Summary
Magnaporthe oryzae is an important fungal pathogen of both rice and wheat. However, how M. oryzae effectors modulate plant immunity is not fully understood. Previous studies have shown that the M. oryzae effector AvrPiz‐t targets the host ubiquitin‐proteasome system to manipulate plant defence. In return, two rice ubiquitin E3 ligases, APIP6 and APIP10, ubiquitinate AvrPiz‐t for degradation. To determine how lysine residues contribute to the stability and function of AvrPiz‐t, we generated double (K1,2R‐AvrPiz‐t), triple (K1,2,3R‐AvrPiz‐t) and lysine‐free (LF‐AvrPiz‐t) mutants by mutating lysines into arginines in AvrPiz‐t. LF‐AvrPiz‐t showed the highest protein accumulation when transiently expressed in rice protoplasts. When co‐expressed with APIP10 in Nicotiana benthamiana, LF‐AvrPiz‐t was more stable than AvrPiz‐t and was less able to degrade APIP10. The avirulence of LF‐AvrPiz‐t on Piz‐t:HA plants was less than that of AvrPiz‐t, which led to resistance reduction and lower accumulation of the Piz‐t:HA protein after inoculation with the LF‐AvrPiz‐t‐carrying isolate. Chitin‐ and flg22‐induced production of reactive oxygen species (ROS) was higher in LF‐AvrPiz‐t than in AvrPiz‐t transgenic plants. In addition, LF‐AvrPiz‐t transgenic plants were less susceptible than AvrPiz‐t transgenic plants to a virulent isolate. Furthermore, both AvrPiz‐t and LF‐AvrPiz‐t interacted with OsRac1, but the suppression of OsRac1‐mediated ROS generation by LF‐AvrPiz‐t was significantly lower than that by AvrPiz‐t. Together, these results suggest that the lysine residues of AvrPiz‐t are required for its avirulence and virulence functions in rice.
Keywords: effector, lysine residue, protein stability, reactive oxygen species, rice immunity
Introduction
Plant defence is a multi‐layered immune network consisting of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI). It relies on strategies involving a range of spatiotemporally regulated plasma membrane receptors (referred to as pattern recognition receptors, PRRs) or structurally conserved nucleotide‐binding and leucine‐rich repeat (NB‐LRR) proteins (Bent and Mackey, 2007; Jones and Dangl, 2006; Jones et al., 2016). The recognition of microbial PAMPs by plant PRRs leads to the first layer of defence, PTI, which confers non‐specific resistance against pathogen infection (Boller and Felix, 2009; Zipfel, 2008). Although pathogens can evade PTI by secreting effectors into plant cells, the host may possess specific NB‐LRR proteins that sense these virulence factors, resulting in a race‐specific immune response, called ETI (Białas et al., 2017; Eitas and Dangl, 2010).
The plant ubiquitination system is involved in a variety of biological processes and is essential for immunity (Banfield, 2015; Li et al., 2014). It regulates one of the most important post‐translational modifications, in which substrate proteins are covalently linked to a ubiquitin chain via the lysine residues and are subjected to 26S proteasome‐dependent degradation (Ciechanover, 1998; Santner and Estelle, 2010). By exploiting the host ubiquitination machinery, pathogen effectors modify immune‐related proteins by altering their stability, enzymatic activity, relocalization or interaction with other proteins (Angot et al., 2007; Spallek et al., 2009). Pseudomonas syringae HopM1, for example, suppresses Arabidopsis PTI by destabilizing the immunity‐related proteins AtMIN2, AtMIN7 and AtMIN10 via the 26S proteasome degradation system (Nomura et al., 2006, 2011). The RXLR‐type effector Avr3a from Phytophthora infestans stabilizes the host E3 ligase CMPG1 from 26S proteasome‐dependent degradation and thereby prevents CMPG‐activated host cell death during biotrophic colonization (Bos et al., 2010). Instead of reprogramming host proteins for degradation, some pathogen effectors can mimic host E3 ligases to ubiquitinate and degrade target proteins. The P. syringae effector AvrPtoB, for example, harbours a C‐terminal E3 ligase domain that can suppress programmed cell death (PCD) (Abramovitch et al., 2006; Janjusevic et al., 2006). By interacting and ubiquitinating immunity‐related proteins, such as Fen (Ntoukakis et al., 2009; Rosebrock et al., 2007), FLS2 (Göhre et al., 2008), CERK1 (Gimenez‐Ibanez et al., 2009) and NPR1 (Chen et al., 2017), AvrPtoB suppresses plant PTI and systemic acquired resistance (SAR) through several signalling pathways.
The importance of the plant ubiquitination system in plant–pathogen interactions has been highlighted further by the discovery of the rice blast effector AvrPiz‐t of Magnaporthe oryzae (Li et al., 2009), which targets two rice RING‐type E3 ligases, APIP6 and APIP10 (Park et al., 2012, 2016). AvrPiz‐t can be translocated into a plant cell, where it interacts with APIP6 and APIP10, and promotes their degradation via the 26S proteasome system. Silencing of either APIP6 or APIP10 in Nipponbare plants (NPB, with a non‐functional Piz‐t) reduces the PTI response, confirming that APIP6 and APIP10 are positive regulators of PTI. However, the silencing of APIP10 in NPB‐Piz‐t:HA transgenic rice (NPB wild‐type plants harbouring a functional Piz‐t:HA gene) causes spontaneous cell death and activates R gene‐mediated resistance, which suggests that APIP10 acts as a negative regulator of ETI. The dual function of APIP10 suggests that AvrPiz‐t achieves its virulence and avirulence function through APIP10. By acting as the substrate of APIP10, AvrPiz‐t was ubiquitinated in vitro and was degraded through the 26S proteasome system in Nicotiana benthamiana (Park et al., 2016). How the proteasomal degradation of AvrPiz‐t affects its avirulence and virulence function in rice cells is unknown.
To reveal the role of AvrPiz‐t lysine residues in ETI and PTI, we generated a lysine‐free AvrPiz‐t mutant (LF‐AvrPiz‐t) and tested its avirulence and virulence functions in this study. We found that the lysine residues in AvrPiz‐t are critical for the stability of both AvrPiz‐t and APIP10 and are important for the avirulence function of AvrPiz‐t on Piz‐t and on Piz‐t accumulation during infection. In addition, the lysine residues affect the suppression of reactive oxygen species (ROS) generation triggered by PAMPs and OsRac1. Together, our results show that the lysine residues of AvrPiz‐t are required for its function in both Piz‐t‐mediated resistance and basal defence in rice.
