females: F(1, 57) = 13.92; P < 0.001]. Two of the four groups of F3 mice, CC and CB, displayed the expected dishabituation: increased investigation directed toward the novel mouse in trial 9. Juveniles whose dams were from the BPA lineage (BC and BB) failed to show enhanced investigation toward a novel mouse (Fig. 2C and 2D).
Figure 2.
Mean ± SEM time in seconds spent investigating an adult ovariectomized mouse during a social recognition task. (A and B) Time juvenile F3 (A) male and (B) female mice spent investigating the stimulus mouse on each trial, including the last trial, when a novel stimulus mouse was presented. (C and D) Time spent by (C) males and (D) females investigating a novel stimulus in trial 9 minus the time spent investigating a familiar mouse in trial 8. *Significant main effect of maternal diet history, P < 0.05.
Gene and protein expression
We present these results in chronological order beginning with analysis of the E18.5 tissues. We probed up to four genes; in some cases, we had limited RNA; thus, not all of the target genes were assessed in all experimental groups.
Embryonic (E18.5) brain, F3 in C57BL/6J mice: RNA and protein
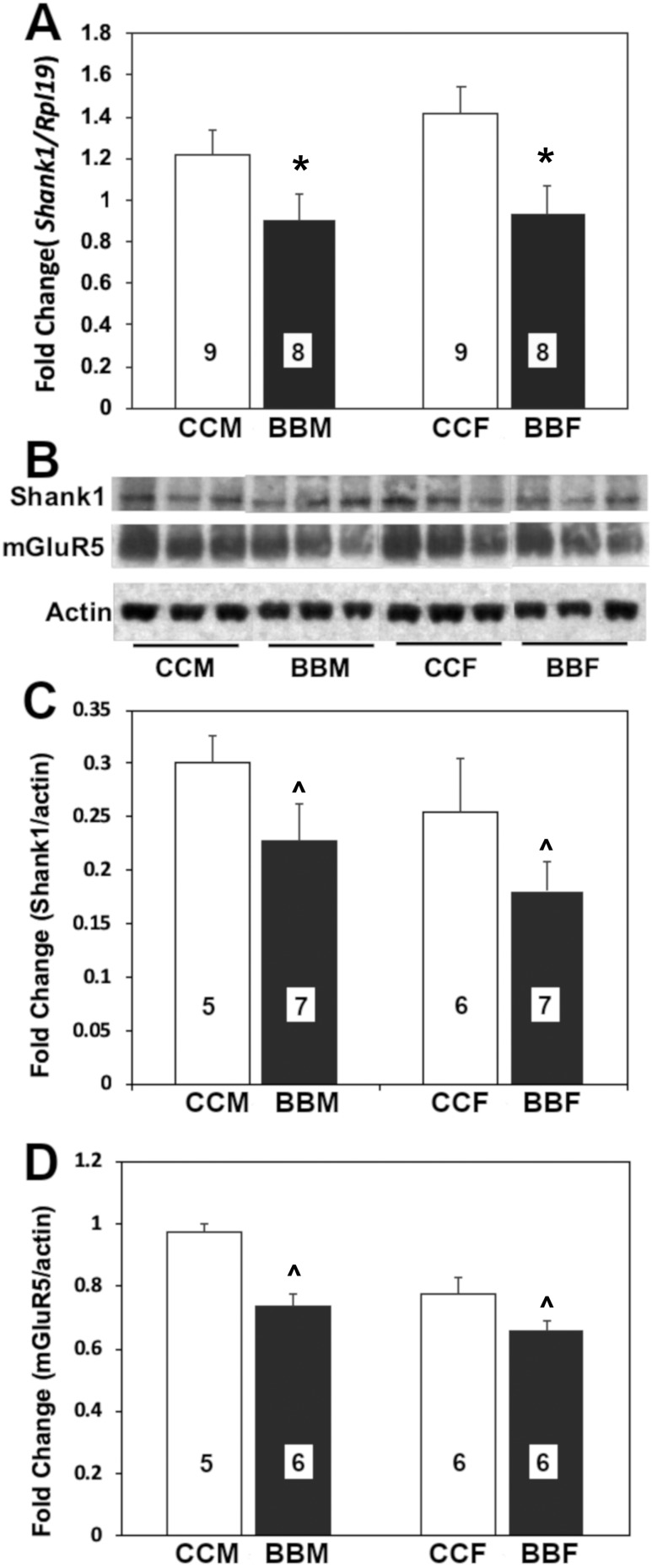
Embryos from the BPA lineage had significantly less transcript of Shank1 [F(1, 33) = 9.50; P < 0.004] (Fig. 3A) than controls. No effect of sex was found, nor was there an interaction.
Figure 3.
Mean + SEM fold-changes in (A) Shank1 gene expression, (B and C) SHANK1 protein, and (B and D) MGLUR5 from F3 embryonic (E18.5) male (M) and female (F) mice from control or BPA lineage. (A) Control male data were set to 1.0, and other values were normalized; tissue assayed was the hypothalamus. (B‒D) Tissue was from the amygdala. Numbers per group are given in the histograms. ^Trend for a difference by lineage. *Significant difference by lineage, P < 0.05. BBF, BPA lineage female; BBM, BPA lineage male; CCF, control female; CCM, control male.
Embryos from the BPA lineage tended toward less SHANK1 protein expression [F(1, 24) = 4.07; P = 0.057] (Fig. 3B and 3C) than controls. No effect of sex was found, nor was there any interaction. Protein levels of MGLUR5 also tended to be lower in the BPA than control E18.5 amygdala [F(1, 22) = 1.92; P = 0.182] (Fig. 3B and 3D). There was no effect of sex, nor was an interaction present between lineage and sex.
