Summary
RXLR effectors encoded by Phytophthora species play a central role in pathogen–plant interactions. An understanding of the biological functions of RXLR effectors is conducive to the illumination of the pathogenic mechanisms and the development of disease control strategies. However, the virulence function of Phytophthora parasitica RXLR effectors is poorly understood. Here, we describe the identification of a P. parasitica RXLR effector gene, PPTG00121 (PpE4), which is highly transcribed during the early stages of infection. Live cell imaging of P. parasitica transformants expressing a full‐length PpE4 (E4FL)‐mCherry protein indicated that PpE4 is secreted and accumulates around haustoria during plant infection. Silencing of PpE4 in P. parasitica resulted in significantly reduced virulence on Nicotiana benthamiana. Transient expression of PpE4 in N. benthamiana in turn restored the pathogenicity of the PpE4‐silenced lines. Furthermore, the expression of PpE4 in both N. benthamiana and Arabidopsis thaliana consistently enhanced plant susceptibility to P. parasitica. These results indicate that PpE4 contributes to pathogen infection. Finally, heterologous expression experiments showed that PpE4 triggers non‐specific cell death in a variety of plants, including tobacco, tomato, potato and A. thaliana. Virus‐induced gene silencing assays revealed that PpE4‐induced cell death is dependent on HSP90, NPK and SGT1, suggesting that PpE4 is recognized by the plant immune system. In conclusion, PpE4 is an important virulence RXLR effector of P. parasitica and recognized by a wide range of host plants.
Keywords: cell death, haustoria, Phytophthora parasitica, RXLR effector, virulence
Introduction
Phytophthora parasitica shares the main features of most Phytophthora species; it is a soil‐borne pathogen with a wide host range (Meng et al., 2014). It causes tobacco black shank and is listed as one of the top 10 oomycete pathogens because of its scientific and economic importance (Kamoun et al., 2015). P. parasitica serves as a model oomycete pathogen, and its compatible interaction with the model plant Arabidopsis thaliana has been established (Attard et al., 2010; Wang Y et al., 2011). There have been fewer functional analyses of P. parasitica genes (Chang et al., 2015; Evangelisti et al., 2013; Gaulin et al., 2002; Khatib et al., 2004; Meng et al., 2015; Zhang et al., 2012), and these studies are far from sufficient to fully understand the biology, pathogenesis and plant interaction mechanisms of P. parasitica.
During the war between pathogens and hosts, plants have evolved two immune systems to defend against invaders: pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI), mediated by pattern recognition receptors (PRRs), and effector‐triggered immunity (ETI), mediated by resistance (R) proteins that recognize avirulence (AVR) effectors (Dodds and Rathjen, 2010; Jones and Dangl, 2006). On perception of non‐self signals (PAMPs or effectors) from pathogens, plant cells activate a complicated signal transduction network. Although the signal transduction pathways implicated in PTI and ETI are different, the downstream cellular events are similar, including a series of cellular responses and also cell death (Dodds and Rathjen, 2010; Pedley and Martin, 2005; Peng et al., 2018). Although cell death induced by a number of Phytophthora RXLR effectors occurs independently of known R proteins, it is probably the result of plant recognition and related to components of the PTI or ETI pathway. Many genes involved in plant immune signalling are required for effector‐induced cell death. For example, MEK2 is required for Avh238‐triggered cell death (Yang et al., 2017), MEK2 and WIPK are involved in Avh241‐induced cell death (Yu et al., 2012), suppressor of G2 allele of skp1 (SGT1) is required for the cell death activity of PexRD2 (Oh et al., 2009) and PITG_22798 (Wang H et al., 2017), a specific mitogen‐activated protein kinase (MAPK) cascade is responsible for Pi_23226‐induced cell death (Lee et al., 2018), and SGT1, HSP90, RAR1 and MAPK cascades are required for PvRXLR16‐induced cell death (Xiang et al., 2017).
During the infection and colonization of plants, pathogens secrete numerous effectors to manipulate plant physiological processes and thereby suppress plant immunity and enhance plant susceptibility. Effectors usually possess dual activities, facilitating infection and triggering plant immunity during plant–microbial interactions (Kamoun, 2006; Kjemtrup et al., 2000; van’t Slot & Knogge, 2002). For example, the glycoside hydrolase 12 protein XEG1 is required for Phytophthora sojae virulence, but is also recognized as a PAMP and triggers cell death and plant immunity (Ma et al., 2015). Necrosis and ethylene‐inducing peptide 1 (Nep1)‐like proteins (NLPs), which are conserved virulence factors widespread in bacterial, oomycete and fungal pathogens, trigger host cell damage‐associated plant immunity and are also recognized as PAMPs (Bohm et al., 2014; Fellbrich et al., 2002; Ottmann et al., 2009; Qutob et al., 2006). Another classic example is the triggering of an R gene‐mediated hypersensitive response (HR) by AVR effectors that typically exert their virulence function on R gene‐absent plants (Kamoun, 2006). In addition to known AVR effectors, a few RXLR effectors, such as PsAvh241 (Yu et al., 2012), PsAvh238 (Wang Q et al., 2011; Yang et al., 2017) and PITG_22798 (Wang H et al., 2017), possess virulence functions even though they induce immune response‐related cell death in plants.
RXLR effectors, which exist by the hundreds in each oomycete genome, are amongst the best‐characterized oomycete effectors (Baxter et al., 2010; Haas et al., 2009; Jiang et al., 2008; Tyler et al., 2006). In recent years, a large number of studies have been carried out to elucidate the biological functions of RXLR effectors from Phytophthora infestans, P. sojae and Hyaloperonospora arabidopsidis (Anderson et al., 2015; Sharpee and Dean, 2016; Wang Q et al., 2011; Whisson et al., 2016; Zheng et al., 2014). However, little is known about RXLR effectors from P. parasitica, except PSE1, which has been reported to alter the auxin content and to promote infection (Evangelisti et al., 2013). In addition, 172 candidate RXLR effectors have been identified recently in the P. parasitica genome, three of which suppress INF1‐induced cell death and enhance P. parasitica virulence (Dalio et al., 2017).
In this study, we investigated the virulence function of the P. parasitica RXLR effector gene PpE4. We found that PpE4 is highly expressed during the early stages of infection and is secreted from haustoria. To evaluate the role of PpE4 in P. parasitica pathogenicity, PpE4‐silenced transformants were created and analysed. These transformants showed a reduced ability to infect plants, and transient expression of PpE4 in Nicotiana benthamiana restored pathogenicity. To further examine its contribution in the promotion of pathogen colonization, an inoculation assay was performed after transient or induced in planta expression of PpE4. Plants expressing PpE4 were more susceptible to P. parasitica infection. PpE4 also triggered non‐specific cell death in a variety of plants in an HSP90‐, NPK‐ and SGT1‐dependent manner, which suggests that PpE4 is recognized by the plant immune system. Based on these results, we conclude that PpE4 is a virulence RXLR effector of P. parasitica and is recognized by a wide variety of host plants.
Results
PpE4 encodes a secreted RXLR effector and is highly expressed during the early phase of infection
Using previous RNA‐sequencing (RNA‐seq) data (Jia et al., 2017), we identified a putative P. parasitica RXLR effector gene PPTG_00121, named PpE4, which was the most highly expressed RXLR effector gene during the infection of Arabidopsis roots (Fig. S1A, see Supporting Information). Over 70% of the total RXLR effector transcripts corresponded to PpE4, and the FPKM (fragments per kilobase million) value was over 8000 at 3–6 h post‐inoculation (hpi) of P. parasitica zoospores (Fig. S1A). To validate the expression profile of PpE4 during development and plant infection, reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) (Bustin et al., 2009) was performed. The expression pattern of P. parasitica PpE4 during A. thaliana root infection initiated with zoospores was consistent with RNA‐seq (Fig. S1B). During infection of N. benthamiana leaves, PpE4 transcripts were rapidly and strongly up‐regulated from 3 to 24 hpi, and then declined and became barely detectable at 48 hpi, which is similar to the observations in vegetative hyphae, zoospores, cysts and germinated cysts before infection (Fig. 1). Biotrophic growth of P. parasitica in N. benthamiana leaves was dominant before 24 hpi, followed by necrotrophic growth with significant cell death (Fig. S2, see Supporting Information). In conclusion, PpE4 transcripts are strongly induced and predominantly accumulated during the biotrophic phase, at levels hundreds of times higher than those in the mycelium.
Figure 1.
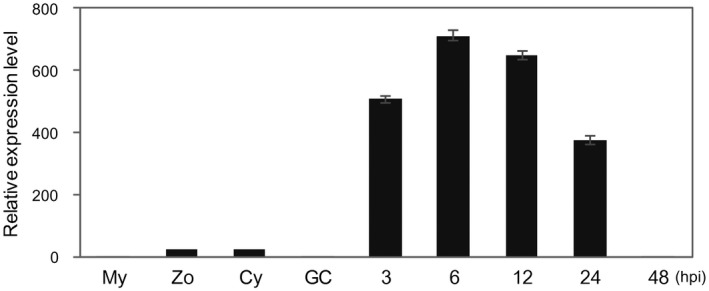
The Phytophthora parasitica RXLR effector gene PpE4 is highly expressed during early plant infection. Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) was used to quantify the relative PpE4 transcript levels during different stages of P. parasitica development and infection. Nicotiana benthamiana leaves inoculated with P. parasitica zoospores were harvested at different hours post‐inoculation (hpi). Cy, cysts; GC, germinated cysts; My, P. parasitica mycelium grown in carrot broth; Zo, zoospores. The relative expression level of PpE4 in mycelia was given a value of unity. Error bars represent the standard deviation (SD) of three biological replicates.
To monitor the secretion of PpE4 during infection, a full‐length PpE4 with its native signal peptide (E4FL)‐mCherry fusion construct was transformed into P. parasitica strain 1121, which stably expresses cytoplasmic green fluorescent protein (GFP), via polyethylene glycol (PEG)–CaCl2‐mediated transformation (Bottin et al., 1999). Six transformants showing a stable red fluorescence signal and one without were chosen for RT‐qPCR and western blot assays. High levels of PpE4 transcripts and fusion proteins accumulated in vegetative mycelia of transformants, whereas no accumulation was observed in E4MC3N4 and strain 1121 (Fig. S3, see Supporting Information). The observation of transformant E4MC4A2 with a strong red fluorescence signal showed that the red fluorescence was evenly distributed in mycelia cultured in vitro (Fig. 2A), whereas it was highly enriched in haustoria during the infection of N. benthamiana leaves at 24 hpi (Fig. 2B). Further detailed observations and fluorescence intensity analyses of E4MC4A2 (Fig. 2C,D) and E4MC4A6 (Fig. S4, see Supporting Information) showed that the mCherry fluorescence signal accumulated outside the GFP fluorescent haustoria, mainly distributed around the haustorial neck, indicating that E4FL‐mCherry accumulates in the extrahaustorial matrix (EHMx) on secretion from haustoria. By contrast, there was no mCherry fluorescence in strain 1121, and GFP fluorescence was distributed evenly in vegetative and infection hyphae, without specific accumulation at haustoria (Fig. 2). This result is consistent with previous studies of P. infestans effectors AVR3a (Whisson et al., 2007), AVR2 (Gilroy et al., 2011), Pi04314 (Wang S et al., 2017), AVR4 and AVRblb1 (van Poppel, 2009), and P. sojae effector Avr1b (Liu et al., 2014).
