Summary
Cell wall peroxidases and plasma membrane‐localized NADPH oxidases are considered to be the main sources of the apoplastic oxidative burst in plants attacked by microbial pathogens. In spite of this established doctrine, approaches attempting a comparative, side‐by‐side analysis of the functions of extracellular reactive oxygen species (ROS) generated by the two enzymatic sources are scarce. Previously, we have reported the role of Arabidopsis NADPH oxidase RBOHD (respiratory burst oxidase homologue D) in plants challenged with the necrotrophic fungus Alternaria brassicicola. Here, we present results on the activity of apoplastic class III peroxidases PRX33 (At3g49110) and PRX34 (At3g49120) investigated in the same Arabidopsis–Alternaria pathosystem. ROS generated by Arabidopsis peroxidases PRX33 and PRX34 increase the necrotic symptoms and colonization success of A. brassicicola. In addition, the knockdown of PRX33 and PRX34 transcript levels leads to a reduced number of host cells showing an extracellular burst of ROS after inoculation with A. brassicicola. Our results also reveal an age‐dependent transcript distribution of ROS‐producing peroxidase and NADPH oxidase enzymes, and some potential new components of the RBOHD, PRX33 and PRX34 signalling networks.
Keywords: Alternaria, Arabidopsis, cell wall peroxidase, ERECTA, JAR1, NHO1, VIGS
Introduction
Plant cells respond to invading microbial pathogens with a series of changes in the activity of their various metabolic pathways. One of the most apparent cellular reactions on pathogen attack is a timely, coordinated accumulation of reactive oxygen species (ROS) in the apoplast and in some cellular compartments, which is often referred to as the ‘oxidative burst’. ROS that are released during the course of the oxidative burst include the free radicals superoxide (O2 •¯), hydroxyl radicals (•OH) and nitric oxide (•NO), with unpaired valence electrons, as well as the non‐radicals hydrogen peroxide (H2O2) and singlet oxygen (1O2) (Groß et al., 2013; Mignolet‐Spruyt et al., 2016; Mittler, 2017). Several enzymatic systems have been recognized as cellular sources of the oxidative burst in the apoplast of plant cells. These include NADPH oxidases, cell wall peroxidases (class III secretory plant peroxidases), amine and polyamine oxidases (AOs and PAOs), oxalate oxidases and quinone reductases (Kärkönen and Kuchitsu 2015). Endeavours to decipher the roles of NADPH oxidases and cell wall peroxidases in plant diseases and immunity have led to significant discoveries, but the functions of AOs, oxalate oxidases and quinone reductases are still largely elusive.
The most extensively studied sources of extracellular ROS in plants challenged by pathogenic microbes are respiratory burst oxidase homologue (RBOH) NADPH oxidases localized in the cellular plasma membrane (Frederickson Matika and Loake, 2013; Liu and He, 2016). The genome of Arabidopsis thaliana (hereafter referred to as Arabidopsis) encodes 10 RBOH proteins. Of these 10 isoforms, RbohD and RbohF are transcribed at the highest levels in root and leaf tissues (Torres et al., 1998). The contribution of Arabidopsis NADPH oxidases RBOHD and RBOHF to the apoplastic oxidative burst, host immunity and/or cell death regulation has been reported in a wide range of Arabidopsis–pathogen interactions. These interactions include virulent or avirulent bacterial, oomycete or necrotrophic and biotrophic fungal pathogens of Arabidopsis (Marino et al., 2012). Concerning the significance of apoplastic Arabidopsis class III peroxidases in plant–pathogen interactions, PRX33 and PRX34 (encoded by loci At3g49110 and At3g49120, respectively) have been described as sources of an extracellular oxidative burst when plants/cell suspensions are challenged by avirulent strains of Pseudomonas syringae (Bindschedler et al., 2006) or treated with microbe‐associated molecular pattern (MAMP) elicitors (Daudi et al., 2012; O’Brien et al., 2012). The proportions of ROS released by NADPH oxidases and peroxidases were evaluated by a pharmacological approach. In Arabidopsis cell suspensions treated with various MAMP elicitors, at least 50% of the H2O2 produced could be credited to peroxidases, predominantly to PRX33 and PRX34, and the remaining 50% or less was attributed to NADPH oxidases and intracellular sources (O’Brien et al., 2012). Peroxidases PRX33 and PRX34 were found to contribute to resistance against virulent and avirulent strains of P. syringae, as well as against infections caused by Botrytis cinerea and powdery mildew fungi (Bindschedler et al., 2006). Interestingly, an oxidative burst triggered by the treatment of Arabidopsis plants with a cell wall elicitor prepared from the fungus Fusarium oxysporum could be abolished by knocking down the transcript levels of PRX33 and PRX34 through the transgenic expression of a French bean class III peroxidase cDNA (FBP1) in an antisense orientation (Daudi et al., 2012). This genotype has been referred to as asFBP1.1. The depletion of the PRX33 mRNA level in an Arabidopsis T‐DNA insertion line, however, resulted in reduced susceptibility to F. oxysporum, indicating that PRX33 promotes susceptibility to this particular fungus (Lyons et al., 2015).
Peroxidases PRX33 and PRX34 have also been shown to act in salicylic acid (SA) signalling, as asFBP1.1 plants are impaired in the expression of several SA‐responsive genes and in SA‐induced NPR1 monomerization (Mammarella et al., 2015). The formation of NPR1 monomers is dependent on the cellular redox state and is also a crucial component of the SA‐mediated signalling pathway (Mou et al., 2003; Tada et al., 2008). Two studies on the cell wall proteome of Arabidopsis leaves also drew attention to PRX33 or PRX34 in responses to pathogen infection and redox imbalances. PRX34 is over‐represented in the apoplastic proteome of Verticillium longisporum‐infected leaves (Floerl et al., 2012), and PRX33 and PRX34 are both more abundant in cell wall protein extracts of ascorbate‐deficient Arabidopsis leaves relative to those of wild‐type plants (Sultana et al., 2015).
In addition to NADPH oxidases and apoplastic peroxidases, AOs and PAOs have been proposed as alternative sources of extracellular ROS accumulation. Polyamines are catabolized by copper‐containing AOs (CuAOs) and FAD‐dependent PAOs. PAOs catalyse the oxidation of spermine (Spm), spermidine (Spd) and/or their acetylated derivatives at the secondary amino groups (Angelini et al., 2010). In Arabidopsis, five PAO isoforms (AtPAO1–AtPAO5) have been identified, and the main physiological role of these proteins has been linked to compartment‐specific H2O2 synthesis in different phases of development and differentiation, as well as in the course of defence mechanisms against pathogens and abiotic stress (Andronis et al., 2014; Fincato et al., 2012; Takahashi et al., 2010). Arabidopsis plants inoculated with an avirulent strain of P. syringae pv. tomato DC3000 (avrRpm1) accumulate polyamines, and infiltration of polyamines into the apoplast of leaves causes an oxidative burst and subsequent cell death (Yoda et al., 2009).
The Arabidopsis thaliana–Alternaria brassicicola pathosystem has been widely used to study plant defence and fungal pathogenesis strategies (Schenk et al., 2003; Su’udi et al., 2011). We have reported previously that the functional Arabidopsis NADPH oxidase RBOHD (respiratory burst oxidase homologue D) is required for an extracellular oxidative burst when plants are spray inoculated with a conidial suspension of the necrotrophic fungus A. brassicicola. An rbohd knockout line showed reduced susceptibility to the pathogen and increased spread of cell death, indicating that RBOHD contributes to the success of the colonization process and is also engaged in the regulation of A. brassicicola‐induced host cell death. Finally, an interplay between RBOHD, ethylene and SA was demonstrated, and diverse roles of RBOHD‐dependent ROS were suggested between cells that were affected directly by the fungus and cells that were in neighbouring positions (Pogány et al., 2009).
The necrotrophic fungus A. brassicicola causes black spot disease and is an economically important pathogen of Brassicaceae species (Thomma, 2003). The interaction between A. brassicicola strain MUCL 20297 and Arabidopsis thaliana ecotype Col‐0 (both used throughout this work) is considered to be incompatible (Narusaka et al., 2005; Thomma et al., 1998). Nevertheless, our experimental conditions (which were excessively favourable for the fungus) enabled considerable pathogen growth (Pogány et al., 2009).
The various enzymatic sources of extracellular ROS in diseased (or wounded) plants are typically examined separately and in different pathosystems. Sometimes, when these sources are investigated together, enzyme inhibitors are used to distinguish between NADPH oxidase‐, class III peroxidase‐ or PAO‐derived ROS (Dmochowska‐Boguta et al., 2013; O’Brien et al., 2012; Roach et al., 2015). As an extension of our work on the cellular functions of the NADPH oxidase RBOHD (Pogány et al., 2009), in this study, we provide results on the contribution of peroxidases PRX33 and PRX34 to a pathogen‐induced apoplastic oxidative burst. The experiments are performed with the same Arabidopsis–Alternaria pathosystem as in the RBOHD study, using knockout and knockdown insertion mutant and gene‐silenced plant samples. Some likely candidates of the RBOHD, PRX33 and PRX34 signalling networks are also exposed.
Results
Elicitation of Arabidopsis leaf cells by A. brassicicola activates PRX33 and PRX34 transcription
To explore the contribution of apoplastic peroxidases and PAOs in the oxidative burst, we monitored mRNA abundance for peroxidase genes PRX33 and PRX34 and PAO isoforms PAO1, PAO2, PAO3, PAO4 and PAO5 in our pathosystem. Expression of the two cell wall peroxidases was clearly induced after 12 h, and the highest expression levels were observed at 24 h after inoculation (hai) for both PRX genes in comparison with mock‐inoculated plants (Fig. 1). In contrast, transcription of PAO1, PAO2, PAO3 and PAO4 remained unaffected and PAO5 mRNA levels were repressed by elicitation with A. brassicicola at 24 hai, indicating that they are not involved in the A. brassicicola‐induced oxidative burst and other host responses (Fig. S1, see Supporting Information).
Figure 1.