Day-of-birth hypothalamus, F3 in C57BL/6J mice
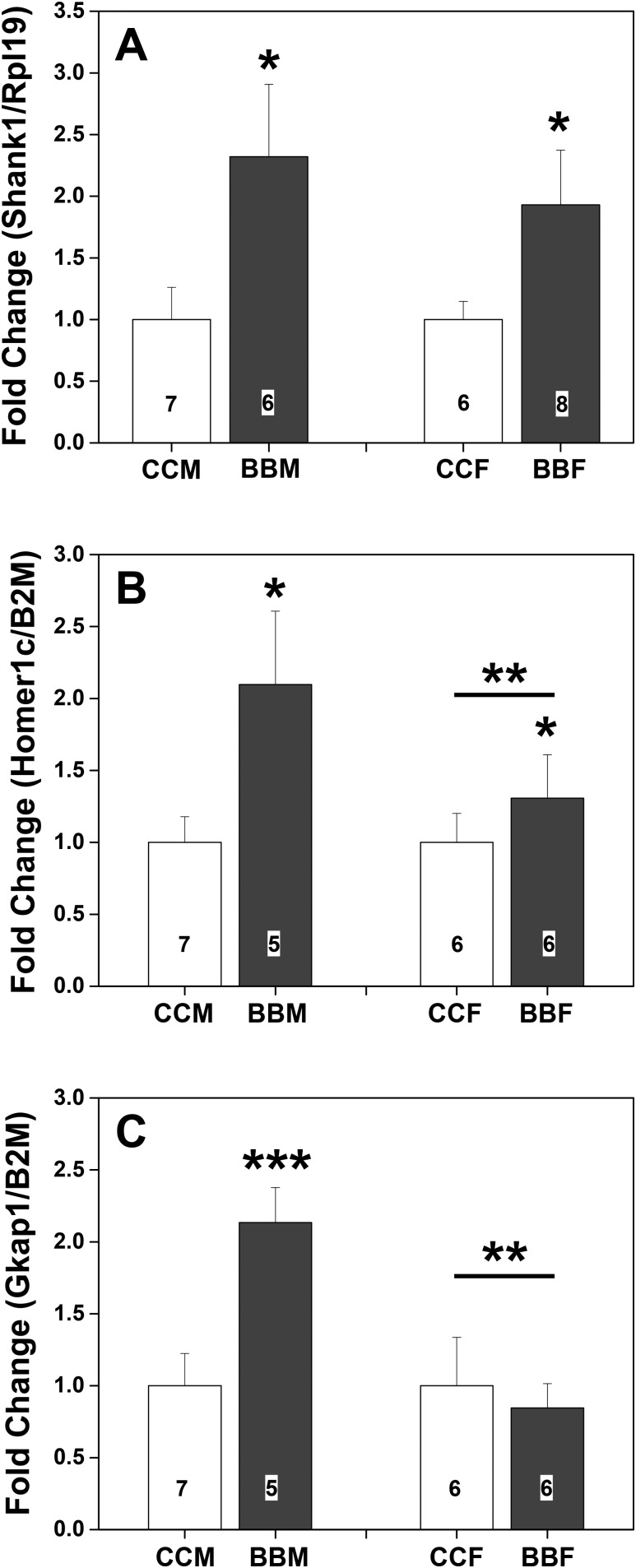
Hypothalami from PN0, BPA-lineage, F3 C57BL/6J males and females were evaluated for Shank1 [F(1, 26) = 7.89; P < 0.01], Homer1c [F(1, 21) = 9.87; P < 0.006], and Gkap [F(1, 22) = 3.88; P = 0.06] (Fig. 4) mRNA. In all cases, BPA history was reflected in higher levels of expression (Fig. 4). Significant sex differences in Homer1c [F(1, 21) = 11.75; P < 0.003] and Gkap [F(1, 22) = 7.67; P < 0.01] were produced by higher levels of mRNA in male than in female brains. A trend for an interaction in Homer1c [F(1, 21) = 4.10; P = 0.058] and a significant interaction for Gkap [F(1, 22) = 6.50; P < 0.02] were caused by greater gene expression in hypothalami from males with a history of BPA exposure. No other effects of sex, diet history, or interactions were noted.
Figure 4.
Mean + SEM fold-changes in gene expression in hypothalami from day of birth (PN0) in F3 male (M) and female (F) mice from control or BPA lineage. (A) Levels of Shank1 RNA. (B) Levels of Homer1c RNA. (C) Levels of Gkap RNA. Control male data were set to 1.0. Numbers per group are given in the histograms. *Significantly different from control lineage. **Significant sex difference. ***Significantly different from all other groups, P < 0.05. BBF, BPA lineage female; BBM, BPA lineage male; CCF, control female; CCM, control male.
Day-of-birth whole brain, F1 in C57BL/6J mice
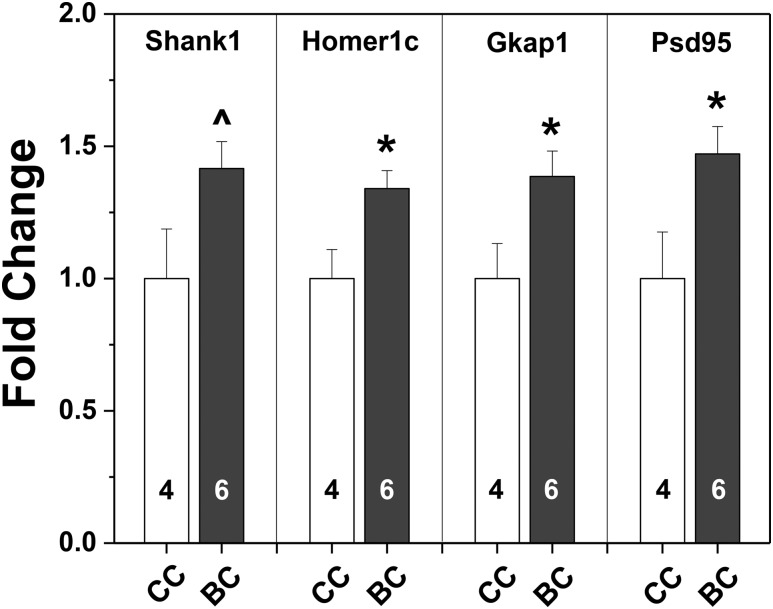
Two groups of whole male brains from the day of birth revealed elevated expression of all four scaffolding protein genes after BPA exposure in utero (Fig. 5). Significant differences based on two-tailed t tests were found for Homer1c, Gkap, and Psd95 [t (9) = 2.80, 2.42, and 2.48, respectively; P < 0.04 at least]. Shank1 expression levels were significant only when a one-tailed t test was used [t (9) = 2.14; P < 0.03].
Figure 5.