Results
The lysine residues of AvrPiz‐t affect its protein stability in plant cells
As lysine residue modification affects target proteins by changing their stability, enzymatic activities, cellular translocation and protein–protein interaction (Ciechanover, 1998; Walsh et al., 2005), we hypothesized that the lysine residues of AvrPiz‐t, as the potential ubiquitin acceptor sites, are essential for AvrPiz‐t stability and function. As shown in Fig. 1a, AvrPiz‐t contains six lysine residues. To determine the effect of the lysine residues, we used a site‐directed mutagenesis method to generate a series of AvrPiz‐t mutants harbouring double (K1,2R‐AvrPiz‐t), triple (K1,2,3R‐AvrPiz‐t) and all six (LF‐AvrPiz‐t) lysine to arginine mutations. Compared with the wild‐type AvrPiz‐t:GFP, transient expression of K1,2R‐AvrPiz‐t:GFP, K1,2,3R‐AvrPiz‐t:GFP and LF‐AvrPiz‐t:GFP showed a higher protein accumulation in rice protoplasts (Fig. 1b). Among them, LF‐AvrPiz‐t was the most stable in terms of protein stability, suggesting that, although the mutation of individual lysine residues can increase AvrPiz‐t protein stability, mutation of all six lysine residues can lead to maximum protein stability in rice cells. Therefore, we selected LF‐AvrPiz‐t for subsequent experiments. When co‐expressed with Myc‐tagged E3 ligase APIP10 in N. benthamiana, the GFP:LF‐AvrPiz‐t:HA protein was more stable than the wild‐type GFP:AvrPiz‐t:HA (Fig. 1c, first panel). Meanwhile, the protein levels of Myc:APIP10 were higher when it was co‐expressed with LF‐AvrPiz‐t than when it was co‐expressed with wild‐type AvrPiz‐t (Fig. 1c, second panel). These results suggest that the lysine residues of AvrPiz‐t are essential for its stability and, consequently, for its ability to degrade APIP10 in plant cells.
Figure 1.
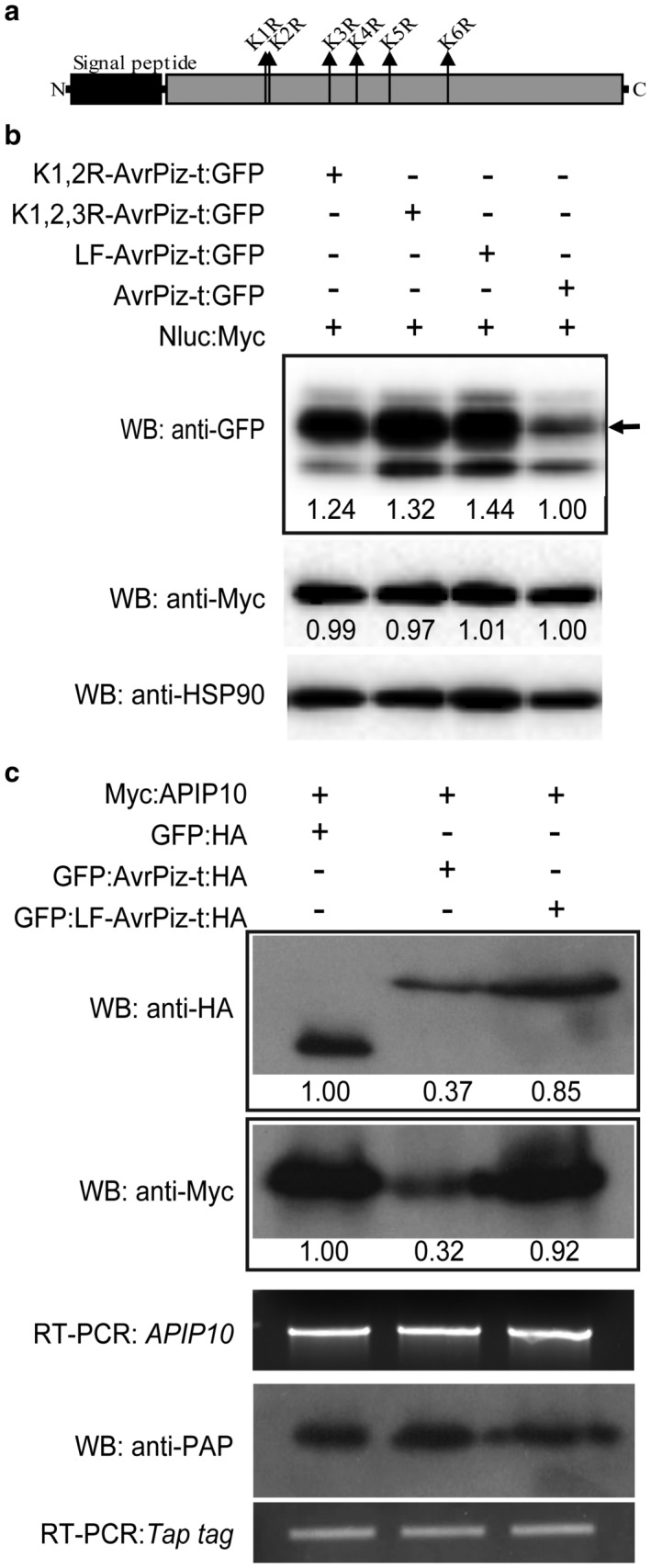
Protein stability of double, triple and lysine‐free mutants of AvrPiz‐t and APIP10 in plant cells. (a) Schematic structure of the AvrPiz‐t protein; the locations of the six lysine residues are labelled as K1–K6 according to their order in the AvrPiz‐t protein. (b) Plasmids of K1,2R‐AvrPiz‐t:GFP, K1,2,3R‐AvrPiz‐t:GFP, LF‐AvrPiz‐t:GFP and AvrPiz‐t:GFP were separately expressed in Nipponbare rice protoplasts, and Nluc:Myc was used as an internal transfection control. Immunoblots with anti‐GFP, anti‐Myc and anti‐HSP90 were conducted to detect the protein levels of different AvrPiz‐t mutants, Nluc‐Myc and endogenous rice HSP90, respectively. (c) Co‐expression of Myc:APIP10 with GFP:LF‐AvrPiz‐t:HA and GFP:AvrPiz‐t:HA in Nicotiana benthamiana. GFP:HA was used as the infiltration control, and the Tap tag was used as an internal control. Semi‐real‐time polymerase chain reaction (semi‐RT‐PCR) was conducted for APIP10 and Tap transcripts. Protein levels were measured by Image Lab software on the basis of the band intensity and were then normalized with the internal control.
The lysine residues of AvrPiz‐t are required for the activation of Piz‐t‐mediated resistance
To investigate the functional role of lysine residues in the recognition by Piz‐t, we generated M. oryzae RB22 transformants harbouring the AvrPiz‐t or LF‐AvrPiz‐t constructs under the control of the native promoter. The M. oryzae RB22 wild‐type isolate that lacks a functional AvrPiz‐t was used for fungal transformation. We obtained six independent transformants of each construct after polymerase chain reaction (PCR) screening and selected two per construct for the following experiments. We then spray inoculated NPB plants with the RB22‐AvrPiz‐t and RB22‐LF‐AvrPiz‐t transformants and found that their pathogenicity did not differ from that of the RB22 wild‐type isolate (Fig. 2a, right panel; Fig. S1, see Supporting Information). To determine whether the lysine mutations in AvrPiz‐t disrupt its recognition of Piz‐t, we spray inoculated NPB‐Piz‐t:HA plants with RB22‐AvrPiz‐t and RB22‐LF‐AvrPiz‐t transformants. Inoculation with RB22‐AvrPiz‐t resulted in a resistance phenotype with no lesions, whereas inoculation with RB22‐LF‐AvrPiz‐t resulted in a susceptible phenotype with merged, necrotic lesions, a phenotype similar to that resulting from inoculation with the RB22 wild‐type isolate (Fig. 2a, left panel; Fig. S1). Results with punch inoculation of NPB‐Piz‐t:HA plants were consistent with the results of spray inoculation, i.e. disease was more significant on plants inoculated with the RB22‐LF‐AvrPiz‐t transformant than with the RB22‐AvrPiz‐t transformant, but the lesions were smaller with the RB22‐LF‐AvrPiz‐t transformant than with the RB22 wild‐type (Fig. 2b). These results suggest that mutations of the lysine residues of AvrPiz‐t partially abolish its avirulence function, thus leading to the failure of Piz‐t‐mediated resistance.