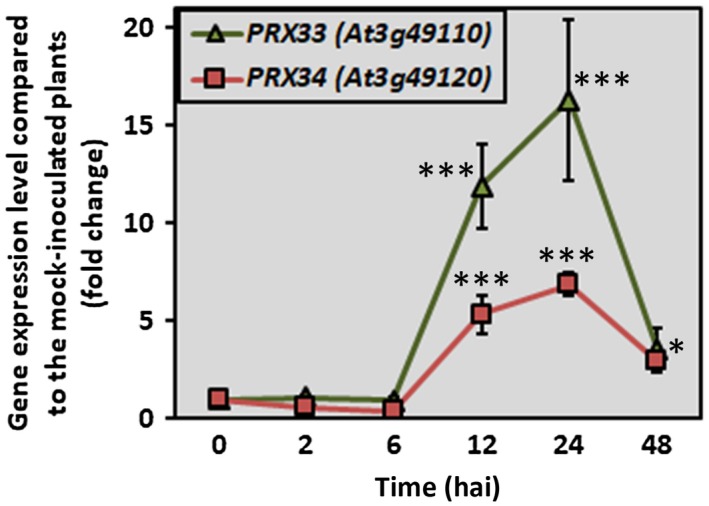
Alternaria brassicicola‐induced activation of PRX33 and PRX34 genes in wild‐type Arabidopsis plants. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with A. brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. Data represent the mean of three independent biological samples with three technical replicates each. Statistical analysis was performed using Mann–Whitney U‐test. Asterisks indicate statistically significant differences (*α = 0.05, ***α = 0.001).
Knockdown of PRX33 and PRX34 transcript levels reduces symptoms and colonization success of A. brassicicola
In a process to select Arabidopsis genotypes with adequate PRX33 and PRX34 knockdown mRNA levels, T‐DNA insertion mutants prx33 (SALK_062314C) and prx34 (SALK_051769C) (Passardi et al., 2006), together with the line asFBP1.1 expressing a French bean class III peroxidase in an antisense orientation (Bindschedler et al., 2006), were initially included in this work. Transcript levels of peroxidase gene PRX33 were significantly reduced in the prx33 insertion line and in asFBP1.1, but mRNA levels for the peroxidase gene PRX34 were not sufficiently suppressed in our hands in the prx34 insertion line or in the asFBP1.1 line. Therefore, we created a Tobacco rattle virus (TRV) vector‐based virus‐induced gene silencing (VIGS) construct (Hayward et al., 2011) that provided Arabidopsis plants with markedly reduced PRX34 (and PRX33) transcript levels. This TRV‐GFP‐PRX VIGS construct is hereafter abbreviated as TRV‐PRX. Diminished activity of PRX33 and PRX34 genes in the prx33 insertion line and in plants treated with the TRV‐PRX VIGS construct is shown in Figs S2 and S3 (see Supporting Information). All subsequent pathogen and ROS detection assays aimed at the characterization of extracellular PRX33 and PRX34 functions in Arabidopsis during fungal pathogenesis were conducted with the prx33 (SALK_062314C) line and with plants carrying the TRV‐PRX VIGS construct. A previously characterized rbohd NADPH oxidase knockout line (SALK_070610C) was also included in some experiments (Pogány et al., 2009).
Down‐regulation of PRX33 and PRX34 yielded reduced fungal symptoms on Arabidopsis leaves. The percentage of leaf chlorosis and necrosis triggered by A. brassicicola in plants with diminished PRX33 and PRX34 transcript levels was only 32%–36% of that in plants showing wild‐type PRX33/PRX34 transcript levels (Fig. 2). The fungal biomass of A. brassicicola was also lower in prx33/prx34 knockdown plants, representing 68% (prx33) and 51% (TRV‐PRX) of that of corresponding inoculated control plants with wild‐type PRX33/PRX34 transcript levels (Fig. 3).
Figure 2.
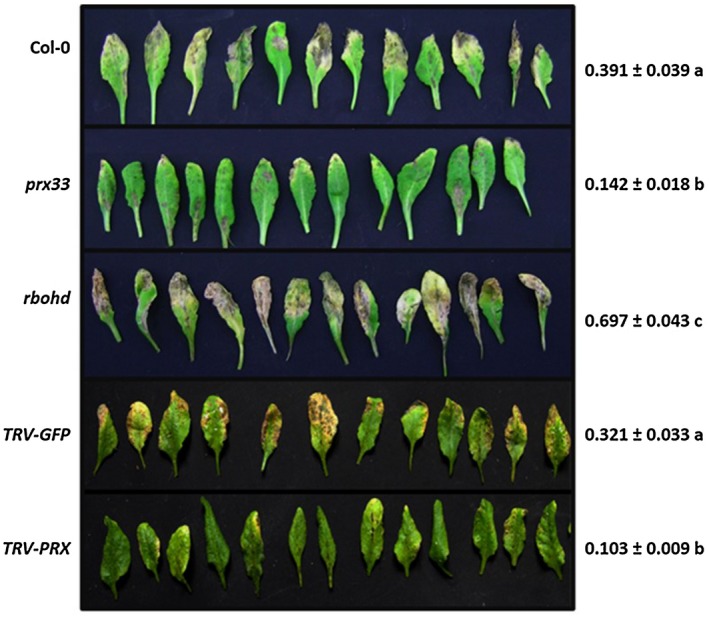
Symptoms of Alternaria brassicicola infection are suppressed in Arabidopsis plants with reduced PRX33/PRX34 mRNA levels. Leaves of A. brassicicola‐infected Col‐0, prx33 knockdown mutant, rbohd knockout mutant, TRV‐GFP and TRV‐PRX plants are shown 10 days after inoculation. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with A. brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. Detached leaves in middle positions (leaf levels 5–8) were evaluated. Results are presented as the chlorotic and necrotic leaf area compared with the total surface area of leaf blades analysed by ImageJ (1 is equal to 100% leaf surface), and represent the means of two experiments (n = 30 for each genotype/treatment) ± standard error (SE). Different letters indicate statistically significant differences between genotypes/treatments using Tukey’s post hoc test. Increased symptoms of A. brassicicola on leaves of the rbohd mutant are included as a reference.
Figure 3.
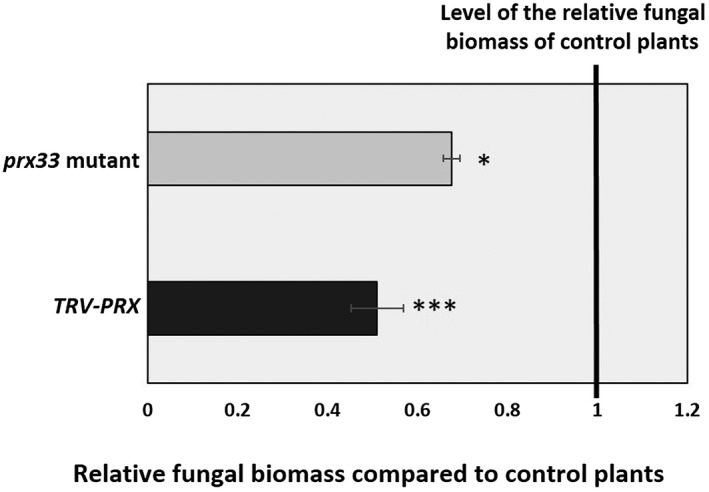
Alternaria brassicicola biomass in prx33 T‐DNA insertion line and in gene‐silenced (TRV‐PRX) plants. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with A. brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. The results show the average of two experiments, each comprising three biological samples (each sample composed of a pool of three Arabidopsis rosettes), analysed in three technical replicates. Statistical analysis was performed using Student’s t‐test. Asterisks indicate statistically significant differences between PRX knockdown Arabidopsis plants (prx33, TRV‐PRX) and their controls (Col‐0, TRV‐GFP) at 10 days after inoculation with A. brassicicola (*α = 0.05, ***α = 0.001).
H2O2 accumulation
H2O2 was detected by a non‐fluorescent 3,3′‐diaminobenzidine (DAB) histochemical staining procedure, which is based on an H2O2‐dependent peroxidase‐catalysed polymerization reaction (Thordal‐Christensen et al., 1997), and a fluorescent 2′,7′‐dichlorofluorescein diacetate (DCFH‐DA) method. DCFH‐DA also reacts with H2O2 in the presence of peroxidases, yielding a fluorescent DCF product (Bozsó et al., 2005). Inoculated leaves were observed microscopically for the DAB‐stained samples and visualized with UV light for the DCFH‐DA‐stained leaves, where the intensity of the fluorescence signal released by individual leaves was detected.
Knockdown of PRX33 and PRX34 transcript levels led to decreased H2O2 formation in Arabidopsis leaves challenged with A. brassicicola (Fig. 4). This altered response was quantitative because the apoplastic burst was not completely abolished, but the number of cells exhibiting a burst of extracellular H2O2 was reduced (Fig. 4C). This is in contrast with the fully inhibited apoplastic ROS accumulation in the rbohd mutant (Pogány et al., 2009). We quantified the number of cells showing extracellular H2O2 accumulation at 2 days after inoculation with A. brassicicola for 20 randomly selected infection sites, observing leaves from two independent experiments. This evaluation revealed that infection sites in wild‐type (Col‐0) plants were composed of an average of 42.6 ± 5.7 cells showing ROS accumulation relative to 16.0 ± 2.5 cells in prx33 plants. Likewise, infection sites in TRV‐GFP plants with normal PRX33/PRX34 transcript levels comprised an average of 34.4 ± 3.8 cells exhibiting H2O2 accumulation relative to 17.0 ± 1.2 cells in prx33/prx34 under‐producing TRV‐PRX plants.
Figure 4.