Mean + SEM fold-changes in gene expression in the whole brains of F1 male mice collected on the day of birth (PN0) from control (CC white bars) or BPA-exposed dams (BC black bars). Control male data were set to 1.0. Numbers per group are given in the histograms. *Significantly different from controls (BC), P < 0.05. ^Significantly different from controls using one-tailed t test, P < 0.03.
Neonatal (P4) whole brains in FVB mice
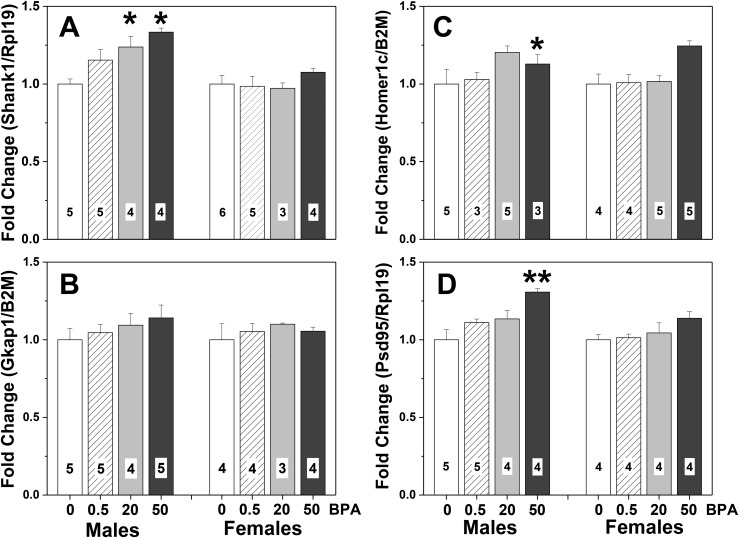
Maternal diet history was a significant variable for all the scaffolding protein genes except Gkap (Fig. 6). Whole brains from FVB F3 BC pups had significantly increased mRNA for Shank1 [F(1, 38) = 4.99; P < 0.006], Homer1c [F(1, 37) = 4.66; P < 0.01], and Psd95 [F(1, 33) = 8.40; P < 0.0005] (Fig. 6). In all cases, mice from control dams had lower levels of gene expression than mice in the highest-dose BPA lineage (P < 0.05). A trend for an effect of sex on Psd95 mRNA levels [F(1, 33) = 3.66] was noted, with the effect for males greater than that for females. Another trend was present for an interaction between dose history and sex for Homer1c [F(1, 37) = 2.58; P = 0.07]. No other effects of sex, diet history, or interactions were noted.
Figure 6.
Means + SEM fold changes in gene expression in whole brains of F3 4-day-old (PN4) FVB mice from one of four BPA dose lineages. (A) Expression levels of Shank1. (B) Expression levels of Gkap. (C) Expression levels of Homer1c. (D) Expression levels of PSD95. Control (CC) male data were set to 1.0. White bars represent data from mice with no BPA exposure, control dams and sires (CC). Striped bars show data from F3 mice produced by dams from the 0.5 μg/kg/d BPA dose lineage; sires were unexposed to BPA. Gray bars represent data from F3 mice from the medium (20 μg/kg/d) BPA dose lineage and control sires (BC). Black bars show data from F3 mice produced by dams from the high-dose lineage (50 μg/kg/d) and control sires. Numbers per group are given in the histograms. *Significantly different from control-lineage males, P < 0.05. **Significantly different from all other doses, same sex, P < 0.05.
Juvenile (P27) lateral septum in C57BL/6J mice
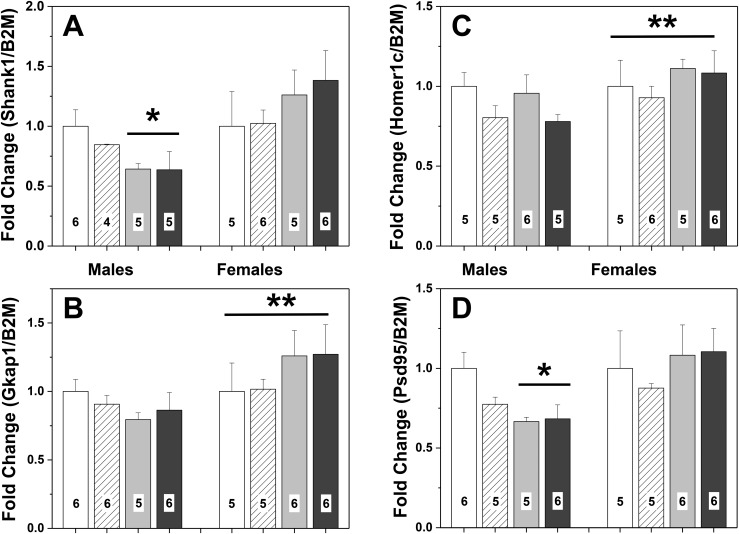
Sex and maternal diet history had pronounced effects on gene expression in the lateral septum. Interactions between the diet history of the dam and the sex of the F3 offspring were found for Shank1 [F(1, 41) = 6.78; P < 0.015] and Psd95 [F(1, 43) = 5.23; P < 0.03], with a trend noted for Gkap [F(1, 44) = 3.65; P = 0.064] expression. For both significant effects, control male brains had greater gene expression than control females or males with maternal BPA history (P < 0.05). Sex differences, males greater than females, were present for Homer1c [F(1, 42) = 12.56; P < 0.001] and Gkap [F(1, 44) = 14.62; P < 0.0005]. In addition, Homer1c expression was higher in mice sired by males from the control than from the BPA lineage [F(1, 42) = 5.15; P < 0.03] (Fig. 7). No other significant main effects or interactions were found. The mice used for qPCR were not tested for behavior, but were littermates of tested animals.
Figure 7.
Mean + SEM fold-changes in gene expression in the lateral septum of F3 mice (PN27) from one of four lineages. (A) Expression levels of Shank1. (B) Expression levels of Gkap. (C) Expression levels of Homer1c. (D) Expression levels of PSD95. Control (CC) male data were set to 1.0. White bars represent data from males and females with control dams and sires (CC). Striped histograms show data from F3 mice that had dams from the control lineage and sires from the BPA lineage (CB). Gray histograms represent data from F3 mice with BPA-lineage dams and control sires (BC). Black bars show data from F3 mice with both parents from the BPA lineage (BB). Numbers per group are given in the histograms. *Significant effect of maternal lineage in males. BC and BB < CC and CB. BC and BB lineages are less than males from CC and CB lineages, P < 0.05. **Significant sex difference, P < 0.03.