Figure 2.
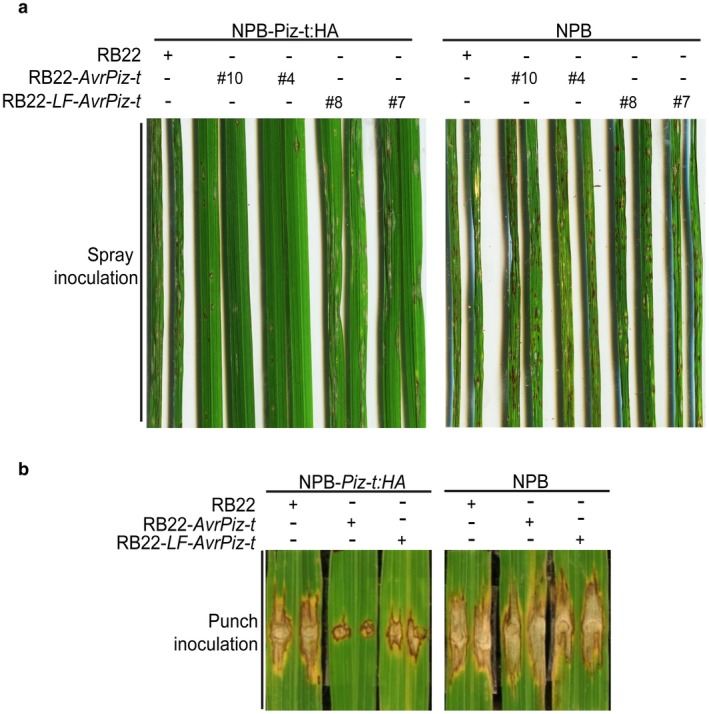
Effects of the lysine residues of AvrPiz‐t on the activation of Piz‐t‐mediated resistance. (a) Effects of spray inoculation of NPB‐Piz‐t:HA transgenic rice and NPB wild‐type rice with different Magnaporthe oryzae RB22 isolates. Two transformants of the M. oryzae isolate RB22 harbouring the LF‐AvrPiz‐t or AvrPiz‐t constructs were used for inoculation, whereas the non‐transformed M. oryzae RB22 was used as a control. Infected leaves were assessed at 6 days post‐inoculation (dpi). (b) Effects of punch inoculation of NPB‐Piz‐t:HA transgenic rice and NPB wild‐type rice with different M. oryzae RB22 isolates. Magnaporthe oryzae RB22‐LF‐AvrPiz‐t or RB22‐AvrPiz‐t transformants from the spray inoculation were used. Six‐week‐old plants were inoculated with an M. oryzae spore suspension. Infected leaves were assessed at 9 dpi.
LF‐AvrPiz‐t fails to induce Piz‐t accumulation during M. oryzae infection
Previous studies have shown that AvrPiz‐t is translocated from M. oryzae into rice cells, where it induces a high level of Piz‐t protein accumulation during M. oryzae infection (Park et al., 2012, 2016). Given the altered stability and avirulence function of LF‐AvrPiz‐t, we speculated that the lysine residue mutations of AvrPiz‐t might affect Piz‐t accumulation. To obtain a comprehensive profile of Piz‐t protein accumulation during M. oryzae infection, we spray inoculated NPB‐Piz‐t:HA transgenic rice with the M. oryzae RB22‐LF‐AvrPiz‐t transformant, the RB22‐AvrPiz‐t transformant and the RB22 wild‐type, respectively. Infected tissues were collected at 0, 24, 48, 96 and 144 h post‐inoculation (hpi). Immunoblot analysis showed that the protein levels of Piz‐t in RB22‐LF‐AvrPiz‐t‐inoculated plants were slightly higher than those in RB22‐AvrPiz‐t‐inoculated plants at 24 hpi (Fig. 3). However, the protein levels of Piz‐t in RB22‐LF‐AvrPiz‐t‐inoculated plants were lower than those in RB22‐AvrPiz‐t‐inoculated plants at 48, 96 and 144 hpi (Fig. 3). The profiles of Piz‐t protein accumulation were consistent with the inoculation results, confirming the importance of lysine residues for the avirulence function of AvrPiz‐t.
Figure 3.
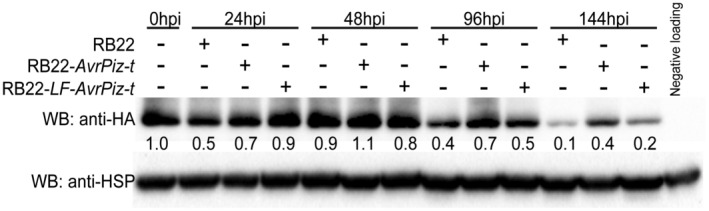
Effects of the lysine residues on AvrPiz‐t‐induced Piz‐t protein accumulation during Magnaporthe oryzae infection. NPB‐Piz‐t:HA transgenic plants were inoculated with M. oryzae isolate RB22, RB22‐AvrPiz‐t or RB22‐LF‐AvrPiz‐t. Immunoblots with anti‐HA were conducted to detected Piz‐t protein accumulation at different time points after inoculation (hpi, hours post‐inoculation). Endogenous rice HSP90 detected by anti‐HSP was used as an internal control, and NPB wild‐type samples served as a negative control (last lane). Protein levels were measured by Image Lab software on the basis of the band intensity and were then normalized with the internal control.
The lysine residues of AvrPiz‐t are necessary for the suppression of basal resistance
In the absence of Piz‐t recognition, AvrPiz‐t suppresses host PTI responses by acting as a virulence effector (Park et al., 2012). To examine whether mutation of the lysine residues of AvrPiz‐t impairs its virulence function, we generated stable transgenic rice by expressing LF‐AvrPiz‐t:HA and AvrPiz‐t:HA under the maize ubiquitin promoter in NPB plants [NPB‐LF‐AvrPiz‐t:HA in this study and NPB‐AvrPiz‐t:HA in Park et al. (2012)]. Quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis showed high transcriptional levels of both LF‐AvrPiz‐t:HA and AvrPiz‐t:HA in transgenic plants (Fig. S2, see Supporting Information). By using luminol‐based chemiluminescence, we quantified the chitin‐ and flg22‐induced ROS production in NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA transgenic plants. Consistent with the results of Park et al. (2012), ectopic expression of AvrPiz‐t in NPB plants suppressed the production of ROS elicited by chitin or flg22 treatment, but the suppression ability was partially reduced in NPB‐LF‐AvrPiz‐t:HA plants (Fig. 4a). In addition to the ROS response, we also measured the transcriptional changes of the plant defence marker genes PAL4 and NAC4 caused by chitin or flg22 treatment of NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA plants. qRT‐PCR analysis showed that the expression of PAL4 in chitin‐treated NPB‐LF‐AvrPiz‐t:HA plants was completely recovered as in NPB plants, but, in flg22‐treated NPB‐LF‐AvrPiz‐t:HA plants, the expression of PAL4 was only partially recovered (Fig. 4b, left panel). The expression of NAC4, however, did not differ significantly between NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA transgenic plants (Fig. 4b, right panel).