Figure 2.
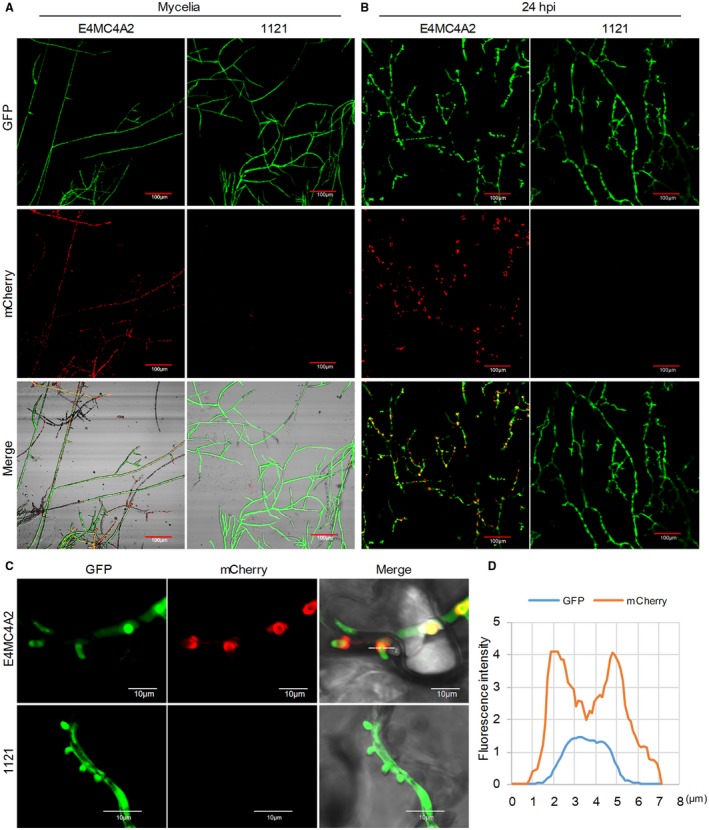
PpE4 accumulates around haustoria after secretion during Phytophthora parasitica infection. (A) Confocal images of mycelia cultured on 5% carrot juice agar medium. The red fluorescence was distributed throughout the mycelial cytoplasm of E4MC4A2 [a transformant expressing cytoplasmic green fluorescent protein (GFP) and full‐length PpE4 (E4FL)‐mCherry], but was not detected in strain 1121 (stably expressing cytoplasmic GFP). (B) Nicotiana benthamiana leaves infected with E4MC4A2 and 1121 were observed by confocal microscopy at 24 h post‐inoculation (hpi). A strong red fluorescence signal was highly accumulated in haustoria, but not in hyphae, during E4MC4A2 infection, whereas GFP fluorescence was evenly distributed in hyphae. No red fluorescence was observed in strain 1121. (C) A magnified lateral view of haustoria showing red fluorescence focused on the outside of the haustoria base and the GFP signal distributed throughout hyphae and haustoria. (D) The fluorescence intensities of GFP and mCherry across the haustorium indicated by the white line labelled ‘2’ in (C). Identical images were obtained from more than 10 haustoria in three independent biological replicates. [Colour figure can be viewed at wileyonlinelibrary.com]
Silencing of PpE4 attenuates the pathogenicity of P. parasitica
Inoculation analysis revealed that constitutive expression of E4FL‐mCherry reduces colonization by P. parasitica (Fig. S5, see Supporting Information). To investigate the potential virulence function of PpE4 in P. parasitica pathogenesis, we generated co‐silencing transformants as described previously (Meng et al., 2015). A hairpin structure derived from a segment of GFP fused with a segment of PpE4 was constructed and introduced into P. parasitica strain 1121 (Fig. S6A, see Supporting Information). Because both GFP and PpE4 were targeted, PpE4 expression was more likely to be decreased in transformants with a significantly reduced GFP signal. A total of 173 independent transformants were generated, and 19 with normal colony morphology showed decreased GFP fluorescence, a frequency consistent with previous reports (Meng et al., 2015; Zhang et al., 2012). RT‐qPCR experiments revealed that five of the 19 candidate transformants had obviously reduced PpE4 expression at 24 hpi compared with strain 1121 (Fig. S6). Further pathogenicity analysis showed that three silenced lines (E4S2A6, E4S2B2 and E4S2F5) produced significantly smaller lesions and less hyphal biomass compared with strain 1121, whereas the virulence of the other two lines (E4S2C4 and E4S2G5) was almost unaffected (Fig. 3A–C). We further confirmed the expression of PpE4 in these transformants after a series of subcultures. The results showed that the expression level of PpE4 in infected N. benthamiana leaves at 15 hpi and 24 hpi remained silenced in transformants E4S2A6, E4S2B2 and E4S2F5, but partially recovered in E4S2C4 and totally recovered in E4S2G5 (Fig. 3D), which was consistent with the results of the pathogenicity assay. Therefore, stable silencing of PpE4 led to the attenuated pathogenicity of P. parasitica and restored target gene expression, suggesting that PpE4 is important to P. parasitica.
Figure 3.
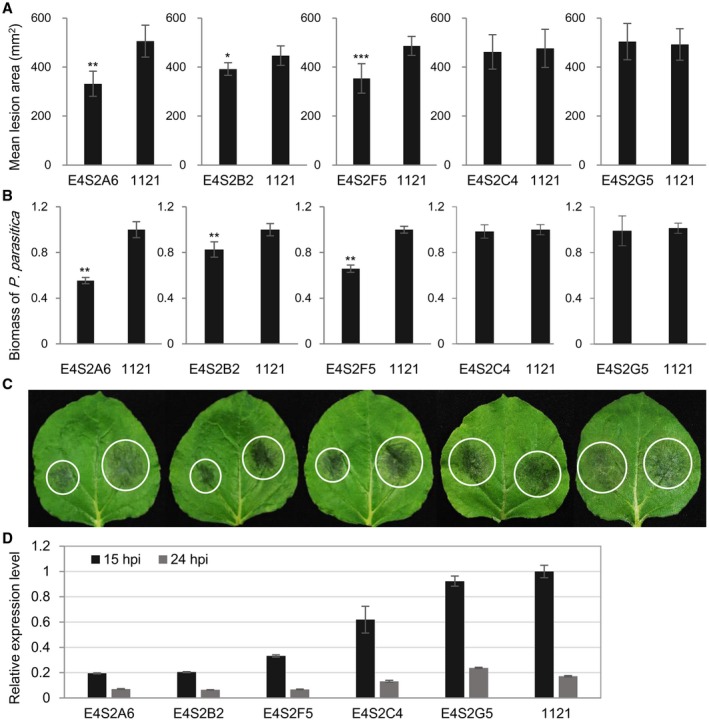
PpE4‐silenced Phytophthora parasitica transformants exhibit reduced pathogenicity. (A) Mean lesion areas of Nicotiana benthamiana leaves inoculated with PpE4‐silenced transformants and the control strain 1121 at 48 h post‐inoculation (hpi). The transformant and the control strain were inoculated on opposite halves of an N. benthamiana leaf. Error bars represent the standard deviation (SD) of 15 leaves, and asterisks denote significant differences from control strain 1121 (two tailed t‐test: *P < 0.05; **P < 0.01; ***P < 0.001). (B) Biomass of P. parasitica on N. benthamiana leaves determined by quantitative polymerase chain reaction (qPCR). Bars represent PpUBC levels relative to NbF‐box levels with SD of three biological replicates. Asterisks denote significant differences from control strain 1121 (two tailed t‐test: **P < 0.01). (C) Representative inoculated leaves. White circles outline the water‐soaked lesions. Similar results were obtained from more than three independent experiments with about 15 leaves for each experiment. (D) PpE4 expression was restored in two transformants E4S2C4 and E4S2G5, whose virulence was not reduced. The subcultured transformants were inoculated onto N. benthamiana leaves and sampled at 15 and 24 hpi. Reverse transcription (RT)‐qPCR was used to determine the PpE4 silencing level. The expression level of PpE4 in strain 1121 sampled at 15 hpi was given a value of unity. Error bars represent the SD of three biological replicates. Two independent experiments were performed with similar results. [Colour figure can be viewed at wileyonlinelibrary.com]
Transient expression of PpE4 in planta restores the pathogenicity of PpE4‐silenced transformants
To verify that the virulence attenuation of the PpE4‐silenced lines is caused by PpE4 silencing, three silenced lines were inoculated onto mature PpE4‐expressing (intracellular expression without signal peptide) or GFP‐expressing leaves, and strain 1121 was inoculated onto GFP‐expressing leaves. The PpE4‐silenced lines inoculated onto PpE4‐expressing leaves formed significantly larger lesions than those inoculated onto GFP‐expressing leaves, whereas there was no difference between the size of the lesions on PpE4‐expressing leaves and those of the control group (1121 inoculated on GFP‐expressing leaves) (Fig. 4A–C). Western blot showed that PpE4 and GFP proteins were stably accumulated under low agroinfiltration concentration [optical density at 600 nm (OD600) = 0.01] (Fig. 4D). This indicates that in planta expression of PpE4 is able to restore the virulence of PpE4‐silenced lines to wild‐type levels. In conclusion, PpE4 positively contributes to the pathogenicity of P. parasitica.
Figure 4.