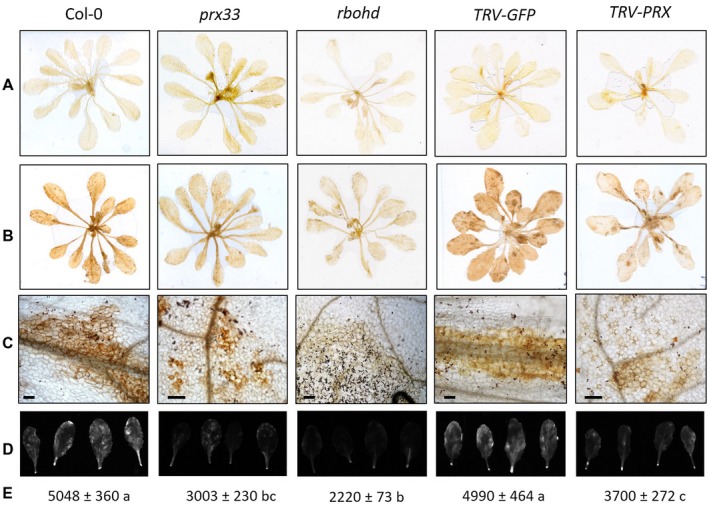
Hydrogen peroxide (H2O2) production in Arabidopsis plants with reduced cell wall peroxidase and NADPH oxidase activity. The accumulation of H2O2 was detected in wild‐type (Col‐0), prx33 and rbohd T‐DNA insertion lines and in TRV‐GFP and TRV‐PRX gene‐silenced Arabidopsis plants by 3,3′‐diaminobenzidine (DAB) (A–C) and 2′,7′‐dichlorofluorescein diacetate (DCFH‐DA) (D, E) staining methods. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with Alternaria brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. The stainings were carried out with mock‐inoculated (A) and Alternaria brassicicola‐infected (B–E) plants (2 days after inoculation). Bars, 50 µm. The intensity of the fluorescence signal emitted by DCFH‐DA‐stained leaves (D) was quantified under UV light using an AlphaImager Mini gel documentation system (E). Results are presented as an average pixel fluorescence intensity and represent the means of two experiments (n = 30 leaves in the middle position for each genotype/treatment) ± standard error (SE). Statistical analysis was performed using one‐way analysis of variance (ANOVA) and Tukey’s post hoc test. Different letters indicate statistically significant results. prx33 and TRV‐PRX plants accumulate less H2O2 than the corresponding control plants. Microscopic observation of the patterns of H2O2 accumulation reveals that prx33 and TRV‐PRX plants show apoplastic H2O2 accumulation in a reduced number of cells in comparison with the corresponding control plants after inoculation with A. brassicicola. Leaves of the rbohd knockout line lacking apoplastic H2O2 accumulation after A. brassicicola infection are shown as a reference. Decreased H2O2 production in rbohd plants was also confirmed by the DCFH‐DA staining method. The mean level of base fluorescence for mock‐inoculated Col‐0 plants was 1240 ± 34.
The adaptation of a quantifiable, fluorescent, ROS detection method suitable for whole leaves gave us similar results on the accumulation of ROS in our A. brassicicola‐infected genotypes/treatments, confirming the lower levels of ROS in prx33/prx34 under‐producing Arabidopsis plants (Fig. 4D). ROS formation, represented by the intensity of fluorescence signals, in leaves of infected prx33 plants was 59% of that in leaves of the corresponding infected wild‐type plants, and in leaves of infected TRV‐PRX plants was 74% of that in leaves of infected TRV‐GFP plants, measured 2 days after inoculation with A. brassicicola. The fluorescence signal intensity in leaves of NADPH oxidase mutant rbohd plants was also detected as a reference, and a marked reduction in ROS accumulation was observed. In rbohd plants, the ROS‐related fluorescence signal intensity was 44% of that of wild‐type Col‐0 plants.
Leaf senescence clearly affects RbohD, PRX33 and PRX34 mRNA levels
The results of several studies have indicated functional interactions between the Arabidopsis NADPH oxidase RBOHD and the senescence hormone ethylene or its precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) (Bouchez et al., 2007; Pogány et al., 2009; Mersmann et al., 2010; Yao et al., 2017). This observation prompted us to investigate the effect of leaf age on the activity of genes (RbohD, PRX33 and PRX34) encoding crucial ROS‐generating proteins in Arabidopsis, before and after elicitation with A. brassicicola. Sampling was performed by dividing true leaves growing on one plant evenly into three age groups: oldest leaves (in leaf positions 1–4), middle leaves (in leaf positions 5–8) and youngest leaves (in leaf positions 9–12). The basal level of transcript abundance examined in mock‐inoculated wild‐type plants was highest in the oldest bottom leaves for all three ROS‐producing proteins, and non‐senescent or younger leaves exhibited moderate or lower basal transcript abundance for RbohD, PRX33 and PRX34 genes (Fig. 5). Twenty‐four hours after inoculation with A. brassicicola, however, non‐senescent middle leaves responded to fungal infection with the highest induction of RbohD, PRX33 and PRX34 transcript levels (Fig. 5).
Figure 5.
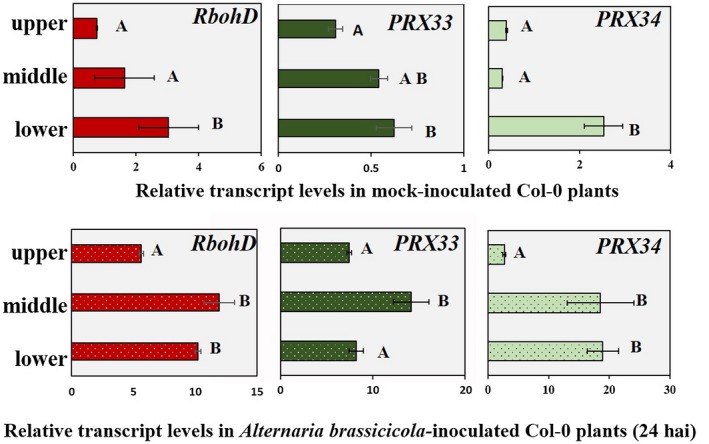
Transcript abundance of RbohD, PRX33 and PRX34 genes at three leaf levels. Gene expression was analysed in leaves representing three consecutive physiological states (upper, middle and lower). Five‐ to six‐week‐old wild‐type Arabidopsis plants (whole rosettes) were spray inoculated with Alternaria brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. Mock‐inoculated and A. brassicicola‐infected [24 h after inoculation (hai)] plants were assayed. Relative gene expression was calculated using the comparative 2–ΔΔCT method and At4g26410 as a reference gene. The results show the average of two experiments, each comprising three biological samples (each sample composed of a pool of the corresponding leaves of three Arabidopsis rosettes), analysed in three technical replicates. True leaves growing on one plant were divided evenly into three age groups: lower leaves (typically in positions 1–4), middle leaves (in positions 5–8) and upper leaves (in positions 9–12). Different letters represent relative transcript values significantly different at P < 0.05. Statistical analysis was performed using one‐way analysis of variance (ANOVA) and Tukey’s post hoc test.
Searching for new components in the PRX33/PRX34 and RBOHD signalling networks
In a pursuit to find unknown elements of the RBOHD or PRX33/PRX34 cellular interaction networks, two approaches were employed. First, T‐DNA insertion lines with perturbed activity of key Arabidopsis immune regulators were inoculated with A. brassicicola. Mutants corresponding to seven crucial determinants [BIK1, NPR1, MPK6, NIA2, EIN2, JAR1 (JASMONATE RESISTANT1) and NHO1 (NON‐HOST RESISTANCE TO P. SYRINGAE PV. PHASEOLICOLA 1)] showed altered host responses (in comparison with wild‐type plants) after inoculation with A. brassicicola. Six were further investigated. BIK1 was excluded because its role in the regulation of RBOHD activity has been confirmed (Kadota et al., 2014; Li et al., 2014). Mutants affected in the activity of two of these pivotal plant immunity factors, JAR1 and NHO1, exhibited consistently altered PRX33 and PRX34 transcript levels (Fig. 6), but the other four mutants showed wild‐type transcript levels for PRX33 and PRX34 genes. Interestingly, no apparent differences in RbohD mRNA levels could be detected between any of the immune mutants and the wild‐type, before or after inoculation with A. brassicicola.
Figure 6.
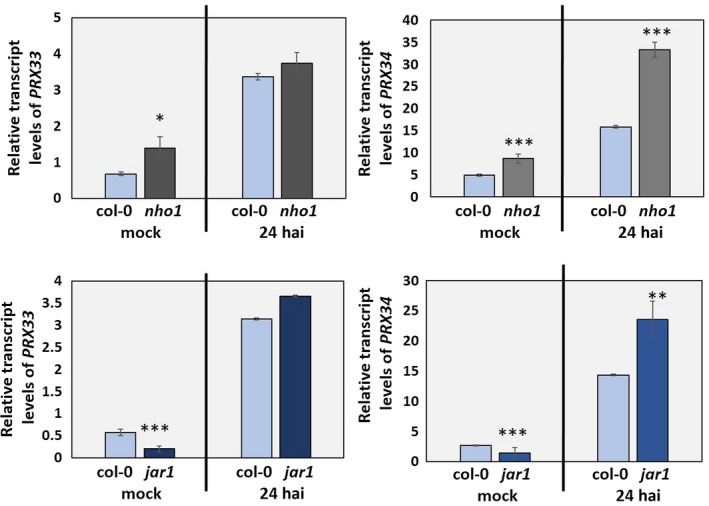
Transcript levels of PRX33 and PRX34 genes in nho1 and jar1 T‐DNA insertion lines relative to wild‐type plants. The level of gene expression was analysed before and after inoculation with Alternaria brassicicola [24 h after inoculation (hai)]. Dysfunctions in NHO1 and JAR1 activity affect PRX33 and PRX34 mRNA levels. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with A. brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. The results show the average of two experiments, each comprising three biological samples (each sample composed of a pool of three Arabidopsis rosettes), analysed in three technical replicates. Statistical analysis was performed using Student’s t‐test. Asterisks indicate statistically significant differences (*α = 0.05, **α = 0.01, ***α = 0.001).
In a second approach, T‐DNA insertion lines affected in the activity of 11 selected target proteins were analysed. Earlier, these proteins had been reported to establish a physical interaction with RBOHD (Geisler‐Lee et al., 2007; Jones et al., 2014). Mutants with associated insertions in the following genes were included: UDP‐Galactose Transporter 3, Annexin 1, Thioredoxin H‐type 7, Calmodulin 4, Glutathione S‐Transferase TAU 19, Quantitative Resistance to Plectosphaerella 1, Cell Elongation Protein, ADP‐Ribosylation Factor C1, Membrane‐Associated Progesterone Binding Protein 3, At4g37445 without functional prediction and NDR1/HIN1‐like 3 (Table S1, see Supporting Information). Two mutants in which the insertions could be linked to the same locus, At2g26330 encoding ERECTA (Quantitative Resistance to Plectosphaerella 1), gave spreading cell death phenotypes on inoculation with A. brassicicola, similar to those seen on the rbohd mutant (Fig. 7). The disease responses of the mutants representing the other 10 RBOHD interactors were the same as in the wild‐type. One of the erecta T‐DNA insertion lines, SALK_066455C, was also assayed for ROS formation after inoculation with A. brassicicola. The accumulation of H2O2, visualized by DAB staining, was highly compromised in whole leaves of the erecta mutant at 48 hai with A. brassicicola (Fig. 8).