Figure 4.
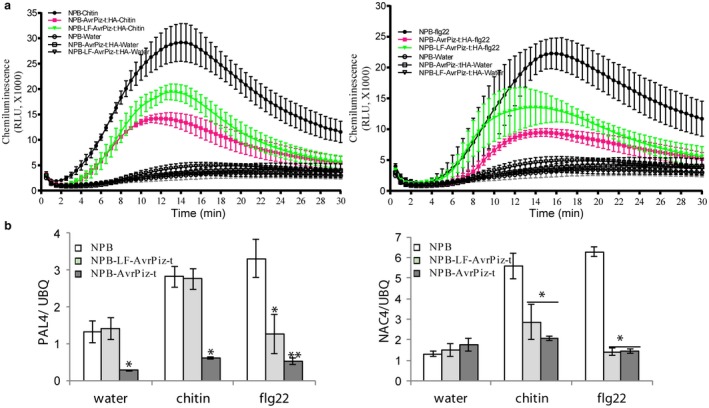
Reactive oxygen species (ROS) and defence gene expression in NPB‐LF‐AvrPiz‐t plants. (a) Pathogen‐associated molecular pattern (PAMP)‐induced ROS production in NPB‐LF‐AvrPiz‐t:HA transgenic plants, NPB‐AvrPiz‐t:HA transgenic plants and NPB wild‐type plants. Values are means ± standard error (SE) (n = 3). RLU, relative light unit. (b) PAMP‐induced marker gene expression in NPB‐LF‐AvrPiz‐t:HA transgenic plants, NPB‐AvrPiz‐t:HA transgenic plants and NPB wild‐type plants. The transcriptional levels of two defence‐related marker genes, PAL4 and NAC4, at 3 h after chitin or flg22 treatment were measured by quantitative real‐time polymerase chain reaction (qRT‐PCR). Values are means ± SE (n = 3). One‐way analysis of variance (ANOVA) test with Tukey’s method was conducted. Significant differences at P < 0.05 and P < 0.01 are indicated by ‘*’ and ‘**’, respectively. UBQ, ubiquitin.
Given the differences in ROS generation and defence gene expression between NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA transgenic plants, we reasoned that the lysine residue mutations in AvrPiz‐t might result in a change in the disease phenotype of NPB‐LF‐AvrPiz‐t:HA plants. Using punch inoculation with the virulent isolate RB22, we quantified disease development in transgenic plants and NPB plants. On the basis of lesion size and fungal biomass, disease symptoms were fewer in NPB‐LF‐AvrPiz‐t:HA plants than in NPB‐AvrPiz‐t:HA plants (Fig. 5a,b). These results, together with the reduced ability of NPB‐LF‐AvrPiz‐t:HA to suppress ROS generation and PAL4 expression, suggest that the lysine residues of AvrPiz‐t are essential for the suppression of plant basal defence.
Figure 5.
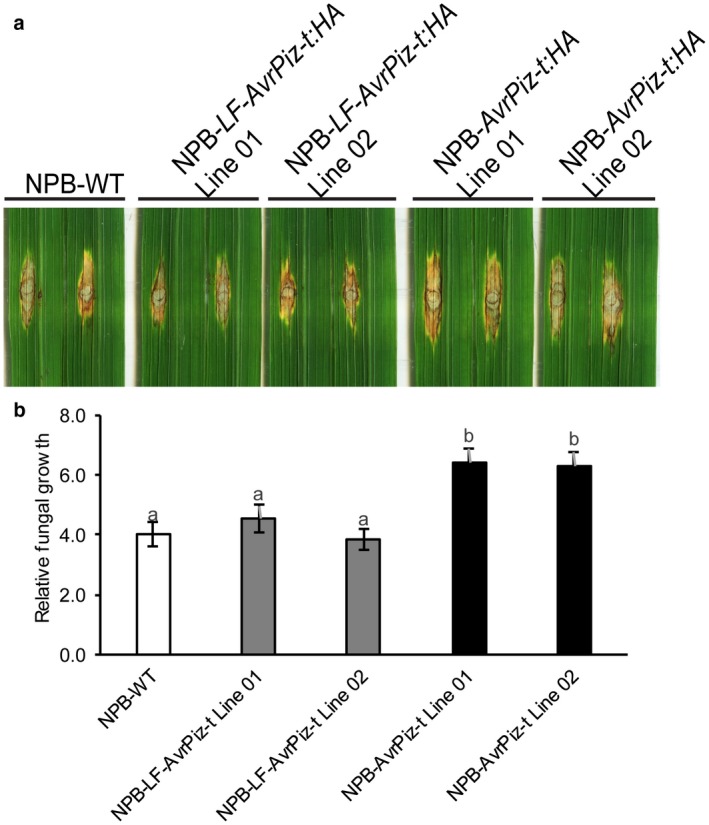
Effects of the lysine residues of AvrPiz‐t on the suppression of rice basal resistance. (a) Effects of punch inoculation of NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA transgenic plants with Magnaporthe oryzae isolate RB22. Two independent plants of each transgenic line were used. NPB wild‐type plants served as a control. Six‐week‐old plants were inoculated with an M. oryzae spore suspension of 5 × 105 spores/mL. Phenotype leaves were assessed at 9 days post‐inoculation (dpi). (b) Relative fungal growth in the photographed leaves was measured by DNA‐based real‐time PCR to quantify the amount of DNA of M. oryzae using two sets of primers specific for M. oryzae Pot2 and rice Ubiquitin. Values are means ± standard error (SE) (n = 3). One‐way analysis of variance (ANOVA) test with Tukey’s method was conducted. Means followed by different letters are significantly different (P < 0.05).
AvrPiz‐t interacts with Rac1 and suppresses Rac1‐mediated ROS accumulation in N. benthamiana, and lysine residues are required for this suppression
OsRac1, a rice homologue of human small GTPase, is an essential regulator of defence‐related ROS production and cell death in rice (Kawasaki et al., 1999; Ono et al., 2001). With the hypothesis that OsRac1 might act as an effector target used by the pathogen to manipulate host ROS production, we tested the interaction between OsRac1 and AvrPiz‐t using the luciferase complementation imaging (LCI) assay. Strong luminescence signals were observed for the interactions of OsRac1 with both AvrPiz‐t and LF‐AvrPiz‐t, respectively (Fig. 6a). The average relative luciferase activities from three replications of the AvrPiz‐t/OsRac1 and LF‐AvrPiz‐t/OsRac1 co‐infiltrations were similar to each other, but higher than the negative controls (Fig. 6b), suggesting that lysine mutations do not affect the AvrPiz‐t and OsRac1 interaction.