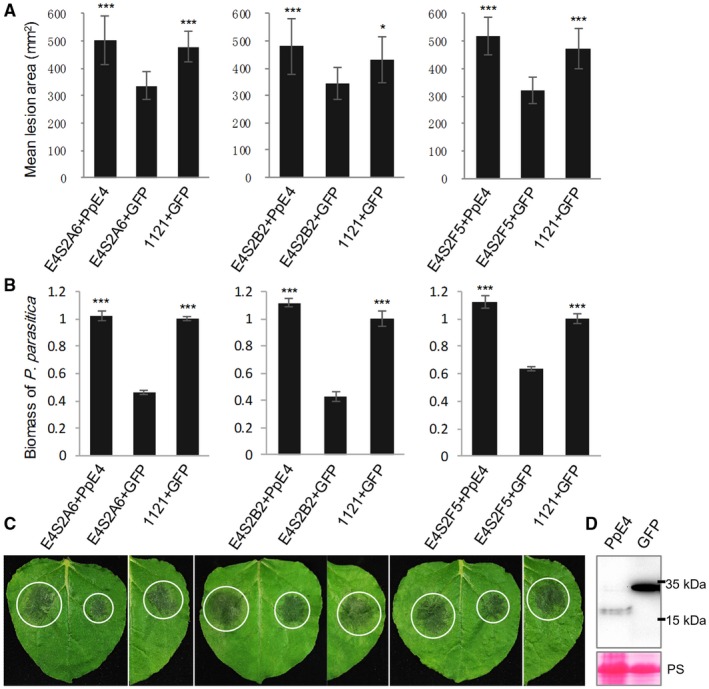
The pathogenicity of PpE4‐silenced Phytophthora parasitica lines is restored by the transient expression of PpE4 in planta. PpE4 and GFP were transiently expressed by agroinfiltration in Nicotiana benthamiana leaves 1 day before inoculation [optical density at 600 nm (OD600) = 0.01]. (A) The lesions formed after the inoculation of PpE4‐silenced lines onto PpE4‐expressing leaves were almost the same size as those formed after the inoculation of 1121 onto GFP‐expressing leaves, whereas the lesions formed by silenced lines inoculated onto GFP‐expressing leaves were significantly smaller. Error bars represent the standard deviation (SD) of 15 leaves, and asterisks denote significant differences from the control group (two‐tailed t‐test: *P < 0.05; ***P < 0.001). (B) Biomass of P. parasitica on N. benthamiana leaves was determined by quantitative polymerase chain reaction (qPCR). Bars represent PpUBC levels relative to NbF‐box levels with SD of three biological replicates. Asterisks denote significant differences from silenced lines inoculated onto GFP‐expressing leaves (two‐tailed t‐test: ***P < 0.001). (C) Representative inoculated leaves. White circles outline the water‐soaked lesions. (D) Protein accumulation detected by western blot using anti‐Flag antibody. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from three independent experiments with more than 15 leaves inoculated for each group in each experiment. [Colour figure can be viewed at wileyonlinelibrary.com]
PpE4 enhances plant susceptibility to P. parasitica
To further determine whether PpE4 contributes to P. parasitica colonization in planta, inoculation was performed onto N. benthamiana leaves expressing mature PpE4 on one half and GFP on the other. The lesions and P. parasitica biomass on the PpE4‐expressing halves were significantly larger than those on the control (Fig. 5A–C). Stable accumulation of PpE4 and GFP proteins in planta was detected by western blot (Fig. 5D). These results suggest that the transient expression of PpE4 renders N. benthamiana more susceptible to P. parasitica.
Figure 5.
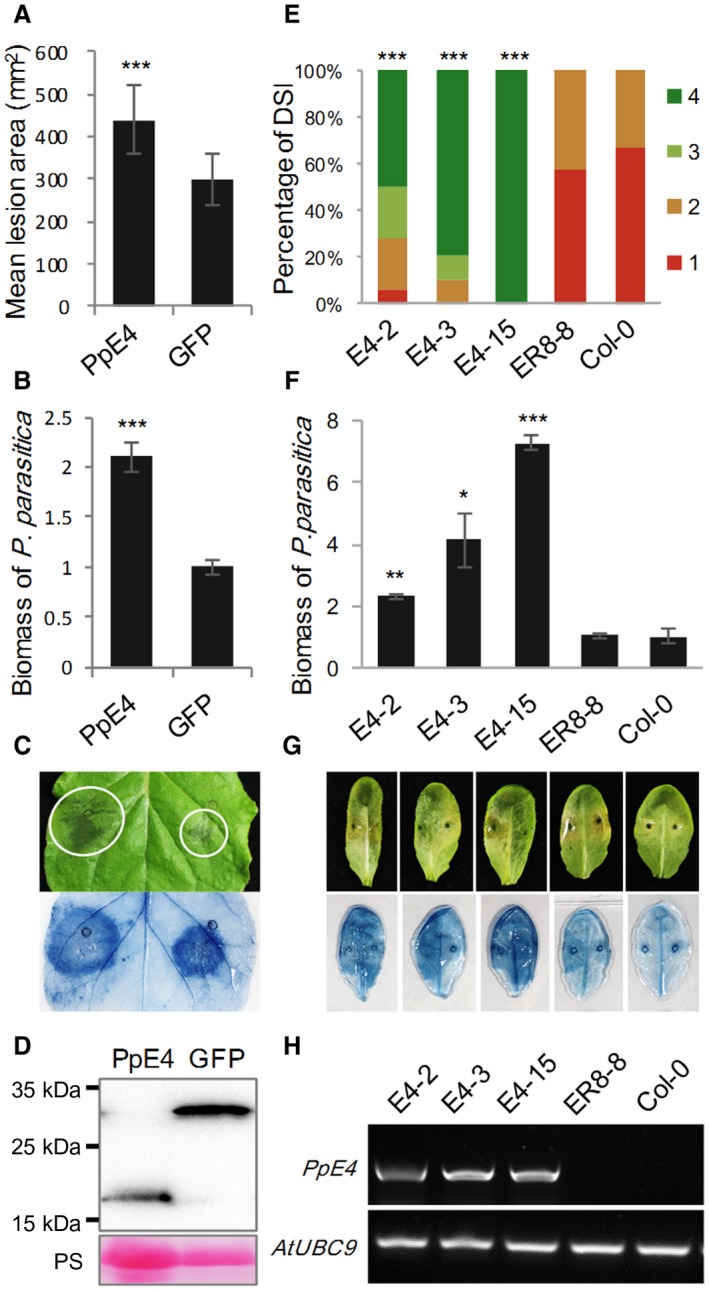
Heterologous expression of PpE4 renders Nicotiana benthamiana and Arabidopsis more susceptible to Phytophthora parasitica infection. (A) Mean lesion areas were measured at 48 h post‐inoculation (hpi). Agrobacterium tumefaciens strains carrying PpE4 or GFP [optical density at 600 nm (OD600) = 0.01] were infiltrated into different sides of the same leaf, 1 day before inoculation of strain 1121. Error bars represent the standard deviation (SD) of 15 leaves, and asterisks denote significant differences from the green fluorescent protein (GFP) control (two‐tailed t‐test: ***P < 0.001). (B) Quantification of P. parasitica biomass in infected N. benthamiana leaves. Bars represent PpUBC levels relative to NbF‐box levels with SD of three biological replicates. Asterisks denote significant differences from the GFP control (two‐tailed t‐test: ***P < 0.001). (C) A typical leaf photographed and stained by trypan blue. White circles outline the water‐soaked lesions. (D) Protein accumulation was determined at 3 days post‐infiltration (dpi) by western blot using anti‐Flag antibody. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from three independent experiments with about 15 leaves for each experiment. (E) Disease severity index (DSI) from grade 1 to grade 4 was recorded at 48 hpi. Homozygous transgenic plants expressing β‐estradiol‐inducible 3×Flag‐PpE4 (E4‐2, E4‐3 and E4‐15), an empty vector pER8 transgenic plant (ER8‐8) and wild‐type Col‐0 were injected with 10 μM 17‐β‐estradiol, 12 h before inoculation of strain 1121. Asterisks represent significant differences from Col‐0 (Wilcoxon rank‐sum test: ***P < 0.001). (F) Biomass of P. parasitica on Arabidopsis leaves. Bars represent PpUBC levels relative to AtUBC levels with SD of five biological replicates. Asterisks denote significant differences from Col‐0 (two‐tailed t‐test: *P < 0.05; **P < 0.01; ***P < 0.001). (G) Disease symptoms of representative leaves. Trypan blue stain was used to highlight the infection hyphae in colonized leaves. (H) Verification of PpE4 expression 12 h after injection of 10 μM 17‐β‐estradiol using semi‐quantitative polymerase chain reaction (PCR). Similar results were obtained from three independent experiments with about 25 leaves for each experiment. [Colour figure can be viewed at wileyonlinelibrary.com]
We also examined the contribution of PpE4 in the Arabidopsis–P. parasitica pathosystem. Chemically inducible transgenic Arabidopsis lines in which the expression of mature PpE4 is strictly regulated by estradiol were constructed (Zuo et al., 2000). Wild‐type Col‐0 and empty vector pER8 transgenic plants were used as controls. The rosette leaves from three homozygous transgenic lines expressing PpE4 and control plants were infiltrated with 10 μM 17‐β‐estradiol to induce PpE4 expression, 12 h before inoculation of P. parasitica zoospores. The disease index statistic indicated that the transgenic plants expressing PpE4 were more susceptible than the controls to P. parasitica infection, and the pathogen biomass in these plants was significantly higher than that in the control plants (Fig. 5E–G). Semi‐quantitative PCR showed that PpE4 was expressed on injection of 17‐β‐estradiol, whereas there was no PpE4 expression in control plants receiving the same treatment (Fig. 5H). These results demonstrate that PpE4 facilitates P. parasitica infection.
PpE4 triggers cell death in various plants
When PpE4 was intracellularly expressed in N. benthamiana leaves by agroinfiltration, it triggered cell death at 3 days post‐infiltration (dpi) (Fig. 6A). To investigate whether this cell death is species specific, PpE4 was transiently expressed in several Solanaceae plants, including three tobacco species, tomato and potato. We found that PpE4 triggered cell death in all tested Solanaceae plants (Fig. 6A). No Arabidopsis transgenic plants were recovered when the 35S promoter was used to drive PpE4 expression, implying that PpE4 is lethal to Arabidopsis cells. Using estradiol‐inducible transgenic plants, we found that PpE4 triggered cell death in Arabidopsis leaves 4 days after induction by 17‐β‐estradiol, whereas no cell death occurred in the control plants (empty vector pER8 transgenic plants and Col‐0) (Fig. 6B). These results indicate that PpE4 triggers non‐specific cell death in a variety of plants.
Figure 6.