Discussion
Here, we replicated previously reported transgenerational actions of BPA on social recognition (22, 23). In addition, we determined that hyperinvestigation during habituation was restricted to F3 mice with both parents from a BPA ancestry. The lack of dishabituation noted in F3 BPA-lineage mice was caused by maternal BPA transmission. Furthermore, we found that transgenerational exposure to BPA produced significant differences in several genes that are functionally and structurally related to excitatory postsynaptic synapses. Within this data set, we also showed that our data generalized to other mouse strains, which experienced different BPA administration paradigms. Because F1 brains exposed to BPA have comparable differences to F3 brains, we hypothesize that the epigenetic modifications produced by BPA directly are propagated over generations. Our results support the hypothesis that environmental chemicals may contribute to increases in social behavior abnormalities and disorders in humans (45–47).
Previously, we reported that F3 offspring descended from F1 mice originally exposed to BPA during gestation were more active in juvenile interaction tests than controls (22). In a social recognition test, similarly generated mice were slower to habituate, investigating the stimulus mice longer during the repeated presentations than control mice did (23). Using four combinations of mating pairs in the second generation (F2), we determined that enhanced activity (delayed habituation) was restricted to progeny from maternal and paternal BPA lineages. Increased investigation times during habituation may reflect increased activity; in fact, F3 BPA-lineage offspring have increased locomotor activity in the open field compared with control-lineage offspring (23). A dose-response study, also conducted in C57BL/6J mice and using a diet similar to ours, found that F1 mice exposed to BPA in utero and during lactation had increased activity and higher metabolism than controls (5). In other studies of F1 BPA-exposed rodents, effects of BPA on activity were inconsistent, varying with sex, species, and dose (48–51). Several studies in children reported a strong association between attention-deficit/hyperactivity disorder and levels of BPA in children’s urine (12, 13).
The other facet of social recognition is dishabituation, scored in the final trial when a novel mouse was presented. Only mice from maternal BPA lineages failed to discriminate a novel mouse from the familiar mouse in the final trial. This failure to dishabituate cannot be attributed to olfactory deficits because we tested F3 BPA and control mice previously for olfactory discrimination with mouse urine (23). Although we know of no behavioral data on BPA-exposed FVB mice, we hypothesized that their social recognition behavior would parallel that of the C57BL/6J mice with maternal BPA history, as the FVB mice were bred through the maternal lineage only.
To date, two other EDCs have been assessed for multigenerational or transgenerational actions on rodent behavior. The Crews/Skinner laboratories tested rats in the F3 generation; exposure to the pesticide vinclozolin was given via injection to F0 dams in gestation (51–54). Of note, mate choice (assessed by time spent near the other rat) was altered in F3 females, but not in males. Some aspects of anxiety were affected in F3 rats, particularly in females, and stress reactivity was changed in both sexes. In these studies, the behavioral outcomes were inherited via the sires. Recently, a lower dose of vinclozolin or a low dose of the polychlorinated bisphenyl A1221 was injected during gestation, and F2 adults from maternal or paternal lineage were tested for ultrasonic vocalizations or sexual behaviors (54). Of note, behavioral effects were produced only in the paternal lineage. Vocalizations in males were reduced by gestational exposure to vinclozolin, and a principle component analysis also revealed an effect of A1221. Male intromission frequencies were increased by both EDCs, and ejaculation latency was significantly reduced by A1221. Small but significant effects were also noted on female sexual behaviors. In toto, these data show that several classes of EDCs produce long-term behavioral abnormalities.
To date, no other laboratory has used the social recognition task in rodents to assess behavioral actions of BPA, although a similar “social preference or choice” task (55, 56) is considered the gold standard for social behavior. The social recognition test in mice has good validity for humans, as it is similar to facial recognition studies that reported deficits in facial (but not object) memory in autism spectrum disorders (ASDs) (57) probands compared with controls (58). Higher levels of socially relevant autistic traits, poorer face recognition, and less eye tracking toward stimulus photographs are correlated with differences in electroencephalographic recordings when individuals with autism are compared with normally developing individuals (59, 60). These results from humans, along with the current data, suggest a relationship between BPA exposure and ASD.
To move toward a mechanism for these transgenerational behavioral effects, we examined differentially expressed genes related to synapse formation and function from F3 RNA-seq data (20). One gene in particular was noteworthy: Shank1, which codes for one of the scaffolding proteins in the PSD. This protein is associated physically with a set of other PSD proteins, including PSD95, GKAP, and HOMER1. Shank1 gene mutations have been identified in patients with ASDs (34, 61–63), and one single-nucleotide polymorphism in PSD95 has been associated with ASDs (64). Postmortem brains of some patients with schizophrenia have mutations in the DLG4 promoter (PSD95) (65–67). As a class, PSD gene mutations are well documented in schizophrenia, ASDs, and other associated phenotypes (67).
Shank1 knockout (KO) mice were reported to have normal social and olfactory behavior (68). However, ultrasonic vocalizations in pups were reduced in Shank1 KO mice compared with wild-type controls (69). In adults, the pattern of calls and scent marking in response to female urine differed between male KO and control mice. These results indicate communication deficits; however, in the radial maze cognitive test, male Shank1 KO mice performed better than controls (70). Application of short hairpin RNA to reduce Shank1 expression blocked synaptic plasticity (spine formation) during learning in forebrain-dependent tasks (71). Both sexes of PSD95 heterozygous mice displayed hypersocial behavior (72).
At all three ages examined (E18.4, PN0, and PN27) and in both regions (hypothalamus and lateral septum), BPA-lineage F3 C57BL/6J mice had expression levels of Shank1 that were different from those of controls. An exciting aspect of these results is that the direction of the differences varied with the age of the mice. E18.5 BPA-lineage mice had less gene expression than controls, and on PN0 they had higher mRNA; on PN27, BPA-lineage brains again had less Shank1 expression. At the two older ages assayed, PSD95 was also differentially expressed in the same direction as Shank1. Synapse production varies over development; first synapses are overproduced, followed by pruning and reduction. We speculate that the timing of these events is disrupted by BPA. In rodents exposed to BPA during gestation, neurogenesis and neuronal migration can be accelerated (73, 74) or delayed (8, 75).