Figure 6.
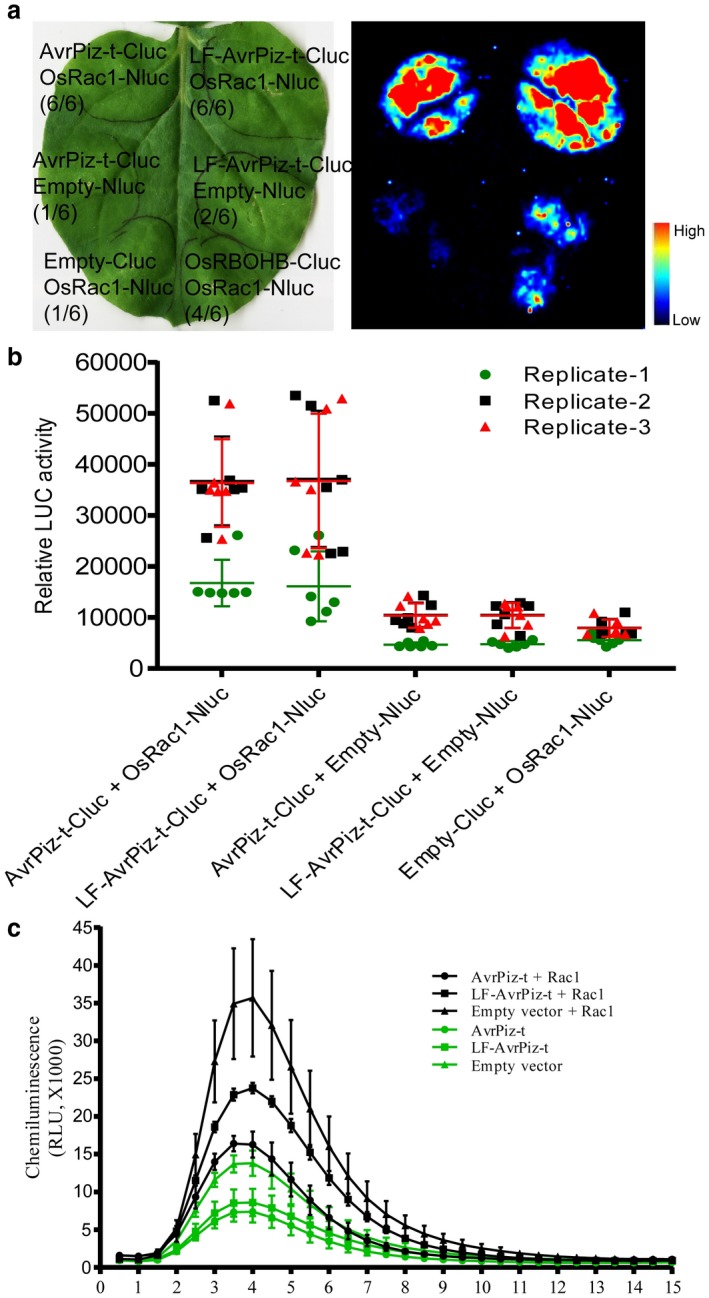
Interaction between OsRac1 and AvrPiz‐t/LF‐AvrPiz‐t and suppression of Rac1‐mediated reactive oxygen species (ROS) accumulation by AvrPiz‐t and LF‐AvrPiz‐t in Nicotiana benthamiana. (a) Luciferase complementation imaging (LCI) assay for the interaction between OsRac1 and AvrPiz‐t/LF‐AvrPiz‐t. OsRac1‐Nluc was co‐expressed with AvrPiz‐t‐Cluc or LF‐AvrPiz‐t‐Cluc, whereas Nluc‐empty vector/AvrPiz‐t‐Cluc, Nluc‐empty vector/LF‐AvrPiz‐t‐Cluc and OsRac1‐Nluc/Cluc‐empty vector were used as negative controls. OsRac1‐Nluc/OsRBOHB‐Cluc was used as a positive control. The numbers in parentheses indicate the number of leaf spots with positive luminescence signals among all of the infiltrated leaves. (b) Relative luciferase (LUC) activity of the interaction between OsRac1 and AvrPiz‐t/LF‐AvrPiz‐t. Three biological replicates with 18 N. benthamiana leaves were measured for the luciferase activity using the Image Lab quantity tool. (c) Rac1‐mediated ROS production in N. benthamiana leaves expressing AvrPiz‐t, LF‐AvrPiz‐t or empty vector with OsRac1. Leaf discs of each infiltrated spot on the same leaves were treated with 8 nm chitin. ROS accumulation was measured using luminol‐based chemiluminescence for 15 min. Values are means ± standard error (SE) (n = 3). RLU, relative light unit.
To assess whether AvrPiz‐t interferes with OsRac1‐mediated ROS accumulation and whether the lysine residues in AvrPiz‐t are required for this interference, we co‐expressed OsRac1 and AvrPiz‐t/LF‐AvrPiz‐t in N. benthamiana and measured chitin‐induced ROS production by luminol‐based chemiluminescence. AvrPiz‐t/LF, AvrPiz‐t and the empty vector (EV) were agroinfiltrated 1 day before OsRac1 in the same leaves. Consistent with the reduced ROS induction in NPB‐AvrPiz‐t:HA transgenic rice (Fig. 4a), the levels of ROS accumulation were suppressed by transient expression of either AvrPiz‐t or LF‐AvrPiz‐t in N. benthamiana (Fig. 6c). However, the suppressive ability of LF‐AvrPiz‐t was partially impaired. These results suggest that AvrPiz‐t suppresses OsRac1‐mediated ROS production in planta and that the lysine residues of AvrPiz‐t are required for this suppression.
Discussion
Pathogen effectors are essential for the infection of host plants. Although many effectors have been characterized in different pathogens, little is known about the role of individual domains or amino acids in effector functions because most effectors lack a conserved motif or domain. AvrPiz‐t is a small secreted peptide with 108 predicted amino acids (Li et al., 2009). Structural analysis has revealed that two cysteine residues, Cys62 and Cys75, in AvrPiz‐t form a disulfide bond, which is responsible for its avirulence function (Zhang et al., 2013). Our previous studies have shown that AvrPiz‐t is ubiquitinated by two rice E3 ligases, APIP6 and APIP10 (Park et al., 2012, 2016). The role of the lysine residues of AvrPiz‐t in APIP6/APIP10‐mediated ubiquitination, and its interaction with host immunity, is unclear. In this study, we used genetic, biochemical, and transient and stable plant expression approaches to determine the function of lysine residues in AvrPiz‐t. Transient expression of LF‐AvrPiz‐t in planta increases its protein accumulation, suggesting that the lysine residues of AvrPiz‐t are essential for the stability of the protein. It is possible that LF‐AvrPiz‐t shows lesser binding affinity to APIP6, APIP10 or other unknown host proteins, thus reducing the degradation rate in rice cells. Another possibility is that the protein folding change in LF‐AvrPiz‐t increases its stability in planta.