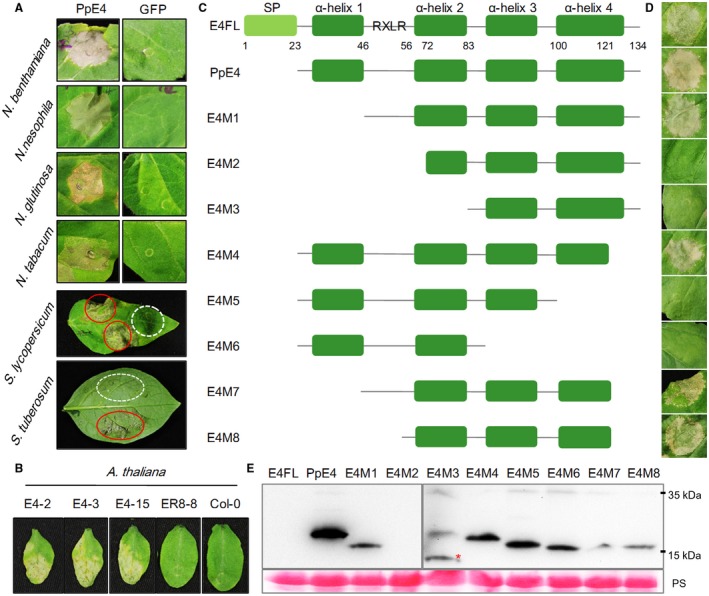
Analysis of cell death triggered by PpE4. (A) Cell death phenotype induced by PpE4 in Solanaceae plants. Agrobacterium tumefaciens carrying PpE4 [optical density at 600 nm (OD600) = 0.4] was infiltrated into the leaves of Nicotiana benthamiana, N. nesophila, N. glutinosa, N. tabacum cv. Florida 301, Solanum lycopersicum and S. tuberosum. Photographs were taken at 5 days post‐infiltration (dpi) for Nicotiana species and 8 dpi for Solanum species. Red circles represent the PpE4‐expressing areas, and white broken circles represent the GFP‐expressing areas. (B) Cell death symptoms triggered by PpE4 in Arabidopsis. Leaves of transgenic Arabidopsis plants harbouring pER8::3×Flag‐PpE4 or the empty vector and Col‐0 were injected with 10 μM 17‐β‐estradiol. Photographs were taken after 5 days. (C) Schematic diagrams of the protein secondary structures of the PpE4 deletion mutants. (D) Cell death symptoms in N. benthamiana leaves expressing PpE4 deletion mutants. Photographs were taken at 5 dpi. (E) Western blot detection of PpE4 deletion proteins using anti‐Flag antibody. The red asterisk indicates a protein band of the correct size. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from three independent experiments. [Colour figure can be viewed at wileyonlinelibrary.com]
According to the protein secondary structure predicted by Phyre2 (Kelley et al., 2015), PpE4 contains four α‐helices downstream of the signal peptide (Fig. S7, see Supporting Information). To identify which domains are crucial for cell death‐inducing activity, we successively deleted the α‐helices to construct a series of deletion mutants and transiently expressed them in N. benthamiana (Fig. 6C–E). E4FL with its native signal peptide could also induce cell death. However, cell death occurred more slowly and more weakly in comparison with that of the mature protein. As shown in Fig. 6, E4M1, E4M4, E4M7 and E4M8 still maintained cell death‐inducing activity, which indicates that the first α‐helix, the RXLR motif and the last 13 amino acids are not required for cell death induction. Deletion of the RXLR‐DEER domain as well as the second α‐helix abolished cell death‐inducing ability. As the DEER motif is in α‐helix 2, its deletion may destroy the structure of α‐helix 2. Moreover, E4M5 and E4M6 were unable to induce cell death, which indicates that α‐helix 3 and part of α‐helix 4 are necessary for cell death induction (Fig. 6C,D). In short, E4M8, with residues 56–121, is sufficient to maintain the integrity of the protein tertiary structure and to trigger cell death.
PpE4‐induced cell death requires HSP90, NPK and SGT1
Cell death induced by a number of pathogen effectors is considered to be the outcome of recognition by the plant immune system, either PTI or ETI, involving a variety of receptors and signal transduction pathways (Lee et al., 2018; Wang H et al., 2017; Wang Q et al., 2011; Xiang et al., 2017; Yang et al., 2017; Yu et al., 2012). To determine which signalling pathway is involved in PpE4‐induced cell death, virus‐induced gene silencing (VIGS) was used to silence a series of genes in N. benthamiana, including genes responsible for R protein function, such as HSP90, SGT1 and RAR1 (Kanzaki et al., 2003; Takahashi et al., 2003; Zhang et al., 2004), genes associated with the activation of the TIR‐NB‐LRR (Toll/interleukin‐1 receptor, nucleotide binding and leucine‐rich repeat) and CC‐NB‐LRR (coiled coil, nucleotide binding and leucine‐rich repeat) R proteins, EDS1 and NDR1 (Knepper et al., 2011; Oh et al., 2014), respectively, the receptor‐like kinases BAK1 and SOBIR1 (Chaparro‐Garcia et al., 2011; Liebrand et al., 2013, 2014), the transcription factors MYB1 and WRKY3, and the MAPK cascade genes NPK, MEK1, MEK2 and SIPK (Jin et al., 2002; Liu et al., 2004b). Cell death was scored after transient expression of PpE4 in these silenced plants. PpE4‐induced cell death was almost abolished in HSP90‐ and NPK‐silenced plants, and significantly attenuated in SGT1‐silenced plants, compared with GFP‐silenced plants (Fig. 7A,B). Western blot assay showed that the PpE4 protein was stably accumulated in the silenced plants (Fig. 7D). In addition, cell death was slightly, but significantly, compromised in BAK1‐silenced plants, whereas cell death was not affected in plants with silenced expression of the other genes (Fig. S8A,B, see Supporting Information). RT‐qPCR assays confirmed that there was a significant reduction in the transcript levels of the targeted genes in silenced plants compared with the levels in GFP‐silenced plants (Figs 7C and S8C). In summary, HSP90, NPK and SGT1 are required for PpE4‐induced cell death.
Figure 7.
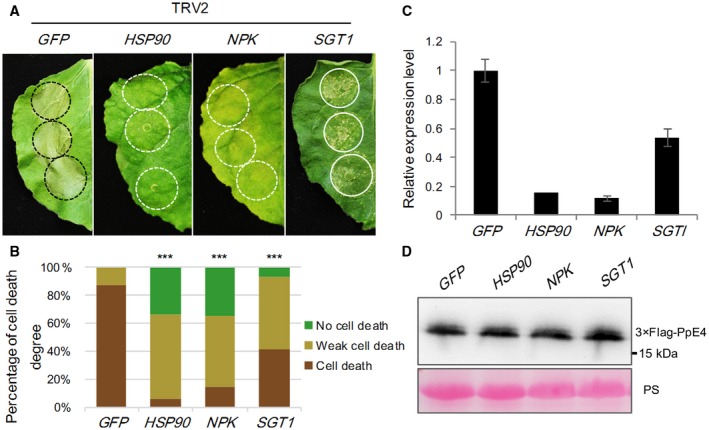
HSP90, NPK and SGT1 are involved in PpE4‐induced cell death. (A) Representative images of PpE4‐induced cell death in silenced Nicotiana benthamiana leaves at 5 days post‐infiltration (dpi). PpE4 was transiently expressed in the upper leaves of silenced plants at 16–20 dpi of TRV constructs. (B) Quantification of cell death in N. benthamiana leaves scored at 5 dpi. The degree of cell death was divided into three levels: no cell death, weak cell death and complete cell death. Asterisks indicate significant differences from GFP‐silenced plants (Wilcoxon rank‐sum test: ***P < 0.001). (C) Relative expression levels of HSP90, NPK and SGT1 transcripts in corresponding virus‐induced gene silencing (VIGS)‐treated plants determined by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Error bars represent the standard deviation (SD) of three biological replicates. (D) Detection of PpE4 protein accumulation in silenced leaves using the anti‐Flag antibody. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from more than three independent experiments with 10 plants for each TRV construct. [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
PpE4 is highly induced during infection and secreted from haustoria
As reported previously, functionally important RXLR effectors are usually induced during infection stages, but only a few RXLR effectors, which contribute the vast majority of RXLR effector transcripts, are considered to be crucial for pathogen pathogenicity (Wang Q et al., 2011). Here, we found that the effector gene PpE4 is highly up‐regulated, accounting for more than 70% of the total RXLR effector transcripts during the early stages of infection (Figs 1 and S1). The relative expression of PpE4 in the transformants was hundreds of times higher than that in strain 1121 in vegetative mycelia. However, only three transformants exhibited a slightly higher expression level than strain 1121 at 36 hpi (Fig. S3B,C), indicating that PpE4 is extremely highly transcribed during infection and is difficult to be over‐expressed artificially by Hsp70 or Ham34 promoter during infection. This implies that PpE4 plays a critical role during P. parasitica infection. RXLR effectors usually accumulate in the EHMx and are especially concentrated at the haustorial neck after secretion (Gilroy et al., 2011; Liu et al., 2014; van Poppel, 2009; Wang S et al., 2017; Whisson et al., 2007). In this study, we demonstrated that E4FL‐mCherry fusion protein accumulates substantially in the EHMx after secretion from haustoria during infection, even though its expression is driven by constitutive promoters (Figs 2, S3 and S4).
PpE4 contributes to infection even though it triggers cell death in plants
In virulence assays of PpE4‐silenced transformants, three stable silenced lines showed attenuated pathogenicity that could be restored by the transient expression of PpE4 in planta. However, the virulence of two other unstable transformants, E4S2C4 and E4S2G5, was not attenuated, with PpE4 expression being initially silenced, but restored after a series of subcultures (Figs 3 and 4). These results indicate that stable silencing of PpE4 affects the pathogenicity of P. parasitica. The restored expression of PpE4 in the silenced transformants and its low frequency of co‐silencing with GFP suggest its importance and tightly regulated expression in P. parasitica, similar to a previous report (Meng et al., 2015). We also found that E4FL‐mCherry‐expressing transformants displayed decreased virulence (Fig. S5). The virulence reduction may be attributed to PpE4 recognition by the plant immune system. Considering that PpE4 is transcribed in small amounts in mycelia and zoospores, it is likely that the constitutive overexpression of PpE4 during the pre‐infection stage disrupts its original expression pattern and affects its pathogenicity. Similarly, the premature expression of Avh238 also affects the ability of P. sojae to infect plants; thus, the timing of effector expression is crucial for pathogenicity (Wang Q et al., 2011).
In addition to its contribution to infection, we also found that PpE4 triggers non‐specific cell death in two major eudicots (Fig. 6). Cell death plays a vital, but ambiguous, role in plant–pathogen interactions, especially those involving biotrophic and hemibiotrophic pathogens. Hemibiotrophs require living cells to establish colonization, and cell death is not preferred at the early stages of infection. During rapid expansion in plants, the invaders induce host cell death to facilitate the transition from biotrophy to necrotrophy (Qutob et al., 2002). However, cell death induced by the recognition of PAMPs or AVR effectors emerging at the very beginning of infection usually abolishes pathogen invasion. Therefore, the timing and intensity of cell death are under sophisticated regulation during plant–pathogen interactions: that which controls cell death wins (Coll et al., 2011; Kabbage et al., 2013).