Figure 7.

Alternaria brassicicola‐infected Col‐0, rbohd, erecta SALK_04410 and erecta SALK_066455C mutant Arabidopsis plants. Fungal symptoms were photographed at 7 days after inoculation. T‐DNA insertion lines with mutations in the genomic sequence of RbohD (At5g47910) and ERECTA (At2g26330) genes were used. Necrotic fungal symptoms are more intense in the two erecta mutants, as in the rbohd mutant, in comparison with the Col‐0 wild‐type. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with A. brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water.
Figure 8.
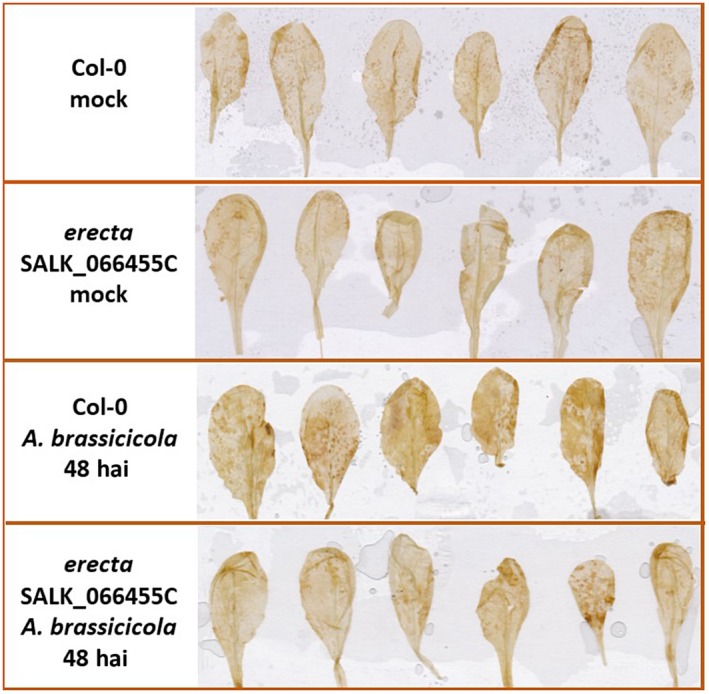
Hydrogen peroxide (H2O2) production in wild‐type (Col‐0) and erecta T‐DNA insertion line Arabidopsis leaves. The accumulation of H2O2 was visualized by 3,3′‐diaminobenzidine (DAB) staining. Five‐ to six‐week‐old Arabidopsis plants (whole rosettes) were spray inoculated with Alternaria brassicicola conidial suspension at a concentration of 5 × 105 conidia/mL distilled water. The staining was carried out with mock‐inoculated and A. brassicicola‐infected plants [48 h after inoculation (hai)]. Leaves of the A. brassicicola‐infected erecta mutant show markedly reduced accumulation of H2O2. The detection was repeated twice using 15 plants for each treatment. Representative leaves are shown.
Discussion
Alternaria brassicicola infection activates PRX33 and PRX34 genes
The mRNA expression of genes encoding ROS‐producing proteins was examined in A. brassicicola‐infected Arabidopsis plants. PRX33 and PRX34 mRNA levels were elevated even at 12 hai, reaching their climax at 24 hai. The analysis of NADPH oxidase RbohD transcript levels, investigated in the same pathosystem, revealed a similar transcriptional response (Pogány et al., 2009). The induction of PRX34 gene expression was also reported in Sclerotinia sclerotiorum‐infected Brassica napus plants at 12–48 hai (Yang et al., 2007). PAO1, PAO2, PAO3, PAO4 and PAO5 encoding genes, however, did not show A. brassicicola‐induced activation at 24 hai with the fungus (Figs 1 and S1).
These data suggest that the apoplastic peroxidases PRX33 and PRX34 play important roles in the response of Arabidopsis to A. brassicicola, but the significance of PAO1, PAO2, PAO3, PAO4 and PAO5 was not proven.
PRX33 and PRX34 contribute markedly to the A. brassicicola‐induced accumulation of H2O2
Impaired RBOHD function leads to the abolishment of apoplastic ROS accumulation in Alternaria‐infected Arabidopsis cells, whereas down‐regulation of PRX33/PRX34 activity results in a reduced number of leaf cells exhibiting the burst of extracellular ROS (Fig. 4). These results reveal that the RBOHD and PRX33/PRX34 ROS‐generating systems contribute simultaneously to the Alternaria‐induced extracellular oxidative burst in Arabidopsis. The reduced number of leaf cells showing H2O2 accumulation indicates a role of the apoplastic peroxidases PRX33/PRX34 in the spread of intercellular ROS signalling initiated by RBOHD (Gilroy et al., 2014, 2016).
Some crucial Arabidopsis stomatal responses mediated by the plant hormone cytokinin have been recently linked to the activity of peroxidases PRX33 and PRX34, together with certain other apoplastic peroxidases (Arnaud et al., 2017). It was shown that cytokinin‐induced stomatal closure, ROS accumulation in guard cells and stomatal immunity to coronatine‐deficient P. syringae pv. tomato DC3000 bacteria were dependent on the activity of the extracellular peroxidases PRX33 and PRX34.
Necrotic symptoms of the fungal infection are enhanced by PRX33/PRX34‐derived ROS
Down‐regulation of PRX33 and PRX34 led to reduced necrotic fungal symptoms on Arabidopsis leaves (Fig. 2) and the relative fungal biomass of A. brassicicola was lower in prx33/prx34 knockdown plants (Fig. 3). These results suggest that, similar to RBOHD, functional PRX33 and PRX34 and their ROS products support the growth of the fungus. However, PRX33/PRX34 activity leads to increased tissue necrosis, in contrast with functional RBOHD, which suppresses tissue necrosis.
It is unclear whether the reduced necrotic symptoms in our prx33/prx34 under‐producers are the result of partially compromised cell death induction or simply the consequence of limited necrotrophic pathogen growth. Mammarella et al. (2015) reported that asFBP1.1 prx knockdown plants retained the ability to mount a hypersensitive cell death response to attacks by avirulent P. syringae strains, suggesting that the apoplastic peroxidases PRX33/PRX34 are dispensable in pathogen‐induced cell death in Arabidopsis.
The inhibition of RBOHD activity in a T‐DNA mutant resulted in more intense tissue necrosis (increased spread of cell death) after inoculation with A. brassicicola. Fungal colonization, however, was suppressed in rbohd knockout plants relative to the wild‐type. Thus, RBOHD regulates host cell death on A. brassicicola infection and functional RBOHD supports the colonization of Arabidopsis by this necrotrophic fungus (Pogány et al., 2009).
Leaf senescence modulates the transcript levels of ROS‐producing enzymes
Leaf senescence has been associated with elevated ROS levels and reduced antioxidant capacity (Barna et al., 2012). In our studies, senescence was connected with an increased basal transcript level of pivotal ROS‐producing enzymes, but also with a reduced ability to develop a full transcriptional response of RbohD, PRX33 and PRX34 genes following the attack of a necrotrophic fungus (Fig. 5).
This is in accord with our microscopic observations, where cells in older leaves of Arabidopsis plants exhibited a mild, but uniform, induction of apoplastic basal H2O2 accumulation, but younger (non‐senescent) leaves responded to A. brassicicola infection with a more pronounced accumulation of extracellular H2O2. Characteristic spatio‐temporal mRNA expression patterns of NADPH oxidase isoforms RbohD and RbohF have been reported recently in unchallenged and pathogen‐elicited Arabidopsis plants (Morales et al., 2016).
Quest for new PRX33/34 and RBOHD interactors
In a search for new components in the PRX33/PRX34 and RBOHD signalling networks, numerous Arabidopsis mutants were tested.
T‐DNA insertion lines jar1 and nho1 consistently exhibited altered PRX33/PRX34 transcript abundance, both before and after inoculation with A. brassicicola (Fig. 6). JAR1 encodes a jasmonic acid‐amido synthetase which catalyses the formation of a biologically active jasmonyl‐isoleucine (JA‐Ile) conjugate. This amino acid is necessary for optimal signalling in some jasmonate responses in Arabidopsis (Staswick and Tiryaki, 2004; Staswick et al., 2002). Synthesis of the JA‐Ile conjugate by JAR1 and related JA‐conjugating enzymes is required for the plant immune system (Ryu et al., 2004; Staswick et al., 1998) and for various abiotic stress responses (Rao et al., 2000). Functional JAR1 alleviates the oxidative burst in Arabidopsis seedlings triggered by the bacterial MAMP elicitor flagellin (Yi et al., 2014). An interaction between JAR1 and ROS has been reported during cell wall damage‐induced lignin synthesis in Arabidopsis (Denness et al., 2011).
NHO1 protein is a glycerol kinase, which converts glycerol to glycerol 3‐phosphate and performs a rate‐limiting step in glycerol metabolism (Kang et al., 2003). This protein is also known as GLI1 or GLYCEROL‐INSENSITIVE 1 (Eastmond, 2004). Arabidopsis NHO1 transcript levels are induced by non‐host Pseudomonas bacterial strains, and functional NHO1 is required for resistance against non‐host and avirulent Pseudomonas strains, as well as against the necrotrophic fungal pathogen Botrytis cinerea. However, the NHO1 gene is ineffective against virulent P. syringae pv. tomato DC3000, and its transcription is suppressed by the DC3000 strain. Interestingly, the JA signalling pathway is specifically required for NHO1 suppression by DC3000 bacteria (Kang et al., 2003; Li et al., 2005; Lu et al., 2001). Although the fact that NHO1 is a significant component of the immune system of Arabidopsis is well established, its specific functions in immune signalling are still largely elusive (Lu et al., 2010; Maeda et al., 2010). Nonetheless, a connection between ROS production and NHO1 activity is emerging (Wang et al., 2014). Perturbation of the cellular redox status in plants, triggered by catalase or glycolate oxidase deficiency or by treatment with the ROS‐generating compound methylviologen, significantly alters the transcript level of NHO1 (Chaouch et al., 2010; El‐Maarouf‐Bouteau et al., 2015; Rojas et al., 2012). Conversely, plants with disturbed glycerol metabolism or with impaired NHO1 gene expression have been shown to exhibit elevated H2O2 formation (Hu et al., 2014; Li et al., 2016). Our results, revealing enhanced PRX33/PRX34 gene activity in the nho1 knockout (in comparison with wild‐type plants), before or after inoculation with A. brassicicola, indicate that these extracellular peroxidases may be partly responsible for the observed interaction between ROS and NHO1 (Fig. 6).