Several lines of convergent evidence show that direct BPA exposure changes the density and/or structure of spines and synapses (26–28, 76). Synapse-related proteins are altered by BPA in cultured fetal rat hypothalamic cells. Also, at a variety of ages, BPA elevated synapse-related protein levels in mouse brains (77). Even a single BPA treatment 10 days after birth decreased synaptophysin in adult brains (78). Bisphenol A exposure weakened electrophysiology in glutamatergic synapses (29). Relevant to our data, adult male rats exposed to BPA during gestation had reduced mRNA for synaptophysin, PSD95, spinophilin, GluR1, and NMDAR1 (79). Male offspring of Swiss-Webster dams treated daily with BPA from E7 until weaning had depressed PSD95 protein in the cortex and hippocampus at 3 and 8 weeks of age (33). Another F1 study used Institute of Cancer Research mice treated with BPA for the same period with three doses of BPA [all higher than the dose we used here (32)]. In general, BPA changed synaptic morphology by reducing the thickness of the PSD and the synaptic active zone, which should reduce effectiveness of receptor activity at the synapse. At PN14, the lowest and largest doses of BPA decreased numeric spine densities in BPA-exposed male offspring. Our findings suggest that synapse modifications persist without additional BPA contact over three generations.
Although we have no supporting data in humans, our data do generalize to other mouse strains, even when the timing and doses of BPA were different. In the FVB experiment, three BPA doses were used; dams were given BPA orally via voluntary consumption for the second half of pregnancy, and whole brains collected on PN4. When these data were compared with hypothalamic gene expression in PN0 C57BL/6J mice, both Shank1 and PSD95 were significantly increased in males from both inbred lines. In the FVB mice, Homer1c RNA was significantly elevated in the BPA males, and in the C57BL/6J study, expression of Gkap was higher in the equivalent group. Thus, there is good agreement, despite several major differences in study design.
In line with the F3 FVB gene expression results, we found a maternal lineage effect in the C57BL/6J lateral septum of PN27 mice. In this study, we used mice from all four F3 crosses, but differential expression for Shank1 and PSD95 was noted only in groups with maternal BPA ancestry (BB and BC groups). These data match the social recognition results, demonstrating that only mice with maternal BPA-lineage exposure failed to show dishabituation. Curiously, gene expression results in all of the studies, except in embryos, were sexually dimorphic, with BPA effects limited to males. However, the behavioral studies reported here, as well as our previous transgenerational work, did not show sex differences (22, 23). This may be due to the age at which we tested the mice, before puberty and the onset of elevated circulating sex hormones. Interestingly, Shank1 microdeletions are present in some male patients with autism; however, in females the same microdeletion is associated with anxiety, not ASD (64). Perhaps these data speak to differences in male and female autistic symptomology, and these gene-by-environment interactions have sex-specific effects.
In agreement with the literature reviewed previously, we found that F1 day-of-birth brains had elevated expression of the PSD genes. Thus, initial gestational exposure to BPA alters expression of these genes, and their expression remains dysregulated over the course of three generations. Most mammalian models for transgenerational modifications emphasize changes in DNA methylation (80, 81). Some genes that are differentially expressed by BPA in transgenerational tissues are good candidates for DNA methylation; however, in the brain, the methylation status of neither Shank1 nor PSD95 changed after incubation with methionine (82). Another potential mechanism is histone modifications, and histone-modifying enzymes are affected transgenerationally by BPA in Caenorhabditis elegans germline (83). Recently in rats, three generations after DDT exposure, male germ cells revealed several differences in genomic regions containing H3K27me3 modifications (84). Even more striking were modifications in small and long noncoding RNAs in sperm from F1 vs F3 lineages.
In summary, our findings show transgenerational BPA actions on one of the genes associated with autism in humans. This unique gene-by-environment relationship is correlated with a social behavioral deficiency in mice that has relevance to autistic behavior. Because these effects are retained for generations after BPA exposure, even an immediate prohibition on use of BPA in plastics, epoxy resins, thermal paper, and other products will not provide remediation.
Acknowledgments
Financial Support: This work was funded by National Institutes of Health (NIH) Grant ES022759 (to E.F.R.), the James Madison University and University of Virginia 4-VA programs (to A.D.H.), NIH Grant P01 ES 022848 (to J.A.F.), and Environmental Protection Agency Grant RD83 543401 (to J.A.F.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ASD
autism spectrum disorder
- BB
bisphenol A lineage pair
- BC
dam from BPA lineage and sire from control lineage
- BPA
bisphenol A
- CB
dam from control lineage and sire from bisphenol A lineage
- CC
control lineage pair
- E
embryonic day
- EDC
endocrine-disrupting compound
- F1
first familial generation
- F2
second familial generation
- F3
third familial generation
- Gkap
DLG associated protein 1
- Homer1c
Homer scaffolding protein 1c
- KO
knockout
- mGluR
metabotropic glutamate receptor
- OVX
ovariectomized
- PN
postnatal day
- PSD
postsynaptic density
- PSD95
discs large MAGUK scaffold protein 4
- qPCR
quantitative polymerase chain reaction
- Shank1
SH3 and multiple ankyrin repeat domains 1
References and Notes
- 1. Song S, Duan Y, Zhang T, Zhang B, Zhao Z, Bai X, Xie L, He Y, Ouyang JP, Huang X, Sun H. Serum concentrations of bisphenol A and its alternatives in elderly population living around e-waste recycling facilities in China: associations with fasting blood glucose. Ecotoxicol Environ Saf. 2019;169:822–828. [DOI] [PubMed] [Google Scholar]