Many RXLR effectors have been demonstrated to promote pathogen colonization when transiently expressed in plants, such as AVR1 (Du et al., a,b), PexRD2 (King et al., 2014), PITG_22798 (Wang H et al., 2017), Avh241 (Yu et al., 2012) and Avh238 (Yang et al., 2017). However, these effectors could also induce HR when detected by corresponding R proteins or recognized by unknown mechanisms in plants. To eliminate the influence of cell death during the inoculation process, we reduced the concentration of the Agrobacterium tumefaciens suspension to an OD600 value of 0.01 to delay and weaken cell death (Wang H et al., 2017). As a result, PpE4 enhanced infection when transiently or stably expressed in plants, in spite of its cell death‐inducing activity (Figs 5 and 6). Three hypotheses may explain this result. First, although PpE4 is highly transcribed at the early stages of infection, the accumulation of PpE4 protein in plant cells via translocation from the pathogen may be insufficient to induce cell death under natural conditions. Second, considering the cases of Avh241 and Avh238, where cell death could be suppressed by other immediate‐early expressed effectors (Wang Q et al., 2011), we suspect that PpE4‐induced cell death may be suppressed by other cooperative effectors. Thus, PpE4 possibly manifests its virulence function and enhances colonization when its cell death activity is blocked. Finally, it is possible that cell death occurs just in time to promote the transition into necrotrophy, enabling an earlier occurrence of the necrotrophic phase. In this situation, cell death triggered by its intracellular expression is beneficial to pathogen infection, making it a virulence factor. In any case, PpE4 exhibits dual functions: it contributes to P. parasitica virulence, whilst triggering recognition‐related cell death in the host plant.
PpE4‐triggered cell death may be related to plant recognition
In this study, we applied VIGS technology to demonstrate that PpE4‐induced cell death requires HSP90, NPK and SGT1 (Fig. 7). As reported previously, HSP90 often forms a complex with its co‐chaperones RAR1 and SGT1 to maintain the function of NB‐LRR proteins (Kadota et al., 2010; Shirasu, 2009). In addition to being involved in R3a‐AVR3a‐mediated HR and INF1‐triggered cell death (Bos et al., 2006; Chapman et al., 2014; Kanzaki et al., 2003), HSP90 and SGT1 are required in both N‐ and Rx‐mediated defence responses against viruses (Boter et al., 2007; Liu et al., 2004a; Lu et al., 2003). However, only SGT1 is required for PITG_22798‐ and Rpiblb2‐AVRblb2‐triggered HR (Oh et al., 2014; Wang H et al., 2017). NPK1 is the Nicotiana homologue of human MEKK1 and encodes a MAP kinase kinase kinase that is involved in responses mediated by the resistance genes N, Bs2 and Rx (Jin et al., 2002; Liu et al., 2004b; Soyano et al., 2003). The fact that HSP90, NPK and SGT1 are involved in either R gene‐ or PRR‐mediated immune signalling suggests that PpE4‐triggered cell death is possibly the consequence of plant recognition. However, this recognition is not mediated by either BAK1‐ or SOBIR1‐associated cell surface receptors, or by EDS1‐ or NDR1‐associated R proteins, because cell death was only slightly affected in BAK1‐silenced plants and not significantly affected in SOBIR1‐, EDS1‐ or NDR1‐silenced plants (Fig. S8). According to a previous study, PpE4 shows moderate sequence similarity to P. sojae effector Avh238 (Yang et al., 2017). Although they are significantly divergent in the C‐terminal region, both PpE4 and Avh238 trigger non‐specific cell death in various plants, but the cell death mechanism may be distinct, as different genes are responsible for the mediation of cell death induced by each effector (Yang et al., 2017). Different components of the PTI or ETI pathways have been reported to be specifically involved in cell death induced by different RXLR effectors, which indicates that there are distinct recognition mechanisms and complicated signalling pathways responsible for each effector (Bos et al., 2006; Lee et al., 2018; Oh et al., 2009; Wang H et al., 2017; Xiang et al., 2017; Yang et al., 2017; Yu et al., 2012). However, it is still unclear how these effectors are recognized in plants.
It is worthwhile to elucidate the cell death induction mechanisms of early‐induced RXLR effectors. Studies of the biological functions and host targets of RXLR effectors are conducive to the illumination of the pathogenic mechanisms and the development of disease control strategies against pathogens, such as P. parasitica, which have a broad host range.
Experimental Procedures
Plant and Phytophthora cultivation
Arabidopsis thaliana seeds were sterilized and sown on 0.8% agar plates containing half‐strength Murashige and Skoog nutrient solution, followed by a 1‐week incubation in a growth chamber, as described previously (Wang Y et al., 2011). The seedlings were then transferred to a matrix containing soil and vermiculite, and grown in a 22–25 °C climate chamber with a photoperiod of 14 h light, 10 h dark and 70% relative humidity for 4 weeks. Nicotiana benthamiana, tobacco and tomato seeds, and potato tubers, were routinely cultured in a matrix in a climate chamber for about 5–6 weeks under the same conditions as used for the growth of Arabidopsis. The P. parasitica strain and transformants were cultured on 5% (v/v) carrot juice agar (CA) medium with 0.01% (w/v) CaCO3 and 0.002% (w/v) β‐sitosterol for 4 days at 23 °C. Then, 5% CA plugs with fresh mycelia were cultured in carrot broth for 4 days. To produce sporangia, carrot broth was replaced with Petri solution [Ca(NO3)2, 0.4 g/L; KH2PO4, 0.15 g/L; Mg(NO3)2, 0.15 g/L; CaCl2, 0.06 g/L], and the culture was cultivated for another 5 days. Zoospores were released by chilling and recovery as described previously (Wang Y et al., 2011).
Total RNA extraction and RT‐qPCR analyses
Total RNA of different samples was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the product manuals. RT‐qPCR was performed using 5 μL of a 1 : 10 dilution of first‐strand cDNA and SYBR Green mix (CWBio, Beijing, China) on a QuantStudio™ 3 Real‐Time PCR System (Thermo Scientific, Waltham, MA, USA). Gene‐specific primers were designed online (http://sg.idtdna.com/PrimerQuest/Home/Index), and the specificity was examined by performing dissociation curve assays. The previously described internal controls were chosen as follows: ubiquitin‐conjugating enzyme (PpUBC) and 40S ribosomal protein S3A (PpWS21) genes for P. parasitica (Yan and Liou, 2006); the AtUBC9 gene for A. thaliana; and the β‐actin gene for N. benthamiana. For the biomass assay, primers specific to PpUBC, AtUBC9 and NbF‐box were used for qPCR.
Vector construction
All the primers and vectors used in this study are listed in Table S1 (see Supporting Information). The gene fragments were amplified using PrimeStar polymerase (TaKaRa) and digested using appropriate restriction endonucleases (Promega, Madison, WI, USA), followed by ligation into vectors using T4 DNA ligase (Promega). The PpE4 and GFP co‐silencing hairpin vector pTH210::E4S was constructed with reference to a previous study (Meng et al., 2015). First, the SpeI‐ and ClaI‐digested GFP fragment and ClaI‐digested kanamycin resistance gene linker were ligated into SpeI‐linearized pBluescript II KS to generate the GFP‐linker‐GFP hairpin structure. Then, the BamHI‐ and SpeI‐digested PpE4 fragment and the SpeI‐released GFP‐linker‐GFP fragment were ligated into BamHI‐linearized pBluescript II KS to generate the PpE4‐GFP‐linker‐GFP‐PpE4 co‐silencing hairpin structure. Finally, the co‐silencing hairpin structure, which was blunt‐ended by Pfu DNA Polymerase (Promega) after being digested by BamHI, was inserted into SmaI‐linearized plasmid pTH210 (Judelson et al., 1991). To construct the overexpression vector, E4FL was inserted into pMCherryH (Ham34 promoter) after being digested with AgeI and NheI, or fused with mCherry by overlapping PCR and then inserted into pTH210 (Hsp70 promoter) after being digested with ApaI and KpnI. The signal peptide of PpE4 was predicted using the SignalP 4.1 online server (http://www.cbs.dtu.dk/services/SignalP/) (Nielsen, 2017). E4FL, mature PpE4 without a signal peptide and its deletion mutants were ligated into pCAMBIA1307‐3×Flag and pER8 vector. For the VIGS assay, primers were designed with reference to previous studies: EDS1 and RAR1 (Liu et al., 2002); SGT1, HSP90 and NPK (Jin et al., 2002); NDR1, MEK2, SIPK, MEK1, MYB1 and WRKY3 (Liu et al., 2004b); BAK1 (Yang et al., 2017); and SOBIR1 (Liebrand et al., 2013). Fragments amplified from N. benthamiana cDNA were cloned into the binary vector pTRV2. All constructs were sequenced by Genscript (Nanjing, China).
Agrobacterium tumefaciens‐mediated transient expression
Agrobacterium tumefaciens GV3101 strains carrying the respective constructs were cultured in Luria–Bertani medium supplemented with the appropriate antibiotics at 28 °C for 1 day, and then harvested and suspended in infiltration buffer [10 mM 2‐(N‐morpholine)‐ethane sulfonic acid (MES), 10 mM MgCl2, pH 5.6, and 200 μM acetosyringone] to an appropriate concentration. For the inoculation of P. parasitica after transient expression, infiltrations were performed at a final OD600 of 0.01; otherwise, an OD600 value of 0.4 was used. After incubation for 1 h at 28 °C, the A. tumefaciens suspensions were infiltrated into plant leaves using needleless syringes (Meng et al., 2015). Cell death was observed at 3–5 dpi in N. benthamiana and tobacco species, and at 5–8 dpi in Solanum lycopersicum and S. tuberosum. For western blot analysis, proteins were extracted at 2 dpi. All experiments were repeated at least three times.
VIGS assay in N. benthamiana
Agrobacterium tumefaciens GV3101 strains carrying different pTRV2 constructs were mixed with pTRV1 in equal ratios to a final OD600 of 0.25. pTRV2::GFP was used as a control, and pTRV2::PDS was used to visualize the silencing process. The lower leaves of four‐leaf stage N. benthamiana plants were infiltrated as described previously (Liu et al., 2002; Ratcliff et al., 2001), and the degree of cell death and gene silencing efficiency were analysed in the upper leaves at 16–20 dpi.