In a different set of experiments, Arabidopsis plants carrying mutations in the ERECTA gene were shown to display spreading necrosis symptoms on A. brassicicola infection, very similar to those observed on the rbohd mutant (Fig. 7). Inoculated leaves of the erecta mutant (SALK_066455C) also exhibited reduced H2O2 accumulation (Fig. 8). ERECTA has been reported previously to establish a physical interaction with RBOHD (Geisler‐Lee et al., 2007; Jones et al., 2014).
ERECTA (QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1, AGI locus code: At2g26330) has been connected to thermotolerance. It belongs to the protein kinase superfamily and to the serine/threonine (Ser/Thr) protein kinase family, which includes proteins that are associated with development, pathogen defence and phytohormone perception (Sánchez‐Rodríguez et al., 2009; Torii et al., 1996). ERECTA plays a role in the immune response of Arabidopsis because erecta mutant plants are more susceptible than wild‐type plants to a variety of pathogens (Magnaporthe oryzae, Plectosphaerella cucumerina, Verticillium longisporum) (Haffner et al., 2014; Llorente et al., 2005; Takahashi et al., 2016). Fusarium graminearum infection apparently down‐regulates mRNA abundance for the ERECTA gene in Arabidopsis pistils (Mondragon‐Palomino et al., 2017). Results in our study demonstrate that ERECTA is also required for ROS accumulation and the suppression of necrotic symptoms caused by A. brassicicola, a necrotrophic fungal pathogen. ERECTA not only physically interacts with RBOHD (one of the major cellular sources of apoplastic ROS) in Arabidopsis, but has been shown to be part of a signalling pathway responsible for ROS sensing and redox‐mediated cortex proliferation in the roots (Cui et al., 2014). Recently, the ROS responsivity of the ERECTA gene has been exposed in methylviologen‐treated Arabidopsis rosettes (Han et al., 2014).
Taken together, we consider JAR1 and NHO1 to be potential members of the PRX33/PRX34 cellular signalling system and ERECTA to be an expected component of the RBOHD interaction network in Arabidopsis leaf cells when plants are challenged by the necrotrophic fungus A. brassicicola.
In this study, we have provided information on the simultaneous and comparable contribution of class III peroxidases PRX33 and PRX34 and NADPH oxidase RBOHD to the apoplastic oxidative burst investigated in the same Arabidopsis–Alternaria pathosystem. We have also presented microscopic images of the altered pattern of ROS accumulation in Arabidopsis plants with reduced PRX33 and PRX34 activity following inoculation with A. brassicicola. In contrast with the ROS‐producing NADPH oxidase RBOHD, which surprisingly inhibits the spread of cell death in pathogen‐infected Arabidopsis tissues (Pogány et al., 2009), functional cell wall peroxidases PRX33 and PRX34 apparently enhance the development of necrotic symptoms triggered by infection with A. brassicicola. Fungal growth, however, is similarly stimulated by both RBOHD and the two apoplastic peroxidases. Finally, a senescence‐dependent distribution of RbohD, PRX33 and PRX34 transcripts is reported. As a conclusion of this study, ROS generated by the extracellular peroxidases PRX33 and PRX34 function in Arabidopsis as susceptibility factors when plants are challenged by the necrotrophic fungal pathogen A. brassicicola.
Experimental Procedures
Plant materials and growth conditions
Wild‐type (Col‐0), transgenic, mutant and gene‐silenced Arabidopsis plants were grown in a peat growing medium (Pindstrup Plus Orange, Pindstrup Mosebrug, Ryomgaard, Denmark) at 22 °C in a growth chamber under a 14‐h light/10‐h dark cycle with 80 µmol/m2/s irradiation. Seeds of the Arabidopsis line asFBP1.1 expressing a French bean (Phaseolus vulgaris) peroxidase sequence in antisense orientation (Bindschedler et al., 2006) were received from Professor F. M. Ausubel (Department of Genetics, Harvard Medical School, Boston, MA, USA). Arabidopsis T‐DNA insertion lines were ordered from the European Arabidopsis Stock Center. The list of Arabidopsis lines used in this study is provided in Table S1. Characterization of the prx33 insertion line (SALK_062314C) is shown in Fig. S4 (see Supporting Information), whereas lines prx34 (SALK 051769C) and rbohd (SALK_070610C) have been described previously (Passardi et al., 2006; Pogány et al., 2009). An Arabidopsis line expressing a green fluorescent protein coding mGFP‐ER sequence (Haseloff et al., 1997), used for all VIGS experiments, was generated by a modified floral dip method (Logemann et al., 2006).
Cultivation of fungal strain and inoculation
Alternaria brassicicola strain MUCL20297, isolated from cabbage (Thomma et al., 1998), was grown on potato dextrose agar medium at room temperature in the dark. The conidial suspension contained 5 × 105 conidia/ml distilled water, and conidia were harvested from 1‐week‐old cultures. The inoculum was sprayed onto the leaf surface of 5–6‐week‐old Arabidopsis plants. The infected plants were incubated in 100% relative humidity in translucent plastic boxes under the same growing conditions as before. For mock‐inoculated plants, distilled water was sprayed onto the leaves and they were kept under the same conditions as the inoculated plants.
Gene expression analysis
For total RNA extraction, Arabidopsis leaves were frozen in liquid nitrogen and ground with a mortar and pestle; 100 mg of plant material was used according to the manufacturer’s instructions (Total RNA Extraction Miniprep Kit, Viogene, Taipei, Taiwan). The RNA content and purity were analysed with a NanoDrop‐1000 (Thermo Fisher Scientific, Waltham, MA, USA) spectrophotometer.
Prior to cDNA synthesis all samples were subjected to DNase treatment (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), DNA‐free™ DNA Removal Kit).
First‐strand cDNA was synthesized with a Thermo Scientific First Strand cDNA Synthesis Kit and used as a template for real‐time polymerase chain reaction (PCR) analysis in ten‐fold dilution. Real‐time PCRs were prepared using a Bioline (Taunton, MA, USA) SensiFAST SYBR® No‐ROX Kit. The reactions were performed in a total volume of 15 μL containing 7.5 μL 2X Bioline SensiFAST qPCR Master Mix, 1.5–1.5 μL forward and reverse primers (10 μm), 2.5 μL template and 2 μL PCR‐grade water. The list of primer pairs used in this work is provided in Table S2 (see Supporting Information). All real‐time PCRs were carried out in a C1000 Touch Thermal Cycler equipped with a CFX96 Real‐Time PCR System (Bio‐Rad, Hercules, CA, USA). The following cycling conditions were selected: 95 °C for 3 min, then 40 cycles of 95 °C for 20 s, followed by 60 °C for 60 s. Finally, a melt curve analysis was performed to determine amplicon specificity with temperature increases from 65 to 95 °C in steps of 0.5 °C. Relative quantification analysis was performed using the comparative 2−ΔΔCt method (Livak and Schmittgen, 2001). To evaluate the level of gene expression, the results were normalized using Ct values from the cDNA amplification of the constitutively expressed Arabidopsis At4g26410 gene (Czechowski et al., 2005).
Determination of fungal biomass
Genomic DNA was isolated using the method of Brouwer et al. (2003) with some modifications. Spray‐infected rosettes of Arabidopsis plants (10 days after inoculation with A. brassicicola) were ground in liquid nitrogen with a mortar and pestle. To 100 mg frozen plant material, 300 µL lysis buffer [2.5 m LiCl, 50 mm Tris‐HCl, 62.5 mm Na2‐ethylenediaminetetraacetic acid (EDTA), 4% Triton X‐100, pH 8.0] and an equal volume of phenol–chloroform–isoamyl alcohol (25 : 24 : 1, v/v) were added, and samples were thoroughly vortexed. After centrifugation (5 min, 16 000 g) at room temperature, the supernatant was recovered and genomic DNA was precipitated by the addition of two volumes of 100% ethanol, incubation for 15 min at −20 °C and another round of centrifugation. The DNA pellet was washed with 70% ethanol, air dried and resuspended in nuclease‐free water. DNA quality and concentration were analysed with a NanoDrop spectrophotometer. Fungal biomass content was determined from total genomic DNA extracts by quantitative real‐time PCR analysis, and the ratio of A. brassicicola to Arabidopsis genomic DNA was assessed using the same platform and conditions as for the gene expression analyses described above. Primer sequences specific for A. brassicicola (Pogány et al., 2009) were derived from the ribosomal ITS region of the fungus (forward, 5′‐TCTCCAGTTTGCTGGAGACT‐3′; reverse, 5′‐GGATGCTGACCTTGGCTGGA‐3′), and a primer pair specific for the sequence of Arabidopsis At4g26410 was used to determine plant biomass content in the samples.
VIGS
TRV, a bipartite virus‐based silencing system, was used for VIGS experiments. TRV1 contains the viral replicase, the RNA‐dependent RNA polymerase and the movement protein, whereas TRV2 contains the coat protein and a multiple cloning site to insert host plant‐derived fragment(s) of target sequences (Hayward et al., 2011). pTRV1 (STOCK: CD3‐1039) and pTRV2 in pCAMBIA3301 (STOCK: CD3‐1043) Agrobacterium plasmids were obtained from the Arabidopsis Biological Resource Center.