The importance of lysine residues in AvrPiz‐t is also indicated by the reduced ability of LF‐AvrPiz‐t to degrade APIP10 relative to the wild‐type AvrPiz‐t in N. benthamiana. Our previous study demonstrated how APIP10 connects AvrPiz‐t to its cognate Piz‐t for immune activation (Park et al., 2016). In the absence of pathogen invasion, APIP10 maintains the homeostasis of the R protein Piz‐t at an optimal level by the 26S proteasome degradation system. Once AvrPiz‐t is translocated into rice cells during pathogen infection, AvrPiz‐t promotes the degradation of APIP10, thus removing the suppression effects of APIP10 on the accumulation of Piz‐t. In this study, we found that the RB22‐LF‐AvrPiz‐t isolate failed to cause Piz‐t accumulation and Piz‐t‐mediated resistance. These results demonstrate that the lysine residues in AvrPiz‐t are required for the activation of Piz‐t‐mediated resistance to M. oryzae. However, how the reduction in APIP10 levels by LF‐AvrPiz‐t affects Piz‐t accumulation remains to be elucidated.
In addition to failing to activate ETI, LF‐AvrPiz‐t partially lost its virulence function in the suppression of PTI responses. Previous research has reported that many microbial effectors can suppress PTI and, in some cases, suppression results from reprogramming of the fate of basal defence‐related proteins (Banfield, 2015; Duplan and Rivas, 2014). AvrPiz‐t interferes with the ubiquitination activities of the rice E3 ligase proteins APIP6 and APIP10, which are positive regulators of PTI in the non‐Piz‐t background. In this study, susceptibility to M. oryzae was lower with the overexpression of LF‐AvrPiz‐t than with the overexpression of AvrPiz‐t in the NPB wild‐type, which is consistent with the impaired ability of LF‐AvrPiz‐t to suppress ROS production and defence‐related gene expression in response to PAMP treatments. These results suggest that the lysine residues of AvrPiz‐t are essential for its virulence function, and that the underlying mechanism could involve reduced interference of APIP6 or APIP10 activity by LF‐AvrPiz‐t. Because LF‐AvrPiz‐t retains some ability to suppress PTI, it is likely that LF‐AvrPiz‐t retains part of its virulence activity without these residues.
The ROS burst is a hallmark of plant defence which is associated with PCD and defence activation. OsRac1 is activated for chitin‐triggered ROS production by interaction with the plasma membrane complex OsCEBiP–OsCERK1–OsRacGEF1 (Akamatsu et al., 2013). We demonstrated that OsRac1 interacts with both AvrPiz‐t and LF‐AvrPiz‐t, suggesting that OaRac1 might serve as an effector target for the manipulation of ROS production in host cells. Previous studies have shown that AvrPiz‐t suppresses BAX‐induced PCD in N. benthamiana (Li et al., 2009) and inhibits the transcriptional activity of a bZIP transcriptional factor APIP5, thus enhancing APIP5‐mediated necrosis, which is beneficial for the necrotrophic growth of M. oryzae in rice cells (Li et al., 2009; Wang et al., 2016). The involvement of AvrPiz‐t in cell death suggests that it could play a role in the regulation of OsRac1‐mediated ROS production and defence activation. It will be interesting to determine whether the lysine mutations in AvrPiz‐t affect OsRac1 activity or its interaction with the defence complex OsCEBiP–OsCERK1–OsRacGEF1 in the future.
Experimental Procedures
Rice cultivation, M. oryzae inoculation and disease evaluation
The rice plants used in this study were germinated on half‐strength Murashige and Skoog (½MS) medium (24 h light at 25 °C) for 1 week, and seedlings of a uniform size were transplanted into soil. For germination, rice seeds with removed husks were sterilized by immersion in 75% ethanol for 1 min and then in 2% sodium hypochlorite (Clorox bleach) for 30 min (Park et al., 2012). After they had been rinsed with sterile water, the seeds were placed on ½MS medium and kept at 25 °C with 24 h light for 1 week. Seedlings of a uniform size were then transplanted into soil. Plants used for fungal inoculation or ROS assays were kept in a growth chamber (Conviron, model BDR16, Winnipeg, Canada) with 12 h of light at 26 °C, 12 h of darkness at 20 °C and 80% relative humidity.
The M. oryzae isolates were cultured on oatmeal agar medium (30 g of ground Quaker Oats oatmeal, 15 g of agar, increased to 1 l with water) in a dark, 28 °C incubator for 1 week, and were then exposed to fluorescent light in a 28 °C incubator for a further week. The first week in the dark supported hyphal growth, and the second week with fluorescent light supported sporulation. For the standardized inoculation of rice with M. oryzae (Ono et al., 2001), the conidia of M. oryzae isolates were collected (0.1% Tween‐20) and their concentration was adjusted with a haemocytometer to 2 × 105 spores/mL for spray inoculation or to 5 × 105 spores/mL for punch inoculation.
For spray inoculation (Park et al., 2012; Qu et al., 2006), 3‐week‐old rice plants were placed in a plastic bag (to provide high humidity) and the leaves were evenly sprayed with fungal spore suspensions. Spray‐inoculated plants were kept in the plastic bag in the dark for 24 h before they were returned to the growth chamber. Six days after spray inoculation, disease reactions of inoculated plants were scored using a 0–5 scale, where ‘0’ represents a resistance phenotype with no lesions and ‘5’ represents a susceptible phenotype with coalesced lesions. For punch inoculation, 6‐week‐old plants were used. The second leaf from the top of each plant was wounded, but not cut through, with a mouse ear punch, and the wound was then inoculated with a droplet (10 µL) of a spore suspension. The inoculated wound was wrapped with Scotch tape so that the droplet remained on the wound. The inoculated leaves were photographed at 9 days post‐inoculation (dpi) and then subjected to cetyltrimethylammonium bromide (CTAB)‐based DNA extraction (Murray and Thompson, 1980). To quantify the relative fungal growth, the ratio of fungal DNA to plant DNA was determined by qRT‐PCR as described previously (Park et al., 2012).
Rice and M. oryzae transformation
Agrobacterium‐mediated rice transformations for NPB‐LF‐AvrPiz‐t:HA and NPB‐AvrPiz‐t:HA overexpression plants were conducted with the modified protocol as described previously (Hiei et al., 1994; Nishimura et al., 2006). Rice embryonic calli induced from mature seeds of NPB rice were co‐cultivated with agrobacteria LBA4404 harbouring the pCXUN‐LF‐AvrPiz‐t:HA construct or the pCXUN‐AvrPiz‐t:HA construct. Positive transformants were selected on hygromycin medium and transferred onto regenerating medium for plantlet formation. To obtain homozygous lines, we selected the next generation of transgenic plants on hygromycin medium and performed a PCR genotyping. Polyethylene glycol (PEG)‐mediated M. oryzae transformations for RB22‐LF‐AvrPiz‐t:HA and RB22‐AvrPiz‐t:HA isolates were generated as described previously (Shirsekar, 2013). Magnaporthe oryzae isolate RB22, which lacks a functional AvrPiz‐t, was used for the fungal transformations.