- 2. Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect. 2008;116(1):39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nahar MS, Soliman AS, Colacino JA, Calafat AM, Battige K, Hablas A, Seifeldin IA, Dolinoy DC, Rozek LS. Urinary bisphenol A concentrations in girls from rural and urban Egypt: a pilot study. Environ Health. 2012;11:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ye X, Wong LY, Kramer J, Zhou X, Jia T, Calafat AM. Urinary concentrations of bisphenol A and three other bisphenols in convenience samples of U.S. adults during 2000-2014. Environ Sci Technol. 2015;49(19):11834–11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson OS, Peterson KE, Sanchez BN, Zhang Z, Mancuso P, Dolinoy DC. Perinatal bisphenol A exposure promotes hyperactivity, lean body composition, and hormonal responses across the murine life course. FASEB J. 2013;27(4):1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox KH, Gatewood JD, Howeth C, Rissman EF. Gestational exposure to bisphenol A and cross-fostering affect behaviors in juvenile mice. Horm Behav. 2010;58(5):754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gioiosa L, Palanza P, Parmigiani S, Vom Saal FS. Risk evaluation of endocrine-disrupting chemicals: effects of developmental sxposure to low doses of bisphenol A on behavior and physiology in mice (Mus musculus). Dose Response. 2015;13(4):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jang YJ, Park HR, Kim TH, Yang WJ, Lee JJ, Choi SY, Oh SB, Lee E, Park JH, Kim HP, Kim HS, Lee J. High dose bisphenol A impairs hippocampal neurogenesis in female mice across generations. Toxicology. 2012;296(1-3):73–82. [DOI] [PubMed] [Google Scholar]
- 9. Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci USA. 2015;112(22):6807–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gore AC, Krishnan K, Reilly MP. Endocrine-disrupting chemicals: effects on neuroendocrine systems and the neurobiology of social behavior. Horm Behav. 2019;111:7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Braun JM. Early-life exposure to EDCs: role in childhood obesity and neurodevelopment. Nat Rev Endocrinol. 2017;13(3):161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tewar S, Auinger P, Braun JM, Lanphear B, Yolton K, Epstein JN, Ehrlich S, Froehlich TE. Association of bisphenol A exposure and attention-deficit/hyperactivity disorder in a national sample of U.S. children. Environ Res. 2016;150:112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y, Zhang H, Kuang H, Fan R, Cha C, Li G, Luo Z, Pang Q. Relationship between bisphenol A exposure and attention-deficit/ hyperactivity disorder: a case-control study for primary school children in Guangzhou, China. Environ Pollut. 2018;235:141–149. [DOI] [PubMed] [Google Scholar]
- 14. Stein TP, Schluter MD, Steer RA, Guo L, Ming X. Bisphenol A exposure in children with autism spectrum disorders. Autism Res. 2015;8(3):272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown JS., Jr Effects of bisphenol-A and other endocrine disruptors compared with abnormalities of schizophrenia: an endocrine-disruption theory of schizophrenia. Schizophr Bull. 2009;35(1):256–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eladak S, Grisin T, Moison D, Guerquin MJ, N’Tumba-Byn T, Pozzi-Gaudin S, Benachi A, Livera G, Rouiller-Fabre V, Habert R. A new chapter in the bisphenol A story: bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil Steril. 2015;103(1):11–21. [DOI] [PubMed] [Google Scholar]
- 17. Kinch CD, Ibhazehiebo K, Jeong JH, Habibi HR, Kurrasch DM. Low-dose exposure to bisphenol A and replacement bisphenol S induces precocious hypothalamic neurogenesis in embryonic zebrafish. Proc Natl Acad Sci USA. 2015;112(5):1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rochester JR, Bolden AL. Bisphenol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ Health Perspect. 2015;123(7):643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horan TS, Pulcastro H, Lawson C, Gerona R, Martin S, Gieske MC, Sartain CV, Hunt PA. Replacement bisphenols adversely affect mouse gametogenesis with consequences for subsequent generations. Curr Biol. 2018;28(18):2948–2954.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drobná Z, Henriksen AD, Wolstenholme JT, Montiel C, Lambeth PS, Shang S, Harris EP, Zhou C, Flaws JA, Adli M, Rissman EF. Transgenerational effects of bisphenol A on gene expression and DNA methylation of imprinted genes in brain. Endocrinology. 2018;159(1):132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lombó M, Fernández-Díez C, González-Rojo S, Navarro C, Robles V, Herráez MP. Transgenerational inheritance of heart disorders caused by paternal bisphenol A exposure. Environ Pollut. 2015;206:667–678. [DOI] [PubMed] [Google Scholar]
- 22. Wolstenholme JT, Edwards M, Shetty SR, Gatewood JD, Taylor JA, Rissman EF, Connelly JJ. Gestational exposure to bisphenol A produces transgenerational changes in behaviors and gene expression. Endocrinology. 2012;153(8):3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolstenholme JT, Goldsby JA, Rissman EF. Transgenerational effects of prenatal bisphenol A on social recognition. Horm Behav. 2013;64(5):833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ziv-Gal A, Wang W, Zhou C, Flaws JA. The effects of in utero bisphenol A exposure on reproductive capacity in several generations of mice. Toxicol Appl Pharmacol. 2015;284(3):354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacLusky NJ, Hajszan T, Leranth C. The environmental estrogen bisphenol A inhibits estradiol-induced hippocampal synaptogenesis. Environ Health Perspect. 2005;113(6):675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hajszan T, Leranth C. Bisphenol A interferes with synaptic remodeling. Front Neuroendocrinol. 2010;31(4):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elsworth JD, Jentsch JD, Vandevoort CA, Roth RH, Redmond DE Jr, Leranth C. Prenatal exposure to bisphenol A impacts midbrain dopamine neurons and hippocampal spine synapses in non-human primates. Neurotoxicology. 2013;35:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimura E, Matsuyoshi C, Miyazaki W, Benner S, Hosokawa M, Yokoyama K, Kakeyama M, Tohyama C. Prenatal exposure to bisphenol A impacts neuronal morphology in the hippocampal CA1 region in developing and aged mice. Arch Toxicol. 2016;90(3):691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu F, Li T, Gong H, Chen Z, Jin Y, Xu G, Wang M. Bisphenol A impairs synaptic plasticity by both pre- and postsynaptic mechanisms. Adv Sci (Weinh). 2017;4(8):1600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinson A, Bourguignon JP, Parent AS. Exposure to endocrine disrupting chemicals and neurodevelopmental alterations. Andrology. 2016;4(4):706–722. [DOI] [PubMed] [Google Scholar]