First, a marker GFP silencing construct (TRV2‐GFP) was created to check the efficiency of silencing. For this purpose, a 256‐bp mGFP fragment was amplified from pEarlyGate 103 (Earley et al., 2006) with GFP‐specific primer pairs (forward, 5′‐CGCTCTAGAATGCCTGAGGGATACGTGCAG‐3′; reverse, 5′‐CGCTCTAGATTCGATGTTGTGGCGGGTCTT‐3′) and inserted into a pGEM®‐T Easy vector (Promega, Madison, WI, USA). The GFP insert from the purified pGEM®‐T Easy vector (Nucleospin Plasmid, Macherey‐Nagel, Düren, NRW, Germany) was digested with EcoRI (Thermo Fisher Scientific, Waltham, MA, USA), and the gel‐purified GFP fragment (Agarose gel DNA extraction kit, Roche) was ligated into the EcoRI‐digested pTRV2 plasmid.
For the TRV2‐GFP‐PRX VIGS vector, a 261‐bp cDNA fragment from the PRX33 gene was amplified (forward, 5′‐CGCGGATCCGCTGATGGCACACAAACATTC‐3′; reverse, 5′‐ GCGGGATCCAATACAATCTGCTCCTGCTCAA‐3′), which shows 31% sequence similarity with PRX34 cDNA. The target sequence specificity of silencing and avoidance of off‐target gene silencing were controlled and confirmed by the siRNA tool (http://bioinfo2.noble.org/RNAiScan.htm) (Xu et al., 2006) searching its Arabidopsis thaliana DCFCI Gene Index V 15 release on 23/04/2010, Arabidopsis thaliana TAIR 9 V 9 release on 19/06/2009 and Arabidopsis thaliana mRNA (from TIGR Ath1 5). The amplified fragment was cloned into the BamHI restriction site of the TRV2‐GFP vector.
Afterwards, VIGS plasmids were introduced into Escherichia coli DH5α by the heat shock method (Tu et al., 2005). The identity of the clones was confirmed by DNA sequence determination. VIGS plasmids were finally purified from E. coli (Nucleospin Plasmid, Macherey‐Nagel) and introduced into Agrobacterium tumefaciens MOG301 (Hood et al., 1993) by electroporation. For VIGS, Agrobacterium strains containing pTRV1 and pTRV2 vectors were grown at 28 °C overnight on LB (Lysogeny Broth) medium supplemented with the appropriate antibiotics (kanamycin sulfate, 30 µg/mL; rifampicin, 50 µg/mL). For inoculum preparation, bacterial cells were suspended in Agrobacterium incubation buffer (1.95 g MES (2‐[N‐morpholino]ethanesulfonic acid), 2 g MgCl2 .6H2O in 1 L distilled water, pH 5.6) and supplemented with acetosyringone (final concentration of 150 mm). Bacterial cell densities were adjusted with a spectrophotometer to an optical density at 600 nm (OD600) = 1.5. After a 3‐h incubation at room temperature, the bacterial suspensions (TRV1 and TRV2), mixed in a ratio of 1 : 1 (v/v), were infiltrated into two lower leaves of 2–3‐week‐old, GFP‐expressing transgenic Arabidopsis plants, which were grown at 22 °C in a growth chamber under a 16‐h light/8‐h dark cycle (Burch‐Smith et al., 2006).
Three weeks after agroinfiltration, the effect of gene silencing was observed under UV light and gene‐silenced plants were selected and used for mechanical inoculation as viral inoculum on new 2–3‐week‐old, GFP‐expressing transgenic Arabidopsis seedlings which were kept under a 14‐h light/10‐h dark cycle. Mechanical inoculation was chosen instead of agroinfiltration because the 16‐h light/8‐h dark photoperiod condition required for agroinoculation did not provide Arabidopsis plants appropriate for pathological experiments with A. brassicicola (plants grown under these conditions did not develop useful rosettes). Fourteen days after mechanical inoculation, the effect of gene silencing was tested on entire plants under UV light and by real‐time PCR from the UV‐selected plants. The mRNA contents of PRX33 and PRX34 were calculated using the same platform and conditions as for the real‐time PCR assays above (PRX33: forward, 5′‐AAATTCAGCCCGAGGATTTC‐3′; reverse, 5′‐GAGCAGCAATGGTGAGCATA‐3′; PRX34: forward, 5′‐CGAGAAACCATTGTAAATGAGT‐3′; reverse, 5′‐CCGAGCCGAATTTGCG‐3′).
H2O2 detection
Mock‐inoculated and A. brassicicola‐infected Arabidopsis plants (2 days after inoculation) were vacuum infiltrated with DAB (Sigma‐Aldrich, St. Louis, MO, USA) dissolved in distilled water (1 mg/mL), incubated in the light for 2 h, decolorized with plant clearing solution (80% ethanol, 20% chloroform, 0.15% trichloroacetic acid) and mounted in 50% glycerol solution, as described previously (Pogány et al., 2004). Stained plants were examined by light microscopy.
The fluorescent dye DCFH‐DA was also used to detect H2O2. Detached Arabidopsis leaves in the middle position (in positions 5–8) were vacuum infiltrated with 0.4 mm DCFH‐DA in 10 mm sodium phosphate buffer (pH 7.4), as described by Bozsó et al. (2005). After a 10‐min incubation period in the dark, leaves were photographed under 302–365‐nm UV light illumination by an AlphaImager Mini gel documentation system (ProteinSimple, Santa Clara, CA, USA). For each genotype or treatment, 30 leaves (in the middle position) were assayed and, for each leaf, an average pixel fluorescence intensity was calculated by AlphaView software of AlphaImager® Systems using the Multiplex Band Analysis function.
Statistical analysis
Experimental data were analysed using Student’s t‐test, the non‐parametric Mann–Whitney U‐test or one‐way analysis of variance (ANOVA) and subsequent Tukey’s honestly significant difference test. Statistical analyses were performed with IBM SPSS Statistics 20 Software (Armonk, New York, USA).
Supporting information
Fig. S1 Transcript levels of five polyamine oxidase (PAO)‐encoding genes of wild type (Col‐0) Arabidopsis thaliana. Five to 6 weeks old Arabidopsis plants (whole rosettes) were spray‐inoculated with A. brassicicola conidium suspension used in a concentration of 5 x 105 conidia in 1 mL distilled water. Samples of mock‐inoculated and A. brassicicola‐infected plants (24 hai) were analyzed. Transcript levels of the five PAO isoforms are not increased as a result of the fungal infection. Data represent the mean of three independent biological samples with three technical replicates for each. Statistical analysis was performed using Student's t‐test. Asterisks indicate statistically significant difference (**α = 0.01).
Fig. S2 Transcript level of apoplastic class III peroxidase gene PRX33 (At3g49110) is reduced in the Arabidopsis prx33 knock‐down T‐DNA insertion line (SALK_062314C). Transcript levels were quantified in untreated Arabidopsis plants by real‐time RT‐PCR. The results show the average of two experiments each comprising three biological samples (each sample composed as a pool of 3 Arabidopsis rosettes) analyzed in three technical replicates. Statistical analysis was performed using Student's t‐test. Asterisks indicate statistically significant difference (***α = 0.001).
Fig. S3 Transcript levels of apoplastic class III peroxidase genes PRX33 (At3g49110) and PRX34 (At3g49120) are reduced in Col‐0 Arabidopsis plants that were treated with a VIGS construct (TRV‐PRX) that targets PRX33 and PRX34 mRNA sequences. Plants labelled as TRV‐GFP were treated with the control construct that targets only mRNA sequences of the GFP marker gene. Silencing of transgene GFP transcript levels and a subsequent abolishment of GFP‐derived green fluorescence in VIGS‐treated plants was used to detect the occurrence of successful gene silencing events. Five to 6 weeks old Arabidopsis plants (whole rosettes) were spray‐inoculated with A. brassicicola conidium suspension used in a concentration of 5 x 105 conidia in 1 mL distilled water. PRX33 and PRX34 mRNA levels were monitored in VIGS‐treated Arabidopsis plants before (A) and after (B) inoculation with A. brassicicola (24 hai) by real‐time RT‐PCR. The results show the average of two experiments each comprising three biological samples (each sample composed as a pool of 3 Arabidopsis rosettes) analyzed in three technical replicates. Statistical analysis was performed using Student’s t‐test. Asterisks indicate statistically significant differences (*α = 0.05, **α = 0.01, ***α = 0.001).
Fig. S4 Confirmation of T‐DNA insertion by genotyping prx33 (SALK_062314C) Arabidopsis line. PCR analysis to confirm the presence of T‐DNA was performed using genomic DNA of the Arabidopsis insertion line prx33 (SALK_062314C) and of wild type Col‐0. The first 4 lanes show results of PCR amplifications, where primers specific for PRX33 sequence (forward 5’ ‐ATTATAGTTGTTGTCAGCATTAGCA‐3’, reverse 5’‐ACCATTTGTTCCTCTGAAGCA‐3’) were used with Col‐0 and prx33 genomic DNA extracts as templates. The last three lanes exhibit PCR results where a T‐DNA left border primer (LBa1:5’‐ TGGTTCACGTAGTGGGCCATCG ‐3’) was combined with PRX33 sequence specific forward primer using prx33 insertion line genomic DNA extract confirming the location of T‐DNA insertion. No template controls were included. PCR products were fractionated in 1% agarose gel and DNA was visualized by staining with GelRed (Fremont, California, USA).
Table S1 Arabidopsis T‐DNA insertion lines used in this work.
Table S2 Primer sequences used in this work.
Acknowledgements
Arabidopsis thaliana line asFBP1.1 and A. brassicicola strain MUCL 20297 were kindly provided by Professors Frederick M. Ausubel (Department of Genetics, Harvard Medical School, Boston, MA, USA) and Bruno Cammue (Centre of Microbial and Plant Genetics, University of Leuven, Leuven, Belgium). Funding from the Hungarian National Research, Development and Innovation Fund (OTKA: K 104730, K 124131) and from the Bolyai Scholarship (BO_609_12) is also gratefully acknowledged.
The authors have no conflicts of interest to declare.