Rice protoplast isolation and transfection
Rice protoplasts were isolated and transfected by following published protocols with minor modification (Yoo et al., 2007; Zhang et al., 2011). Ten‐day‐old etiolated rice seedlings were grown on ½MS medium in the dark at 28 °C. The sheath and stem parts of the seedlings were cut into 0.5‐mm fragments with a razor blade and immediately submerged into cell wall digestion buffer [1.5% Cellulase RS, 0.75% Macerozyme R‐10, 0.6 m mannitol, 10 mm 2‐(N‐Morpholino)ethanesulfonic acid hydrate, 4‐Morpholineethanesulfonic acid (MES) (Sigma Cat. No. 2933, St. Louis, MO, USA), 10 mm CaCl2 and 0.1% bovine serum albumin (BSA), pH 5.7]. The digestion was conducted under vacuum infiltration (20 kPa) for 30 min, and the digested tissue was then incubated for another 5–6 h in the dark with gentle shaking (60–80 rpm). After digestion, rice residues were washed three times with W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 2 mm MES and 0.5% glucose, pH 5.7) and filtered through 40‐μm nylon mesh. Protoplasts were harvested by centrifugation of the filtered solution at 1500 rpm for 3 min. The pelleted protoplasts were washed three times with W5 solution and then resuspended in MMG solution (0.4 m mannitol, 15 mm MgCl2 and 4 mm MES, pH 5.7) for transfection. PEG‐mediated protoplast transfections were conducted using 200 μL of protoplasts per 5 μg of plasmids in a 2‐mL microfuge tube. An equal volume of PEG solution [40% (w/v) PEG 4000 (Sigma Cat. No. 81240), 0.2 m mannitol and 0.1 m CaCl2] was added and mixed well. The mixture was incubated at room temperature for 15 min in the dark. Then, two volumes of W5 solution were added to terminate PEG effects. Protoplasts were collected by centrifugation, and the pellet was resuspended in W5 solution and incubated in the dark for 16 h.
Luciferase complementation imaging assay
In vivo protein–protein interactions were detected via luciferase complementation imaging assay, as described previously (Chen et al., 2008). OsRac1 was fused with N‐terminal luciferase (Nluc), whereas LF‐AvrPiz‐t and AvrPiz‐t were fused with C‐terminal luciferase (Cluc). Agrobacterium strain GV3101, carrying Nluc or Cluc constructs, was mixed and infiltrated into leaves of N. benthamiana. The pair of OsRac1 and OsRBOHB served as a positive interaction control. Three days after agroinfiltration, leaves co‐expressing different constructs were treated with 1 mm luciferin substrate and subjected to luminescence intensity measurement via a Bio‐Rad ChemiDoc XRS+ imaging system (Bio‐Rad, Hercules, CA, USA).
ROS quantification by luminol‐based chemiluminescence
A luminol‐based chemiluminescence method was used to quantify ROS accumulation in rice and N. benthamiana leaves in response to PAMP treatment (Schwacke and Hager, 1992; Smith and Heese, 2014). In brief, discs of 6‐week‐old rice leaves (the third leaf from the top) or of agroinfiltrated N. benthamiana leaves were placed in sterile water overnight and then submerged in reaction mixture [100 µL of Immunstar‐HRP substrate from Bio‐Rad (Cat. No. 170‐5040) and 1 µL of peroxidase‐streptavidin from Jackson Immunoresearch (Cat. No. 016‐030‐084)] with 8 nm chitin (Toronto Research Chemicals, Cat. No. H290750, N,N′,N′′,N′′′,N′′′′,N′′′′′‐hexaacetylchitohexaose, Toronto, Canada) or 100 nm flg22. All reaction mixtures were immediately subjected to luminescence measurement in a Glomax 20/20 Luminometer (one measurement per 10 s for 15–30 min) (Madison, WI, USA).
qRT‐PCR analysis
To quantify the expression levels of marker genes (PAL4 and NAC4) after PAMP treatments, leaf discs of different transgenic rice plants were immersed in water containing either 100 nm flg22 or 8 nm chitin for 3 h, as described previously (Park et al., 2012). Total RNA was extracted with Trizol reagent (Invitrogen, Cat. No. 15596, Carlsbad, CA, USA) and subjected to first‐strand cDNA synthesis using a Promega RT‐PCR kit (Madison, WI, USA). qRT‐PCR was performed using an iQ5 real‐time PCR detection system (Bio‐Rad). The transcriptional levels were calculated by the relative 2−ΔCT method with the rice ubiquitin gene as an endogenous reference for normalization.
Constructs and primer sequences used in this study
The plasmid constructs, primers and their purposes are listed in Table S1 (see Supporting Information). The primer sequences for qRT‐PCR are listed in Table S2 (see Supporting Information).
Supporting information
Fig. S1 Rice blast scores of the NPB‐Piz‐t:HA and NPB plants spray inoculated with the AvrPiz‐t‐ and LF‐AvrPiz‐t‐carrying isolates of Magnaporthe oryzae. The phenotype results are presented in Fig. 2a. Disease reactions of 30 seedlings per isolate were scored using a 0–5 scale system. Infected leaves were photographed at 6 days post‐inoculation (dpi) as shown in Fig. 2. Values are means ± standard error (SE) (n = 30). One‐way analysis of variance (ANOVA) test with Tukey's method was conducted. Means with different letters are significantly different (P < 0.05).
Fig. S2 Transcriptional expression of LF‐AvrPiz‐t:HA and AvrPiz‐t:HA in transgenic rice. Leaf tissues of the transgenic lines used for punch inoculation were sampled before inoculation. The quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis was conducted with the gene‐specific primers to both LF‐AvrPiz‐t:HA and AvrPiz‐t:HA. Values are means ± standard error (SE) (n = 30). One‐way analysis of variance (ANOVA) test with Tukey's method was conducted. Means with different letters are significantly different (P < 0.05).
Table S1 List of plasmid constructs and primers used in this study.
Table S2 List of primers used for quantitative real‐time polymerase chain reaction (qRT‐PCR).
Acknowledgements
We thank OARDC Graduate Research Enhancement Competitive Grants for financial support. We are grateful to Dr Mingzhe Shen for his suggestions on this project.