- 31. Alavian-Ghavanini A, Lin PI, Lind PM, Risén Rimfors S, Halin Lejonklou M, Dunder L, Tang M, Lindh C, Bornehag CG, Rüegg J. Prenatal bisphenol A exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci Rep. 2018;8(1):11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu X, Xie L, Hong X, Ruan Q, Lu H, Zhang Q, Zhang G, Liu X. Perinatal exposure to bisphenol-A inhibits synaptogenesis and affects the synaptic morphological development in offspring male mice. Chemosphere. 2013;91(8):1073–1081. [DOI] [PubMed] [Google Scholar]
- 33. Kumar D, Thakur MK. Anxiety like behavior due to perinatal exposure to bisphenol-A is associated with decrease in excitatory to inhibitory synaptic density of male mouse brain. Toxicology. 2017;378:107–113. [DOI] [PubMed] [Google Scholar]
- 34. Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18(3):147–157. [DOI] [PubMed] [Google Scholar]
- 35. Goldsby JA, Wolstenholme JT, Rissman EF. Multi- and transgenerational consequences of bisphenol A on sexually dimorphic cell populations in mouse brain. Endocrinology. 2017;158(1):21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang W, Hafner KS, Flaws JA. In utero bisphenol A exposure disrupts germ cell nest breakdown and reduces fertility with age in the mouse. Toxicol Appl Pharmacol. 2014;276(2):157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imwalle DB, Scordalakes EM, Rissman EF. Estrogen receptor α influences socially motivated behaviors. Horm Behav. 2002;42(4):484–491. [DOI] [PubMed] [Google Scholar]
- 38. Tejada LD, Rissman EF. Sex differences in social investigation: effects of androgen receptors, hormones and test partner. J Neuroendocrinol. 2012;24(8):1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bielsky IF, Hu SB, Ren X, Terwilliger EF, Young LJ. The V1a vasopressin receptor is necessary and sufficient for normal social recognition: a gene replacement study. Neuron. 2005;47(4):503–513. [DOI] [PubMed] [Google Scholar]
- 40. Lim MM, Bielsky IF, Young LJ. Neuropeptides and the social brain: potential rodent models of autism. Int J Dev Neurosci. 2005;23(2-3):235–243. [DOI] [PubMed] [Google Scholar]
- 41. Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6(6):e21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. RRID:AB_2187584, https://scicrunch.org/resolver/RRID:AB_2187584.
- 43. RRID:AB_1523944, https://scicrunch.org/resolver/RRID:AB_1523944.
- 44. RRID:AB_306371, https://scicrunch.org/resolver/RRID:AB_306371.
- 45. Carter CJ, Blizard RA. Autism genes are selectively targeted by environmental pollutants including pesticides, heavy metals, bisphenol A, phthalates and many others in food, cosmetics or household products. Neurochem Int. 2016;101:83–109. [DOI] [PubMed] [Google Scholar]
- 46. Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Curr Probl Pediatr Adolesc Health Care. 2014;44(10):277–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sealey LA, Hughes BW, Sriskanda AN, Guest JR, Gibson AD, Johnson-Williams L, Pace DG, Bagasra O. Environmental factors in the development of autism spectrum disorders. Environ Int. 2016;88:288–298. [DOI] [PubMed] [Google Scholar]
- 48. Sullivan AW, Beach EC, Stetzik LA, Perry A, D’Addezio AS, Cushing BS, Patisaul HB. A novel model for neuroendocrine toxicology: neurobehavioral effects of BPA exposure in a prosocial species, the prairie vole (Microtus ochrogaster). Endocrinology. 2014;155(10):3867–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson SA, Painter MS, Javurek AB, Ellersieck MR, Wiedmeyer CE, Thyfault JP, Rosenfeld CS. Sex-dependent effects of developmental exposure to bisphenol A and ethinyl estradiol on metabolic parameters and voluntary physical activity. J Dev Orig Health Dis. 2015;6(6):539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Esterik JC, Dollé ME, Lamoree MH, van Leeuwen SP, Hamers T, Legler J, van der Ven LT. Programming of metabolic effects in C57BL/6JxFVB mice by exposure to bisphenol A during gestation and lactation. Toxicology. 2014;321:40–52. [DOI] [PubMed] [Google Scholar]
- 51. Gillette R, Miller-Crews I, Nilsson EE, Skinner MK, Gore AC, Crews D. Sexually dimorphic effects of ancestral exposure to vinclozolin on stress reactivity in rats. Endocrinology. 2014;155(10):3853–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008;3(11):e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci USA. 2007;104(14):5942–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krishnan K, Mittal N, Thompson LM, Rodriguez-Santiago M, Duvauchelle CL, Crews D, Gore AC. Effects of the endocrine-disrupting chemicals, vinclozolin and polychlorinated biphenyls, on physiological and sociosexual phenotypes in F2 generation Sprague-Dawley rats. Environ Health Perspect. 2018;126(9):097005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3(5):287–302. [DOI] [PubMed] [Google Scholar]
- 56. Moy SS, Nadler JJ, Young NB, Nonneman RJ, Grossman AW, Murphy DL, D’Ercole AJ, Crawley JN, Magnuson TR, Lauder JM. Social approach in genetically engineered mouse lines relevant to autism. Genes Brain Behav. 2009;8(2):129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pascual B, de Bot ST, Daniels MR, França MC Jr, Toro C, Riverol M, Hedera P, Bassi MT, Bresolin N, van de Warrenburg BP, Kremer B, Nicolai J, Charles P, Xu J, Singh S, Patronas NJ, Fung SH, Gregory MD, Masdeu JC. “Ears of the lynx” MRI sign is associated with SPG11 and SPG15 hereditary spastic paraplegia. AJNR Am J Neuroradiol. 2019;40(1):199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arkush L, Smith-Collins AP, Fiorentini C, Skuse DH. Recognition of face and non-face stimuli in autistic spectrum disorder. Autism Res. 2013;6(6):550–560. [DOI] [PubMed] [Google Scholar]