References
- Andronis, E.A. , Moschou, P.N. , Toumi, I. and Roubelakis‐Angelakis, K.A. (2014) Peroxisomal polyamine oxidase and NADPH‐oxidase cross‐talk for ROS homeostasis which affects respiration rate in Arabidopsis thaliana. Front. Plant Sci. 5, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini, R. , Cona, A. , Federico, R. , Fincato, P. , Tavladoraki, P. and Tisi, A. (2010) Plant amine oxidases ‘‘on the move’’: an update. Plant Physiol. Biochem. 48, 560–564. [DOI] [PubMed] [Google Scholar]
- Arnaud, D. , Lee, S. , Takebayashi, Y. , Choi, D. , Choi, J. , Sakakibara, H. and Hwang, I. (2017) Cytokinin‐mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in Arabidopsis . Plant Cell, 29, 543–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna, B. , Fodor, J. , Harrach, B.D. , Pogány, M. and Király, Z. (2012) The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 59, 37–43. [DOI] [PubMed] [Google Scholar]
- Bindschedler, L.V. , Dewdney, J. , Blee, K.A. , Stone, J.M. , Asai, T. , Plotnikov, J. , Denoux, C. , Hayes, T. , Gerrish, C. , Davies, D.R. , Ausubel, F.M. and Bolwell, G.P. (2006) Peroxidase‐dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez, O. , Huard, C. , Lorrain, S. , Roby, D. and Balagué, C . (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1 . Plant Physiol. 145, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozsó, Z. , Ott, P.G. , Szatmári, A. , Czelleng, G. , Varga, G. , Besenyei, E. , Sárdi, É. , Bányai, É. and Klement, Z. (2005) Early detection of Bacterium‐induced basal resistance in tobacco leaves with diaminobenzidine and dichlorofluorescein diacetate. J. Phytopathol. 153, 596–607. [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , Van den Ackerveken, G. , Cammue, B.P.A. and Thomma, B.P.H.J. (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Schiff, M. , Liu, Y. and Dinesh‐Kumar, S.P. (2006) Efficient virus‐induced gene silencing in Arabidopsis . Plant Physiol. 142, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch, S. , Queval, G. , Vanderauwera, S. , Mhamdi, A. , Vandorpe, M. , Langlois‐Meurinne, M. , Van Breusegem, F. , Saindrenan, P. and Noctor, G. (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength‐related manner. Plant Physiol. 153, 1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Kong, D. , Wei, P. , Hao, Y. , Torii, K.U. , Lee, J.S. and Li, J. (2014) SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox‐mediated cortex proliferation in the Arabidopsis root. Mol. Plant, 7, 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.‐R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi, A. , Cheng, Z. , O’Brien, J.A. , Mammarella, N. , Khan, S. , Ausubel, F.M. and Bolwell, G.P. (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern‐triggered immunity. Plant Cell, 24, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denness, L. , McKenna, J.F. , Segonzac, C. , Wormit, A. , Madhou, P. , Bennett, M. and Hamann, T. (2011) Cell wall damage‐induced lignin biosynthesis is regulated by a reactive oxygen species‐ and jasmonic acid‐dependent process in Arabidopsis . Plant Physiol. 156, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochowska‐Boguta, M. , Nadolska‐Orczyk, A. and Orczyk, W. (2013) Roles of peroxidases and NADPH oxidases in the oxidative response of wheat (Triticum aestivum) to brown rust (Puccinia triticina) infection. Plant Pathol. 62, 993–1002. [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J. (2004) Glycerol‐insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J., 37, 617–625. [DOI] [PubMed] [Google Scholar]
- El‐Maarouf‐Bouteau, H.A.Y.A.T. , Sajjad, Y. , Bazin, J. , Langlade, N. , Cristescu, S.M. , Balzergue, S. and Bailly, C. (2015) Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ. 38, 364–374. [DOI] [PubMed] [Google Scholar]
- Fincato, P. , Moschou, P.N. , Ahou, A. , Angelini, R. , Roubelakis‐Angelakis, K.A. , Federico, R. and Tavladoraki, P. (2012) The members of Arabidopsis thaliana PAO gene family exhibit distinct tissue‐ and organ‐specific expression pattern during seedling growth and flower development. Amino Acids, 42, 831–841. [DOI] [PubMed] [Google Scholar]
- Floerl, S. , Majcherczyk, A. , Possienke, M. , Feussner, K. , Tappe, H. , Gatz, C. , Feussner, I. , Kües, U. and Polle, A. (2012) Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana . PLoS ONE, 7, e31435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson Matika, D.E. and Loake, G.J. (2013) Redox regulation in plant immune function. Antioxid. Redox Signal. 21, 1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler‐Lee, J. , O'Toole, N. , Ammar, R. , Provart, N.J. , Millar, A.H. and Geisler, M. (2007) A predicted interactome for Arabidopsis . Plant Physiol. 145, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S. , Białasek, M. , Suzuki, N. , Górecka, M. , Devireddy, A.R. , Karpiński, S. and Mittler, R. (2016) ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 171, 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S. , Suzuki, N. , Miller, G. , Choi, W.G. , Toyota, M. , Devireddy, A.R. and Mittler, R. (2014) A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19, 623–630. [DOI] [PubMed] [Google Scholar]
- Groß, F. , Durner, J. and Gaupels, F. (2013) Nitric oxide, antioxidants and prooxidants in plant defence responses. Front. Plant Sci. 4, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner, E. , Karlovsky, P. , Splivallo, R. , Traczewska, A. and Diederichsen, E. (2014) ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum . BMC Plant Biol. 14, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H.J. , Peng, R.H. , Zhu, B. , Fu, X.Y. , Zhao, W. , Shi, B. and Yao, Q.H. (2014) Gene expression profiles of Arabidopsis under the stress of methyl viologen: a microarray analysis. Mol. Biol. Rep. 41, 7089–7102. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. , Siemering, K.R. , Prasher, D.C. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, A. , Padmanabhan, M. and Dinesh‐Kumar, S.P. (2011) Virus‐induced gene silencing in Nicotiana benthamiana and other plant species. Plant Rev. Genet. 678, 55–63. [DOI] [PubMed] [Google Scholar]
- Hood, E.E. , Gelvin, S.B. , Melchers, L.S. and Hoekema, A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. [Google Scholar]
- Hu, J. , Zhang, Y. , Wang, J. and Zhou, Y. (2014) Glycerol affects root development through regulation of multiple pathways in Arabidopsis . PLoS ONE, 9, e86269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.A. , Xuan, Y. , Xu, M. , Wang, S.‐R. , Ho, H.‐C. , Lalonde, S. , You, H.C. , Sardi, I.M. , Parsa, A.S. , Smith‐Valley, E. , Su, T. , Frazer, A.K. , Pilot, G. , Pratelli, R. , Grossmann, G. , Acsaraya, R.B. , Hu, C.‐H. , Engineer, C. , Villers, F. , Ju, C. , Takeda, K. , Su, Z. , Dong, Q. , Assmann, M.S. , Chen, J. , Kwak, M.J. , Schroeder, I.J. , Albert, R. , Rhee, Y.S. and Frommer, B.F. (2014) Border control—a membrane‐linked interactome of Arabidopsis . Science, 344, 711–716. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, G.D.J. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kang, L. , Li, J. , Zhao, T. , Xiao, F. , Tang, X. , Thilmony, R. , He, S. and Zhou, J.M. (2003) Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. USA, 100, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkönen, A. and Kuchitsu, K. (2015) Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry, 112, 22–32. [DOI] [PubMed] [Google Scholar]
- Li, L. , Li, M. , Yu, L. , Zhou, Z. , Liang, X. , Liu, Z. , Cai, G. , Gao, L. , Zhang, X. , Wang, Y. and Chen, S. (2014) The FLS2‐associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe, 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Li, X. , Lin, H. , Zhang, W. , Zou, Y. , Zhang, J. , Tang, X. and Zhou, J.M. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA, 102, 12 990–12 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Song, N. , Zhao, C. , Li, F. , Geng, M. , Wang, Y. , Liu, W. , Xie, C. and Sun, Q. (2016) Application of glycerol for induced powdery mildew resistance in Triticum aestivum L. Front. Physiol. 7, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. and He, C. (2016) Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 35, 995–1007. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Llorente, F. , Alonso‐Blanco, C. , Sanchez‐Rodriguez, C. , Jorda, L. and Molina, A. (2005) ERECTA receptor‐like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant J. 43, 165–180. [DOI] [PubMed] [Google Scholar]
- Logemann, E. , Birkenbihl, R.P. , Ülker, B. and Somssich, I.E. (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods, 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Zou, Y. and Feng, N. (2010) Overexpression of AHL20 negatively regulates defenses in Arabidopsis . J. Integr. Plant Biol. 52, 801–808. [DOI] [PubMed] [Google Scholar]
- Lu, M. , Tang, X. and Zhou, J.‐M. (2001) Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell, 13, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, R. , Stiller, J. , Powell, J. , Rusu, A. , Manners, J.M. and Kazan, K. (2015) Fusarium oxysporum triggers tissue‐specific transcriptional reprogramming in Arabidopsis thaliana . PLoS ONE, 10, e0121902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, K. , Houjyou, Y. , Komatsu, T. , Hori, H. , Kodaira, T. and Ishikawa, A. (2010) Nonhost resistance to Magnaporthe oryzae in Arabidopsis thaliana . Plant Signal. Behav. 5, 755–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammarella, N.D. , Cheng, Z. , Fu, Z.Q. , Daudi, A. , Bolwell, G.P. , Dong, X. and Ausubel, F.M. (2015) Apoplastic peroxidases are required for salicylic acid‐mediated defense against Pseudomonas syringae . Phytochemistry, 112, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, D. , Dunand, C. , Puppo, A. and Pauly, N. (2012) A burst of plant NADPH oxidases. Trends Plant Sci. 17, 9–15. [DOI] [PubMed] [Google Scholar]
- Mersmann, S. , Bourdais, G. , Rietz, S. and Robatzek, S. (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet‐Spruyt, L. , Xu, E. , Idänheimo, N. , Hoeberichts, F.A. , Mühlenbock, P. , Brosché, M. , Breusegem, V.F. and Kangasjärvi, J. (2016) Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 67, 3831–3844. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2017) ROS are good. Trends Plant Sci. 22, 11–19. [DOI] [PubMed] [Google Scholar]