Transformation of A. thaliana
Agrobacterium tumefaciens carrying the empty vector pER8 or pER8::3×Flag‐PpE4 was cultured and suspended in a solution of 5% sucrose and 0.02% Silwet L‐77 (GE Healthcare, Uppsala, Sweden). Arabidopsis ecotype Col‐0 was transformed by dipping in the suspension as described previously (Clough and Bent, 1998). The kanamycin‐resistant seedlings were screened on selective medium and planted in soil. Then, the expression level of PpE4 in transgenic plants after induction by 17‐β‐estradiol was determined by semi‐quantitative PCR (Zuo et al., 2000).
Transformation of P. parasitica
To generate silencing and overexpressing transformants, P. parasitica protoplasts were transformed using the PEG–CaCl2‐mediated method as described previously (Bottin et al., 1999; Meng et al., 2015). The silencing and overexpression plasmids (pTH210::E4S, pMCherryH::E4FL and pTH210::E4FL‐mCherry) were linearized by BamHI and separately co‐transformed with linearized pTH209 into protoplasts of strain 1121, which stably expresses hyphal cytoplasmic GFP. The transformed protoplasts were regenerated overnight, and the recovered mycelia were selected on 5% CA medium with 4 µg/mL geneticin and 100 µg/mL hygromycin. After 3–7 days, the primary transformants were transferred to new selective medium in six‐well plates and named sequentially and maintained for subsequent analyses.
Fluorescence microscopy
To identify PpE4 and GFP co‐silencing transformants, transformants with attenuated GFP signal were identified using an Olympus BX‐51TRF fluorescence microscope (Olympus, Tokyo, Japan) with the GFP filter (BP450‐480). For E4FL‐mCherry‐expressing lines, the putative transformants were observed under the mCherry filter (BP520‐550). Images of vegetative mycelia and infection hyphae in N. benthamiana leaves were captured on an Olympus IX83‐FV1200 confocal microscope with 488 nm excitation and a 500–530 nm emission spectrum for GFP. For mCherry, the emission spectrum was acquired between 595 and 625 nm under 559 nm excitation to eliminate potential autofluorescence from P. parasitica hyphae and cell damage. The detached N. benthamiana leaves inoculated with the transformants and the control strain were incubated at 23 °C for 12–48 h to allow the penetration and formation of intercellular hyphae with haustoria. The control strain and transformants were observed under the same conditions.
Inoculation of P. parasitica
Nicotiana benthamiana leaves were detached 24 h after agroinfiltration and kept in a plastic tray covered with moist filter paper. The petioles were wrapped with wet cotton; the leaves were inoculated with 1000 zoospores of P. parasitica strain 1121 and incubated in a growth chamber at 23 °C. For pathogenicity assays of P. parasitica transformants, fresh mycelia of transformants and the control strain grown on 5% CA plugs were inoculated on each side of detached N. benthamiana leaves. More than 15 leaves were used in each assay. At 36–48 hpi, the hyphal expansion was marked under a fluorescence microscope to measure the lesion diameter. Total DNA was extracted from identical areas on each side of the leaf, and the biomass was calculated by the DNA ratio of P. parasitica in infected tissues using qPCR (Meng et al., 2015). The rosette leaves of wild‐type Arabidopsis Col‐0 and T3 homozygous pER8 and pER8::3×Flag‐PpE4 transgenic plants were injected with 10 μM 17‐β‐estradiol (Zuo et al., 2000). After 12 h, the treated leaves were detached and placed in a plastic tray with wet cotton covering the petioles. Then, 2000 zoospores were dropped onto the abaxial surface of each leaf. About 25 leaves from more than 15 plants of each line were analysed for each assay. The disease severity index (DSI) was recorded at 48 hpi, with grade 1 being no visible symptoms and few hyphae colonized on the leaf surface, grade 2 being the development of restricted water‐soaked lesions with a diameter of less than 2 mm, grade 3 being the development of water‐soaked lesions smaller than the inoculation sites with abundant hyphae colonized, and grade 4 being the development of large lesions with massive hyphae spreading beyond the inoculation sites. The expansion of P. parasitica hyphae was visualized by trypan blue staining and the P. parasitica biomass was determined in equal amounts of inoculated leaves by qPCR.
Western blot analysis
Mycelia or plant leaves were ground into a powder in liquid nitrogen and vigorously mixed with a double volume of precooled RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% TritonX‐100, 1% sodium deoxycholate, 0.1% sodium dodecylsulfate (SDS), 5 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM ethylenediaminetetraacetic acid (EDTA), 10 mM dithiothreitol (DTT), 1% (w/v) protease inhibitor cocktail (Sigma, St. Louis, MO, USA)]. After 20 min of incubation on ice, the sample was centrifuged at 20 000 g for 15 min to obtain the supernatant. After the addition of loading buffer and boiling for 5 min, total proteins were separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). Then, the proteins were transferred to poly(vinylidene difluoride) (PVDF) membranes (Roche, Basel, Switzerland), followed by blocking in 10% skimmed milk (BD, Sparks, MD, USA) dissolved in Tris‐buffered saline (TBS; pH 7.2). Mouse anti‐Flag monoclonal antibody (Abbkine, Redlands, CA, USA) and mouse anti‐mCherry monoclonal antibody (Abbkine) were used at 1 : 2000 dilution to detect the corresponding fusion proteins. The membranes were washed and incubated with a goat anti‐mouse antibody (Abbkine). The protein bands were visualized by chemiluminescence using an eECL Western blot kit (CWBio), and photographs were taken under a ChemiDOC™ XRS+ imaging system (Bio‐Rad Laboratories, Hercules, CA).
Supporting information
Fig. S1 Expression pattern of PpE4 during Phytophthora parasitica infection of Arabidopsis. (A) FPKM (fragments per kilobase million) value of PpE4 and other RXLR effector genes from RNA‐sequencing (RNA‐seq) data. The sums of the FPKM values of all the 76 RXLR effector genes detected (FPKM value larger than unity) during infection of Arabidopsis roots were calculated. (B) Relative PpE4 transcript levels during different stages of P. parasitica infection quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Arabidopsis roots inoculated with P. parasitica zoospores were harvested at different hours post‐inoculation (hpi). My, P. parasitica mycelia grown in carrot broth. The relative expression level of PpE4 in mycelia was given a value of unity. Error bars represent the standard deviation (SD) of three pooled samples.
Fig. S2 The infection process of Phytophthora parasitica on Nicotiana benthamiana. Biotrophic growth was dominant before 24 h post‐inoculation (hpi), followed by a rapid switch to necrotrophic growth with large‐scale cell death. Nicotiana benthamiana leaves infected with zoospores of strain 1121 [stably expresses hyphal cytoplasmic green fluorescent protein (GFP)] were observed under a fluorescence microscope at 3, 6, 12, 24 and 48 hpi. The green fluorescence represents infection hyphae; the red fluorescence is the chloroplast autofluorescence of healthy leaf cells, which turns black when cell death occurs in the leaves. At 3 hpi, the cysts germinated and colonized on the epidermal cells, and extensive hyphae formed at 6 hpi. Cell death occurred at the inoculation sites at 12 hpi. Together with the spread of abundant hyphae, cell death occurred at the whole inoculation sites at 24 hpi. At 48 hpi, cell death occurred in large areas, with sporangia developing at the inoculation sites. CD, cell death; S, sporangia. Bars, 100 μm.
Fig. S3 Generation of Phytophthora parasitica transformants expressing the E4FL‐mCherry fusion protein. (A) Schematic diagram of the fusion protein constructs in vector pTH210 or pMCherryH. Expression of E4FL (full‐length PpE4 with its own signal peptide) fused with mCherry at its C‐terminus was driven by the constitutive Ham34 or Hsp70 promoter. Relative expression level of PpE4 in vegetative mycelia (B) and in infected Nicotiana benthamiana leaves at 36 h post‐inoculation (hpi) (C) quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Expression of E4FL‐mCherry in E4MC4A2 is driven by the Ham34 promoter, whereas, in other transformants it is driven by the Hsp70 promoter. The expression level of PpE4 in strain 1121 was given a value of unity. Error bars represent the standard deviation (SD) of three biological replicates. (D) Accumulation of E4FL‐mCherry fusion proteins in vegetative mycelia was confirmed by western blot using mCherry antibody. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from three independent experiments.
Fig. S4 The localization of E4FL‐mCherry in transformant E4MC4A6 during infection. (A) Confocal image showing the accumulation of E4FL‐mCherry outside the haustoria after secretion at 24 h post‐inoculation (hpi). (B) The fluorescence intensities of green fluorescent protein (GFP) and mCherry across the haustoria are indicated by the white lines labelled ‘1’ and ‘2’ in (A). Identical images were obtained from more than 10 haustoria in three independent biological replicates.
Fig. S5 Attenuated pathogenicity of E4FL‐mCherry‐expressing Phytophthora parasitica transformants. Fresh mycelial plugs of transformants (E4MC4A2, E4MC4A6, E4MC4B2 and E4MC3N4) and control strain 1121 were inoculated on the left and right sides of Nicotiana benthamiana leaves, respectively, and the lesion diameters were measured at 48 h post‐inoculation (hpi). (A) Lesions caused by E4FL‐mCherry‐expressing transformants were significantly smaller than those caused by the 1121 strain and E4MC3N4. Error bars represent the standard deviation (SD) of 15 leaves. Asterisks denote significant differences from the control strain 1121 (two tailed t‐test: **P < 0.01; ***P < 0.001). (B) Representative inoculated leaves. Similar results were obtained from more than three independent experiments.
Fig. S6 Generation of Phytophthora parasitica PpE4‐silencing transformants. (A) Diagram of the PpE4 and GFP co‐silencing hairpin structure construct. The kanamycin‐resistant gene (kanR) was used as the linker sequence. (B) Green fluorescent protein (GFP) signals in mycelia of five PpE4‐silenced transformants and strain 1121. (C) Relative expression level of PpE4 in five GFP signal‐decreased transformants sampled at 24 h post‐inoculation (hpi) on Nicotiana benthamiana leaves was quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). The expression level of PpE4 in strain 1121 was given a value of unity. Error bars represent the standard deviation (SD) of three biological replicates. Two independent experiments were performed with similar results.
Fig. S7 Secondary structure of the PpE4 protein predicted by Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?xml:id=index).