- 59. Black MH, Chen NTM, Iyer KK, Lipp OV, Bölte S, Falkmer M, Tan T, Girdler S. Mechanisms of facial emotion recognition in autism spectrum disorders: insights from eye tracking and electroencephalography. Neurosci Biobehav Rev. 2017;80:488–515. [DOI] [PubMed] [Google Scholar]
- 60. Davis J, McKone E, Zirnsak M, Moore T, O’Kearney R, Apthorp D, Palermo R. Social and attention-to-detail subclusters of autistic traits differentially predict looking at eyes and face identity recognition ability. Br J Psychol. 2017;108(1):191–219. [DOI] [PubMed] [Google Scholar]
- 61. Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, O’Connor I, Russell C, Drmic IE, Hamdan FF, Michaud JL, Endris V, Roeth R, Delorme R, Huguet G, Leboyer M, Rastam M, Gillberg C, Lathrop M, Stavropoulos DJ, Anagnostou E, Weksberg R, Fombonne E, Zwaigenbaum L, Fernandez BA, Roberts W, Rappold GA, Marshall CR, Bourgeron T, Szatmari P, Scherer SW. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet. 2012;90(5):879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78(1):P8–P27. [DOI] [PMC free article] [PubMed]
- 63. Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, Giuliano F, Stordeur C, Depienne C, Mouzat K, Pinto D, Howe J, Lemière N, Durand CM, Guibert J, Ey E, Toro R, Peyre H, Mathieu A, Amsellem F, Rastam M, Gillberg IC, Rappold GA, Holt R, Monaco AP, Maestrini E, Galan P, Heron D, Jacquette A, Afenjar A, Rastetter A, Brice A, Devillard F, Assouline B, Laffargue F, Lespinasse J, Chiesa J, Rivier F, Bonneau D, Regnault B, Zelenika D, Delepine M, Lathrop M, Sanlaville D, Schluth-Bolard C, Edery P, Perrin L, Tabet AC, Schmeisser MJ, Boeckers TM, Coleman M, Sato D, Szatmari P, Scherer SW, Rouleau GA, Betancur C, Leboyer M, Gillberg C, Delorme R, Bourgeron T. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10(9):e1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang J, Li L, Shao SS, He Z, Chen YL, Kong R, Zhang XH, Gong JH, Song RR. Association analysis of genetic variant of rs13331 in PSD95 gene with autism spectrum disorders: a case-control study in a Chinese population. J Huazhong Univ Sci Technolog Med Sci. 2016;36(2):285–288. [DOI] [PubMed] [Google Scholar]
- 65. Cheng MC, Lu CL, Luu SU, Tsai HM, Hsu SH, Chen TT, Chen CH. Genetic and functional analysis of the DLG4 gene encoding the post-synaptic density protein 95 in schizophrenia. PLoS One. 2010;5(12):e15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Soler J, Fañanás L, Parellada M, Krebs MO, Rouleau GA, Fatjó-Vilas M. Genetic variability in scaffolding proteins and risk for schizophrenia and autism-spectrum disorders: a systematic review. J Psychiatry Neurosci. 2018;43(4):223–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Balan S, Yamada K, Hattori E, Iwayama Y, Toyota T, Ohnishi T, Maekawa M, Toyoshima M, Iwata Y, Suzuki K, Kikuchi M, Yoshikawa T. Population-specific haplotype association of the postsynaptic density gene DLG4 with schizophrenia, in family-based association studies. PLoS One. 2013;8(7):e70302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wöhr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011;6(6):e20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28(7):1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Collins SM, Galvez R. Neocortical SHANK1 regulation of forebrain dependent associative learning. Neurobiol Learn Mem. 2018;155:173–179. [DOI] [PubMed] [Google Scholar]
- 72. Winkler D, Daher F, Wüstefeld L, Hammerschmidt K, Poggi G, Seelbach A, Krueger-Burg D, Vafadari B, Ronnenberg A, Liu Y, Kaczmarek L, Schlüter OM, Ehrenreich H, Dere E. Hypersocial behavior and biological redundancy in mice with reduced expression of PSD95 or PSD93. Behav Brain Res. 2018;352:35–45. [DOI] [PubMed] [Google Scholar]
- 73. Komada M, Asai Y, Morii M, Matsuki M, Sato M, Nagao T. Maternal bisphenol A oral dosing relates to the acceleration of neurogenesis in the developing neocortex of mouse fetuses. Toxicology. 2012;295(1-3):31–38. [DOI] [PubMed] [Google Scholar]
- 74. Nakamura K, Itoh K, Sugimoto T, Fushiki S. Prenatal exposure to bisphenol A affects adult murine neocortical structure. Neurosci Lett. 2007;420(2):100–105. [DOI] [PubMed] [Google Scholar]
- 75. Komada M, Itoh S, Kawachi K, Kagawa N, Ikeda Y, Nagao T. Newborn mice exposed prenatally to bisphenol A show hyperactivity and defective neocortical development. Toxicology. 2014;323:51–60. [DOI] [PubMed] [Google Scholar]
- 76. Bowman RE, Luine V, Diaz Weinstein S, Khandaker H, DeWolf S, Frankfurt M. Bisphenol-A exposure during adolescence leads to enduring alterations in cognition and dendritic spine density in adult male and female rats. Horm Behav. 2015;69:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Iwakura T, Iwafuchi M, Muraoka D, Yokosuka M, Shiga T, Watanabe C, Ohtani-Kaneko R. In vitro effects of bisphenol A on developing hypothalamic neurons. Toxicology. 2010;272(1-3):52–58. [DOI] [PubMed] [Google Scholar]
- 78. Viberg H, Lee I. A single exposure to bisphenol A alters the levels of important neuroproteins in adult male and female mice. Neurotoxicology. 2012;33(5):1390–1395. [DOI] [PubMed] [Google Scholar]
- 79. Wang C, Niu R, Zhu Y, Han H, Luo G, Zhou B, Wang J. Changes in memory and synaptic plasticity induced in male rats after maternal exposure to bisphenol A. Toxicology. 2014;322:51–60. [DOI] [PubMed] [Google Scholar]
- 80. Rissman EF, Adli M. Minireview: transgenerational epigenetic inheritance: focus on endocrine disrupting compounds. Endocrinology. 2014;155(8):2770–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nilsson E, King SE, McBirney M, Kubsad D, Pappalardo M, Beck D, Sadler-Riggleman I, Skinner MK. Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS One. 2018;13(8):e0202662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beri S, Tonna N, Menozzi G, Bonaglia MC, Sala C, Giorda R. DNA methylation regulates tissue-specific expression of Shank3. J Neurochem. 2007;101(5):1380–1391. [DOI] [PubMed] [Google Scholar]
- 83. Camacho J, Allard P.. Histone modifications: epigenetic mediators of environmental exposure memory. Epigenet Insights. 2018;11:2516865718803641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, Klukovich R, Xie Y, Tang C, Yan W. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin. 2018;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]