- Mondragon‐Palomino, M. , John‐Arputharaj, A. , Pallmann, M. and Dresselhaus, T. (2017) Similarities between reproductive and immune pistil transcriptomes of Arabidopsis species. Plant Physiol. 174, 1559–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, J. , Kadota, Y. , Zipfel, C. , Molina, A. and Torres, M.‐A. (2016) The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67, 1663–1676. [DOI] [PubMed] [Google Scholar]
- Mou, Z. , Fan, W. and Dong, X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell, 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Narusaka, Y. , Narusaka, M. , Seki, M. , Ishida, J. , Shinozaki, K. , Nan, Y. , Park, P. , Shiraisi, T. and Kobayashi, M. (2005) Cytological and molecular analyses of non‐host resistance of Arabidopsis thaliana to Alternaria alternata . Mol. Plant Pathol. 6, 615–627. [DOI] [PubMed] [Google Scholar]
- O’Brien, J.A. , Daudi, A. , Finch, P. , Butt, V.S. , Whitelegge, J.P. , Souda, P. , Ausubel, M.F. and Bolwell, G.P. (2012) A peroxidase‐dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP‐elicited defense. Plant Physiol. 158, 2013–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi, F. , Tognolli, M. , De Meyer, M. , Penel, C. and Dunand, C. (2006) Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta, 223, 965–974. [DOI] [PubMed] [Google Scholar]
- Pogány, M. , Koehl, J. , Heiser, I. , Elstner, E.F. and Barna, B. (2004) Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to Tobacco necrosis virus and confers tolerance to oxidative stress. Physiol. Mol. Plant Pathol. 65, 39–47. [Google Scholar]
- Pogány, M. , von Rad, U. , Grün, S. , Dongó, A. , Pintye, A. , Simoneau, P. , Bahnweg, G. , Kiss, L. , Barna, B. and Durner, J. (2009) Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis‐Alternaria pathosystem. Plant Physiol. 151, 1459–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M.V. , Lee, H.‐I. , Creelman, R.A. , Mullet, J.E. and Davis, K.R. (2000) Jasmonic acid signaling modulates ozone‐induced hypersensitive cell death. Plant Cell, 12, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach, T. , Colville, L. , Beckett, R.P. , Minibayeva, F.V. , Havaux, M. and Kranner, I. (2015) A proposed interplay between peroxidase, amine oxidase and lipoxygenase in the wounding‐induced oxidative burst in Pisum sativum seedlings. Phytochemistry, 112, 130–138. [DOI] [PubMed] [Google Scholar]
- Rojas, C.M. , Senthil‐Kumar, M. , Wang, K. , Ryu, C.M. , Kaundal, A. and Mysore, K.S. (2012) Glycolate oxidase modulates reactive oxygen species‐mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis . Plant Cell, 24, 336–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.M. , Murphy, J.F. , Mysore, K.S. and Kloepper, J.W. (2004) Plant growth promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1‐independent and jasmonic acid‐dependent signaling pathway. Plant J. 39, 381–392. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Rodríguez, C. , Estévez, J.M. , Llorente, F. , Hernández‐Blanco, C. , Jordá, L. , Pagán, I. , Berrocal, M. , Marco, Y. , Somerville, S. and Molina, A. (2009) The ERECTA receptor‐like kinase regulates cell wall‐mediated resistance to pathogens in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 22, 953–963. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Manners, J.M. , Anderson, P.J. , Simpson, S.R. , Wilson, W.I. , Somerville, C.S. and Maclean, J.D. (2003) Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. and Tiryaki, I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis . Plant Cell, 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. , Tiryaki, I. and Rowe, M.L. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole‐3‐acetic acids in an assay for adenylation. Plant Cell, 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. , Yuen, G.Y. and Lehman, C.C. (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare . Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Su’udi, M. , Kim, M.G. , Park, S.‐R. , Hwang, D.‐J. , Bae, S.‐C. and Ahn, I.‐P. (2011) Arabidopsis cell death in compatible and incompatible interactions with Alternaria brassicicola . Mol. Cells, 31, 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana, N. , Florance, H.V. , Johns, A. and Smirnoff, N. (2015) Ascorbate deficiency influences the leaf cell wall glycoproteome in Arabidopsis thaliana . Plant Cell Environ. 38, 375–384. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T. , Shibuya, H. and Ishikawa, A. (2016) ERECTA contributes to non‐host resistance to Magnaporthe oryzae in Arabidopsis . Biosci. Biotechnol. Biochem. 80, 1390–1392. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y. , Cong, R. , Sagor, G.H. , Niitsu, M. , Berberich, T. and Kusano, T. (2010) Characterization of five polyamine oxidase isoforms in Arabidopsis thaliana . Plant Cell Rep. 29, 955–965. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. , Eggermont, K. , Penninckyx, I.A.M.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P.A. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Torii, N. , Mitsukawa, T. , Oosumi, Y. , Matsuura, R. , Yokayama, R.F. , Whittier, Y. and Komeda, Y. (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine‐rich repeats. Plant Cell, 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. , Onouchi, H. , Hamada, S. , Machida, C. , Hammond‐Kosack, K.E. and Jones, J.D.G. (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. [DOI] [PubMed] [Google Scholar]
- Tu, Z. , He, G. , Li, X.K. , Chen, J.M. , Chang, J. , Chen, L. , Yao, O. , Liu, P.D. , Ye, H. , Shi, J. and Wu, X. (2005) An improved system for competent cell preparation and high efficiency plasmid transformation using different Escherichia coli strains. Electron. J. Biotechnol. 8, 113–120. [Google Scholar]
- Wang, C. , El‐Shetehy, M. , Shine, M.B. , Yu, K. , Navarre, D. , Wendehenne, D. , Kachroo, A. and Kachroo, P. (2014) Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. [DOI] [PubMed] [Google Scholar]
- Xu, P. , Zhang, Y. , Kang, L. , Roossinck, M.J. and Mysore, K.S. (2006) Computational estimation and experimental verification of off‐target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Srivastava, S. , Deyholos, M.K. and Kav, N.N. (2007) Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum . Plant Sci. 173, 156–171. [Google Scholar]
- Yao, Y. , He, R.J. , Xie, Q.L. , Zhao, X.H. , Deng, X.M. , He, J.B. , Song, L. , He, J. , Marchant, A. , Chen, X.‐Y. and Wu, A.‐M. (2017) ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)‐dependent mechanism in response to different stresses in Arabidopsis . New Phytol. 213, 1667–1681. [DOI] [PubMed] [Google Scholar]
- Yi, S.Y. , Shirasu, K. , Moon, J.S. , Lee, S.‐G. and Kwon, S.‐Y. (2014) The activated SA and JA signaling pathways have an influence on flg22‐triggered oxidative burst and callose deposition. PLoS ONE, 9, e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda, H. , Fujimura, K. , Takahashi, H. , Munemura, I. , Uchimiya, H. and Sano, H. (2009) Polyamines as a common source of hydrogen peroxide in host‐ and nonhost hypersensitive response during pathogen infection. Plant Mol. Biol. 70, 103–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Transcript levels of five polyamine oxidase (PAO)‐encoding genes of wild type (Col‐0) Arabidopsis thaliana. Five to 6 weeks old Arabidopsis plants (whole rosettes) were spray‐inoculated with A. brassicicola conidium suspension used in a concentration of 5 x 105 conidia in 1 mL distilled water. Samples of mock‐inoculated and A. brassicicola‐infected plants (24 hai) were analyzed. Transcript levels of the five PAO isoforms are not increased as a result of the fungal infection. Data represent the mean of three independent biological samples with three technical replicates for each. Statistical analysis was performed using Student's t‐test. Asterisks indicate statistically significant difference (**α = 0.01).
Fig. S2 Transcript level of apoplastic class III peroxidase gene PRX33 (At3g49110) is reduced in the Arabidopsis prx33 knock‐down T‐DNA insertion line (SALK_062314C). Transcript levels were quantified in untreated Arabidopsis plants by real‐time RT‐PCR. The results show the average of two experiments each comprising three biological samples (each sample composed as a pool of 3 Arabidopsis rosettes) analyzed in three technical replicates. Statistical analysis was performed using Student's t‐test. Asterisks indicate statistically significant difference (***α = 0.001).
Fig. S3 Transcript levels of apoplastic class III peroxidase genes PRX33 (At3g49110) and PRX34 (At3g49120) are reduced in Col‐0 Arabidopsis plants that were treated with a VIGS construct (TRV‐PRX) that targets PRX33 and PRX34 mRNA sequences. Plants labelled as TRV‐GFP were treated with the control construct that targets only mRNA sequences of the GFP marker gene. Silencing of transgene GFP transcript levels and a subsequent abolishment of GFP‐derived green fluorescence in VIGS‐treated plants was used to detect the occurrence of successful gene silencing events. Five to 6 weeks old Arabidopsis plants (whole rosettes) were spray‐inoculated with A. brassicicola conidium suspension used in a concentration of 5 x 105 conidia in 1 mL distilled water. PRX33 and PRX34 mRNA levels were monitored in VIGS‐treated Arabidopsis plants before (A) and after (B) inoculation with A. brassicicola (24 hai) by real‐time RT‐PCR. The results show the average of two experiments each comprising three biological samples (each sample composed as a pool of 3 Arabidopsis rosettes) analyzed in three technical replicates. Statistical analysis was performed using Student’s t‐test. Asterisks indicate statistically significant differences (*α = 0.05, **α = 0.01, ***α = 0.001).
Fig. S4 Confirmation of T‐DNA insertion by genotyping prx33 (SALK_062314C) Arabidopsis line. PCR analysis to confirm the presence of T‐DNA was performed using genomic DNA of the Arabidopsis insertion line prx33 (SALK_062314C) and of wild type Col‐0. The first 4 lanes show results of PCR amplifications, where primers specific for PRX33 sequence (forward 5’ ‐ATTATAGTTGTTGTCAGCATTAGCA‐3’, reverse 5’‐ACCATTTGTTCCTCTGAAGCA‐3’) were used with Col‐0 and prx33 genomic DNA extracts as templates. The last three lanes exhibit PCR results where a T‐DNA left border primer (LBa1:5’‐ TGGTTCACGTAGTGGGCCATCG ‐3’) was combined with PRX33 sequence specific forward primer using prx33 insertion line genomic DNA extract confirming the location of T‐DNA insertion. No template controls were included. PCR products were fractionated in 1% agarose gel and DNA was visualized by staining with GelRed (Fremont, California, USA).
Table S1 Arabidopsis T‐DNA insertion lines used in this work.
Table S2 Primer sequences used in this work.