Fig. S8 PpE4‐triggered cell death is not compromised in Nicotiana benthamiana plants with silenced expression of several genes involved in plant immune signalling. Nicotiana benthamiana leaves were infiltrated with pTRV2 constructs targeting EDS1, NDR1, MEK1, MEK2, SIPK, MYB1, WRKY3, EDS1, NDR1, BAK1 and SOBIR1; pTRV2::GFP was used as a control. Agrobacterium tumefaciens carrying PpE4 was infiltrated into the upper leaves of silenced plants at 16–20 days post‐infiltration (dpi). (A) Cell death photographed at 5 dpi. (B) Quantification of cell death on N. benthamiana leaves. The degree of cell death was divided into three levels: no visible cell death, weak cell death and complete cell death. Asterisk represents a significant difference from the control (Wilcoxon rank‐sum test: *P < 0.05). (C) Relative expression levels of silenced genes in corresponding virus‐induced gene silencing (VIGS)‐treated plants determined by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Error bars represent the standard deviation (SD) of three biological replicates. The experiments were repeated three times with more than 10 plants for each TRV construct.
Table S1 Primers and vectors used in this study.
Acknowledgements
We thank Professor Brett Tyler (Oregon State University, Corvallis, OR, USA), Professor Francine Govers (Wageningen University, Wageningen, the Netherlands), Professor Gary Loake (University of Edinburgh, Edinburgh, UK), Professor Patrick Schäfer (University of Warwick, Coventry, UK) and Dr Ruth Eichmann (University of Warwick, Coventry, UK) for helpful discussions and useful suggestions. This work was supported by funding from the National Natural Science Foundation of China (#31561143007), National Key R&D Program of China (#2017YFD0200602‐2). China Agriculture Research System (#CARS‐09), the Programme of Introducing Talents of Innovative Discipline to Universities (Project 111) from the State Administration of Foreign Experts Affairs (#B18042).
References
- Anderson, R.G. , Deb, D. , Fedkenheuer, K. and McDowell, J.M. (2015) Recent progress in RXLR effector research. Mol. Plant–Microbe Interact. 28, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Attard, A. , Gourgues, M. , Callemeyn‐Torre, N. and Keller, H. (2010) The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 187, 449–460. [DOI] [PubMed] [Google Scholar]
- Baxter, L. , Tripathy, S. , Ishaque, N. , Boot, N. , Cabral, A. , Kemen, E. , Thines, M. , Ah‐Fong, A. , Anderson, R. and Badejoko, W. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science, 330, 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm, H. , Albert, I. , Oome, S. , Raaymakers, T.M. , Van den Ackerveken, G. and Nurnberger, T. (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern‐induced immunity in Arabidopsis . PLoS Pathog. 10, e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, J.I. , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. , Armstrong, M.R. , Birch, P.R. and Kamoun, S. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Boter, M. , Amigues, B. , Peart, J. , Breuer, C. , Kadota, Y. , Casais, C. , Moore, G. , Kleanthous, C. , Ochsenbein, F. , Shirasu, K. and Guerois, R. (2007) Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell, 19, 3791–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottin, A. , Larche, L. , Villalba, F. , Gaulin, E. , Esquerré‐Tugayé, M.‐T. and Rickauer, M. (1999) Green fluorescent protein (GFP) as gene expression reporter and vital marker for studying development and microbe–plant interaction in the tobacco pathogen Phytophthora parasitica var. nicotianae . FEMS Microbiol. Lett. 176, 51–56. [DOI] [PubMed] [Google Scholar]
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M.W. , Shipley, G.L. , Vandesompele, J. and Wittwer, C.T. (2009) The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clin. Chem. 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Chang, Y.H. , Yan, H.Z. and Liou, R.F. (2015) A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol. Plant Pathol. 16, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro‐Garcia, A. , Wilkinson, R.C. , Gimenez‐Ibanez, S. , Findlay, K. , Coffey, M.D. , Zipfel, C. , Rathjen, J.P. , Kamoun, S. and Schornack, S. (2011) The receptor‐like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana . PLoS One, 6, e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, S. , Stevens, L.J. , Boevink, P.C. , Engelhardt, S. , Alexander, C.J. , Harrower, B. , Champouret, N. , McGeachy, K. , Van Weymers, P.S. , Chen, X. , Birch, P.R. and Hein, I. (2014) Detection of the virulent form of AVR3a from Phytophthora infestans following artificial evolution of potato resistance gene R3a . PLoS One, 9, e110158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. and Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalio, R.J.D. , Maximo, H.J. , Oliveira, T.S. , Dias, R.O. , Breton, M.C. , Felizatti, H. and Machado, M. (2017) Phytophthora parasitica effector PpRxLR2 suppresses Nicotiana benthamiana immunity. Mol. Plant–Microbe Interact. 31, 481–493. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Berg, J. , Govers, F. and Bouwmeester, K. (2015a) Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 207, 735–747. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Mpina, M.H. , Birch, P.R. , Bouwmeester, K. and Govers, F. (2015b) Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti, E. , Govetto, B. , Minet‐Kebdani, N. , Kuhn, M.L. , Attard, A. , Ponchet, M. , Panabieres, F. and Gourgues, M. (2013) The Phytophthora parasitica RXLR effector penetration‐specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol. 199, 476–489. [DOI] [PubMed] [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. , Felix, G. , Kemmerling, B. , Krzymowska, M. and Nurnberger, T. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis . Plant J. 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Gaulin, E. , Jauneau, A. , Villalba, F. , Rickauer, M. , Esquerré‐Tugayé, M.‐T. and Bottin, A. (2002) The CBEL glycoprotein of Phytophthora parasitica var‐nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115, 4565–4575. [DOI] [PubMed] [Google Scholar]
- Gilroy, E.M. , Breen, S. , Whisson, S.C. , Squires, J. , Hein, I. , Kaczmarek, M. , Turnbull, D. , Boevink, P.C. , Lokossou, A. , Cano, L.M. , Morales, J. , Avrova, A.O. , Pritchard, L. , Randall, E. , Lees, A. , Govers, F. , van West, P. , Kamoun, S. , Vleeshouwers, V.G. , Cooke, D.E. and Birch, P.R. (2011) Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2‐like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grunwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M. , Jones, J.D. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L. , Maclean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J. , Morgan, W. , Morris, P.F. , Munro, C.A. , O’Neill, K. , Ospina‐Giraldo, M. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W. , Meyers, B.C. , van West, P. , Ristaino, J. , Govers, F. , Birch, P.R. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Jia, J. , Lu, W. , Zhong, C. , Zhou, R. , Xu, J. , Liu, W. , Gou, X. , Wang, Q. , Yin, J. , Xu, C. and Shan, W. (2017) The 25–26 nt small RNAs in Phytophthora parasitica are associated with efficient silencing of homologous endogenous genes. Front. Microbiol. 8, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Axtell, M.J. , Dahlbeck, D. , Ekwenna, O. , Zhang, S. , Staskawicz, B. and Baker, B. (2002) NPK1, an MEKK1‐like mitogen‐activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell, 3, 291–297. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. , Tyler, B.M. and Michelmore, R.W. (1991) Transformation of the oomycete pathogen, Phytophthora infestans . Mol. Plant–Microbe Interact. 4, 602–607. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota, Y. , Shirasu, K. and Guerois, R. (2010) NLR sensors meet at the SGT1‐HSP90 crossroad. Trends Biochem. Sci. 35, 199–207. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O.J. , Jones, J.D.G. , Judelson, H.S. , Ali, G.S. , Dalio, R.J.D. , Roy, S.G. , Schena, L. , Zambounis, A. and Panabieres, F. (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki, H. , Saitoh, H. , Ito, A. , Fujisawa, S. , Kamoun, S. , Katou, S. , Yoshioka, H. and Terauchi, R. (2003) Cytosolic HSP90 and HSP70 are essential components of INF1‐mediated hypersensitive response and non‐host resistance to Pseudomonas cichorii in Nicotiana benthamiana . Mol. Plant Pathol. 4, 383–391. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. and Sternberg, M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib, M. , Lafitte, C. , Esquerré‐Tugayé, M.T. , Bottin, A. and Rickauer, M. (2004) The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510. [Google Scholar]
- King, S.R. , McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Bukharova, T. , Sukarta, O. , Win, J. , Kamoun, S. , Birch, P.R. and Banfield, M.J. (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell, 26, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjemtrup, S. , Nimchuk, Z. and Dangl, J.L. (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Knepper, C. , Savory, E.A. and Day, B. (2011) The role of NDR1 in pathogen perception and plant defense signaling. Plant Signal. Behav. 6, 1114–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Lee, S.E. , Oh, S. , Seo, E. and Choi, D. (2018) HSP70s enhance a Phytophthora infestans effector‐induced cell death via a MAPK cascade in Nicotiana benthamiana . Mol. Plant‐Microbe Interact. 31, 356–362. [DOI] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , van den Berg, G.C.M. , Zhang, Z. , Smit, P. , Cordewener, J.H.G. , America, A.H.P. , Sklenar, J. , Jones, A.M.E. , Tameling, W.I.L. , Robatzek, S. , Thomma, B.P.H.J. and Joosten, M.H.A.J. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA, 110, 10 010–10 015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W. , van den Burg, H.A. and Joosten, M.H. (2014) Two for all: receptor‐associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Song, T. , Zhang, X. , Yuan, H. , Su, L. , Li, W. , Xu, J. , Liu, S. , Chen, L. and Chen, T. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Burch‐Smith, T. , Schiff, M. , Feng, S. and Dinesh‐Kumar, S.P. (2004a) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2004b) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N‐mediated resistance to tobacco mosaic virus. Plant J. 38, 800–809. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. and Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D.C. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z. , Song, T. , Zhu, L. , Ye, W. , Wang, Y. , Shao, Y. , Dong, S. , Zhang, Z. , Dou, D. , Zheng, X. , Tyler, B.M. and Wang, Y. (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell, 27, 2057–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Zhang, Q. , Ding, W. and Shan, W. (2014) Phytophthora parasitica: a model oomycete plant pathogen. Mycology, 5, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Zhang, Q. , Zhang, M. , Gu, B. , Huang, G. , Wang, Q. and Shan, W. (2015) The protein disulfide isomerase 1 of Phytophthora parasitica (PpPDI1) is associated with the haustoria‐like structures and contributes to plant infection. Front. Plant Sci. 6, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H. (2017) Predicting Secretory Proteins with SignalP. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- Oh, S.K. , Kwon, S.Y. and Choi, D. (2014) Rpi‐blb2‐mediated hypersensitive cell death caused by Phytophthora infestans AVRblb2 requires SGT1, but not EDS1, NDR1, salicylic acid‐, jasmonic acid‐, or ethylene‐mediated signaling. Plant Pathol. J. 30, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.K. , Young, C. , Lee, M. , Oliva, R. , Bozkurt, T.O. , Cano, L.M. , Win, J. , Bos, J.I. , Liu, H.‐Y. and van Damme, M. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2. Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann, C. , Luberacki, B. , Kufner, I. , Koch, W. , Brunner, F. , Weyand, M. , Mattinen, L. , Pirhonen, M. , Anderluh, G. , Seitz, H.U. , Nurnberger, T. and Oecking, C. (2009) A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. USA, 106, 10 359–10 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.F. and Martin, G.B. (2005) Role of mitogen‐activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541–547. [DOI] [PubMed] [Google Scholar]