References
- Abramovitch, R.B. , Janjusevic, R. , Stebbins, C.E. and Martin, G.B. (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA, 103, 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu, A. , Wong, H.L. , Fujiwara, M. , Okuda, J. , Nishide, K. , Uno, K. , Imai, K. , Umemura, K. , Kawasaki, T. , Kawano, Y. and Shimamoto, K . (2013) An OsCEBiP/OsCERK1‐OsRacGEF1‐OsRac1 module is an essential early component of chitin‐induced rice immunity. Cell Host Microbe, 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Angot, A. , Vergunst, A. , Genin, S. and Peeters, N. (2007) Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathogens, 3, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield, M.J. (2015) Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell. Microbiol. 17, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Białas, A. , Zess, E.K. , De la Concepcion, J.C. , Franceschetti, M. , Pennington, H.G. , Yoshida, K. , Upson, J.L. , Chanclud, E. , Wu, C.H. , Langner, T. and Maqbool, A. (2017) Lessons in effector and NLR biology of plant–microbe systems. Mol. Plant–Microbe Interact. 31, 34–45. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bos, J.I. , Armstrong, M.R. , Gilroy, E.M. , Boevink, P.C. , Hein, I. , Taylor, R.M. , Zhendong, T. , Engelhardt, S. , Vetukuri, R.R. , Harrower, B. , Dixelius, C. , Bryan, G. , Sadanandom, A. , Whisson, S.C. , Kamoun, S. and Birch, P.R.J. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA, 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Chen, J. , Li, M. , Chang, M. , Xu, K. , Shang, Z. , Zhao, Y. , Palmer, I. , Zhang, Y. , McGill, J. and Alfano, J.R. (2017) A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe, 22(777–788), e777. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. and Zhou, J.M. (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover, A. (1998) The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan, V. and Rivas, S. (2014) E3 ubiquitin‐ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas, T.K. and Dangl, J.L. (2010) NB‐LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Göhre, V. , Spallek, T. , Häweker, H. , Mersmann, S. , Mentzel, T. , Boller, T. , de Torres, M. , Mansfield, J.W. and Robatzek, S. (2008) Plant pattern‐recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Janjusevic, R. , Abramovitch, R.B. , Martin, G.B. and Stebbins, C.E. (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science, 311, 222–226. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. , Vance, R.E. and Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science, 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T. , Henmi, K. , Ono, E. , Hatakeyama, S. , Iwano, M. , Satoh, H. and Shimamoto, K. (1999) The small GTP‐binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA, 96, 10 922–10 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Lu, D. and Shan, L. (2014) Ubiquitination of pattern recognition receptors in plant innate immunity. Mol. Plant Pathol. 15, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Wang, B. , Wu, J. , Lu, G. , Hu, Y. , Zhang, X. , Zhang, Z. , Zhao, Q. , Feng, Q. , Zhang, H. and Wang, Z. (2009) The Magnaporthe oryzae avirulence gene AvrPiz‐t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz‐t . Mol. Plant–Microbe Interact. 22, 411–420. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A. , Aichi, I. and Matsuoka, M. (2006) A protocol for Agrobacterium‐mediated transformation in rice. Nat. Protoc. 1, 2796–2802. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , DebRoy, S. , Lee, Y.H. , Pumplin, N. , Jones, J. and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science, 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , Mecey, C. , Lee, Y.‐N. , Imboden, L.A. , Chang, J.H. and He, S.Y. (2011) Effector‐triggered immunity blocks pathogen degradation of an immunity‐associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA, 108, 10 774–10 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis, V. , Mucyn, T.S. , Gimenez‐Ibanez, S. , Chapman, H.C. , Gutierrez, J.R. , Balmuth, A.L. , Jones, A.M. and Rathjen, J.P. (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science, 324, 784–787. [DOI] [PubMed] [Google Scholar]
- Ono, E. , Wong, H.L. , Kawasaki, T. , Hasegawa, M. , Kodama, O. and Shimamoto, K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA, 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.‐H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. and Valent, B. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.H. , Shirsekar, G. , Bellizzi, M. , Chen, S. , Songkumarn, P. , Xie, X. , Shi, X. , Ning, Y. , Zhou, B. , Suttiviriya, P. , Wang, M. , Umemura, K. and Wang, G.‐L. (2016) The E3 ligase APIP10 connects the effector AvrPiz‐t to the NLR receptor Piz‐t in rice. PLoS Pathogens, 12, e1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S. , Liu, G. , Zhou, B. , Bellizzi, M. , Zeng, L. , Dai, L. , Han, B. and Wang, G.L. (2006) The broad‐spectrum blast resistance gene Pi9 encodes a nucleotide‐binding site–leucine‐rich repeat protein and is a member of a multigene family in rice. Genetics, 172, 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock, T.R. , Zeng, L. , Brady, J.J. , Abramovitch, R.B. , Xiao, F. and Martin, G.B. (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature, 448, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner, A. and Estelle, M. (2010) The ubiquitin‐proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R. and Hager, A. (1992) Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein‐kinase activity. Planta, 187, 136–141. [DOI] [PubMed] [Google Scholar]
- Shirsekar, G.S. (2013) Ubiquitination in Innate Immunity of Rice (Oryza sativa). Columbus, OH: The Ohio State University. [Google Scholar]
- Smith, J.M. and Heese, A. (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leaf tissue in response to living Pseudomonas syringae . Plant Methods, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek, T. , Robatzek, S. and Göhre, V. (2009) How microbes utilize host ubiquitination. Cell. Microbiol. 11, 1425–1434. [DOI] [PubMed] [Google Scholar]
- Walsh, C.T. , Garneau‐Tsodikova, S. and Gatto, G.J. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. 44, 7342–7372. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Ning, Y. , Shi, X. , He, F. , Zhang, C. , Fan, J. , Jiang, N. , Zhang, Y. , Zhang, T. , Hu, Y. , Bellizzi, M. and Wang, G.‐L. (2016) Immunity to rice blast disease by suppression of effector‐triggered necrosis. Curr. Biol. 26, 2399–2411. [DOI] [PubMed] [Google Scholar]
- Yoo, S.‐D. , Cho, Y.‐H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. , Li, Y. , Liu, B. , Feng, D. , Wang, J. and Wang, H. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.‐M. , Zhang, X. , Zhou, Z.‐R. , Hu, H.‐Y. , Liu, M. , Zhou, B. , and Zhou, J. (2013) Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz‐t. J. Biomol. NMR, 55, 219–223. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. (2008) Pattern‐recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Rice blast scores of the NPB‐Piz‐t:HA and NPB plants spray inoculated with the AvrPiz‐t‐ and LF‐AvrPiz‐t‐carrying isolates of Magnaporthe oryzae. The phenotype results are presented in Fig. 2a. Disease reactions of 30 seedlings per isolate were scored using a 0–5 scale system. Infected leaves were photographed at 6 days post‐inoculation (dpi) as shown in Fig. 2. Values are means ± standard error (SE) (n = 30). One‐way analysis of variance (ANOVA) test with Tukey's method was conducted. Means with different letters are significantly different (P < 0.05).
Fig. S2 Transcriptional expression of LF‐AvrPiz‐t:HA and AvrPiz‐t:HA in transgenic rice. Leaf tissues of the transgenic lines used for punch inoculation were sampled before inoculation. The quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis was conducted with the gene‐specific primers to both LF‐AvrPiz‐t:HA and AvrPiz‐t:HA. Values are means ± standard error (SE) (n = 30). One‐way analysis of variance (ANOVA) test with Tukey's method was conducted. Means with different letters are significantly different (P < 0.05).
Table S1 List of plasmid constructs and primers used in this study.
Table S2 List of primers used for quantitative real‐time polymerase chain reaction (qRT‐PCR).