- Peng, Y. , van Wersch, R. and Zhang, Y. (2018) Convergent and divergent signaling in PAMP‐triggered immunity and effector‐triggered immunity. Mol. Plant–Microbe Interact. 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kemmerling, B. , Brunner, F. , Kuefner, I. , Engelhardt, S. , Gust, A.A. , Luberacki, B. , Seitz, H.U. , Stahl, D. , Rauhut, T. , Glawischnig, E. , Schween, G. , Lacombe, B. , Watanabe, N. , Lam, E. , Schlichting, R. , Scheel, D. , Nau, K. , Dodt, G. , Hubert, D. , Gijzen, M. and Nuernberger, T. (2006) Phytotoxicity and innate immune responses induced by Nep1‐like proteins. Plant Cell, 18, 3721–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F. , Martin‐Hernandez, A.M. and Baulcombe, D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Sharpee, W.C. and Dean, R.A. (2016) Form and function of fungal and oomycete effectors. Fungal Biol. Rev. 30, 62–73. [Google Scholar]
- Shirasu, K. (2009) The HSP90‐SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. [DOI] [PubMed] [Google Scholar]
- Soyano, T. , Nishihama, R. , Morikiyo, K. , Ishikawa, M. and Machida, Y. (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK‐mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 17, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A. , Casais, C. , Ichimura, K. and Shirasu, K. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2‐mediated disease resistance in Arabidopsis . Proc. Natl. Acad. Sci. USA, 100, 11 777–11 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. and Beynon, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- van Poppel, P.M. (2009) The Phytophthora infestans avirulence gene PiAvr4 and its potato counterpart R4 . PhD thesis, University of Wageningen, Wageningen, the Netherlands. [Google Scholar]
- van’t Slot, K.A. and Knogge, W. (2002) A dual role for microbial pathogen‐derived effector proteins in plant disease and resistance. Crit. Rev. Plant Sci. 21, 229–271. [Google Scholar]
- Wang, H. , Ren, Y. , Zhou, J. , Du, J. , Hou, J. , Jiang, R. , Wang, H. , Tian, Z. and Xie, C. (2017) The cell death triggered by the nuclear localized RxLR effector PITG_22798 from Phytophthora infestans is suppressed by the effector AVR3b. Int. J. Mol. Sci. 18, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. , Kale, S.D. , Gu, B. , Sheng, Y. , Sui, Y. , Wang, X. , Zhang, Z. , Cheng, B. , Dong, S. , Shan, W. , Zheng, X. , Dou, D. , Tyler, B.M. and Wang, Y. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Boevink, P.C. , Welsh, L. , Zhang, R. , Whisson, S.C. and Birch, P.R.J. (2017) Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 216, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Meng, Y. , Zhang, M. , Tong, X. , Wang, Q. , Sun, Y. , Quan, J. , Govers, F. and Shan, W. (2011) Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 12, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Wang, S. and Birch, P.R. (2016) The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, J. , Li, X. , Yin, L. , Liu, Y. , Zhang, Y. , Qu, J. and Lu, J. (2017) A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana . BMC Plant Biol. 17, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H.Z. and Liou, R.F. (2006) Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genet. Biol. 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Wang, Q. , Jing, M. , Guo, B. , Wu, J. , Wang, H. , Wang, Y. , Lin, L. , Wang, Y. , Ye, W. , Dong, S. and Wang, Y. (2017) Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214, 361–375. [DOI] [PubMed] [Google Scholar]
- Yu, X. , Tang, J. , Wang, Q. , Ye, W. , Tao, K. , Duan, S. , Lu, C. , Yang, X. , Dong, S. , Zheng, X. and Wang, Y. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196, 247–260. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Meng, Y. , Wang, Q. , Liu, D. , Quan, J. , Hardham, A.R. and Shan, W. (2012) PnPMA1, an atypical plasma membrane H(+)‐ATPase, is required for zoospore development in Phytophthora parasitica . Fungal Biol. 116, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Dorey, S. , Swiderski, M. and Jones, J.D. (2004) Expression of RPS4 in tobacco induces an AvrRps4‐independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , McLellan, H. , Fraiture, M. , Liu, X. , Boevink, P.C. , Gilroy, E.M. , Chen, Y. , Kandel, K. , Sessa, G. and Birch, P.R. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22‐triggered immunity. PLoS Pathog. 10, e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J. , Niu, Q.‐W. and Chua, N.‐H. (2000) An estrogen receptor‐based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression pattern of PpE4 during Phytophthora parasitica infection of Arabidopsis. (A) FPKM (fragments per kilobase million) value of PpE4 and other RXLR effector genes from RNA‐sequencing (RNA‐seq) data. The sums of the FPKM values of all the 76 RXLR effector genes detected (FPKM value larger than unity) during infection of Arabidopsis roots were calculated. (B) Relative PpE4 transcript levels during different stages of P. parasitica infection quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Arabidopsis roots inoculated with P. parasitica zoospores were harvested at different hours post‐inoculation (hpi). My, P. parasitica mycelia grown in carrot broth. The relative expression level of PpE4 in mycelia was given a value of unity. Error bars represent the standard deviation (SD) of three pooled samples.
Fig. S2 The infection process of Phytophthora parasitica on Nicotiana benthamiana. Biotrophic growth was dominant before 24 h post‐inoculation (hpi), followed by a rapid switch to necrotrophic growth with large‐scale cell death. Nicotiana benthamiana leaves infected with zoospores of strain 1121 [stably expresses hyphal cytoplasmic green fluorescent protein (GFP)] were observed under a fluorescence microscope at 3, 6, 12, 24 and 48 hpi. The green fluorescence represents infection hyphae; the red fluorescence is the chloroplast autofluorescence of healthy leaf cells, which turns black when cell death occurs in the leaves. At 3 hpi, the cysts germinated and colonized on the epidermal cells, and extensive hyphae formed at 6 hpi. Cell death occurred at the inoculation sites at 12 hpi. Together with the spread of abundant hyphae, cell death occurred at the whole inoculation sites at 24 hpi. At 48 hpi, cell death occurred in large areas, with sporangia developing at the inoculation sites. CD, cell death; S, sporangia. Bars, 100 μm.
Fig. S3 Generation of Phytophthora parasitica transformants expressing the E4FL‐mCherry fusion protein. (A) Schematic diagram of the fusion protein constructs in vector pTH210 or pMCherryH. Expression of E4FL (full‐length PpE4 with its own signal peptide) fused with mCherry at its C‐terminus was driven by the constitutive Ham34 or Hsp70 promoter. Relative expression level of PpE4 in vegetative mycelia (B) and in infected Nicotiana benthamiana leaves at 36 h post‐inoculation (hpi) (C) quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Expression of E4FL‐mCherry in E4MC4A2 is driven by the Ham34 promoter, whereas, in other transformants it is driven by the Hsp70 promoter. The expression level of PpE4 in strain 1121 was given a value of unity. Error bars represent the standard deviation (SD) of three biological replicates. (D) Accumulation of E4FL‐mCherry fusion proteins in vegetative mycelia was confirmed by western blot using mCherry antibody. Protein loading is indicated by Ponceau stain (PS). Similar results were obtained from three independent experiments.
Fig. S4 The localization of E4FL‐mCherry in transformant E4MC4A6 during infection. (A) Confocal image showing the accumulation of E4FL‐mCherry outside the haustoria after secretion at 24 h post‐inoculation (hpi). (B) The fluorescence intensities of green fluorescent protein (GFP) and mCherry across the haustoria are indicated by the white lines labelled ‘1’ and ‘2’ in (A). Identical images were obtained from more than 10 haustoria in three independent biological replicates.
Fig. S5 Attenuated pathogenicity of E4FL‐mCherry‐expressing Phytophthora parasitica transformants. Fresh mycelial plugs of transformants (E4MC4A2, E4MC4A6, E4MC4B2 and E4MC3N4) and control strain 1121 were inoculated on the left and right sides of Nicotiana benthamiana leaves, respectively, and the lesion diameters were measured at 48 h post‐inoculation (hpi). (A) Lesions caused by E4FL‐mCherry‐expressing transformants were significantly smaller than those caused by the 1121 strain and E4MC3N4. Error bars represent the standard deviation (SD) of 15 leaves. Asterisks denote significant differences from the control strain 1121 (two tailed t‐test: **P < 0.01; ***P < 0.001). (B) Representative inoculated leaves. Similar results were obtained from more than three independent experiments.
Fig. S6 Generation of Phytophthora parasitica PpE4‐silencing transformants. (A) Diagram of the PpE4 and GFP co‐silencing hairpin structure construct. The kanamycin‐resistant gene (kanR) was used as the linker sequence. (B) Green fluorescent protein (GFP) signals in mycelia of five PpE4‐silenced transformants and strain 1121. (C) Relative expression level of PpE4 in five GFP signal‐decreased transformants sampled at 24 h post‐inoculation (hpi) on Nicotiana benthamiana leaves was quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). The expression level of PpE4 in strain 1121 was given a value of unity. Error bars represent the standard deviation (SD) of three biological replicates. Two independent experiments were performed with similar results.
Fig. S7 Secondary structure of the PpE4 protein predicted by Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?xml:id=index).
Fig. S8 PpE4‐triggered cell death is not compromised in Nicotiana benthamiana plants with silenced expression of several genes involved in plant immune signalling. Nicotiana benthamiana leaves were infiltrated with pTRV2 constructs targeting EDS1, NDR1, MEK1, MEK2, SIPK, MYB1, WRKY3, EDS1, NDR1, BAK1 and SOBIR1; pTRV2::GFP was used as a control. Agrobacterium tumefaciens carrying PpE4 was infiltrated into the upper leaves of silenced plants at 16–20 days post‐infiltration (dpi). (A) Cell death photographed at 5 dpi. (B) Quantification of cell death on N. benthamiana leaves. The degree of cell death was divided into three levels: no visible cell death, weak cell death and complete cell death. Asterisk represents a significant difference from the control (Wilcoxon rank‐sum test: *P < 0.05). (C) Relative expression levels of silenced genes in corresponding virus‐induced gene silencing (VIGS)‐treated plants determined by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Error bars represent the standard deviation (SD) of three biological replicates. The experiments were repeated three times with more than 10 plants for each TRV construct.
Table S1 Primers and vectors used in this study.


